SUMMARY
Innate lymphoid cells (ILCs), strategically positioned throughout the body, undergo population declines over time. A solution to counteract this problem is timely mobilization of multipotential progenitors from the bone marrow. It remains unknown what triggers the mobilization of bone marrow ILC progenitors (ILCPs). We report that ILCPs are regulated by the circadian clock to emigrate and generate mature ILCs in the periphery. We found that circadian-clock-defective ILCPs fail to normally emigrate and generate ILCs. We identified circadian-clock-controlled endocrine and cytokine cues that, respectively, regulate the retention and emigration of ILCPs at distinct times of each day. Activation of the stress-hormone-sensing glucocorticoid receptor upregulates CXCR4 on ILCPs for their retention in the bone marrow, while the interleukin-18 (IL-18) and RORα signals upregulate S1PR1 on ILCPs for their mobilization to the periphery. Our findings establish important roles of circadian signals for the homeostatic efflux of bone marrow ILCPs.
In brief
Liu et al. report that common ILC progenitors in the bone marrow highly express genes that regulate the circadian clock and that the mobilization of these progenitors from the bone marrow occurs in a circadian-clock-dependent manner. Mechanistically, circadian signals regulate the bone marrow retention and emigration receptors for these ILC progenitors.
Graphical Abstract

INTRODUCTION
The innate immune system detects pathogens and generates signals to mount immediate immune responses.1 The importance of innate lymphoid cells (ILCs) for tissue innate immunity and barrier integrity has been well established.2–4 While wide-spread, ILCs are strategically localized in the alimentary tract, skin, urogenital tract, lungs, and certain non-barrier tissues, such as fat and secondary lymphoid tissues. Activation of ILCs is triggered by tissue-derived cytokines, such as interleukin-12 (IL-12), IL-15, IL-18, IL-33, IL-25, thymic stromal lymphopoietin (TSLP), IL-1b, and IL-23, in a subset-specific manner.5 ILCs produce effector molecules and cytokines that lyse infected cells and tumor cells or activate various cell types, such as epithelial cells, endothelial cells, fibroblasts, macrophages, granulocytes, and lymphocytes to fight infections, repair damaged tissues, and regulate immune responses.2,6–8
Tissue ILCs undergo population attrition over time during aging and pathological conditions.9–14 ILC attrition can lead to peripheral ILC deficiency, which can compromise not only the innate immunity but also the integrity of barrier tissues. Unlike their largely sessile mature forms in peripheral tissues,15 ILC progenitors in humans and animals are highly migratory.16,17 This feature gives them the potential to counteract peripheral ILC loss in the steady state and under pathological conditions. Among ILC progenitors, the multipotential ILC progenitors (ILCPs) that make ILC1s, ILC2s, and ILC3s, are the major population circulating in the blood of humans and mice.16–18 ILCPs in the bone marrow (BM) are largely divided into parenchyma and sinusoid ILCP subsets, and the ILCPs in the sinusoid niche are preferentially mobilized.16 The mobilization of BM ILCPs is controlled by the coordinated expression and functions of several chemoattractant and adhesion receptors, such as CXCR4, Itg-α4, and sphingosine-1-phosphate (S1P) receptor 1 (S1PR1).16 However, the factors that control the reciprocal regulation of these BM retention and emigration receptors for ILCPs have not been established.
Biological processes are generally synchronized to achieve high levels of efficiency and homeostasis.19 The circadian clock is one of the most important cues that control biological synchronization.20 The circadian clock controls the activity of tissue and immune cells, including epithelial cells, endothelial cells, neutrophils, B cells, T cells, and dendritic cells. A phenomenon important for immunological homeostasis is the release of hematopoietic progenitor cells and mature leukocytes in the 24-h oscillation pattern.21 The circadian clock also influences lymphocyte migration and activation in lymphoid tissues.22,23 It has been reported that, within the ILC system, the circadian clock regulates intestinal ILC3s to support and control the intestinal barrier along with its microbiota.24–27 It remains unknown, however, whether the circadian clock regulates the distribution and activity of ILCPs.
We conducted the current study to identify signals that control the mobilization and ILC-producing activity of BM ILCPs. We report that the mobilization of ILCPs from the BM is cooperatively regulated by circadian-clock-regulated cytokine and endocrine cues. A cytokine and transcription factor pathway that induces both the emigration and ILC-producing activity of ILCPs in the circadian fashion has been identified, along with an endocrine signal that induces the expression of the BM retention receptor CXCR4 to suppress ILCP mobilization at different zeitgeber times (ZTs). Our findings indicate that circadian signals control the synchronized mobilization of BM ILCPs to the periphery.
RESULTS
ILCPs highly express clock genes and undergo ZT-dependent fluctuations in the blood circulation
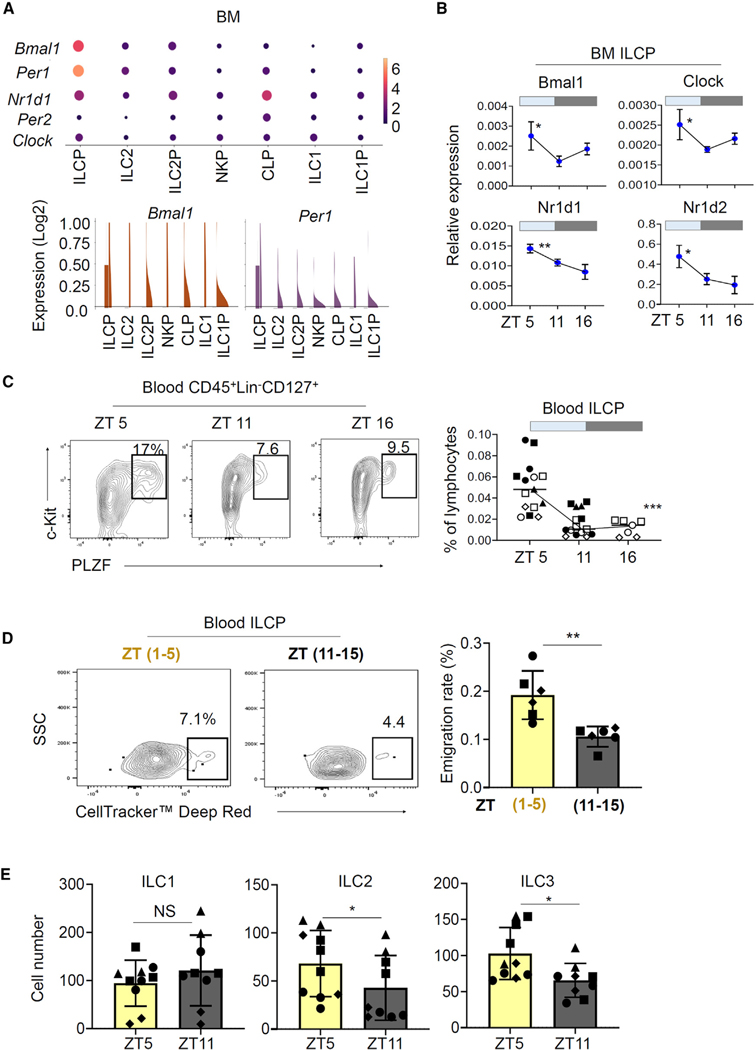
The circadian clock is composed of the master clock in the suprachiasmatic nuclei of the anterior hypothalamus and of individual clocks in cells and tissues throughout the body, which cooperatively generate synchronized and coordinated circadian outputs to control many biological processes.28 The circadian locomotor output cycles kaput (Clock) and basic-helix-loop-helix arnt-like 1 (BMAL1) complex transactivates E box-containing-clock-controlled genes.29 Single-cell RNA sequencing (scRNA-seq) data on ILCP subsets indicate that BM and blood ILCPs express the major clock genes, such as Bmal1, Per1, or Nr1d1 (Figures 1A and S1A). ILC2s and ILC2Ps also expressed the clock genes, albeit at relatively lower levels compared to that of ILCPs (Figure 1A). Interestingly, the expression of Bmal1, Clock, Nr1d1, and Nr1d2 in BM ILCPs changed at different ZTs, which refer to hours after the onset of light for animals in the 12-h light-12-h dark cycle (Figure 1B). In general, the expression of clock genes was higher at ZT 5 compared to ZT 11 and 16. Given that ILCPs are the major ILCPs circulating in the blood of humans and mice,16,17 the expression of clock genes by ILCPs raises the question whether the mobilization of BM ILCPs is regulated by the circadian clock.
Figure 1. BM ILCPs highly express clock genes and emigrate to the blood in a ZT-dependent manner.
(A) Expression of circadian genes in BM ILCPs. The data were retrieved from publicly available scRNA-seq data (GSE193835) on ILCPs from C57BL/6 mice.
(B) Expression of core clock genes at the mRNA level in BM ILCPs at different ZTs. RT-qPCR was performed on sorted BM ILCPs.
(C) Changes in ILCP frequency in the blood at different ZTs.
(D) Emigration rates of BM ILCPs during the ZT 1–5 vs. 11–15 periods. CellTracker Deep Red was injected into the bones of mice before they were euthanized 4 h later.
(E) In vitro differentiation potential of BM ILCPs harvested at different ZTs.
Sorted BM Lin−CD127+c-Kit+ PD-1+ ILCPs were examined in (B) and (E). Rag1−/− (B–D) and PlzfGFP-Cre (E; also called PlzfCre) mice were used. Pooled data obtained from at least three different experiments (n = 6–15) are shown with SEM. p values from one-way ANOVA (B and C) and two-tailed/paired Student’s t test (D and E) are shown. Not significant (NS).
A major consequence of circadian regulation on the immune system is the day-night fluctuation of circulating immune cells and efflux of BM hematopoietic stem and progenitor cells (HSPCs).30,31 In mice, the acrophase (the peak) of BM HSPC efflux to the blood circulation occurs at ZT 5, and the bathyphase (the nadir) is found between ZT 11 and 17.22 To determine this possibility for ILCP progenitors, we examined the number of circulating ILCPs at distinct ZTs (ZT 5, 11, and 16) and found that the number and frequency of blood ILCPs were high at ZT 5 but low at ZT 11 and 16 in mice (Figure 1C), without significant changes in numbers of BM ILCPs (Figure S1B). The fluctuation of ILC2Ps numbers was relatively small and insignificant (Figure S1C). We performed a BM fluorescent dye labeling experiment16,32,33 to track and assess the emigration rate of BM ILCPs at two different time periods that encompass the acrophase and bathyphase of BM HSPC efflux. We detected increased emigration of ILCPs during the ZT 1–5 period compared to the ZT 11–15 period (Figure 1D), indicating time-dependent emigration of ILCPs mirroring that of HSPCs. Emigrated BM ILCPs were frequently found in the intestines and lymphoid tissues but less frequently in lung and fat tissues, indicating a preference for their migration (Figure S1D). In contrast, BM ILC2P preferentially migrated to lung over other tissues.
