Abstract
The nonnucleoside reverse transcriptase (RT) inhibitor (NNRTI) delavirdine (DLV) selects in vitro for the human immunodeficiency virus type 1 (HIV-1) RT mutation P236L, which confers high-level resistance to DLV but not other NNRTIs. Unexpectedly, P236L has developed infrequently in HIV-1 isolates obtained from patients receiving DLV; K103N is the predominant resistance mutation observed in that setting. We characterized the replication fitness of viruses derived from pNL4-3 containing P236L or K103N in both H9 and primary human peripheral blood mononuclear cell cultures infected in parallel with the two mutants. In the absence of DLV, p24 production by wild-type virus occurred more rapidly and to higher levels than with either mutant; P236L consistently demonstrated a two- to threefold decrease in p24 relative to K103N. At low levels of DLV, growth of wild-type virus was severely inhibited, and K103N replicated two- to threefold more efficiently than P236L. At high concentrations of DLV, P236L replication and K103N replication were both inhibited. Recombinant RTs containing K103N or P236L were analyzed for DNA polymerization on heteropolymeric RNA templates and RNase H degradation of RNA-DNA hybrids. Neither mutant demonstrated defects in polymerization. K103N demonstrated normal RNA 5′-end-directed RNase H cleavage and slowed DNA 3′-end-directed RNase H cleavage compared to wild-type RT. P236L demonstrated slowing of both DNA 3′-end- and RNA 5′-end-directed RNase H cleavage, consistent with its reduced replication efficiency relative to K103N. These data suggest that NNRTI resistance mutations can lead to reductions in the efficiency of RNase H cleavage, which may contribute to a reduction in the replication fitness of HIV-1.
Major advances have been made recently in the therapy of human immunodeficiency virus type 1 (HIV-1) infection, which is estimated to affect more than 30 million people worldwide (54). Currently recommended combination antiretroviral regimens contain at least two reverse transcriptase (RT) inhibitors (RTIs) and either a protease inhibitor or a nonnucleoside RTI (NNRTI) (14). Nucleoside RTIs (nRTIs) compete with deoxynucleoside triphosphates (dNTPs) during polymerization and act as premature chain terminators upon incorporation (2). These compounds include zidovudine (AZT [or ZDV]), didanosine (ddI), lamivudine (3TC), and stavudine (d4T).
The NNRTIs are noncompetitive inhibitors that bind to a hydrophobic pocket adjacent to the polymerase active site of RT (16, 23, 31, 47). This binding causes an allosteric change of the polymerase active site which inhibits DNA polymerization (22, 23, 31, 47, 48). Nevirapine, delavirdine (DLV), and efavirenz (DMP-266) are NNRTIs that have been approved for clinical use. Recent reports suggest that combination regimens containing one NNRTI and two nRTIs provide levels of viral load suppression similar to that offered by regimens containing a protease inhibitor combined with two nRTIs (49).
HIV-1 resistance to antiretroviral drugs is a major factor limiting the efficacy of antiretroviral therapy (15, 26). HIV-1 resistance to RT inhibitors is mediated by specific mutations in the viral RT. A number of studies have shown that there are high rates of virus and CD4 cell turnover in HIV-1-infected patients (27, 52). Because of the error-prone nature of reverse transcription and these high replication rates, it is estimated that all possible single resistance mutations can be randomly generated within 1 day in an infected individual (15). Selective pressure introduced through drug therapy can result in rapid shifts in the relative replicative fitness of these mutants, leading to dramatic changes in the relative prevalence of different genotypes within a patient’s HIV-1 quasispecies (15, 20). Theoretical analyses have shown that a replicative advantage of as little as 2% can lead to the emergence of a low-frequency mutant as the dominant population over many replication cycles (15). Therefore, characterization of the relative fitness of drug-resistant mutants under different selective pressures will lead to a better understanding of how specific drug resistance mutations emerge during therapy.
Recent studies have explored how nRTI resistance mutations affect HIV-1 replication fitness. There have been no consistent correlations between replication fitness of nRTI-resistant mutants and their prevalence in clinical isolates, although the increased replication fitness of an AZT-resistant mutant (13) has been postulated to explain the persistence of AZT resistance in certain patients after discontinuation of AZT (10, 13).
The most common nevirapine resistance mutations are K103N and Y181C, which confer cross-resistance to other NNRTIs (26, 44). However, passage of HIV-1 in vitro in the presence of DLV selects for a unique P236L mutation (21). P236L confers a higher level of resistance to DLV than K103N or Y181C, but isolates with P236L remain sensitive to other NNRTIs (9, 21). This observation suggested that, unlike treatment with other NNRTIs, therapy with DLV might not always result in NNRTI cross-resistance. However, only 6% of DLV-resistant HIV isolates from patients receiving DLV monotherapy contained the P236L mutation, whereas the K103N mutation was detected in over 80% of these isolates (17). The effects of NNRTI resistance mutations such as P236L and K103N on HIV-1 replication are not known, but significant differences in the replication fitness of these two mutants might account for the unexpected predominance of K103N during therapy.
Recent studies have also examined how resistance mutations may affect HIV-1 RT biochemical function. HIV-1 RT is a heterodimer composed of a 66-kDa subunit (p66), which contains the polymerase and RNase H active sites, and a 51-kDa subunit (p51), which is composed of the first 440 amino acid residues of p66. HIV-1 RT exhibits a number of biochemical activities, including RNA-directed DNA polymerization, DNA-directed DNA polymerization, and RNase H activity (53). Cleavage of RNA-DNA hybrids by HIV-1 RNase H has been shown to be accomplished through two different modes of RT binding (25, 42). The first mode of cleavage is directed by the 3′ end of the elongating plus-strand DNA and is thought to occur in concert with DNA polymerization. A second mode of RNase H cleavage, directed by the 5′ end of the RNA in the RNA-DNA hybrid, presumably allows further degradation of small RNA fragments that remain annealed to the newly synthesized plus DNA strand after DNA 3′-end-directed RNase H cleavage (19, 24, 38–40, 43).
Biochemical studies of drug-resistant HIV-1 RT mutants have focused on RNA- and DNA-dependent DNA polymerization, RT fidelity (the accuracy with which RT incorporates nucleotides during polymerization) (8), and processivity (the number of nucleotides that a polymerase incorporates in a single binding event) (7). Thus, the nRTI-resistant mutant L74V demonstrates alterations in fidelity (45), and M184V has abnormalities in both processivity and fidelity (4–6, 28, 36, 41, 50, 51). However, these mutants develop readily during therapy with nRTIs, suggesting that some detectable abnormalities in RT function do not result in negative selection during therapy in patients.
Little is known about the effects of naturally occurring RT resistance mutations on RNase H function. One would expect that known resistance mutations to nRTIs and NNRTIs, which are quite distant from the RNase H active site, should not affect RNase H function. However, previous work has shown that mutations at certain residues in the highly conserved primer grip region of the polymerase domain (codons 227 to 235) produce selective defects in RNase H activity without affecting polymerization (40). It has been postulated that certain primer grip mutations interfere with RNase H activity by altering the positioning of the HIV-1 RT during cleavage of the RNA-DNA hybrid. Some primer grip residues (F227, W229, L234, and H235) also line the NNRTI binding pocket (47), raising the question of whether other NNRTI-resistant mutants might affect RNase H activity through a similar mechanism.
In this study, we demonstrate that the NNRTI resistance mutation P236L, when introduced into an HIVNL43 background, is replication-defective relative to wild-type and K103N viruses in both a T-cell line and in primary human peripheral blood mononuclear cells (PBMCs). This observation suggests that the high level of DLV resistance conferred by P236L is insufficient to compensate for its decreased replication fitness relative to other NNRTI resistance mutants having less DLV resistance, and it may explain why P236L is not commonly selected for during therapy with DLV. In addition, we show that P236L has normal DNA polymerase activity and processivity, but is defective in both DNA 3′-end-directed and RNA 5′-end-directed RNase H activities.
MATERIALS AND METHODS
Reagents.
DLV was obtained from Pharmacia and Upjohn (Kalamazoo, Mich.). It was stored as a 2 mM stock in 95% ethanol at −80°C. The DLV stock was diluted immediately before each experiment in the appropriate tissue culture medium. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases: the infectious molecular clone pNL4-3 was obtained from Malcolm Martin, and the HeLa-CD4-LTR-β-Gal cell line was obtained from Michael Emmerman.
Cell culture.
