Abstract
The biological activity of the human immunodeficiency virus type 1 (HIV-1) Tat (Tat1) transcriptional activator requires the recruitment of a Tat1-CyclinT1 (CycT1) complex to the TAR RNA target encoded within the viral long terminal repeat (LTR). While other primate immunodeficiency viruses, such as HIV-2 and mandrill simian immunodeficiency virus (SIVmnd), also encode Tat proteins that activate transcription via RNA targets, these proteins differ significantly, both from each other and from Tat1, in terms of their ability to activate transcription directed by LTR promoter elements found in different HIV and SIV isolates. Here, we show that CycT1 also serves as an essential cofactor for HIV-2 Tat (Tat2) and SIVmnd Tat (Tat-M) function. Moreover, the CycT1 complex formed by each Tat protein displays a distinct RNA target specificity that accurately predicts the level of activation observed with a particular LTR. While Tat2 and Tat-M share the ability of Tat1 to bind to CycT1, they differ from Tat1 in that they are also able to bind to the related but distinct CycT2. However, the resultant Tat-CycT2 complexes fail to bind TAR and are therefore abortive. Surprisingly, mutation of a single residue in CycT2 (asparagine 260 to cysteine) rescues the ability of CycT2 to bind Tat1 and also activates not only TAR binding by all three Tat-CycT2 complexes but also Tat function. Therefore, the RNA target specificity of different Tat-CycT1 complexes is modulated by natural sequence variation in both the viral Tat transcriptional activator and in the host cell CycT molecule recruited by Tat. Further, the RNA target specificity of the resultant Tat-CycT1 complex accurately predicts the ability of that complex to activate transcription from a given LTR promoter element.
Human immunodeficiency virus type 1 (HIV-1) encodes a transcriptional transactivator, here termed Tat1, that can dramatically enhance the level of expression of genes linked to the viral long terminal repeat (LTR) promoter element (for a review, see references 6 and 16). Unusual attributes of Tat1 include the fact that it acts predominantly at the level of transcription elongation, rather than initiation, and the finding that the cis-acting target for Tat1 is an RNA sequence rather than a DNA sequence (7, 9, 16, 17, 21). This RNA target, here termed TAR1, is a 57-nucleotide RNA stem-loop structure that serves to present two critical protein binding sites, i.e., a terminal hexanucleotide loop and a small U-rich bulge located immediately 5′ to this loop (Fig. 1). It has been demonstrated that the bulge serves as a binding site for Tat1, while the terminal loop is important for the recruitment to TAR1 of an essential Tat1 cofactor (7, 10, 24, 27).
FIG. 1.
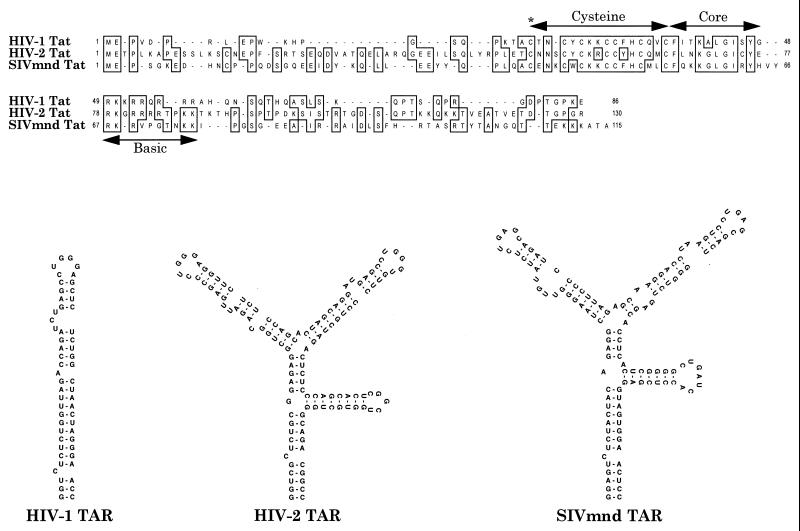
Comparison of primate immunodeficiency virus Tat proteins and TAR elements. A sequence alignment of the HIV-1 (strain IIIB), HIV-2 (strain ROD), and SIVmnd Tat proteins is shown (8, 26). While very different in terms of length, and in the more amino- and carboxyl-terminal sequences, all three proteins contain recognizable cysteine-rich, core, and basic domains, as indicated. A critical cysteine residue (C22 in Tat1) that is required for biological activity is indicated by an asterisk. The predicted secondary structures of the viral TAR elements suggest the presence of one Tat target site in HIV-1 TAR and duplicated target sites in both HIV-2 and SIVmnd TAR. The critical functional elements in TAR are the terminal hexanucleotide loops and the 5′ proximal bulges.
Mutational analysis of the HIV-1 Tat protein has identified two functional domains that are critical for transactivation of the HIV-1 LTR (18, 20). The Tat1 basic domain (Fig. 1) is important for TAR1 binding, while the Tat1 activation domain, which includes amino-terminal, cysteine, and core motifs, is critical for the recruitment of a cellular Tat1 cofactor (19, 20, 24, 27). Recently, it has become apparent that this cofactor is cyclin T1 (CycT1), a component of the cellular transcription elongation factor P-TEFb (22, 28, 30, 31). It is now known that Tat1 and CycT1 form a heterodimer that is then recruited to TAR1 (4, 13, 28). Once bound to TAR1, CycT1 and its associated proteins, particularly the CDK9 kinase, are then believed to modify the initiated RNA polymerase II (pol II) transcription complex to a more elongation-competent state by phosphorylation of the C-terminal domain of pol II (6, 14, 16, 28, 29, 31).
Evidence validating CycT1 as a critical cofactor for Tat1 function includes the demonstration that CycT1 can specifically bind to the Tat activation domain both in vitro and in vivo and that the resultant heterodimer has a far-higher affinity for TAR1 than either Tat1 or CycT1 alone (4, 13, 28). Indeed, evidence suggests that neither protein is able to bind TAR1 in vivo on its own (4, 20, 28). Convincing genetic evidence for the critical importance of CycT1 has come from the demonstration that expression of human CycT1 (hCycT1) in mouse cells can effectively rescue Tat1 function in these normally nonpermissive cells (4, 13, 28). Analysis of the related mouse CycT1 (mCycT1) protein has demonstrated that mCycT1 can bind Tat1 effectively but that the resultant mCycT1-Tat1 complex is only very inefficiently bound by TAR1, thus explaining the lack of Tat function in mouse cells (4). Remarkably, the substitution of a single residue in mCycT1 with the human equivalent (tyrosine 261 to cysteine) fully rescues both TAR binding and Tat function in mouse cells (4, 13). The inability of mouse cells to support Tat1 function in the absence of a coexpressed hCycT1 derivative has also allowed a preliminary definition of sequences in hCycT1 that are required to support Tat1 function. This analysis has demonstrated that the first 272 amino acids (aa) of the 726-aa hCycT1 protein are fully sufficient to support Tat function and have further defined a region in CycT1, located between residues 250 and 262, that is critical for Tat1 and/or TAR1 binding in vitro and that has been termed the Tat-TAR recognition motif (TRM) (13).
