Abstract
The recently discovered hepatitis G virus (HGV) or GB virus C (GBV-C) is widely distributed in human populations, and homologues such as HGV/GBV-CCPZ and GBV-A are found in a variety of different primate species. Both epidemiological and phylogenetic analyses support the hypothesis that GB viruses coevolved with their primate hosts, although their degree of sequence similarity appears incompatible with the high rate of sequence change of HGV/GBV-C over short observation periods. Comparison of complete coding sequences (8,500 bases) of different genotypes of HGV/GBV-C showed an excess of invariant synonymous sites (at 23% of all codons) compared with the frequency expected by chance (10%). To investigate the hypothesis that RNA secondary-structure formation through internal base pairing limited sequence variability at these sites, an algorithm was developed to detect covariant sites among HGV/GBV-C sequences of different genotypes. At least 35 covariant sites that were spatially associated with potential stem-loop structures were detected, whose positions correlated with positions in the genome that showed reductions in synonymous variability. Although the functional roles of the predicted secondary structures remain unclear, the restriction of sequence change imposed by secondary-structure formation provides a mechanism for differences in net rate of accumulation of nucleotide substitutions at different sites. However, the resulting disparity between short- and long-term rates of sequence change of HGV/GBV-C violates the assumptions of the “molecular clock.” This places a major restriction on the use of nucleotide or amino acid sequence comparisons to calculate times of divergence of other viruses evolving under the same structural constraints as GB viruses.
Although the flavivirus hepatitis G virus (HGV) or GB virus C (GBV-C) is newly discovered (17, 19), several observations suggest that it may have always infected humans and originated through coevolution with its primate hosts. First, it is widely distributed in human populations, with frequencies of active or past infection ranging from 5 to 15%, and its distribution extends even to highly isolated populations, such as indigenous tribes in Papua New Guinea and Central and South America. Second, although infection is frequently persistent and associated with high levels of circulating viremia, no evidence links HGV/GBV-C to any identifiable hepatic or nonhepatic disease, consistent with a process of mutual adaptation. Third, the geographical distribution of HGV/GBV-C variants reflects that of ancient human migrations (14). For example, sequences from the Far East are almost invariably genotype 3, and this genotype is otherwise found only in native inhabitants of North and South America. In contrast, Caucasian and other populations from India westward including Northern Africa are infected with genotype 2. Genotype 1 is confined to sub-Saharan Africa and shows the greatest overall sequence diversity (22, 25); particularly divergent variants have been recovered from Pygmy and other African populations (27a, 28). These genotype distributions can potentially be mapped to the emergence and migration of modern humans out of Africa 100,000 years ago (7, 28).
Finally, viruses closely related to HGV/GBV-C have been found in a variety of Old World and New World primate species, and their phylogenetic relationships mirror those of their hosts. HGV/GBV-C variants in wild-caught chimpanzees from Central and West Africa show around 27% nucleotide (15% amino acid) sequence divergence from HGV/GBV-C (1, 3). Distinct variants of HGV/GBV-CCPZ, recovered from different subspecies of chimpanzees, differed at 19% of nucleotide sites (9.5% of amino acid sites) in NS5, a level of diversity greater than that found between the most divergent genotypes of HGV/GBV-C in humans (11% nucleotide and 3.3% amino acid divergence) and consistent with their likely greater population age. Even more divergent homologues of HGV/GBV-C, described as GBV-A, have been recovered from several species of New World primates, with 42% nucleotide (38% amino acid) sequence divergence in the NS5 region from homologous sequences of HGV/GBV-C from humans and chimpanzees (4, 16). Again mirroring host relationships, genetic variants of GBV-A differing from each other by around 25% are closely associated with different new world primate species (4, 5, 16). Observations of congruent sequence relationships between GB viruses and their primate host species are consistent with their coevolution.
However, this hypothesis is difficult to reconcile with the high short-term rate of sequence change of HGV/GBV-C, estimated at 3.9 × 10−4 site/year over the whole genome (23), a rate comparable to that of other RNA viruses (e.g., 4 × 10−4 in hepatitis C virus NS5 [26]). This rate appears incompatible with the lack of sequence diversity between HGV/GBV-C isolates in human populations, if the current distribution of genotypes derives from the migration of modern humans out of Africa. The rate of sequence change is also inconsistent with sequence relationships between human GB viruses and those found in other primate species. However, if GB viruses did not coevolve with their hosts, their current distribution can have originated only through multiple cross-racial and cross-species transmissions over the last few hundred years. This latter hypothesis does not accord with the geographical distribution of HGV/GBV-C genotypes in humans or with the congruent virus and host phylogenies. This explanation is also inconsistent with the recently described species barriers (GBV-A cannot be transmitted to chimpanzees, and HGV/GBV-C cannot be transmitted to New World primates [4a]).
A more radical explanation of the current data is that there are major differences between GB viruses and higher organisms in the constraints operating on sequence change. For vertebrates and other eukaryotes, there is a close concordance between fossil-based estimates of the times of species separation and the extent of divergence of nucleotide and amino acid sequences in a wide variety of nuclear genes (15). In contrast, restrictions on variability at certain sites in the HGV/GBV-C genome may prevent the accumulation of substitutions that limit the extent to which HGV/GBV-C can diversify over time. In the present study, we have compared sequences of different genotypes of HGV/GBV-C to identify such restrictions and to investigate whether these can be explained by mechanisms such as RNA secondary-structure formation.
MATERIALS AND METHODS
Nucleotide sequences.