BM ILCPs, with heterogeneous ILC-poietic activity and the trafficking receptor phenotype, are produced at distinct ZTs
We examined the possibility that BM ILCPs at different ZTs would be different from each other in their differentiation potential. We found that the BM ILCPs at ZT 5 were more efficient than those at ZT 11 in generating ILC2s and ILC3s (Figure 1E). Thus, the ILC-making capacity of BM ILCPs also changes throughout the day, in addition to their efflux to the blood circulation.
The emigration of BM ILCPs is largely mediated by the three trafficking receptors CXCR4, S1PR1, and α4β1 (VLA-4),16 which play distinct roles. CXCR4 is the BM retention receptor, whereas S1PR1 is the BM emigration receptor for ILCPs. The integrin α4β1 promotes the interaction between developing hematopoietic progenitor cells and stromal cells in the arterial niche of the central zone in BM.34 For efficient emigration of ILCPs, coordinated up- and downregulation of these three receptors is required. We found increased expression of the BM retention receptor CXCR4 on ILCPs and ILC2Ps at ZT 11 over ZT 5 but no significant change for the expression of Itg-α4 or Itg-β1 at ZT 5 or 11 (Figures 2A and S2). In contrast, S1PR1 expression was increased at ZT 5 compared to ZT 11 and 16 (Figure 2B). To determine their chemotactic behavior toward BM and blood chemoattractants, we performed a chemotaxis assay for the BM ILCPs at distinct ZTs in a CXCL12(−)/S1P(+) chemotactic gradient, mimicking the natural chemotactic environment of BM.35,36 In line with the CXCR4 and S1PR1 expression, the chemotactic activity of BM ILCPs to S1P and away from CXCL12 was higher at ZT 5 than ZT 16 (Figure 2C). However, we could not find significant differences in the expression and activity of S1PR1 for BM ILC2Ps between ZT5 and ZT 11 (Figures 2B and 2C). Overall, these results indicate the presence of ZT-dependent regulation of BM retention and emigration receptors for ILCPs.
Figure 2. The expression of CXCR4 and S1PR1 on ILCPs changes at different ZTs.
(A) Expression of cell surface CXCR4, Itg-α4, and Itg-β1 by BM ILCPs at different ZTs.
(B) Expression of cell-surface S1PR1 by BM ILCPs at different ZTs.
(C) Chemotaxis of BM ILCPs harvested at different ZTs to CXCL12 (100 ng/mL) in a negative gradient and S1P (100 nM) in a positive gradient. BM common lymphoid progenitors (CLPs, Lin−CD127+Flt3+CD25−), CHILP (Lin−CD127+c-Kit+ PLZF−CD25−), ILCP (Lin−CD127+c-Kit+PLZF+), and ILC2P (Lin−CD127+CD25+c-Kit–) cells were examined.
Both Rag1−/− and PlzfGFP-Cre mice were used in (A) and (B), and Rag1−/− mice were used in (C). Pooled data obtained from at least three different experiments (n = 4–23) are shown with SEM. p values from two-tailed/paired Student’s t test (A and C) and one-way ANOVA with Tukey multiple comparison test (B) are shown. Not significant (NS).
The circadian clock of ILCPs is required for optimal emigration from the BM and generation of intestinal ILCs
To study circadian-clock-defective ILCPs, we created PlzfCre Bmal1f/f mice. While there was no difference in the numbers of BM ILCPs between control and PlzfCre Bmal1f/f animals, a significant decrease in blood-circulating ILCPs was observed in the clock-deficient mice (Figure 3A). Also decreased were ILCPs in peripheral tissues, such as the colon and the small intestine (Figure 3B). Consistently, the emigration of BM ILCPs was decreased in these animals (Figure 3C). However, the numbers of BM and blood ILC2Ps were not significantly changed (Figures S3A and S3B). To understand the molecular basis of such a defect in ILCP emigration, we examined the expression of CXCR4 and S1PR1. Bmal1-deficient ILCPs had decreased expression of S1PR1 but largely intact expression of CXCR4 (Figure 3D). Thus, Bmal1 and the circadian clock regulate the expression of S1PR1 and the emigration of ILCPs from the BM.
Figure 3. Bmal1 deficiency in ILCPs decreases the emigration of BM ILCPs.
(A) Frequency of BM and blood ILCPs in control Bmal1f/f versus PlzfCre Bmal1f/f mice.
(B) Numbers of ILCPs in the intestines of Bmal1f/f versus Plzfcre Bmal1f/f mice.
(C) Emigration rates of BM ILCPs in Bmal1f/f versus PlzfCre Bmal1f/f mice.
(D) Expression of cell-surface S1PR1 and CXCR4 by BM ILCPs at ZT 5.
Pooled data obtained from four different experiments (n = 8) are shown with SEM. p values from Student’s t test (two-tailed and unpaired) are shown. The ILCPs were examined at ZT 5.
Because ILCPs generate mature ILCs, their defective emigration would have an impact on mature ILC populations in peripheral tissues. We examined the numbers of mature ILCs in peripheral tissues in control and Bmal1-deficient animals. We found that the numbers of ILC2s and ILC3s, but not ILC1s, were significantly decreased in the colon and small intestine of PlzfCre Bmal1f/f animals (Figures 4A and S4A). However, the numbers of mature ILCs and ILCPs in the lungs were not significantly altered (Figure S4B). Unlike their BM counterparts (Figure 1A), lung ILCP clusters expressed circadian-related genes, such as Nr3c1, Rora, Nr1d1, and Nr1d2, at relatively low levels (Figure S4C). Also decreased in the colon of PlzfCre Bmal1f/f mice were the numbers of effector cytokine-producing ILC2s and ILC3s (Figure 4B). Because the ILC deficiency in the intestines could be due to a differentiation problem of Bmal1-deficient ILCPs, we also examined their differentiation potential to mature ILC subsets in vitro on OP9-DL1 cells. Bmal1-deficient ILCPs have decreased function in making ILC2s and ILC3s compared to their wild-type (WT) counterparts (Figure 4C). Thus, the circadian clock controls not only the emigration of BM ILCPs but also their differentiation to mature ILC2 and ILC3 subsets.
Figure 4. Bmal1 deficiency in ILCPs leads to mature ILC deficiency in the intestine.
(A) Numbers of ILC1, ILC2, and ILC3 subsets in the colon and small intestine of control Bmal1f/f versus PlzfCre Bmal1f/f mice.
(B) Frequency of ILC cells producing effector cytokines in the colon and small intestinal tissues of Bmal1f/f versus PlzfCre Bmal1f/f mice. ILC1s (Lin−CD127+CD90+GATA3−RORγt−Tbet+), ILC2s (Lin−CD127+CD90+GATA3+RORγt−), and ILC3s (Lin−CD127+CD90+GATA3−RORγt+) were examined.
(C) Differentiation potential of BM ILCPs from Bmal1f/f versus PlzfCre Bmal1f/f mice.
Pooled data obtained from four different experiments (n = 6–8) are shown with SEM. p values from Student’s t test (two-tailed and unpaired) are shown.
Glucocorticoid receptor functions to enhance the expression of the BM retention receptor CXCR4 on ILCPs
The central clock in the brain generates various signals to control peripheral circadian events. A group of hormones produced in a circadian manner include the stress alert hormones, glucocorticoids (GCs), which are implicated in regulating CXCR4 expression on hematopoietic stem cells and lymphocytes.37–40 We found that the Nr3c1 gene that encodes GC receptor (GR) was highly expressed in BM ILCPs compared with other ILCP subsets, including ILC2Ps and common helper-like ILCPs (CHILPs) (Figure 5A). Interestingly, Nr3c1 was expressed more highly in parenchyma ILCPs over sinusoid ILCPs. It was also highly expressed in ILCPs at the protein level (Figure 5B). While GCs are produced in the circadian fashion at the beginning of night in nocturnal animals,38 the GR expression by ILCPs did not change at ZT 5 vs. 11 (Figure 5B). The GR stimulation by the synthetic ligand dexamethasone (Dex) increased CXCR4 expression and the chemotaxis of ILCPs to CXCL12, which was blocked by RU486, a GR antagonist (Figures 5C and S5A). However, neither Dex nor RU486 regulated the S1PR1 expression on ILCPs and ILC2Ps. In vivo experiments showed that CXCR4 expression was increased after Dex treatments (at ZT 5) but decreased by RU486 (at ZT 11) (Figures 5D, S5C, and S5D). For ILC2P, we found a positive effect of Dex on CXCR4 expression in vitro but not in vivo (Figures 5C and 5D).
Figure 5. GR activation increases the expression of the BM retention receptor CXCR4 on ILCPs.
(A) Expression of Nr3c1 in BM ILCPs. The data were retrieved from publicly available scRNA-seq data (GSE141330 and GSE193835) on ILCPs from C57BL/6 mice.
(B) Expression of intracellular GR by BM ILCPs.
(C) Effects of Dex and/or RU486 on the expression of CXCR4 and S1PR1 by cultured BM ILCPs. Lin− BM cells were cultured with cytokines (SCF and IL-7 at 20 ng/mL) on OP9-DL1 cells for 24 h, and ILCPs and ILC2Ps were examined for cell-surface expression of the trafficking receptors.
(D) Regulation of CXCR4 expression by RU486 at ZT 11 and Dex at ZT 5 on ILCPs in the BM of Rag1−/− mice. Rag1−/− mice were intraperitoneally injected with dexamethasone (Dex, 500 μg/kg) for 3 days or RU486 (20 mg/kg) for 2 days.
(E) Binding of GR to the promoter region of the Cxcr4 gene. ChIP sequencing (ChIP-seq) analysis on Lin− BM cells was performed.
(F) Effects of RU486 and Dex on numbers of blood ILCPs at ZT 11 and 5 respectively, in Rag1−/− mice.
(G) Effects of RU486 and Dex on the emigration of BM ILCPs at ZT 11 and 5, respectively, in Rag1−/− mice. When indicated, Rag1−/− mice were intraperitoneally administered Dex (500 μg/kg) for 3 days or RU486 (20 mg/kg) for 2 days in a row.
