Abstract
A nef gene is present in all primate lentiviruses, including human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus of macaque monkeys (SIVmac). However, the nef genes of HIV-1 and SIVmac exhibit minimal sequence identity, and not all properties are shared by the two. Nef sequences of SIVmac239 were replaced by four independent nef alleles of HIV-1 in a context that was optimal for expression. The sources of the HIV-1 nef sequences included NL 4-3, a variant NL 4-3 gene derived from a recombinant-infected rhesus monkey, a patient nef allele, and a nef consensus sequence. Of 16 rhesus monkeys infected with these SHIVnef chimeras, 9 maintained high viral loads for prolonged periods, as observed with the parental SIVmac239, and 6 have died with AIDS 52 to 110 weeks postinfection. Persistent high loads were observed at similar frequencies with the four different SIV recombinants that expressed these independent HIV-1 nef alleles. Infection with other recombinant SHIVnef constructions resulted in sequence changes in infected monkeys that either created an open nef reading frame or optimized the HIV-1 nef translational context. The HIV-1 nef gene was uniformly retained in all SHIVnef-infected monkeys. These results demonstrate that HIV-1 nef can substitute for SIVmac nef in vivo to produce a pathogenic infection. However, the model suffers from an inability to consistently obtain persisting high viral loads in 100% of the infected animals, as is observed with the parental SIVmac239.
Experimental infection of rhesus monkeys with simian immunodeficiency virus (SIV) is one of the most widely used animal models for the study of AIDS pathogenesis. A nef gene is present in all primate lentiviruses, including SIVmac and human immunodeficiency virus type 1 (HIV-1), but nef is not necessary for the ability of these viruses to replicate. Evidence for the importance of the nef gene has been derived from the study not only of SIV in monkeys but also of HIV-1 in people. Rhesus monkeys infected with SIVmac239 containing a deletion in the nef gene typically maintain low viral loads (20) and only rarely progress to AIDS (47). In addition, individual humans have been found in the United States (21) and in Australia (8) who harbor only nef-deleted forms of HIV-1. These patients have maintained low viral loads for more than a decade in the absence of antiretroviral intervention, a phenotype likely due at least in part to the absence of an intact nef gene.
The nef genes of SIVmac and HIV-1 are correspondingly located at the 3′ ends of their genomes. However, SIVmac nef and HIV-1 nef share little amino acid homology and do not share all properties. For example, it has been observed that Nef of SIVmac, SIVsm, and HIV-2 interact with the zeta chain of the T-cell receptor, whereas none of five independent HIV-1 nef alleles tested shares this activity (16). In addition, HIV-1 nef expresses a highly conserved SH3 binding element, PXXPXXP, which is required for its ability to bind efficiently to Src family kinases (27). SIVmac nef, on the other hand, contains only a single PXXP motif and binds to Src family kinases less efficiently than does HIV-1 nef (11, 37).
Recently, the three-dimensional structure of HIV-1 nef has been elucidated (28). The structure determination revealed that HIV-1 nef contains a long, relatively unstructured leader sequence (amino acids 1 to 75 in HIV-1SF2 nef) which is linked to a highly structured core (amino acids 76 to 207 in HIV-1SF2 nef). Within its leader sequence, HIV-1 nef shares no detectable similarity with SIVmac nef. However, the sequences of the core structural region of HIV-1 nef shares limited homology with SIVmac nef. Despite only limited homology, it is likely that within the HIV-1 nef core are structural and functional motifs that are common between SIVmac nef and HIV-1 nef. In support of this contention, it has been demonstrated that the nef genes of SIVmac and HIV-1 downregulate the surface expression of CD4 (2, 6, 13). It has also been observed that SIVmac nef and HIV-1 nef similarly associate with a complex containing a serine kinase (38). In addition, SIVmac nef and HIV-1 nef are similarly capable of causing lymphocyte activation to enhance SIVmac replication (4). Both SIV nef and HIV-1 nef have also been found to downregulate major histocompatibility complex class 1 expression (39) and to enhance viral infectivity (3, 7, 26, 31, 45). The maintenance of these common activities despite limited overall homology indicates that these activities may contribute to the ability of SIVmac and HIV-1 to cause pathogenic infections in susceptible hosts. Identification of a recombinant SIV containing HIV-1 nef sequences that is pathogenic in monkeys could facilitate the dissection of the diverse activities for their relative importance and contributions in vivo.
In this report, we demonstrate that HIV-1 nef is able to substitute for SIVmac nef to cause pathogenic infection in rhesus monkeys. However, recombinant constructs containing HIV-1 nef were not as efficient as the parental SIVmac239 for consistently yielding high virus loads and for inducing AIDS.
MATERIALS AND METHODS
Plasmid construction.
All mutations in this study were engineered by using either a splice overlap extension (15) or a modified recombinant-PCR (RT-PCR) (12) technique. In all constructions the SIVmac nef sequences were deleted from the 3′ extent of the SIVmac env gene to 120 bases 5′ of the NF-κB binding site in SIVmac239 (35). The NL 4-3 nef allele was obtained from the infectious NL 4-3 clone (1), the RULDA nef allele was derived from a patient isolate obtained from J. Sullivan and T. Greenough, and the consensus nef allele (40) was acquired from R. Swanstrom. Thorough DNA sequence analysis verified the proper sequences for all mutants; recombinant clones selected for study were shown to contain the exactly desired sequence.
SHIVnef replication assays.
All stocks of SHIVnef recombinants described here were generated by DEAE-dextran transfection (32) of the cell line CEMx174. Recombinant constructs representing SHIVnef were transfected into CEMx174 cells, and virus in the cell-free supernatant was harvested at or near the peak of virus production as previously described (14). Transfected or infected CEMx174 cells were grown in RPMI 1640 (Gibco-BRL, Grand Island, N.Y.) that was supplemented with 10% fetal calf serum (Gibco-BRL). The levels of p27 viral protein that were produced from transfections or infections or contained within viral stocks were quantified by using a SIV core antigen kit (Coulter, Hialeah, Fla.).
Experimental infection of rhesus monkeys.
Virus diluted to contain 50 ng of p27 antigen was inoculated intravenously into juvenile rhesus monkeys (Macaca mulatta). At various time points postinoculation, blood samples were collected as previously described (14). Animals that became moribund were sacrificed, and complete necropsies were performed.
Determination of viral RNA and infectious cell loads.
Cell-associated virus loads were determined by quantitative cocultivation of peripheral blood mononuclear cells (PBMC) with CEMx174 cells (9). PBMC were purified, counted in a hemocytometer, and cocultured with CEMx174 cells in various numbers. On day 21, the presence of SIV p27 antigen was determined and the numbers of PBMC needed to recover SIV was calculated. The results described here represent averages of duplicate determinations. Virion-associated SIV RNA in plasma samples was quantified by RT-PCR assay on an Applied Biosystems Prism 7700 sequence detection system (Perkin-Elmer Cetus, Norwalk, Conn.) (46).
PCR amplification of recombinant SIV sequences recovered from PBMC.
PBMC (5 × 106) isolated from animals inoculated with SHIVnef recombinants were lysed in 0.5 ml of lysis buffer (10 mM Tris, pH 8.2; 0.4 M NaCl; 2 mM EDTA; pH 8.2) that was supplemented with 33 μl of 10% sodium dodecyl sulfate (SDS) and 10 μl of proteinase K (10 mg/ml) for 1 h at 56°C. After lysis, 160 μl of saturated NaCl was added, and the tube was inverted to mix the reagents. The mixture was then centrifuged at 14,000 rpm in a microcentrifuge for 10 min. The clear supernatant was removed and placed in a fresh tube, and 700 μl of isopropanol was added. The mixture was inverted and centrifuged for 10 min at 14,000 rpm. The supernatant was then removed, and the pellet was washed with 70% ethanol and then air dried for 1 h. One microgram of cellular DNA served as the template for PCR amplifications of the nef region of the recombinant SHIVnef genomes. Primers that annealed to the envelope 5′-GCCGTCTGGAGATCTGCGACAG-3′ and 3′ long terminal repeat (LTR) regions 5′-GCAGAGCGACTGAATACAGAGCGAAA-3′ were used to amplify the region spanning nef. The resulting fragments were then treated with T4 DNA polymerase (New England Biolabs, Beverly, Mass.) and inserted into a SmaI-digested puc18 vector (Promega, Madison, Wis.). The DNA sequence of the inserted fragment was then determined by using an ABI 377 DNA sequencer (Perkin-Elmer Cetus). All sequence data presented here represent a consensus of two independent clones from each of two independent PCR amplifications.
