Abstract
We have used phage-displayed peptide libraries to identify novel ligands to the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein gp120. Screening of libraries of random 12-mers, 7-mers, and cyclic 9-mers produced two families of gp120 binding peptides. Members of a family with the prototype sequence RINNIPWSEAMM (peptide 12p1) inhibit the interaction between gp120 and both four-domain soluble CD4 (4dCD4) and monoclonal antibody (MAb) 17b, a neutralizing antibody that covers the chemokine receptor binding surface on gp120. Peptide 12p1 inhibits the interaction of 4dCD4 with gp120 from three different HIV strains, implying that it binds to a conserved site on gp120. Members of a second family of peptides, with the prototype sequence TSPYEDWQTYLM (peptide 12p2), bind more weakly to gp120. They do not detectably affect its interaction with 4dCD4, but they enhance its binding to MAb 17b. A common sequence motif in the two peptide families and cross-competition for gp120 binding suggest that they have overlapping contacts. Their divergent effects on the affinity of gp120 for MAb 17b may indicate that their binding stabilizes distinct conformational states of gp120. The functional properties of 12p1 suggest that it might be a useful lead for the development of inhibitors of HIV entry.
Chemical ligands to virus surface proteins can inhibit entry by trapping intermediate conformational states. Entry of enveloped viruses is initiated through recognition of cellular receptors by envelope (Env) glycoproteins. The Env glycoprotein of human immunodeficiency virus type 1 (HIV-1) consists of two noncovalently associated polypeptide chains, gp120 and gp41, derived by cleavage of a single precursor (gp160). gp120 is the surface subunit responsible for binding of the virus to the target cells, and gp41 is the transmembrane subunit involved in the membrane fusion step. Binding of gp120 to cellular CD4 is thought to induce a conformational change in gp120 (24, 25), thereby allowing it to interact with a chemokine receptor on the cell surface (16, 29, 32, 39). The interaction of gp120 with these cellular receptors in turn releases gp41 from a metastable conformation into a fusion-active state (4, 30, 33). A triggered release of the fusogenic conformation of the viral envelope protein appears to be a general mechanism of infection by enveloped viruses (2, 3, 10).
The multiple conformational changes that gp120 and gp41 undergo upon binding to the cellular receptors provide various targets for the development of antiviral ligands. Recently, it has been shown that peptide ligands that bind to gp41 have antiviral activity and can reduce viral loads in humans (12, 14, 34–36). It is proposed that the inhibitory activity of these peptides derives from their ability to trap an early fusion-inactive conformation of gp41, preventing it from developing a fully fusion-competent complex (8). The search for ligands that bind specifically to gp120 and strongly affect the viral entry process has been less successful. The interaction between CD4 and gp120 has been the prime target for discovery of antiviral gp120 ligands (9, 11, 15, 21). Extensive mutagenesis studies have mapped the gp120 binding site on the C′C"D strands of domain 1 of CD4 and identified Phe43 and Arg59 as key side chains (1, 17, 18, 23, 26, 27, 38), but neither linear nor cyclic peptides that include and extend the sequence of the C′C"D strands have significant inhibitory activity (9, 11). The crystal structure (13) of a ternary complex of gp120, CD4, and the Fab from monoclonal antibody (MAb) 17b (31), an HIV neutralizing MAb against a CD4-induced site on gp120, helps explain this failure to design inhibitory peptides. The structure shows that the C" ridge of CD4 indeed contacts gp120 and that the critical Phe43 side chain inserts into the mouth of a hydrophobic cavity. In addition, the exposed edge of the C" strand of CD4 has β-sheet-like hydrogen bonds with a strand in gp120 and Arg59 at the end of the D strand has salt bridges with conserved acid residues on the viral glycoprotein. Thus, there appears to be an extended set of contacts with defined spatial relationships, which are difficult to incorporate into designed ligands based on inspection of CD4 alone. Moreover, the snapshot of the CD4-bound conformational state seen in the crystal structure suggests that significant conformational rearrangements may accompany CD4 binding and that the contacts just described are probably not possible with gp120 in its unliganded form. One of the likely consequences of the conformational rearrangements is exposure of the 17b epitope, which corresponds roughly to the chemokine receptor site (22).
As an alternative approach to discovery of ligands to a CD4-unbound state of gp120, we have turned to screening of random peptide libraries. In particular, we have used phage-displayed peptide libraries (28) to isolate peptide ligands that interact specifically with gp120. These peptides fall into two sets: one that inhibits CD4 and MAb 17b binding, another that apparently does not affect the CD4 interaction while enhancing affinity for 17b. We have further characterized one member of the former set, which might provide a useful beginning for the design of molecules that inhibit HIV attachment.
MATERIALS AND METHODS
General.
CD41-371 (4dCD4), CD41-183 (2dCD4), HXB2-gp120, and MAb 803-15.6 were from Procept, Inc., and were prepared as described elsewhere (6, 7). SF2-gp120 and ADA-gp100 were gifts from Ellis Reinherz’s laboratory at the Dana Farber Cancer Institute. MAb 17b (originally from the laboratory of James Robinson at Tulane University) was a gift from Joseph Sodroski’s laboratory at the Dana Farber Cancer Institute. Peptides were synthesized by standard solid-phase 9-fluorenylmethoxycarbonyl (Fmoc) methods on a PAL support (Perseptive Biosystems, Framingham, Mass.), which yields peptide amides, using a 431 Applied Biosystems peptide synthesizer. Fmoc-amino acids were purchased from Peptides International (Louisville, Ky.) and Perseptive Biosystems. Fmoc-benzoyl phenylalanine (Fmoc-Bpa) was purchased from Bachem Bioscience (King of Prussia, Pa.). 6-Biotinoyl-hexanoic N-succinimide anhydride (biotin-ONSu) was from Molecular Probes (Eugene, Oreg.). Peptides were purified by high-pressure liquid chromatography on a C18 reversed-phase column, eluting with a gradient of H2O-acetonitrile with 0.1% trifluoroacetic acid. The identity of the peptides was confirmed by fast atom bombardment mass spectrometry (Mass Spectrometry Facility, Department of Chemistry and Biological Chemistry, Harvard University). Single-stranded DNA from phage M13 was sequenced at the Biopolymers Facility, Harvard Medical School.
Phage selection.