While the essential role of CycT1 in mediating HIV-1 Tat function is now established, it has remained unclear whether CycT1 plays a similar role in mediating the activity of Tat proteins derived from other primate immunodeficiency viruses. Of these other Tat proteins, HIV-2 Tat (Tat2) has been most intensively investigated and like Tat1, has been shown to form a specific complex with CycT1 that can bind TAR2 with high affinity in vitro (28). Like Tat1, Tat2 has also been shown to be associated with CDK9 in expressing cells (14, 29). While the roles of Tat1 and Tat2 in the viral life are therefore clearly similar, significant mechanistic differences do exist (2, 3, 8, 11). Thus, the HIV-2 TAR element (TAR2) differs from TAR1 in that it displays two functional Tat2 binding sites (Fig. 1), each of which contains a U-rich bulge located 5′ to a conserved terminal loop (3, 11). While Tat1 is fully able to activate transcription via TAR2, Tat2 is only partially active on HIV-1 TAR (2, 3, 8, 11). This difference, which may be in part due to a lower affinity of Tat2 for the TAR1 RNA target (23), remains to be fully explained. Tat proteins from simian immunodeficiency viruses (SIV) have thus far received relatively little attention. In Fig. 1, we show the sequence of the Tat protein (Tat-M) and TAR element (TAR-M) from mandrill simian immunodeficiency virus (SIVmnd), a virus that is highly divergent from both HIV-1 and HIV-2 (26). While TAR-M appears to have in common with TAR2 the property of having two Tat target sites, it also displays potentially significant differences in sequence in both the terminal loops, which differ from the TAR1/TAR2 consensus (5′-CUGGGX-3′), and in the sequence context of the flanking RNA bulges.
In this article we report an analysis of the interaction of the HIV-1, HIV-2, and SIVmnd Tat proteins and TAR elements with CycT1 and show that CycT1 is a critical cofactor for all three transactivators. However, the CycT1-Tat complexes formed by these diverse Tat proteins differ significantly in terms of their TAR RNA sequence specificity, thus explaining their differential abilities to activate heterologous LTR promoters. We demonstrate that the sequences in CycT1 required to bind and support the function of Tat1, Tat2, and Tat-M are very similar but not identical. Indeed, Tat2 and Tat-M proved able to interact not only with CycT1 but also with CycT2A and CycT2B, two cyclin partners for CDK9 that are not bound by Tat1 (4, 22). However, these latter interactions were found to be nonproductive in that none of these CycT2-Tat complexes proved able to bind to TAR. Surprisingly, however, mutation of asparagine 260 to cysteine (N260C) in CycT2B was found to permit not only a specific interaction between Tat1 and CycT2B but also TAR binding and hence the rescue of Tat function in murine cells by all three primate immunodeficiency virus Tat proteins. Overall, these data reveal that CycT1 has been conserved as a critical Tat cofactor during primate immunodeficiency evolution and demonstrate that the ability of specific CycT-Tat complexes to be recruited to a viral TAR RNA target is both modulated by natural sequence variation in Tat and highly dependent on a critical cysteine residue that is present in hCycT1 but lacking in mCycT1 and hCycT2.
MATERIALS AND METHODS
Plasmid construction.
The mammalian expression plasmids pBC12/CMV and derivatives expressing hCycT1, mCycT1, hCycT2A, hCycT2B, or a single-amino-acid mutant form of mCycT1 (Y261C) have been described previously (4). The reciprocal hCycT1 mutant (C261Y) was constructed by recombinant PCR, as were the N260C mutant of CycT2B and a series of scanning mutants of hCycT1 (X1-X8 [Fig. 6]) in which groups of two or three amino acids located between hCycT1 residues 238 and 276 were changed to alanine. Derivatives of the indicator plasmid pBC12/HIV/CAT (4) were constructed by replacing the HIV-1IIIB LTR promoter with LTRs derived from HIV-2ROD or SIVmnd (8, 11, 26). HIV-2ROD and SIVmnd Tat cDNA expression plasmids, similar to the previously described HIV-1 Tat expression plasmid pcTat (4), were constructed by insertion of the relevant Tat cDNA sequence into pBC12/CMV. Mutant Tat expression plasmids have either been described previously (Tat1-C22S [4]) or were constructed by recombinant PCR (Tat2-C50S, Tat-M-C37S). The integrity of each of these wild-type and mutant clones was confirmed by DNA sequence analysis.
FIG. 6.
The hCycT2B(N260C) mutant can support the function of divergent lentiviral Tat proteins. Murine LmTK− cells were transfected with the indicated viral LTR-based reporter and Tat expression plasmid along with plasmids expressing the indicated hCycT proteins, as described for Fig. 4.
Derivatives of the yeast expression plasmid pVP16, encoding wild-type or mutant CycT variants fused to the herpes simplex virus VP16 activation domain, have been or were constructed as previously described (4). pGBT9 derivatives expressing GAL4-Tat fusion proteins and pPGK derivatives expressing wild-type or mutant Tat proteins in yeast were constructed by substitution of Tat cDNA sequences from HIV-2ROD or SIVmnd, amplified from mammalian expression plasmids, in place of the Tat1 gene. A plasmid expressing an MS2-TAR1 hybrid RNA molecule in yeast has been described previously (4). Similar plasmids expressing MS2-TAR2 and MS2-TAR-M hybrid RNAs were derived by insertion of the relevant TAR sequence into pIII/MS2. Yeast two-hybrid assays with GAL4-Tat and VP16-CycT fusion proteins were performed as previously described, as were three-hybrid assays with MS2-TAR hybrid RNAs, authentic Tat proteins, and VP16-CycT fusions (4, 12, 25).
Assay of Tat function in mammalian cells.
Murine LmTK− cells were transfected with DEAE dextran (5) with a pBC12/LTR/CAT indicator plasmid and pBC12/CMV/lacZ in the presence or absence of an HIV-1, HIV-2, or SIVmnd Tat expression plasmid and a pBC12-based plasmid expressing an intact or mutant CycT1 protein, as described previously (4).
Human 293T cultures were transfected by using calcium phosphate coprecipitation (5) with 100 ng of a pBC12/LTR/CAT indicator plasmid, 50 ng of pBC12/CMV/lacZ, and various quantities (0 to 100 ng) of a Tat expression plasmid. In all transfection experiments, the total amount of DNA was equalized by using the parental pBC12/CMV plasmid. CAT activity in cell lysates was determined 48 h posttransfection (4) and normalized for minor variations in transfection efficiency as determined by β-galactosidase activity in the same lysates.
