Abstract
Noncoding DNA undergoes widespread context-dependent transcription to produce noncoding RNAs. In recent decades, tremendous advances in genomics and transcriptomics have revealed important regulatory roles for noncoding DNA elements and the RNAs that they produce. Enhancers are one such element that are well-established drivers of gene expression changes in response to a variety of factors such as external stimuli, cellular responses, developmental cues, and disease states. They are known to act at long distances, interact with multiple target gene loci simultaneously, synergize with other enhancers, and associate with dynamic chromatin architectures to form a complex regulatory network. Recent advances in enhancer biology have revealed that upon activation, enhancers transcribe long noncoding RNAs, known as enhancer RNAs (eRNAs), that have been shown to play important roles in enhancer-mediated gene regulation and chromatin-modifying activities. In the brain, enhancer dysregulation and eRNA transcription has been reported in numerous disorders from acute injuries to chronic neurodegeneration. Because this is an emerging area, a comprehensive understanding of eRNA function has not yet been achieved in brain disorders, however the findings to date have illuminated a role for eRNAs in activity-driven gene expression and phenotypic outcomes. In this review, we highlight the breadth of the current literature on eRNA biology in brain health and disease and discuss the challenges as well as focus areas and strategies for future in-depth research on eRNAs in brain health and disease.
Keywords: enhancer, noncoding RNA, eRNA, lncRNA, epigenetics, brain
Introduction:
The advent of whole-genome sequencing has revealed the incredible breadth of noncoding genomic regions, which accounts for approximately 98% of the human genome (Consortium, 2012; Djebali et al., 2012). The biological significance of these regions was once controversial, but the regulatory role of the noncoding genome has now been widely studied and is well established. Among the various noncoding regulatory elements, enhancers are a major group of cis-acting elements that are dispersed throughout the genome and modulate the expression of target genes. Enhancers often engage with their target genes via long-range interactions, which often, but not always, involves complex looping of intermediary chromatin. These interactions can span hundreds of kilobases, and by some estimates the average distance between an enhancer and its target genes is approximately 120 kb in humans (Jin et al., 2013; Mifsud et al., 2015; Sanyal et al., 2012). The gene regulatory activity of enhancers is thought to be in large part responsible for the heterogeneity in cell-type specific gene expression (Andersson et al., 2014; Heintzman et al., 2009; Heinz et al., 2015), as well as gene expression responses to stimuli, cues, injuries, and disease (Corradin & Scacheri, 2014). Thus, understanding enhancer biology and their gene regulatory networks is an important area of study.
Despite the importance of enhancer elements, we do not have a complete understanding of their functions or their interactions with target genes. The recent discovery of widespread, activity-dependent transcription of noncoding RNAs from enhancer elements, known as enhancer RNAs (eRNAs), has revealed a new facet of enhancer functionality in eukaryotes and suggests that not just the enhancer elements, but also the eRNAs, have important gene regulatory roles (De Santa et al., 2010; Kim et al., 2010). Functional characterization of eRNAs has revealed diverse roles, which often include chromatin remodeling (Bose et al., 2017; Mousavi et al., 2013), orchestrating enhancer-promoter interactions (Li et al., 2013; Tsai et al., 2018; Xiang et al., 2014), facilitating the scaffolding of regulatory proteins at target loci (Bose et al., 2017; Hsieh et al., 2014), and regulating RNA polymerase II (Pol II) activity at target genes (Schaukowitch et al., 2014; Zhao et al., 2016). These studies have been reviewed elsewhere (Arnold et al., 2019; Han & Li, 2022) and will not be covered in this review.
In the brain, several early studies have presented strong evidence of eRNA expression and associations with disease progression and outcomes for various disorders. Because this is an emerging area of research, the functions of eRNAs and their precise mechanisms in the brain have not yet been fully elucidated. We anticipate that a greater focus on eRNA biology will help answer outstanding questions in the field and illuminate the therapeutic potential of these molecules. In this article, we review the current literature across the spectrum of neurological disorders with the goal of providing a broad overview of the current knowledge on eRNAs in brain health and disease.
eRNAs in Neuronal Activity:
The first study to report enhancer transcription was conducted in primary mouse cortical neurons wherein transcriptional responses to KCl-induced membrane_-depolarization and Ca2+ signaling were studied (Kim et al., 2010). In this study, the authors evaluated two established predictive features of enhancers - H3K4me1 deposition, and genomic binding of the CREB-binding protein (CBP) - to identify active enhancer loci. Upon KCl-induced membrane depolarization of the neurons, they observed a dramatic increase in CBP binding from a baseline of approximately 1,000 bound sites in untreated cells to 28,000 bound sites post-treatment, including at 11,835 putative neuronal enhancers sites. Several of these enhancers were proximal to immediate-early genes (IEGs) such as c-Fos, Rgs, Nr4a2, and Arc/Arg3.1. Using the Arc promoter as a model, they evaluated the ability of select enhancers to induce expression in an Arc promoter-driven luciferase reporter system. They confirmed that six out of the seven enhancers examined were able to induce luciferase expression via the Arc promoter in response to KCl treatment. Further analysis showed that several of the enhancers produced eRNAs whose expression corresponded with the mRNA expression of the most proximal genes, suggesting that the expression of eRNAs from active enhancers may be linked to the transcription of their target genes.