Determination of the percentage of CD4+ cells in blood of SHIVnef-inoculated animals.
Whole blood was drawn from SHIVnef-inoculated animals at various times postinoculation and stained with OKT4, a fluorescein isothiocyanate (FITC)-conjugated murine monoclonal antibody that reacts with rhesus macaque CD4. The stained samples were analyzed with a FACSscan flow cytometer (Becton Dickinson, San Jose, Calif.).
Cell line 221 and Western blot assays.
Cell line 221 cells (106) that were grown in RPMI 1640 medium (Gibco BRL) that was supplemented with 20% fetal calf serum (Gibco-BRL) and 10% interleukin-2 (IL-2) (60 to 90 U/ml; Hemagen, Waltham, Mass.) were washed and resuspended in RPMI medium that was supplemented with 5% serum in a 48-well tissue culture plate (Corning CoStar, Cambridge, Mass.) as previously described (4). SIV or SHIVnef containing 10 ng of p27 antigen was used for each infection.
For Western blot analysis, 3 × 106 CEMx174 cells were infected with 50 ng of either SIV or SHIVnef that was derived from the cell-free supernatant of CEMx174-transfected cells. Seven days postinfection, the cells were pelleted and lysed with 500 μl of lysis buffer (0.5% Nonidet P-40, 50 mM HEPES [pH 7.5], 150 mM NaCl) containing 2 mM NaCO3, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, and 1% aprotinin (Sigma Chemical Co., St. Louis, Mo.). The cell lysates were centrifuged at 13,000 × g for 30 min at 4°C. Then, 10 μl of the lysates was boiled for 5 min with an equal volume of Laemmli sample buffer prior to SDS-polyacrylamide gel electrophoresis (PAGE) through a 10% gel. The proteins were electroblotted onto an Immobilon-P membrane (Millipore, Bedford, Mass.), which was then blocked with 8% skim milk in phosphate-buffered saline (PBS)–005% Tween 20 (PBST) for 1 h, which was followed by an incubation with a 1:500 dilution of the anti-HIV-1 NL 4-3 nef-specific monoclonal antibody EH (kindly provided by J. Hoxie) in the same blocking solution for 1 h at room temperature. Primary antibodies were removed by washing the membranes three times with PBST at room temperature. The dilution of the secondary antibody and the detection of nef protein were performed according to the protocol of the enhanced chemiluminescence system (Amersham, Chicago, Ill.).
RESULTS
Introduction to recombinant constructions.
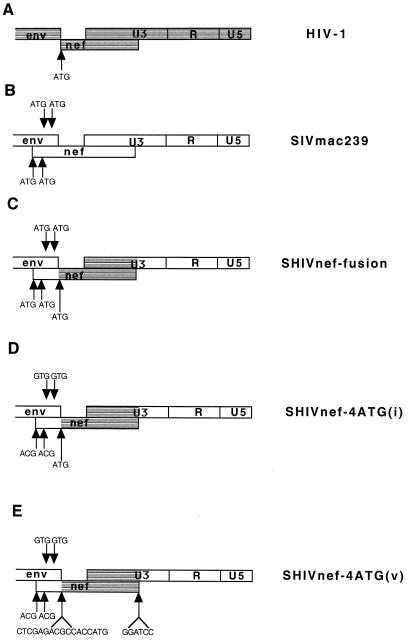
In the HIV-1 genome, the env and nef genes are located adjacent to each other, and no overlap of sequence exists between the two genes (Fig. 1A). In contrast, env and nef overlap by 167 bases in the SIVmac genome (Fig. 1B). The SIVmac nef sequences in the region of overlap are not homologous to HIV-1 nef sequences, and their importance for the function of SIV nef is unknown. The 120 bases immediately 5′ of the NF-κB site in SIVmac contain transcriptional control elements as well as nef coding sequences (17, 34). In animals infected with SIV containing a 182-bp deletion in the region that is uniquely nef, additional deletions accumulate over time in nef (22); however, these 120 bases of U3/nef sequence are consistently retained, which is consistent with a role in transcription (17, 22, 34). We thus created a SHIVnef-fusion construct in which NL 4-3 nef coding sequences were fused in frame to the upstream SIVmac nef sequences and were inserted such that 120 bases of SIVmac 3′ LTR U3 sequences located immediately 5′ of the NF-κB binding site were retained in the SHIVnef genome (Fig. 1C).
FIG. 1.
Diagrams of HIV-1, SIVmac, and SHIVnef genomes. The gray areas represent HIV-1 sequences, and the white areas represent SIVmac sequences. The arrows indicate the sequence contained in different SHIVnef recombinants. The arrows below the nef gene indicate ATGs contained in the nef reading frame, and the arrows above the nef gene indicate ATGs contained in the non-env/non-nef reading frame in the region of overlap containing both SIV env and SIV nef. The genes or genetic elements depicted in this figure are not drawn to scale.
Results of infection with SHIVnef-fusion.
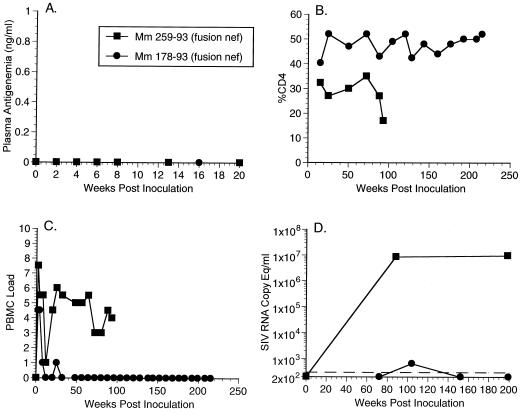
In the first set of experiments (AE542, Table 1), two juvenile rhesus monkeys (Mm178-93 and Mm259-93) were inoculated intravenously with SHIVnef-fusion containing 50 ng of p27 antigen. We monitored plasma antigenemia, CD4 cell counts, numbers of infectious cells in PBMC, and viral RNA loads with blood samples obtained at intervals after experimental infection of the monkeys. In the initial weeks postinoculation (p.i.), plasma antigenemia was not detected in samples from these two animals (Fig. 2A and Table 1). In addition, Mm178-93 did not maintain consistently measurable numbers of infectious cells in PBMC or RNA loads in a detectable range (Fig. 2C and D). This animal was monitored until week 208 p.i. with undetectable RNA loads and a normal CD4 percentage level (CD4%) (Fig. 2 and Table 1). Conversely, Mm259-93 displayed persisting high numbers of infectious cells in PBMC and RNA loads and died from AIDS 93 weeks p.i. (Fig. 2 and Table 1). In the weeks preceding death, the CD4% in this animal dropped from 34 to 16% (Fig. 2B).
TABLE 1.