Pooled data obtained from at least three different experiments (n = 5–6) are shown with SEM. p values from two-tailed/paired Student’s t test (B, E, and G) or one- or two-way ANOVA (B–D) are shown.
To gain insights into the mechanism behind the upregulation of CXCR4 expression by Dex, we searched for and found several GC response elements (GREs) in the regulatory region before the coding exon of the Cxcr4 gene and observed GR binding activities to regions containing the GREs by a chromatin immunoprecipitation (ChIP) assay (Figures 5E and S5E). In vivo administration of RU486 increased the number of circulating ILCPs, while Dex administration had an opposite effect on blood ILCPs (Figures 5F and S5F). Importantly, RU486 increased, while Dex decreased, the emigration of BM ILCPs (Figures 5G, S5F, and S5G). However, we did not detect any significant effect of the treatments on the emigration of BM ILC2Ps (Figure S5H). These results indicate that the GC-GR axis preferentially promotes the time-dependent retention of BM ILCPs by increasing CXCR4 expression.
Regular and inverse RORα agonists bidirectionally regulate the expression of S1PR1 and the emigration of BM ILCPs
RORα expression is induced by the transactivation activity of the BMAL1-CLOCK complex, and it is a key mediator of circadian-induced gene expression.41 We observed that Rora is highly expressed by ILCPs and ILC2Ps among the ILCPs in BM (Figure 6A) and blood (Figure S6A). Rora expression was high at ZT 5 but low at ZT 11 and 16 in BM ILCPs (Figure 6B). The expression of Rora was significantly reduced in the Bmal1-deficient ILCPs (Figure 6C). We found a RORα response element (RORE) in the promoter region of the mouse S1pr1 gene (Figure 6D). The ChIP assay detected significant RORα binding activity in this region (Figure 6D). Interestingly, the binding of RORα to this site was higher at ZT 5 than 11. RORα binding to the Bmal1 gene was detected at both ZT 5 and 11 at similar levels. We performed a luciferase reporter assay containing the S1pr1 promoter harboring the S1pr1 RORE motif. The RORα agonist neoruscogenin (Neo) moderately increased the transcriptional activity of the S1pr1 promoter (Figure 6E). Neo also increased S1pr1 protein expression and chemotactic activity of BM ILCPs (Figures 6F and 6G). In contrast, the RORα inverse agonist SR3335 decreased the expression and activity of S1PR1 on ILCPs. The regular (Neo) and inverse (7α-hydroxycholesterol [OHC] and SR3335) RORα agonists did not affect the expression of CXCR4 on ILCPs (Figure S6B). Thus, RORα triggering increased the expression of S1PR1 by ILCPs.
Figure 6. RORα activation upregulates the emigration receptor S1PR1 on BM ILCPs.
(A) Expression of Rora at mRNA level in BM ILCPs from C57BL/6 mice based on publicly available scRNA-seq data (GSE193835).
(B) Expression of Rora at the mRNA level in sorted BM ILCPs at different ZTs. RT-qPCR was performed.
(C) Rora mRNA expression was determined at around ZT 5 using RT-qPCR in sorted BM Lin−CD127+ cells of Bmal1f/f versus Plzfcre Bmal1f/f mice.
(D) RORα binding to the S1pr1 and Bmal1 genes in BM Lin−CD127+ ILC-lineage cells isolated from mice sacrificed at ZT 5 and 11.
(E) The transcriptional activity of the S1pr1 promoter region was determined with a luciferase reporter assay. The reporter plasmid was transfected into pre-cultured Lin− BM cells for 10 h with or without regular or inverse RORα agonists and assayed for luciferase activity.
(F) Expression of cell surface S1PR1 by in vitro cultured BM ILCPs with/without regular or inverse RORα agonists.
(G) Chemotaxis of the cultured BM ILCPs with regular or inverse RORα agonists. BM ILCPs were cultured with or without regular or inverse RORα agonists (10 μM) and cytokines (SCF and IL-7 at 20 ng/mL) on OP9-DL1 cells as the feeder layer for 5 days. Cells from Rag1−/− mice were used unless indicated otherwise. Pooled data obtained from at least three different experiments (n = 3–6) are shown with SEM. p values from one- or two-way ANOVA (B and D–G) and two-tailed/paired Student’s t test (C) are shown.
Circadian-controlled IL-18 increases the emigration of BM ILCPs by enhancing RORα expression and function
BM ILCPs highly express the α chain of the IL-18 receptor.42–44 IL-18, while not required for the development of ILCPs,45 is an effective mobilizer of BM ILCPs to the blood circulation.16 IL-18 expression is also regulated in a circadian fashion in certain tissues, such as intestines and liver,46,47 and, therefore, the function of IL-18 for the circadian regulation of ILCP mobilization remains to be established. First, we examined whether IL-18 expression is similarly regulated in the steady-state BM. We found that IL-18 is produced in a diurnal manner in the mouse BM at both mRNA and protein levels (Figures S7A and S7B). Because Il18r1 or IL-18Rα are highly expressed in ILCPs in both the BM and blood (Figure S7C),16 we examined the possibility that IL-18 stimulates ILCPs for regulation of clock genes. In cultures of sorted BM ILCPs, we found that IL-18 induces the expression of Rora (Figure 7A). Also increased were Bmal1 and Clock genes. We also employed synchronized ILCPs utilizing a horse serum-induced synchronization approach.48 IL-18 increased the expression of Rora and Bmal1 genes as well as that of S1PR1 in the synchronized ILCPs (Figure S7D). However, no impact of IL-18 on CXCR4 expression was observed (Figure S7E).
Figure 7. IL-18 enhances the RORα function in promoting the emigration of ILCPs.
(A) Enhanced expression of Rora, Bmal1, and Clock transcripts by IL-18. RT-qPCR on cultured ILCPs with IL-18 (20 ng/mL) for 5 days in the presence of IL-7 and SCF was performed.
(B) The effect of IL-18 neutralization on the frequency of blood circulating ILCPs at different ZTs in Rag1−/− mice. Mice were injected intraperitoneally with a neutralizing anti-IL-18 (YIGIF74–1G7) or isotype control antibody (Ab) on day 1 and 4 and sacrificed at the indicated ZTs on day 6.
(C) S1PR1 expression on BM ILCPs cultured for 5 days with regular or inverse RORα agonists in the presence and absence of IL-18.
(D) Expression of S1PR1 by BM ILCPs treated with or without 7α-OHC and/or IL-18.
(E) Effects of regular or inverse RORα agonists on the frequency of BM and blood ILCPs.
(F) Effects of regular or inverse RORα agonists on the emigration of BM ILCPs.
(E and F) Rag1−/− mice were treated with or without IL-18 (daily for 4 days) and Neo or SR3335 (2 times in 4 days) and examined at around ZT 5.
(G) Impact of the circadian clock on the response of cultured BM ILCPs to RORα ligands and IL-18. BM ILCPs were cultured with SCF and IL-7 on OP9-DL1 cells for 5 days in the presence or absence of the indicated factors. Cells from Rag1−/− mice were used unless indicated otherwise.
Pooled data obtained from at least three different experiments (n = 4–6) are shown with SEM. p values from two-tailed/paired Student’s t test (A) and one- or two-way ANOVA (B–G) are shown.
Blocking IL-18 with a neutralizing antibody decreased the numbers of blood ILCPs, particularly at ZT 5 (Figure 7B). Furthermore, IL-18 increased both the basal and Neo-induced expression of S1PR1 by ILCPs in vitro (Figure 7C). 7α-OHC is a cholesterol metabolite that acts as a natural RORα inverse agonist.49 7α-OHC had a negative effect on the expression of S1PR1 (Figure 7D), and this change was observed only in the presence of IL-18. We found that SR3335 (an inverse RORα agonist) increased the number of BM ILCPs but decreased that of blood ILCPs and the emigration of BM ILCPs (Figures 7E, 7F, and S7F). IL-18 treatment enhanced the effect of Neo on ILCP mobilization. Thus, IL-18 enhances the expression of RORα and its effect on inducing the expression of S1PR1. Finally, we examined whether the response of ILCPs in the upregulation of the emigration receptor S1PR1 to RORα ligands and IL-18 is affected by the circadian clock (Figure 7G). Bmal1-deficient ILCPs expressed S1PR1 at a relatively low level and had a reduced response to IL-18 and Neo compared to WT ILCPs. Thus, the circadian clock is required for normal expression of S1PR1 in response to RORα ligands and IL-18.
DISCUSSION
We investigated the role of the circadian clock in regulating the emigration and ILC-producing activity of BM ILCPs. We found that the homeostatic mobilization of BM ILCPs is not constant throughout the day but synchronized in a ZT-dependent manner. Our study identified a function of the circadian clock in replenishing the peripheral ILC activity in the gut through the timed and synchronized release of the BM ILCPs. A key feature of this process is that the expression of trafficking receptors that control the retention versus emigration by ILCPs is under the control of the circadian clock. Two circadian cues that reciprocally regulate the BM retention and emigration receptors at different ZTs have been identified in this study.
We found that the emigration of BM ILCPs is synchronized, occurring at an elevated level at around ZT 5 in mice. Behind this synchronization of ILCP emigration is the coordinated regulation of the expression of the chemokine receptor CXCR4 and the lysosphingolipid receptor S1PR1. However, the circadian clock does not have a significant effect on the expression of the integrin α4β1, which mediates cell adhesion to BM stromal cells.50 We previously reported that the expression of α4β1 is high on the parenchyma ILCPs and low on sinusoid ILCPs.16 Thus, as parenchyma ILCPs become sinusoid ILCPs, this integrin is developmentally downregulated. CXCR4 senses the BM chemokines CXCL12 and CXCL14 for cell retention within BM. Downregulation or functional blocking of CXCR4 facilitates the emigration of ILCPs.16 On the other hand, S1PR1 should be upregulated to allow ILCPs to emigrate via the BM sinusoids. Our findings in this study indicate that the circadian clock downregulates CXCR4 but upregulates S1PR1 at around ZT 5 in mice for the synchronized emigration of BM ILCPs in mice.