293 human primary embryonal kidney cells (American Type Culture Collection; Manassas, Va.) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum, l-glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 U/ml). H9 human lymphoma cells (American Type Culture Collection) were grown in RPMI supplemented with 20% (vol/vol) fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 U/ml). HeLa-CD4-LTR (long terminal repeat)-β-Gal (β-galactosidase) cells were grown in DMEM supplemented with 10% (vol/vol) fetal bovine serum, G418 (200 μg/ml), and hygromycin (100 μg/ml). Primary human PBMCs isolated from HIV-negative donors by Ficoll-Hypaque density gradient centrifugation were grown in RPMI 1640 supplemented with 20% (vol/vol) fetal bovine serum, penicillin (100 U/ml), gentamicin (50 μg/ml), and 5% interleukin 2. All cells were propagated in 5% CO2 at 37°C.
Site-directed mutagenesis.
pRHA1 was created by subcloning the pol gene of pNL43 (1) into the phagemid vector pTZ18U (Bio-Rad, Hercules, Calif.) by using SphI and EcoRI restriction endonuclease sites. The NNRTI resistance mutations P236L and K103N were introduced separately into the RT coding sequence of pRHA1 by the method of Kunkel et al. (32). The mutagenic oligonucleotides were 5′-TTAAAACAGAACAAATCAGTAACA-3′ (K103N) and 5′-TGAACTCCATCTTGATAAATG-3′ (P236L). Individual clones, pRHA1(K103N) and pRHA1(P236L), were isolated and sequenced in both directions to verify the absence of other mutations (Perkin-Elmer/Applied Biosystems, Foster City, Calif.).
Generation of NNRTI-resistant virus stocks.
NNRTI resistance mutations were separately subcloned from pRHA1(K103N) and pRHA1(P236L) into pNL43 by using SpeI and AgeI. Individual clones of pNL43(K103N) and pNL43(P236L) were isolated and sequenced to verify the integrity of the cloning sites and the presence of the appropriate RT resistance mutation. 293 cells were transiently transfected with either pNL43, pNL43(K103N), or pNL43(P236L) by lipofection (SuperFect; Qiagen, Santa Clarita, Calif.). After 48 h, H9 cells were added to increase the titer. Supernatant was harvested 48 h after addition of H9 cells, clarified by centrifugation at 500 × g, and stored at −80°C. Virus stocks were analyzed for p24 antigen concentration by an enzyme-linked immunosorbent assay kit (Dupont, Boston, Mass.). In addition, the relative infectivities of virus stocks were determined by titration in HeLa-CD4-LTR-β-Gal cells, as previously described (30).
Viral replication kinetics assays.
A total of 0.1 × 106 H9 cells or 0.2 × 106 PBMCs were washed with PBS and infected separately at a multiplicity of infection of 0.001 in a final volume of 200 μl for 1 h in 5% CO2 at 37°C. Infected cells were then washed and cultured in a final volume of 200 μl with or without DLV. Infections were performed in triplicate. At least three independently generated virus stocks were used in separate experiments. On days 0, 2, 4, 6, and 8 after infection, 100-μl aliquots of clarified supernatant were removed to assay for p24 antigen and replaced with fresh medium. DLV was included in the replacement medium, as required. At day 8, cultured cells were centrifuged at 500 × g and the pellet was stored at −80°C. Analysis of cell viability was performed by trypan blue exclusion; no evidence of cellular toxicity was detected at the doses of DLV used in these experiments (data not shown). Results of replication kinetics assays were reproduced by using amounts of virus stocks corresponding to 5 ng of p24 antigen per ml of culture.
Expression of heterodimeric RT.
We used pRSETB (IBI, Rochester, N.Y.) for expression of each RT subunit in vitro. pRSETB is a T7 promoter-driven expression vector that encodes an amino-terminal hexahistidine (His6) tag to allow purification by metal affinity chromatography. Immediately following the His6 tag is an enterokinase cleavage site that allows the removal of the His6 tag.
The regions of pRHA1 encoding the p66 and p51 subunits of RT were amplified by PCR with the proofreading thermostable polymerase Pfu (Stratagene, La Jolla, Calif.) and separately cloned into pRSETB. 5′ p66/Gly(pRSETB) (5′-GGTCCCATTAGTCCTATTGAGACTG) was the amino-terminal sense primer for amplification of both subunits. This primer was engineered to introduce a BamHI restriction endonuclease site for subsequent cloning and a glycine residue immediately upstream of the amino-terminal proline of RT. This was required since enterokinase will not cleave upstream of a proline residue. 3′ p66 (5′-CTATACTTTCCTGATTCCAGCACTG) was the carboxy-terminal antisense primer for amplification of the p66 subunit, and 3′ p51 (5′-TTAGAAAGTTTCTGCTCCTATTATGGG) was used for PCR of the p51 subunit. These primers were engineered to introduce an EcoRI restriction endonuclease site for subsequent cloning and a translation termination codon immediately downstream of the last codon of each respective subunit. PCR products were treated with BamHI and EcoRI and cloned into pRSETB separately to make the respective p66 and p51 expression vectors pRSETB/NL43p66 and pRSETB/NL43p51. Sequences of constructs, including the cloning sites, were verified on both strands to confirm the absence of extraneous mutations that may have been introduced by PCR.
NNRTI resistance mutations that were originally introduced into pRHA1 were subcloned into these constructs with MscI and AgeI. The p66 and p51 expression vectors were transfected separately into BL21(DE3)pLysS (Stratagene), an Escherichia coli strain possessing a T7 RNA polymerase gene under the control of the lac promoter. Upon addition of isopropyl-β-d-thiogalactopyranoside (IPTG), T7 RNA polymerase is expressed, which then induces expression of the p66 or p51 subunits. Induced cells were harvested 4 h after the addition of IPTG and lysed with a French pressure cell (3). Crude bacterial cell lysates were purified by metal affinity chromatography (Talon; Clontech, Palo Alto, Calif.) followed by DEAE-Sepharose and SP-Sepharose chromatography, as previously described (33). Purified RT subunits were visualized on Coomassie- or silver-stained sodium dodecyl sulfate (SDS)-polyacrylamide gels. The molarity of each protein preparation was determined according to a modification of the Bradford method (12) (Bio-Rad). Heterodimeric RTs were made by mixing equimolar amounts of the p66 and p51 subunits in 50 mM Tris (pH 7.0), 25 mM NaCl, 1 mM EDTA, 0.5 mM dithiothreitol (DTT) and 25% glycerol. Mutant RT heterodimers contained the respective NNRTI resistance mutation in both p66 and p51. All RT preparations were assayed for the absence of contaminating exo- and endonucleases (data not shown).
Generation of RNA templates.
All RNA templates used for DNA polymerization and RNase H assays were synthesized by T7 RNA polymerase-directed runoff transcription. The 142-nucleotide (nt)-long RNA (5′GGGCGAAUUA GCUUUUGUUCCCUUUAGUGAGGGUUAAUUCCGAGCUUGGCGUAAUCAUGGUCAUAGCUGUUUCCUGUGUGAAAUUGUUAUCCGCUCACAAUUCCACACAACAUACGAGCCGGAACCAUAAAGUGUAAAGCCU) and the 41-nt-long RNA (5′-GGGCGAAUUCGAGCUCGGUACCCGGGGAUCCUCUAGAGTCG) were derived from the plasmid pBS+(Δ) digested with either BstNI or AccI, respectively. A 545-nt-long RNA homologous to the 5′-end region of HIV-1 was derived from pSP-PBS following digestion by XmnI (35). Purification, quantitation, and 5′-end labeling with [γ-32P]ATP were carried out as previously described (37).
Analysis of RT activity.
The relative activity for DNA polymerization of each RT preparation was determined by measuring the rate of incorporation of [α-32P]dTTP into a poly(rA)-oligo(dT) template-primer. Relative activities determined by using this homopolymeric template were verified by using the 142- and 545-nt-long heteropolymeric RNAs primed by an appropriate DNA oligonucleotide, as described below.
Assays of RNA-dependent DNA polymerization by HIV-1 RT.
Substrates were prepared by hybridizing the appropriate DNA primer to the RNA template at a 2:1 molar ratio (40 nM primer, 20 nM template) in 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 50 mM NaCl, and 1 mM DTT at 65°C for 10 min, followed by cooling to room temperature for 90 min.