Summary of animal infections with SHIVnef recombinantsa
| Expt no. | Animal no. | HIV-1 nef allele | Construct specifics | Viral loads:
|
Death with AIDS | Time to death (wks) | ||
|---|---|---|---|---|---|---|---|---|
| 3–10 mos | 10 mos | Late | ||||||
| AE542 | 178-93 | NL 4-3 | SHIVnef-fusion | Low | Low | Low | No | |
| AE542 | 259-93 | NL 4-3 | SHIVnef-fusion | High | High | High | Yes | 93 |
| AE576 | 119-94 | NL 4-3 | SHIVnef-4ATG(i) | High | High | High | Yes | 80 |
| AE576 | 144-94 | NL 4-3 | SHIVnef-4ATG(i) | High | High | High | No | |
| AE585 | 125-94 | NL 4-3 | SHIVnef-4ATG(v) | Low | Low | Low | No | |
| AE585 | 211-94 | NL 4-3 | SHIVnef-4ATG(v) | Low | Low | Low | No | |
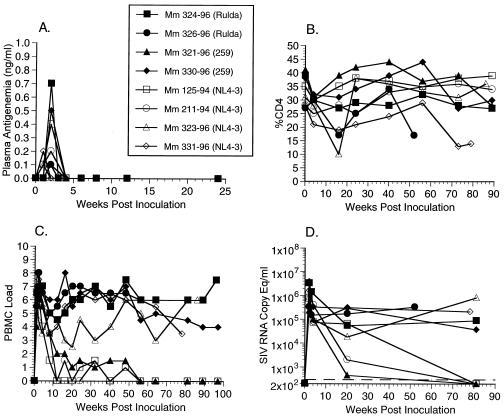
| AE585 | 323-96 | NL 4-3 | SHIVnef-4ATG(v) | Med | Med | High | Yes | 87 |
| AE585 | 331-96 | NL 4-3 | SHIVnef-4ATG(v) | High | High | High | Yes | 78 |
| AE585 | 321-96 | 259 | SHIVnef-4ATG(v) | Low | Low | Low | No | |
| AE585 | 330-96 | 259 | SHIVnef-4ATG(v) | High | High | High | Yes | 110 |
| AE585 | 324-96 | RULDA | SHIVnef-4ATG(v) | High | High | High | Yes | 96 |
| AE585 | 326-96 | RULDA | SHIVnef-4ATG(v) | High | High | High | Yes | 52 |
| AE598 | 37-97 | Consensus | SHIVnef-4ATG(v) | Med | High | High | No | |
| AE598 | 40-97 | Consensus | SHIVnef-4ATG(v) | Low | Low | Low | No | |
| AE598 | 41-97 | Consensus | SHIVnef-4ATG(v) | Med | High | High | No | |
| AE598 | 45-97 | Consensus | SHIVnef-4ATG(v) | Low | Low | Low | No | |
| AE601 | 168-96 | Consensus | EGFPiresnef | High | High | High | Yes | 51 |
| AE603 | 354-96 | RULDA | SHIVnef-4ATG(v) | Low | Low | ND | No | |
| AE603 | 360-96 | RULDA | SHIVnef-4ATG(v) | Low | Low | ND | No | |
Column 1 indicates the independent animal experiments described in this report. Column 2 lists the individual animals that were used in each experiment. Column 3 specifies the HIV-1 nef allele that was expressed in the SHIVnef inoculum for each animal. The NL 4-3 nef allele is derived from the commonly used laboratory adapted strain of the same name; the 259 nef allele was isolated from Mm259-93 at the time of death and contained 14 amino acid changes from the NL 4-3 nef (Fig. 4A); the RULDA nef allele was isolated from an HIV-1-infected patient that progressed rapidly to AIDS, and the consensus allele represents a compilation of sequences derived from a cohort of HIV-1-infected patients (40). Column 4 shows the nature of the recombinant constructions. The SHIVnef-fusion expresses a SIV–HIV-1 nef chimeric gene (Fig. 1C), whereas SHIVnef-4ATG(i) and SHIVnef-4ATG(v) express HIV-1 nef as an independent gene in an optimal context for expression (Fig. 1D and E). EGFPiresnef was engineered to express the EGFP gene, the IRES element of encephalomyocarditis virus, and HIV-1 nef as previously described (5). Columns 5, 6, and 7 represent the viral loads observed for individual animals at the times indicated. A designation of a high load denotes a burden that is consistent with what is observed for SIVmac239-infected animals, whereas a designation of a low load denotes a burden that is consistent with what is observed for SIVΔnef-infected animals. A medium (Med) load designation indicates an intermediate phenotype, and ND indicates that the viral load data has not been acquired as yet for this time frame. Column 8 specifies the animals in this study that have died to date with AIDS, and column 9 specifies the number of weeks from inoculation to death for these animals.
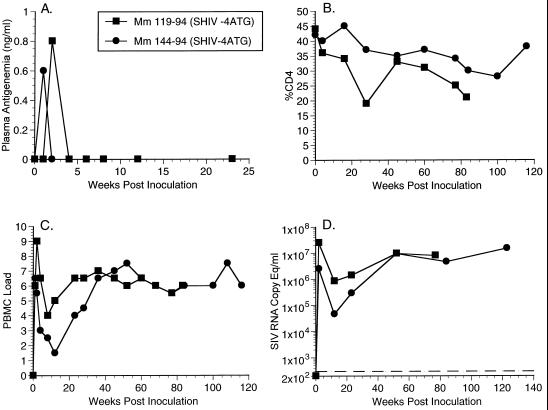
FIG. 2.
Plasma antigenemia, CD4%, PBMC load, and RNA copy eq/ml measurements for animal experiment 542 (AE542, Table 1). (A) Plasma antigenemia in SHIVnef-infected rhesus monkeys. p27 concentrations in plasma were determined at the time points indicated. The limit of detection is approximately 0.05 ng/ml. Week 0 is a preinfection sample taken immediately before inoculation with SHIVnef. (B) CD4% in SHIVnef-infected rhesus monkeys. Whole blood was drawn from SHIVnef-inoculated animals at various times p.i. and stained with OKT4, an FITC-conjugated murine monoclonal antibody that was raised against rhesus macaque CD4 (American Type Culture Collection). The stained samples were analyzed with a FACSscan flow cytometer (Becton Dickinson). (C) Frequency of infectious cells in PBMC of SHIVnef-infected rhesus macaques. Viral loads were graded on a scale from 0 to 10 indicating the number of PBMC needed to recover SIV. A “0” indicates that no virus was recovered with 106 cells, a “1” indicates successful virus recovery from 106 cells, and values 2 to 10 indicate successful virus recovery from 333,333, 111,111, 37,037, 12,345, 4,115, 1,371, 457, 152, or 51 cells, respectively. (D) Plasma SIV RNA levels at the indicated weeks p.i. for animals infected with SHIVnef recombinants. The dashed lines indicate the threshold sensitivity of the assay (300 copy eq/ml). Prior to week 72, plasma was not stored appropriately for plasma RNA measurements of animal experiment 542 (AE542). A value of 0 was assumed for week 0.
