Abstract
We recently identified three nuclear proteins (p45, p39, and p26) that bind to a 91-nucleotide (nt) RNA element between nt 1243 and 1333 in hepatitis B virus (HBV) RNA, and we showed that these proteins and HBV RNA are regulated coordinately by gamma interferon and tumor necrosis factor alpha. Purification and sequence analysis of tryptic peptides obtained from p39 revealed sequence homology to the mouse La protein. Immunoprecipitation experiments showed that p45, p39, and p26 were recognized by anti-La-specific antiserum, indicating that p45 is the full-length La protein and that p39 and p26 are likely to be proteolytic La cleavage products. Furthermore, in competition experiments we found that all three La proteins bind, in a phosphorylation-dependent manner, to the same predicted stem-loop structure located between nt 1275 and 1291 of HBV, with Kds of approximately 1.0 nM. Collectively, these results support the notion that the La protein may contribute to HBV RNA stability, constitutively and in response to inflammatory cytokines.
RNA-protein interactions regulate gene expression by controlling the processing (62), export (43, 51, 62), translation (65), intracellular localization (67), and degradation (39, 58) of mRNA. mRNA stability is regulated by hormones and cytokines (72) that induce the phosphorylation or dephosphorylation of RNA-binding proteins (17, 26, 46) and modulate RNase activities (39a, 41, 57). Furthermore, prokaryotic multiprotein complexes consisting of RNase E, polynucleotide phosphorylase (11, 55), ATP-dependent RNA helicases (56), heat shock-chaperone proteins GroEL and DnaK, and the glycolytic enzyme endolase (49) are thought to coordinate the stabilization or destabilization of certain mRNAs. In addition, it was shown that these complexes also contain cellular RNAs (49).
The hepatitis B virus (HBV) is a DNA virus that replicates through an RNA intermediate and encodes four unspliced, overlapping messages that terminate at a common polyadenylation signal (61). The vigor and kinetics of the cytotoxic T-lymphocyte (CTL) response to HBV determine the outcome of infection (19). By the use of transgenic mice that express some (20, 21, 30) or all of the viral proteins and replicate the virus (35), many of the host-virus interactions responsible for viral clearance and disease pathogenesis during HBV infection have been defined (3, 4, 18, 50). Recently, we have shown that inflammatory cytokines, especially gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α), induced by adoptively transferred HBV-specific CTLs or during lymphocytic choriomeningitis virus and murine cytomegalovirus infections, can abolish hepatic HBV gene expression and replication in the livers of these animals (12, 13, 29–34). Importantly, the cytokines suppress HBV gene expression by a posttranscriptional mechanism (36, 69), presumably reflecting destabilization of HBV mRNA.
Recently, we demonstrated that liver nuclear extracts from untreated, CTL-injected, lymphocytic choriomeningitis virus-infected, and murine cytomegalovirus-infected HBV transgenic mice contain three proteins (p45, p39, and p26) that bind a 91-nucleotide (nt) in vitro transcript of HBV (40). This transcript is located in the 5′ region of the HBV posttranscriptional regulatory element (between nt 1200 and 1650), which is thought to mediate the nuclear export of HBV RNA (23, 42). A tight correlation was observed among the downregulation of HBV RNA, the disappearance of p45, and the appearance of p26, suggesting that these proteins might contribute to the regulation of HBV mRNA stability by the cytokines (40). Furthermore, we showed that the elimination of p45 and the induction of p26 are coupled events that require IFN-γ and TNF-α, suggesting that cytokine-induced signal transduction pathways might regulate the RNA-binding activity of these proteins (40).
The goal of the current study was the purification and molecular identification of p45, p39, and p26 and the mapping of the RNA target element(s) to which they bind. In this report, we demonstrate that all three proteins are recognized by anti-La antibodies and that they bind HBV RNA in a phosphorylation-dependent manner. Furthermore, we showed that they bind to a predicted stem-loop in the 91-nt transcript with high affinity and that the stem structure and specific loop nucleotides are required for binding. Collectively, these results are consistent with the notion that the La protein, especially p45, may be part of a complex mechanism that controls HBV RNA stability, constitutively and in response to inflammatory cytokines.
MATERIALS AND METHODS
HBV transgenic mice.
The HBV transgenic mouse lineages 1.3.32 (official designation, Tg{HBV 1.3 genome}Chi32) and 1.3.46 (official designation, Tg{HBV 1.3 genome}Chi46) used in this study have been described previously (35). Lineages 1.3.32 and 1.3.46 express all of the HBV transcripts under the control of their respective promoters and replicate HBV at high levels in the liver and kidney without any evidence of cytopathology (35). Mice were matched for age (8 to 10 weeks), sex (male), and serum hepatitis B e antigen concentration by a commercially available solid-phase radioimmunoassay (Sorin Biomedica, Saluggia, Italy).
HBsAg-specific CTLs.
Ld-restricted, CD3+ CD4− CD8+ hepatitis B surface antigen (HBsAg)-specific CTL clones that recognize an epitope located between residues 28 and 39 of HBsAg (HBsAg28–39) and secrete IFN-γ and TNF-α upon antigen recognition (3) were used for the studies. In all experiments, 107 CTLs were injected intravenously into transgenic mice 5 days after in vitro stimulation with irradiated P815 cells that stably express the HBV large envelope protein (50). CTL-induced liver disease was monitored by measuring serum alanine aminotransaminase levels at various time points after CTL injection. Liver tissue obtained at autopsy was either processed for histological analysis or snap frozen for subsequent molecular analyses.
RNA analyses.
Snap-frozen (liquid nitrogen) liver tissues were mechanically pulverized, and total genomic RNA was isolated for Northern blot analyses exactly as previously described (35).
Preparation of liver nuclear and cytosolic extracts from HBV transgenic mice.
Frozen liver tissue (0.2 to 0.5 g) was thawed and homogenized in a fivefold volume of ice-cold homogenization buffer containing 10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 2.5 mM MgCl2, 1 mM EDTA (buffer A) containing 0.5 mM dithiothreitol (DTT), and a 1/25 volume of proteinase inhibitor mix (Boehringer Mannheim, Indianapolis, Ind.) by five strokes in a glass homogenizer with a loose-fitting motor-driven (50 rpm) Teflon pestle. The homogenate was centrifuged at 2,000 × g for 20 min, and the resulting supernatant was stored at −80°C. The pellet was resuspended in 6 ml of buffer A containing 0.88 M sucrose (buffer B), loaded on a 7-ml cushion of buffer B, and centrifuged at 10,000 × g for 30 min. The supernatant was discarded, and the pellet was dissolved in 5 ml of buffer A containing 2.0 M sucrose (buffer C). The slurry was loaded on a 7-ml cushion of buffer C and centrifuged at 180,000 × g for 70 min. The supernatant was discarded, and the nuclei was resuspended in 100 μl of storage buffer containing 20 mM Tris-HCl (pH 8.0), 75 mM NaCl, 2.5 mM MgCl2, 0.5 mM EDTA, 50% glycerol, 0.5 mM DTT, and a 1/10 volume of proteinase inhibitor mix (Boehringer Mannheim). Nuclei were counted by light microscopy and lysed by adding 5× lysis buffer containing 100 mM Tris-HCl (pH 8.0), 2.1 M NaCl, 7.5 mM MgCl2, 1.0 mM EDTA, and 25% glycerol to final concentrations of 33 mM Tris-HCl (pH 8.0), 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 5% glycerol, and 0.5 mM DTT, and a 1/10 volume of proteinase inhibitor mix (Boehringer Mannheim). The viscous lysate was transferred into dialysis tubes (molecular weight cutoff, 6,000 to 8,000; Spectro/Por; Spectrum Companies, Gardena, Calif.) and dialyzed three times against 500 ml of dialysis buffer F containing 10 mM Tris-HCl (pH 7.4), 100 mM NaCl, 3 mM MgCl2, 0.5 mM EDTA, 10% glycerol, 0.5 mM DTT, and proteinase inhibitor mix (Boehringer Mannheim). The dialyzed nuclear extract was cleared by centrifugation for 10 min at 24,000 × g, and the protein content was determined by the Bradford dye-binding procedure, with a commercial kit (Bio-Rad Laboratories, Hercules, Calif.).