Phage libraries were purchased from New England Biolabs (Beverly, Mass.). Peptides are fused to the N terminus of the protein of gene III, with a GGS spacer. For phage selection, Immulon-2 microplates (Dynatech Inc., Chantilly, Va.) were coated with MAb 803-15.6 (0.150 ml, 100 μg/ml in phosphate-buffered saline [PBS]) for 3 h at 25°C. Microplates were blocked with PBSTB (PBS, 0.05% Tween 20, 10 mg of bovine serum albumin per ml [BSA]) for 2 h at 4°C, and HXB2-gp120 (0.15 ml, 10 μg/ml in PBSTB) was then added for 16 h at 4°C. The plates were washed with cold PBST (PBS, 0.05% Tween 20) and incubated for 1 h at 4°C with the phage suspension (0.15 ml containing 1011 PFU) in the presence of 1 mM gp160(502–516) (APTKAKRRVVQREKR). Unbound phage were removed by washing 10 times with cold PBST (4°C). In the first round of selection, bound phage were eluted with 0.2 M glycine (pH 2.2)–10 mg of BSA per ml (0.15 ml) for 10 min at 25°C. Eluted phage were immediately neutralized in 20 μl of 2 M Tris (pH 9.2). In subsequent rounds of selection, phage were eluted with 0.15 ml of a 1 mM solution of gp160(502–516) in PBSTB (30 min at 37°C). This solution was then warmed to 60°C for 10 min to release gp120-bound phage. Eluted phage were immediately amplified by infecting a mid-log-phase 298F′ Escherichia coli cell culture and incubating it for 4.5 h at 37°C with vigorous shaking. Phage were obtained by double precipitation with a polyethylene glycol-NaCl solution and dissolved in PBS to a final concentration of ∼1013 PFU/ml.
Competition ELISA for binding of gp120 to 4dCD4.
Immulon-2 microplates were coated with 4dCD4 (0.1 ml, 1 μg/ml in PBS) for 3 h at 25°C. Control plates were coated with PBS alone. The plates were then blocked with 0.2 ml of PBSTB for 16 h at 4°C. The gp120 (15 ng/ml for HXB2 and SF2 and 250 ng/ml for ADA, in PBSTB) and soluble 2dCD4 (0 to 100 nM in PBSTB) or peptide inhibitor (0 to 500 μM in PBSTB) (final total volume, 0.2 ml in PBSTB) were then added to the plates and incubated for 3 h at 25°C. After the plates were washed five times with cold PBST, bound gp120 was probed with MAb 803-15.6 (0.1 ml, 50 ng/ml for HXB2 and SF2, and 1 μg/ml for ADA, in PBSTB) for 2 h at 4°C. Bound MAb was detected by incubation with alkaline phosphatase secondary antibody (0.1 ml, 1/400 to 1/2,000 anti-mouse AP-IgG1 [Southern Biotechnologies] in PBSTB) for 1 h at 4°C. After another PBST wash, the enzyme-linked immunosorbent assay (ELISA) product was developed using 0.1 ml of alkaline phosphatase substrate solution (Bio-Rad).
SPR competition assay for binding of gp120 to MAb 17b.
Surface plasmon resonance (SPR) binding assays were performed on a Biacore (Piscataway, N.J.) instrument at 25°C in HBSTB buffer (10 mM HEPES [pH 7.4] containing 0.15 M NaCl, 3.4 mM EDTA, 0.1% Tween 20, and 1 mg of BSA per ml) at a constant flow rate of 10 μl/min. The MAb 17b was coupled to a CM5 sensor chip through protein amino groups by using the standard amino coupling kit (Biacore Inc.). MAb 17b was dissolved in 10 mM acetic acid-sodium acetate buffer (pH 4.9). The surface was regenerated twice with 5 μl of 4.5 M MgCl2 after gp120 binding. Sequential injections (20 μl) of HXB2-gp120 (200 nM) with peptide at increasing concentrations (0.2 to 200 μM) were made over a surface with 1,500 RU of coupled MAb 17b. The amount of gp120 bound (expressed as surface plasmon resonance units [RU]) was measured 10 s after the end of the injection and by subtracting RUs bound for the same solution on a blank mock-coupled surface. Results are given as the percentage of gp120 bound relative to gp120 bound in the absence of peptide: 100 + 100 × [(RU − RUgp120)/RUgp120].
UV cross-linking of Bpa-peptide to gp120.
12p1(Arg1/Bpa) was obtained by automated peptide synthesis. Biotin-ONSu was coupled to the N terminus with diisopropylethylamine in dimethylformamide overnight at room temperature. The activity of biotin-12p1(Arg1/Bpa) peptide was confirmed by the CD4-gp120 competition ELISA described above (data not shown). Cross-linking reactions were carried out in Falcon 3912 assay microplates (Becton Dickinson, Oxnard, Calif.). In a typical cross-linking reaction, 2 μl of a 7 μM Bpa-peptide solution in PBS, 2 μl of a 2 μM SF2-gp120 solution in PBS, and 2 μl of competitor solution in PBS, were mixed in the dark and incubated for 10 min at room temperature. The solution was then frozen on dry ice in the dark and irradiated for 12 min with a long-wavelength UV source (for the method, see reference 20); 2 μl of reducing sodium dodecyl sulfate loading buffer was then added, and the solution was boiled for 3 min to stop the reaction. Western blotting was performed by standard procedures, as follows. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% polyacrylamide) under reducing conditions and transferred in 10 mM CAPS (pH 11.0), at 150 mA for 1.5 h, to Immobilon-P polyvinylidene difluoride membranes (Millipore Corp., Bedford, Mass.) by using a Mini Trans-Blot cell (Bio-Rad). The membranes were blocked with PBSTB for 2 h at 25°C and incubated with a 1/204 dilution streptavidin-horseradish peroxidase (Pierce) in PBSTB for 1 h at 25°C. Bound streptavidin was detected with the enhanced chemiluminescence substrate (Pierce).
RESULTS
Screening of phage-displayed peptide libraries against gp120.
Three random peptide libraries, a linear (X)7, a cyclic Cys(X)7Cys, and a linear (X)12 were screened against gp120. The peptide libraries were expressed at the N terminus of the protein of gene III of the M13 phage. There are up to five copies of the protein at one end of each phage particle. Initially, the peptide libraries were screened against microplates coated directly with gp120, using a pH 2.2 solution to elute, but after eight rounds of selection this screening protocol did not yield any enrichment above background (determined by screening against BSA-coated plates in a parallel experiment). The screening strategy was therefore changed to favor multivalent interactions. Multivalent interactions between the phage and the protein on the surface should allow for the selection of weak binders (Kd is above the micromolar range). A sandwich format similar to that used to obtain ligands to the erythropoietin receptor (37) was developed to maximize the amount of CD4 binding to gp120 on the microplate and to make it possible to elute gp120-bound phage specifically. In our sandwich format, gp120 is attached to the plate through a MAb (MAb 803-15.6) directed to the gp120 C terminus (C5 region) (7), a site that is located on the side opposite the CD4 and chemokine binding regions. The principle of using anti-C5 antibodies to capture gp120 onto a solid-phase support has been described previously, in the context of an ELISA to test gp120 binding to CD4 and other anti-gp120 antibodies (19). By using the sandwich format, the amount of CD4-binding gp120 on the plate is maximized (data not shown). Moreover, we expected that two gp120 molecules could attach to a single antibody molecule and thus be close enough that one phage particle could interact simultaneously with more than one protein molecule.
