SUMMARY
Terminal differentiation requires massive restructuring of the transcriptome. During intestinal differentiation, the expression patterns of nearly 4,000 genes are altered as cells transition from progenitor cells in crypts to differentiated cells in villi. We identify dynamic occupancy of RNA polymerase II (Pol II) to gene promoters as the primary driver of transcriptomic shifts during intestinal differentiation in vivo. Changes in enhancer-promoter looping interactions accompany dynamic Pol II occupancy and are dependent upon HNF4, a pro-differentiation transcription factor. Using genetic loss-of-function, chromatin immunoprecipitation sequencing (ChIP-seq), and immunoprecipitation (IP) mass spectrometry, we demonstrate that HNF4 collaborates with chromatin remodelers and loop-stabilizing proteins and facilitates Pol II occupancy at hundreds of genes pivotal to differentiation. We also explore alternate mechanisms that drive differentiation gene expression and find that pause-release of Pol II and post-transcriptional mRNA stability regulate smaller subsets of differentially expressed genes. These studies provide insights into the mechanisms of differentiation in renewing adult tissue.
Graphical Abstract

In brief
Vemuri et al. demonstrate the critical role of dynamic RNA polymerase II activity in driving transcriptome shifts during intestinal differentiation. The intestinal transcription factor HNF4 sculpts a favorable chromatin landscape and is required for dynamic polymerase recruitment driving differentiation. Their study offers insights into transcriptional mechanisms driving tissue renewal.
INTRODUCTION
Transcriptomic shifts are a fundamental prerequisite for cellular differentiation across various tissues and organs. These shifts involve a dynamic reprogramming of gene regulatory patterns, where specific sets of genes are activated or suppressed to guide a cell’s transformation from a multipotent state into a highly specialized and functional cell type. The intestinal epithelium is a prime example of rapid and perpetual differentiation. In the mucosa of the small intestine, stem cells and actively dividing progenitor cells are predominantly located within the crypts of Lieberkühn.1 In contrast, differentiated, non-dividing cells responsible for vital absorptive and secretory functions are found along the protruding villi. The entire lifespan of a stem cell from the base of the crypt until it differentiates as it migrates to the tip of the villus is 3–5 days.2,3 Differentiation of the intestinal epithelium from crypts onto villi underlies many areas of intestinal health, encompassing everything from nutrient absorption to immune defense.4-6
A fundamental aspect of crypt-villus differentiation lies in the precise control of gene expression, ensuring that cells along this axis maintain distinct identities and functions. Studies centering on cis-regulatory elements have provided valuable insights into chromatin patterns and gene regulatory events occurring in the intestinal epithelium. During murine embryogenesis, significant chromatin modifications at distant regulatory elements lay the groundwork for the subsequent activation of enterocyte-specific genes during differentiation.7 Subsequently, multiple studies have highlighted distinct chromatin accessibility patterns among various intestinal epithelial cell populations during cell fate specification, especially with regard to secretory lineages.8-11 The coordination between transcription factors CDX2 and HNF4α at enhancer sites helps in maintaining an open chromatin state in differentiated villus cells.12 Profiling of H3K27ac, H3K4me2, and DNase I in crypt and villus cell populations revealed that this state of accessible chromatin is universal across both cell types, which facilitates lineage specification of differentiated cell types based on the available set of transcription factors.13,14 The upregulation of differentiation specific genes such as those associated with enterocyte metabolism also corresponds with a loss of DNA methylation from the stem to the differentiated cell state, along with an increase in enhancer activity, and CDX2 and HNF4 binding.12,14,15 This cascade of epigenomic events culminates in a dynamic transcriptome as cells progress from crypts to villi, resulting in the differential regulation of nearly 4,000 genes.16-18 Nevertheless, it remains unclear how upstream regulators intersect with basic trans-regulatory mechanisms, such as the recruitment and regulation of RNA polymerase II (Pol II) at gene promoters.