Protein purification.
All the following steps were performed on ice or at 4°C. Nuclear extract was brought to 35% saturation with ammonium sulfate and stirred for 30 min. After centrifugation of the slurry for 30 min at 20,000 × g, the supernatant was brought to 65% saturation with ammonium sulfate and stirred for 30 min. The slurry was centrifuged at 20,000 × g for 30 min, the pellet was suspended in dialysis buffer F (see above), and the extract was dialyzed overnight. The next purification steps were performed with fast protein liquid chromatography (Pharmacia, Piscataway, N.J.). The extract was loaded (1 ml/min) on a heparin column (Pharmacia) which was preequilibrated with wash buffer containing 10 mM Tris-HCl (pH 7.4), 3 mM MgCl2, 0.5 mM EDTA, and 200 mM NaCl. The column was washed until protein was no longer detectable in the flowthrough. Bound protein was eluted in a linear gradient ranging from 200 mM to 2 M NaCl at a flow rate of 1 ml/min. Fractions (1 ml) were assayed for p45, p39, and p26 by UV cross-linking as described below. RNA-binding-protein-containing fractions were pooled, concentrated by ultrafiltration on Centricon 10 (Millipore, Bedford, Mass.), and loaded (1 ml/min) onto a source 30Q column (Pharmacia), and the flowthrough was further run through a source 30S column (Pharmacia). The final flowthrough was tested for p45, p39, and p26 by UV cross-linking; concentrated by ultrafiltrations as described above; and subsequently loaded (0.7 ml/min) onto a Superdex 75-pg gel filtration column (Pharmacia) equilibrated with 10 mM Tris-HCl (pH 7.4)–3 mM MgCl2–0.5 mM EDTA–100 mM NaCl. Elution was performed in equilibration buffer at a flow rate of 0.17 ml/min, and 0.51-ml fractions were collected and assayed for p45, p39, and p26 by UV cross-linking (ranging of fractions 13 to 26). For molecular mass determinations, the Superdex column was calibrated with standard proteins in a gel filtration calibration kit (Sigma). The gel filtration fractions containing p45, p39, and p26 (ranging of fractions 15 to 20) were separately concentrated and subjected to sodium dodecyl phosphate–14% polyacrylamide gel electrophoresis (SDS-PAGE). To determine the exact positions of p45, p39, and p26, UV cross-linking samples were loaded onto the same gel, and the gel was stained with Coomassie blue (Bio-Rad), destained, and exposed to Kodak Biomax (Kodak, Rochester, N.Y.) overnight at 4°C. The autoradiogram was matched with the gel, and a dominant protein band corresponding to the position of the p39 UV cross-linking signal was cut out. The protein band was subsequently submitted for tryptic in-gel digestion, and high-pressure liquid chromatography (HPLC) fractionation of tryptic peptides obtained from p39 and N-terminal sequencing of several peptides were performed by the Scripps Protein and Nucleic Acid Core Facility.
Immunoprecipitation and Western blotting.
One hundred milligrams of protein A-coupled Sepharose CL-4B beads (Pharmacia) was swollen in 1.5 ml of TS buffer (15 mM Tris-HCl [pH 7.4], 150 mM NaCl) for 4 h at room temperature. The swollen gel was collected by centrifugation and washed three times with TS buffer. The final pellet was resuspended in a total volume of 600 μl of TS buffer. Three hundred microliters was combined with 100 μl of control human serum or with anti-La-positive human serum (Centers for Disease Control and Prevention prototype, kindly provided by E. Chan, The Scripps Research Institute [14]). The mixture was incubated overnight at 4°C and subsequently for 4 h at room temperature. The slurry was washed three times with TS buffer and resuspended in 300 μl of TS buffer. One hundred microliters from this mixture was incubated with 20 μg of nuclear extracts prepared from untreated or CTL-injected mice and rotated for 6 h at 4°C. Precipitates were collected by centrifugation at 14,000 × g. The supernatant was taken off, and 5 μg of protein was analyzed by UV cross-linking and Western blotting. For Western blotting (semidry procedure), the SDS gel was incubated in 25 mM Tris-HCl (pH 9.0)–20% methanol at room temperature for 20 min. The nitrocellulose membrane was equilibrated in 25 mM Tris-HCl (pH 10.6)–20% methanol. The transfer was performed at 1.2 mA/cm2 for 90 min. After the transfer, the membrane was incubated in blocking buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.001% Tween 20, and 10% milk powder) at room temperature for 45 min. The blocking solution was changed, and anti-La-positive human serum (Centers for Disease Control and Prevention prototype, dilution 1:600) was added and further incubated overnight at 4°C. Subsequently, the membrane was washed six times for 5 to 10 min at room temperature with washing buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.001% Tween 20) and replaced by blocking buffer. Peroxidase-conjugated rabbit anti-human immunoglobulin A (IgA), IgG, IgM, and kappa and lambda antibodies (DAKOPATTS, Glostrup, Denmark) were added in a 1:1,000 dilution and incubated for 1 h at 37°C. The membrane was washed six times for 5 to 10 min at room temperature with washing buffer followed by detection with the ECL detection system (Amersham, Arlington Heights, Ill.) according to the manufacturer’s instructions.
In vitro transcription.
A plasmid containing the entire HBV genome (ayw subtype) was used for the production of DNA templates for the generation of HBV transcripts. Two primers were used. Primer 1 (5′-CCATCGAT-TAATACGACTCACTATAG-3′) contained a restriction site for ClaI (shown in italics), the T7 RNA polymerase promoter sequence (shown in boldface), and the sense HBV ayw DNA sequences (28) spanning nt 1243 to 1261 (5′-GAACCTTTTCGGCTCCTCT-3′). Primer 2 contained antisense HBV sequences from nt 1312 to 1333 (5′-GTCCCGATAATGTTTGCTCCAG-3′, RNA.B), 1317 to 1294 (5′-CTCCAGACCTGCTGCGAGCAAAAC-3′, RNA.C), and 1293 to 1276 (5′-AAGCGGCTAGGAGTTCCG-3′, RNA.D). For the generation of templates with mutations in the binding region of the RNA-binding proteins (stem-loop 2; see Fig. 1B and 6), the following primers containing antisense HBV sequences (nt 1293 to 1276) and nucleotide changes (shown in boldface and underlined) were used: 5′-AAGCGGATCTTAGTTCCG-3′ (RNA.D-M1), 5′-AAGCTTCTAGGAGTTCTT-3′ (RNA.D-M2), 5′-AAGCGGATAGGAGTTCCG-3′ (RNA.D-M3), 5′-AAGCGACTAGGAGTTCCG-3′ (RNA.D-M4), 5′-AAGCGGCTAGGAGTTCCG-3′ (RNA.D-M5), and 5′-AAGCGGCTAGGCGTTCCG-3′ (RNA.D-M6). RNA.E is a synthetic oligoribonucleotide spanning HBV nt 1243 to 1281 (5′-GAACCUUUUCGGCUCCUCUGCCGAUCCAUACUGCGGAAC-3′, produced by Oligos Etc., Wilsonville, Oreg.). The mouse β-actin template was generated by using the following primers. Primer 1 included a ClaI site and a T7 RNA polymerase promoter followed by β-actin-specific DNA sequences spanning nt 27 to 45 (5′-GGGCCGCTCTAGGCACCAA-3′) (2). Primer 2 contained antisense β-actin-specific sequence between nt 121 and 140 (5′-TGTTCAATGGGGTACTTCAG-3′) (2). The mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) template was generated by using the following primers. Primer 1 included a ClaI site and a T7 RNA polymerase promoter followed by GAPDH-specific DNA sequences spanning nt 383 to 401 (5′-GGAGCCAAACGGGTCATCA-3′) (60). Primer 2 contained antisense GAPDH-specific sequence between nt 478 and 497 (5′-TGCAGGATGCATTGCTGACA-3′) (60). The human immunodeficiency virus (HIV) Rev response element (RRE) template was generated by using the following primers. Primer 1 included a ClaI site and a T7 RNA polymerase promoter followed by RRE-specific DNA sequences spanning nt 1565 to 1583 (5′-GAGCAGTGGGAATAGGAGC-3′) (25); primer 2 contained antisense RRE-specific sequence spanning nt 1826 to 1819 (5′-TCCCTAGGAGCTGTTGAT-3′) (25). The Mason-Pfizer virus constitutive transport element (CTE) (52) template was generated by using the following primers. Primer 1 included a ClaI site and a T7 RNA polymerase promoter followed by Mason-Pfizer virus-specific DNA sequences spanning nt 8007 to 8025 (5′-CCTCCCCTCTGAGCTAGAC-3′) (64). Primer 2 contained antisense Mason-Pfizer virus-specific sequence between nt 8238 and 8221 (5′-AAGACATCATCCGGGCAG-3′) (64). PCRs for HBV, RRE, and CTE templates were produced with 1 ng of plasmid (plasmids containing specific RRE and CTE sequences were a generous gift from T. Hope). For the production of the mouse β-actin and GAPDH templates reverse-transcribed mouse liver RNA, the mixture contained 80 pmol of each primer in 1× PCR buffer; 0.2 mM GTP, ATP, TTP, and CTP; and 2.5 U of Taq DNA polymerase (Boehringer Mannheim). PCR was performed as follows: 5 min at 95°C, followed by 35 cycles of 1 min at 95°C, 1 min at 56°C, 1 min at 72°C, and 5 min at 72°C. The PCR products were purified by size exclusion with a commercial kit (PCR purification kit; Boehringer Mannheim), ethanol precipitated, and used as templates to generate transcripts. Transcription reactions were carried out with 0.5 to 1.0 μg of PCR product in a final volume of 20 μl in transcription buffer (Promega, Madison, Wis.) containing 0.31 mM ATP, CTP, and GTP; 7.5 mM [α-32P]UTP (800 Ci/mmol) (NEN, Boston, Mass.); 5 mM DTT; and 20 U of RNasin (Promega). The reaction was started by addition of 20 U of T7 RNA polymerase (Promega). After incubation for 45 min at 37°C, another 20 U of T7 RNA polymerase was added and the reaction was continued for 45 min at 37°C, another 20 U of T7 RNA polymerase was added and the reaction was continued for 45 min at 37°C. The reaction was terminated by adding 10 μg of yeast tRNA and 1 U of DNase I (Promega), and the reaction mixture incubated for 15 min at 37°C. After phenol-chloroform extraction and ethanol precipitation, transcripts were dissolved in 10 mM Tris-HCl (pH 7.4)–diethyl pyrocarbonate-treated water.
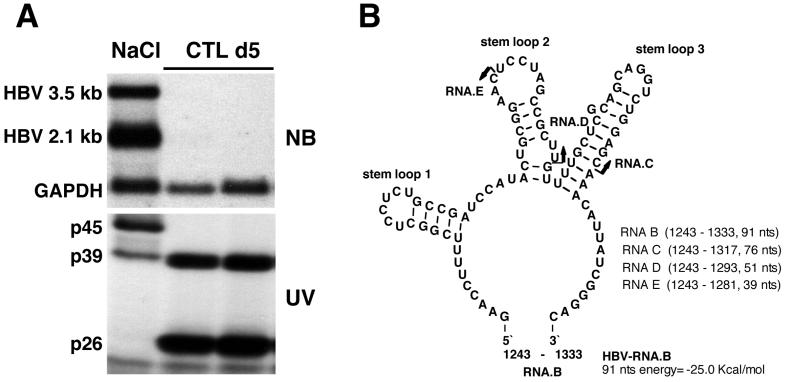
FIG. 1.
HBV RNA-binding proteins p45 and p39 are detectable in liver nuclear extracts from NaCl-injected mice, while p39 and p26 are detectable in CTL-injected mice. (A) Northern blotting and UV cross-linking analysis of 20 μg of total liver RNA or 5 μg of liver nuclear extract prepared from the same liver were performed as described in Materials and Methods. Sex and serum HBsAg-matched mice (lineage 1.3.32) were intravenously injected with 107 CTLs or with saline and sacrificed on day 5 after CTL administration. The upper panel shows the Northern blot analysis, and the lower panel shows the UV cross-linking analysis of nuclear extracts. (B) Predicted secondary structure of HBV in vitro transcript RNA.B used in this study. The secondary structure was calculated with the program MFOLD version 3 by Zuker and Turner available on the MFOLD server (71, 74). Arrows indicate the 3′ ends of in vitro transcripts RNA.C and RNA.D and of an oligoribonucleotide, RNA.E. The positions for all RNAs are shown according to the HBV ayw subtype sequence.
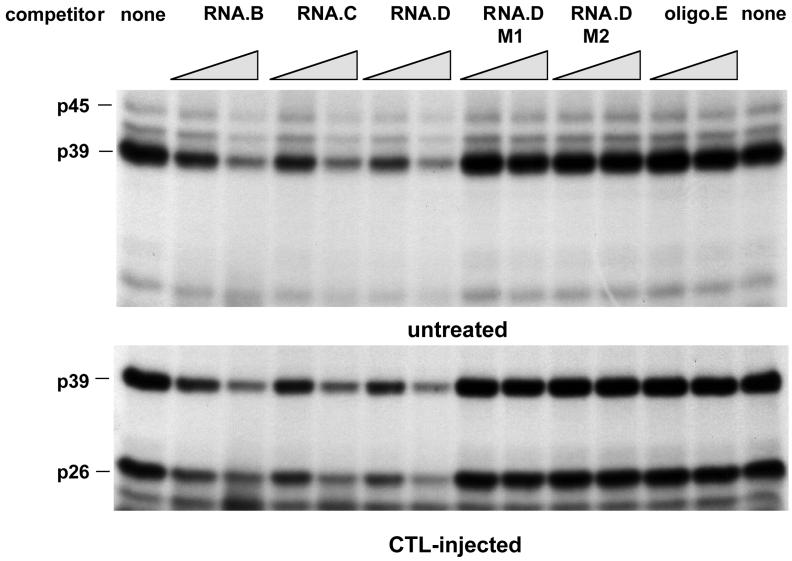
FIG. 6.
Mapping of RNA.B for a sequential-structural element recognized by p45, p39, and p26. UV cross-linking experiments were performed with liver nuclear extracts from untreated or CTL-injected mice under standard conditions described in Materials and Methods. A 30- and a 60-fold molar excess of unlabeled in vitro transcripts RNA.B, RNA.C, RNA.D, and RNA.E (Fig. 1B) were added into the binding reaction mixture prior to the addition of 40 fmol of 32P-labeled RNA.B.
UV cross-linking experiments.
Standard binding reactions were carried out in a final volume of 40 μl with 5 μg of total nuclear protein and 40 fmol of the 32P-labeled transcripts in binding buffer containing 10 mM Tris-HCl (pH 7.4), 3 mM MgCl2, 1.5 mM EDTA, 450 mM NaCl, 0.01% Triton X-100, 20 μg of yeast tRNA, and 6 μg of heparin for 20 min at room temperature. The reaction mixtures were incubated on ice, irradiated for 10 min with UV light (254 nm) in a Stratalinker (Stratagene, La Jolla, Calif.) approximately 3 cm under the bulbs, and then digested with 40 μg of RNase A and 100 U of RNase T1 for 45 min at 37°C. Forty microliters of SDS sample buffer (2% SDS, 5% mercaptoethanol, 63 mM Tris-HCl [pH 6.8], 10% glycerol, and 0.01% bromophenol blue) was added, and samples were boiled for 5 min, placed on ice, and resolved on an SDS–12.5% PAGE gel. After electrophoresis, the gels were stained with Coomassie blue, destained, dried, and exposed to Kodak Biomax overnight at −80°C. Competition experiments were carried out by addition of excess cold competitor to the binding reaction 3 min before or after the addition of the labeled transcript.