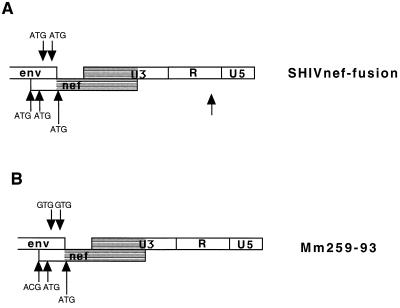
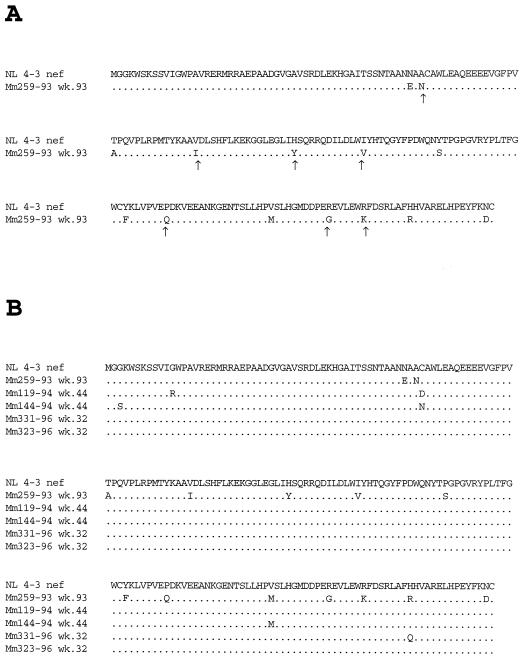
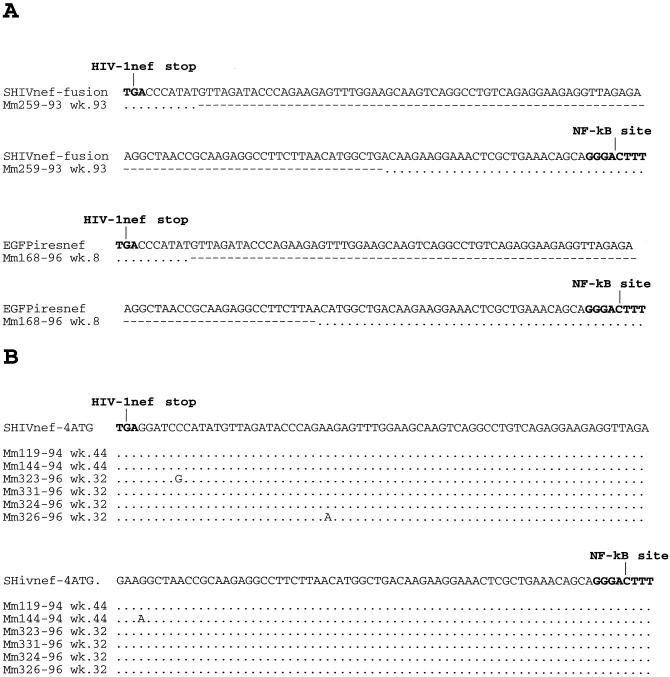
We investigated whether changes had occurred in the SHIVnef-fusion genome in Mm259-93 by the time of death. Sequence analysis indicated that three of the four ATGs in the region of overlap containing both SIV env and SIV nef sequences had been mutated without affecting the predicted SIVmac env amino acid sequence (Fig. 3). The unchanged ATG in this region was in a sequence context that has been reported to be suboptimal for utilization as an initiating methionine for the expression of downstream sequences (23–25) (Fig. 3). These observations suggested that sequence changes within Mm259-93 led to the expression of the HIV-1 nef open reading frame independent of the upstream SIV nef sequences to which it was fused. We also examined the NL 4-3 nef sequences isolated from Mm259-93 to determine whether changes were introduced into the nef coding region which also may have contributed to the development of higher loads in this animal. At the time of death, the HIV-1 nef in Mm259-93 had acquired 14 amino acid changes in the NL 4-3 nef sequences (Fig. 4A). Seven of these changes produced the same amino acid present at the corresponding position of SIVmac nef (Fig. 4A).
FIG. 3.
Sequence changes in the region of overlap containing both SIV env and SIV nef. SHIVnef sequences present in PBMC of Mm259-93 at the time of death are compared with the SHIVnef fusion construct that was used for infection. One of the preserved ATGs represents the initiating ATG of HIV-1 nef. The other preserved ATG in the region of overlap is not in an appropriate consensus (5′-TCCATGA-3′) context for the initiation of translation (a purine at the −3 position and a guanosine at the +4 position relative to the adenosine of the ATG) (23–25). These data represent a consensus sequence of two clones from each of two independent PCRs.
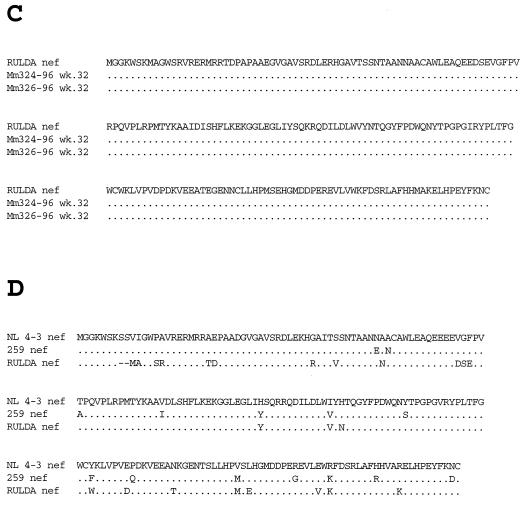
FIG. 4.
Analysis of HIV-1 nef sequences contained in SHIVnef recombinants recovered from infected monkeys. The sequences represent a consensus of two clones from each of two independent PCRs. (A) Amino acid changes observed in NL 4-3 nef after infection of Mm259-93 with SHIVnef-fusion. The arrows indicate where nucleotide changes resulted in the same amino acid that is present at the corresponding position of SIVmac nef. (B) A comparison of HIV-1 nef amino acid sequences after passage in animals that were infected with SHIVnef that expressed NL 4-3 nef sequences and maintained high viral loads. (C) Same as in panel B except with RULDA nef sequences. (D) An alignment of the amino acids expressed by NL 4-3 nef, 259 nef which was passaged through Mm259-93, and RULDA nef which was isolated from a rapid-progressor HIV-1 patient. In all panels, dots denote sequence identity and the dashes denote deletions. The sequences analyzed were isolated from these animals at the indicated number of weeks p.i.
Infection of monkeys with optimized SHIVnef constructions.
We next created recombinants capable of expressing HIV-1 nef as an independent open reading frame. As described above, SIVmac contains two ATG codons in the nef reading frame in the 167 base region of env-nef overlap; the 5′ methionine acts as the start codon for SIVmac nef translation. These two ATGs were mutated to ACGs, which did not affect the predicted amino acid sequence in the env reading frame (Fig. 1D). Two additional ATGs are present in the SIVmac genome in the region of env-nef overlap in the non-env/non-nef reading frame (Fig. 1C). These triplets were mutated to GTGs which again did not affect the predicted amino acid sequence of env (Fig. 1D). These ATGs were eliminated to ensure optimal expression of HIV-1 nef sequences which were inserted downstream (23–25). NL 4-3 nef coding sequences were inserted immediately 3′ of the SIVmac env coding sequences to create SHIV-4ATG(i) (Fig. 1D). In order to facilitate the insertion of independent HIV-1 nef alleles into the SIVmac nef locus, an XbaI restriction site was introduced immediately downstream of the SIVmac env stop codon and a BamHI site was introduced 120 bases upstream of the SIVmac NF-κB site as previously described (SIVΔnefXESAB) (4). HIV-1 nef alleles were engineered to contain a consensus translation initiation motif ACGCCCACC (25) immediately 5′ of the start codons and were then inserted into these XbaI (CTCGAG) and BamHI (GGATCC) sites [SHIVnef-4ATG(v), Fig. 1E]. The nef alleles for insertion were derived from the infectious molecular clone NL 4-3 (1), a rapidly progressing AIDS patient (coded RULDA), a consensus nef sequence (40), and the variant NL 4-3 sequence that evolved in Mm259-93 (Fig. 4A). The nef alleles were inserted into the SIVΔnefXESAB vector to produce the recombinants SHIVnef NL 4-3v, SHIVnef RULDA, SHIVnef consensus, and SHIVnef 259, respectively.
We inoculated two animals, Mm119-94 and Mm144-94 (AE576, Table 1), with SHIVnef-4ATG(i) which expressed NL 4-3 nef sequences (Fig. 1D). Plasma antigenemia was detected in Mm119-94 and Mm144-94 in the initial weeks p.i. (Fig. 5A), and these animals maintained persisting high cell-associated and RNA loads (Fig. 5C and D and Table 1). The CD4% values in Mm119-94 were declining up until the time of death with AIDS at 80 weeks p.i. (Fig. 5B). Mm144-94 has remained healthy for 116 weeks p.i. despite the continued persistence of high viral loads.
FIG. 5.
Plasma antigenemia, CD4%, PBMC load, and RNA copy eq/ml measurements for animal experiment 576 (AE576, Table 1). See legend to Fig. 2 for details.
We infected four additional animals with SHIVnef NL 4-3v (Fig. 1E). We also inoculated two animals with SHIVnef259 which expressed the HIV-1 nef allele which had acquired 14 amino acid changes from NL 4-3 nef through infection in Mm259-93 (Fig. 4A) and two animals which expressed an HIV-1 nef allele which was isolated from a patient who progressed rapidly to AIDS (RULDA) (AE585, Table 1). All eight animals in these experiments displayed detectable antigenemia in the initial weeks p.i. (Fig. 6A). However, only five maintained persisting high viral and RNA loads (Fig. 6C and D), and all five died with AIDS 52 to 110 weeks p.i. (Table 1). Only one of two animals inoculated with SHIVnef259 maintained high loads and died with AIDS (Mm330-96). Of the four animals inoculated with SHIVnef NL 4-3(v) (Mm125-96, Mm211-96, Mm323-96, and Mm331-96 [Table 1]), two (Mm323-96 and 331-96) maintained persisting high viral burdens and died with AIDS; two (Mm125-96 and Mm211-96) have maintained low loads and remain healthy 102 weeks p.i.