Activity-regulated IEGs in neurons, such as the ones identified in the aforementioned study encompass the earliest gene expression response to signal transduction events and serve important roles in processes such as memory formation, behavioral responses, and synaptic plasticity (Flavell & Greenberg, 2008; Minatohara et al., 2015; Tischmeyer & Grimm, 1999). In addition, induction of IEG expression is linked to several neuropathologies, including neurodegenerative diseases (Hendrickx et al., 2014), cerebral ischemia (Akins et al., 1996), and neuropsychiatric conditions (Gallitano, 2020; Marballi & Gallitano, 2018). Therefore, studying the gene regulatory relationship between IEGs and the upstream enhancers/eRNAs that modulate their expression has been a focus of several studies. One group characterized transcription at the enhancers corresponding to the IEGs Fos, Fosb, and Nr4a1 in embryonic cortical rat neurons following KCl-induced membrane depolarization (Carullo et al., 2020). While evaluating temporal patterns of eRNA expression, they observed a significant upregulation of transcription at two highly conserved Fos enhancers (Fos E1 and Fos E3) within 7.5 minutes of 25 mM KCl treatment, whereas the Fos mRNA was not upregulated until 15 minutes following KCl treatment. This indicated that eRNA transcription preceded Fos mRNA transcription in response to KCl-treatment. This apparent temporal pattern is in line with previous studies showing that eRNA transcription precedes the transcription of its target genes (Arner et al., 2015; Kim et al., 2015; Schaukowitch et al., 2014). Further, knockdown of the Fos E1 eRNA using antisense oligonucleotides (ASOs) led to a marked decrease in Fos mRNA expression, whereas the knockdown of Fos mRNA had no effect on any Fos eRNA expression, demonstrating that the transcription of the Fos mRNA is dependent on the transcription of the eRNA, but not vice-versa. Exogenous expression of the Fos E1 eRNA using vectors did not lead to Fos mRNA induction, indicating that native transcription of eRNAs from the cis-enhancer element is necessary for target gene engagement. This was further confirmed through CRISPR-activation (a CRISPR-based method that facilitates recruitment of transcriptional activators to genomic loci to induce their overexpression) in which induction of both the endogenous Fos enhancers successfully induced Fos mRNA transcription. Notably, simultaneous induction of both the Fos enhancers had an additive effect on Fos mRNA expression, however the enhancers themselves appeared to be uncoupled in that the activation of one of the enhancers led to eRNA transcription from only that enhancer and not the other enhancer. Next, the authors investigated whether the eRNA transcripts themselves or merely the act of transcription at the enhancers facilitated induction of the Fos gene. To study this, they employed a technique called CRISPR-display (Shechner et al., 2015), using a nuclease-deficient Cas9 to target a highly conserved domain of the Fos E1 eRNA to its endogenous enhancer locus, and investigated the sufficiency of the eRNA in inducing Fos gene expression. They found that the display of the E1 functional domain at the E1 locus alone (but not another enhancer locus, or an unrelated intronic region) was sufficient to induce expression of the Fos mRNA. This demonstrates that, in some cases, the presence of the eRNA transcript at the appropriate enhancer site alone is sufficient to induce target gene expression.
Other studies investigating eRNA activity in neurons found that some eRNAs can also elicit target gene activation by binding and sequestering transcriptional inhibitors. For example, one study investigating Pol II engagement with IEGs in activity-induced neurons found a functional role for eRNAs in Pol II pause-release at target genes through interactions with a protein called Negative Elongation Factor (NELF) (Schaukowitch et al., 2014). Pausing of RNA Pol II proximal to gene promoters is a phenomenon that plays an important role in priming the loci for rapid expression, such as at IEGs, bypassing several slower initiation steps (Adelman & Lis, 2012). NELF, along with another protein DREB-sensitivity inducing factor (DSIF), binds to Pol II to form a paused elongation complex (PEC) proximal to the promoter. One of the NELF subunits, NELF-E, contains an RNA-recognition motif (RRM), which is thought to bind nascent mRNA to stabilize the PEC at gene loci, to pause transcription at those loci in a ‘poised state’ (Aoi et al., 2020; Wu et al., 2003; Yamaguchi et al., 2002). The authors hypothesized that eRNAs from enhancers linked with the IEGs might compete with the nascent mRNA transcripts to bind and sequester NELF away from the gene loci, thereby relieving the inhibitory effects of NELF on Pol II and releasing it to resume transcription at the gene loci. Following this logic, inhibiting eRNA expression would prevent the sequestration of NELF by the eRNA, thus maintaining the paused state of Pol II at the gene locus. To test this, they used ASOs to knockdown eRNAs associated with several IEG enhancers (such as Arc, c-Fos, Gadd45b, etc.) and observed an increase in NELF occupancy at their gene promoters, suggesting that eRNAs do in fact play a role in sequestering NELF away from gene loci to activate gene transcription via Pol II release. This was corroborated by a concomitant decrease in the levels of elongating Pol II at the target gene loci upon eRNA knockdown. Further, they confirmed that the interaction of NELF with the eRNAs is mediated by the same RRM motif that is known to bind nascent mRNAs, further supporting the hypothesis that eRNAs compete with the nascent mRNAs to bind NELF.
The features of eRNAs that drive such specific interactions with regulatory proteins are not fully understood. A follow-up study was conducted on several eRNAs arising from the enhancers of the IEGs Nr4a1, Arc, Junb, c-Fos, and Fosb to evaluate whether secondary structures played a role in NELF binding (Gorbovytska et al., 2022). Using SHAPE-MaP, they biochemically resolved the secondary structures of the 5’ ends of these eRNAs under the premise that this region would be most likely to exhibit domains critical to eRNA function, considering that the 3’ end is prone to rapid exosomal decay due to the lack of polyadenylation. The SHAPE-MaP results indicated that some eRNAs, such as those associated with Fos, Junb, and Arc enhancers were highly structured, whereas others such those associated with Fosb and Gadd45b were relatively unstructured. An examination of structural motifs did not identify any common motifs between the eRNAs, indicating that secondary structures do not underlie the binding of the eRNAs to NELF. Next, they evaluated whether the length of the eRNAs played a role in NELF binding. To test this, they reconstituted the PEC (Pol II, NELF, and DSIF) in vitro and conducted electrophoretic mobility shift assays (EMSA) against fragments of the 5’ regions of Arc and Nr4a1 eRNAs. Using full-length 5’ fragments (200 nu) of the Arc and Nr4a1 eRNAs, NELF was successfully dissociated from the PEC. Mutant forms of these eRNA fragments that disrupted their structural domains retained the ability to successfully dissociate NELF from the PEC, confirming that this interaction is not dependent on secondary structures. Interestingly, reducing the length of the fragments to 100 nu and 50 nu significantly attenuated the dissociative effect in a length-dependent manner, however eRNA length was determined not to be a key driver of the interaction with NELF and the authors posited that the length-dependence could be explained by the necessity of eRNAs to span multiple RNA-binding sites on NELF subunits to induce NELF dissociation from the PEC, though this requires further validation. Given that the effect of eRNAs on NELF dissociation did not appear to be tied to shared secondary structure or eRNA length, the question remained as to what feature of the eRNAs was responsible for their binding to NELF. To investigate further, they conducted EMSA experiments using simple oligonucleotide repeats to determine if nucleotide repeats were the driving factor behind this interaction. Interestingly, they found that poly(GA) and poly(GU) repeat-containing RNAs were able to induce robust dissociation of NELF from the PEC, even at low RNA concentrations, indicating that dispersed unpaired guanosines may underpin the ability of RNAs to induce NELF dissociation from the PEC. They confirmed this through generation of G-to-C and G-to-A mutants of the Nr4a1 eRNA, which exhibited a greatly diminished dissociative effect on NELF as compared to the wild-type sequences. Upon evaluation of other activity-induced eRNAs, they found that the sequences were indeed enriched with guanosines in their 5’ ends, however the enrichment of guanosines is not a general feature of eRNAs, suggesting that such a mechanism is not conserved across the wider population of eRNAs and may be a specific feature of the eRNA-NELF relationship.