Dephosphorylation of nuclear proteins.
In the dephosphorylation reaction, 5 to 10 μg of nuclear extract was treated with 0.5 to 3 U of calf intestine alkaline phosphatase (CIAP) (Ambion, Austin, Tex.) for 30 min at 37°C in 20 μl of 1× dephosphorylation buffer (Ambion). Control assays were performed in the presence of reaction buffer but without CIAP; in addition, we showed that the strength of the RNA-protein complex signal after UV cross-linking was unchanged by CIAP. To rule out the possibility that CIAP dephosphorylates the bound 32P-labeled in vitro transcript RNA.B, we performed the following control experiment. After UV irradiation of the binding reaction mixture, the samples were digested with RNase and then treated with CIAP for 30 min at 37°C, and we showed that the strength of the RNA-protein complex signal after UV cross-linking was unchanged by CIAP treatment (data not shown).
RESULTS
Characteristics of nuclear HBV RNA-binding activities.
As shown in Fig. 1A, nuclear extracts prepared from HBV transgenic mouse liver contain three proteins that form RNA-protein complexes with a 91-nt 32P-labeled HBV transcript (designated RNA.B, shown in Fig. 1B) with apparent molecular masses of 45 kDa (p45), 39 kDa (p39), and 26 kDa (p26) in UV cross-linking experiments. In addition, an RNA-protein complex with an apparent molecular mass of 42 kDa was occasionally detected in variable amounts in all extracts containing p45. Note that p45 activity is constitutively present in the liver, as are the overlapping 3.5- and 2.1-kb HBV transcripts, both of which contain RNA.B, and it disappears following CTL injection, concomitant with the appearance of p26 and the disappearance of HBV RNA. In contrast, p39 is present under both conditions, appearing to be induced following CTL injection in this experiment. The predicted secondary structure of RNA.B is shown in Fig. 1B.
Molecular characterization of p45, p39, and p26.
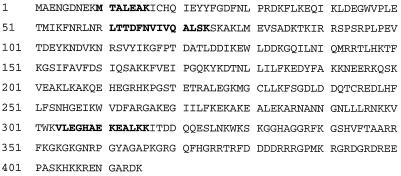
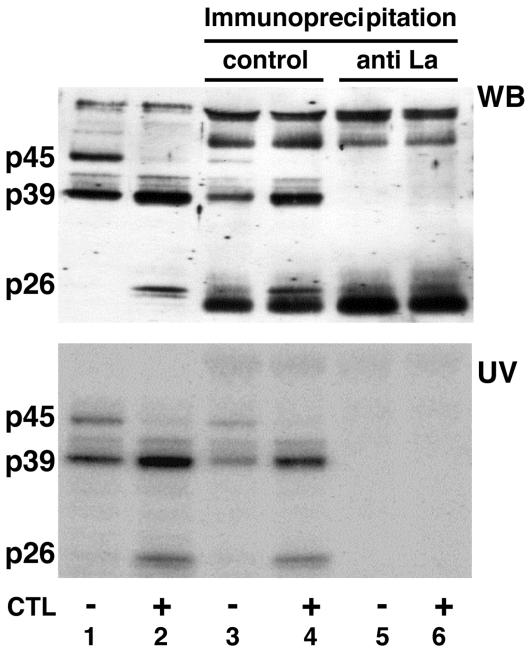
In order to characterize the HBV RNA-binding proteins at the molecular level, we subjected them to ammonium sulfate precipitation and heparin-affinity, ion-exchange, and molecular-exclusion chromatography (see Materials and Methods). Duplicate samples of gel filtration fractions containing p39 were separated by SDS-PAGE and subjected to UV cross-linking and Coomassie blue staining to locate the precise position of p39. The Coomassie blue-stained protein band corresponding to the mobility of p39 detected by UV cross-linking was cut out of the gel and digested with trypsin. After HPLC separation of the tryptic peptides, several peptides were subjected to N-terminal sequencing in the Scripps Research Institute Molecular Biology Core Facility. Three peptide sequences showed striking homology to the mouse La protein sequence (Fig. 2), identifying p39 as La protein (68). The mouse La protein has an apparent molecular mass of 47.7 kDa (68), and it is known that the La protein prepared from rabbit thymus and calf thymus was protease sensitive and displayed distinct 39- and 26-kDa cleavage products after repeated freezing and thawing or incubation of the samples at 37°C (14, 16, 37). Hence, we assayed whether p45 might be the full-length La protein while p39 and p26 might be proteolytic cleavage products of La, by attempting to deplete them from liver nuclear extracts with an anti-La antiserum. Figure 3 shows the Western blot (top) and UV cross-linking (bottom) results obtained with immunodepleted nuclear extracts analyzed on the same membrane. Lanes 1 and 2 demonstrate that similar patterns of proteins are detected by Western blotting and by UV cross-linking, with the exception of two high-molecular-weight protein bands that were detected only by Western blotting. Importantly, all three HBV RNA-binding proteins were depleted from the nuclear extracts by the anti-La antiserum (Fig. 3, lanes 5 and 6) but not by the control serum (Fig. 3, lanes 3 and 4). These results suggest that p45 is the full-length La protein while p39 is a constitutively detectable proteolytic product and p26 is a La proteolytic product that was induced following CTL injection.
FIG. 2.
Sequence analysis of tryptic peptides obtained from p39 revealed 100% homology to the mouse La protein. p39 was purified and processed as described in Materials and Methods. The sequence tags observed by N-terminal sequencing of three tryptic peptides obtained from p39 are shown in boldface.
FIG. 3.
HBV RNA-binding proteins are recognized by anti-La-positive human serum. Five micrograms of liver nuclear extract prepared from untreated or CTL-injected mice (lanes 1 and 2) and 5 μg of liver nuclear extract from untreated or CTL-injected mice after immunoprecipitation with control human serum (lanes 3 and 4) or with anti-La-positive human serum (lanes 5 and 6) were incubated with 40 fmol of in vitro-labeled RNA.B. The UV cross-linking reaction was performed as described in Materials and Methods. The gel was transferred to a nitrocellulose membrane and analyzed for La protein by Western blotting (WB) and by autoradiography to detect p45, p39, and p26 as described in Materials and Methods.
Binding of p45, p39, and p26 to HBV RNA is phosphorylation dependent.
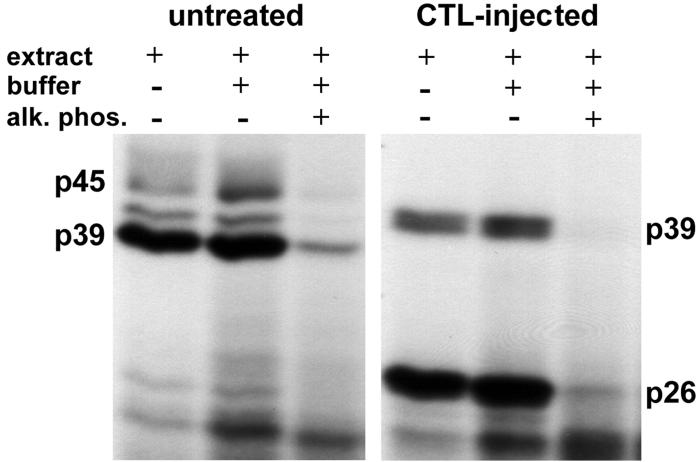
Since IFN-γ and TNF-α, induced in the liver following CTL injection and other intrahepatic inflammatory processes (12, 13, 34), mediate the switch from p45 to p26 (40), and since these cytokines are known to activate cellular kinases at a proximal step in their signal transduction cascade, we assayed whether the phosphorylation status of these proteins might influence their RNA-binding activity by treating the nuclear extracts with CIAP before the binding reaction was performed. As shown in Fig. 4, the RNA-binding activity of p45, p39, and p26 was strongly reduced after dephosphorylation. Since CIAP can dephosphorylate nucleic acids, control experiments were performed to see if the UV cross-linked labeled RNA was dephosphorylated by the phosphatase, thereby artifactually reducing the signal from the RNA-protein complex. No change in signal intensity was observed in this control experiment (data not shown). While these results suggest that the RNA-binding activity of these proteins is regulated by phosphorylation and dephosphorylation, it remains to be determined whether specific protein phosphatases and/or kinases actually regulate the RNA-binding activity of p45, p39, and p26 in vivo.