HIV-1 RT was prebound to the substrate for 3 min at 37°C. Reactions were initiated with a mixture of magnesium acetate and dNTPs to obtain final reaction conditions of 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 1 mM DTT, 50 mM NaCl, 6 mM magnesium acetate, and 50 μM (each) dNTP in a final volume of 12.5 μl. Trap reaction mixtures also contained 50 ng of heparin. DNA polymerization reaction mixtures were incubated for 16 min at 37°C and terminated with 12.5 μl of a 2× termination solution (90% formamide [vol/vol], 10 mM EDTA [pH 8.0], 0.1% [wt/vol] each xylene cyanole and bromphenol blue).
For time course analyses, reactions were scaled up to allow for removal of the appropriate number of aliquots. At various intervals, 12.5 μl of the reaction solution was removed and added to a separate tube containing 12.5 μl of the 2× termination solution. Products were resolved by denaturing polyacrylamide gel electrophoresis followed by autoradiography.
Determination of the degree of susceptibility to DLV.
The 142-nt-long RNA was hybridized to a 5′-end-32P-radiolabeled DNA primer, CP-3 (5′-CTCGTATGTTGTGTGGAATTGTGAGCGGAT). A DLV titration was performed with a fivefold serial dilution of DLV in separate reactions run in parallel. Reaction conditions were as described above for RNA-dependent DNA polymerization, with the addition of DLV stock solution to final concentrations ranging from 0.0256 to 2,000 μM. Reaction mixtures were incubated at 37°C for 16 min after addition of RT. They were terminated and resolved as described above.
RT processivity.
Two separate substrates were generated for the analysis of RNA-dependent DNA polymerase processivity. The first was made by annealing 40 fmol of a 5′-end-32P-labeled DNA oligonucleotide, CP-3, hybridized to 20 fmol of the 142-nt-long RNA. The second was made by annealing 40 fmol of a 5′-end-32P-labeled DNA oligonucleotide, PBS-1281 (5′-TCGCTTTCAAGTCCCTGTT), hybridized to 20 fmol of the viral 545-nt-long RNA. Reactions were carried out in the presence of a heparin trap and sampled at times ranging from 15 s to 16 min.
RT RNase H activity.
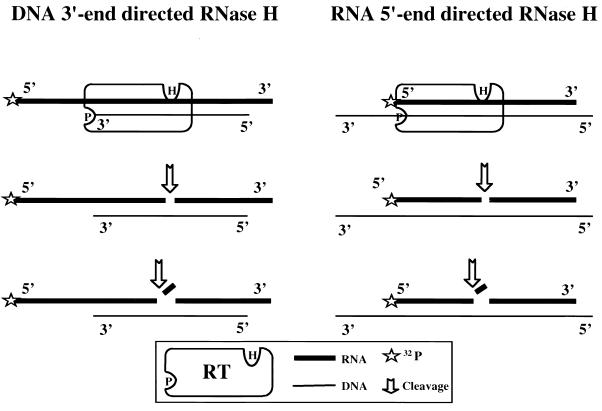
For analysis of DNA 3′-end-directed RNase H activity, 100 fmol of the DNA oligonucleotide PG003 (5′-AGAGGATCCCCGGGTACCGAGCTCGA) was hybridized to 80 fmol of the 5′-end-32P-labeled 41-nt-long RNA per reaction. For RNA 5′-end-directed RNase H activity analysis, 100 fmol of the DNA oligonucleotide MEW-3 (5′-TGCATGCCTGCAGGTCGACT CTAGAGGATCCCCGGGTACCGAGCTCGAATTCGCCCTATAGTGAGTCGTATTACAAT) was hybridized to 80 fmol of the 5′-end 32P-labeled 41-nt-long RNA. The amount of RT used for all RNase H activity measurements was normalized based on the relative activity of DNA polymerization. On a per weight basis, the amount of RT added to each reaction mixture ranged from 50 to 100 fmol of heterodimer. Reaction conditions were the same as for the DNA polymerization experiments, except that dNTPs were omitted from the reaction mixture. All experiments included E. coli RNase H and no-enzyme control reaction mixtures.
RESULTS
Effects of NNRTI resistance mutations on viral replication kinetics in the absence and presence of DLV.
The P236L mutation is an uncommon occurrence in patients receiving DLV monotherapy (17). To explain this observation, we hypothesized that the predominant drug-resistant mutant selected during DLV therapy would be the one which replicates most efficiently under these conditions. The likelihood of selecting a specific drug-resistant mutant would reflect a combination of the mutant’s level of drug resistance (which is mediated by a reduction in drug binding) and its replication fitness. We postulated that, since the occurrence of P236L relative to that of K103N is uncommon, despite conferring a higher degree of DLV resistance, P236L must replicate less efficiently than K103N. The balance between the degree of drug resistance and replication fitness would also determine the ability of other DLV-resistant mutants to compete with each other and presumably would vary under different selective conditions.
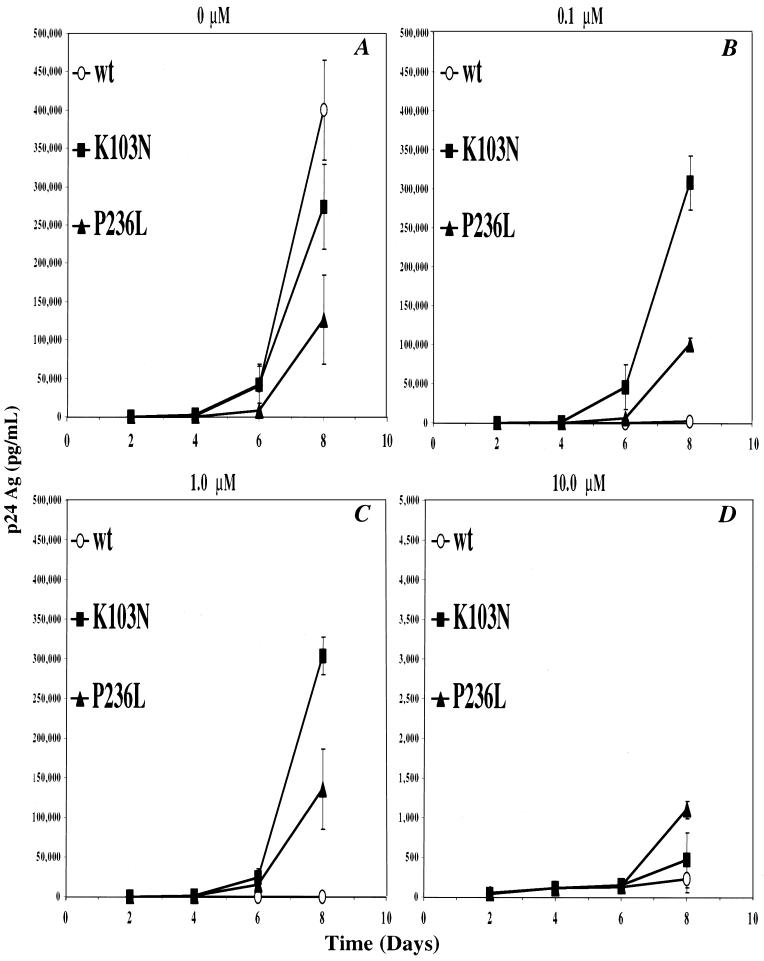
To test these predictions, we generated separate virus stocks of either HIVNL43, HIVNL43(K103N), or HIVNL43(P236L) derived from the corresponding mutagenized pNL43 clones (see Materials and Methods). To determine if HIVNL43(P236L) replicates less efficiently than HIVNL43(K103N) and HIVNL43, parallel cultures of H9 cells or primary human PBMCs were infected with each virus stock separately at a multiplicity of infection of 0.001 and grown over a period of 8 days (Fig. 1 and 2). In H9 cells, the P236L mutant demonstrated a three- to fourfold-lower culture p24 antigen concentration relative to wild-type virus and a two- to threefold-lower culture p24 antigen concentration relative to K103N in the absence of DLV at day 8 after infection (Fig. 1A). In the presence of either 0.1 or 1.0 μM DLV, there was a dramatic inhibition of wild-type virus, but there was little effect on the replication of P236L or K103N (Fig. 1B and C). At these DLV concentrations, K103N still demonstrated significantly higher p24 antigen concentrations than P236L. At 10 μM DLV, there was significant inhibition of both P236L and K103N viruses, with the P236L mutant demonstrating a two-fold-greater level of replication than K103N (Fig. 1D).
FIG. 1.