Initial screening by using the sandwich format with acid elution yielded only peptides that bound to MAb 803-15.6, even though a prescreening step with antibody alone on the plate was carried out in the screening protocol. To improve the specificity of the enrichment toward gp120, phage were screened in the presence of the peptide gp160(502–516) (APTKAKRRVVQREKR), which corresponds to the MAb 803-15.6 epitope (7), to block any antibody sites that remained free of gp120. This step was carried out at low temperature (4°C), so that the peptide did not induce the dissociation of gp120 from the antibody. Specific elution of gp120-bound phage was achieved by releasing gp120 by adding gp160(502–516) peptide and warming the mixture to 37°C. To dissociate the bound phage from gp120, a final incubation at 60°C for 10 min was added just before infection of the cell culture.
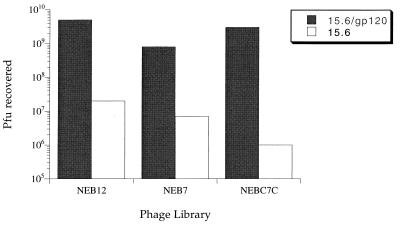
The three libraries were screened against gp120 in the sandwich format by using gp160(502–516) peptide blocking and either peptide or acid elution (see Materials and Methods). The library of 12-mers was enriched >100-fold above background after three to four rounds of selection. After five or six rounds of selection, both the library of 7-mers and the library of cyclic 9-mers were also enriched >100-fold above background levels. The specificity of the enriched phage toward gp120 was determined by screening against MAb 803-15.6 alone. In all cases, the number of phage recovered decreased by at least 100-fold (Fig. 1). Phage from individual plaques were amplified, and the single-stranded DNA was extracted by NaI-polyethylene glycol precipitation and sequenced. The resulting peptide sequences are shown in Table 1.
FIG. 1.
PFU recovered when 1011 PFU of phage solutions, obtained after four or five rounds of enrichment against MAb 803-15.6/gp120, were screened against mAb 803-15.6/gp120 or mAb 803-15.6 alone. For each of the three phage-displayed peptide libraries tested, PFU recovered from mAb 803-15.6/gp120 plates was at least 100-fold higher than from MAb 803-15.6 plates, indicating that the phage libraries had been specifically enriched for interaction with gp120.
TABLE 1.
Amino acid sequences of the phage-displayed peptides isolated by screening against HXB2-gp120
| Peptidea | Sequence | Frequency of isolatesb |
|---|---|---|
| 12p1 | RINNIPWSEAMM | 8 (13)c |
| 12p2 | TSPYEDWQTYLM | 5 (13)c/3 (8)d |
| 12p3 | DTHRWPWYISQE | 1 (8)d |
| 12p4 | SSVDESTAQVNW | 1 (8)d |
| 12p5 | SPWYSDYTRYLW | 1 (8)d |
| 12p6 | SDPYKLWASYMY | 1 (8)d |
| 12p7 | QPASNMTLGRGL | 1 (8)d |
| 7p1 | SWPWDTW | 3 (7)c |
| 7p2 | FWHPMHD | 4 (7)c |
| C7Cp1 | CHPLTPWEC | 6 (6)c |
Peptides are designated by the number of amino acids in the sequence, followed by the clone number.
Numbers in parentheses refer to the total number of plaques sequenced.
Peptide elution.
Acid elution.
Binding specificity of the peptides.
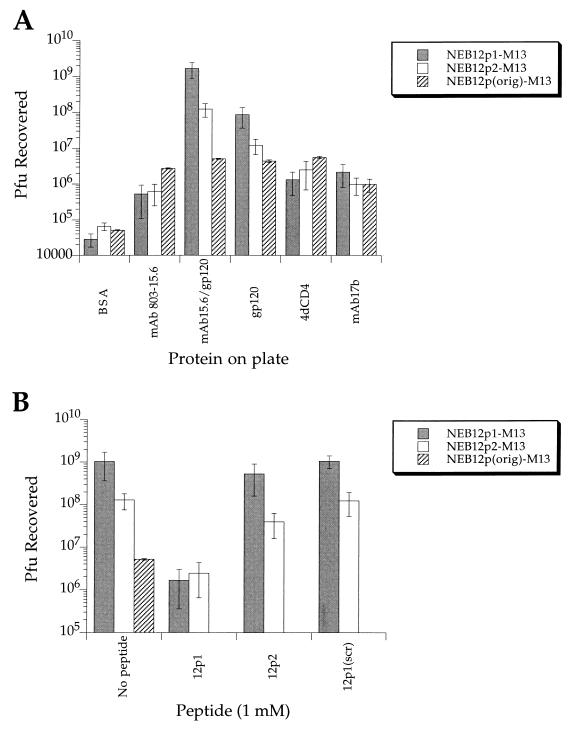
The binding specificity of the phage carrying the peptide sequences isolated was tested by enriching individual phage isolates and measuring the amount of phage adsorbed to a variety of proteins immobilized on microplates. The results for NEB12p1-M13 and NEB12p2-M13 are shown in Fig. 2A. Single plaques were amplified to ∼1013 PFU/ml, and aliquots containing 2 × 1011 PFU were screened against immobilized BSA, MAb 803-15.6 (100 μg/ml), gp120 (10 μg/ml), MAb 803-15.6 (100 μg/ml) plus gp120 (10 μg/ml), 4dCD4 (10 μg/ml), and MAb 17b (100 μg/ml). To account for nonspecific binding of the phage particles to the proteins tested, we measured the phage recovered when the original nonenriched library (2 × 1011 PFU) was tested with the same proteins. The results of screening the original nonenriched library show significant (10-fold above the BSA values) nonspecific binding of the phage particles to MAb 803-15.6, gp120, 4dCD4, and MAb 17b. Taking this background into account, the results indicate that phage carrying 12p1 have very high specificity (1,000-fold over background) for gp120. This is also true for phage carrying 12p2, although in this case the total number of phage recovered from MAb 803-15.6–plus–gp120 and gp120 plates was 10-fold smaller than the recovery of NEB12p1-M13, probably because 12p2 is a weaker binder. Phage carrying the peptide sequences 12p3, 12p4, 12p5, 12p6, and 12p7, which were obtained by screening with blocking and acid elution at all steps, were also amplified and tested for specific binding (data not shown). We found that phage NEB12p3, NEB12p5, and NEB12p6 bound specifically to gp120, but we did not detect significant binding of NEB12p4-M13 or NEB12p7-M13, either to gp120 or to any of the other proteins. These last two are likely to be irrelevant phage isolated when nonspecific acid elution was used in the screening method.