The downregulation of CXCR4 on ILCPs would decrease the chemotactic activity of ILCPs to CXCL12, which is mainly expressed by BM stromal cells such as LepR+ CXC chemokine ligand (CXCL) 12-abundant reticular (CAR) cells.51 This allows developing ILCPs to move away from the BM stromal cells that produce hematopoietic factors for lymphoid lineage cells, such as stem cell factor (SCF) and IL-7.52–56 We found that CXCR4 is upregulated on BM ILCPs when GR is triggered by its ligands. Corticosteroids, such as cortisol, which bind and trigger GR activation, is produced from the cortex of adrenal glands under the regulation of adrenocorticotropic hormone, which is released from the anterior pituitary in mammals.57 Endogenous levels of GCs peak around the time of the sleep-wake transition. GR expression is high in ILCPs, making them sensitive to GCs. We observed that BM parenchyma ILCPs expressed GR at levels higher than that of sinusoid ILCPs. This implies that parenchyma ILCPs are a major target of GCs to upregulate their CXCR4 expression, thereby promoting their retention in BM. In this regard, the function of GCs in increasing CXCR4 expression on ILCPs appears to promote ILCP retention in the BM. At the molecular level, this induction of CXCR4 expression is likely to be mediated by GR binding to the Cxcr4 gene.
While the cortisol-GR axis upregulates CXCR4, we found that S1PR1 expression on ILCPs is upregulated by RORα activation. There are many RORα ligands, such as cholesterol, cholesterol sulfate, 7α-OHC, 7β-OHC, 7-keto-cholesterol, and 24S-OHC, which act as either agonists or inverse agonists.49 For example, 7α-OHC, which negatively regulates S1PR1 expression, acts as a RORα inverse agonist (or inhibitor), and others function as agonists. Interestingly, the production of these ligands also oscillates in the 24-h circadian cycle.58,59 Thus, it is plausible that these physiological RORα ligands bidirectionally regulate the expression of S1PR1 by ILCPs to control ILCP emigration at different ZTs. Neo, used as an RORα agonist in this study, is naturally found in plants,60 and, therefore, the emigration of ILCPs and other cells could be controlled pharmacologically or dietetically.
We identified IL-18 as a circadian-controlled cytokine that enhances the function of RORα in supporting ILCP emigration. This is presumably mediated by upregulation of RORα by IL-18 in ILCPs. We found that the somewhat moderate effect of RORα ligands can be boosted by IL-18. This synergistic relationship between IL-18 and RORα provides an important control point for ILCP mobilization. Because IL-18 is produced in the circadian fashion, IL-18 has the potential to synergistically affect the circadian regulation of ILCP emigration by RORα ligands. We observed that the IL-18 level was increased at ZT 5 in the steady state within BM, when S1PR1 expression by ILCPs was also increased. Moreover, elevated production of IL-18 in inflammatory conditions as an NLRP3 inflammasome product61,62 implies that inflammatory IL-18 may cooperate with the circadian cues for regulation of ILCP mobilization. It also has the potential to override the timing of the homeostatic emigration of ILCPs.
The phenotype of clock-defective (i.e., Bmal1-deficient) ILCPs was largely in line with the observed function of the circadian clock in the emigration of BM ILCPs. Bmal1-deficient ILCPs were abnormally low in S1PR1 expression compared to their WT counterparts. The reason for the decreased S1PR1 expression is explained by low RORα expression by Bmal1-deficient ILCPs. In line with their low S1PR1 expression, the emigration efficiency of Bmal1-deficient ILCPs was significantly decreased compared with their WT counterparts. The defective circadian clock in ILCPs was sufficient to reduce the numbers of mature ILC2s and ILC3s in the intestine. This result, however, should be interpreted with caution because the gene deletion at the ILCP level is permanent, affecting not only ILCPs but also single potential ILCPs and mature ILCs. In this regard, several groups reported that deletion of a clock gene in RORγt-expressing cells can cause functional ILC3 deficiency in the intestines.24,25,63 Another caveat is that the promyelocytic leukaemia zinc finger protein (PLZF) is also expressed by cells other than ILCPs, such as natural killer T (NKT) cells,64 but we do not expect that NKT cells would indirectly affect the circadian mobilization of ILCPs.
We found that there is a considerable heterogeneity among BM ILCPs in the regulation of their emigration. More specifically, ILCPs and ILC2Ps were different in their emigration response to circadian cues and GR ligands. ILCPs change the expression of CXCR4 and S1PR1 at different ZTs, but this does not clearly occur for ILC2Ps. In PlzfCre Bmal1f/f mice, ILCPs were abnormal in the expression of S1PR1, but its expression on ILC2Ps was not significantly affected. This may be explained by the fact that ILC2Ps are mainly present in the parenchyma of BM and are relatively inefficient, compared with ILCPs, in circulating the blood in the steady state.16 Moreover, ILC2Ps do not express IL-18Rα and only express S1PR1 at a moderate level. ILC2P emigration appears to be regulated not by the circadian clock but by other extrinsic factors. In this regard, ILC2P mobilization is mainly induced by IL-33, where CXCR4 downregulation is important for their mobilization.65
We also observed that the circadian clock moderately controls the ILC-producing ability of BM ILCPs. The ILCPs that were mobilized at ZT 1–5 were more efficient in making mature ILCs than those mobilized later, in ZT 11–15. Interestingly, ILCP differentiation to ILC2s and ILC3s, but not to ILC1s, was dependent on ZTs. We also observed that the Bmal1-deficient ILCPs were reduced in their capacity to generate ILC2s and ILC3s. Overall, our data indicate that the circadian clock is not only important for increasing the numbers of circulating ILCPs but also for their ability to make mature ILC2s and ILC3s in peripheral tissues. A potential mechanism for the observed phenomenon is the regulation of RORα expression by the circadian clock. More specifically, we found that its expression was downregulated in Bmal1-deficient ILCPs. Because RORα supports the development of ILC2s and ILC3s,66–68 the observed downregulation of RORα can cause reduced production of ILC2s and ILC3s.
Finally, we found significant differences among organs (e.g., intestines vs. lung) in terms of ILCP migration. Some of the emigrated BM ILCPs were found in the small and large intestinal tissues. Thus, the intestinal tract is a sensitive site to host the emigrated ILCPs. In contrast, the lungs do not appear to efficiently take the emigrated BM ILCPs. In this regard, the numbers of Bmal1-deficient ILCPs in the small and large intestines were decreased, but their presence in lung tissues was not affected. Moreover, the numbers of mature ILC2s and ILC3s in the intestinal tract, but not in the lungs, of PlzfCre Bmal1f/f mice were significantly reduced. The lungs could have their own autonomous ILCP and ILC2 populations that sustain the ILC2-rich condition,42,69,70 whereas the intestinal tract may rely on the continuous supply of ILCPs from the BM through the circadian regulation of their emigration. Additionally, the gut is rich in host and bacterial metabolites that can potentially function as RORα ligands to stimulate ILCPs, whereas lung tissues are relatively devoid of these metabolites, leading to smaller effects of the circadian regulation. More studies are required in this regard.
In sum, we found that the circadian clock is a major regulator of the mobilization of BM ILCPs in the steady state. We identified two circadian cues that reciprocally control the BM retention and emigration receptors at different ZTs, allowing the operation of these two opposing mechanisms at distinct times of a day so that the overall effects on ILCP mobilization are synergistic rather than canceling each other. These are the GC-GR signal for CXCR4 expression and the RORα-IL-18 axis for S1PR1 expression, which, respectively, induce ILCP retention and emigration from the BM. In addition, we demonstrated that the circadian clock of ILCPs is required for normal levels of tissue ILCs in the gut. The results emphasize that the circadian clock is required to sustain peripheral ILCs by regulating the mobilization of the common ILCPs and their ILC production capacity. These findings are expected to have significant ramifications in ILC homeostasis.
Limitations of the study
First, we used several different methods based on PLZF or PD-1 expression along with other surface antigens, such as c-Kit and α4β7, to identify rare ILCPs in the blood circulation and peripheral tissues. A caveat is that these approaches may include additional cell subsets, such as CHILPs and a minor subset of ILC2Ps, despite the use of exclusion markers such as CD25 and other lineage markers. It is also possible that the methods may identify somewhat different ILCPs, assuming the presence of significant heterogeneity in marker gene expression among ILCPs. Second, we were able to measure the emigration of ILCPs and the expression of trafficking receptors only at 2 or 3 different ZTs when changes were clearly detectable rather than continuously or at more time points to establish the full circadian rhythmicity. This approach, while a practical choice to increase feasibility based on conventional efflux behavior of BM hematopoietic progenitors, may miss potential changes that could additionally occur at other times. Third, because of the study focus on BM ILCPs, we did not study the impact of the circadian clock on ILCPs in additional tissues, such as thymus, which also hosts ILC2 progenitors.10,71 Further studies should be performed to overcome the limitation and gain more broad insights. Fourth, we did not confirm the lack of functional Bmal1 expression in ILCPs versus other cell types in PlzfCre Bmal1f/f mice due to technical difficulties associated with the rarity of ILCPs and animal availability. The PlzfCre and Bmal1f/f lines have been characterized well in many different applications, and, therefore, the chance that these mice do not behave as intended is low. Fifth, this study did not aim to determine potential differences between sexes using statistically enough male and female animals. While we do not expect significant differences between the sexes, further studies with more animals are needed to address this point. Finally, minor perturbation of the dark cycle at ZT 11 could have happened because we did not intentionally use a red light while working on animals in this study. Because most of the data were obtained at ZT 5 and 11 during the light cycle, and the mice were handled to avoid direct exposure to light sources whenever possible in this study, the impact of any minor light perturbation on the results is expected to be low.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests should be directed to the lead contact, Chang Kim (chhkim@umich.edu).
Materials availability
All unique reagents generated in this study are available via the lead contact with a completed Materials Transfer Agreement.
Data and code availability
This paper does not report original high throughput data.
This paper does not report original codes.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Animals
All animal experiments conform to the relevant regulatory standards and the animal protocol approved by the Animal Care and Use Committees at University of Michigan (PRO00009958). This study used C57BL/6 (Taconic Biosciences), CD45.1 (Jackson Laboratory, stock 006584), PlzfGFP-cre (B6(SJL)-Zbtb16tm1.1(EGFP/cre)Aben/J (also called PlzfCre, Jackson Laboratory, stock 024529), Rag1 −/− (Jackson Laboratory, stock 002216), and Rag2 −/−IL2rg −/− mice (Taconic Biosciences, 4111-F). Bmal1f/f mice (Jackson Laboratory 007668) were crossed with PlzfGFP-cre mice. All mice were kept under a specific pathogen–free condition on a regular rodent chow ad libitum and exposed to the standard 12 h light/12 h dark cycle. Most experiments were performed on 6- to 8-week-old male and female mice.