Together, these studies laid the groundwork for our understanding of eRNA functions and interactions in neurons and showed that eRNAs can act through diverse mechanisms. In the next sections of this article, we summarize the current literature on the expression, functions, and significance of eRNAs in the context of neurodevelopment and neurological disorders.
eRNAs in Neurodevelopment:
Neurodevelopment is characterized by intricate, finely tuned gene regulatory processes that drive cell fate and phenotype. Enhancers are known to play a central role in these events (Choi et al., 2021; de la Torre-Ubieta et al., 2018; Frank et al., 2015; Nord et al., 2013). A recent report investigated transcribed enhancers in the cerebellum of mice at multiple stages of development (Ramirez et al., 2023), building on a previous study which demonstrated that unique signatures of active enhancers distinguished the various stages of cerebellar development (Ramirez et al., 2022). Using the FANTOM5 atlas of bidirectionally_-transcribed enhancers in combination with ChIP-Seq data for the enhancer marks H3K27ac and H3K4me1, they identified a total of 1,664 high-confidence actively transcribed enhancers that produce eRNAs during various stages of cerebellar development ranging from embryonic day 11 (E11) to postnatal day 9 (P9). K-means clustering revealed that these transcribed enhancers could be grouped into three distinct clusters that peak in expression at different timepoints: E12, E14, and P9. Additionally, they found that they were significantly enriched in cerebellar samples versus other tissue types in the FANTOM5 consortium. This suggested that the identified enhancers likely regulate timepoint-specific and region-specific gene expression profiles in the developing brain. Through correlation of transcription at these enhancers and transcription at proximal genes within the same conserved topologically associated domains (TADs), they were able to identify 964 putative target genes whose expression was significantly correlated to that of these enhancers. Using Gene Ontology analysis, they found that these genes had functions fundamental to the developing brain, including axonogenesis, glial cell differentiation, and neuronal differentiation. Notably, putative target genes of the transcribed enhancers that peaked during embryonic development were associated with stem cell proliferation and neuronal differentiation, whereas those of transcribed enhancers that peaked at P9 were associated with neurotransmitter transport and secretion. This suggests that the transcription of eRNAs from developmentally regulated enhancers at specific time_-points is tightly correlated with the maturation and patterning of the cerebellar tissue. Target genes for non-transcribed enhancers, on the other hand, were associated with constitutive cellular functions conserved across all cell-types, such as RNA splicing and DNA repair. In total, 44.32% of the high-confidence transcribed enhancers were predicted to regulate multiple genes and 24.6% of target genes were associated with multiple high-confidence transcribed enhancers, indicating a degree of synergism, which is a hallmark of enhancer function. Together, this study provides promising evidence for a link between time point-specific eRNA expression and important stage-specific gene expression profiles in the developing brain. Because this is an emerging area of research, mechanistic evidence confirming these correlational observations has not yet been reported but is an important focus area for further studies.
eRNAs in Neurodegeneration:
Neurodegenerative pathologies involve an array of chronic symptoms that are driven by molecular changes over a number of years. Aberrant enhancer activity and enhancer variants have been reported in these pathologies (Achour et al., 2015; Francelle et al., 2017; Kikuchi et al., 2019; Li et al., 2019; McClymont et al., 2018; Soldner et al., 2016). Notably, enhancer variants in Alzheimer’s Disease (AD) have been found to disrupt chromatin looping (Kikuchi et al., 2019), which is an epigenetic mechanism previously shown to be regulated by eRNA binding (Hsieh et al., 2014; Li et al., 2013; Tsai et al., 2018). While a potential role for eRNAs in the neurodegenerative gene regulatory environment is intriguing, this area is largely understudied, leaving major gaps in knowledge. The few studies currently published in the literature are reviewed in this section.
In one study investigating methylated enhancer sites in AD, the authors characterized transcription at differentially methylated enhancers in human tissue. Out of 2,563 identified enhancer loci with detectable transcription in the human prefrontal cortex, 36 eRNAs were differentially expressed in the AD samples (n=25) as compared to healthy controls (Li et al., 2019). Of these, 32 showed modest, but statistically significant, upregulation (log fold-change ranging 0.150 to 0.363) and four showed downregulation (log fold-change ranging from −0.186 to −0.314). They found that these differentially expressed eRNAs were enriched at AD-relevant enhancer loci. Although this study did not evaluate eRNA function, the differential expression of eRNAs in AD tissues is interesting and might suggest an involvement of eRNAs in shaping the AD pathophysiology. Another study investigating the AD-relevant APOE gene in skin fibroblasts found that it was regulated by the eRNA AANCR, which when partially transcribed, paused Pol II by forming R-loops to prevent its own complete transcription and the transcription of APOE and APOC1 (Watts et al., 2022). Extending this observation further to a osmotic stress model using renal proximal tubule cells, the authors found that when the cells were exposed to hypertonic stress, the R-loops were resolved and the AANCR eRNA was transcribed completely, which in turn activated APOE transcription as expected. This suggests that this pause mechanism allows the APOE locus to remain poised for rapid transcription in response to external stimuli such as stress. Previously, single-nucleotide polymorphisms (SNPs) in the APOE enhancer (and thus the corresponding AANCR eRNA) have been associated with Alzheimer’s risk (Bullido et al., 1998; Wang et al., 2000), but whether these SNPs disrupt APOE expression through disruption of AANCR eRNA expression is unknown and is yet to be confirmed in an AD model.
In Huntington’s Disease (HD), a study using the R6/1 transgenic mouse model evaluated eRNA expression in the striatum and its association with dysregulated strial gene expression (Le Gras et al., 2017). They identified 677 downregulated and 335 upregulated eRNAs in the striatum as compared to wild-type controls. To correlate these changes with differential gene expression, the authors considered genes neighboring the enhancer loci as putative target genes. For the enhancers that showed a downregulation in eRNA expression, they observed that the putative target genes were also largely downregulated. Confirming this observation, using qPCR they found that for a selected set of eRNAs that were downregulated, the expression of the corresponding putative target genes was also downregulated. Gene Ontology analysis showed that these genes represented important neuronal components such as voltage gated potassium channel complexes, post-synaptic density, and dendritic spine, feeding into processes such as long-term synaptic depression, regulation of synaptic transmission, and cognition, to name a few. Notably, they also found that the genomic loci of the downregulated eRNAs were enriched in Srf binding sites, a transcription factor that is significantly downregulated in the HD mouse striatum. This provides a potential mechanism by which decreased transcription factor binding may drive decreases in eRNA expression, resulting in the downregulation of the enhancers’ target genes. Together, these results suggest that the apparently concomitant downregulation of eRNAs and their putative target genes may represent a gene regulatory network that is associated with the changes in the HD brain. Notably though, a similarly broad correlation was not found between upregulated eRNAs and their adjacent genes. An important consideration when interpreting this data is that these are merely correlational observations that may not necessarily constitute gene regulatory relationships between the enhancers and their targets given that enhancers often facilitate long-range interactions with genes beyond those in the immediate vicinity. Further studies will be necessary to biochemically validate these networks before they can be confidently implicated in the etiology of HD. Other than these select few studies, we did not find additional reports in the literature investigating eRNA expression or function in AD, HD, or other neurodegenerative diseases. Thus, the field is wide open for further investigation.