FIG. 2.
(A) PFU recovered when 1011 PFU of phage carrying peptide sequences 12p1, 12p2, and irrelevant 12-mer sequences (original nonenriched 12-mer library) were screened against proteins immobilized on microplates. PFU recovered from phage carrying irrelevant sequences indicates nonspecific binding of the phage particle to the proteins (background binding). (B) PFU recovered when 1011 PFU of phage carrying peptide sequences 12p1 and 12p2 were screened against MAb 803-15.6/gp120 microplates in the absence or presence of 1 mM solution of peptides 12p1, 12p2, and 12p1[Scrambled].
Peptides 12p1 and 12p2 bind to interacting sites.
To study whether 12p1 and 12p2 bind to independent sites on gp120, we measured the ability of the free peptides to inhibit the binding of phage-displayed peptides to gp120. Figure 2B shows the PFU recovered when ∼2 × 1011 PFU of NEB12p1-M13 or NEB12p2-M13 were screened against gp120, immobilized through MAb 803-15.6 on microplates, in the absence or presence of 1 mM 12p1, 12p2, or 12p1[Scrambled]. The results show that 12p1 can efficiently compete the binding of both 12p1- and 12p2-displaying phage, indicating either that there is some binding-site overlap or that the two sites are in allosteric contact. 12p2 is a much weaker competitor but still reduces the PFU recovered for both 12p1 and 12p2 carrying phage. A control with a 12p1 scrambled sequence showed no decrease in recovered PFU, confirming that the effect observed for 12p1 is sequence specific.
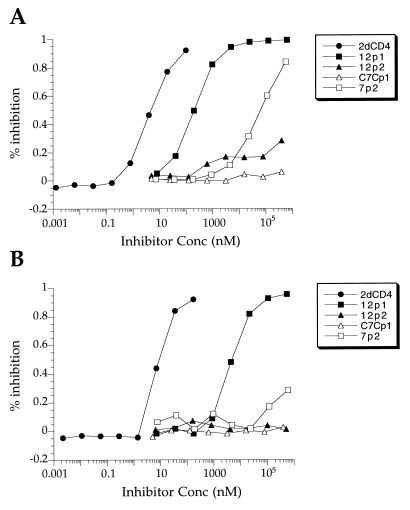
Effect of the peptides on the interaction between 4dCD4 and gp120.
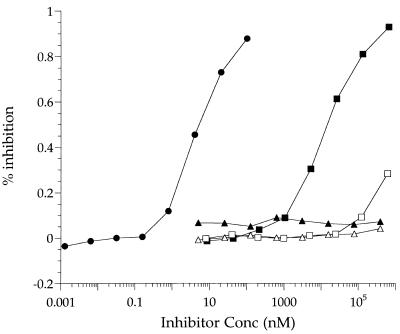
The gp120 binding peptides were tested for inhibition of CD4-gp120 binding by using a 4dCD4/HXB2-gp120 competition ELISA (Table 2 and Fig. 3). In this assay, 2dCD4 had a 50% inhibitory concentration (IC50) of 9 nM, a value that agrees with the Kd of 10 nM measured for 4dCD4 and HXB2-gp120 by SPR (7a). Two of the dodecamers, 12p1 and 12p3, had IC50 of 26 and 500 μM, respectively. 12p2, 12p5, and 12p6 did not have any inhibitory activity. The sequence specificity of inhibition by 12p1 was tested by using a scrambled version. This peptide had no inhibitory activity at concentrations up to 500 μM. The two heptamers, 7p1 and 7p2, reduced the binding of gp120 and CD4 by about 30% at 130 and 500 μM, respectively, the highest concentrations tested. A unique sequence was isolated from the library of cyclic peptides, but it did not inhibit the interaction between CD4 and gp120.
TABLE 2.
IC50s for inhibition of the 4dCD4-gp120 interaction as measured by competition ELISA
| Peptide | Peptide sequence | IC50a (μM)
|
||
|---|---|---|---|---|
| T-tropic gp120
|
M-tropic gp120
|
|||
| HXB2 | SF2 | ADA | ||
| 2dCD4 | 8.7 ± 2.1 nM (13) | 6.1 ± 1.6 nM (4) | 9.4 ± 0.5 nM (2) | |
| 12p1 | RINNIPWSEAMM | 25.7 ± 7.3 (13) | 0.33 ± 0.13 (6) | 4.2 ± 0.7 (2) |
| 12p1[2–8] | INNIPWS | 610 ± 130 (2) | 12 ± 3 (2) | NDb |
| 12p1[4–10] | NIPWSEA | >500 | >500 | ND |
| 12p1[6–12] | PWSEAMM | >500 | >500 | ND |
| 12p1[Scrambled] | IPAIMENMWNRS | >500 | >500 | >500 |
| 12p2 | TSPYEDWQTYLM | >500 | >500 | >500 |
| 12p3 | DTHRWPWYISQE | 500 | 259 ± 12 (2) | 348 ± 3 (2) |
| 12p5 | SPWYSDYTRYLW | >140c | >140c | ND |
| 12p6 | SDPYKLWASYMY | >100c | >100c | ND |
| 7p1 | SWPWDTW | >130c | 21 (1) | ND |
| 7p2 | FWHPMHD | >500 | 68 ± 4 (2) | >500 |
| C7Cp1 | CHPLTPWEC | >500 | >500 | >500 |
Values are means ± standard errors; the number of experiments is given in parentheses.
ND, not determined.
Low solubility prevented higher concentrations from being attained.
FIG. 3.
4dCD4/HXB2-gp120 competition ELISA. Immulon-2 microplates were coated with 4dCD4 and incubated for 3 h at 25°C with HXB2-gp120 in the presence of inhibitor. Bound gp120 was detected by incubating first with MAb 803-15.6 and then with AP-IgG1 and developing with alkaline phosphatase substrate. Solid circles, 2dCD4; solid squares, 12p1; solid triangles, 12p2; open triangles, C7Cp1; open squares, 7p2.
Effect of the peptides on the interaction between MAb 17b and gp120.