METHOD DETAILS
Treatments of animals
Mice were treated with IL-18 (200 ng per mouse, i.p. once a day for 4 days), anti-IL-18 (0.1 mg per mouse, i.p., on day 1 and 4 and sacrificed on day 6), neoruscogenin (Neo, 2 mg/kg, i.v., every other day for 2 times), SR3335 (10 mg/kg, i.v., every other day for 2 times) and/or dexamethasone (500 μg/kg, daily for 3 days) at ZT 5, and with RU486 (20 mg/kg, daily for 2 days) at ZT 11. Mice were sacrificed when indicated or at around ZT 5. Mice were handled to avoid exposure to direct light sources whenever possible.
Isolation of BM ILC progenitors
Single cell suspensions were obtained from various tissues such as BM, intestine, lung, and lymph node tissues at ZT 5. BM cells were flushed from femurs and tibias as previously described.16 Lin− CD127+ c-Kit+ PLZF-GFP (or PD-1)+ ILCPs were sorted (~95% pure) from the BM with a flow cytometry sorter (FACSMelody, BD Biosciences). Cell isolation from intestine tissues was performed after removing Peyer’s patches for the small intestine by longitudinal opening, cold PBS washing, and epithelial extraction with Hanks’ balanced salt solution containing 1 mM EDTA, 2% HEPES, and NaHCO3 (0.35 g/L). Intestinal lamina propria (LP) cell suspensions were obtained from digestion with collagenase IV (1.5 mg/mL; Worthington, Lakewood, NJ) containing 10% newborn calf serum for 45 min at 37°C. Lung tissues were cut into small pieces and digested with collagenase IV (1.5 mg/mL; Worthington). Digested tissues were homogenized by passing through iron meshes and treated with a red blood cell lysis buffer.
Flow cytometry
Gating information for ILC progenitor subsets and mature ILCs (NK, ILC1, ILC2, and ILC3) has been previously described (22). ILC progenitors in this study were identified by the following phenotype: BM Lin−CD127+Flt3+CD25− CLP, Lin−CD127+c-Kit+ PLZF−CD25− CHILP, Lin−CD127+c-Kit+ PLZF/PD-1+ ILCP, and Lin−CD127+CD25+ c-Kit– ILC2P. Cells were stained with antibodies to lineage-specific molecules (CD3ε, CD4, CD8, CD11b, CD11c, CD19, B220, Gr-1, NK1.1, and TER119) and ILC progenitor-expressed antigens such as CD25 (clone 3C7), CD45.2 (104), CD45.1 (A20), CD90.2 (53–2.1), CD127 (A7R34), SCA-1 (D7), KLRG1 (2F1), FLT3 (A2F10), c-Kit (ACK2), CD122 (5H4), PD-1 (29F.1A12), and α4β7 (DATK32).
Anti-NK1.1 was omitted from the lineage cocktail for ILC1P and NKP cells. Expression of trafficking receptors such as S1PR1 (713412), CXCR4 (L276F12), Itg-α4 (R1–2), and Itg-β1 (HMβ1–1) was assessed by a three-step staining with primary rat antibody, biotinylated anti-rat IgG 2a/b/c (clone MRG2a-83/2b-85/2c-67), and fluorescent dye–conjugated streptavidin. Cells were fixed and permeabilized with the Transcription Factor Staining Buffer Kit (Tonbo Biosciences) for further staining of intracellular antigens such as T-BET (eBio-4B10), GATA3 (TWAJ), RORγt (AFKJS-9), PLZF (9E12), and GR (D6H2L). For intracellular staining of cytokines, cells were stained for surface markers, followed by activation with phorbol 12-myristate 13-acetate (50 ng/mL; Sigma-Aldrich), ionomycin (1 μg/mL; Sigma-Aldrich), and monensin (2 μM, Sigma-Aldrich) for 3 to 4 h. Cells were fixed with 1% paraformaldehyde for at least 2 h, then permeabilized with saponin buffer, and stained with antibodies for IL-22 (Poly5164), IFNγ (XMG1.2), IL-17 (TC11–18H10.1), IL-10 (TRFK5), and IL-13 (W17010B). Stained cells were analyzed on a NovoCyte Flow Cytometer (ACEA Biosciences Inc.).
Emigration of BM ILC progenitors
For labeling BM cells with Cell-Tracker Deep Red (Thermo Fisher Scientific), the skin above the skull or tibia was cut open, and the bones were drilled with a 30-gauge needle to make holes for injection of Cell-Tracker Deep Red (3 μL) was slowly injected into each site using a custom 34-gauge blunt needle (Hamilton) attached to 5-μL syringe (#65 Hamilton Co., Reno, NV, USA), which is described previously.32 The labeling was generally performed at ZT 5 but was performed at ZT 1 or 11 for the experiments in Figure 1D. Mice were euthanized 4, 24 or 48 h after the injection, and BM and blood cells were examined for numbers and frequencies of Cell-Tracker Deep Red-labeled ILCPs. Emigration rate (%) was calculated based on the formula: [A: Frequency of Cell-Tracker Deep Red-labeled cells of CD45+ cells in the blood or colon]/[Frequency of Cell-Tracker Deep Red-labeled cells of CD45+ cells in the BM + A].
Chemotaxis of ILC progenitors in a BM-mimicking chemoattractant gradient
The in vitro chemotaxis assay was performed using 24-well Transwell plates with a pore size of 5.0 μm (Corning, Corning, NY). In brief, Lin− CD127+ BM cells were freshly sorted from Rag1−/− mice euthanized at ZT5 or 16 and used at 5 × 105 per well. The cells were placed in the top chamber of the Transwells along with murine CXCL12 (100 ng/mL, BioLegend), and S1P (100 nM; Cayman Chemical) was added to the bottom chambers. After 3 h chemotaxis, the migrated ILC progenitors in the bottom chamber were identified and quantified by flow cytometry.
Chromatin immunoprecipitation (ChIP) for DNA binding of RORα and GR
Putative RORα binding sites on the S1pr1 genes promoter were identified based on the following RORE consensus motif (WWRNTRGGTCA). Putative GR binding sites on the Cxcr4 genes were identified based on the following sequences: GRE(KGYACMNNNTGTYCTK) according to a defined motif definition (https://motifmap.ics.uci.edu/and https://www.bioinformatics.org/sms/iupac.html). ChIP PCR primers were designed for the putative binding motifs corresponding to the known binding peaks in public ChIP-seq data (GSE146745 for RORα and GSM788651 for GR) visualized with Integrated Genome Browser (Bioviz). ChIP PCR primers were designed to cover the putative RORα binding motifs corresponding to the known RORα binding peaks (GSE146745). ChIP was performed with the SimpleChIP Kit (Cell Signaling Technology, Danvers, MA) on Lin− CD127+ BM cells sorted from Rag1−/− mice sacrificed at ZT 5 or 11. Immunoprecipitation was performed using rabbit control immunoglobulin G (IgG), GR (Cell Signaling Technology, D6H2L) or RORα (Santa Cruz Biotechnology sc-28612). qPCR analysis was performed using SYBR Green PCR Master Mix (Thermo Fisher Scientific).
Luciferase promoter reporter assay
The 1.7 kb S1pr1 gene promoter was PCR-amplified with the primers described in the key resources table and cloned into pGL3 (Promega, Madison, WI). Lineage-depleted BM cells (3 × 106), isolated from Rag1−/− mice at ZT 5, were pre-cultured for 2 days with SCF and IL-7 (20 ng/mL each) before transfection of the reporter plasmid using the Mouse T cell Nucleofector kit (Lonza, Allendale, NJ). Transfected cells were cultured for 10 h in complete DMEM medium with or without regular (Neo) or inverse (SR3555) RORα agonists, and luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Anti-mouse CD25 (Clone 3C7) | BioLegend | Cat# 101910; RRID: AB_2280288 |
| Anti-mouse CD45.2 (Clone 104) | BioLegend | Cat# 109838; RRID: AB_2650900 |
| Anti-mouse CD45.1 (Clone A20) | BioLegend | Cat# 110706; RRID: AB_313495 |
| Anti-mouse CD90.2 (Clone 53–2.1) | BioLegend | Cat# 140310; RRID: AB_10643586 |
| Anti-mouse CD127 (Clone A7R34) | BioLegend | Cat# 135040; RRID: AB_2566161 |
| Anti-mouse Sca-1 (Clone D7) | BioLegend | Cat# 108120; RRID: AB_493273 |
| Anti-mouse KLRG-1 (Clone 2F1/KLRG1) | BioLegend | Cat# 138421; RRID: AB_2563800 |
| Anti-mouse α4β7 (Clone DATK32) | BioLegend | Cat# 120606; RRID: AB_493267 |
| Anti-mouse Flt3 (Clone A2F10) | BioLegend | Cat# 135310; RRID: AB_2107050 |
| Anti-mouse c-Kit (Clone ACK2) | BioLegend | Cat# 135108; RRID: AB_2028407 |
| Anti-mouse Nkp46 (Clone 29A1.4) | BioLegend | Cat# 137646; RRID: AB_2876479 |
| Anti-mouse IL-23R (Clone 12B2B64) | BioLegend | Cat# 150906; RRID: AB_2687346 |
| Anti-mouse CD122 (Clone 5H4) | BioLegend | Cat# 105906; RRID: AB_2125736 |
| Anti-mouse PD-1 (Clone 29F.1A12) | BioLegend | Cat# 135210; RRID: AB_2159183 |
| Anti-mouse CD159a (Clone 16A11) | BioLegend | Cat# 142804; RRID: AB_10965542 |
| Anti-mouse Ly6C (Clone HK1.4) | BioLegend | Cat# 128006; RRID: AB_1186135 |
| Anti-mouse CD28 (Clone 37.51) | BioLegend | Cat# 102110; RRID: AB_312875 |
| Anti-mouse S1pr1 (Clone 713412) | R&D systems | Cat# MAB7089; RRID: AB_10994183 |
| Anti-mouse CXCR4 (Clone L276F12) | BioLegend | Cat# 146508; RRID: AB_2562785 |