FIG. 4.
RNA-binding activity of p45, p39, and p26 depends on their phosphorylation status. Two micrograms of liver nuclear extract from untreated or CTL-injected mice was treated with 0.5 and 1.0 U of CIAP (alk. phos.) prior to addition of 40 fmol of 32P-labeled in vitro transcript RNA.B. The dephosphorylation reaction was performed in 20 μl of 1× reaction buffer for 30 min at 37°C. The binding reaction was performed and analyzed as described in Materials and Methods.
Affinity and specificity of protein binding to RNA.
Experiments were performed to determine the binding affinity and specificity of the RNA-binding proteins for RNA.B. First, we added increasing amounts of nuclear extract to two different concentrations of RNA.B to determine the optimal protein concentration required for the titration of RNA.B. UV cross-linking results were analyzed by phosphorimaging with arbitrary units to quantitate the ribonucleoprotein complexes formed at varying nuclear extract concentrations. We observed a linear increase in RNA binding at nuclear extract concentrations between 0.5 and 4 μg per reaction mixture (data not shown). Based on these results, 0.5 and 2.0 μg of nuclear extract were used to measure the binding affinity of p45, p39, and p26 at increasing RNA.B concentrations. UV cross-linking results were analyzed by phosphorimaging to quantitate RNA-protein complex formation at increasing RNA.B concentrations. The apparent Kd was calculated according to the mass action equation (44): Kd = [r] [p]/[c], where [r], [p], and [c] are the molar concentrations of free RNA, protein, and complex, respectively, assuming that the total RNA concentration (Rt) is much higher than the total protein concentration (Pt) (Rt − [c]) ≈ Rt. The final equation was 1/[c] = (Kd/Pt) (1/Rt) + (1/Pt). Plotting 1/[c] (1/arbitrary unit) versus 1/Rt (1/RNA.B concentration) generated a straight line with the apparent Kd defined as the point of intersection with the x axis. By this method, we determined the apparent Kds as 1.4 to 1.5 nM for p45, 0.4 to 1.1 nM for p39, and 0.9 to 1.0 nM for p26.
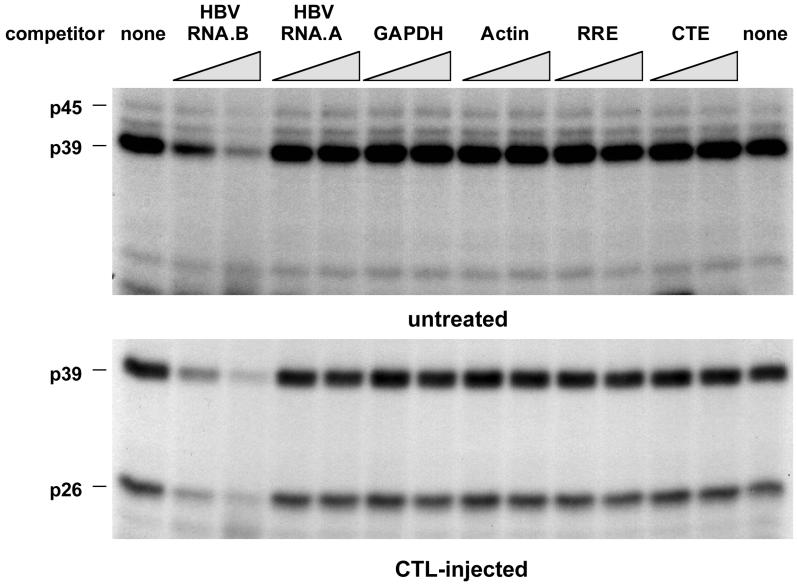
In order to establish the binding specificity of the RNA-binding proteins, competition experiments were performed with unlabeled transcripts, including RNA.B and several other transcripts derived from an AU-rich region of HBV (nt 767 to 870) designated RNA.A or from mouse GAPDH (nt 383 to 497 [60]), mouse β-actin (nt 27 to 140 [2]), the HIV RRE (nt 1565 to 1826 [25]; a generous gift from T. Hope), and the Mason-Pfizer monkey virus CTE (nt 8007 to 8238 [64], also a generous gift from T. Hope). All experiments were done with a 10- and a 30-fold molar excess of unlabeled competitors. As shown in Fig. 5, unlabeled RNA.B inhibited the binding of p45, p39, and p26 to labeled RNA.B in a concentration-dependent manner while the other transcripts did not, indicating that the binding interaction is specific.
FIG. 5.
Competition of various in vitro transcripts with RNA.B for binding of p45, p39, and p26. UV cross-linking experiments were performed with liver nuclear extracts from untreated or CTL-injected mice under standard conditions described in Materials and Methods. A 30- and a 60-fold molar excess of unlabeled in vitro transcripts RNA.B, GAPDH, actin, RRE, and CTE were added into the binding reaction mixture prior to the addition of 40 fmol of 32P-labeled RNA.B.
To map the La-binding domain within the 91-nt HBV RNA.B element more precisely, additional competition experiments were performed with in vitro transcripts RNA.C (nt 1243 to 1317) and RNA.D (nt 1243 to 1293) and an RNA oligonucleotide (RNA.E, nt 1243 to 1281) representing 3′ deletions of RNA.B (Fig. 1B). As shown in Fig. 6, RNA.B, RNA.C, and RNA.D inhibited the binding of p45, p39, and p26 to labeled RNA.B in a concentration-dependent manner, while RNA.E did not compete. These results suggest that a sequence or structural element between nt 1275 and 1291 (i.e., stem-loop 2 in Fig. 1B) was recognized by these RNA-binding proteins.
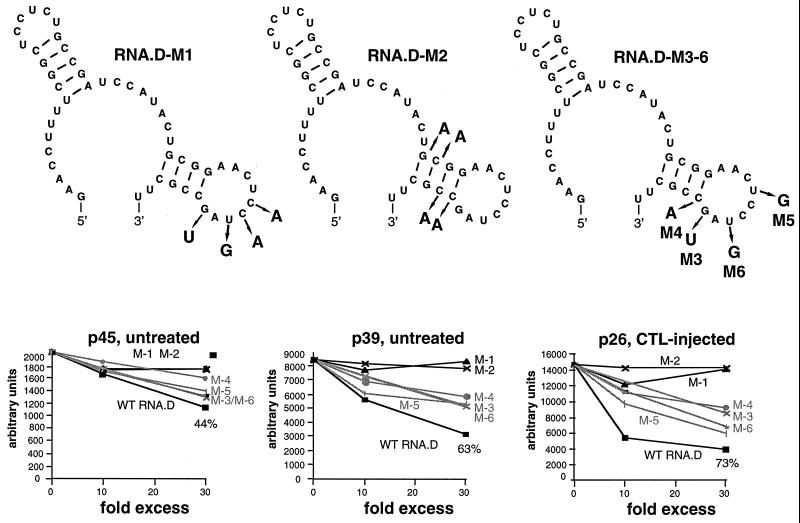
The secondary structure of RNA.D (nt 1243 to 1293) predicted by using MFOLD version 3 by Zuker and Turner (71, 74) is shown in Fig. 7. The 3′ stem-loop was of interest since it is included in RNA.C and RNA.D, both of which compete for the binding of RNA.B, but not in RNA.E, which does not. Therefore, we introduced two sets of mutations into the template for the in vitro transcription of RNA.D, designated RNA.D-M1 (containing mutations in the predicted loop) and RNA.D-M2 (containing mutations in the predicted stem) in Fig. 6 and 7. As shown in Fig. 6, neither of these mutant homologues of RNA.D was able to inhibit the binding of p45, p39, or p26 to RNA.B. Finally, we introduced single mutations into the RNA.D template, creating RNA.D-M3, -M4, -M5, and -M6, shown in Fig. 7. The ability of a 10- and a 30-fold molar excess of unlabeled RNA.D and mutant RNA.D homologues to inhibit the binding of p45, p39, and p26 to labeled RNA.B was assessed in competitive UV cross-linking experiments and analyzed by phosphorimaging. As shown in Fig. 7, the relative signal intensities of p45, p39, and p26 (expressed in arbitrary units) plotted against the relative concentrations of competitor indicate that single nucleotide substitutions in RNA.D-M3, -M4, -M5, and -M6 reduced their ability to inhibit the binding of p26, p39, and p45. These results suggest that both the structure and the sequence of stem-loop 2 are probably important for its recognition by the RNA-binding proteins.