RESULTS
In Fig. 1, we present a comparison of the Tat proteins and TAR elements encoded by HIV-1, HIV-2, and SIVmnd. These sequences are representative of three of the five known families of primate immunodeficiency viruses and evolutionarily are approximately equidistant from one another (15, 26). While the Tat proteins encoded by each of these viruses retain recognizable cysteine, core, and basic motifs, they are nevertheless quite divergent, not least in terms of their size.
HIV-2 and SIVmnd Tat, unlike HIV-1 Tat, can bind to both CycT1 and CycT2.
Previously, it has been demonstrated that Tat1 function in mouse cells can be rescued by expression of hCycT1 or of a mutant mCycT1 containing cysteine in place of tyrosine at position 261 (Y261C) (4, 13). In contrast, Tat1 activity in mouse cells was not rescued by coexpression of CycT2A or CycT2B, alternative cyclin partners of CDK9 (22), or by wild-type mCycT1. These data demonstrate compellingly that hCycT1, but not hCycT2, is an essential cellular cofactor for transactivation by Tat1. Nevertheless, given the sequence diversity among primate lentivirus Tat proteins of different phylogenetic lineages, we considered that divergent Tat proteins and TAR elements might possibly vary in their CycT binding specificity. Notably, the related hCycT2A and hCycT2B proteins, as well as mCycT1, are highly homologous to hCycT1 within their respective N-terminal 280 amino acids, the hCycT1 domain that is both necessary and sufficient to support Tat1 function (13). In particular, the proposed Tat/TAR recognition motif present in CycT1 (13) is largely conserved in both versions of the hCycT2 protein (see below). We therefore attempted to determine whether the Tat2 and Tat-M proteins would be able to target mCycT1 or hCycT2 proteins in addition to or instead of hCycT1.
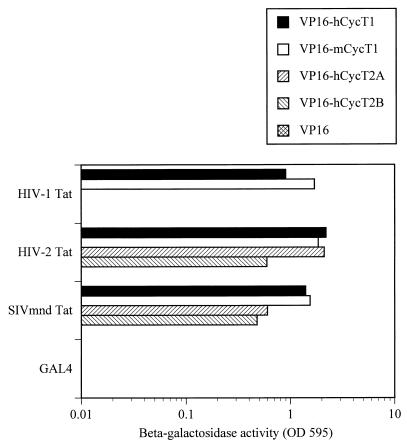
Previously, we have used two- and three-hybrid assays in yeast cells to demonstrate that Tat1 is able to specifically interact with both hCycT1 and mCycT1 in vivo but that the resultant Tat1-mCycT1 heterodimer, unlike the Tat1-hCycT1 complex, is then unable to effectively bind to TAR1 (4). In fact, this is the predicted result for the bona fide Tat cofactor, in that Tat1 is unable to activate the wild-type HIV-1 LTR in murine cells but is fully active when targeted to an RNA target substituted for TAR1 (1, 20). We first examined the ability of Tat2 and Tat-M proteins to bind CycT1 and CycT2 proteins in vivo, using the yeast two-hybrid protein-protein interaction assay (12) (Fig. 2). As previously observed for Tat1 (4), both Tat2 and Tat-M proved able to interact efficiently with both hCycT1 and mCycT1. However, Tat2 and Tat-M also proved able to bind hCycT2A and hCycT2B, a property that is not possessed by Tat1 (4).
FIG. 2.
Interactions between Tat proteins and cyclins in vivo. The ability of the indicated lentiviral Tat proteins and human and murine cyclins to interact was assayed by yeast two-hybrid analysis, as previously described (4). Tat proteins were expressed as GAL4 fusions, whereas CycT variants were expressed fused to the VP16 transcription activation domain.
It was important to demonstrate that each of these CycT-Tat interactions is, in fact, dependent on the functional integrity of the respective Tat cofactor binding domain. The well-characterized Tat1 activation domain point mutant C22S has been previously shown to be defective for hCycT1 binding and, hence, unable to transactivate viral gene expression (4, 20). Therefore, to test the specificity and potential functional relevance of the observed interaction between hCycT2 and Tat-2 or Tat-M, mutants precisely analogous to C22S (Fig. 1), termed Tat2(C50S) and Tat-M(C37S), were generated. In each case, despite protein expression levels found to be equivalent to their wild-type counterparts (data not shown), these point mutant Tat proteins proved to be highly attenuated in their ability to interact with any form of CycT (Table 1) or to transactivate reporter gene expression directed by a cognate LTR in human cells (data not shown). Thus, the observed interactions between Tat1, Tat2, or Tat-M and various CycT proteins are all specific in that they are dependent on a functionally intact Tat activation domain.
TABLE 1.
Tat-CycT interactions require an intact Tat activation domain
| Fusion protein | % β-galactosidase activity observeda
|
|||||
|---|---|---|---|---|---|---|
| Tat1 | Tat1 (C22S) | Tat2 | Tat2 (C50S) | Tat-M | Tat-M (C37S) | |
| VP16-hCycT1 | 100 | <1 | 100 | <1 | 100 | 11 ± 1.0 |
| VP16-mCycT1 | 165 ± 5.0 | <1 | 93 ± 18 | <1 | 111 ± 11 | 4.9 ± 0.8 |
| VP16-hCycT1 (C261Y) | 6.4 ± 2.7 | <1 | 84 ± 17 | <1 | 100 ± 5.0 | 6.5 ± 0.9 |
| VP16-hCycT2B | <1 | <1 | 23 ± 4 | <1 | 34 ± 5.0 | <1 |
| VP16 | <1 | <1 | <1 | <1 | <1 | <1 |
Values given are percentages of the β-galactosidase activities observed in yeast Y190 cells expressing the relevant wild-type GAL4-Tat and VP16-hCycT1 fusion proteins and the averages of three independent experiments.
The C261Y mutant of hCycT1, which bears the converse of the Y261C mutation that rescues the ability of mCycT1 to support Tat function, has previously been reported (13) to be attenuated for Tat1 binding in vitro. In Table 1, we confirm this observation in vivo by the yeast two-hybrid assay. Surprisingly, however, the hCycT1(C261Y) mutant proved able to interact efficiently with the wild-type Tat2 and Tat-M proteins (Table 1). No interaction was observed with any of the activation domain mutant Tat proteins. Thus, the requirement for Cys 261 in mediating efficient Tat-CycT interactions is clearly context dependent; while it is critical for hCycT1 to interact with Tat1, it is absent in mCycT1, which nevertheless binds efficiently to Tat1 in vivo, and can be replaced in hCycT1 without significantly affecting binding to Tat2 or Tat-M.