Currently available complete genomic sequences of HGV/GBV-C sequences of genotypes 1 to 3 (GenBank/EMBL accession numbers in parentheses) include the type 1 sequence, GBV-C (U36380); the type 2a sequences PNF2161 (U44402), R10291 (U45966), HGV-Iw (D87255), HGV-1539 (AF031829), GT110 (D90600), and CG01BD (AB003289); the type 2b sequence GBV-C(EA) (U63715); the type 3 sequences GT230 (D90601), CG07BD (AB003290), HGV-IM71 (AB008342), GSI85 (D87262), G13HC (AB003293), HCV-GD (AF006500), HGVCN (U94695), BG1HC (AB003288), HGVC-964 (U75356), and D87708 to D87715 (13); and sequences of unclassified genotype, i.e., CG12LC (AB003291) and G05BD (AB003292). Sequences were numbered from the start of the coding region after alignment. HGV/GBV-C homologues in primate sequences were from chimpanzees (AF068910 to AF068913 [3]) and the New World primate species Sanguinis mystax, S. labiatus, and Aotus trivirgatus (U94421, AF023424, and AF023425).
Coding sequences of serum albumin were obtained from the following mammalian species (GenBank accession numbers in parentheses): cow (Y17769), sheep (X17055), cat (X84842), gerbil (AB006197), horse (X74045), human (V00494), macaque (M90463), rabbit (U18344), and rat (V01222). Alpha globin sequences compared included gibbon (M94634), human (V00493), rhesus (J04495), baboon (X05289), lemur (M29648), horse (M17902), rabbit (J00658), seal (M73996), mouse (L75940), rat (M17083), hamster (X57029), and sheep (X70213).
Generation of simulated sequence data sets.
The expected stochastic frequencies of invariant and variable codons and of covariant substitutions were determined by using control sequence data sets containing the same number of sequences generated in the following ways.
(i) Control set A.
Nucleotide changes were introduced at random positions into a representative HGV/GBV-C sequence (R10291) for the 17 representative sequences of types 1 to 3, and GT320 as a control for type 3 sequences) at a prespecified frequency (nucleotide divergence × length of sequence). The introduced substitutions reproduced the relative frequency of synonymous and nonsynonymous substitutions (30:1), the relative frequency of transitions and transversions (2:1), and the base composition at synonymous sites (12.9% A, 31.7% C, 35.8% G, and 19.9% U) observed upon comparison of 17 representative HGV/GBV-C sequences. Retention of the observed transition/transversion ratio was essential because two of the four transitions preserve base pairing through the possibility of G-U base pairs, whereas all transversions disrupt base pairing. All distances (Jukes-Cantor [J-C], synonymous [dS] and nonsynonymous [dN], transitions [Ts], transversions [Tv]) and base composition at synonymous sites in the simulated data set were within 10% of the values of the data set to be emulated. Control set A was used to determine whether there was an excess frequency of invariant synonymous positions and of potential covariant sites among HGV/GBV-C sequences.
(ii) Control set B.
The nucleotide identity of all bases in the representative and type 3 data sets was changed (G→C→U→A→G). The resulting sequences retain the phylogenetic relationships of the original sequences and the distribution of variability across the genome and within codons, but all base pairings (G-C, A-U, and G-U) contributing to secondary structure were disrupted. However this control could not be used to explore the expected frequency of covariant sites since the G+C content of synonymous sites was reduced from 67 to 52%, resulting in a reduction from 40.0 to 33.7% in the number of potential base pairings in the shifted data set that occur by chance.
(iii) Control set C.
Each codon within the virus data sets was randomly assigned to a new position. Control set C retains the phylogenetic relationships between sequences and does not alter dN, dS, Ts, or Tv distances or the number of covariant sites. The data set was used to evaluate the stochastic association between covariant sites and surrounding stem-loop base pairing. Since this control data set disrupts potential base pairings, it allowed an evaluation of the association between covariant sites and the size and position of potential stem-loops.
(iv) Control set D.
The order of nucleotides in representative sequences was randomized to determine the contribution of sequence order to the free energy on folding in the program RNADraw, v. 1.0 (provided by O. Matzura).
Measurement of codon variability.
Aligned data sets of nucleotide sequences were compared codon by codon. At each site, only sequences coding for the most frequent amino acid were analyzed for synonymous variability. At each site, the mean number of synonymous differences between codons was calculated and normalized by correcting for multiple substitution so that sites at equilibrium would have a mean divergence of 1.0. For two-, three-, four-, and sixfold-degenerate sites, the mean distances were therefore corrected by factors of 2, 1.5, 1.333, and 1.2, respectively. Variability across the genome was calculated by using the mean diversity of sites in a sliding window of 50 codon positions. Control data set A was used to estimate the frequency of invariant synonymous sites arising by chance.
Detection of covariance.
Each base position in sequence alignments of HGV/GBV-C was screened for covariance by comparison with each downstream base. Sequences were scanned for downstream potential base pairings to each variable nucleotide position in the alignment, in which nucleotide substitutions in one sequence were matched and contributed to base pairing with the complementary site. G · U pairing were scored as 0.8 of Watson-Crick pairing, and the identification of a match required a mean score of 0.84 per sequence. After identification of complementary covariant sites, the maximum number of base pairings between nucleotides surrounding the site was scored using a sliding window of 7 bases from positions −7 to +7 from the site. Five or more consecutive complementary sites could potentially form stem-loop structures.
Sequence software.
All sequence randomization and nucleotide distance measurements were performed with the Simmonic 2000 package. Programs for measurement of synonymous variability and for covariance screening are available from the authors.
RESULTS
Relationship between rate of sequence change and divergence.