| Anti-mouse CCR2 (Clone QA18A56) | BioLegend | Cat# 160102; RRID: AB_2876563 |
| Anti-mouse CXCR6 (Clone SA051D1) | BioLegend | Cat# 151102; RRID: AB_2566544 |
| Anti-mouse CD3ε (Clone 145–2C11) | BioLegend | Cat# 100326; RRID: AB_893317 |
| Anti-mouse CD8 (Clone 53–6.7) | BioLegend | Cat# 100732; RRID: AB_893423 |
| Anti-mouse CD4 (Clone GK1.5) | BioLegend | Cat# 100432; RRID: AB_893323 |
| Anti-mouse CD11c (Clone N418) | BioLegend | Cat# 117326; RRID: AB_2129643 |
| Anti-mouse CD11b (Clone M/70) | BioLegend | Cat# 101230; RRID: AB_2129374 |
| Anti-mouse CD19 (Clone 605) | BioLegend | Cat# 115532; RRID: AB_2072926 |
| Anti-mouse B220 (Clone RA3–6B2) | BioLegend | Cat# 103234; RRID: AB_893353 |
| Anti-mouse Gr-1 (Clone RB6–8C5) | BioLegend | Cat# 108426; RRID: AB_893557 |
| Anti-mouse NK1.1 (Clone PK136) | BioLegend | Cat# 108726; RRID: AB_2132707 |
| Anti-mouse Ter-119 (Clone TER-119) | BioLegend | Cat# 116226; RRID: AB_893635 |
| Anti-mouse Itg-a4 (Clone R1–2) | BioLegend | Cat# 103622; RRID: AB_ 2565777 |
| Anti-mouse Itg-b1 (Clone HMβ1–1) | BioLegend | Cat# 102212; RRID: AB_ 492829 |
| Anti-mouse T-bet (Clone eBio4B10) | ThermoFisher | Cat# 12582582; RRID: AB_925761 |
| Anti-mouse GATA3 (Clone TWAJ) | ThermoFisher | Cat# 50996642; RRID: AB_10596663 |
| Anti-mouse RORgt (Clone Q31–378) | BD Biosciences | Cat# 562894; RRID: AB_2687545 |
| Anti-mouse PLZF (Clone 9E12) | BioLegend | Cat# 145806; RRID: AB_2566165 |
| Anti-mouse GR(D6H2L) | Cell Signaling Technology | Cat# 12041S; RRID: AB_2631286 |
| Anti-mouse RORa (Clone H-65) | Santa Cruz Biotechnology | Cat# sc-28612; RRID: AB_2265518 |
| Anti-mouse IL-22 (Clone Poly5164) | BioLegend | Cat# 516404; RRID: AB_2124255 |
| Anti-mouse IFNg (Clone XMG1.2) | BioLegend | Cat# 505808; RRID: AB_315402 |
| Anti-mouse IL-17 (Clone TC11–18H10.1) | BioLegend | Cat# 506940; RRID: AB_2565781 |
| Anti-mouse IL-10 (Clone JES5–16E3) | BioLegend | Cat# 505009; RRID: AB_315363 |
| Anti-mouse IL-13 (Clone W17010B) | BioLegend | Cat# 159405; RRID: AB_2910327 |
| Anti-mouse IL-18 (Clone YIGIF741G7) | BioXcell | Cat# BE0237; RRID: AB_ 2687719 |
|
| ||
| Experimental models: Cell Lines | ||
|
| ||
| OP9/DL1 | Cells were kindly provided by M. Chieng |
Ref. 72 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6 | Taconic Biosciences | C57BL/6NTac |
| Mouse: CD45.1/Ly5.1 | Jackson Laboratory | JAX stock #006584 |
| Mouse: Rag1−/− | Jackson Laboratory | JAX stock #002216 |
| Mouse: Plzfgfp/cre | Jackson Laboratory | JAX stock #024529 |
| Mouse: Rag2−/−gc−/− | Taconic Biosciences | Taconic, 4111-F |
| Mouse: B6.129S4(Cg)-Bmal1tm1Weit/J | Jackson Laboratory | JAX stock #007668 |
| Mouse: Plzfgfp/cre× B6.129S4(Cg)-Bmal1tm1Weit/J | This paper | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| Forward primer for qRT-PCR Bmal1: CAGTGCCACTGACTACCAAGA Reverse primer for qRT-PCR Bmal1: GCTGAACAGCCATCCTTAGCA |
This paper | N/A |
| Forward primer for qRT-PCR Clock: AGCGATCATGACAGATGGAAG Reverse primer for qRT-PCR Clock: TATTCATAGGTGGATGGCTCC |
This paper | N/A |
| Forward primer for qRT-PCR Nr1d1: CCTACAGTCTGCAGGAGCTC Reverse primer for qRT-PCR Nr1d1: GACAGCAGCTTCTCGGAATG |
This paper | N/A |
| Forward primer for qRT-PCR Nr1d2: GAGGTGGTGGAATTTGCAAAGA Reverse primer for qRT-PCR Nr1d2: ACAACTGCTGTGAACAAGCTCA |
This paper | N/A |
| Forward primer for qRT-PCR Per1: GGAACTGGGTGCTGTGCACT Reverse primer for qRT-PCR Per1: TCCATGGCAGAGTCCTGAGA |
This paper | N/A |
| Forward primer for qRT-PCR Per2: GTGACACAAGTCACACCAGC Reverse primer for qRT-PCR Per2: GACCCACGGATGAACCTCTC |
This paper | N/A |
| Forward primer for qRT-PCR Rora: GTGGCTTCAGGAAAAGGT Reverse primer for qRT-PCR Rora: GCGCGACATTTACCCAT |
This paper | N/A |
| Forward primer for qRT-PCR Il18: CCTGACATCTTCTGCAACCT Reverse primer for qRT-PCR Il18: TTCCGTATTACTGCGGTTGT |
This paper | N/A |
| Forward primer for Reporter plasmid S1pr1: TTAAGCTAGCGATATGGATGC ATAGCTAAGTG Forward primer for Reporter plasmid S1pr1: TTAACTCGAGCAGCAGAGAG AAGGGAAGAG |
This paper | N/A |
| Forward primer for ChIP-PCR S1pr1: | This paper | N/A |
| TGATGGCCACTGTCCCATGAA | ||
| Reverse primer for ChIP-PCR S1pr1: | ||
| TGTAAGGAGTCCAAGTGCAGA | ||
| Forward primer for ChIP-PCR Bmal1: | This paper | N/A |
| AGCGGATTGGTCGGAAAGT | ||
| Reverse primer for ChIP-PCR Bmal1: | ||
| ACCTCCGTCCCTGACCTACT | ||
| Forward primer for ChIP-PCR Cxcr4a: | This paper | N/A |
| CCTGGCTGGCATCCTGCT | ||
| Reverse primer for ChIP-PCR Cxcr4a: | ||
| GTCCAAGAGCCACTGCTG | ||
| Forward primer for ChIP-PCR Cxcr4b: | This paper | N/A |
| CATAAGTCTGTACCATGATGCT | ||
| Reverse primer for ChIP-PCR Cxcr4b: | ||
| CTTGCACCTGTTCATCCCTCCG | ||
| Forward primer for ChIP-PCR Cxcr4c: | This paper | N/A |
| CTGCAGGATGGGAAGCGAC | ||
| Reverse primer for ChIP-PCR Cxcr4c: | ||
| TTATCACTGGGTGGATCTGG | ||
| Forward primer for ChIP-PCR Cxcr4d: | This paper | N/A |
| AATTGGTTCCACTAAAGAGG | ||
| Reverse primer for ChIP-PCR Cxcr4d: | ||
| GCTGGTTAAGGTCCAGAGAG | ||
|
| ||
| Software and algorithms | ||
|
| ||
| SeqGeq v9.0 | FlowJo, LLC | https://docs.flowjo.com/seqgeq/ |
| Prism v.8.0. | GraphPad | https://www.graphpad.com |
| FlowJo v.10.5.0 | FlowJo | https://www.flowjo.com |
| Seurat | Satija Lab | https://satijalab.org/seurat/ |
In vitro differentiation of ILCPs
Sorted Lin− CD127+ c-Kit+ PD-1+ BM ILCPs at ZT 5 were cocultured with OP9-DL1 cells72 for 10 days in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS in the presence of murine SCF (mSCF) and IL-7. The OP9-DL1 cells were pretreated with mitomycin C (50 ng/mL) for 25 min to stop cell division prior to cell culture. Half of the culture medium was replaced every 3 days, and cultured cells were harvested on indicated days. When indicated, recombinant IL-18 (20 ng/mL, BioLegend) and dexamethasone (10 nM), RU486 (500 nM) or RORα ligands (SR3335 and Neo at 10 mM; 7a-OHC at 20 mM, Cayman Chemical) were added to the culture.
In vitro synchronization of ILCPs in a 50% horse serum
BM ILCPs were synchronized as previously described.48 In brief, an equal volume of horse serum (Sigma-Aldrich, h1270) was pre-warmed and diluted in DMEM. Sorted Lin−CD127+PD-1+ ILCPs (for circadian genes expression) or Lin−BM cells (for the expression of S1PR1 and CXCR4 on ILCP and ILC2P) at ZT 5 were incubated in the 50% horse serum-containing DMEM for 2 h at 37 °C under 5% CO2 for serum shock, and the cells were washed and cultured further in complete DMEM (10% FBS) in the presence of mIL-7 and mSCF with or without mIL-18 (all at 20 ng/mL) for 24 or 30h.
Quantitative PCR for RNA expression
For qRT-PCR analysis, total RNA from freshly sorted or cultured BM ILCPs (1–2×104 cells) at indicated ZTs was extracted using the RNeasy Micro Kit (QIAGEN) and reverse-transcribed with the Sensiscript RT Kit (QIAGEN). Gene expression was analyzed by qRT-PCR with SYBR Green PCR Master Mix (Thermo Fisher Scientific). The PCR primers used in this study are listed in the key resources table.
scRNA-seq data analysis
The bone marrow scRNA-seq data (GSE193835) were analyzed by using SeqGeq (version 9.0, FlowJo, LLC). Other data were also obtained from Gene Expression Omnibus (GEO: GSE193835, GSE176357 and GSE141330). Dimensional reduction with principal components analysis (PCA), cell clustering, and ViolinBox plugin applications were performed to show the expression of circadian genes in BM ILC progenitors.16 The scRNA-seq dataset for blood ILC subsets (GSE176357) was analyzed using Seurat V4.3. The data files (matrix.mtx, features.tsv, and barcodes.tsv) were loaded into the R studio (V4.2). QC was performed with the PercentageFeatureSet function in Seurat. Normalization and variance stabilization of count data were performed based on LogNormalize (https://satijalab.org/seurat/reference/lognormalize). Linear dimensional reduction (PCA) and cell clustering with FindNeighbors and FindClusters functions based on Louvain algorithm, and non-linear dimensional reduction using UMAP was performed. The expression data for circadian genes in ILC progenitor clusters were visualized using the VlnPlot function. All codes and software, including the R package, were obtained from Github (https://satijalab.org/seurat/articles/pbmc3k_tutorial.html). The Nr3c1 expression data from BM scRNA-seq data (GSE141330) were visualized using the Shiny application (http://murine-ilc-atlas.ie-freiburg.mpg.de).