Replication kinetics of wild-type (wt), and K103N and P236L mutants in H9 cells. Separate cultures of H9 cells were infected with HIVNL43 (○), HIVNL43K103N (■), or HIVNL43P236L (▴) as described in Materials and Methods. Viral replication was monitored over a period of 8 days by measuring the HIV-1 p24 antigen (Ag) concentration. Cultures were grown in the absence of DLV (A) or with 0.1 μM DLV (B), 1 μM DLV (C), or 10 μM DLV (D). Error bars are the standard deviation of the mean p24 antigen concentration.
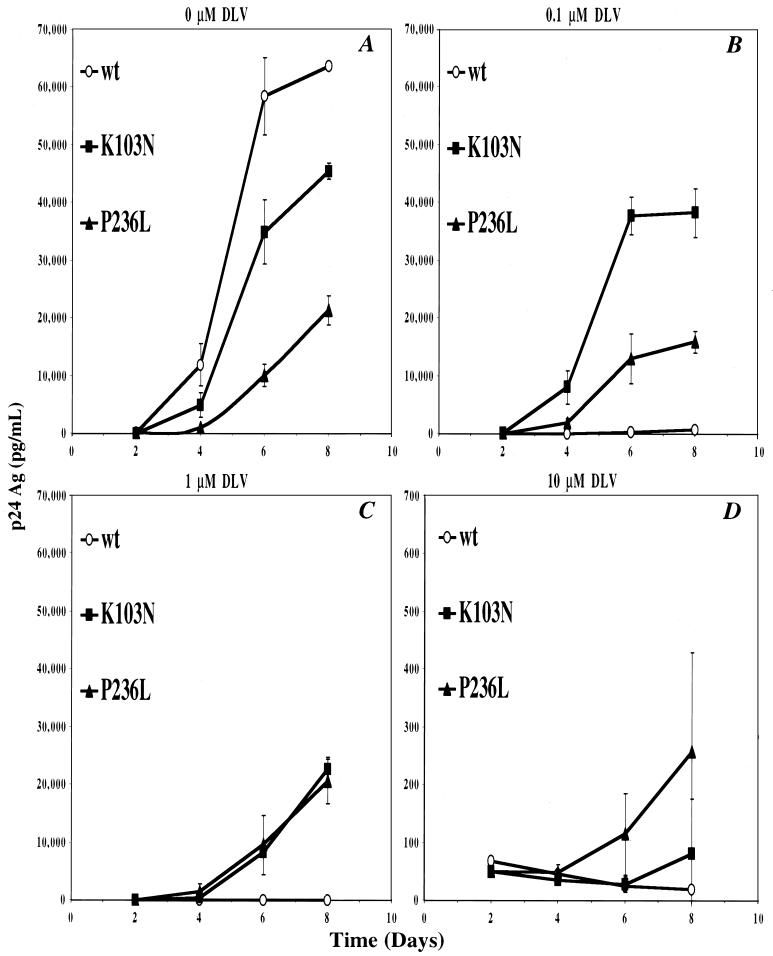
FIG. 2.
Replication kinetics of wild-type (wt) and K103N and P236L mutants in PBMCs. Separate cultures of PMBCs were infected with HIVNL43 (○), HIVNL43K103N (■), or HIVNL43P236L (▴) as described in Materials and Methods. Viral replication was monitored over a period of 8 days by measuring the HIV-1 p24 antigen (Ag) concentration. Cultures were grown in the absence of DLV (A) or with 0.1 μM DLV (B), 1 μM DLV (C), or 10 μM DLV (D). Error bars are the standard deviation of the mean p24 antigen concentration.
In human PBMCs, there was an overall reduction in p24 antigen content relative to H9 cells. By 4 days after infection in the absence of DLV, the P236L mutant demonstrated a 10-fold-lower culture p24 antigen concentration relative to wild-type virus and a 5-fold decrease relative to the K103N mutant (Fig. 2A). After 6 days, P236L demonstrated fivefold and threefold decreases in p24 antigen concentration relative to the wild type and K103N, respectively. At 0.1 μM DLV, replication of HIVNL43 was inhibited by 1,000-fold relative to replication in the absence of drug. Replication kinetics of HIVNL43(K103N) and HIVNL43(P236L) were minimally affected by this low concentration of DLV, and the K103N virus replicated more efficiently than P236L (Fig. 2B). At 1 μM DLV, the replication of HIVNL43(P236L) was still unaffected relative to the no-drug control. In contrast, the replication of HIVNL43(K103N) was inhibited at this intermediate DLV concentration but still remained similar to that of P236L (compare Fig. 2B and C). When the concentration of DLV was raised to 10 μM, replication of all three genotypes was so markedly inhibited that no significant differences in replication efficiencies were detected (Fig. 2D).
Similar results were obtained with three independently generated sets of wild-type and mutant virus stocks. The consistency of our observations suggests that the introduction of a spurious mutation during generation of these virus stocks is unlikely to account for the observed replication defect of HIVNL43(P236L). In addition, a similar reduction in the replication fitness of P236L relative to those of the wild-type and K103N viruses was seen when virus input was based on p24 antigen concentration (data not shown).
DLV susceptibilities of wild-type and NNRTI-resistant RTs.
In order to determine the biochemical basis for the replication defect of P236L, we generated recombinant wild-type, K103N, and P236L heterodimeric RTs. The p66 and p51 sequences from pNL43 were cloned into pRSETB to generate the RT subunit expression vectors pRSETB/NL43p66 and pRSETB/NL43p51, into which NNRTI resistance mutations were subcloned from pRHA1. RT subunits were expressed in E. coli separately and purified to greater than 95% homogeneity (data not shown).
Relative activities for DNA polymerization were determined by enzyme titration with a homopolymeric poly(rA)-oligo(dT) substrate (data not shown). RT preparations were normalized by this measurement for all subsequent experiments. To facilitate comparison of wild-type and mutant RT catalytic functions on various substrates, each experimental assay contained an amount of RT producing equivalent synthetic activity, as measured on the standard homopolymeric substrate. Relative activities were also determined in DNA polymerization reactions by using two different heteropolymeric RNA templates, which yielded results similar to those obtained with a homopolymeric template (data not shown). Preparations of the P236L and the K103N mutant RTs had polymerization-specific activities indistinguishable from that of the wild type. This indicates that the mutations did not significantly alter the ability of the mutant RTs to carry out primer elongation under these experimental conditions.
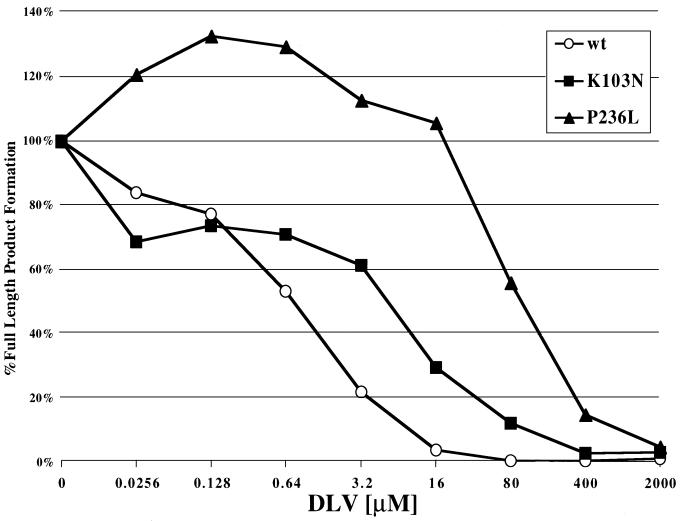
To determine the relative DLV susceptibilities of each recombinant RT, RNA-dependent DNA polymerization was measured in the presence of different concentrations of DLV (Fig. 3). Wild-type RT was most susceptible to DLV, with 50% inhibition occurring at approximately 0.64 μM. P236L RT was the most resistant RT to DLV, with 50% inhibition occurring at greater than 80 μM DLV. Interestingly, polymerization by P236L appeared to be stimulated by low levels of DLV (Fig. 3). K103N RT exhibited an intermediate level of DLV resistance, with 50% inhibition occurring between 3.2 and 16 μM. No differences in the distribution of extension products were seen in the absence or presence of DLV (data not shown). The relative differences in DLV susceptibilities among these RT preparations are compatible with previous findings (21) and are consistent with our studies of DLV inhibition of these mutants in cell culture (Fig. 1 and 2).
FIG. 3.