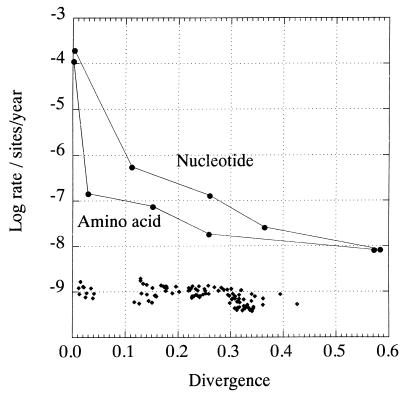
If HGV/GBV-C and related viruses in primates have evolved in concert with their hosts, then rates of sequence change could be calculated from known times of human population movements and from paleontological estimates of the timing of primate species divergence. However, these inferred rates differ markedly for different estimated times of divergence. For example, the accumulated rate of sequence change of 5.6 × 10−6 nucleotide substitution per site per year over 100,000 years (0.1 Myr) between different human populations was approximately 100 times lower than the rate measured over 8.4 years (4 × 10−4 per year [23]). Rates of synonymous change showed a similar decline over this period (3.2 × 10−6 per year compared with 6.5 × 10−4 per year over 8.4 years). Even greater differences were observed when more distantly related species were compared (Fig. 1). These changes in net rate occur even though neither the nucleotide divergence, corrected for multiple substitutions between human and chimpanzee GB viruses (0.36), nor the synonymous distances between human isolates (0.64) approached saturation.
FIG. 1.
Relationship of rates of nucleotide and amino acid sequence change to sequence divergence, using the hypothesis that HGV/GBV-C and related viruses in primates have cospeciated with their hosts. Datum points (●) from left to right represent the following divergence times: 8.4 years (time course in HGV/GBV-C-infected individual) (23), 100,000 years (divergence of modern humans), 1.6 Myr (divergence of troglodytes and verus subspecies of chimpanzees) (Pan troglodytes [21]), 7 Myr (divergence of humans and chimpanzees 12, 21), and 35 Myr (divergence of Old World [human and chimpanzees] and New world [Sanguinis mystax, S. labiatus, and Aotus trivirgatus] primates [12]). The sequences compared were from the NS5 region of the genome (amino acid positions 2498 to 2561 in sequence PNF2161 [U44402]), with divergences and rates based on J-C distances. For comparison, rates of substitution of nucleotide sequences of alpha globin of mammals, placentals, and birds are plotted (⧫), using times of divergence estimated from paleontological records (15).
In contrast to HGV/GBV-C, little change in the rate of nucleotide substitution was evident upon comparison of eukaryotic genes. For example, the rate predicted from comparison of human and chimpanzee alpha globin sequences (1.1 × 10−9 substitution per site per year) was similar to rates calculated from more distantly related animals (e.g., human and duck, 0.55 × 10−9/year). Overall, pairwise comparisons between 16 mammalian, placental, and avian species produced rates ranging from 0.47 to 1.7 × 10−9 per year. These rates were calculated over a range of corrected nucleotide sequence distances (0.012 to 0.42) in which the rate of sequence change of HGV/GBV-C declined by at least 100,000-fold (Fig. 1) (15).
Frequency and distribution of variability at synonymous sites.
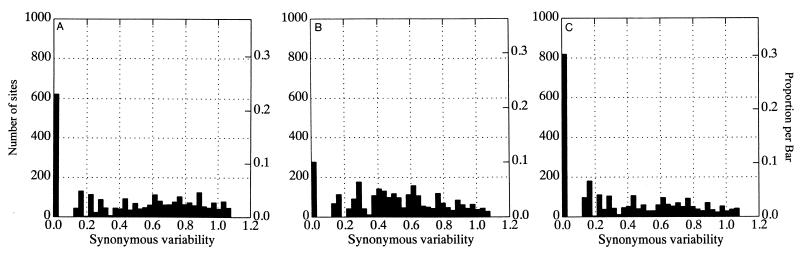
Hypotheses to explain differences in the rate of sequence change of HGV/GBV-C would not only have to account for the much greater constraint on the encoded amino acid sequences but also provide a mechanism that would inhibit the accumulation of substitutions at synonymous sites which are usually regarded as selectively neutral. Evidence for a restriction in variability at synonymous sites was provided by analysis of the distribution of variable and invariant nucleotide positions in HGV/GBV-C sequences (Fig. 2A). Among 17 HGV/GBV-C sequences of genotypes 1, 2, and 3, a total of 622 codons were invariant at synonymous sites (23%) compared with a mean of 277 (10.0% ± 0.6%) between 10 independently generated simulated data sets of descendants of the HGV/GBV-C sequence, R10291 (control set A; Fig. 2B). These sequences had the same overall degree of divergence (J-C distance, 0.139; dN, 0.017; dS, 0.62; Ts/Tv ratio, 2.0) and had similar codon usage and base compositions biases at the codon positions 1, 2, and 3. In a second dataset of 17 genotype 3 sequences, the mean variability was lower (J-C distance, 0.10; dN, 0.016; dS, 0.43) and 30.3% codons were invariant at synonymous controls (Fig. 2C), 2.3 times the value in its matched control sequence data set. In contrast, comparisons of the mammalian coding sequences of alpha globin and albumin produced no evidence for a significant excess of invariant codon positions with respect to that expected by chance (ratios of 1.15 and 1.26, respectively, with respect to values of randomized control sequences).
FIG. 2.
Frequency histograms of variability at synonymous sites in 17 HGV/GBV-C sequences of genotypes 1, 2, and 3 (A), of expected distribution of synonymous variability arising by chance (control data set A) (B), and of variability in 17 HGV/GBV-C sequences of genotype 3 (C).
Unequal codon usage does not explain the excess number of invariant synonymous sites in HGV/GBV-C sequences, because such biases are frequently more extreme in eukaryotic gene sequences such as alpha globin. For example, 36% of third-base sites among HGV/GBV-C sequences were G and 32% were C (total G+C content, 68%), compared with the more biased frequencies in alpha globin sequences of 32 and 51% (G+C content, 83%).