QUANTIFICATION AND STATISTICAL ANALYSIS
Sample numbers for the experiments were determined based on power calculation and our previous experience. Statistical significance was calculated by paired or unpaired two-tailed t test with Prism (version 8.0, GraphPad Software) to compare two experimental groups. One and two-way analysis of variance (ANOVA) was used for testing differences among data series and data with multiple variables. ANOVA analyses were followed by a Tukey multiple comparisons test to obtain p values for specific comparisons among the data groups. The data from PlzfCre Bmal1f/f animals were plotted in relative to those of control mice to normalize inter-experimental variations. In this case, the values from the control mice were always one after normalization. The Mann Whitney U test was used for these data. *, **, and *** indicate a significance at p values of <0.05, <0.01, and <0.001, respectively. Sample numbers are listed in each figure legend. Error bars indicate SEM. “NS” indicates not significant in figures. Graphs that do not have either * or NS did not yield statistically significant differences. In most figures, distinct symbols were used to distinguish data from repeated independent experiments.
Supplementary Material
Highlights.
Emigration of bone marrow ILCPs is dependent on zeitgeber times
Circadian signals differentially regulate the efflux of bone marrow ILCPs
IL-18 and glucocorticoid signals distinctly regulate S1PR1 or CXCR4
ACKNOWLEDGMENTS
The authors thank J. Suan for maintaining animals used for the study and R. Kostlan for general assistance. We also thank the members of the Mary H. Weiser Food Allergy Center and the Immunology program thesis committee (N. Lukacs, N. Kamada, V. Keshamouni, and Y. Laouar) for critical input. This study was supported in part by grants from the NIH (R01AI173179, R56AI173179, R21AI148898, and R01AI121302). C.H.K. has been supported by the Kenneth and Judy Betz Professorship in food allergy research at the University of Michigan. Core facility services were supported in part by P30CA046592.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114200.
REFERENCES
- 1.Remick BC, Gaidt MM, and Vance RE (2023). Effector-Triggered Immunity. Annu. Rev. Immunol. 41, 453–481. 10.1146/annurev-immunol-101721-031732. [DOI] [PubMed] [Google Scholar]
- 2.Meininger I, Carrasco A, Rao A, Soini T, Kokkinou E, and Mjösberg J. (2020). Tissue-Specific Features of Innate Lymphoid Cells. Trends Immunol. 41, 902–917. 10.1016/j.it.2020.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Branzk N, Gronke K, and Diefenbach A. (2018). Innate lymphoid cells, mediators of tissue homeostasis, adaptation and disease tolerance. Immunol. Rev. 286, 86–101. 10.1111/imr.12718. [DOI] [PubMed] [Google Scholar]
- 4.Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, et al. (2018). Innate Lymphoid Cells: 10 Years On. Cell 174, 1054–1066. 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 5.Colonna M. (2018). Innate Lymphoid Cells: Diversity, Plasticity, and Unique Functions in Immunity. Immunity 48, 1104–1117. 10.1016/j.immuni.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geremia A, Arancibia-Cá rcamo CV, Fleming MPP, Rust N, Singh B, Mortensen NJ, Travis SPL, and Powrie F. (2011). IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J. Exp. Med. 208, 1127–1133. 10.1084/jem.20101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K, Hreggvidsdottir HS, Heinsbroek SE, Legrand N, Buskens CJ, et al. (2013). Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 14, 221–229. 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- 8.Coccia M, Harrison OJ, Schiering C, Asquith MJ, Becher B, Powrie F, and Maloy KJ (2012). IL-1beta mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4(+) Th17 cells. J. Exp. Med. 209, 1595–1609. 10.1084/jem.20111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darboe A, Nielsen CM, Wolf AS, Wildfire J, Danso E, Sonko B, Bottomley C, Moore SE, Riley EM, and Goodier MR (2020). Age-Related Dynamics of Circulating Innate Lymphoid Cells in an African Population. Front. Immunol. 11, 594107. 10.3389/fimmu.2020.594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudakov JA, Mertelsmann AM, O’Connor MH, Jenq RR, Velardi E, Young LF, Smith OM, Boyd RL, van den Brink MRM, and Hanash AM (2017). Loss of thymic innate lymphoid cells leads to impaired thymopoiesis in experimental graft-versus-host disease. Blood 130, 933–942. 10.1182/blood-2017-01-762658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh A, Kazer SW, Roider J, Krista KC, Millar J, Asowata OE, Ngoepe A, Ramsuran D, Fardoos R, Ardain A, et al. (2020). Innate Lymphoid Cell Activation and Sustained Depletion in Blood and Tissue of Children Infected with HIV from Birth Despite Antiretroviral Therapy. Cell Rep. 32, 108153. 10.1016/j.celrep.2020.108153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider C, Lee J, Koga S, Ricardo-Gonzalez RR, Nussbaum JC, Smith LK, Villeda SA, Liang HE, and Locksley RM (2019). Tissue-Resident Group 2 Innate Lymphoid Cells Differentiate by Layered Ontogeny and In Situ Perinatal Priming. Immunity 50, 1425–1438.e5. 10.1016/j.immuni.2019.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Cheng L, Zhao J, Li G, Zhang L, Chen W, Nie W, Reszka-Blanco NJ, Wang FS, and Su L. (2015). Plasmacytoid dendritic cells promote HIV-1-induced group 3 innate lymphoid cell depletion. J. Clin. Invest. 125, 3692–3703. 10.1172/JCI82124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazenberg MD, Haverkate NJE, van Lier YF, Spits H, Krabbendam L, Bemelman WA, Buskens CJ, Blom B, and Shikhagaie MM (2019). Human ectoenzyme-expressing ILC3: immunosuppressive innate cells that are depleted in graft-versus-host disease. Blood Adv. 3, 3650–3660. 10.1182/bloodadvances.2019000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasteiger G, Fan X, Dikiy S, Lee SY, and Rudensky AY (2015). Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 350, 981–985. 10.1126/science.aac9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q, Lee JH, Kang HM, and Kim CH (2022). Identification of the niche and mobilization mechanism for tissue-protective multipotential bone marrow ILC progenitors. Sci. Adv. 8, eabq1551. 10.1126/sciadv.abq1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim AI, Li Y, Lopez-Lastra S, Stadhouders R, Paul F, Casrouge A, Serafini N, Puel A, Bustamante J, Surace L, et al. (2017). Systemic Human ILC Precursors Provide a Substrate for Tissue ILC Differentiation. Cell 168, 1086–1100.e10. 10.1016/j.cell.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 18.Bar-Ephraim YE, Koning JJ, Burniol Ruiz E, Konijn T, Mourits VP, Lakeman KA, Boon L, Bögels M, van Maanen JP, Den Haan JMM, et al. (2019). CD62L Is a Functional and Phenotypic Marker for Circulating Innate Lymphoid Cell Precursors. J. Immunol. 202, 171–182. 10.4049/jimmunol.1701153. [DOI] [PubMed] [Google Scholar]
- 19.Pérez-García S, García-Navarrete M, Ruiz-Sanchis D, Prieto-Navarro C, Avdovic M, Pucciariello O, and Wabnik K. (2021). Synchronization of gene expression across eukaryotic communities through chemical rhythms. Nat. Commun. 12, 4017. 10.1038/s41467-021-24325-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palomino-Segura M, and Hidalgo A. (2021). Circadian immune circuits. J. Exp. Med. 218, e20200798. 10.1084/jem.20200798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheiermann C, Kunisaki Y, and Frenette PS (2013). Circadian control of the immune system. Nat. Rev. Immunol. 13, 190–198. 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Méndez-Ferrer S, Lucas D, Battista M, and Frenette PS (2008). Haematopoietic stem cell release is regulated by circadian oscillations. Nature 452, 442–447. 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- 23.Druzd D, Matveeva O, Ince L, Harrison U, He W, Schmal C, Herzel H, Tsang AH, Kawakami N, Leliavski A, et al. (2017). Lymphocyte Circadian Clocks Control Lymph Node Trafficking and Adaptive Immune Responses. Immunity 46, 120–132. 10.1016/j.immuni.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Robinette ML, Billon C, Collins PL, Bando JK, Fachi JL, Sécca C, Porter SI, Saini A, Gilfillan S, et al. (2019). Circadian rhythm-dependent and circadian rhythm-independent impacts of the molecular clock on type 3 innate lymphoid cells. Sci. Immunol. 4, eaay7501. 10.1126/sciimmunol.aay7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godinho-Silva C, Domingues RG, Rendas M, Raposo B, Ribeiro H, da Silva JA, Vieira A, Costa RM, Barbosa-Morais NL, Carvalho T, and Veiga-Fernandes H. (2019). Light-entrained and brain-tuned circadian circuits regulate ILC3s and gut homeostasis. Nature 574, 254–258. 10.1038/s41586-019-1579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talbot J, Hahn P, Kroehling L, Nguyen H, Li D, and Littman DR (2020). Feeding-dependent VIP neuron-ILC3 circuit regulates the intestinal barrier. Nature 579, 575–580. 10.1038/s41586-020-2039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks JF 2nd, Behrendt CL, Ruhn KA, Lee S, Raj P, Takahashi JS, and Hooper LV (2021). The microbiota coordinates diurnal rhythms in innate immunity with the circadian clock. Cell 184, 4154–4167.e12. 10.1016/j.cell.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reppert SM, and Weaver DR (2001). Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 63, 647–676. 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 29.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, and Weitz CJ (1998). Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569. 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 30.Ross DD, Pollak A, Akman SA, and Bachur NR (1980). Diurnal variation of circulating human myeloid progenitor cells. Exp. Hematol. 8, 954–960. [PubMed] [Google Scholar]
- 31.Haus E, Lakatua DJ, Swoyer J, and Sackett-Lundeen L. (1983). Chronobiology in hematology and immunology. Am. J. Anat. 168, 467–517. 10.1002/aja.1001680406. [DOI] [PubMed] [Google Scholar]