Surprisingly, the genomic distribution of Pol II along the crypt-villus axis remains unexplored, underscoring the gap in our knowledge of gene regulation during intestinal differentiation. The recruitment of Pol II to target gene promoters is intricately linked to three-dimensional intrachromosomal contacts within topologically associated domains.19,20 The spatial organization of chromatin loops brings distal enhancers into close proximity with gene promoters, facilitating the recruitment of transcription factors and the assembly of the transcriptional machinery, including Pol II.21 In the context of the intestine, the pro-differentiation factor HNF4 exerts a significant influence on this process, as evidenced by loss of enhancer-promoter looping events (measured by H3K4me3 HiChIP-seq, which identifies long range interactions associated with H3K4me3 marked promoters) in villus-specific genes when HNF4 is depleted.17
Within the Pol II transcription process itself, multiple regulatory checkpoints influence gene expression through a variety of factors and mechanisms. At the initiation checkpoint, the assembly of the transcription initiation complex and the recruitment of Pol II at the gene promoter mark the commencement of gene transcription. Variations in Pol II recruitment to the gene promoter can lead to substantial alterations in gene expression.22 Additionally, promoter-proximal pausing of Pol II (hereafter referred to as “Pol II pausing”) has emerged as a prominent regulatory mechanism influencing gene expression.23-26 This phenomenon involves the temporary halting of Pol II shortly after transcription initiation, poised to rapidly respond to environmental cues. The dynamic equilibrium between paused and actively elongating Pol II has far-reaching implications for the spatial and temporal control of gene expression patterns. Additionally, fluctuations in the stability of transcribed mRNA after splicing can influence the patterns of gene expression during differentiation.27,28
In this study, we delve into the intricate regulatory network involving Pol II occupancy, Pol II pausing, chromatin looping, transcription factor binding, and post-transcriptional regulation, shedding light on the sophisticated mechanisms governing regionalized gene expression in the intestinal epithelium. Additionally, we zoom in on the impact of transcription factors such as HNF4 on Pol II occupancy, elucidating its role in bridging the intestinal epigenome with gene expression.
RESULTS
Dynamic Pol II occupancy drives differential gene expression during intestinal differentiation
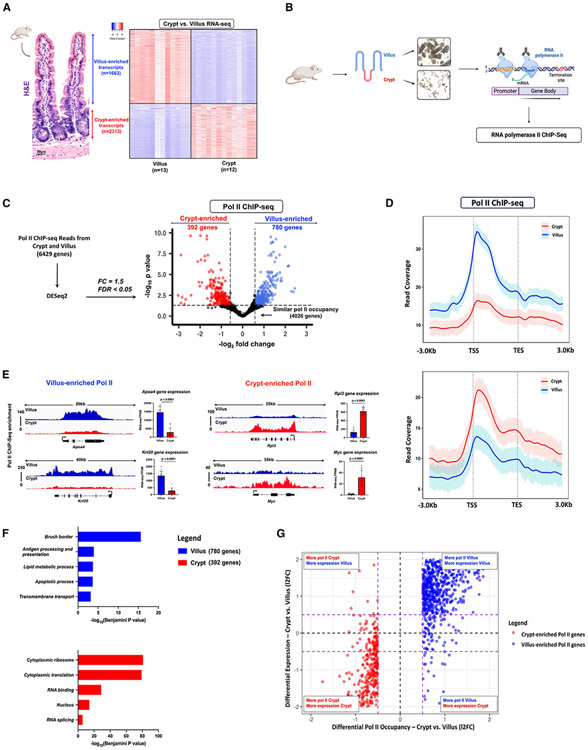
Thousands of genes are differentially expressed in the context of intestinal differentiation, as evidenced by RNA sequencing (RNA-seq) data acquired from isolated crypt and villus epithelium.18 Our analysis of this data has identified 1,663 transcripts that displayed enrichment in villi and 2,313 transcripts exhibiting enrichment in crypts (log2 fold change [l2FC] > 1 or < −1, false discovery rate [FDR] < 0.05, Fragments Per Kilobase of transcript per Million mapped reads [FPKM] >1) (Figure 1A). However, the contribution of RNA polymerase II (Pol II) dynamics during intestinal differentiation has not been systematically explored. Changes in Pol II recruitment and occupancy, pause-release, or downstream mRNA stability could all contribute to differentiation-specific gene expression. To address this knowledge gap, we conducted chromatin immunoprecipitation sequencing (ChIP-seq) analysis in isolated duodenal villus or crypt cells, using an antibody-targeting total RNA polymerase II (Figure 1B). ChIP replicates from each cell type exhibited a high degree of correlation (Pearson coefficient > 0.9) (Figure S1A). There were 6,429 genes that exhibited detectable Pol II signal-over-noise (Figures S1C-S1E; Table S1), as determined using size-matched random regions to calculate background signal (Figure S1B). Individual replicates across the 6,429 genes demonstrated a comparable distribution pattern across the gene bodies, wherein enhanced signal was generally observed at transcriptional start sites (TSS). This signal gradually decreased toward the transcriptional end sites (TES), and its strength further decreased upon entering intergenic regions (Figures S1D-S1F), consistent with other reports of Pol II distribution.29,30 Genomic browser tracks of housekeeping genes, Actb and Gapdh, provided visual validation of the signal quality and robustness inherent in the ChIP experiment (Figure S1F).