eRNAs in Neuropsychiatric Disorders
A growing body of evidence suggests that polymorphisms and mutations in intronic and intergenic DNA regions (including enhancers) underpin an array of neuropsychiatric pathologies, including alcoholism (Davidson et al., 2011; Dong et al., 2018), autism (Inoue & Inoue, 2016; Yao et al., 2015), and schizophrenia (Bharadwaj et al., 2013; Eckart et al., 2016) among others. The first study investigating eRNAs in addiction was conducted in an adolescent intermittent ethanol (AIE) exposure model in rats (Kyzar et al., 2019). AIE is known to induce lifelong epigenetic changes in the amygdala. This study focused on Arc gene expression in the amygdala, which is known to be attenuated following AIE. The Arc eRNA originates from the Arc synaptic activity response element (Arc SARE). It regulates Arc gene expression via the NELF_-dissociation mechanism described earlier (Kyzar et al., 2019; Schaukowitch et al., 2014). Here, the authors evaluated the effects of AIE-induced epigenetic changes on Arc eRNA regulation. In adult rats that underwent AIE, they found that Arc eRNA and Arc mRNA levels were decreased as compared to saline-exposed controls (AIS). Simultaneously, there was an increase in H3K27me3 levels, a repressive epigenetic modification, along the Arc promoter and Arc SARE, along with decreased mRNA levels of a well-known histone de-methylator, KDM6B. Considering that acute ethanol exposure in adulthood has been shown to reverse characteristic anxiety-like behaviors in AIE rats, the authors investigated the effects of such exposure on these epigenetic and gene expression patterns. Interestingly, following acute ethanol exposure of AIE rats in adulthood, KDM6B mRNA levels returned to normal and there was a concomitant increase in the levels of Arc eRNA and mRNA as compared to adult AIE rats exposed to saline only. This apparent trend in normalization following acute ethanol exposure was also observed for a variety of epigenetic marks at the Arc SARE, including a decrease in the repressive mark H3K27me3 and increases in the activating marks H3K27ac and CBP. These findings demonstrated that acute ethanol exposure in adult AIE rats reversed many of the long-term changes induced by AIE, including many that modulate Arc eRNA expression. Evaluating whether KDM6B plays a role in epigenetic changes regulating Arc eRNA expression in the amygdala of control rats, siRNA-induced ablation of KDM6B led to increased methylation at the Arc promoter and Arc SARE, as well as decreased expression of the Arc eRNA. This indicates that KDM6B regulates histone methylation at these loci to modulate Arc eRNA expression. In addition, KDM6B and Arc eRNA knockdown in the amygdala both induced anxiety-like behavior in control rats. This suggests that the anxiety-relieving effects of acute ethanol exposure in adult AIE rats may be driven by the normalization of KDM6B levels, and thus Arc eRNA levels. Another study provided additional evidence for the role of the epigenetic regulation of Arc SARE and Arc eRNA in anxiety-like behaviors using CRISPR-based epigenomic editing in rats (Bohnsack et al., 2022). Deposition of H3K27ac using dCas9-P300 at the Arc SARE to activate it was sufficient to reduce anxiety-like and excessive drinking behaviors in AIE rats. In contrast, deposition of H3K27me3 using dCas9-KRAB at Arc SARE to repress it was sufficient to induce anxiety-like behaviors and excessive drinking in control rats. Together these studies provided important insights connecting epigenetic reprogramming and eRNA expression to alcohol use disorder.
Another study evaluated eRNA expression in schizophrenia patient samples (n=258) and control patient samples (n=279) (Hauberg et al., 2019). Using RNA-Seq combined with ATAC-Seq in neuronal and glial cells in combination with publicly available cap end gene expression (CAGE) data from the FANTOM5 Project, they identified 118 differentially expressed eRNAs and 1,647 differentially expressed protein-coding genes. Using Weighted Gene Co-expression Network Analysis (WGCNA) to delineate co-expression patterns of genes and eRNAs, they identified a module that exhibited significant association with schizophrenia (that included enrichment with differentially expressed transcripts, previously established schizophrenia-related genes, and co-regulation differences between schizophrenia and controls). The most significantly altered eRNA in this module - enh3256 - was predicted to regulate the Golgi phosphoprotein3 (GOLPH3L) gene, which was also present in this module. They validated this regulatory relationship through a luciferase reporter assay in which siRNA-induced knockdown of enh3256 in human embryonic kidney cells (HEK-293) resulted in the inhibition of GOLPH3L expression. While this is the only study in the literature to report a potential connection between eRNAs and schizophrenia-associated gene dysregulation, it underscores the importance of further exploring eRNAs in the etiology of schizophrenia and can serve as a framework for future clinical investigations.
eRNAs in Cerebral Ischemia and Reperfusion Injury:
The noncoding transcriptome has been widely studied in ischemic stroke and ischemia-reperfusion injury over the last two decades. Early studies on gene expression in stroke provided a comprehensive view of mRNA expression (Raghavendra Rao et al., 2002; Schmidt-Kastner et al., 2002; Tang et al., 2002) and noncoding RNA expression (Dharap et al., 2009; Dharap et al., 2011, 2012; Liu et al., 2010; Lusardi et al., 2014; Tan et al., 2009) gene expression in the post-stroke brain, however a limitation of these studies was the use of microarrays, which is an inherently biased technique that relies on probes against annotated sequences rather than an unbiased transcriptome-wide mapping of RNAs by techniques such as RNA-seq. With the advent of RNA-seq, a much wider and deeper understanding of gene expression in the post-stroke brain was possible, which revealed the differential expression of hundreds of previously undiscovered noncoding RNAs and mRNAs (Bhattarai et al., 2019; Bhattarai et al., 2017; Wei et al., 2021; Zheng et al., 2022). As part of these efforts, our group recently published the first reports on eRNA expression and functions in the post-stroke mouse cortex during the acute reperfusion window (Bhattarai et al., 2021; Ruiz et al., 2021).