- 32.Herisson F, Frodermann V, Courties G, Rohde D, Sun Y, Vandoorne K, Wojtkiewicz GR, Masson GS, Vinegoni C, Kim J, et al. (2018). Direct vascular channels connect skull bone marrow and the brain surface enabling myeloid cell migration. Nat. Neurosci. 21, 1209–1217. 10.1038/s41593-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Q, Kim MH, Friesen L, and Kim CH (2020). BATF regulates innate lymphoid cell hematopoiesis and homeostasis. Sci. Immunol. 5, eaaz8154. 10.1126/sciimmunol.aaz8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hurley RW, McCarthy JB, and Verfaillie CM (1995). Direct adhesion to bone marrow stroma via fibronectin receptors inhibits hematopoietic progenitor proliferation. J. Clin. Invest. 96, 511–519. 10.1172/JCI118063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golan K, Kollet O, and Lapidot T. (2013). Dynamic Cross Talk between S1P and CXCL12 Regulates Hematopoietic Stem Cells Migration, Development and Bone Remodeling. Pharmaceuticals 6, 1145–1169. 10.3390/ph6091145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim CH, and Broxmeyer HE (1998). In vitro behavior of hematopoietic progenitor cells under the influence of chemoattractants: stromal cell-derived factor-1, steel factor, and the bone marrow environment. Blood 91, 100–110. [PubMed] [Google Scholar]
- 37.Guo B, Huang X, Cooper S, and Broxmeyer HE (2017). Glucocorticoid hormone-induced chromatin remodeling enhances human hematopoietic stem cell homing and engraftment. Nat. Med. 23, 424–428. 10.1038/nm.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olejniczak I, Oster H, and Ray DW (2022). Glucocorticoid circadian rhythms in immune function. Semin. Immunopathol. 44, 153–163. 10.1007/s00281-021-00889-2. [DOI] [PubMed] [Google Scholar]
- 39.Cain DW, Bortner CD, Diaz-Jimenez D, Petrillo MG, Gruver-Yates A, and Cidlowski JA (2020). Murine Glucocorticoid Receptors Orchestrate B Cell Migration Selectively between Bone Marrow and Blood. J. Immunol. 205, 619–629. 10.4049/jimmunol.1901135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimba A, Cui G, Tani-Ichi S, Ogawa M, Abe S, Okazaki F, Kitano S, Miyachi H, Yamada H, Hara T, et al. (2018). Glucocorticoids Drive Diurnal Oscillations in T Cell Distribution and Responses by Inducing Interleukin-7 Receptor and CXCR4. Immunity 48, 286–298.e6. 10.1016/j.immuni.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Jetten AM (2009). Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl. Recept. Signal. 7, e003. 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeis P, Lian M, Fan X, Herman JS, Hernandez DC, Gentek R, Elias S, Symowski C, Knöpper K, Peltokangas N, et al. (2020). In Situ Maturation and Tissue Adaptation of Type 2 Innate Lymphoid Cell Progenitors. Immunity 53, 775–792.e9. 10.1016/j.immuni.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu W, Cherrier DE, Chea S, Vosshenrich C, Serafini N, Petit M, Liu P, Golub R, and Di Santo JP (2019). An Id2(RFP)-Reporter Mouse Re-defines Innate Lymphoid Cell Precursor Potentials. Immunity 50, 1054–1068.e3. 10.1016/j.immuni.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chea S, Schmutz S, Berthault C, Perchet T, Petit M, Burlen-Defranoux O, Goldrath AW, Rodewald HR, Cumano A, and Golub R. (2016). Single-Cell Gene Expression Analyses Reveal Heterogeneous Responsiveness of Fetal Innate Lymphoid Progenitors to Notch Signaling. Cell Rep. 14, 1500–1516. 10.1016/j.celrep.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 45.Xie M, Zhang M, Dai M, Yue S, Li Z, Qiu J, Lu C, and Xu W. (2022). IL-18/IL-18R Signaling Is Dispensable for ILC Development But Constrains the Growth of ILCP/ILCs. Front. Immunol. 13, 923424. 10.3389/fimmu.2022.923424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S, Lin Y, Yuan X, Li F, Guo L, and Wu B. (2018). REV-ERBalpha integrates colon clock with experimental colitis through regulation of NF-kappaB/NLRP3 axis. Nat. Commun. 9, 4246. 10.1038/s41467-018-06568-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin Y, Wang S, Gao L, Zhou Z, Yang Z, Lin J, Ren S, Xing H, and Wu B. (2021). Oscillating lncRNA Platr4 regulates NLRP3 inflammasome to ameliorate nonalcoholic steatohepatitis in mice. Theranostics 11, 426–444. 10.7150/thno.50281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balsalobre A, Damiola F, and Schibler U. (1998). A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937. 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 49.Solt LA, and Burris TP (2012). Action of RORs and their ligands in (patho)physiology. Trends Endocrinol. Metab. 23, 619–627. 10.1016/j.tem.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papayannopoulou T, Craddock C, Nakamoto B, Priestley GV, and Wolf NS (1995). The VLA4/VCAM-1 adhesion pathway defines contrasting mechanisms of lodgement of transplanted murine hemopoietic progenitors between bone marrow and spleen. Proc. Natl. Acad. Sci. USA 92, 9647–9651. 10.1073/pnas.92.21.9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Omatsu Y, Sugiyama T, Kohara H, Kondoh G, Fujii N, Kohno K, and Nagasawa T. (2010). The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity 33, 387–399. 10.1016/j.immuni.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 52.Cordeiro Gomes A, Hara T, Lim VY, Herndler-Brandstetter D, Nevius E, Sugiyama T, Tani-Ichi S, Schlenner S, Richie E, Rodewald HR, et al. (2016). Hematopoietic Stem Cell Niches Produce Lineage-Instructive Signals to Control Multipotent Progenitor Differentiation. Immunity 45, 1219–1231. 10.1016/j.immuni.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding L, and Morrison SJ (2013). Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495, 231–235. 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding L, Saunders TL, Enikolopov G, and Morrison SJ (2012). Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462. 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greenbaum A, Hsu YMS, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, and Link DC (2013). CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 495, 227–230. 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Méndez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma’ayan A, Enikolopov GN, and Frenette PS (2010). Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466, 829–834. 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uchoa ET, Aguilera G, Herman JP, Fiedler JL, Deak T, and de Sousa MBC (2014). Novel aspects of glucocorticoid actions. J. Neuroendocrinol. 26, 557–572. 10.1111/jne.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petrenko V, Sinturel F, Riezman H, and Dibner C. (2023). Lipid metabolism around the body clocks. Prog. Lipid Res. 91, 101235. 10.1016/j.plipres.2023.101235. [DOI] [PubMed] [Google Scholar]
- 59.Jones PJ, and Schoeller DA (1990). Evidence for diurnal periodicity in human cholesterol synthesis. J. Lipid Res. 31, 667–673. [PubMed] [Google Scholar]
- 60.Helleboid S, Haug C, Lamottke K, Zhou Y, Wei J, Daix S, Cambula L, Rigou G, Hum DW, and Walczak R. (2014). The identification of naturally occurring neoruscogenin as a bioavailable, potent, and high-affinity agonist of the nuclear receptor RORalpha (NR1F1). J. Biomol. Screen 19, 399–406. 10.1177/1087057113497095. [DOI] [PubMed] [Google Scholar]
- 61.Arend WP, Palmer G, and Gabay C. (2008). IL-1, IL-18, and IL-33 families of cytokines. Immunol. Rev. 223, 20–38. 10.1111/j.1600-065X.2008.00624.x. [DOI] [PubMed] [Google Scholar]
- 62.Pourcet B, Zecchin M, Ferri L, Beauchamp J, Sitaula S, Billon C, Delhaye S, Vanhoutte J, Mayeuf-Louchart A, Thorel Q, et al. (2018). Nuclear Receptor Subfamily 1 Group D Member 1 Regulates Circadian Activity of NLRP3 Inflammasome to Reduce the Severity of Fulminant Hepatitis in Mice. Gastroenterology 154, 1449–1464.e20. 10.1053/j.gastro.2017.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teng F, Goc J, Zhou L, Chu C, Shah MA, Eberl G, and Sonnenberg GF (2019). A circadian clock is essential for homeostasis of group 3 innate lymphoid cells in the gut. Sci. Immunol. 4, eaax1215. 10.1126/sciimmunol.aax1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, and Bendelac A. (2008). The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403. 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stier MT, Zhang J, Goleniewska K, Cephus JY, Rusznak M, Wu L, Van Kaer L, Zhou B, Newcomb DC, and Peebles RS Jr. (2018). IL-33 promotes the egress of group 2 innate lymphoid cells from the bone marrow. J. Exp. Med. 215, 263–281. 10.1084/jem.20170449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Halim TYF, MacLaren A, Romanish MT, Gold MJ, McNagny KM, and Takei F. (2012). Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity 37, 463–474. 10.1016/j.immuni.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 67.Lo BC, Canals Hernaez D, Scott RW, Hughes MR, Shin SB, Underhill TM, Takei F, and McNagny KM (2019). The Transcription Factor RORalpha Preserves ILC3 Lineage Identity and Function during Chronic Intestinal Infection. J. Immunol. 203, 3209–3215. 10.4049/jimmunol.1900781. [DOI] [PubMed] [Google Scholar]
- 68.Ferreira ACF, Szeto ACH, Heycock MWD, Clark PA, Walker JA, Crisp A, Barlow JL, Kitching S, Lim A, Gogoi M, et al. (2021). ROR-alpha is a critical checkpoint for T cell and ILC2 commitment in the embryonic thymus. Nat. Immunol. 22, 166–178. 10.1038/s41590-020-00833-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim CH, Hashimoto-Hill S, and Kim M. (2016). Migration and Tissue Tropism of Innate Lymphoid Cells. Trends Immunol. 37, 68–79. 10.1016/j.it.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Puttur F, Denney L, Gregory LG, Vuononvirta J, Oliver R, Entwistle LJ, Walker SA, Headley MB, McGhee EJ, Pease JE, et al. (2019). Pulmonary environmental cues drive group 2 innate lymphoid cell dynamics in mice and humans. Sci. Immunol. 4, eaav7638. 10.1126/sciimmunol.aav7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qian L, Bajana S, Georgescu C, Peng V, Wang HC, Adrianto I, Colonna M, Alberola-Ila J, Wren JD, and Sun XH (2019). Suppression of ILC2 differentiation from committed T cell precursors by E protein transcription factors. J. Exp. Med. 216, 884–899. 10.1084/jem.20182100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holmes R, and ZúñIiga-Pflücker JC (2009). The OP9-DL1 system: generation of T-lymphocytes from embryonic or hematopoietic stem cells in vitro. Cold Spring Harb. Protoc. 2009, pdb prot5156. 10.1101/pdb.prot5156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper does not report original high throughput data.
This paper does not report original codes.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.