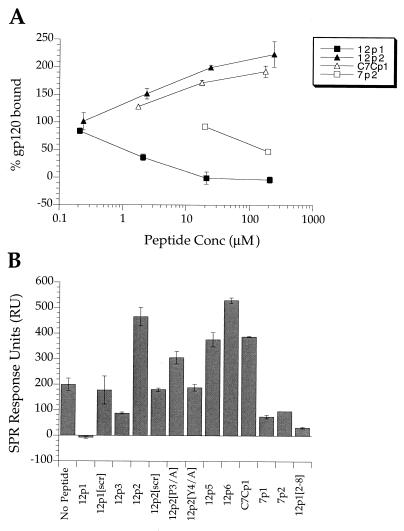
MAb 17b is an HIV neutralizing monoclonal antibody with a binding site on gp120 that overlaps the chemokine receptor binding surface (22). The affinity of 17b for gp120 is enhanced in a CD4-gp120 complex (31). The crystal structure of the ternary complex of gp120, CD4, and the 17b antigen-binding fragment (Fab) shows that CD4 and 17b bind on opposite sides of the so-called bridging sheet of gp120, suggesting that this structural element might play a critical role in the conformational changes that gp120 undergoes upon CD4 binding (13). It also suggests that 17b might to some extent serve as a “surrogate” for the chemokine receptor. The allosteric interaction between the CD4 and MAb 17b binding sites prompted us to study whether the peptides isolated as described above were able to affect the affinity of gp120 for 17b in the same way that CD4 does. The peptides were tested for inhibition of the interaction between gp120 and MAb 17b by using an SPR competition assay (Fig. 4). The results show that 12p1, 12p3, 7p1, and 7p2 inhibit the binding of gp120 to immobilized MAb 17b while 12p2, 12p5, 12p6, and C7Cp1 actually enhance this association. We note that peptides that inhibit the interaction between MAb 17b and gp120 also inhibit the binding of CD4 and gp120 whereas those that increase the affinity of gp120 for MAb 17b do not appear to inhibit the association of CD4 and gp120. The enhancement by 12p2 is sequence specific, since a scrambled version with the sequence PQMTYSDYLWTE (12p2[Scrambled]) does not have activity (Fig. 4B). All the peptide sequences isolated that bind to gp120 contain a proline followed by a hydrophobic residue. We investigated whether this motif is critical for 12p2 enhancement of MAb 17b-gp120 affinity. Substitution of Ala for Pro3 (12p2[P3/A]) or Tyr4 (12p2[Y4/A]) in 12p2 diminished activity significantly, indicating that these amino acid side chains are important for binding of the peptide to gp120 (Fig. 4B). The importance of the Pro-hydrophobic amino acid motif on the activity of 12p1 was studied in more detail by alanine scan and truncation experiments (see below).
FIG. 4.
(A) MAb 17b/HXB2-gp120 competition assay by SPR. Sequential injections of HXB2-gp120 (at 200 nM) with peptide at increasing concentrations (0.2 to 200 μM) were made over a surface with 1,500 RU of coupled MAb 17b. The results are given as percentage of gp120 bound relative to gp120 bound in the absence of peptide. (B) gp120 bound to 1,500 RU of coupled MAb 17b when 200 nM solutions of gp120 were injected after mixing with 12p1 (210 μM), 12p1[Scrambled] (200 μM), 12p3 (202 μM), 12p2 (240 μM), 12p2[Scrambled] (272 μM), 12p2[P3/A] (220 μM), 12p2[Y4/A] (277 μM), 12p5 (50 μM), 12p6 (50 μM), C7Cp1 (176 μM), 7p1 (204 μM), 7p2 (196 μM), and 12p1[2–8] (301 μM).
Effect of the peptides on the interaction between 4dCD4 and gp120 from different viral strains.
The functional properties of 12p1 suggest that this peptide could be used as a potential lead for the development of HIV inhibitors. We further explored whether the inhibitory effect of 12p1 on CD4-gp120 binding depends on the viral strain. The inhibitory capacities of the identified peptides were tested with gp120 from an additional T-tropic strain (SF2) and an M-tropic strain (ADA) (Table 2 and Fig. 5). 12p1 inhibited the interaction of CD4 with gp120 from ADA with an IC50 of 5 μM and was an even stronger inhibitor of the SF2 strain (IC50, 0.3 μM). The remarkable inhibitory effect with the SF2 strain was also observed for the heptapeptides. The different IC50s obtained for different gp120 strains are not likely to reflect the different affinity of each gp120 for 4dCD4, because in each case 2dCD4 inhibits gp120 binding with similar IC50. Thus, 12p1 can inhibit the interaction between CD4 and gp120 from three different HIV strains, although with different IC50s, suggesting that it binds to a largely conserved site on gp120.
FIG. 5.
4dCD4/SF2-gp120 (A) and ADA-gp100 (B) competition ELISA, carried out as described in the legend to Fig. 3.
Identification of residues in peptide 12p1 that are critical for inhibition of CD4-gp120 binding.
We performed an alanine scan of 12p1 to define the residues that are critical for inhibition of the CD4-gp120 interaction (Table 3). Although the length of the peptide increases the inhibitory activity by more than 100-fold (compare the IC50s of 12p1 and 7p1 or 7p2 using SF2-gp120 in Table 2), the alanine scan shows that a core of six residues (NNIPWS) around the Pro-Trp motif are critical for inhibition. The rest of the amino acids in the sequence can be exchanged for alanine without much loss of inhibitory capacity. Three heptapeptides spanning the 12p1 sequence from Ile2 to Met12 were synthesized and tested for CD4-gp120 inhibitory activity. A peptide with the sequence INNIPWS (12p1[2–8]) had an IC50 of 12 μM in the 4dCD4/SF2-gp120 ELISA, showing that it was about twice as strong an inhibitor as 7p1. The remaining two, NIPWSEA (12p1[4–10]) and PWSEAMM (12p1[6–12]), showed no inhibitory activity. This result highlights the importance of Asn3 for inhibition. Peptide 12p3, which has similar functional properties to 12p1, has His in position 3, which potentially could mimic interactions made by an Asn. Residues at the C terminus of 12p1 might interact with gp120 through backbone contacts, because their side chains can be replaced by alanine without loss of inhibition (Table 3), even though some polypeptide chain is needed for tight binding, as indicated by the weaker activity of 12p1[2–8]. The core sequence, INNIPWS, in 12p1 is also sufficient for inhibiting the interaction of gp120 and MAb 17b. Figure 4B shows that the short peptide, 12p1[2–8], retains most of the ability of intact 12p1 to reduce the binding of gp120 to MAb 17b.
TABLE 3.