We previously identified 77 high-confidence eRNAs that were upregulated in the adult cerebral cortex of mouse following a 1 h middle cerebral artery occlusion (MCAO) and 6, 12, or 24 h of reperfusion (Bhattarai et al., 2021). These eRNAs were highly expressed by 6 h of reperfusion, and 28 of them remained significantly upregulated through the later reperfusion timepoints of 12 h and 24 h. Of these, 37 eRNAs were intergenic in origin and the remaining 40 originated proximal to protein-coding genes (i.e., within 5 kb of the annotated genes). Further, we found that 55 of the 77 eRNAs were uniquely expressed in response to stroke and not detected in the sham cortices. All of these transcripts were unspliced, nonadenylated and relatively short with a median length of 350 nucleotides. In addition, probing randomly selected eRNAs using cell fractionation and qPCR, we found that they were localized to the chromatin (Bhattarai et al., 2021), which is a hallmark of eRNAs (Gayen & Kalantry, 2017; Werner et al., 2017). To determine their significance in the post-stroke brain, we delivered antisense oligos in vivo targeting two robustly expressed stroke-induced eRNAs (eRNA_93384 & eRNA_06347) and found that in both cases, the perturbation of the eRNAs significantly increased the mean infarct volumes at 24 h of reperfusion as compared to animals receiving non-targeting oligos (Bhattarai et al., 2021). This suggested a potential neuroprotective role for these eRNAs in the post-stroke brain. When we evaluated eRNA expression in females, we noted sex-dependent as well as sex-independent expression levels and temporal patterns between the sexes (Ruiz et al., 2021). To determine whether perturbation of eRNAs had the same effects in males and females, we knocked down the expression of eRNA_06347 (which showed a similar spatiotemporal expression in both the sexes) in the female brain and evaluated the effects on infarct volume. Like males, the female brains exhibited significantly larger infarct volumes upon eRNA knockdown as compared to controls, and the increased volumes were consistent with that seen in males (Ruiz et al., 2021). Together, these studies demonstrated that eRNAs are expressed very early after the onset of stroke, are robustly expressed across the acute window of reperfusion, show sex-dependent as well as sex-independent expression patterns, and at least some of them have a neuroprotective role in the cortex. Additional functional studies and cell-type specific investigations are needed to gain a more comprehensive understanding of eRNAs in stroke and evaluate their potential as therapeutic targets.
Future Directions
Over the past couple of decades, tremendous advancements have been made in mapping noncoding RNAs and understanding their functions. Along this timescale, the discovery of eRNAs is relatively recent. Therefore, a complete picture of their expression, mechanisms, and functions is yet to be achieved. Our review of the literature on eRNAs in the brain revealed that while substantial progress has been made in probing eRNA expression in brain health and disease, there is still a long road ahead in terms of determining their functions and mechanisms relevant to disease etiology. Additionally, there are numerous disorders for which no literature on eRNAs exists, such as Parkinson’s Disease, multiple sclerosis, substance use disorder, and traumatic brain injury to name a few. In the following section, we discuss some challenges in the field and present strategies to help address them and advance eRNA research in neuroscience.
A fundamental challenge in eRNA research is the accurate and high-confidence mapping of eRNAs. Unlike mRNAs or most other noncoding eRNAs, the discovery of eRNAs requires a combination of high-throughput RNA-seq together with appropriate epigenetic and chromatin annotations that can identify enhancer domains with high confidence. Combining genome-wide datasets, such as GRO-Seq, RNA-Seq, H3K27ac/H3K4me1 ChIP-Seq, and/or ATAC-Seq is a robust strategy for eRNA identification but has been performed in very few studies to date. High-throughput identification, especially in bulk tissues, introduces the significant challenge of deconstructing the cell-type and tissue-based localization of eRNAs. The brain consists of a transcriptional landscape that is highly region-specific and cell-type specific given the inherent complexity of the brain’s architecture (Darmanis et al., 2015). For example, a study evaluating three neuronal populations derived from either the hippocampus, cortex, or striatum found that 776, 390, and 898 transcriptionally active putative enhancers were specific to neurons from each of these regions, respectively (Carullo et al., 2020). These differences might be relevant for the region-specific or cell-type-specific gene regulatory networks that modulate context-dependent cellular outcomes. Approaches such as single-molecule RNA fluorescence in-situ hybridization (smFISH) in combination with IHC for cell-type markers can be applied to resolve the localization of eRNAs at the cellular and subcellular level. By incorporating more advanced technologies such as spatial biology and spatial transcriptomics, the precise expression and three-dimensional localization of eRNAs can be determined in a massively parallel, high-throughput manner, enabling large scale analyses.
Another challenge stems from the lack of knockout cell lines or animal models, and the need for robust in vitro models that represent in vivo eRNA dynamics accurately. Yao et al. note that the cellular context of the brain may play a vital role in shaping eRNA expression; in their study, only 57 of the 103 transcribed enhancers enriched in the human brain were detected in FANTOM5 expression data from in vitro neuronal and astrocyte cultures (Yao et al., 2015). This raises an important caveat to consider regarding representation and compatibility of the in vitro models to the in vivo eRNA response. That said, because a deeper understanding of enhancer/eRNA mechanisms and functions ultimately requires genomic perturbations, often at multiple interlinked loci, the feasibility of developing knockout animal models for the thousands of enhancers in the mammalian genome is limited, which underscores the importance of developing in vitro models for a more tractable and high-throughput platform for biochemical and mechanistic investigations.
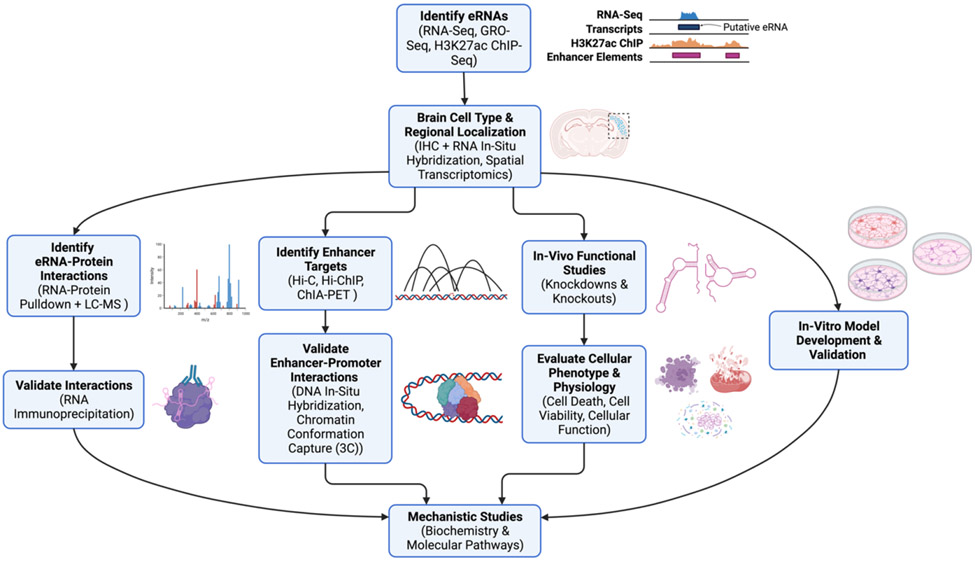
Finally, there remains a need for high-throughput approaches for functional characterization of eRNA biochemistry and molecular interactions. Identifying enhancer-gene pairs is a continuous challenge in the field. To our knowledge, no studies in the area of neurological disorders have used techniques such as chromatin conformation capture (3C) or Hi-C to map higher-order interactions in the 3D chromatin topology for eRNA-centric evaluations. Such an analysis is important considering the complexity and fluidity of the chromatin landscape, which governs the crosstalk between enhancers and their target genes. In addition, identifying the binding partners of eRNAs, especially regulatory proteins, is important to help elucidate the functional complexes underlying eRNA activity and the potential involvement of disease-relevant factors. Such relationships may be driven by a variety of eRNA motifs including nucleotide repeats, specific sequence signatures, or secondary structures which are not yet fully understood, thus complicating and limiting the precision of in silico predictions. The use of RNA-protein pulldown assays in combination with liquid-chromatography mass spectrometry (LC-MS) is a good starting point to begin mapping the diverse protein binding partners of eRNAs, with the added advantage that it can be performed on both in vivo and in vitro samples and can be scaled in a high-throughput manner. An experimental framework for eRNA identification and characterization is presented in Fig. 1.