FIG. 6.
Plasma antigenemia, CD4%, PBMC load, and RNA copy eq/ml measurements for animal experiment 585 (AE585, Table 1). See legend to Fig. 2 for details.
We next investigated whether the changes that were observed in the NL 4-3 nef allele after passage in Mm259-93 were also observed in the NL 4-3 nef allele after passage in Mm119-94, Mm144-94, Mm323-96, and Mm331-96 which also maintained persisting high viral loads (Table 1). Sequence analysis revealed that changes in NL 4-3 nef seen in Mm259-93 were rarely seen in these four animals (Fig. 4B). The exception was the Val168→Met mutation, which was observed in both Mm144-94 and Mm259-93 (Fig. 4B). We also examined nef sequences from Mm324-96 and Mm326-96 which were inoculated with SHIVnef RULDA and maintained high viral loads throughout the course of infection (Table 1). At week 32 p.i., no amino acid changes were observed in the sequences of RULDA nef isolated from these animals (Fig. 4C). The original RULDA nef allele did contain Y102, V114, M168, and K184 (the numbers correspond to NL 4-3 nef), which were the amino acids created at these positions after sequence changes in Mm259-93 (Fig. 4D).
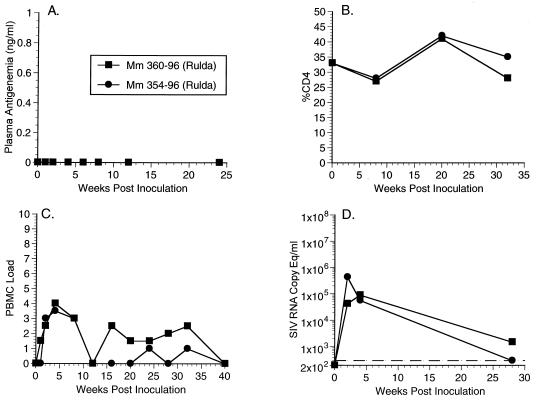
Since both Mm324-96 and Mm326-96, which were inoculated with SHIVnef RULDA maintained high viral loads and died from AIDS and no changes were observed in the RULDA nef sequences in the first 32 weeks p.i., we inoculated two additional animals, Mm354-96 and Mm360-96 with the same dose of the same stock of SHIVnef RULDA in animal experiment 603 (AE603, Table 1). In contrast to our earlier experiment with this recombinant, neither monkey was antigenemic in the initial weeks p.i. (Fig. 7A). In addition, the peaks in SIV load in the PBMC in these animals in the initial weeks of infection were lower than what was observed for other SHIVnef-infected animals (Fig. 7C). However, in Mm354-96, a peak plasma SIV RNA concentration of 430,000 copy equivalents (copy eq)/ml was reached at week 2 p.i. (Fig. 7D), which is similar to the concentrations observed in animal experiment 585 (Fig. 5). However, by week 28 p.i. the SIV RNA concentration had dropped to only 300 copy eq/ml (Fig. 7D), and at week 40 p.i. viral loads were undetectable in this animal (Fig. 7C). In the case of Mm360-96 the peak in SIV RNA expression was significantly lower (89,000 copy eq/ml) than was observed for other SHIVnef-infected animals (Fig. 7D). At 28 weeks p.i. Mm360-96 expressed low but detectable levels of SIV RNA in plasma (Fig. 7D), and at week 40 p.i. undetectable levels of SIV in the PBMC were detected (Fig. 7C). These two animals have maintained stable percentages of CD4 cells and have remained healthy (Fig. 7B and Table 1).
FIG. 7.
Plasma antigenemia, CD4%, PBMC load, and RNA copy eq/ml measurements for animal experiment 603 (AE603, Table 1). See legend to Fig. 2 for details.
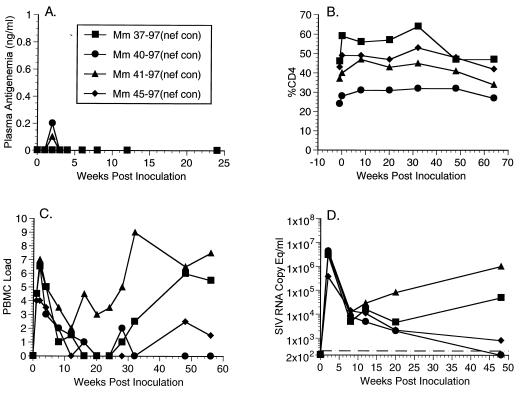
We created one, final recombinant (SHIVnef consensus) in the hopes of establishing a SHIVnef that would yield consistently high viral loads and would be 100% pathogenic in rhesus monkeys. The sequences expressed in the nef consensus allele represent a compilation of sequences derived from a cohort of HIV-1-infected patients (40). Of the four animals inoculated with SHIVnef consensus (AE598, Table 1), two (Mm40-97 and Mm41-97) expressed detectable levels of plasma antigenemia in the initial weeks p.i. (Fig. 8A). Mm37-97 initially maintained low cell-associated viral loads but these cell loads rose dramatically by 48 weeks p.i. and were maintained in subsequent weeks (Fig. 8C and Table 1). SIV RNA concentrations also increased in the interval from 20 to 48 weeks p.i. in this animal (Fig. 8D). Mm41-97, on the other hand, maintained high viral loads (Fig. 8C and Table 1) and high plasma RNA concentrations (Fig. 8D) throughout the course of infection. Conversely, Mm40-97 and Mm45-97 maintained low SIV burdens (Fig. 8C and Table 1) and low or undetectable RNA concentrations at late times p.i. (Fig. 8D). All four animals in this experiment have maintained stable percentages of CD4 cells and remain healthy (Fig. 8B and Table 1).
FIG. 8.
Plasma antigenemia, CD4%, PBMC load, and RNA copy eq/ml measurements for animal experiment 598 (AE598, Table 1). See legend to Fig. 2 for details.
Growth kinetics of SHIVnef recombinants.
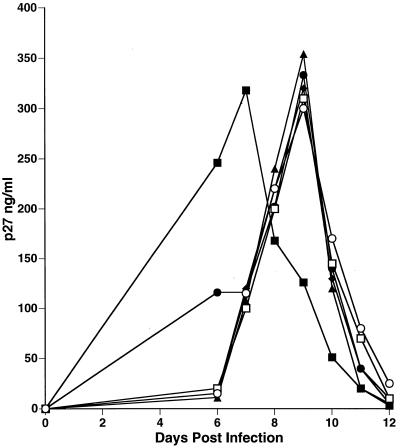
SHIVnef stocks containing 1 ng of p27 antigen were used to infect CEMx174 cells. Supernatants were collected starting at day 6 postinfection and assayed for p27 antigen concentration. SIVmac239 production peaked at day 7, and SHIVnef recombinant production peaked at day 9 postinfection (Fig. 9). Despite the slight delay, peak yields were similar for the SHIVnef recombinants and the parental SIVmac239.
FIG. 9.
Infection of CEMx174 cells with SIVmac239 and various SHIVnef recombinants. A 1-ng portion of p27 antigen that was derived from the cell-free supernatant of transfected CEMx174 cells was used for each infection. Depicted are the SIV p27 concentrations in the cell-free supernatants of the infected cells. Symbols: ■, SIVmac239; ●, SHIVnef NL 4-3v; ▴, SHIVnef RULDA; ⧫, SHIVnef259; □, SHIVnef consensus; ○, SHIVnef fusion.
Expression of HIV-1 nef.