Susceptibilities of wild-type (wt), K103N, and P236L RTs to DLV. The substrate used for the DLV titration was made by annealing the 5′-end-32P-labeled DNA oligonucleotide CP-3 to a 142-nt-long RNA. The titration was performed with fivefold serial dilutions of DLV in parallel separate reactions. Reaction conditions and resolution of products of synthesis are described in Materials and Methods.
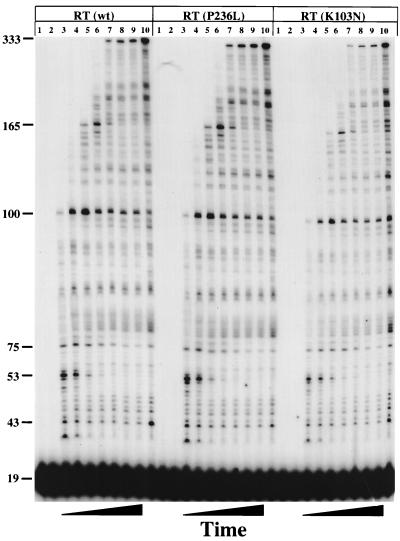
Effects of NNRTI resistance mutations on RT processivity.
The processivity of a polymerase is defined as the number of nucleotides incorporated in a single binding event. Processivity relates to the movements of the RT during polymerization and is a measure of the efficiency of synthesis. Since this value depends on the dissociation rate of the RT during synthesis and on the nucleotide addition rate, it is sensitive to conditions and mutations that change any of many steps in the cycle of binding, primer elongation, and dissociation. We initially hypothesized that the P236L RT would demonstrate abnormalities in polymerization detectable by a change in processivity. This expectation was based on the location of this mutation relative to the polymerization active site and its close proximity to the primer grip region. To make the measurement, equal amounts of wild-type, K103N, or P236L RT (normalized for their relative specific activities) were prebound to a 545-nt-long heteropolymeric, viral RNA template-DNA primer substrate. The processivity of each RT was measured over time after initiation of polymerization with Mg+2, dNTPs, and heparin. Heparin, a trapping polymer, was used to bind and sequester RT dissociated from the primer-template. A trap control, in which heparin was added during prebinding of RT to the primer-template, demonstrated that the amount of heparin used was sufficient to sequester all RTs after dissociation (Fig. 4, lane 1). This confirmed that all extension products represent synthesis resulting from a single binding event between RT and the primer-template. Finally, a no-trap control was used to verify that equal specific activities of each mutant RT were used (Fig. 4, lane 10). No significant differences in the size distribution or kinetics of accumulation of extension products were seen when comparing wild-type, P236L, and K103N RTs on this template-primer. We obtained similar results by using a 142-nt-long RNA template derived from the plasmid pBS+(Δ) (data not shown). These results show that these two DLV resistance mutations do not alter processivity of primer elongation, suggesting that the overall polymerization cycle is unaltered.
FIG. 4.
Processivities of wild-type (wt), K103N, and P236L RTs. The substrate used for processivity analysis was made by annealing 40 fmol of a 5′-end-32P-labeled DNA oligonucleotide, PBS-1281, to a 545-nt-long RNA. The amount of RT used was normalized based upon relative activities of DNA polymerization. Reactions were performed and resolved as described in Materials and Methods. Lanes 1 to 10 for each mutant represent the same reaction conditions. Lane 1 was a trap control in which heparin was added before the addition of RT. Lanes 2 through 9 were reactions in which heparin was added after RT was prebound. Reaction mixtures were allowed to polymerize for the following amounts of time (by lane): 2, 0 s; 3, 15 s; 4, 30 s; 5, 1 min; 6, 2 min; 7, 4 min; 8, 8 min; and 9, 16 min. Lane 10 indicates reactions in which polymerization was allowed to proceed for 16 min in the absence of a heparin trap.
Effects of NNRTI resistance mutations on DNA 3′-end-directed RNase H cleavage by RT.
Previous studies have shown that alanine substitutions at a number of residues in the primer grip region impair RNase H activity (40). P236L, in addition to lining the NNRTI binding pocket, is also immediately adjacent to the primer grip region (23). To determine whether the replication defect of HIVNL43(P236L) can be explained by an abnormality in DNA 3′-end-directed RNase H cleavage, we performed an RNase H time course analysis in the absence of polymerization by using a recessed DNA primer hybridized to a 5′-end-labeled 41-nt-long RNA (Fig. 5). Wild-type, P236L, and K103N RTs were prebound to the substrate in separate reactions before the addition of Mg+2. The amount of each RT used was again adjusted to an equivalent activity for polymerization on the homopolymer template. A sample of each reaction mixture was removed at time intervals ranging from 15 s to 16 min, and the degradation products were resolved by electrophoresis on denaturing polyacrylamide gels (Fig. 6). E. coli RNase H was used as a positive control to demonstrate that the RNA-DNA hybrid was annealed and could be cleaved (data not shown). A control without added RT demonstrated the integrity of the RNA substrate and the absence of contaminating RNases (data not shown).
FIG. 5.
Substrates used to measure DNA 3′-end- and RNA 5′-end-directed RNase H cleavage. For analysis of DNA 3′-end-directed RNase H activity, a recessed DNA primer was hybridized to a 5′-end-32-P-labeled 41-nt-long RNA. RT binding is directed by the 3′ end of the DNA as shown. RNase H cleavage of the RNA initiates at approximately 18 nt upstream of the DNA 3′ end. Additional cleavage occurs as shown. This substrate emulates cleavage that occurs during minus strand synthesis. For analysis of RNA 5′-end-directed RNase H activity, the same 5′-end-32P-labeled 41-nt-long RNA was used, but now hybridized to a 77-nt-long DNA. In this case, the RNA, rather than the DNA, was recessed. Initial cleavage is about 18 nt from the RNA 5′ end, with additional cleavage as shown. This substrate emulates an RNA fragment left behind from minus strand synthesis. RNase H cleavage is represented by an arrow. The postulated positions of the polymerase (P) and RNase H (H) catalytic sites of RT relative to the 5′ and 3′ ends of the nucleic acids are shown diagramatically.
FIG. 6.
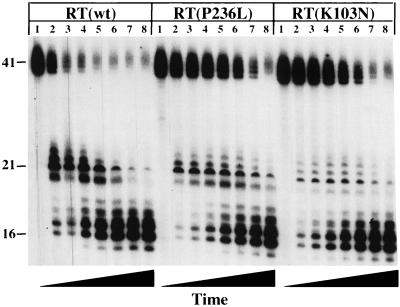
DNA 3′-end-directed RNase H activity of wild-type (wt), K103N, and P236L RTs. The substrate used was made by annealing the DNA oligonucleotide PG003 to a 5′-end-labeled 41-nt-long RNA such that the 3′ end of the DNA primer was recessed relative to the RNA 5′ end. Reactions were performed in the absence of dNTPs and resolved as described in Materials and Methods. Reactions were allowed to proceed for the following amounts of time (by lane): 1, 0 s; 2, 15 s; 3, 30 s; 4, 1 min; 5, 2 min; 6, 4 min; 7, 8 min; 8, 16 min. Disappearance of full-length product was quantitated by PhosphorImaging (data not shown).
Both P236L and K103N demonstrated lower rates of DNA 3′-end-directed RNase H cleavage relative to that of wild-type RT. This difference was evident when monitoring either the degradation of the 5′-end-labeled RNA substrate or the accumulation of degradation products. Wild-type RT significantly degraded the starting material after 2 min [Fig. 6, RT(wt), lane 5], whereas an equivalent amount of degradation required 16 min for both K103N and P236L [Fig. 6, RT(P236L) and RT(K103N), lanes 8]. The reductions in DNA 3′-end-directed RNase H activities of the P236L and K103N mutant RTs relative to that of the wild type were observed in six and four independent experiments, respectively. Thus, both the K103N and P236L mutants are defective in DNA 3′-end-directed RNase H activity, despite their different replication efficiencies in cell culture.
Effects of NNRTI resistance mutations on RNA 5′-end-directed RNase H activity.