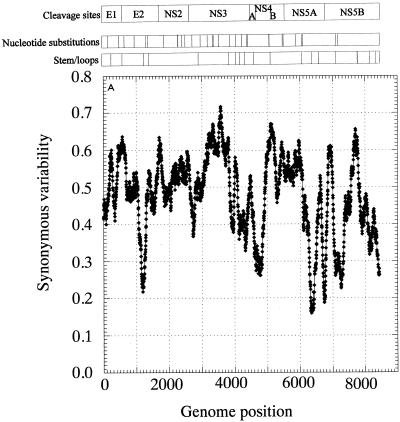
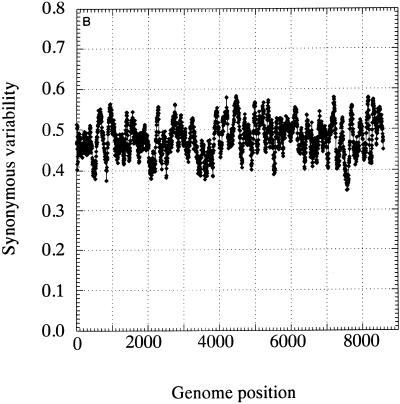
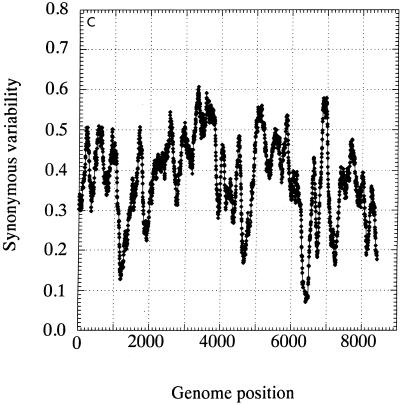
The distribution of invariant synonymous sites in the genome of HGV/GBV-C was not random (Fig. 3). Mean synonymous diversity over a sliding window of 50 codons ranged from 0.159 to 0.716, with extreme suppression of variability around nucleotide positions 1300, 4600, 6300, and 6700 (Fig. 3A). A much more restricted range of variability was observed in a matched simulated data set (control set A; range, 0.363 to 0.564 [Fig. 3B]). The pattern of variability across the genome was reproduced in a separate sequence data set of type 3 sequences, even though the underlying degree of sequence variability was lower (Fig. 3C). There was no evidence for the existence of evolutionarily conserved potential coding sequences (containing initiating methionine and stop codons) in either the +2 or +3 reading frames of the positive-sense genome or the +1 or +3 frames of the antisense genome, either in the parts of the genome showing low variability at synonymous sites or elsewhere (data not shown).
FIG. 3.
Mean synonymous diversity over HGV/GBV-C genome, averaged over a sliding window of 50 codons in 17 HGV/GBV-C sequences of genotypes 1, 2, and 3 (A), control data set A (B), and 17 HGV/GBV-C sequence of genotype 3 (C). Predicted or experimentally determined sites of cleavage of HGV/GBV-C polyprotein (2, 18), sites of nucleotide substitution over 8.4 years (23), and positions of predicted stem-loop binding predicted by covariance algorithm are shown above panel A.
Secondary structure of RNA.
The genomes of many RNA viruses, including HGV/GBV-C and HCV, contain regions that form internally base-paired stem-loop structures that play a role in RNA replication and translation through ribosomal binding to an internal ribosome entry site. While current descriptions of structured regions of viral genomes are generally confined to the untranslated regions at the extreme ends of viral genomes, there is also evidence for a series of stem-loop structures in the core gene of HCV (27), which may play a direct role in the translation of the HCV polyprotein (24). Secondary structures in the HCV core gene would also account for the marked suppression of synonymous substitutions in this region of the genome in the absence of convincing evidence for an overlapping reading frame (10, 24). More extensive internal base pairing might therefore underlie the greater than expected frequency of invariant synonymous sites observed in the HGV/GBV-C genome.
Conventional methods for predicting RNA secondary structures are computationally intensive and are often based upon the calculation of minimum free energies (greatest internal base pairing) for each possible configuration of an RNA sequence. These secondary-structure predictions are sometimes independently supported by the occurrence of covariance, where a nucleotide substitution of an internally base-paired sequence is matched by a substitution in the paired sequence that preserves binding. However, to date, formal statistical methods to analyze the contribution of covariance detection in the identification of particular secondary-structure prediction have not been described, although recently a method was described in which thermodynamic structure prediction was combined with phylogenetic information to search for conserved structures in human immunodeficiency virus and HCV (9).
The length of the coding region of the HGV/GBV-C genome (8,500 bases) clearly poses problems for methods based only upon free-energy calculation because the number of possible internal base-paired combinations would be extremely large and similar in free energy. However, the existence of multiple divergent genomic sequences without substantial phylogenetic structure suggested to us that covariance might be used in place of free-energy calculations as the primary method to detect internal base pairings. An algorithm was used in which alignments of HGV/GBV-C sequences were systematically scanned for potential base pairings between variable sites and scored by the number of paired substitutions from the upstream and downstream consensus bases that maintained base pairing (covariance score). Candidate covariant sites were additionally scored on the length of sequence either side of the site that was capable of forming a stem-loop structure (Table 1). Analysis of a representative data set of 17 HGV/GBV-C sequences of all genotypes identified a numerical excess of sites (approximately 35 sites) with high combined covariance and stem-loop scores over the number detected in control sequences whose codon positions were randomly reordered to break any stem-loop structure (control set C), or sequences with randomly introduced variability at an equivalent level to that between epidemiologically unlinked HGV/GBV-C sequences (control set A). A separate analysis of a second data set containing 17 type 3 sequences also showed a similar excess of sites (30 sites) with high covariance and stem-loop values over control sequences (data not shown).
TABLE 1.
Association of covariance scores with stem-loop length
| Sequence | Covariance valuea | Stem-loop length (/7)b
|
||
|---|---|---|---|---|
| 5 | 6 | 7 | ||
| HGV/GBV-C genotypes 1 to 3 | 1 | 1,196 | 176 | 9 |
| 2 | 196 | 30 | 1 | |
| 3 | 57 | 13 | 3 | |
| 4 | 35 | 7 | 1 | |
| 5 | 21 | 6 | 2 | |
| 6 | 17 | 1 | ||
| 7 | 4 | 3 | ||
| 8 | 3 | 8 | ||
| Control A (simulated diversity) | 1 | 1,382 | 173 | 13 |
| 2 | 352 | 50 | ||
| 3 | 70 | 6 | ||
| 4 | 15 | 1 | ||
| 5 | ||||
| 6 | ||||
| 7 | ||||
| 8 | ||||
| Control C (codon order randomization) | 1 | 1,164 | 132 | 7 |
| 2 | 219 | 24 | 2 | |
| 3 | 57 | 8 | ||
| 4 | 40 | 5 | ||
| 5 | 18 | 2 | ||
| 6 | 11 | |||
| 7 | 1 | |||
| 8 | 1 | 1 | ||
Number of nucleotide substitutions from the consensus base contributing to base pairing at each site (maximum, 8).