FIG. 7.
Sequential and/or structural features of stem-loop 2 are substantive for the binding of p45, p39, and p26. UV cross-linking experiments were performed with liver nuclear extracts from untreated or CTL-injected mice under standard conditions described in Materials and Methods. A 10- and a 30-fold molar excess of unlabeled in vitro transcripts RNA.D-M1 to RNA.D-M6 were added into the binding reaction mixture prior to the addition of 40 fmol of 32P-labeled RNA.B. The decreases in signal intensity for p45, p39, and p26 were separately analyzed by phosphorimaging. The upper panel shows the predicted structure of RNA.D with nucleotide changes indicated by the arrows. The lower panel shows the plot of complex formation (arbitrary units) versus competitor concentrations (10- and 30-fold). WT, wild type.
DISCUSSION
Intrahepatic inflammatory processes characterized by the production of IFN-γ and TNF-α downregulate HBV gene expression in the livers of transgenic mice (12, 13, 29–34, 36, 69) by a posttranscriptional mechanism (69) that could contribute to viral clearance during HBV infection. To begin to define this mechanism, we have recently shown that liver cell nuclei contain a family of three proteins that bind to a 91-nt element (RNA.B) located at the 5′ end of the HBV posttranscriptional regulatory element (40). Furthermore, we suggested that these proteins might contribute to HBV RNA stability, since they are regulated by the same cytokines that destabilize the viral transcripts and because their relative abundance is tightly linked to the presence or absence of HBV RNA (40).
In the studies reported herein, we demonstrate that the HBV RNA-binding proteins p45, p39, and p26 are recognized and depleted from nuclear extracts by anti-La antibodies. These results strongly suggest that p45 is the full-length mouse La protein, that p39 is a constitutive proteolytic cleavage product of p45, and that p26 is generated from p45 in an IFN-γ- and/or a TNF-α-dependent manner. The La protein is a well-described RNA-binding protein (15, 16, 45, 70) that binds to poly(U)-rich elements in RNA polymerase III transcripts (tRNA precursors and 5S RNA) as well as several other cellular and viral RNAs (1, 66, 70). La appears to be necessary for the processing of tRNA (73); stimulates translation of poliovirus (10, 48) and hepatitis C virus RNA (1); has helicase activity (8); and seems to be translocated from the nucleus to the cytoplasm during cell stress secondary to virus infection (5, 6), transformation (59), and UV irradiation (7). Recently, the La protein was reported to stabilize histone mRNA (47). In addition, IFN-γ and TNF-α have been shown to induce membrane expression of La in cells (22, 24). The La protein carries a nuclear localization sequence and a nuclear retention signal (63), and it coprecipitates with certain viral RNAs (38). La is a phosphoprotein (9, 27, 53, 54) that binds RNA in a phosphorylation-dependent manner in some (9, 53) but not all (27) of the systems in which it has been studied. In the present study, we demonstrated that the ability of all three La proteins to bind HBV RNA is phosphorylation dependent. Since these experiments were performed with crude nuclear extracts, we do not know whether phosphorylation of La itself or that of other accessory molecules is required for the binding interaction to occur. This should be clarified in future experiments with purified recombinant La proteins instead of nuclear extracts. Such studies will also allow more precise measurement of the binding affinity of La for HBV RNA in the absence of possible cellular cofactors.
It is important to note that the RNA-binding activity of p45, p39, and p26 La depends on the phosphorylation status of the nuclear extracts used (Fig. 4) and that the disappearance of p45 and the appearance of p26 are regulated by IFN-γ and TNF-α (40). Therefore, the activation of signal transduction pathways by IFN-γ and TNF-α following CTL injection could reflect a phosphorylation-dependent proteolytic cleavage of p45 into a 26-kDa RNA-binding fragment and one or more fragments that are unable to bind HBV RNA. Others have shown that La is sensitive to proteolytic cleavage, yielding several cleavage products similar in size to p39 and p26 (14, 16, 37).
In this study, the specificity of the interaction of p45, p39, and p26 with the 91-nt HBV RNA.B target element was confirmed in the competition experiments shown in Fig. 5 to 7. Importantly, mutational analysis of RNA.B revealed that all three RNA-binding proteins recognize a single 17-bp target element, located between nt 1275 and 1291 (Fig. 7). Interestingly, this element displays a predicted stem-loop structure that appears to be recognized by all three proteins, since mutational disruption of the predicted stem (RNA.D-M2) abolished their ability to bind the RNA. Similarly, binding was reduced by point mutations in the loop (RNA.D-M1), suggesting that the proteins display sequence specificity as well as structural specificity for their substrate. Additional experiments with recombinant La protein and mutant RNA substrates containing single nucleotide mutations in the loop and other substrates containing compensatory mutations that maintain the structure of the stem will be necessary to further define the nature of the binding site(s) within the substrate.
In summary, we have previously shown that a close relationship exists between the presence of three HBV RNA-binding proteins (p45, p39, and p26) and the abundance of HBV RNA in the livers of HBV transgenic mice (40). The current results demonstrate that all three proteins are related to the cellular nucleoprotein, La, and that p45 is probably the full-length protein while p39 and p26 are constitutive and cytokine-inducible proteolytic cleavage products, respectively. We also demonstrate that all three La isoforms bind the same predicted stem-loop in HBV RNA between nt 1275 and 1291 with high affinity in a phosphorylation-dependent manner. These results suggest that conditions, such as inflammation, that alter the content, metabolism, and distribution of La in the hepatocyte may contribute to the posttranscriptional control of the steady-state content of HBV RNA and, thereby, influence the outcome of HBV infection.
ACKNOWLEDGMENTS
We thank Edward K. Chan (The Scripps Research Institute, La Jolla, Calif.) for providing anti-La human antiserum; Joel Gottesfeld (The Scripps Research Institute) for consultations and advice; Thomas J. Hope (Salk Institute, La Jolla, Calif.) for providing plasmids carrying the HIV RRE and Mason-Pfizer monkey virus CTEs; the Scripps Molecular Biology Core Facility for the production of oligonucleotides; the Scripps Protein and Nucleic Acid Core Facility for tryptic digestion of the proteins, HPLC purification, and N-terminal sequencing of peptides; and Jennifer Newmann for help with manuscript preparation.
This work was supported by grants R37-CA40489 and R01-AI40696 from the National Institutes of Health.
Footnotes
Paper no. 11631-MEM from The Scripps Research Institute.