Alanine scan on peptide 12p1
| Peptide sequence | IC50 (μM)a |
|---|---|
| 2dCD4 | 8.7 ± 2.1 nM (13) |
| R-I-N-N-I-P-W-S-E-A-M-M | 25.7 ± 7.3 (13) |
| A-I-N-N-I-P-W-S-E-A-M-M | 9 |
| R-A-N-N-I-P-W-S-E-A-M-M | 300 |
| R-I-A-N-I-P-W-S-E-A-M-M | >500 |
| R-I-N-A-I-P-W-S-E-A-M-M | >500 |
| R-I-N-N-A-P-W-S-E-A-M-M | >500 |
| R-I-N-N-I-A-W-S-E-A-M-M | >500 |
| R-I-N-N-I-P-A-S-E-A-M-M | >500 |
| R-I-N-N-I-P-W-A-E-A-M-M | >500 |
| R-I-N-N-I-P-W-S-A-A-M-M | 43 |
| R-I-N-N-I-P-W-S-E-A-A-M | 15 |
| R-I-N-N-I-P-W-S-E-A-M-A | 55 |
Values are means ± standard errors; the number of experiments is given in parentheses.
Direct binding of 12p1 to gp120 detected by UV cross-linking experiments.
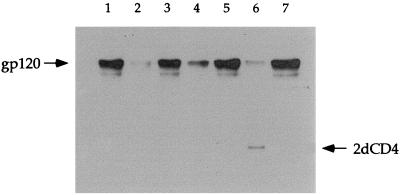
Direct binding of 12p1 to gp120 was studied by UV cross-linking with a Bpa peptide derivative (5). A 12p1 derivative was synthesized with Arg1 replaced by Bpa and with biotin coupled to the N terminus of the peptide sequence. The inhibitory activity of this 12p1 derivative was confirmed by using the 4dCD4-gp120 ELISA (data not shown). The biotin-12p1(R1/Bpa) peptide (2 μM) was incubated with SF2-gp120 (0.8 μM) for 10 min in the dark, frozen with dry ice, and UV irradiated for 12 min. To control for the specificity of the cross-linking reaction, 12p1 (430 μM), 12p1[Scrambled] (260 μM), 12p2 (1.2 mM), and 12p2[Scrambled] (1.1 mM), as well as 2dCD4 (23 μM) and MAb 803-15.6 (5 μM), were used as competitors. The protein cross-linked with biotinylated-Bpa peptide was detected by Western blotting with streptavidin-horseradish peroxidase and enhanced chemiluminescence substrate. The Western blot of the cross-linking experiment is shown in Fig. 6. There is a strong band for gp120 cross-linked with Bpa-peptide in the absence of competitor peptide 12p1 (lane 1). Cross-linking between gp120 and biotin-12p1(R1/Bpa) peptide was inhibited by 12p1 (lane 2), an unlabelled competitor. The inhibitory effect of 12p1 was sequence specific, since the scrambled version, 12p1[Scrambled], was unable to reduce cross-linking (lane 3). 12p2 was also able to inhibit cross-linking (lane 4), but its effect was much weaker than that observed for 12p1. The inhibitory effect of 12p2 was also sequence specific, as tested with a scrambled version, 12p2[Scrambled] (lane 5). Thus, 12p1 and 12p2 compete with each other, indicating that the site for 12p2 overlaps the site for 12p1. The inability of 12p2 to inhibit the binding of phage NEB12p1-M13 to gp120 plates (Fig. 2B) can probably be ascribed to multimeric interactions, which enhance the affinity of the phage for immobilized gp120. Cross-linking of gp120 and biotin-12p1(R1/Bpa) peptide is also inhibited by 2dCD4 (Fig. 6, lane 6), confirming that the binding site for 12p1 overlaps the CD4 site on gp120. In contrast, MAb 803-15.6 is unable to inhibit cross-linking (lane 7), as expected, since it binds to the opposite side of gp120 from CD4.
FIG. 6.
Western blot of UV cross-linking of biotin-12p1(Arg1/Bpa) (2 μM) to gp120 (0.8 μM). Lanes: 1, no competitor; 2, in the presence of 12p1 (430 μM); 3, in the presence of 12p1[Scrambled] (260 μM); 4, in the presence of 12p2 (1.2 mM); 5, in the presence of 12p2[Scrambled] (1.1 mM); 6, in the presence of 2dCD4 (23 μM); 7, in the presence of mAb 803-15.6 (5 μM). UV cross-linking was carried out on dry ice for 12 min.
DISCUSSION
We have screened phage-displayed peptide libraries and identified two families of peptide ligands that specifically bind HIV gp120. Table 4 shows the correlation between amino acid sequence and peptide activity. A family of peptides with the prototype sequence RINNIPWSEAMM (12p1) inhibits the interactions between gp120 and both 4dCD4 and MAb 17b. Another family of peptides with the prototype sequence TSPYEDWQTYLM (12p2) does not affect the binding of gp120 to 4dCD4 but enhances the binding of gp120 to MAb 17b.
TABLE 4.
Correlation between amino acid sequence and activity
| Peptide | Sequenceb | Effecta on gp120 binding to:
|
|
|---|---|---|---|
| CD4 | MAb 17b | ||
| 12p1 | R I N N I P W S E A M M | −−−−− | −−−−− |
| 12p3 | D T H R W P W Y I S Q E | − | −− |
| 7p1 | S W P W D T W | −− | −− |
| 7p2 | F W H P M H D | −− | −− |
| 12p2 | T S P Y E D W Q T Y L M | 0 | +++++ |
| 12p5 | S P W Y S D Y T R Y L W | 0 | +++ |
| 12p6 | S D P Y K L W A S Y M Y | 0 | +++++ |
| C7Cp1 | C H P L T P W E C | 0 | +++ |
Minus signs indicate inhibition; plus signs indicate enhancement; 0 indicates no effect.
Underlined amino acids represent the core sequence as identified from alanine-scanning and truncation experiments (see the text).
The crystal structure of the ternary complex of CD4, gp120, and a Fab of MAb 17b shows that CD4 and 17b bind on opposite sides of the bridging sheet at the base of the V1/V2 loop of gp120. We do not yet know the binding sites of our peptides on gp120, but their functional properties suggest that they interact near the bridging sheet and that they might exert their effects by stabilizing particular conformational states of this region of gp120. There is a proline followed by a hydrophobic amino acid in all the gp120 binding sequences we have isolated. This common motif is therefore likely to play an important role in the binding of both families of peptides to gp120, a conclusion that is consistent with the results of the truncation experiments on 12p1 (Tables 2 and 3) and with the reduced effect of 12p2 on MAb 17b-gp120 binding when these amino acids are replaced by alanine (Fig. 4B). The ability of 12p1 to inhibit the binding of phage bearing 12p2 to gp120 (Fig. 2B) and the ability of 12p2 to complete the cross-linking of Bpa-labeled 12p1 to gp120 (Fig. 6) also suggest that the two families of peptides interact with overlapping sites on gp120.