From a sliding window of 7, the excess number of sites over control sequences underlined.
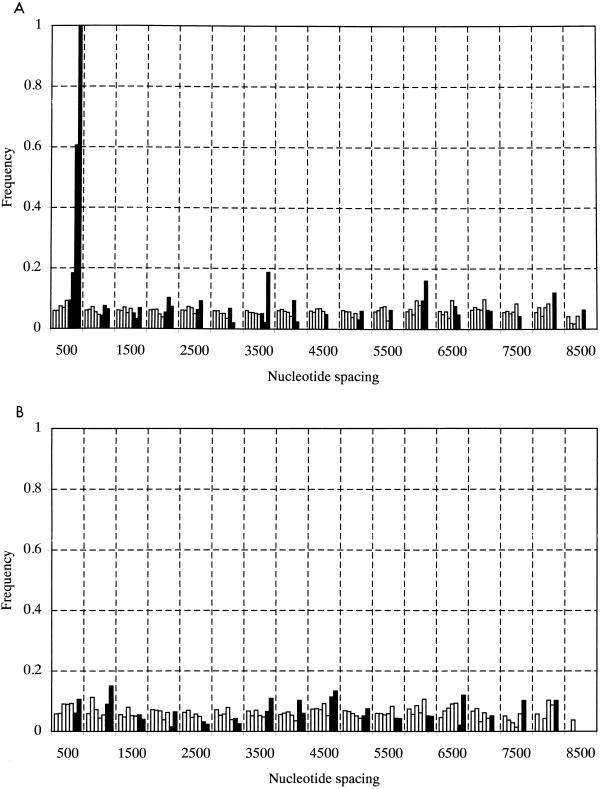
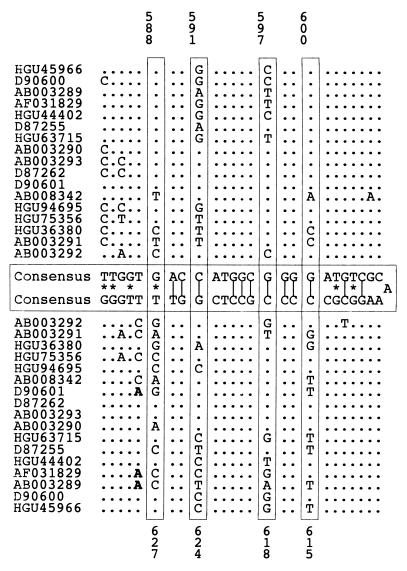
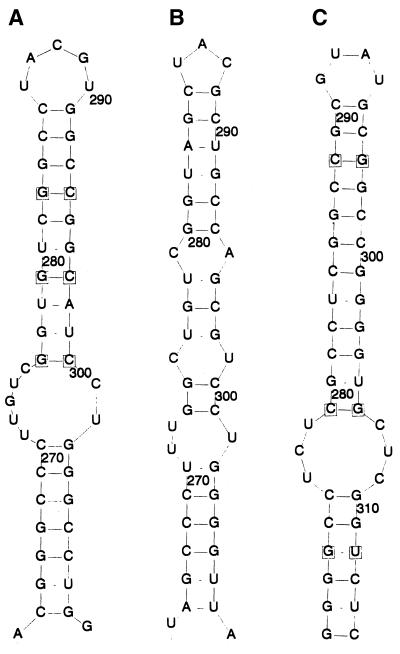
Five lines of evidence argue for the biological reality of these proposed structures. First, the spacing between covariant sites in HGV/GBV-C was nonrandom, since high covariance values were strongly associated with separations between upstream and downstream sites of less than 500 bases (Fig. 4A). The same association between spacing and covariance value was observed in a separate analysis of type 3 HGV/GBV-C sequences (data not shown) but was absent in control data sets A, B, or C (Fig. 4B; data not shown). Second, multiple covariant sites were frequently found in the same stem-loop structure, more often than expected by chance; an example of a stem-loop containing four covariant sites is shown in Fig. 5. Third, proposed structures in HGV/GBV-C sequences were found in homologous positions in the more distantly related HGV/GBV-CTRO and GBV-A sequences by using a minimum-energy algorithm to predict RNA secondary structures between base positions 1 and 1,500 of their coding regions (RNADraw) (Fig. 6). Covariant sites between the available GBV-A sequences were found in this region. These structures were conserved despite the low degree of nucleotide sequence similarity between the GB viruses in this part of the E1 gene (the nucleotide sequence divergence ranged from 43 to 53% between positions 263 and 309 in the R10291 genome). Third-base positions in the predicted stem-loops for human HGV/GBV-C sequences and GBV-A were paired, and all covariant substitutions in the loop were synonymous.
FIG. 4.
Frequency histogram of covariance scores between paired nucleotide sites according to their separation in the genome of 17 HGV/GBV-C sequences of genotypes 1, 2, and 3 (A) and control data set C (B). Solid bars represent frequencies of nucleotide pairs with covariance scores of 6 or greater. Frequencies were adjusted to account for the number of pairs compared.
FIG. 5.
Example of a potential stem-loop structure identified by covariance screening of 17 HGV/GBV-C sequences of genotypes 1 to 3. Covariant sites are indicated by numbering. Analogous structures were detected by analysis of type 3 sequences only (data not shown). All changes from consensus nucleotide sequence were synonymous except for those marked in bold. Vertical lines indicate Watson-Crick base pairing; asterisks indicate GU base pairing.