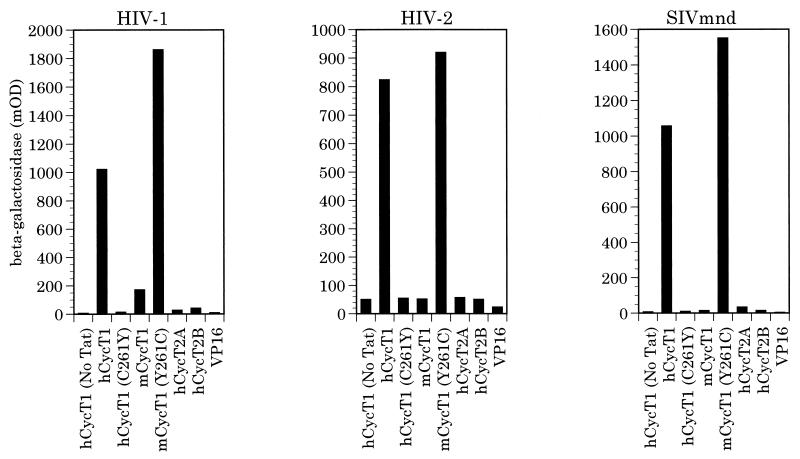
We next used the yeast three-hybrid RNA-protein interaction assay (25) to determine whether the complexes formed between these three distinct Tat proteins and the various forms of CycT would be able to bind to the cognate TAR elements in vivo. As shown in Fig. 3, only hCycT1 and the Y261C mutant of mCycT1 were able to bind to each of these three TAR elements, in each case in a fully Tat-dependent manner. Both the wild-type mCycT1 protein and hCycT1 bearing the C261Y mutation proved unable to mediate TAR binding when complexed with any of these three Tat proteins. Similarly, both CycT2A and CycT2B also failed to bind to TAR, a finding that, in the case of Tat1, is presumably secondary to an inability to bind to Tat1 (Fig. 2). In contrast, for Tat2 and Tat-M, it appears that CycT2A and CycT2B, like mCycT1 and hCycT1(C261Y), can bind these Tat proteins effectively (Fig. 2 and Table 1), but the resulting complexes are then unable to bind to TAR2 or TAR-M (Fig. 3).
FIG. 3.
RNA binding activities of different cyclin T-Tat complexes. The ability of different human and murine cyclins to bind to the indicated primate immunodeficiency virus TAR elements, in the presence or absence of the cognate Tat protein, was assayed by the yeast three-hybrid assay as previously described (4). Cyclins were expressed as VP16 activation domain fusions, as shown in Fig. 2, while Tat proteins were expressed in their wild-type, nonfused form. Induced β-galactosidase activity was measured by light absorption at 595 nm and is expressed in milli-optical density (mOD) units.
Tat2 and Tat-M function is supported by hCycT1 and mCycT1(Y261C) but not by mCycT1 or hCycT2 proteins.
While we are unaware of any previous study examining this question, the inability of the mCycT1-Tat2 and mCycT1-Tat-M complexes to be recruited to the relevant TAR RNA target (Fig. 3) suggests that Tat2 and Tat-M will be weak activators in murine cells. Similarly, the inability of complexes containing hCycT2A, hCycT2B, or hCycT1(C261Y) to be recruited to TAR2 or TAR-M predicts that the coexpression of these CycT proteins in murine cells will not rescue function, even though they can bind Tat2 and Tat-M (Fig. 2). Conversely, the ability of hCycT1 and mCycT1(Y261C) to be recruited to both TAR elements (Fig. 3) predicts that the expression of these proteins in murine cells will rescue Tat2 and Tat-M function, as previously shown for Tat1 (4, 13).
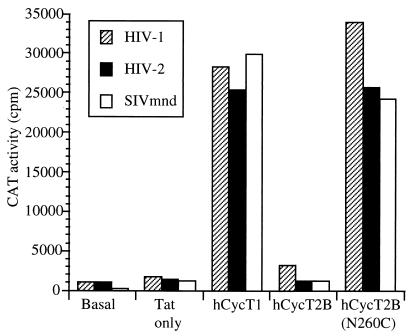
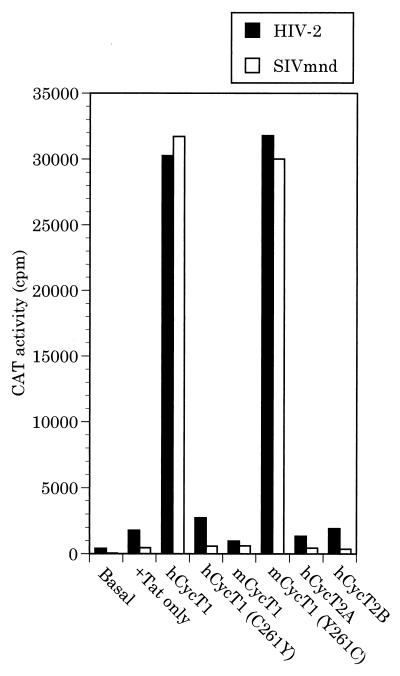
To test this prediction, we examined the ability of Tat2 and Tat-M to transactivate a cognate LTR promoter in murine cells. As shown in Fig. 4, both Tat2 and Tat-M indeed demonstrated only very low levels of transactivation in murine cells. Overexpression of mCycT1, hCycT1(C261Y), hCycT2A, or hCycT2B did not affect the low level of transactivation observed. In contrast, the biological activity of both Tat2 and Tat-M could be rescued by coexpression of either hCycT1 or of the mutant mCycT1(Y261C) protein, both of which are able to bind TAR in the presence of Tat (Fig. 3). Overall, these data demonstrate that formation of a CycT-Tat complex is necessary but not sufficient to support Tat function in vivo. Rather, there is a perfect concordance between the observed ability of a CycT-Tat complex to bind to TAR (Fig. 3) and the ability of this same complex to support transactivation by Tat in otherwise nonpermissive cells (Fig. 4).
FIG. 4.
Rescue of Tat function in murine cells. Murine LmTK− cells were transfected with indicator constructs consisting of the HIV-2 or SIVmnd LTR promoter element linked to the CAT indicator gene together with expression plasmids encoding HIV-2 or SIVmnd Tat and one of the indicated cyclins. At ∼48 h after transfection, CAT expression levels were determined as described previously (4).
A single-amino-acid substitution in hCycT2B results in the formation of Tat-CycT2B complexes able to bind TAR.