Fig. 1: Pipeline for Characterization of eRNAs in Neurological Disorders.
This graphic presents a general strategy for identification and characterization of eRNAs in the brain that can be applied to a variety of models and pathologies. High-throughput identification of eRNAs using next-generation sequencing and cell-type localization of these eRNAs provides an unbiased method of mapping and contextualizing eRNA expression. Subsequent studies can characterize eRNA function by identifying eRNA-protein complexes, mapping enhancer-gene interactions, and applying gain- or loss-of-function strategies. Further mechanistic insights in individual cell-types can be achieved at greater depth using in vitro models.
Overall, eRNA research in neuroscience has gained substantial momentum over the past few years and the discoveries to_-date have established that eRNAs have important roles in neural development and disease. This foundation of early studies presents a compelling case for a greater focus on eRNAs in brain health and disease and illuminates their potential as therapeutic targets.
Acknowledgements:
Figure 1 was created with BioRender.com.
Funding:
This work was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under Award Number R01NS115835 to Ashutosh Dharap.
Footnotes
Competing Interests:
The authors have no relevant financial or non-financial interests to disclose.
References:
- Achour M, Le Gras S, Keime C, Parmentier F, Lejeune FX, Boutillier AL, … Merienne K (2015). Neuronal identity genes regulated by super-enhancers are preferentially down-regulated in the striatum of Huntington's disease mice. Hum Mol Genet, 24(12), 3481–3496. 10.1093/hmg/ddv099 [DOI] [PubMed] [Google Scholar]
- Adelman K, & Lis JT (2012). Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet, 13(10), 720–731. 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akins PT, Liu PK, & Hsu CY (1996). Immediate early gene expression in response to cerebral ischemia. Friend or foe? Stroke, 27(9), 1682–1687. 10.1161/01.str.27.9.1682 [DOI] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, … Sandelin A (2014). An atlas of active enhancers across human cell types and tissues. Nature, 507(7493), 455–461. 10.1038/nature12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoi Y, Smith ER, Shah AP, Rendleman EJ, Marshall SA, Woodfin AR, … Shilatifard A (2020). NELF Regulates a Promoter-Proximal Step Distinct from RNA Pol II Pause-Release. Mol Cell, 78(2), 261–274 e265. 10.1016/j.molcel.2020.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner E, Daub CO, Vitting-Seerup K, Andersson R, Lilje B, Drablos F, … Hayashizaki Y (2015). Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science, 347(6225), 1010–1014. 10.1126/science.1259418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold PR, Wells AD, & Li XC (2019). Diversity and Emerging Roles of Enhancer RNA in Regulation of Gene Expression and Cell Fate. Front Cell Dev Biol, 7, 377. 10.3389/fcell.2019.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj R, Jiang Y, Mao W, Jakovcevski M, Dincer A, Krueger W, … Akbarian S (2013). Conserved chromosome 2q31 conformations are associated with transcriptional regulation of GAD1 GABA synthesis enzyme and altered in prefrontal cortex of subjects with schizophrenia. J Neurosci, 33(29), 11839–11851. 10.1523/JNEUROSCI.1252-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai S, Akella A, Gandhi A, & Dharap A (2021). Modulation of Brain Pathology by Enhancer RNAs in Cerebral Ischemia. Mol Neurobiol, 58(4), 1482–1490. 10.1007/s12035-020-02194-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai S, Aly A, Garcia K, Ruiz D, Pontarelli F, & Dharap A (2019). Deep Sequencing Reveals Uncharted Isoform Heterogeneity of the Protein-Coding Transcriptome in Cerebral Ischemia. Mol Neurobiol, 56(2), 1035–1043. 10.1007/s12035-018-1147-0 [DOI] [PubMed] [Google Scholar]
- Bhattarai S, Pontarelli F, Prendergast E, & Dharap A (2017). Discovery of novel stroke-responsive lncRNAs in the mouse cortex using genome-wide RNA-seq. Neurobiol Dis, 108, 204–212. 10.1016/j.nbd.2017.08.016 [DOI] [PubMed] [Google Scholar]
- Bohnsack JP, Zhang H, Wandling GM, He D, Kyzar EJ, Lasek AW, & Pandey SC (2022). Targeted epigenomic editing ameliorates adult anxiety and excessive drinking after adolescent alcohol exposure. Sci Adv, 8(18), eabn2748. 10.1126/sciadv.abn2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose DA, Donahue G, Reinberg D, Shiekhattar R, Bonasio R, & Berger SL (2017). RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell, 168(1–2), 135–149 e122. 10.1016/j.cell.2016.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullido MJ, Artiga MJ, Recuero M, Sastre I, Garcia MA, Aldudo J, … Valdivieso F (1998). A polymorphism in the regulatory region of APOE associated with risk for Alzheimer's dementia. Nat Genet, 18(1), 69–71. 10.1038/ng0198-69 [DOI] [PubMed] [Google Scholar]
- Carullo NVN, Phillips Iii RA, Simon RC, Soto SAR, Hinds JE, Salisbury AJ, … Day JJ (2020). Enhancer RNAs predict enhancer-gene regulatory links and are critical for enhancer function in neuronal systems. Nucleic Acids Res, 48(17), 9550–9570. 10.1093/nar/gkaa671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Lysakovskaia K, Stik G, Demel C, Soding J, Tian TV, … Cramer P (2021). Evidence for additive and synergistic action of mammalian enhancers during cell fate determination. Elife, 10. 10.7554/eLife.65381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, E. P. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature, 489(7414), 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradin O, & Scacheri PC (2014). Enhancer variants: evaluating functions in common disease. Genome Med, 6(10), 85. 10.1186/s13073-014-0085-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, … Quake SR (2015). A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A, 112(23), 7285–7290. 10.1073/pnas.1507125112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S, Lear M, Shanley L, Hing B, Baizan-Edge A, Herwig A, … MacKenzie A (2011). Differential activity by polymorphic variants of a remote enhancer that supports galanin expression in the hypothalamus and amygdala: implications for obesity, depression and alcoholism. Neuropsychopharmacology, 36(11), 2211–2221. 10.1038/npp.2011.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Stein JL, Won H, Opland CK, Liang D, Lu D, & Geschwind DH (2018). The Dynamic Landscape of Open Chromatin during Human Cortical Neurogenesis. Cell, 172(1-2), 289–304 e218. 10.1016/j.cell.2017.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, … Natoli G (2010). A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol, 8(5), e1000384. 