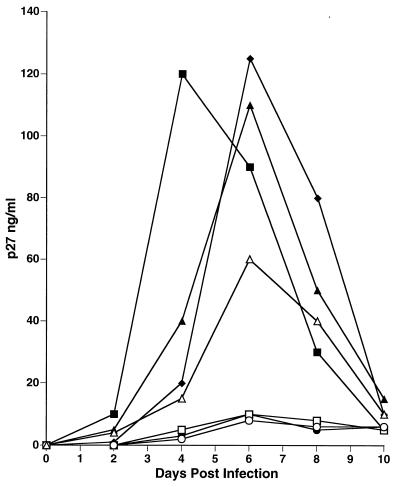
Recently, we demonstrated that the expression of SIVmac nef or HIV-1 nef contributed to activation of the rhesus monkey cell line 221, leading to greatly enhanced replication of SIVmac in the absence of IL-2 (4). We infected 221 cells with SHIVnef recombinants (Table 1) to compare the replication kinetics of these recombinants with those of parental SIVmac. nef-Dependent enhancement of viral replication was not observed with SHIVnef-fusion infections of 221 cells in comparison to a control SIVΔnef infection (Fig. 10). Conversely, SHIVnef-4ATG(i) and SHIVnef NL 4-3v, both of which expressed NL 4-3 nef as an independent gene in an appropriate context for translation, demonstrated significant replication enhancement over a SIVΔnef infection. Levels of p27 production were comparable to SIVmac239 infection (Fig. 10). We tested the influence of inclusion of ATGs on the downstream expression of HIV-1 nef sequences by infecting 221 cells with SHIVnef recombinants that contained an ATG at the 5′ extent of HIV-1 nef sequences in which either the SIV env-nef overlap region was not mutated at all [SHIVnef(i)] or in which only two ATGs in the nef reading frame in this region were mutated to ACGs (SHIVnef-2Met), leaving the two remaining ATGs in this region intact. SHIVnef(i) produced levels of SIV that were similar to those obtained with the SIVΔnef control infection (Fig. 10). SHIVnef-2Met infections produced higher levels of virus than did SHIVnef(i) infections. However, the levels produced by SHIVnef-zATG were significantly lower than those that were observed for SHIV-4ATG(i) and SHIVnef NL 4-3(v) infections (Fig. 10).
FIG. 10.
Infection of unstimulated 221 cells with SHIVnef recombinants. Cell line 221 cells incubated in RPMI medium containing 5% fetal calf serum without exogenous IL-2 were infected with 10 ng of SIV p27 antigen of SHIVnef recombinant virus. p27 antigen was quantitated in the cell-free supernatant at the times indicated. Symbols: ■, SIVmac239; ●, SHIVnef-fusion; ▴, SHIVnef-4ATG(i); ⧫, SHIVnef NL 4-3v; □, SIVmac239Δnef; ○, SHIVnef(i); ▵, SHIVnef-2ATG.
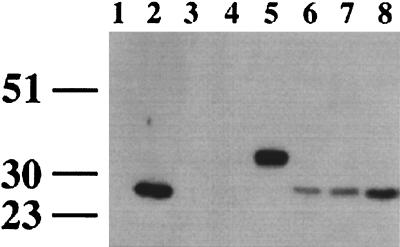
The levels of HIV-1 nef expression by these SHIVnef recombinants were assayed directly by Western blotting. A total of 3 × 106 CEMx174 cells were infected with recombinant virus containing 50 ng of p27 antigen. The cells were lysed 7 days postinfection, and lysates were Western blotted and incubated with a nef-specific monoclonal antibody (EH). HIV-1 nef was detected in cells infected with an HIV-1 NL 4-3 control, SHIVnef-2Met, SHIVnef-4ATG(i), and SHIVnef NL 4-3(v) infections (Fig. 11). As expected, a larger-sized protein was detected in lysates from the SHIVnef-fusion-infected cells (Fig. 11, lane 5). A nef-specific band was not detected when a version of SHIVnef was used with a premature, in-frame stop codon (lane 4) or with SHIVnef(i) (lane 3), thus demonstrating that retention of the upstream ATGs severely limited the expression of downstream HIV-1 nef sequences in SHIVnef(i).
FIG. 11.
Expression of HIV-1 nef in CEMx174 cells infected with SHIVnef recombinants. HIV-1- or SHIVnef-infected cells were harvested at day 7, and proteins from cell lysates were separated by SDS-PAGE and electroblotted onto a membrane filter. HIV-1 nef was detected by using a nef-specific monoclonal antiserum (EH). Lanes: 1, mock infected; 2, HIV-1 NL 4-3 nef; 3, SHIVnef(i); 4, SHIVnef-stop; 5, SHIVnef-fusion; 6, SHIVnef-2Met; 7, SHIVnef-4ATG(i); 8, SHIVnef-4ATG(v).
Genetic analysis of U3 sequences in SHIVnef recombinants.
In all of the SHIVnef constructs described in this study, 120 bases of SIV U3 sequence immediately 5′ of the SIV NF-κB site were retained (Fig. 1). This region has been consistently retained in SIVΔnef-infected animals (22). We analyzed the stability of the SIV U3 sequences in SHIVnef-infected animals that maintained high viral loads. SIV that was isolated at the time of death from Mm259-93 had retained only the 28 bases immediately upstream of the SIV NF-κB site and had deleted 92 bases of SIV U3 sequence (Fig. 12A). Mm168-96, which was inoculated with EGFPiresnef as previously described (5) and which maintained persisting high viral loads and died with AIDS 51 weeks p.i. (Table 1), retained only 24 bases of U3 immediately 5′ of the SIV NF-κB site and had deleted 96 bases of SIV U3 sequence (Fig. 12A). Conversely, SHIVnef isolated from Mm119-94, Mm144-94, Mm323-96, and Mm331-96 (Table 1) had retained the entire 120 base SIV U3 sequence that was contained in the original SHIVnef construction (Fig. 12B).
FIG. 12.
Sequence analysis of SHIVnef U3 sequences before and after passage in rhesus monkeys. Dots denote no change in the U3 sequence from the inoculum, and dashes denote deletions in the U3 sequence after animal infection.
DISCUSSION
In this report we have demonstrated that HIV-1 nef sequences can substitute for SIVmac nef sequences to produce a pathogenic infection. Of the 19 rhesus monkeys described in this study, 11 have maintained persisting high viral loads as is seen in SIVmac239 infections and 8 have died with AIDS 51 to 110 weeks p.i. (Table 1). The frequency of high virus loads and disease development with our SHIVnef constructs is clearly higher than what we have observed with SIVmac239Δnef. Of the 20 rhesus monkeys that we have infected with SIVmac239Δnef, only one developed moderate loads during the first year of observation and only three have shown moderate or high loads and/or evidence of disease progression over the entire period of observation (mean, 5.1 years; longest period, 9.0 years) (47). Alexander et al. (4) have previously demonstrated the ability of HIV-1 nef to substitute for SIV nef in the context of virus in the 221 replication enhancement assay, and Sinclair et al. (41) have previously demonstrated the ability of the HIV-1nef allele to substitute for the SIV nef allele in the context of virus in assays of infectivity enhancement and accelerated replication kinetics.
Despite our use of four distinct HIV-1 nef alleles expressed in an optimal context, infected animals maintained persisting high viral loads with similar frequencies of about 50% regardless of which HIV-1 nef allele was expressed in the recombinant genome (Table 1). This was the case even with infections that used a nef allele (259) that had been passaged through a monkey (Mm259-93) and that had acquired amino acid changes at 14 positions (Fig. 4A). We did not observe a pattern of change to a 259 nef genotype in the SHIVnef NL 4-3v-infected animals that maintained persisting high viral loads (Fig. 4B). If there is selective pressure for such change in rhesus monkeys to a sequence more like SIVmac, it is not sufficiently strong to be seen in all animals.