Figure 1. Dynamic Pol II occupancy correlates with dynamic gene expression during intestinal differentiation.
(A) Hematoxylin and eosin (H&E) staining shows crypt and villus structure from WT mice. Images are representative of three biological replicates. Heatmap of villus-enriched and crypt-enriched transcripts reveals dynamic gene expression patterns during differentiation from crypts onto villi (crypt vs. villus RNA-seq, n = 13 villi and 12 crypts; DESeq2: l2FC > 1 or < −1, FDR < 0.05, FPKM > 1) GEO: GSE133949).
(B) Experimental design for Pol II ChIP sequencing from isolated duodenal crypt and villus cells (n = 3 biological replicates).
(C) Volcano plot of differential Pol II occupancy between villus and crypt cells (n = 3 biological replicates). Significant Pol II occupancy was called with DESeq2 (l2FC > 0.58 or < −0.58; FDR < 0.05) (see Table S3). Genes with significant Pol II enrichment in the villus and crypt were identified with blue and red points, respectively. Points in black represent genes with similar Pol II binding patterns in both cell types. 780 genes in the villus and 392 genes in the crypt exhibit differential Pol II occupancy (see Table S2).
(D) Metagene plots show the average signal profiles of Pol II in genes with villus-enriched or crypt-enriched Pol II (differential Pol II occupancy; DESeq2, l2FC > 0.58 or < −0.58; FDR < 0.05). Confidence bands were generated at 95%.
(E) Functional annotation (DAVID) of villus-enriched Pol II genes and crypt-enriched Pol II genes. p values were calculated using DAVID (see full table in Table S2).
(F) Examples of differential Pol II binding to gene loci as illustrated using merged Pol II ChIP-seq replicate data. Villus tracks are depicted in blue and crypt tracks are depicted in red (demonstrated through merged Pol II ChIP-seq replicate data, n=3 per group). Loci are indicated above, data visualized using IGV. Bar plots show FPKM values for each gene in crypt and villus derived from RNA-seq (GEO: GSE133949). FPKM data are presented as mean ± SEM (n = 13 villi and 12 crypts, two-sided Student’s t test).
(G) Quadrant plots comparing Pol II-enriched genes with crypt versus villus differential expression patterns. The red and blue points indicate crypt- and villus-enriched Pol II gene sets respectively (differential Pol II occupancy; DESeq2, l2FC > 0.58 or < −0.58; FDR < 0.05), which display large magnitude FCs in Pol II occupancy and differential expression (crypt vs. villus RNA-seq, DESeq2 l2FC > 1 or < −1, FDR < 0.05, FPKM > 1; GEO: GSE133949) (see Table S3). The dotted purple lines show a FC cut-off of 1.5.
To test whether Pol II is dynamically recruited to different genes during intestinal differentiation, we employed DESeq2 to quantitatively determine differences between villus and crypt as a function of Pol II ChIP read counts per gene. Using an FDR cut-off of 0.05 and at least a 1.5-fold difference, we identified two subsets of 780 and 392 genes with significantly enriched Pol II in the villus versus crypt, respectively (Figure 1C; Tables S2 and S3). These gene subsets exhibited greater compartment-specific Pol II occupancy both at the TSS and across the gene body (Figures 1D and S2A). Illustrative examples of differentially enriched Pol II genes include Apoa4 and Krt20 in the villus and Rpl3 and Myc in the crypts (Figure 1E). We see in these examples that differential Pol II occupancy also corresponds with differential expression of steady-state mRNA, as evidenced by the significantly elevated transcripts detected by RNA-seq. Functional annotation of villus-enriched Pol II gene sets demonstrated enrichment of mature enterocyte properties, such as brush border and lipid metabolism, whereas genes with enriched Pol II in crypts had functions associated with a proliferative phenotype (Figure 1F; Table S2). These observations suggest that Pol II occupancy is highly distinct and is pivotal in controlling cell-specific intestinal functions. To more broadly examine whether dynamic Pol II occupancy correlated with dynamic gene expression during differentiation, we examined changes in Pol II read counts versus changes in corresponding mRNA transcripts for genes exhibiting dynamic Pol II occupancy during differentiation. We saw a robust association between dynamic Pol II occupancy and steady-state gene expression (Figures 1G and S2B), with nearly 92% of villus-enriched Pol II genes and 85% of crypt-enriched Pol II genes also showing significant mRNA enrichment in their respective compartment (Figure S2C). These results indicate that dynamic Pol II occupancy is a major driver of the extensive transcriptomic changes that occur during intestinal differentiation.