FIG. 6.
(A) Stem-loop structure in the E1 genes of HGV/GBV-C (A) and likely homologues in HGV/GBV-CTRO (B) and GBV-A (C) sequences. Covariant sites in panels A and C are boxed.
Fourth, while potential stem-loop structures detected by the covariance algorithm were distributed throughout the HGV/GBV-C genome (Fig. 3A and C), there was a close correlation between these sites and regions where synonymous site variability was suppressed. For example, the region from positions 3000 to 3950 showed the greatest synonymous variability (peak value, >0.7) and was uninterrupted by detectable covariant sites. This region was bounded by the predicted stem-loop structures between positions 2964 and 3114 and positions 2958 and 3120 (5′ end) and between positions 3894 and 3930 and positions 3897 and 3927 (3′ end), and these coincided with steep declines in mean synonymous variability. There was no obvious association between synonymous variability and sites of cleavage of the HGV/GBV-C polyprotein (Fig. 3A). Together, stem-loop structures predicted from covariant sites detected in both HGV/GBV-C sequence data sets accounted for at least 20% of the HGV/GBV-C genome. The 24 synonymous nucleotide substitutions observed in the longitudinal study of HGV/GBV-C sequence change over 8.4 years (23) were more frequently found in areas of the genome with higher mean synonymous variability (Fig. 3A).
Finally, the proposed extensive RNA secondary structure of the HGV/GBV-C genome was independently supported by the high free energies predicted by RNA folding. Although the formation of RNA secondary structures was favoured by the high G+C content of the genome (32% G and 27% C) and the excess of U over A residues (23 and 18%, respectively), the order of bases also contributed additionally to the high free energy values observed for subgenomic regions. The mean free energy of eight overlapping 1,500-base fragments of the R10291 HGV/GBV-C sequence was 293 kJ/mol lower (range, 207 to 426 kJ/mol) than that of the same sequences in which nucleotides were randomly reordered (control set D). A similar difference in free energy was observed for the chimpanzee HGV/GBV-CTRO sequence (mean reduction, 301 kJ/mol; range, 225 to 399 kJ) or upon comparison of shorter subgenomic regions of R10291 (e.g., mean of 89 kJ/mol and range of 23 to 185 kJ/mol over a sequence length of 500 bases). In contrast, no significant difference in free energy was observed upon randomization of base order for 1,500 base regions of sequences where the identity of nucleotides was altered to change base pairing (G→C→U→A→G [control set B]). Similarly, no significant reduction was observed upon reordering the human gene sequences of alpha globin (−13 kJ/mol over a sequence length of 429 bases) or albumin (+13 kJ/mol over 1,845 bases).
Nucleotide order randomization also lowered the free energy for the reverse complement of the first 1,500 bases of the HGV/GBV-C genome (−119 kJ/mol), but to a lesser extent than that of the sense sequence (−294 kJ/mol), suggesting that the secondary structure of the positive strand was more relevant biologically. Secondary-structure predictions based on free energy provided independent evidence for the majority of the short-range stem-loop structures identified by covariance screening (Fig. 6) but did not support structures in which covariant sites were separated by more than 100 bases.
DISCUSSION
This study provides evidence for marked restrictions on the sequence variability of HGV/GBV-C that may limit the extent to which virus genome sequences can diverge over time. In contrast to mammalian genes, HGV/GBV-C sequences showed an increased frequency of invariant synonymous sites over that expected by chance, suggesting a functional role of certain nucleotide positions that was independent of the coding capacity for the HGV/GBV-C polyprotein and which did not result from the presence of overlapping gene sequences in other reading frames. A similar although less marked excess of invariant synonymous sites was recently reported in a separate analysis of swine vesicular disease virus and foot-and-mouth disease viruses sequences (8). The contribution of secondary-structure formation to the observed restricted variability of HGV/GBV-C was supported not only by the spatial association between covariant sites and stem-loop structures (Fig. 4; Table 1) but also by the contribution of base order to free-energy calculations of RNA folding. Similarly, randomization of the nucleotide order of swine vesicular disease virus 3B/C sequences led to a consistent although proportionately smaller reduction in free energy on folding (mean 7.6% reduction over 641 bases [24a], compared with 16% over an equivalent length of HGV/GBV-C).
In a folded genome, sequence change in regions that are internally base paired would require simultaneous nucleotide substitutions on both sides of potential stem-loop structures to maintain base pairing and would lead to a much lower frequency of sequence change than in unpaired sites. The preferential accumulation of nucleotide changes at nonpaired sites may therefore account for the rapid sequence change of HGV/GBV-C over short periods, while further divergence, such as between different isolates of HGV/GBV-C or between different primate species, may occur only through covariant substitutions, which accumulate more slowly over longer periods (Fig. 1). For example, the substitution rate of HGV/GBV-C over 8.4 years, 4 × 10−4 per site per year (23), predicts a frequency of covariant substitutions less than 1.5 × 10−7, which is comparable to the long-term rate of substitution observed between viruses infecting different primate species (such between HGV/GBV-C and GBV-A sequences [Fig. 1]). An even greater differential frequency (1.1 × 10−5 unpaired and 1 × 10−10 at covariant sites) would occur at nonsynonymous sites, and this greater restriction on nonsynonymous-sequence change may contribute to the extreme dN/dS ratio observed between HGV/GBV-C sequences (0.033) compared with that for coding sequences of other viruses and eukaryotes.
More precise matching of variable and nonvariable sites in the HGV/GBV-C genome with unpaired and paired nucleotides will require the completion of a model of its secondary structure. These investigations would include determining the relationship between secondary structure and the sites of nucleotide substitution in the HGV/GBV-C over short observation periods where high frequencies of sequence change at synonymous sites was observed (Fig. 4) (23). The base pairings predicted by covariance scanning will contribute to future attempts to determine the overall structure of HGV/GBV-C genome, since they restrict the remaining structure prediction by free-energy calculation to relatively short lengths of sequence. Such analyses would be assisted by the observation that all of the sites identified by covariance were relatively closely spaced (<500 bases); this may be a general feature of HGV/GBV-C genomic RNA structure.