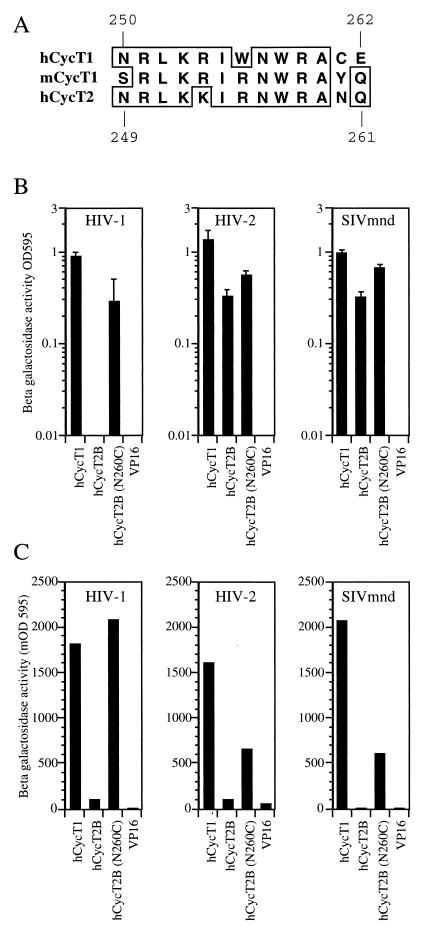
We and others have previously demonstrated that a single amino acid change (Y261C) in the mCycT1 protein allows the Tat-mCycT1 complex to bind TAR and support Tat function (4, 13). Sequences in hCycT1 that are critical for interaction with both Tat1 and TAR in vitro have been mapped to the area between residues 250 and 262 in hCycT1 (13), and these are largely conserved in hCycT2 (Fig. 5A), the notable exception being the critical cysteine residue at position 261 (asparagine 260 in hCycT2). Therefore, we hypothesized that substitution of asparagine 260 with cysteine in CycT2 might restore TAR binding, at least in the case of Tat2 and Tat-M, and thereby result in a protein that would be able to rescue Tat activity in murine cells. Since hCycT2A and hCycT2B differ only in their C-terminal sequences, and the analogous C-terminal sequences in hCycT1 have been shown to be dispensable for Tat function (13), hCycT2B was arbitrarily selected and asparagine 260 was replaced by cysteine (N260C).
FIG. 5.
A single-amino-acid substitution in hCycT2B induces Tat1 binding and permits TAR recruitment by divergent Tat proteins. (A) Sequence comparison between the hCycT1 TRM (residues 250 to 262) and the corresponding sequences in mCycT1 and hCycT2. (B) Interactions between the indicated GAL4-Tat fusion proteins and either VP16-hCycT1 or wild-type or mutant (N260) forms of the VP16-hCycT2B fusion protein measured by the yeast two-hybrid assay. (C) Recruitment of hCycT2B(N260) but not wild-type hCycT2B to TAR. The yeast three-hybrid assay was performed with plasmids expressing Tat proteins and MS2-TAR hybrid RNAs derived from the indicated primate immunodeficiency virus along with the indicated VP16-CycT proteins. OD595, optical density at 595 nm, given in milli-optical density (mOD) units.
We first examined the effect of this mutation on the formation of Tat-hCycT2B complexes by the yeast two-hybrid assay (Fig. 5B). In fact, hCycT2B is already able to bind specifically to the activation domains of Tat2 and Tat-M (Fig. 2 and Table 1), and this mutation had only a minor enhancing effect (∼threefold) on the level of Tat-hCycT2B binding (Fig. 5B). In contrast, while the wild-type hCycT2B protein is unable to bind to Tat1 (Fig. 2), the N260C mutant displayed a significant level of binding. These interactions were again specific in that they were not observed with Tat activation domain mutants (data not shown). We next used the yeast three-hybrid assay to examine whether the complexes formed between the various Tat proteins and hCycT2B(N260C) would be able to bind to a cognate TAR element. As shown in Fig. 5C, all three Tat proteins proved able to recruit hCycT2B(N260C), but not the wild-type hCycT2B protein, to their respective TAR RNA elements. Since hCycT2B(N260C) can form a complex with all three Tat proteins (Fig. 5B), and the resultant complexes can all be subsequently recruited to TAR (Fig. 5C), we predicted that hCycT2B(N260C) should also rescue the activity of these three Tat proteins in murine cells. In fact, as shown in Fig. 6, expression of hCycT2B(N260C) in murine cells dramatically enhanced Tat1, Tat2, and Tat-M function. For each Tat protein, the level of transactivation observed upon coexpression with hCycT2B(N260C) was approximately equivalent to that observed in the presence of hCycT1.
Different hCycT1-Tat complexes display distinct RNA target specificities.
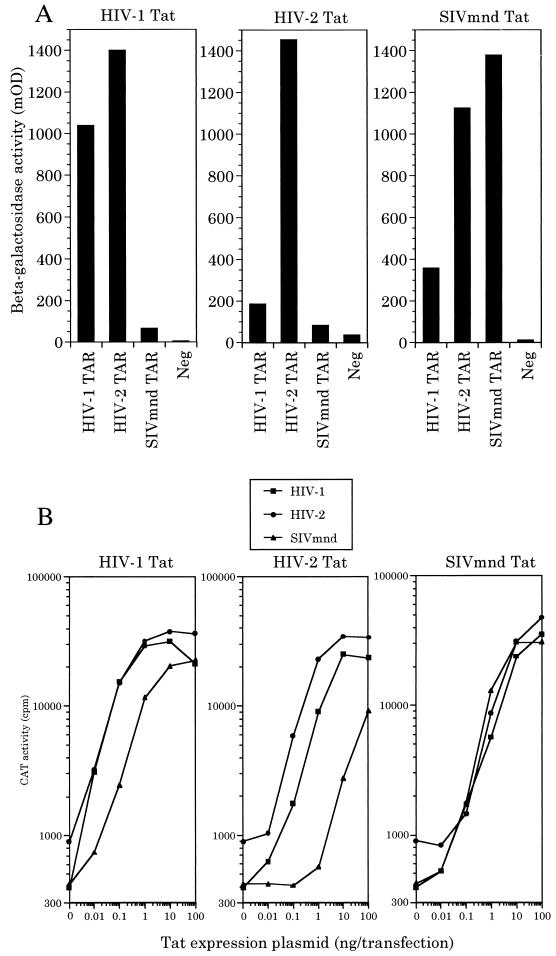
Since these data demonstrate that hCycT1, and not hCycT2, is an essential cofactor for divergent primate lentivirus Tat proteins, we next attempted to determine whether different Tat-CycT1 complexes might possess distinct RNA target specificities. In fact, yeast three-hybrid protein-RNA binding assays (Fig. 7A) revealed a substantial degree of variation in the level of TAR RNA binding by different CycT1-Tat complexes measured in vivo. Specifically, the hCycT1-Tat1 complex bound both TAR1 and TAR2 efficiently but was significantly (10- to 20-fold) attenuated in its ability to bind TAR-M. In contrast, hCycT1-Tat2 bound to TAR1 ∼10-fold-less well than to TAR2 and gave a level of binding to TAR-M that was only slightly above background. Finally, the hCycT1-Tat-M complex was less discriminating in that it bound all three TAR elements effectively, although TAR1 did give a slightly lower activity than either TAR2 or TAR-M (Fig. 7A).