10.1371/journal.pbio.1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Bowen K, Place R, Li LC, & Vemuganti R (2009). Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab, 29(4), 675–687. 10.1038/jcbfm.2008.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Nakka VP, & Vemuganti R (2011). Altered expression of PIWI RNA in the rat brain after transient focal ischemia. Stroke, 42(4), 1105–1109. 10.1161/STROKEAHA.110.598391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Nakka VP, & Vemuganti R (2012). Effect of focal ischemia on long noncoding RNAs. Stroke, 43(10), 2800–2802. 10.1161/STROKEAHA.112.669465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, … Gingeras TR (2012). Landscape of transcription in human cells. Nature, 489(7414), 101–108. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Liao Z, Gritsch D, Hadzhiev Y, Bai Y, Locascio JJ, … Scherzer CR (2018). Enhancers active in dopamine neurons are a primary link between genetic variation and neuropsychiatric disease. Nat Neurosci, 21(10), 1482–1492. 10.1038/s41593-018-0223-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckart N, Song Q, Yang R, Wang R, Zhu H, McCallion AS, & Avramopoulos D (2016). Functional Characterization of Schizophrenia-Associated Variation in CACNA1C. PLoS One, 11(6), e0157086. 10.1371/journal.pone.0157086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, & Greenberg ME (2008). Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci, 31, 563–590. 10.1146/annurev.neuro.31.060407.125631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francelle L, Lotz C, Outeiro T, Brouillet E, & Merienne K (2017). Contribution of Neuroepigenetics to Huntington's Disease. Front Hum Neurosci, 11, 17. 10.3389/fnhum.2017.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CL, Liu F, Wijayatunge R, Song L, Biegler MT, Yang MG, … West AE (2015). Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nat Neurosci, 18(5), 647–656. 10.1038/nn.3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallitano AL (2020). Editorial: The Role of Immediate Early Genes in Neuropsychiatric Illness. Front Behav Neurosci, 14, 16. 10.3389/fnbeh.2020.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayen S, & Kalantry S (2017). Chromatin-enriched lncRNAs: a novel class of enhancer RNAs. Nat Struct Mol Biol, 24(7), 556–557. 10.1038/nsmb.3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbovytska V, Kim SK, Kuybu F, Gotze M, Um D, Kang K, … Kuhn CD (2022). Enhancer RNAs stimulate Pol II pause release by harnessing multivalent interactions to NELF. Nat Commun, 13(1), 2429. 10.1038/s41467-022-29934-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, & Li W (2022). Enhancer RNA: What we know and what we can achieve. Cell Prolif, 55(4), e13202. 10.1111/cpr.13202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauberg ME, Fullard JF, Zhu L, Cohain AT, Giambartolomei C, Misir R, … CommonMind C (2019). Differential activity of transcribed enhancers in the prefrontal cortex of 537 cases with schizophrenia and controls. Mol Psychiatry, 24(11), 1685–1695. 10.1038/s41380-018-0059-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, … Ren B (2009). Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature, 459(7243), 108–112. 10.1038/nature07829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, & Glass CK (2015). The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol, 16(3), 144–154. 10.1038/nrm3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx A, Pierrot N, Tasiaux B, Schakman O, Kienlen-Campard P, De Smet C, & Octave JN (2014). Epigenetic regulations of immediate early genes expression involved in memory formation by the amyloid precursor protein of Alzheimer disease. PLoS One, 9(6), e99467. 10.1371/journal.pone.0099467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang X, … Kantoff PW (2014). Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci U S A, 111(20), 7319–7324. 10.1073/pnas.1324151111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue YU, & Inoue T (2016). Brain enhancer activities at the gene-poor 5p14.1 autism-associated locus. Sci Rep, 6, 31227. 10.1038/srep31227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, … Ren B (2013). A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature, 503(7475), 290–294. 10.1038/nature12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi M, Hara N, Hasegawa M, Miyashita A, Kuwano R, Ikeuchi T, & Nakaya A (2019). Enhancer variants associated with Alzheimer's disease affect gene expression via chromatin looping. BMC Med Genomics, 12(1), 128. 10.1186/s12920-019-0574-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, … Greenberg ME (2010). Widespread transcription at neuronal activity-regulated enhancers. Nature, 465(7295), 182–187. 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YW, Lee S, Yun J, & Kim A (2015). Chromatin looping and eRNA transcription precede the transcriptional activation of gene in the beta-globin locus. Biosci Rep, 35(2). 10.1042/BSR20140126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyzar EJ, Zhang H, & Pandey SC (2019). Adolescent Alcohol Exposure Epigenetically Suppresses Amygdala Arc Enhancer RNA Expression to Confer Adult Anxiety Susceptibility. Biol Psychiatry, 85(11), 904–914. 10.1016/j.biopsych.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gras S, Keime C, Anthony A, Lotz C, De Longprez L, Brouillet E, … Merienne K (2017). Altered enhancer transcription underlies Huntington's disease striatal transcriptional signature. Sci Rep, 7, 42875. 10.1038/srep42875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Marshall L, Oh G, Jakubowski JL, Groot D, He Y, … Labrie V (2019). Epigenetic dysregulation of enhancers in neurons is associated with Alzheimer's disease pathology and cognitive symptoms. Nat Commun, 10(1), 2246. 10.1038/s41467-019-10101-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, … Rosenfeld MG (2013). Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature, 498(7455), 516–520. 10.1038/nature12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DZ, Tian Y, Ander BP, Xu H, Stamova BS, Zhan X, … Sharp FR (2010). Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab, 30(1), 92–101. 10.1038/jcbfm.2009.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusardi TA, Murphy SJ, Phillips JI, Chen Y, Davis CM, Young JM, … Saugstad JA (2014). MicroRNA responses to focal cerebral ischemia in male and female mouse brain. Front Mol Neurosci, 7, 11. 10.3389/fnmol.2014.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marballi KK, & Gallitano AL (2018). Immediate Early Genes Anchor a Biological Pathway of Proteins Required for Memory Formation, Long-Term Depression and Risk for Schizophrenia. Front Behav Neurosci, 12, 23. 10.3389/fnbeh.2018.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClymont SA, Hook PW, Soto AI, Reed X, Law WD, Kerans SJ, … McCallion AS (2018). Parkinson-Associated SNCA Enhancer Variants Revealed by Open Chromatin in Mouse Dopamine Neurons. Am J Hum Genet, 103(6), 874–892. 10.1016/j.ajhg.2018.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, … Osborne CS (2015). Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet, 47(6), 598–606. 10.1038/ng.3286 [DOI] [PubMed] [Google Scholar]