If their sites overlap, why do the two families of peptides have apparently divergent effects? The affinity of the 12p2 family for gp120 can be estimated from the concentrations required to show enhancement of MAb 17b binding (Fig. 4A). The dissociation constant is estimated to be 30 μM, at least 100-fold higher than that of 12p1 and too weak to give detectable inhibition of the CD4 interaction in ELISAs such as those in Fig. 3 and 5. It is therefore likely that the Pro-hydrophobic motif targets the peptides to a common site on gp120 but that there is detectable competition with CD4 binding only when the peptide has sufficiently high affinity. We suggest that the divergent effects of the peptides from the two families on MAb 17b binding are given by the amino acids flanking this motif. In family 2, the Pro-hydrophobic motif is located toward the N terminus of the sequence and is preceded by a polar residue. In family 1, the Pro-hydrophobic motif is positioned in the center of the peptide sequence, with amino acids to the N terminus being the most critical for binding. In particular, three of the four sequences of family 1 have a large hydrophobic residue before the Pro, and the truncation studies demonstrate critical roles for Asn3 in 12p1 and His3 in 12p3. If the binding of family 2 peptides to gp120 partly mimics the effects of CD4, for example by stabilizing the bridging sheet, the observed strengthening of the 17b interaction is a likely outcome, since binding of CD4 to gp120 also increases the affinity of the latter for MAb 17b. It is possible that amino acids in the peptide sequence make adventitious favorable contacts with MAb 17b when both are bound to gp120. The dodecamers of family 2 have a consensus sequence of three hydrophobic amino acids at the C terminus that might have this effect, but C7Cp1 also causes enhancement and does not have these consensus amino acids in its sequence. We therefore believe that enhancement of MAb 17b affinity by direct contact with peptides is unlikely.
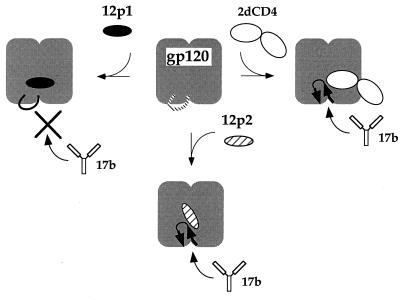
The more strongly binding peptides in family 1 not only compete detectably with CD4 in the ELISAs but also inhibit the association of gp120 with MAb 17b. The inhibitory effect of family 1 on the MAb 17b interaction might be due to either an allosteric effect or steric blocking of the 17b surface on gp120, presumably by the additional contacts responsible for tight binding of the family 1 peptides. A truncated version of 12p1, 12p1[2–8], has a strong inhibitory effect on MAb 17b-gp120 binding (Fig. 4B), indicating that the same core of amino acids is responsible for inhibition of both CD4 and MAb 17b binding to gp120. This result is more consistent with the hypothesis that binding of family 1 peptides favors a conformation of gp120 that does not mimic the CD4-bound state, even though these peptides interact at a site that competes for CD4 binding. That is, we suggest that the two families of peptides bind to overlapping sites but that they stabilize different states of gp120 (Fig. 7).
FIG. 7.
Model for the relationship of peptide, CD4, and MAb 17b binding sites. The diagram is based on the recent crystal structure of the ternary complex of gp120-2dCD4-Fab 17b (13). The two domains of gp120 are represented by the bilobal icon. These domains are linked by the bridging sheet (loop with arrows) in the CD4-bound state, which is probably not a stable structure in the absence of CD4 (dashed line). CD4 binding to gp120 stabilizes the sheet and induces the formation of a site for MAb 17b (top right). Thus, CD4 binding enhances the affinity for this MAb. We propose that peptide 12p1 binds near the CD4 site and inhibits MAb 17b binding by preventing the formation of the bridging-sheet structure (top left) whereas peptide 12p2 binds at a site overlapping that of 12p1 but stabilizes the bridging sheet (bottom). Thus, like CD4, 12p2 enhances the affinity of MAb 17b.
The functional properties of 12p1 and its family of peptides are those of potential anti-HIV compounds: they inhibit binding of the viral envelope protein with CD4 and also with an antibody that covers the site of the chemokine receptor. In addition, 12p1 inhibits the interaction of 4dCD4 with gp120 from three different HIV strains, two T-tropic and one M-tropic, indicating that it could have a broad anti-HIV activity. Peptide 12p1 did not have significant inhibitory activity up to 200 μM in a cell-cell fusion assay, however. There are several reasons that can explain its lack of in vivo activity. The peptide library was screened against soluble monomeric gp120, whereas on the surface of the virus gp120 is a trimer. It is possible that the peptide binding site is conformationally altered or less accessible on the viral trimeric gp120, decreasing the affinity of the peptide for the virus-attached gp120. Efforts are under way in our laboratory to obtain soluble trimeric gp120, which could be used as a more physiologically relevant target for the screening of libraries. Inhibition of viral entry probably requires blocking a large number of gp120 molecules (high occupancy) for fairly long periods. Thus, an inhibitor might need to have both a relatively high affinity and a low off-rate to ensure that enough gp120 molecules remain blocked to prevent attachment to the cell. Finally, it is also possible that the peptide was quickly digested by proteases in the cell culture medium. We are attempting to design stronger inhibitors based on the peptide sequence we have identified.
ACKNOWLEDGMENTS
We thank Marie Rose van Schravendijk, Richard Rickles, Gerhard Niederfellner, Andrea Musacchio, Tom Kirchhausen, Iris Rapoport, Tim Strassmaier, and Don Wiley for valuable help during this work. We also thank colleagues at Procept, Inc., for CD4, HXB2-gp120, and MAb 803-15.6; Ellis Reinherz for SF2-gp120 and ADA-gp100; and Joseph Sodroski and James Robinson for MAb 17b.
This work was supported by NIH grants GM-39589 (to S.C.H.) and AI-27336 (to E. Reinherz). M.F. thanks the Ministry of Education and Science (Spain) for a postdoctoral fellowship. S.C.H. is an investigator in the Howard Hughes Medical Institute.