FIG. 7.
The LTR promoter specificity of diverse Tat proteins is predicted by the TAR binding affinity of the relevant hCycT1-Tat complex. (A) The ability of hCycT1 to bind to the indicated viral TAR elements in the presence of each viral Tat protein was determined by the yeast three-hybrid assay, as shown in Fig. 3. The negative control (Neg) reflects the level of β-galactosidase activity obtained in the absence of any TAR element. (B) Dose response analysis in transfected human 293T cells of the response of CAT-based indicator constructs containing the HIV-1, HIV-2, or SIVmnd LTR promoter to increasing levels of the indicated Tat proteins.
To examine whether these differences in Tat-CycT1 RNA binding activity would correlate with the ability of these three distinct Tat proteins to activate gene expression, we compared the effect of widely different levels of Tat1, Tat2, or Tat-M on the level of expression of a chloramphenicol acetyl transferase (CAT) indicator gene linked to the HIV-1, HIV-2, or SIVmnd LTR promoter element. As may be readily observed (Fig. 7B), these diverse lentiviral regulatory proteins display significant differences in their abilities to activate different LTR promoter targets. In the case of Tat1, we observed efficient and equivalent activation of the HIV-1 and HIV-2 LTR promoter but ∼10- to 20-fold-less activity on the SIVmnd LTR. Tat2 displayed the greatest level of LTR selectivity, being ∼sixfold-less active on the HIV-1 LTR and over 100-fold-less active on the SIVmnd LTR promoter. Finally, Tat-M showed little LTR target specificity, although it did appear to be slightly less active on the HIV-1 LTR promoter. Based on these data, we can conclude that these different hCycT1-Tat complexes do indeed display distinct affinities for different TAR RNA targets in vivo and, further, that these different binding affinities are highly predictive of the level of Tat-induced transcriptional activation seen with each TAR-containing LTR promoter.
Divergent Tat proteins require a similar sequence motif in hCycT1 for biological activity.
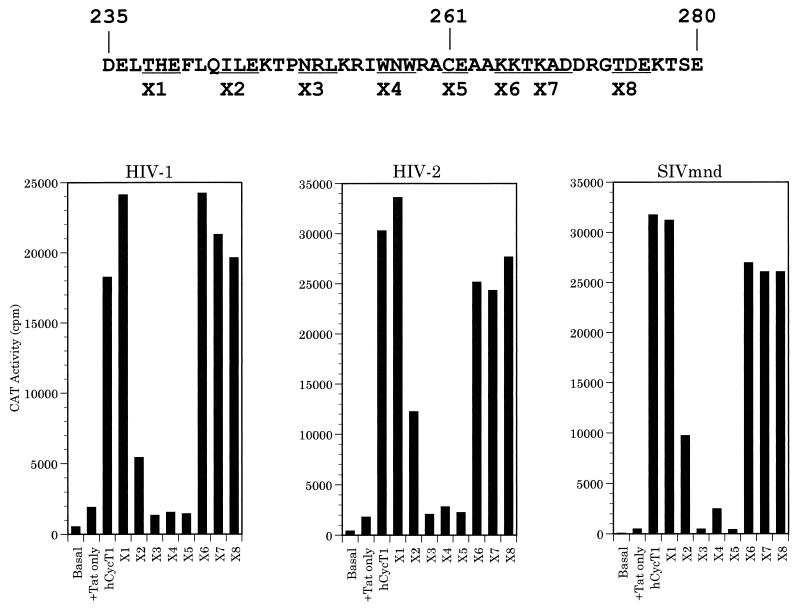
As noted above, Garber et al. (13) recently reported the identification of a motif in hCycT1, termed the TRM sequence, that is critical for Tat1 and/or TAR1 binding in vitro. This motif was reported to extend from approximately residue 250 to 262 in hCycT1 and therefore includes the critical cysteine residue at position 261. The observation that Tat2 and Tat-M differ from Tat1 in being able to bind to hCycT2 (Fig. 2), as well as the finding that Tat1 differs significantly from Tat2 and Tat-M in terms of its ability to bind the hCycT1(C261Y) mutant (Table 1) raised the possibility that Tat2 and Tat-M might display significant differences in their specific binding site on CycT1. To address this issue, we first performed a deletion analysis of hCycT1 to map the extent of sequences required to support each Tat function in murine cells. These data demonstrated that residues 1 to 280 of hCycT1 were sufficient to support the function of Tat1, Tat2, and Tat-M in murine cells, while a protein consisting of residues 1 to 260 was inactive in all three cases, although residues 1 to 260 did retain the ability to bind to CDK9 (data not shown). This result, which is consistent with the recent report that residues 1 to 272, but not residues 1 to 254, of hCycT1 are sufficient to support Tat1 function in murine cells (13), therefore demonstrated that sequences located immediately N-terminal to hCycT1 residue 280 were likely to be critical for the biological activity of all three primate immunodeficiency virus Tat proteins. To more clearly define these sequences, we constructed a set of eight alanine scanning mutations (X1 to X8) between residues 238 and 276 of full-length hCycT1, as shown at the top of Fig. 8, and compared the ability of these mutants to support Tat1, Tat2, or Tat-M function in murine cells. As shown in the lower part of Fig. 8, these hCycT1 mutants gave essentially the same activity with each Tat protein. Specifically, hCycT1 mutants X1, X6, X7, and X8 were all fully active; mutant X2 (alanine inserted in place of residues 244 to 246) was partially active, while mutants X3 (250 to 252), X4 (256 to 258), and X5 (261 and 262) were essentially inactive. These data therefore appear to support the hypothesis that the sequences in hCycT1 required for Tat1, Tat2, and Tat-M function are similar. As expected, analysis of these mutants for their abilities to bind to Tat or TAR, by the yeast two- and three-hybrid assays, demonstrated that the X2, X3, X4, and X5 mutants of hCycT1 are all significantly attenuated in their ability to bind to Tat and/or TAR, while the flanking fully active hCycT1 mutants retain full Tat and TAR binding activity. All these hCycT1 mutants retained the ability to bind to CDK9 effectively, and epitope-tagged versions were found to be equivalently expressed in mammalian cells (data not shown). We therefore conclude that these mutants delineate an hCycT1 sequence, located between residues 244 to 262, that is (i) critical for the in vivo biological activity of all three of these lentiviral Tat proteins and (ii) equivalent to the hCycT1 TRM sequence (residues 250 to 262) previously defined in vitro by Garber et al. (13).