Fusion of upstream SIVmac nef sequences to HIV-1 nef sequences (SHIVnef-fusion; Fig. 1C) resulted in the stable expression of a larger chimeric protein (Fig. 11). However, this chimera did not result in a nef gene that was functional in the 221 cell assay (Fig. 10). In SIV recovered from Mm259-93, three of the four ATGs present in the env-nef overlap region of the SHIVnef-fusion inoculum were mutated (Fig. 3). The unchanged ATG in this region (5′-TCCATGA-3′) was in a context that was not consistent with its use as an initiation codon (a purine at the −3 position and a guanosine at the +4 position relative to the adenosine of the ATG) (23–25) and thus its presence would not be likely to interfere with expression of the downstream HIV-1 nef sequences. In addition, inclusion of upstream ATGs in the region of SIV env-nef overlap appeared to inhibit the expression of the downstream HIV-1 nef sequences in SHIVnef(i) (Fig. 10 and 11). The changes observed in the env-nef region in Mm259-93 therefore rendered the HIV-1 nef sequences open and in a context that was optimal for expression. This is consistent with our previous observation of SIV sequences from Mm168-96 in which the EGFP gene and the IRES element were removed to also render the HIV-1 nef sequences open and in an optimal context for expression (5). It is also consistent with the ease with which extraneous sequences are lost from this region in other recombinant constructs that have been studied previously (22, 29, 48). Conversely, the HIV-1 nef coding sequences were retained intact in all experimentally infected monkeys. In other experiments not shown, a SHIVnef in which a stop codon was introduced into the HIV-1 nef reading frame quickly reverted in infected monkeys to an open nef reading frame (data not shown). These observations not only demonstrate the extreme flexibility in the SIVmac genome in response to selective pressure but also reveal that an optimal context for translation initiation by elimination of the upstream ATGs is necessary for appropriate expression by these SHIVnef chimeras. The retention of HIV-1 nef sequences, selection for an open HIV-1 nef reading frame, and sequence changes to optimize HIV-1 nef translation are all consistent with an advantageous contribution by the HIV-1 nef gene in the context of SIV in rhesus monkeys.
In SIVmac, an important transcriptional enhancer element is located within an 80-bp region of U3 that is immediately upstream of the NF-κB binding site (17, 30, 34). This enhancer element allows SIVmac replication in the complete absence of NF-κB and Sp1 binding sites (17) and is contained within the nef reading frame within U3. In monkeys infected with SIV containing a 182-bp deletion in the region that is uniquely nef, the virus progressively loses much of the remaining nef sequences that are overlapped by U3 (22). However, this nef mutant virus consistently retains regions of nef sequence that contribute to virus replication in a cis-acting fashion; these include the polypurine tract, the U3 terminal sequences and, importantly, the transcriptional enhancer element contained within the 80 bp immediately upstream of NF-κB (18, 33, 36, 42–44). It is thus curious that, in the current study, SIV sequences were lost in Mm168-96 and Mm259-93 to within 27 to 35 nucleotides of the NF-κB site (Fig. 12). It is possible that these 27 to 35 nucleotides retain much or all of the transcriptional enhancer activity. However, the SIV U3 deletions in Mm168-96 and Mm259-93 bring the 3′ carboxy-terminal sequences of HIV-1 nef into much closer proximity to the NF-κB binding element (Fig. 12). This region near the C terminus of the nef coding sequence has not been rigorously studied as a transcriptional control element in HIV-1. However, in a human naturally infected with nef-deleted HIV-1, progressive deletions in U3 over time consistently spared the 80 bp upstream of the NF-κB binding site (21). Thus, movement of HIV-1 transcriptional control elements within nef to closer proximity to the core enhancer element (NF-κB and Sp1) could have contributed to the U3 sequence rearrangements observed in Mm168-96 and Mm259-93.
The SHIVnef constructions described here have brought some cis-acting HIV-1 sequences under the operation of SIV-encoded enzymatic activities. The polypurine tract, located within nef sequences just upstream of the start of U3, serves as the primer for the synthesis of plus-strand DNA by reverse transcriptase and is absolutely essential for replication. In addition, terminal sequences at the 5′ end of U3 are absolutely required for proviral DNA integration by the virus-encoded integrase (10, 19). U-box sequences, just upstream of the polypurine tract, are also required for replication at the RT step (18). The sequences of these HIV-1 cis-acting elements all differ slightly from their SIV counterparts. Although the HIV-1 cis-acting sequences are sufficiently functional with SIV-encoded enzymes to allow for viral replication, they could possibly be responsible for the slight delay in replication seen in Fig. 9.
While HIV-1 nef can clearly substitute for SIVmac nef in this relevant animal model, we were not able to achieve moderate or high virus loads and disease progression in a workable time frame in 100% of the animals. Possible explanations for this result are varied. Since SIVmac nef and HIV-1 nef differ somewhat in the activities that can be measured in vitro, HIV-1 nef may lack one or more functions that contribute to pathogenesis in monkeys. Since all of the functional activities reported for nef involve interactions with host cell proteins, HIV-1 nef may demonstrate less-than-optimal coupling with macaque cellular partners, as opposed to the human cellular partners for which it has evolved. It is also possible that the action of SIV enzymes on cis-acting HIV-1 sequences present in the recombinant constructs has resulted in suboptimal viral replication or that the hybrid constructions are not optimized for cis-acting sequence functions. With regard to the latter possibility, SHIVnef constructions necessarily create hybrid LTRs which may be altered in their transcriptional regulatory elements. The variability in viral loads and in time to develop AIDS can be viewed in an optimistic light as actually similar to the variable viral loads and disease progression observed for HIV-1-infected people. However, practical use of the SHIVnef model for studying the effects of nef mutations or anti-nef drugs would benefit from improvements to the system that would produce a more consistent outcome.
ACKNOWLEDGMENTS
We thank J. Lifson for plasma RNA measurements; D. L. Xia and A. McPhee for technical assistance; P. Sehgal and E. Roberts for animal care, blood sampling, and clinical care; J. Sullivan and T. Greenough for the RULDA isolate; R. Swanstrom for the nef consensus sequences; J. Hoxie for the nef monoclonal antibody (EH); A. Lackner and the New England Regional Primate Research Center Department of Pathology for performance of necropsies; and J. Newton for manuscript preparation.