REFERENCES
- 1.Brodsky M H, Warton M, Myers R M, Littman D R. Analysis of the site in CD4 that binds to the HIV envelope glycoprotein. J Immunol. 1990;144:3078–3086. [PubMed] [Google Scholar]
- 2.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemaglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 3.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 4.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 5.Dormán G, Prestwich G D. Benzophenone photophores in biochemistry. Biochemistry. 1994;33:5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 6.Ferrer M, Godbout K L, Sullivan B J, Austen D A, Sanderson C T, Kelley K C, Osburne M S, Harrison S C, van Schravendijk M R. Construction and characterization of a radio-iodinatable mutant of recombinant human CD4. J Immunol Methods. 1997;210:215–225. doi: 10.1016/s0022-1759(97)00195-6. [DOI] [PubMed] [Google Scholar]
- 7.Ferrer, M., B. J. Sullivan, K. L. Godbout, E. Burke, H. S. Stump, J. Godoy, A. Golden, A. T. Profy, and M. R. van Schravendidjk. Structural and functional characterization of an epitope in the conserved C-terminal region of HIV-1 gp120. J. Pept. Res., in press. [DOI] [PubMed]
- 7a.Ferrer, M. Unpublished results.
- 8.Furuta R A, Wild C T, Weng Y, Weiss C D. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 9.Gizachew D, Moffett D B, Busse S C, Westler W M, Dratz E A, Teintze M. NMR studies on the conformation of the CD4 36-59 peptide bound to HIV-1 gp120. Biochemistry. 1998;37:10616–10625. doi: 10.1021/bi980652o. [DOI] [PubMed] [Google Scholar]
- 10.Hughson F M. Enveloped viruses: a common mode of membrane fusion? Curr Biol. 1997;7:565–569. doi: 10.1016/s0960-9822(06)00283-1. [DOI] [PubMed] [Google Scholar]
- 11.Jameson B A, Rao P E, Kong L I, Hahn B H, Shaw G M, Hood L E, Kent S B H. Location and chemical synthesis of a binding site for HIV-1 on the CD4 protein. Science. 1988;240:1335–1339. doi: 10.1126/science.2453925. [DOI] [PubMed] [Google Scholar]
- 12.Kilby J M, Hopkins S, Venetta T M, DiMassimo B, Cloud G A, Lee J Y, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson M R, Nowak M A, Shaw G M, Saag M S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 13.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawless M K, Barney S, Guthrie K I, Bucy T B, Petteway S R, Merutka G. HIV-1 membrane fusion mechanism: structural studies of the interactions between biologically-active peptides from gp41. Biochemistry. 1996;35:13697–13708. doi: 10.1021/bi9606962. [DOI] [PubMed] [Google Scholar]
- 15.Lifson J D, Hwang K M, Nara P L, Fraser B, Padgett M, Dunlop N M, Eiden L E. Synthetic CD4 peptide derivatives that inhibit HIV infection and cytopathicity. Science. 1988;241:712–716. doi: 10.1126/science.2969619. [DOI] [PubMed] [Google Scholar]
- 16.Littman D R. Chemokine receptors: keys to AIDS pathogenesis? Cell. 1998;93:677–680. doi: 10.1016/s0092-8674(00)81429-4. [DOI] [PubMed] [Google Scholar]
- 17.Moebius U, Clayton L K, Abraham S, Diener A, Yunis J J, Harrison S C, Reinherz E L. Human immunodeficiency virus gp120 binding C′C" ridge of CD4 domain 1 is also involved in interactions with class II major histocompatibility complex molecules. Proc Natl Acad Sci USA. 1992;89:12008–12012. doi: 10.1073/pnas.89.24.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moebius U, Clayton L K, Abraham S, Harrison S C, Reinherz E L. The human immunodeficiency virus gp120 binding site on CD4: delineation by quantitative equilibrium and kinetic binding studies of mutants in conjunction with a high-resolution CD4 atomic structure. J Exp Med. 1992;176:507–517. doi: 10.1084/jem.176.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore J P, Wallace L A, Follett E A C, McKeating J A. An enzyme-linked immunosorbent assay for antibodies to the envelope glycoproteins of divergent strains of HIV-1. AIDS. 1989;3:155–163. doi: 10.1097/00002030-198903000-00006. [DOI] [PubMed] [Google Scholar]
- 20.Rapoport I, Chen Y C, Cupers P, Shoelson S E, Kirchhausen T. Dileucine-based sorting signals bind to the β chain of AP-1 at a site distinct and regulated differently from the tyrosine-based motif-binding site. EMBO J. 1998;17:2148–2155. doi: 10.1093/emboj/17.8.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Repke H, Gabuzda D, Palú G, Emmrich F, Sodroski J. Effects of CD4 synthetic peptides on HIV type 1 envelope glycoprotein function. J Immunol. 1992;149:1809–1816. [PubMed] [Google Scholar]
- 22.Rizzuto C D, Wyatt R, Hernández-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 23.Sattentau Q J, Arthos J, Hanna D K N, Healey D, Beverly P C, Sweet R, Truneh A. Structural analysis of the human immunodeficiency virus-binding domain of CD4. J Exp Med. 1989;170:1319–1334. doi: 10.1084/jem.170.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sattentau Q J, Moore J P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sattentau Q J, Moore J P. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schockmel G A, Somoza C, Davis S J, Williams A F, Healey D. Construction of a binding site for human immunodeficiency virus Type 1 gp120 in rat CD4. J Exp Med. 1992;175:301–304. doi: 10.1084/jem.175.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon J H M, Somoza C, Schockmel G A, Collin M, Davis S J, Williams A F, James W. A rat CD4 mutant containing the gp120-binding site mediates human immunodeficiency virus type 1 infection. J Exp Med. 1993;177:949–954. doi: 10.1084/jem.177.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith G P, Petrenko V A. Phage display. Chem Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan K, Liu J-H, Wang J-H, Shen S, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 33.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 34.Wild C, Dubay J W, Greenwell T, Baird T, Oas T G, McDanal C, Hunter E, Matthews T. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc Natl Acad Sci USA. 1994;91:12676–12680. doi: 10.1073/pnas.91.26.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc Natl Acad Sci USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wild C T, Shugars D C, Greenwell T K, McDanal C B, Matthews T J. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wrighton N C, Farrell F X, Chang R, Kashyap A K, Barbone F P, Mulcahy L S, Johnson D L, Barrett R W, Jolliffe L K, Dower W J. Small peptides as potent mimetics of the protein hormone erythropoietin. Science. 1996;273:458–463. doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]
- 38.Wu H, Myszka D G, Tendian S W, Brouillette C G, Sweet R W, Chaiken I M, Hendrickson W A. Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding. Proc Natl Acad Sci USA. 1996;93:15030–15035. doi: 10.1073/pnas.93.26.15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]