- Minatohara K, Akiyoshi M, & Okuno H (2015). Role of Immediate-Early Genes in Synaptic Plasticity and Neuronal Ensembles Underlying the Memory Trace. Front Mol Neurosci, 8, 78. 10.3389/fnmol.2015.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Dell'orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, … Sartorelli V (2013). eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell, 51(5), 606–617. 10.1016/j.molcel.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, … Visel A (2013). Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell, 155(7), 1521–1531. 10.1016/j.cell.2013.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra Rao VL, Bowen KK, Dhodda VK, Song G, Franklin JL, Gavva NR, & Dempsey RJ (2002). Gene expression analysis of spontaneously hypertensive rat cerebral cortex following transient focal cerebral ischemia. J Neurochem, 83(5), 1072–1086. 10.1046/j.1471-4159.2002.01208.x [DOI] [PubMed] [Google Scholar]
- Ramirez M, Badayeva Y, Yeung J, Wu J, Abdalla-Wyse A, Yang E, … Goldowitz D (2022). Temporal analysis of enhancers during mouse cerebellar development reveals dynamic and novel regulatory functions. Elife, 11. 10.7554/eLife.74207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M, Robert R, Yeung J, Wu J, Abdalla-Wyse A, Consortium, F., & Goldowitz D (2023). Identification and characterization of transcribed enhancers during cerebellar development through enhancer RNA analysis. BMC Genomics, 24(1), 351. 10.1186/s12864-023-09368-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz D, Bhattarai S, & Dharap A (2021). Sex-based eRNA expression and function in ischemic stroke. Neurochem Int, 150, 105149. 10.1016/j.neuint.2021.105149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, & Dekker J (2012). The long-range interaction landscape of gene promoters. Nature, 489(7414), 109–113. 10.1038/nature11279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, & Kim TK (2014). Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell, 56(1), 29–42. 10.1016/j.molcel.2014.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Kastner R, Zhang B, Belayev L, Khoutorova L, Amin R, Busto R, & Ginsberg MD (2002). DNA microarray analysis of cortical gene expression during early recirculation after focal brain ischemia in rat. Brain Res Mol Brain Res, 108(1–2), 81–93. 10.1016/s0169-328x(02)00516-8 [DOI] [PubMed] [Google Scholar]
- Shechner DM, Hacisuleyman E, Younger ST, & Rinn JL (2015). Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods, 12(7), 664–670. 10.1038/nmeth.3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, … Jaenisch R (2016). Parkinson-associated risk variant in distal enhancer of alpha-synuclein modulates target gene expression. Nature, 533(7601), 95–99. 10.1038/nature17939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan KS, Armugam A, Sepramaniam S, Lim KY, Setyowati KD, Wang CW, & Jeyaseelan K (2009). Expression profile of MicroRNAs in young stroke patients. PLoS One, 4(11), e7689. 10.1371/journal.pone.0007689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Lu A, Aronow BJ, Wagner KR, & Sharp FR (2002). Genomic responses of the brain to ischemic stroke, intracerebral haemorrhage, kainate seizures, hypoglycemia, and hypoxia. Eur J Neurosci, 15(12), 1937–1952. 10.1046/j.1460-9568.2002.02030.x [DOI] [PubMed] [Google Scholar]
- Tischmeyer W, & Grimm R (1999). Activation of immediate early genes and memory formation. Cell Mol Life Sci, 55(4), 564–574. 10.1007/s000180050315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PF, Dell'Orso S, Rodriguez J, Vivanco KO, Ko KD, Jiang K, … Sartorelli V (2018). A Muscle-Specific Enhancer RNA Mediates Cohesin Recruitment and Regulates Transcription In trans. Mol Cell, 71(1), 129–141 e128. 10.1016/j.molcel.2018.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Kwon JM, Shah P, Morris JC, & Goate A (2000). Effect of APOE genotype and promoter polymorphism on risk of Alzheimer's disease. Neurology, 55(11), 1644–1649. 10.1212/wnl.55.11.1644 [DOI] [PubMed] [Google Scholar]
- Watts JA, Grunseich C, Rodriguez Y, Liu Y, Li D, Burdick JT, … Cheung VG (2022). A common transcriptional mechanism involving R-loop and RNA abasic site regulates an enhancer RNA of APOE. Nucleic Acids Res, 50(21), 12497–12514. 10.1093/nar/gkac1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Chen J, Chen X, Wu S, Chen Z, Huang Y, … He W (2021). Screening for differentially expressed circRNAs in ischemic stroke by RNA sequencing. BMC Neurol, 21(1), 370. 10.1186/s12883-021-02397-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner MS, Sullivan MA, Shah RN, Nadadur RD, Grzybowski AT, Galat V, … Ruthenburg AJ (2017). Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat Struct Mol Biol, 24(7), 596–603. 10.1038/nsmb.3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, … Gilmour D (2003). NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev, 17(11), 1402–1414. 10.1101/gad.1091403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, … Chen LL (2014). Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res, 24(5), 513–531. 10.1038/cr.2014.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Inukai N, Narita T, Wada T, & Handa H (2002). Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol, 22(9), 2918–2927. 10.1128/MCB.22.9.2918-2927.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao P, Lin P, Gokoolparsadh A, Assareh A, Thang MW, & Voineagu I (2015). Coexpression networks identify brain region-specific enhancer RNAs in the human brain. Nat Neurosci, 18(8), 1168–1174. 10.1038/nn.4063 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Wang L, Ren S, Wang L, Blackburn PR, McNulty MS, … Huang H (2016). Activation of P-TEFb by Androgen Receptor-Regulated Enhancer RNAs in Castration-Resistant Prostate Cancer. Cell Rep, 15(3), 599–610. 10.1016/j.celrep.2016.03.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Lin L, Jiang W, Chen L, Zhang X, Zhang Q, … Hao J (2022). Single-cell RNA-seq reveals the transcriptional landscape in ischemic stroke. J Cereb Blood Flow Metab, 42(1), 56–73. 10.1177/0271678X211026770 [DOI] [PMC free article] [PubMed] [Google Scholar]