Structural constraints on sequence change independent of coding capacity may inhibit sequence change in other viruses with single-stranded genomes. For example, separate genotypes of simian immunodeficiency virus (SIVAGM) are associated with different subspecies of African green monkeys (11), while a similar association between genetic variants of SIVCPZ in different chimpanzee subspecies (Pan troglodytes troglodytes and P. troglodytes schweinfurthii has recently been reported (6). If these associations also represent coevolution of viruses with their hosts, there is a similar discrepancy between their long- and short-term rates of sequence change. In the second example, the 37.7% sequence divergence which accumulated over a period of at least 0.5 million years between SIV variants infecting the troglodytes (SIVCPZ-US and SIVCPZ-GAB) and schweinfurthii (SIVCPZ-ANT) subspecies (21) represents a net rate of sequence change of 3.4 × 10−7 per site per year. This rate is approximately 5,000 times lower than the rate (1.4 × 10−4 per site per year) inferred from comparison of different subtypes in the main (M) group of human immunodeficiency variants, which are considered to have originated over the last 50 years (based on a mean pairwise distance between single representative sequences of types A to J of 14.2% [29]). Despite the genetic and structural differences between flaviviruses and lentiviruses, the absolute values and differential between short- and long-term rates of sequence change of human immunodeficiency virus type 1 and SIVCPZ were remarkably similar to those predicted for HGV/GBV-C.
The functional role of the predicted RNA secondary structures in HGV/GBV-C remains unclear. Among several possibilities that may apply equally to other single-stranded RNA viruses, RNA folding may be required for packaging of the genome into the viral nucleocapsid or to protect the genome from RNA-degrading enzymes or it may be involved in the regulation of transcription or translation in analogous ways to the function of internal ribosome entry site structures in flaviviruses and picornaviruses. Recently, an internal stem-loop structure in the coding part of the human rhinovirus 14 genome sequence, comparable in size and free energy to those detected in HGV/GBV-C, was shown to be essential for human rhinovirus 14 negative-strand transcription (20). The long-range interactions between different genomic regions implied by these observations suggest an organized overall structure of the RNA genome, in which stem-loop structures may play an important structural or catalytic role in virus replication.
Irrespective of its functional significance, the evidence for markedly different substitution frequencies at different sites puts a fundamental limitation on the use of sequence divergence in the timing of virus origins. While the species association of HGV/GBV-C and related viruses in primates provides indirect evidence for their long-term rates of change, such inferences cannot be made for viruses with less close host associations. While it is possible that much of the genetic heterogeneity of RNA viruses originated very recently, our inability to measure long-term rates of sequence change or to understand the restrictions on sequence variability currently leaves open the possibility of very ancient origins of many RNA viruses. These findings suggest that the molecular clock, which has been of major value in the reconstruction of vertebrate and other eukaryotic phylogenies, cannot be simply applied to viruses with more unusual genomic structures.
REFERENCES
- 1.Adams N J, Prescott L E, Jarvis L M, Lewis J C M, McClure M O, Smith D B, Simmonds P. Detection of a novel flavivirus related to hepatitis G virus/GB virus C in chimpanzees. J Gen Virol. 1998;79:1871–1877. doi: 10.1099/0022-1317-79-8-1871. [DOI] [PubMed] [Google Scholar]
- 2.Belyaev A S, Chong S, Novikov A, Kongpachith A, Masiarz F R, Lim M, Kim J P. Hepatitis G virus encodes protease activities which can effect processing of the virus putative nonstructural proteins. J Virol. 1998;72:868–872. doi: 10.1128/jvi.72.1.868-872.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birkenmeyer L G, Desai S M, Muerhoff A S, Leary T P, Simons J N, Montes C C, Mushahwar I K. Isolation of a GB virus-related genome from a chimpanzee. J Med Virol. 1998;56:44–51. doi: 10.1002/(sici)1096-9071(199809)56:1<44::aid-jmv8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 4.Bukh J, Apgar C L. Five new or recently discovered (GBV-A) virus species are indigenous to new world monkeys and may constitute a separate genus of the Flaviviridae. Virology. 1997;229:429–436. doi: 10.1006/viro.1997.8461. [DOI] [PubMed] [Google Scholar]
- 4a.Bukh, J., et al. Unpublished data.