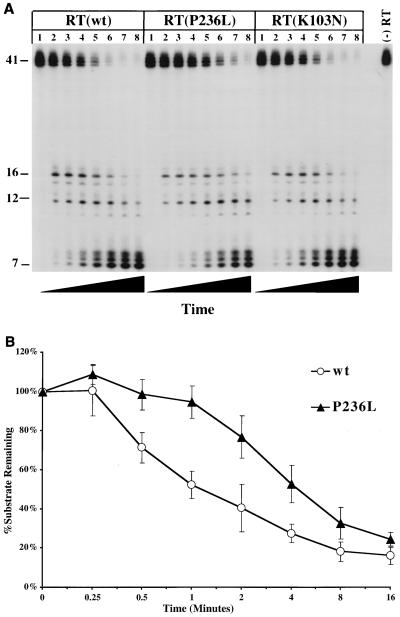
Since there are two modes of RNase H activity, it was important to know whether either of the DLV-resistant mutants also demonstrated abnormalities in RNA 5′-end-directed RNase H cleavage. We performed a time course analysis of RNase H cleavage by using the same 5′-end-radiolabeled 41-nt-long RNA, now hybridized to a 77-nt-long DNA (Fig. 5). In these reactions, the RNA, rather than the DNA, was recessed. This substrate emulates an RNA fragment left behind after completion of minus strand synthesis, which must be removed by RNA 5′-end-directed RNase H activity in order for the RT to complete plus strand DNA synthesis. Wild-type, P236L, and K103N RTs were prebound separately to each substrate, and samples from each reaction mixture were taken at selected time intervals (Fig. 7A). Quantitation of starting material degradation was performed by PhosphorImaging. Both wild-type and K103N RTs cleaved more than 90% of the starting material after 4 min [Fig. 7A, RT(wt) and RT(K103N), lanes 6]. However, the P236L mutant had only degraded 75% of the starting material at this time point and required between 8 and 16 min to degrade more than 90% of the starting material [Fig. 7A, RT(P236L), lanes 7 and 8]. Similar reductions in the rates of RNA 5′-end-directed cleavage by P236L RT were seen in five additional experiments. Figure 7B summarizes data from six independent experiments on the rates of RNA 5′-end-directed RNase H cleavage by wild-type and P236L RTs. No consistent differences in RNA 5′-end-directed cleavage by the K103N mutant were observed in four independent experiments. In contrast to the results obtained from the assays of DNA 3′-end-directed RNase H activity, there were no detectable differences in the abilities of wild-type and K103N RTs to carry out RNA 5′-end-directed RNase H cleavage. Thus, only P236L demonstrated a reduced rate of RNA 5′-end-directed RNase H cleavage.
FIG. 7.
(A) RNA 5′-end-directed RNase H activity of wild-type (wt), K103N, and P236L RTs. The substrate used was made by annealing a DNA oligonucleotide, MEW-3, to a 5′-end-labeled 41-nt-long RNA such that the 5′ end of the RNA was recessed relative to the DNA 3′ end. Reactions were performed in the absence of dNTPs and resolved as described in Materials and Methods. Reactions were allowed to proceed for the following amounts of time (by lane): 1, 0 s; 2, 15 s; 3, 30 s; 4, 1 min; 5, 2 min; 6, 4 min; 7, 8 min; 8, 16 min. (B) Plot of degradation of full-length product by wild-type (open circles) and P236L (solid triangles) RTs, quantitated by PhosphorImaging. The plot represents the average of six experiments. Error bars represent standard deviations.
DISCUSSION
It is important to understand the selective forces that interact to favor the emergence of drug-resistant mutants during antiretroviral therapy. Such an understanding will allow the more rational design of therapeutic strategies to prevent the emergence of resistant HIV-1 variants and lead to a broader understanding of the factors that influence the stability of an HIV-1 quasispecies. HIV-1 resistance to the NNRTI DLV has several characteristics of a good model system to study the interplay of different selective forces exerted on an HIV-1 quasispecies during antiretroviral therapy. The genetic basis for DLV resistance is simple, with only a limited number of single or double mutations conferring high-level phenotypic resistance. The primary mechanism for drug resistance is well understood and involves a reduction in drug binding. DLV resistance can develop through a number of different alternative genetic pathways which have different consequences in terms of cross-resistance to other NNRTIs. Finally, the most common genetic pathway to resistance is not what would be predicted from DLV binding or in vitro passage studies. The P236L mutation appears in cell culture after passage in the presence of DLV, but it is seen in patients only infrequently. The frequent appearance of P236L in cell culture passage experiments may have been influenced by experimental conditions, such as the concentration of drug used or the way in which mutants were detected. Alternatively, the infrequent occurrence of P236L during therapy with DLV could result from features of the P236L mutant RT that make the virus less fit than other DLV-resistant viruses.
Our studies have demonstrated that the P236L mutation confers a replication defect, relative to wild-type virus, in an HIVNL4-3 background. In the absence of DLV, the P236L mutant displayed a consistent decrease in replication fitness, relative to that of wild-type virus, in primary human PBMCs and in a T-cell line. This degree of replication impairment is not commonly seen in RTI-resistant mutants that are recovered from treated patients. The M184V mutant, selected for by 3TC, does not affect replication of the virus in human T-cell lines, but does confer a replication defect in primary human PBMCs, which have lower dNTP pools than T-cell lines (5). The L74V mutation selected during ddI therapy is less fit than wild-type virus in PBMCs (46). The D67N/K70R/T215Y/K219Q mutant selected during AZT therapy actually replicates more efficiently in quiescent human PBMCs than does wild-type virus, although no difference from the wild type was detected in activated PBMCs (13). Although the magnitude of reduction in replication fitness of the P236L mutant appears to be greater than that of M184V or L74V, these studies cannot be directly compared with ours because of methodologic differences in the assessment of replication fitness.
An important feature of our study is that we did not limit our comparison of P236L to wild-type virus, but also compared the fitness of P236L to K103N, the most commonly observed DLV-resistant mutant. The replication defect of P236L relative to K103N was evident in the absence of DLV and persisted at low DLV concentrations. However, at higher levels of DLV in PBMCs, the relative levels of fitness of K103N and P236L were indistinguishable, and at the highest concentration of DLV tested in H9 cells, P236L had a slight replication advantage compared to K103N. Our studies suggest that predictions of the frequency of drug resistance mutations in HIV-1 isolates from treated patients cannot be based solely on the level of drug resistance conferred by these mutants. As can be seen in our experiments with P236L and K103N, under conditions of escalating DLV concentrations, “fitness” of HIV-1 is a relative measure and is highly dependent on the selective forces present. We postulate that the degree of reduction in fitness of the P236L mutant, relative to that of K103N, explains the low frequency with which P236L is detected during therapy with DLV.
Of interest is the demonstration by Iversen and colleagues that the V75I mutation reverses the marked replication defect of the F77L and F77L/Q151M mutants that contribute to multinucleoside drug resistance, and this finding may explain the selection for this combination of mutations in patient isolates (29). Studies are under way in our laboratory to determine if RT polymorphisms seen in clinical isolates with P236L may compensate for the replication defect observed in an HIVNL4-3 background.
Surprisingly, despite the reduced replication rate observed in tissue culture, the P236L mutant displayed normal polymerization activities in vitro. We found no consistent reductions in the polymerization-specific activity of P236L RT preparations relative to wild-type or K103N RTs. In addition, we found no evidence for a decrease in polymerization efficiency or processivity of the P236L mutant on an RNA template. The P236L mutant RT did display significant reductions in both RNA 5′-end- and DNA 3′-end-directed RNase H activities relative to wild-type RT, but only differed from the K103N mutant in its ability to catalyze RNA 5′-end-directed RNase H cleavage. Our studies do not address the question of whether abnormalities in other aspects of RT function, such as fidelity of nucleotide incorporation, DNA-dependent DNA polymerization, tRNA priming, or strand transfer, could contribute to the replication defect of the P236L mutant. In addition, our studies do not address whether the RNase H abnormalities observed persist in the presence of DLV.
Our studies also suggest that a more subtle defect in replication of the K103N mutant may be explained by an impairment in DNA 3′-directed RNase H activity. Clearly, the defect in DNA 3′-end-directed RNase H activity conferred by the K103N mutation does not interfere with its selection during NNRTI therapy in patients. Since the degrees of impairment of DNA 3′-end-directed RNase H cleavage by the K103N and P236L mutants appear to be of a similar magnitude, we postulate that the defect in DNA 3′-directed RNase H cleavage is not sufficient to explain the replication defect of the P236L mutant.