REFERENCES
- 1.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso S, Minty A, Bourlet Y, Buckingham M. Comparison of three actin-coding sequences in the mouse; evolutionary relationships between the actin genes of warm-blooded vertebrates. J Mol Evol. 1986;23:11–22. doi: 10.1007/BF02100994. [DOI] [PubMed] [Google Scholar]
- 3.Ando K, Guidotti L G, Cerny A, Ishikawa T, Chisari F V. CTL access to tissue antigen is restricted in vivo. J Immunol. 1994;153:482–488. [PubMed] [Google Scholar]
- 4.Ando K, Moriyama T, Guidotti L G, Wirth S, Schreiber R D, Schlicht H J, Huang S N, Chisari F V. Mechanisms of class I restricted immunopathology. A transgenic mouse model of fulminant hepatitis. J Exp Med. 1993;178:1541–1554. doi: 10.1084/jem.178.5.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baboonian C, Venables P J, Booth J, Williams D G, Roffe L M, Maini R N. Virus infection induces redistribution and membrane localization of the nuclear antigen La (SS-B): a possible mechanism for autoimmunity. Clin Exp Immunol. 1989;78:454–459. [PMC free article] [PubMed] [Google Scholar]
- 6.Bachmann M, Althoff H, Troster H, Selenka C, Falke D, Muller W E. Translocation of the nuclear autoantigen La to the cell surface of herpes simplex virus type 1 infected cells. Autoimmunity. 1992;12:37–45. doi: 10.3109/08916939209146128. [DOI] [PubMed] [Google Scholar]
- 7.Bachmann M, Chang S, Slor H, Kukulies J, Muller W E. Shuttling of the autoantigen La between nucleus and cell surface after uv irradiation of human keratinocytes. Exp Cell Res. 1990;191:171–180. doi: 10.1016/0014-4827(90)90002-r. [DOI] [PubMed] [Google Scholar]
- 8.Bachmann M, Pfeifer K, Schroder H C, Muller W E. Characterization of the autoantigen La as a nucleic acid-dependent ATPase/dATPase with melting properties. Cell. 1990;60:85–93. doi: 10.1016/0092-8674(90)90718-t. [DOI] [PubMed] [Google Scholar]
- 9.Bachmann M, Schroder H C, Wagner K G, Mayet W J, Pfeifer K, Muller W E. Purification and characterization of the Ro and La antigens. Modulation of their binding affinities to poly(U) by phosphorylation and the presence of ATP. Biol Chem Hoppe-Seyler. 1986;367:671–680. doi: 10.1515/bchm3.1986.367.2.671. [DOI] [PubMed] [Google Scholar]
- 10.Belsham G J, Sonenberg N, Svitkin Y V. The role of the La autoantigen in internal initiation. Curr Top Microbiol Immunol. 1995;203:85–98. doi: 10.1007/978-3-642-79663-0_4. [DOI] [PubMed] [Google Scholar]
- 11.Carpousis A J, Van Houwe G, Ehretsmann C, Krisch H M. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 12.Cavanaugh V J, Guidotti L G, Chisari F V. Inhibition of hepatitis B virus replication during adenovirus and cytomegalovirus infections in transgenic mice. J Virol. 1998;72:2630–2637. doi: 10.1128/jvi.72.4.2630-2637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavanaugh V J, Guidotti L G, Chisari F V. Interleukin-12 inhibits hepatitis B virus replication in transgenic mice. J Virol. 1997;71:3236–3243. doi: 10.1128/jvi.71.4.3236-3243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan E K, Francoeur A M, Tan E M. Epitopes, structural domains, and asymmetry of amino acid residues in SS-B/La nuclear protein. J Immunol. 1986;136:3744–3749. [PubMed] [Google Scholar]
- 15.Chan E K, Sullivan K F, Tan E M. Ribonucleoprotein SS-B/La belongs to a protein family with consensus sequences for RNA-binding. Nucleic Acids Res. 1989;17:2233–2244. doi: 10.1093/nar/17.6.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan E K, Tan E M. The small nuclear ribonucleoprotein SS-B/La binds RNA with a conserved protease-resistant domain of 28 kilodaltons. Mol Cell Biol. 1987;7:2588–2591. doi: 10.1128/mcb.7.7.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen F Y, Amara F M, Wright J A. Mammalian ribonucleotide reductase R1 mRNA stability under normal and phorbol ester stimulating conditions: involvement of a cis-trans interaction at the 3′ untranslated region. EMBO J. 1993;12:3977–3986. doi: 10.1002/j.1460-2075.1993.tb06075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chisari F V. Cytotoxic T cells and viral hepatitis. J Clin Investig. 1997;99:1472–1477. doi: 10.1172/JCI119308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chisari F V, Ferrari C. Hepatitis B virus immunopathology. Springer Semin Immunopathol. 1995;17:261–281. doi: 10.1007/BF00196169. [DOI] [PubMed] [Google Scholar]
- 20.Chisari F V, Filippi P, McLachlan A, Milich D R, Riggs M, Lee S, Palmiter R D, Pinkert C A, Brinster R L. Expression of hepatitis B virus large envelope polypeptide inhibits hepatitis B surface antigen secretion in transgenic mice. J Virol. 1986;60:880–887. doi: 10.1128/jvi.60.3.880-887.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chisari F V, Pinkert C A, Milich D R, Filippi P, McLachlan A, Palmiter R D, Brinster R L. A transgenic mouse model of the chronic hepatitis B surface antigen carrier state. Science. 1985;230:1157–1160. doi: 10.1126/science.3865369. [DOI] [PubMed] [Google Scholar]
- 22.Clark D A, Lamey P J, Jarrett R F, Onions D E. A model to study viral and cytokine involvement in Sjogren’s syndrome. Autoimmunity. 1994;18:7–14. doi: 10.3109/08916939409014674. [DOI] [PubMed] [Google Scholar]
- 23.Donello J E, Beeche A A, Smith G J, Lucero G R, Hope T J. The hepatitis B virus posttranscriptional regulatory element is composed of two subelements. J Virol. 1996;70:4345–4351. doi: 10.1128/jvi.70.7.4345-4351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorner T, Hucko M, Mayet W J, Trefzer U, Burmester G R, Hiepe F. Enhanced membrane expression of the 52 kDa Ro(SS-A) and La(SS-B) antigens by human keratinocytes induced by TNF alpha. Ann Rheum Dis. 1995;54:904–909. doi: 10.1136/ard.54.11.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duchet S, Letourneur F, Loussert-Ajaka I, Chaplain C, Gomas E, Brun-Vezinet F, Simon F, Saragosti S. gag and env sequences of an A/G/H recombinant from a Zairian HIV type 1 isolate. AIDS Res Hum Retroviruses. 1997;13:1351–1354. doi: 10.1089/aid.1997.13.1351. [DOI] [PubMed] [Google Scholar]
- 26.Eisenstein R S, Tuazon P T, Schalinske K L, Anderson S A, Traugh J A. Iron-responsive element-binding protein. Phosphorylation by protein kinase C. J Biol Chem. 1993;268:27363–27370. [PubMed] [Google Scholar]
- 27.Fan H, Sakulich A L, Goodier J L, Zhang X, Qin J, Maraia R J. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell. 1997;88:707–15. doi: 10.1016/s0092-8674(00)81913-3. [DOI] [PubMed] [Google Scholar]
- 28.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 29.Gilles P N, Fey G, Chisari F V. Tumor necrosis factor alpha negatively regulates hepatitis B virus gene expression in transgenic mice. J Virol. 1992;66:3955–3960. doi: 10.1128/jvi.66.6.3955-3960.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guidotti L G, Ando K, Hobbs M V, Ishikawa T, Runkel L, Schreiber R D, Chisari F V. Cytotoxic T lymphocytes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice. Proc Natl Acad Sci USA. 1994;91:3764–3768. doi: 10.1073/pnas.91.9.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guidotti L G, Borrow P, Hobbs M V, Matzke B, Gresser I, Oldstone M B, Chisari F V. Viral cross talk: intracellular inactivation of the hepatitis B virus during an unrelated viral infection of the liver. Proc Natl Acad Sci USA. 1996;93:4589–4594. doi: 10.1073/pnas.93.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guidotti L G, Chisari F V. To kill or to cure: options in host defense against viral infection. Curr Opin Immunol. 1996;8:478–483. doi: 10.1016/s0952-7915(96)80034-3. [DOI] [PubMed] [Google Scholar]
- 33.Guidotti L G, Guilhot S, Chisari F V. Interleukin-2 and alpha/beta interferon down-regulate hepatitis B virus gene expression in vivo by tumor necrosis factor-dependent and -independent pathways. J Virol. 1994;68:1265–1270. doi: 10.1128/jvi.68.3.1265-1270.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guidotti L G, Ishikawa T, Hobbs M V, Matzke B, Schreiber R, Chisari F V. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity. 1996;4:25–36. doi: 10.1016/s1074-7613(00)80295-2. [DOI] [PubMed] [Google Scholar]
- 35.Guidotti L G, Matzke B, Schaller H, Chisari F V. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guilhot S, Guidotti L G, Chisari F V. Interleukin-2 downregulates hepatitis B virus gene expression in transgenic mice by a posttranscriptional mechanism. J Virol. 1993;67:7444–7449. doi: 10.1128/jvi.67.12.7444-7449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Habets W J, den Brok J H, Boerbooms A M, van de Putte L B, van Venrooij W J. Characterization of the SS-B (La) antigen in adenovirus-infected and uninfected HeLa cells. EMBO J. 1983;2:1625–1631. doi: 10.1002/j.1460-2075.1983.tb01636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamelin R, Chan E K, Tan E M, Arlinghaus R B. Antibodies against small nuclear ribonucleoproteins immunoprecipitate complexes containing ts110 Moloney murine sarcoma virus genomic and messenger RNAs. Virology. 1986;152:87–99. doi: 10.1016/0042-6822(86)90374-0. [DOI] [PubMed] [Google Scholar]
- 39.Harford J B, Morris D R. mRNA metabolism & post-transcriptional gene regulation. New York, N.Y: Wiley-Liss; 1997. [Google Scholar]
- 39a.Heise, T. Unpublished data.