FIG. 8.
Mutational definition of an hCycT1 sequence motif required to mediate Tat function. The protein sequence of hCycT1 between residues 235 and 280 is shown, with the critical cysteine 261 indicated. Alanine scanning mutants X1 to X8 were derived by substitution of alanine for the indicated contiguous stretches of two to three residues in the context of the full-length hCycT1 sequence. The bar graph shows the ability of each of these hCycT1 mutants to rescue the function of the indicated primate immunodeficiency virus LTR promoter-Tat combination in mouse cells.
DISCUSSION
The recent identification of hCycT1, a key component of the transcription elongation factor P-TEFb, as a critical cofactor for HIV-1 Tat has led to a molecular explanation for several key aspects of Tat1 function (4, 13, 22, 28). For example, the observation that both the terminal loop and bulge of TAR1 are critical for in vivo function, while only the latter is required for Tat1 binding in vitro, can now be explained by the finding that the high-affinity interaction of the Tat1-hCycT1 heterodimer with TAR1 is dependent on both the terminal loop and the bulge of TAR1 (4, 7, 24, 27, 28). In addition, the earlier finding that Tat1 is unable to activate the wild-type HIV-1 LTR in murine cells but is fully capable of activating this LTR when recruited to a heterologous RNA target substituted in place of TAR1 (1, 20) can also be explained by the finding that mCycT1 effectively binds to Tat1 but that the resultant Tat1-mCycT1 complex is then only poorly able to bind to TAR1 (4). Put another way, these two findings demonstrate that hCycT1 actively participates in mediating recruitment of the Tat1-hCycT1 complex to TAR1 and is a major determinant of the sequence specificity of this complex.
Previously, several groups have provided data demonstrating that the Tat2 protein is only poorly able to activate transcription via the HIV-1 TAR element, although Tat2 is fully active on TAR2 (2, 3, 8, 11). It therefore seemed likely that other primate immunodeficiency virus Tat proteins, such as the highly divergent SIVmnd Tat (Fig. 1), would also display a significant degree of RNA target specificity. We considered two possible explanations for these different RNA target specificities. On the one hand, it seemed possible that the complex formed between each of these immunodeficiency virus Tat proteins and hCycT1 would display a different RNA target specificity due simply to the different Tat component (23). Alternately, it also seemed possible that highly divergent Tat proteins, such as Tat-M, might actually recruit a different form of P-TEFb, such as a CycT2-containing form, and that this would explain the difference in their RNA target specificity.
Different Tat-CycT1 complexes have different RNA binding specificities.
As shown in Fig. 2 and Table 1, we observed that Tat2 and Tat-M were indeed distinct from Tat1 in being able to readily form an in vivo complex, in an activation domain-dependent manner, not only with hCycT1 but also with CycT2A and hCycT2B. However, these latter complexes are functionally abortive in that only hCycT1, and neither form of hCycT2, can be recruited to TAR (Fig. 3) and can therefore rescue Tat2 and Tat-M function in nonpermissive murine cells (Fig. 4). If hCycT1 is indeed the only relevant cofactor for each of these three Tat proteins in human cells, then one would predict that the ability of each Tat protein to activate gene expression driven by a viral LTR promoter containing the TAR1, TAR2, or TAR-M RNA target site would correlate with the relative affinity of the relevant Tat-hCycT1 complex for each TAR element. As shown in Fig. 7, there is indeed a remarkably strong correlation between the LTR promoter target specificity, measured in transfected human cells (Fig. 7B), and the relative efficiency of TAR binding by each Tat-hCycT1 complex, measured by the yeast three-hybrid assay (Fig. 7A). Overall, these data strongly suggest that the observed differences in the LTR promoter target specificity of these three Tat proteins are due largely or entirely to differences in the efficiency of recruitment of the relevant Tat-hCycT1 complex to TAR.
Sequence determinants for CycT binding by Tat.
While hCycT1 is clearly the relevant cellular cofactor for all three Tat proteins examined in the present study, the data demonstrate that the protein determinants on hCycT1 required to bind each Tat protein, while similar (Fig. 8), are not identical. This is most clearly revealed by the ability, noted above, of Tat2 and Tat-M to bind to hCycT2A and hCycT2B, an activity lacking in Tat1 (Fig. 2). In addition, we note that the C261Y mutant of hCycT1 binds to Tat1 poorly, although binding to both Tat2 and Tat-M is comparable to that of wild-type hCycT1 (Table 1). This latter result is interesting for two reasons. First, it is surprising that hCycT1(C261Y) is a relatively poor target for Tat1 binding because mCycT1, which also bears a tyrosine at position 261, binds Tat1 effectively by this in vivo assay system. Second, this result is of interest in that it partly explains the apparent disagreement in the literature over whether cysteine 261 is required only for TAR1 binding or also for Tat1 binding (4, 13). Specifically, we have previously reported that Tat1 binds wild-type mCycT1 at least as effectively as hCycT1 in vivo (see also Fig. 2 and Table 1) but that the resultant complex fails to bind TAR1 (4). However, substitution of tyrosine 261 in mCycT1 with cysteine fully rescues TAR1 binding (Fig. 3). In contrast, Garber et al. (13) have reported that substitution of cysteine 261 in hCycT1 with alanine entirely ablates both Tat1 and TAR1 binding in vitro. Wild-type mCycT1 was reported to bind to Tat1 ∼three- to fourfold less effectively in vitro than either hCycT1 or the mCycT1(Y261C) mutant but was essentially inactive for TAR1 binding. Garber et al. therefore concluded that Cys261 was essential for Tat1 binding in hCycT1 but only contributory in the context of mCycT1 (13). Our data, derived by a different, in vivo assay system, largely confirm this proposal in that mutation of Cys261 to tyrosine in hCycT1 generates a protein that binds Tat1 far less well than mCycT1 (Table 1). However, the finding that mCycT1 binds to Tat1, Tat2, and Tat-M as effectively as hCycT1 in vivo, combined with the observation that the hCycT1(C261Y) mutant binds efficiently to both Tat2 and Tat-M (Fig. 2 and Table 1), is clearly inconsistent with the hypothesis that this cysteine residue invariably plays a critical role in mediating Tat binding. Instead, the relative importance of Cys261 in mediating Tat-CycT interactions appears to be dependent on the particular Tat and CycT protein being tested. One possible interpretation of the observation that Cys261 is critical in some cases (e.g., binding of Tat1 to hCycT1) but not others (e.g., binding of Tat1 to mCycT1) is that Cys261 forms one of a number of contact sites for Tat on CycT. The relative importance of this contact to the formation of a particular Tat-CycT complex would thus vary, depending on the existence and relative affinity of these other Tat-CycT contact sites. In this context, it is of interest that binding of Tat1 to CycT2B was again entirely dependent on the presence of a cysteine residue introduced at position 260, while binding of CycT2B to Tat2 and Tat-M was enhanced only slightly by this same mutation (Fig. 5B).