Our results suggest that the reduction in DNA 3′-end-directed RNase H activity catalyzed by K103N RT explains HIVNL43(K103N)’s subtle replication abnormality relative to wild-type virus and only partially contributes to P236L’s replication defect. This observation suggests that the RNA 5′-end-directed RNase H cleavage defect of the P236L mutant contributes significantly to its slowed replication kinetics in cell culture. Our studies do not exclude the possibility that an abnormality in RNA 5′-end-directed RNase H activity alone can impair the replication fitness of HIV-1. However, we favor the interpretation that it is the combination of defects in both RNA 5′-end and DNA 3′-end-directed RNase H activities that accounts for the replication defect of the P236L mutant.
Our reasoning is supported by the currently accepted view of the mechanism of HIV replication. The newly reverse-transcribed minus strand viral DNA must serve as the template for synthesis of the plus strand DNA. This requires that the original plus strand RNA be completely degraded. Previous work from our laboratory supports the hypothesis that degradation of plus strand RNA occurs in two steps. Polymerization-associated RNase H cleavage, which occurs during synthesis of minus strand DNA, is directed by the 3′ end of the DNA in the RNA-DNA hybrid (DNA 3′-end-directed RNase H cleavage). Our previous studies suggest that this process reduces the plus strand RNA to oligomers, many of which spontaneously dissociate from the minus strand DNA (18). Plus strand RNA fragments that are large enough to remain annealed to the minus strand DNA are cleaved by RTs that rebind and position on the 5′ ends of these RNAs. Additional cleavages completely remove the RNA during this polymerization-independent mode of cleavage, referred to as RNA 5′-end-directed RNase H activity. We postulate that a defect in polymerization-associated RNase H activity (i.e., DNA 3′-end-directed RNase H cleavage) can be compensated for by the later 5′-end-directed cleavage process. However, a defect in both DNA 3′-end- and RNA 5′-end-directed modes of cleavage would adversely affect the rate at which the virus can accomplish complete RNA removal. The combination of defects in both types of RNase H cleavage would result in a significant slowing of the overall replication rate of HIV-1.
We postulate that the effect of P236L on RNase H activity is secondary to abnormalities in positioning of RT on the RNA-DNA hybrid, since it is unlikely that the P236L mutation, which is quite remote from the active site of RNase H, would have a direct effect on the RNase H active site. Of course, the P236L mutation could cause sufficient disruption of the tertiary structure of RT to directly affect the RNase H activity. We believe the latter explanation is less likely, since the effect of P236L on RNase H activity was quite specific, with no discernible impact on DNA polymerization. Previous work by others has demonstrated that modification of a number of residues in the polymerase domain can selectively affect RNase H activity with little effect on RNA- or DNA-dependent DNA polymerization (11, 34). Of interest is that C280S, a mutation which confers resistance to RNase H inhibitors, exerts its effect through both the p66 and p51 subunits (34).
To our knowledge, this is the first demonstration that drug-resistant mutants of HIV-1 that occur clinically have abnormalities in RNase H cleavage. In addition, the defect in RNA 5′-end-directed RNase H cleavage described here can contribute to a reduction in HIV-1 replication fitness. We propose that the efficiency of RNA 5′-end-directed RNase H cleavage can contribute significantly to the replication fitness of HIV-1. In addition, abnormalities of RNase H cleavage may be a property of certain drug-resistant HIV-1 mutants selected for during NNRTI therapy. Studies to determine if other NNRTI-resistant mutants of HIV-1 demonstrate alterations in RNase H cleavage are in progress.
ACKNOWLEDGMENTS
This research was supported in part by grants R01-AI041387, MO1-RR00044-34S2, N01-AI-27658, and R01-GM-49573.
We thank Vicente Planelles for generously providing the pSP-PBS construct. We give special thanks to Michael Chiulli and Luis Berrios for performance of p24 antigen assays and to Michelle Wisnewski, Jin Kim, and Carrie Dykes for helpful discussions.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arts E J, Wainberg M A. Mechanisms of nucleoside analog antiviral activity and resistance during human immunodeficiency virus reverse transcription. Antimicrob Agents Chemother. 1996;40:527–540. doi: 10.1128/aac.40.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Wiley & Sons, Inc.; 1997. pp. 16.8.6–16.8.8. [Google Scholar]
- 4.Avidan O, Hizi A. The processivity of DNA synthesis exhibited by drug-resistant variants of human immunodeficiency virus type-1 reverse transcriptase. Nucleic Acids Res. 1998;26:1713–1717. doi: 10.1093/nar/26.7.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Back N K, Nijhuis M, Keulen W, Boucher C A, Oude Essink B O, van Kuilenburg A B, van Gennip A H, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 6.Back N K T, Berkhout B. Limiting deoxynucleoside triphosphate concentrations emphasize the processivity defect of lamivudine-resistant variants of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1997;41:2484–2491. doi: 10.1128/aac.41.11.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bambara R A, Fay P J, Mallaber L M. Methods of analyzing processivity. Methods Enzymol. 1995;262:270–280. doi: 10.1016/0076-6879(95)62023-0. [DOI] [PubMed] [Google Scholar]
- 8.Bebenek K, Kunkel T A. The fidelity of retroviral reverse transcriptases. Cold Spring Harbor Monogr Ser. 1993;5:85–102. [Google Scholar]
- 9.Been-Tiktak A M, de Haas C J, de Graaf L, Boucher C A, Verhoef J, Borleffs J C, Nottet H S, Schuurman R. In-vitro selection of HIV-1 variants resistant to non-nucleoside reverse transcriptase inhibitors in monocyte-derived macrophages. J Antimicrob Chemother. 1997;40:847–853. doi: 10.1093/jac/40.6.847. [DOI] [PubMed] [Google Scholar]
- 10.Boucher C A B, van Leeuwen R, Kellam P, Schipper P, Tijnagel J, Lange J M A, Larder B A. Effects of discontinuation of zidovudine treatment on zidovudine sensitivity of human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1525–1530. doi: 10.1128/aac.37.7.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyer P L, Ferris A L, Clark P, Whitmer J, Frank P, Tantillo C, Arnold E, Hughes S H. Mutational analysis of the fingers and palm subdomains of human immunodeficiency virus type-1 reverse transcriptase. J Mol Biol. 1994;243:472–483. doi: 10.1006/jmbi.1994.1673. [DOI] [PubMed] [Google Scholar]
- 12.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 13.Caliendo A M, Savara A, An D, DeVore K, Kaplan J C, D’Aquila R T. Effects of zidovudine-selected human immunodeficiency virus type 1 reverse transcriptase amino acid substitutions on processive DNA synthesis and viral replication. J Virol. 1996;70:2146–2153. doi: 10.1128/jvi.70.4.2146-2153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carpenter C C, Fischl M A, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S, Richman D D, Saag M S, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A. Antiretroviral therapy for HIV infection in 1998: updated recommendations of the International AIDS Society—USA Panel. JAMA. 1998;280:78–86. doi: 10.1001/jama.280.1.78. [DOI] [PubMed] [Google Scholar]
- 15.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 16.Cohen K A, Hopkins J, Ingraham R H, Pargellis C, Wu J C, Palladino D E, Kinkade P, Warren T C, Rogers S, Adams J, et al. Characterization of the binding site for nevirapine (BI-RG-587), a nonnucleoside inhibitor of human immunodeficiency virus type-1 reverse transcriptase. J Biol Chem. 1991;266:14670–14674. [PubMed] [Google Scholar]
- 17.Demeter, L. M., R. W. Shafer, P. M. Meehan, J. Holden-Wiltse, M. A. Fischl, W. W. Freimuth, M. Para, and R. C. Reichman. Delavirdine susceptibilities and associated reverse transcriptase mutations in HIV-1 isolates from patients in a phase I/II trial of delavirdine monotherapy (ACTG 260). Submitted for publication. [DOI] [PMC free article] [PubMed]
- 18.DeStefano J J, Buiser R G, Mallaber L M, Myers T W, Bambara R A, Fay P J. Polymerization and RNase H activities of the reverse transcriptases from avian myeloblastosis, human immunodeficiency, and Moloney murine leukemia viruses are functionally uncoupled. J Biol Chem. 1991;266:7423–7431. [PubMed] [Google Scholar]