- 40.Heise T, Guidotti L G, Cavanaugh V J, Chisari F V. Hepatitis B virus RNA-binding proteins associated with cytokine-induced clearance of viral RNA from the liver of transgenic mice. J Virol. 1999;73:474–481. doi: 10.1128/jvi.73.1.474-481.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heise T, Krones A, Nath A, Jungermann K, Christ B. Parallel acelleration of phosphoenolpyruvate carboxykinase mRNA degradation and increase in ribonuclease activity induced by insulin in cultured rat hepatocytes. Biol Chem Hoppe-Seyler. 1998;379:875–883. doi: 10.1515/bchm.1998.379.7.875. [DOI] [PubMed] [Google Scholar]
- 42.Huang J, Liang T J. A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol Cell Biol. 1993;13:7476–7486. doi: 10.1128/mcb.13.12.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Izaurralde E, Mattaj I W. RNA export. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- 44.Lehninger A L. Biochemistry. 2nd ed. New York, N.Y: Worth Publishers; 1975. pp. 1152–1154. [Google Scholar]
- 45.Lerner M R, Boyle J A, Hardin J A, Steitz J A. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science. 1981;211:400–402. doi: 10.1126/science.6164096. [DOI] [PubMed] [Google Scholar]
- 46.Malter J S, Hong Y. A redox switch and phosphorylation are involved in the post-translational up-regulation of the adenosine-uridine binding factor by phorbol ester and ionophore. J Biol Chem. 1991;266:3167–3171. [PubMed] [Google Scholar]
- 47.McLaren R S, Caruccio N, Ross J. Human La protein: a stabilizer of histone mRNA. Mol Cell Biol. 1997;17:3028–3036. doi: 10.1128/mcb.17.6.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miczak A, Kaberdin V R, Wei C L, Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci USA. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moriyama T, Guilhot S, Klopchin K, Moss B, Pinkert C A, Palmiter R D, Brinster R L, Kanagawa O, Chisari F V. Immunobiology and pathogenesis of hepatocellular injury in hepatitis B virus transgenic mice. Science. 1990;248:361–364. doi: 10.1126/science.1691527. [DOI] [PubMed] [Google Scholar]
- 51.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 52.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjold M L, Dahlberg J E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfeifle J, Anderer F A, Franke M. Multiple phosphorylation of human SS-B/LA autoantigen and its effect on poly(U) and autoantibody binding. Biochim Biophys Acta. 1987;928:217–226. doi: 10.1016/0167-4889(87)90124-8. [DOI] [PubMed] [Google Scholar]
- 54.Pizer L I, Deng J S, Stenberg R M, Tan E M. Characterization of a phosphoprotein associated with the SS-B/La nuclear antigen in adenovirus-infected and uninfected KB cells. Mol Cell Biol. 1983;3:1235–1245. doi: 10.1128/mcb.3.7.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Py B, Causton H, Mudd E A, Higgins C F. A protein complex mediating mRNA degradation in Escherichia coli. Mol Microbiol. 1994;14:717–729. doi: 10.1111/j.1365-2958.1994.tb01309.x. [DOI] [PubMed] [Google Scholar]
- 56.Py B, Higgins C F, Krisch H M, Carpousis A J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 57.Rao K S, Sirdeshmukh R, Gupta P D. Modulation of cytosolic RNase activity by endogenous RNase inhibitor in rat vaginal epithelial cells on estradiol administration. FEBS Lett. 1994;343:11–14. doi: 10.1016/0014-5793(94)80597-0. [DOI] [PubMed] [Google Scholar]
- 58.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rother R P, Thomas P S. La/SSB ribonucleoprotein levels increased in transformed cells. Clin Exp Immunol. 1991;83:369–374. doi: 10.1111/j.1365-2249.1991.tb05645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sabath D E, Broome H E, Prystowsky M B. Glyceraldehyde-3-phosphate dehydrogenase mRNA is a major interleukin 2-induced transcript in a cloned T-helper lymphocyte. Gene. 1990;91:185–191. doi: 10.1016/0378-1119(90)90087-8. [DOI] [PubMed] [Google Scholar]
- 61.Schaller H, Fischer M. Transcriptional control of hepadnavirus gene expression. Curr Top Microbiol Immunol. 1991;168:21–39. doi: 10.1007/978-3-642-76015-0_2. [DOI] [PubMed] [Google Scholar]
- 62.Sharp P A. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 63.Simons F H, Broers F J, Van Venrooij W J, Pruijn G J. Characterization of cis-acting signals for nuclear import and retention of the La (SS-B) autoantigen. Exp Cell Res. 1996;224:224–236. doi: 10.1006/excr.1996.0132. [DOI] [PubMed] [Google Scholar]
- 64.Sonigo P, Barker C, Hunter E, Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986;45:375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- 65.Standart N, Jackson R J. Regulation of translation by specific protein/mRNA interactions. Biochimie. 1994;76:867–879. doi: 10.1016/0300-9084(94)90189-9. [DOI] [PubMed] [Google Scholar]
- 66.Stefano J E. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–154. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- 67.St. Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 68.Topfer F, Gordon T, McCluskey J. Characterization of the mouse autoantigen La (SS-B). Identification of conserved RNA-binding motifs, a putative ATP binding site and reactivity of recombinant protein with poly(U) and human autoantibodies. J Immunol. 1993;150:3091–3100. [PubMed] [Google Scholar]
- 69.Tsui L V, Guidotti L G, Ishikawa T, Chisari F V. Posttranscriptional clearance of hepatitis B virus RNA by cytotoxic T lymphocyte-activated hepatocytes. Proc Natl Acad Sci USA. 1995;92:12398–12402. doi: 10.1073/pnas.92.26.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Venrooij W J, Slobbe R L, Pruijn G J. Structure and function of La and Ro RNPs. Mol Biol Rep. 1993;18:113–119. doi: 10.1007/BF00986765. [DOI] [PubMed] [Google Scholar]
- 71.Walter A E, Turner D H, Kim J, Lyttle M H, Muller P, Mathews D H, Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams D L, Sensel M, McTigue M, Binder R. Hormonal and developmental regulation of mRNA turnover. In: Belasco J, Brawermann G, editors. Control of messenger RNA stability. New York, N.Y: Academic Press, Inc.; 1993. pp. 161–197. [Google Scholar]
- 73.Yoo C J, Wolin S L. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]
- 74.Zuker, M., and D. H. Turner. 1995–1999, copyright date. [Online.] MFOLD, version 3. Michael Zuker, Washington University School of Medicine. http://mfold2.wustl.edu/∼mfold/rna/form1.cgi. [6 May 1999, last date accessed.]