This study was supported by PHS grants AI25328, AI38559, and RR00168.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken C, Konner J, Landau N, Lenburg M E, Trono D. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 3.Aiken C, Trono D. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol. 1995;69:5048–5056. doi: 10.1128/jvi.69.8.5048-5056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander L, Du Z, Rosenzweig M, Jung J U, Desrosiers R C. A role for natural simian immunodeficiency virus and human immunodeficiency virus type 1 nef alleles in lymphocyte activation. J Virol. 1997;71:6094–6099. doi: 10.1128/jvi.71.8.6094-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander L, Veazey R S, Czajak S, DeMaria M, Rosenzweig M, Lackner A A, Desrosiers R C, Sasseville V G. Recombinant simian immunodeficiency virus expressing green fluorescent protein identifies infected cells in rhesus monkeys. AIDS Res Hum Retroviruses. 1999;15:11–21. doi: 10.1089/088922299311664. [DOI] [PubMed] [Google Scholar]
- 6.Benson E, Sanfridson A, Ottinger J S, Doyle C, Cullen B R. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus nef prevents viral super infection. J Exp Med. 1993;177:1561–1566. doi: 10.1084/jem.177.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowers M Y, Spina C A, Kwob T J, Fitch N J S, Richman D D, Guatelli J C. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol. 1994;68:2906–2914. doi: 10.1128/jvi.68.5.2906-2914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C, Lawson V A, Crowe S, Maerz A, Sonza S, Learmont J, Sullivan J S, Cunningham A, Dwyer D, Downton D, Mills J. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 9.Desrosiers R C, Lifson J D, Gibson J S, Czajak S C, Howe A Y M, Arthur L O, Johnson R P. Identification of highly attenuated mutants of simian immunodeficiency virus. J Virol. 1998;72:1431–1437. doi: 10.1128/jvi.72.2.1431-1437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du Z, Ilyinskii P O, Lally K, Desrosiers R C, Engelman A. A mutation in integrase can compensate for mutations in the simian immunodeficiency virus att site. J Virol. 1997;71:8124–8132. doi: 10.1128/jvi.71.11.8124-8132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du Z, Lang S M, Sasseville V G, Lackner A A, Ilyinskii P O, Daniel M D, Jung J U, Desrosiers R C. Identification of a nef allele that causes lymphocyte activation and acute disease in macaque monkeys. Cell. 1995;82:665–674. doi: 10.1016/0092-8674(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 12.Du Z, Regier D A, Desrosiers R C. An improved recombinant PCR mutagenesis procedure that uses alkaline denatured plasmid template. BioTechniques. 1995;18:376–378. [PubMed] [Google Scholar]
- 13.Garcia J V, Miller A D. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature. 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 14.Gibbs J S, Lackner A A, Lang S M, Simon M A, Sehgal P K, Daniel M D, Desrosiers R C. Progression to AIDS in the absence of genes for vpr or vpx. J Virol. 1995;69:2378–2383. doi: 10.1128/jvi.69.4.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 16.Howe A Y M, Jung J U, Desrosiers R C. Zeta chain of the T-cell receptor interacts with nef of simian immunodeficiency virus and human immunodeficiency virus type 2. J Virol. 1998;72:9827–9834. doi: 10.1128/jvi.72.12.9827-9834.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilyinskii P O, Desrosiers R C. Efficient transcription and replication of simian immunodeficiency virus in the absence of NF-κB and Sp1 binding elements. J Virol. 1996;70:3118–3126. doi: 10.1128/jvi.70.5.3118-3126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilyinskii P O, Desrosiers R C. Identification of a sequence element immediately upstream of the polypurine tract that is essential for replication of simian immunodeficiency virus. EMBO. 1998;17:3766–3774. doi: 10.1093/emboj/17.13.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katz R A, Skalka A M. The retroviral enzymes. Biochemistry. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 20.Kestler H W, III, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for the development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 21.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 22.Kirchhoff F, Kestler III H W, Desrosiers R C. Upstream U3 sequences in simian immunodeficiency virus are selectively deleted in vivo in the absence of an intact nef gene. J Virol. 1994;68:2031–2037. doi: 10.1128/jvi.68.3.2031-2037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8132. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981;9:5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak M J. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lang S M, Iafrate A J, Stahl-Hennig C, Kuhn E M, Nisslein T, Kaup F-J, Haupt M, Hunsmann G, Skowronski J, Kirchohoff F. Association of simian immunodeficiency virus Nef with cellular serine/threonine kinases is dispensable for the development of AIDS in rhesus macaques. Nat Med. 1997;3:860–865. doi: 10.1038/nm0897-860. [DOI] [PubMed] [Google Scholar]
- 27.Lee C-H, Leung B, Lemmon M A, Zheng J, Cowburn D, Kuriyan J, Saksela K. A single amino acid in the SH3 domain of Hck determines its high affinity and specificity in binding to HIV-1 nef protein. EMBO J. 1995;14:5006–5015. doi: 10.1002/j.1460-2075.1995.tb00183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee C-H, Saksela K, Mirza U A, Chait B T, Kuriyan J. Crystal structure of the conserved core of HIV-1 nef complexed with a Src family SH3 domain. Cell. 1996;85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- 29.Luciw P A, Cheng-Mayer C, Levy J A. Mutational analysis of the human immunodeficiency virus: the orf-B region down-regulates virus replication. Proc Natl Acad Sci USA. 1987;84:1434–1438. doi: 10.1073/pnas.84.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markovitz D M, Smith M J, Hilfinger J, Hannibal M C, Petryniak B, Nabel G J. Activation of the human immunodeficiency virus type 2 enhancer is dependent on purine box and κB regulatory elements. J Virol. 1992;66:5479–5484. doi: 10.1128/jvi.66.9.5479-5484.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller M D, Warmerdam M T, Gaston I, Greene W C, Feinberg M B. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J Exp Med. 1994;179:101–113. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naidu Y M, Kestler III H W, Li Y, Butler C V, Silva D P, Schmidt D K, Troup C D, Sehgal P K, Sonigo P, Daniel M D, Desrosiers R C. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J Virol. 1988;62:4691–4696. doi: 10.1128/jvi.62.12.4691-4696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Omer C A, Resnick R, Faras A J. Evidence for involvement of an RNA primer in initiation of strong-stop plus DNA synthesis during reverse transcription in vitro. J Virol. 1984;50:465–470. doi: 10.1128/jvi.50.2.465-470.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pohlmann S, Floos S, Ilyinskii P O, Stamminger T, Kirchhoff F. Sequences just upstream of the simian immunodeficiency virus core enhancer allow efficient replication in the absence of NF-kappaB and Sp1 binding elements. J Virol. 1998;72:5589–5598. doi: 10.1128/jvi.72.7.5589-5598.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Regier D A, Desrosiers R C. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res Hum Retroviruses. 1990;6:1221–1231. doi: 10.1089/aid.1990.6.1221. [DOI] [PubMed] [Google Scholar]
- 36.Resnick R, Omer C A, Faras A J. Involvement of retrovirus reverse transcriptase-associated RNase H in the initiation of strong-stop (+) DNA synthesis and the generation of the long terminal repeat. J Virol. 1984;51:813–821. doi: 10.1128/jvi.51.3.813-821.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saksela K, Cheng G, Baltimore D. Proline-rich (PxxP) motifs in HIV-1 nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of nef+ viruses but not for down-regulation of CD4. EMBO J. 1995;14:484–491. doi: 10.1002/j.1460-2075.1995.tb07024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawai E T, Baur A, Struble H, Peterlin B M, Levy J A, Cheng-Mayer C. Human immunodeficiency virus type 1 nef associates with a cellular serine kinase in T lymphocytes. Proc Natl Acad Sci USA. 1995;91:1539–1543. doi: 10.1073/pnas.91.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz O, Marechal V, LeGall S, Lemonnier F, Heard J-M. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 nef protein. Nat Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 40.Shugars D C, Smith M S, Glueck D H, Nantermet P V, Seillier-Moiseiwitsch F, Swanstrom R. Analysis of human immunodeficiency virus type 1 nef gene sequences present in vivo. J Virol. 1993;67:4639–4650. doi: 10.1128/jvi.67.8.4639-4650.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinclair E, Barbosa P, Feinberg M B. The nef gene products of both simian and human immunodeficiency virus enhance virus infectivity and are functionally interchangeable. J Virol. 1997;71:3641–3651. doi: 10.1128/jvi.71.5.3641-3651.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith J K, Cywinski A, Taylor J M. Initiation of plus-strand DNA synthesis during reverse transcription of an avian retrovirus genome. J Virol. 1984;49:200–204. doi: 10.1128/jvi.49.1.200-204.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith J K, Cywinski A, Taylor J M. Specificity of initiation of plus-strand DNA by Rous sarcoma virus. J Virol. 1984;52:314–319. doi: 10.1128/jvi.52.2.314-319.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sorge J, Hughes S H. Polypurine tract adjacent to the U3 region of the Rous sarcoma virus genome provides a cis-acting function. J Virol. 1982;43:482–488. doi: 10.1128/jvi.43.2.482-488.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spina C A, Kwoh T J, Chowers M Y, Guatelli J C, Richman D D. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J Exp Med. 1994;179:115–123. doi: 10.1084/jem.179.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suryanarayana K, Wiltrout T A, Vasquez G M, Hirsch V M, Lifson J D. Plasma SIV RNA viral load by determination by real-time quantification of product generation in reverse transcriptase-polymerase chain reaction. AIDS Res Hum Retroviruses. 1998;14:183–189. doi: 10.1089/aid.1998.14.183. [DOI] [PubMed] [Google Scholar]
- 47.Wyand, M., K. Manson, S. Czajak, M. Simon, and R. C. Desrosiers. Unpublished data.
- 48.Yilman T, Hsu D, Jones L, Owens S, Grubman M, Mebus C, Yamanaka M, Dale B. Protection of cattle against rinderpest with vaccinia virus recombinants expressing the HA or F gene. Science. 1988;242:1058–1061. doi: 10.1126/science.3194758. [DOI] [PubMed] [Google Scholar]