- 19.DeStefano J J, Mallaber L M, Fay P J, Bambara R A. Determinants of the RNase H cleavage specificity of human immunodeficiency virus reverse transcriptase. Nucleic Acids Res. 1993;21:4330–4338. doi: 10.1093/nar/21.18.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domingo E, Martinez-Salas E, Sobrino F, de la Torre J C, Portela A, Ortin J, Lopez-Galindez C, Perez-Brena P, Villanueva N, Najera R, et al. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance—a review. Gene. 1985;40:1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- 21.Dueweke T J, Pushkarskaya T, Poppe S M, Swaney S M, Zhao J Q, Chen I S, Stevenson M, Tarpley W G. A mutation in reverse transcriptase of bis(heteroaryl)piperazine-resistant human immunodeficiency virus type 1 that confers increased sensitivity to other nonnucleoside inhibitors. Proc Natl Acad Sci USA. 1993;90:4713–4717. doi: 10.1073/pnas.90.10.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esnouf R, Ren J, Ross C, Jones Y, Stammers D, Stuart D. Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors. Nature Struct Biol. 1995;2:303–308. doi: 10.1038/nsb0495-303. [DOI] [PubMed] [Google Scholar]
- 23.Esnouf R M, Ren J, Hopkins A L, Ross C K, Jones E Y, Stammers D K, Stuart D I. Unique features in the structure of the complex between HIV-1 reverse transcriptase and the bis(heteroaryl)piperazine (BHAP) U-90152 explain resistance mutations for this nonnucleoside inhibitor. Proc Natl Acad Sci USA. 1997;94:3984–3989. doi: 10.1073/pnas.94.8.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuentes G M, Fay P J, Bambara R A. Coordination of DNA synthesis and removal of downstream segments of RNA by human immunodeficiency virus, murine leukemia virus, and avian myeloblastosis virus reverse transcriptases. Nucleic Acids Res. 1996;24:1719–1726. doi: 10.1093/nar/24.9.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furfine E S, Reardon J E. Reverse transcriptase RNase H from the human immunodeficiency virus: relationship of the DNA polymerase and RNA hydrolysis activities. J Biol Chem. 1991;266:406–412. [PubMed] [Google Scholar]
- 26.Hirsch M S, Conway B, D’Aquila R T, Johnson V A, Brun-Vezinet F, Clotet B, Demeter L M, Hammer S M, Jacobsen D M, Kuritzkes D R, Loveday C, Mellors J W, Vella S, Richman D D. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. International AIDS Society—USA Panel. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 27.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 28.Hsu M, Inouye P, Rezende L, Richard N, Li Z, Prasad V R, Wainberg M A. Higher fidelity of RNA-dependent DNA mispair extension by M184V drug-resistant than wild-type reverse transcriptase of human immunodeficiency virus type 1. Nucleic Acids Res. 1997;25:4532–4536. doi: 10.1093/nar/25.22.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iversen A K N, Shafer R W, Wehrly K, Winters M A, Mullins J I, Chesebro B, Merigan T C. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J Virol. 1996;70:1086–1090. doi: 10.1128/jvi.70.2.1086-1090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 32.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 33.Le Grice S F, Naas T, Wohlgensinger B, Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991;10:3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loya S, Gao H-Q, Avidan O, Boyer P L, Hughes S H, Hizi A. Subunit-specific mutagenesis of the cysteine 280 residue of the reverse transcriptase of human immunodeficiency virus type 1: effects on sensitivity to a specific inhibitor of the RNase H activity. J Virol. 1997;71:5668–5672. doi: 10.1128/jvi.71.7.5668-5672.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu Y, Planelles V, Li X, Palaniappan C, Day B, Challita-Eid P, Amado R, Stephens D, Kohn D B, Bakker A, Fay P, Bambara R A, Rosenblatt J D. Inhibition of HIV-1 replication using a mutated tRNALys-3 primer. J Biol Chem. 1997;272:14523–14531. doi: 10.1074/jbc.272.23.14523. [DOI] [PubMed] [Google Scholar]
- 36.Oude Essink B B, Back N K, Berkhout B. Increased polymerase fidelity of the 3TC-resistant variants of HIV-1 reverse transcriptase. Nucleic Acids Res. 1997;25:3212–3217. doi: 10.1093/nar/25.16.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palaniappan C, Fay P J, Bambara R A. Nevirapine alters the cleavage specificity of ribonuclease H of human immunodeficiency virus 1 reverse transcriptase. J Biol Chem. 1995;270:4861–4869. doi: 10.1074/jbc.270.9.4861. [DOI] [PubMed] [Google Scholar]
- 38.Palaniappan C, Fuentes G M, Rodriguez-Rodriguez L, Fay P J, Bambara R A. Helix structure and ends of RNA/DNA hybrids direct the cleavage specificity of HIV-1 reverse transcriptase RNase H. J Biol Chem. 1996;271:2063–2070. [PubMed] [Google Scholar]
- 39.Palaniappan C, Kim J K, Wisniewski M, Fay P J, Bambara R A. Control of initiation of viral plus strand DNA synthesis by HIV reverse transcriptase. J Biol Chem. 1998;273:3808–3816. doi: 10.1074/jbc.273.7.3808. [DOI] [PubMed] [Google Scholar]
- 40.Palaniappan C, Wisniewski M, Jacques P S, Le Grice S F, Fay P J, Bambara R A. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J Biol Chem. 1997;272:11157–11164. doi: 10.1074/jbc.272.17.11157. [DOI] [PubMed] [Google Scholar]
- 41.Pandey V N, Kaushik N, Rege N, Sarafianos S G, Yadav P N, Modak M J. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35:2168–2179. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 42.Peliska J A, Benkovic S J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 43.Rausch J W, LeGrice S F J. Substituting a conserved residue of the ribonuclease H domain alters substrate hydrolysis by retroviral reverse transcriptase. J Biol Chem. 1997;272:8602–8610. doi: 10.1074/jbc.272.13.8602. [DOI] [PubMed] [Google Scholar]
- 44.Richman D D, Havlir D, Corbeil J, Looney D, Ignacio C, Spector S A, Sullivan J, Cheeseman S, Barringer K, Pauletti D, Shih C-K, Myers M, Griffin J. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol. 1994;68:1660–1666. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubinek T, Bakhanashvili M, Taube R, Avidan O, Hizi A. The fidelity of 3′ misinsertion and mispair extension during DNA synthesis exhibited by two drug-resistant mutants of the reverse transcriptase of human immunodeficiency virus type 1 with Leu74→Val and Glu89→Gly. Eur J Biochem. 1997;247:238–247. doi: 10.1111/j.1432-1033.1997.00238.x. [DOI] [PubMed] [Google Scholar]
- 46.Sharma P L, Crumpacker C S. Attenuated replication of human immunodeficiency virus type 1 with a didanosine-selected reverse transcriptase mutation. J Virol. 1997;71:8846–8851. doi: 10.1128/jvi.71.11.8846-8851.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smerdon S J, Jager J, Wang J, Kohlstaedt L A, Chirino A J, Friedman J M, Rice P A, Steitz T A. Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1994;91:3911–3915. doi: 10.1073/pnas.91.9.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spence R A, Anderson K S, Johnson K A. HIV-1 reverse transcriptase resistance to nonnucleoside inhibitors. Biochemistry. 1996;35:1054–1063. doi: 10.1021/bi952058+. [DOI] [PubMed] [Google Scholar]
- 49.Staszewski S, Morales-Ramirez J, Flanigan T, Hardy D, Johnson P, Nelson M, Ruiz N. Proceedings of the 12th World AIDS Conference. 1998. A phase II, multi center, randomized, open-label study to compare the antiretroviral activity and tolerability of efavirenz (EFV) + indinavir (IDV), versus EFV + zidovudine (ZDV) + lamivudine (3TC), versus IDV + ZDV + 3TC at 24 weeks (DMP 266-006), abstr. 22336. [Google Scholar]
- 50.Wainberg M A. Increased fidelity of drug-selected M184V mutated HIV-1 reverse transcriptase as the basis for the effectiveness of 3TC in HIV clinical trials. Leukemia. 1997;11(Suppl. 3):85–88. [PubMed] [Google Scholar]
- 51.Wainberg M A, Drosopoulos W C, Salomon H, Hsu M, Borkow G, Parniak M, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad V R. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]
- 52.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 53.Whitcomb J M, Hughes S H. Retroviral reverse transcription and integration: progress and problems. Annu Rev Cell Biol. 1992;8:275–306. doi: 10.1146/annurev.cb.08.110192.001423. [DOI] [PubMed] [Google Scholar]
- 54.World Health Organization. The current global situation of the HIV/AIDS pandemic. Geneva, Switzerland: World Health Organization; 1998. [Google Scholar]