The N260C mutation permits CycT2B to support TAR binding and Tat function.
While not necessarily critical for Tat-CycT interactions, Cys261 is evidently required in all cases for the interaction of the Tat-CycT complex with TAR. Indeed, the CycT2B protein, which contains an asparagine residue at the equivalent position (260), was rendered permissive for Tat-dependent TAR binding and was able to support Tat1, Tat2, and Tat-M function when Asn 260 was replaced by cysteine. Similarly, we and others have previously reported (4, 13) that mutation of a tyrosine residue to cysteine at the analogous position in mCycT1 (Fig. 5A) also rescues TAR binding and Tat function in murine cells (4, 13). While the critical importance of this cysteine for TAR binding by Tat-CycT complexes is therefore clear, the underlying mechanistic basis for this requirement is not presently known. Shared binding of a zinc atom (13) could conceivably alter the conformation of the Tat-CycT complex such that the affinity of Tat for TAR is increased. Alternatively, Cys261 or even zinc could directly interact with TAR. Clearly, this question must be addressed in structural studies to be unambiguously resolved.
The finding that hCycT1 has been conserved as a uniquely critical Tat cofactor throughout primate immunodeficiency virus evolution suggests that reagents that selectively disrupt the Tat-hCycT1-TAR complex might prove effective as anti-HIV agents. The question of whether such drugs would allow the development of resistant forms of Tat1 or TAR1 is, of course, difficult to address at this stage. The fact that the hCycT1 TRM has been largely but not entirely conserved through primate immunodeficiency virus evolution does, however, suggest that it might be difficult for the virus to effectively escape from reagents that target this interaction. The importance of the Tat-hCycT1-TAR interaction in the HIV-1 life cycle, combined with the absence of any evidence for an equivalent mechanism in the regulation of host cell gene expression, clearly suggests that this is a highly appropriate target for drug screening.
REFERENCES
- 1.Alonso A, Derse D, Peterlin B M. Human chromosome 12 is required for optimal interactions between Tat and TAR of human immunodeficiency virus type 1 in rodent cells. J Virol. 1992;66:4617–4621. doi: 10.1128/jvi.66.7.4617-4621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arya S K, Beaver B, Jagodzinski L, Ensoli B, Kanki P J, Albert J, Fenyo E-M, Biberfeld G, Zagury J F, Laure F, Essex M, Norrby E, Wong-Staal F, Gallo R C. New human and simian HIV-related retroviruses possess functional transactivator (tat) gene. Nature. 1987;328:548–550. doi: 10.1038/328548a0. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout B, Gatignol A, Silver J, Jeang K-T. Efficient trans-activation by the HIV-2 Tat protein requires a duplicated TAR RNA structure. Nucleic Acids Res. 1990;18:1839–1846. doi: 10.1093/nar/18.7.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cullen B R. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 6.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 7.Dingwall C, Ernberg I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A. HIV-1 Tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 1990;9:4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emerman M, Guyader M, Montagnier L, Baltimore D, Muesing M A. The specificity of the human immunodeficiency virus type 1 transactivator is different from that of human immunodeficiency virus type 1. EMBO J. 1987;6:3755–3760. doi: 10.1002/j.1460-2075.1987.tb02710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feinberg M B, Baltimore D, Frankel A D. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc Natl Acad Sci USA. 1991;88:4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng S, Holland E C. HIV-1 Tat transactivation requires the loop sequence within TAR. Nature. 1988;334:165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- 11.Fenrick R, Malim M H, Hauber J, Le S-Y, Maizel J, Cullen B R. Functional analysis of the Tat trans-activator of human immunodeficiency virus type 2. J Virol. 1989;63:5006–5012. doi: 10.1128/jvi.63.12.5006-5012.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields S, Song O-K. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 13.Garber M E, Wei P, KewalRamani V N, Mayall T P, Herrmann C H, Rice A P, Littman D R, Jones K A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrmann C H, Rice A P. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin M J, Hui H, Robertson D L, Müller M C, Barré-Sinoussi F, Hirsch V M, Allan J S, Shaw G M, Sharp P M, Hahn B H. Mosaic genome structure of simian immunodeficiency virus from West African green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones K A. Taking a new TAK on Tat transactivation. Genes Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- 17.Kao S-Y, Calman A F, Luciw P A, Peterlin B M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 18.Kuppuswamy M, Subramanian T, Srinivasan A, Chinnadurai G. Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nucleic Acids Res. 1989;17:3551–3561. doi: 10.1093/nar/17.9.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo Y, Madore S J, Parslow T G, Cullen B R, Peterlin B M. Functional analysis of interactions between Tat and the transactivation response element of human immunodeficiency virus type 1 in cells. J Virol. 1993;67:5617–5622. doi: 10.1128/jvi.67.9.5617-5622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madore S J, Cullen B R. Genetic analysis of the cofactor requirement for human immunodeficiency virus type 1 Tat function. J Virol. 1993;67:3703–3711. doi: 10.1128/jvi.67.7.3703-3711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marciniak R A, Sharp P A. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng J, Zhu Y, Milton J T, Price D H. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhim H, Rice A P. TAR RNA binding properties and relative transactivation activities of human immunodeficiency virus type 1 and 2 Tat proteins. J Virol. 1993;67:1110–1121. doi: 10.1128/jvi.67.2.1110-1121.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy S, Delling U, Chen C-H, Rosen C A, Sonenberg N. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated transactivation. Genes Dev. 1990;4:1365–1373. doi: 10.1101/gad.4.8.1365. [DOI] [PubMed] [Google Scholar]
- 25.Sengupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsujimoto H, Hasegawa A, Maki N, Fukasawa M, Miura T, Speidel S, Cooper R W, Moriyama E N, Gojobori T, Hayami M. Sequence of a novel simian immunodeficiency virus from a wild-caught African mandrill. Nature. 1989;341:539–541. doi: 10.1038/341539a0. [DOI] [PubMed] [Google Scholar]
- 27.Weeks K M, Ampe C, Schultz S C, Steitz T A, Crothers D M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990;249:1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- 28.Wei P, Garber M E, Fang S-M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 29.Yang X, Herrmann C H, Rice A P. The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxyl-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Q, Chen D, Pierstorff E, Luo K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 1998;17:3681–3691. doi: 10.1093/emboj/17.13.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]