- 5.Erker J C, Desai S M, Leary T P, Chalmers M L, Montes C C, Mushahwar I K. Genomic analysis of two GB virus A variants isolated from captive monkeys. J Gen Virol. 1998;79:41–45. doi: 10.1099/0022-1317-79-1-41. [DOI] [PubMed] [Google Scholar]
- 6.Gao F, Bailes E, Robertson D L, Chen Y, Rodenburg C M, Michael S F, Cummins L B, Arthur L O, Peeters M, Shaw G M, Sharp P M, Hahn B H. Origins of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez Perez M A, Norder H, Bergstrom A, Lopez E, Visona K A, Magnius L O. High prevalence of GB virus C strains genetically related to strains with Asian origin in Nicaraguan hemophiliacs. J Med Virol. 1997;52:149–155. [PubMed] [Google Scholar]
- 8.Haydon D, Knowles N, McCauley J. Models for the detection of non-random base substitution in virus genes: models of synonymous nucleotide substitution in picornavirus genes. Virus Genes. 1998;16:253–266. doi: 10.1023/a:1008018403582. [DOI] [PubMed] [Google Scholar]
- 9.Hofacker I L, Fekete M, Flamm C, Huynen M A, Rauscher S, Stolorz P E, Stadler P F. Automatic detection of conserved RNA structure elements in complete RNA virus genomes. Nucleic Acids Res. 1998;26:3825–3836. doi: 10.1093/nar/26.16.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ina Y, Mizokami M, Ohba K, Gojobori T. Reduction of synonymous substitutions in the core protein gene of hepatitis C virus. J Mol Evol. 1994;38:50–56. doi: 10.1007/BF00175495. [DOI] [PubMed] [Google Scholar]
- 11.Jin M J, Hui H, Robertson D L, Muller M C, Barre-Sinoussi F, Hirsch V M, Allan J S, Shaw G M, Sharp P M, Hahn B H. Mosaic genome structure of simian immunodeficiency virus from West African green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones S, Mertin R, Pilbeam D, editors. Human evolution. Cambridge, United Kingdom: Cambridge University Press; 1992. [Google Scholar]
- 13.Katayama K, Kageyama T, Fukushi S, Hoshino F B, Kurihara C, Ishiyama N, Okamura H, Oya A. Full-length GBV-C/HGV genomes from nine Japanese isolates: characterization by comparative analyses. Arch Virol. 1998;143:1063–1075. doi: 10.1007/s007050050356. [DOI] [PubMed] [Google Scholar]
- 14.Katayama Y, Apichartpiyakul C, Handajani R, Ishido S, Hotta H. GB virus C hepatitis G virus (GBV-C/HGV) infection in Chiang Mai, Thailand, and identification of variants on the basis of 5′-untranslated region sequences. Arch Virol. 1997;142:2433–2445. doi: 10.1007/s007050050253. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, Hedges S B. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 16.Leary T P, Desai S M, Erker J C, Mushahwar I K. The sequence and genomic organization of a GB virus A variant isolated from captive tamarins. J Gen Virol. 1997;78:2307–2313. doi: 10.1099/0022-1317-78-9-2307. [DOI] [PubMed] [Google Scholar]
- 17.Leary T P, Muerhoff A S, Simons J N, Pilot-Matias T J, Erker J C, Chalmers M L, Schlauder G S, Dawson G J, Desai S M, Mushahwar I K. Sequence and genomic organisation of GBV-C: a novel member of the Flaviviridae associated with human non-A-E hepatitis. J Med Virol. 1996;48:60–67. doi: 10.1002/(SICI)1096-9071(199601)48:1<60::AID-JMV10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 18.Leary T P, Muerhoff A S, Simons J N, PilotMatias T J, Erker J C, Chalmers M L, Schlauder G G, Dawson G J, Desai S M, Mushahwar I K. Sequence and genomic organization of GBV-C: A novel member of the flaviviridae associated with human non-A-E hepatitis. J Med Virol. 1996;48:60–67. doi: 10.1002/(SICI)1096-9071(199601)48:1<60::AID-JMV10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 19.Linnen J, Wages J, ZhangKeck Z Y, Fry K E, Krawczynski K Z, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih J W K, Young L, Piatak M, Hoover C, Fernandez J, Chen S, Zou J C, Morris T, Hyams K C, Ismay S, Lifson J D, Hess G, Foung S K H, Thomas H, Bradley D, Margolis H, Kim J P. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 20.McKnight K L, Lemon S M. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA. 1998;4:1569–1584. doi: 10.1017/s1355838298981006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morin P A, Moore J J, Chakraborty R, Jin L, Goodall J, Woodruff D S. Kin selection, social structure, gene flow, and the evolution of chimpanzees. Science. 1994;265:1193–1201. doi: 10.1126/science.7915048. [DOI] [PubMed] [Google Scholar]
- 22.Muerhoff A S, Smith D B, Leary T P, Erker J C, Desai S M, Mushahwar I K. Identification of GB virus C variants by phylogenetic analysis of 5′-untranslated and coding region sequences. J Virol. 1997;71:6501–6508. doi: 10.1128/jvi.71.9.6501-6508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakao H, Okamoto H, Fukuda M, Tsuda F, Mitsui T, Masuko K, Lizuka H, Miyakawa Y, Mayumi M. Mutation rate of GB virus C hepatitis G virus over the entire genome and in subgenomic regions. Virology. 1997;233:43–50. doi: 10.1006/viro.1997.8615. [DOI] [PubMed] [Google Scholar]
- 24.Reynolds J E, Kaminski A, Kettinen H J, Grace K, Clarke B E, Carroll A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Simmonds, P., and D. Haydon. Unpublished observations.
- 25.Smith D B, Cuceanu N, Davidson F, Jarvis L M, Mokili J L K, Hamid S, Ludlam C A, Simmonds P. Discrimination of hepatitis G virus/GBV-C geographical variants by analysis of the 5′ non-coding region. J Gen Virol. 1997;78:1533–1542. doi: 10.1099/0022-1317-78-7-1533. [DOI] [PubMed] [Google Scholar]
- 26.Smith D B, Pathirana S, Davidson F, Lawlor E, Power J, Yap P L, Simmonds P. The origin of hepatitis C virus genotypes. J Gen Virol. 1997;78:321–328. doi: 10.1099/0022-1317-78-2-321. [DOI] [PubMed] [Google Scholar]
- 27.Smith D B, Simmonds P. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J Mol Evol. 1997;45:238–246. doi: 10.1007/pl00006226. [DOI] [PubMed] [Google Scholar]
- 27a.Soni, P. N., et al. Unpublished data.
- 28.Tanaka Y, Mizokami M, Orito E, Ohba K, Kato T, Kondo Y, Mboudjeka I, Zekeng L, Kaptue L, Bikandou B, MPele P, Takehisa J, Hayami M, Suzuki Y, Gojobori T. African origin of GB virus C hepatitis G virus. FEBS Lett. 1998;423:143–148. doi: 10.1016/s0014-5793(98)00083-0. [DOI] [PubMed] [Google Scholar]
- 29.Zhu T F, Korber B T, Nahmias A J, Hooper E, Sharp P M, Ho D D. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature. 1998;391:594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]