A subset of genes exhibits dynamic regulation at the post-transcriptional level
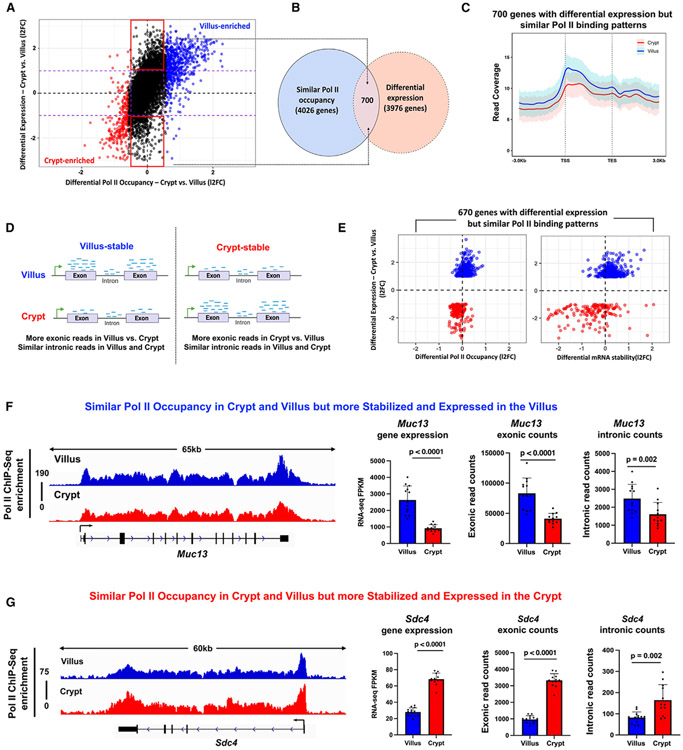
While dynamic Pol II occupancy seemed to explain a large proportion of intestinal gene expression changes, there were 4,026 genes that had detectable Pol II ChIP, signal but did not make our DESeq2 cut-offs (Figure 1C; Table S3). We categorized this gene set as having similar Pol II occupancies between crypt and villus cells, although we observed that most genes in this set still exhibited varying Pol II occupancy at non-significant levels (genes indicated by black dots, Figure 1C). Within the set of 4,026 genes, a subgroup of 700 genes exhibited differential expression (FPKM > 1, 12FC > 1 or < −1, FDR < 0.05) despite sharing similar Pol II occupancy levels, suggesting alternative mechanisms of transcript regulation at these loci (Figures 2A and 2B; Table S2). A metagene analysis on these 700 genes confirmed similar Pol II binding profiles spanning from the TSS to the TES, with nearly overlapping confidence intervals (Figure 2C). Given the uniformity in Pol II occupancies across these genes in both crypts and villi, we became intrigued by the possibility that the distinct expression patterns observed in this subgroup might be attributed to post-transcriptional mechanisms, such as alterations in mRNA stability. We examined changes in mRNA stability using the crypt versus villus RNA-seq data described above. Intronic reads in RNA-seq data are an indicator of newly transcribed, unspliced transcripts, whereas exonic reads should be present in both spliced and unspliced transcripts. We estimated the differences in mRNA stability across the crypt-villus axis using DiffRAC, which is designed to calculate the abundance of de novo transcripts versus mature mRNAs using the relative quantities of intronic reads (pre-mRNA) versus exonic reads (mature mRNA).31 Analyzing the read distribution across gene exons and introns under distinct conditions enables us to discern whether the transcript remains stable in those conditions (Figure 2D). For this analysis, we removed 30 genes from the 700 that either had no introns or had zero intron counts in all replicates across both crypts and villi. In these 670 genes, we observed a wider spread in effect size estimates for differential stability than for differential Pol II (Figure 2E; Table S3). To put this into perspective, nearly 22% of the 670 genes exhibited differential stability at a fold-change of 1.5 compared with none at the same filter for differential Pol II (Figure 2E). However, a similar analysis with the set of genes showing differential Pol II occupancy (1,172 villus- and crypt-enriched Pol II-bound genes) showed a similar fraction (18%) of genes exhibited differential mRNA stability in addition to differential Pol II occupancy (Figure S3A; Table S3). Thus, while dynamic mRNA stability contributes to differential expression during differentiation, it does not appear unique to genes lacking dynamic Pol II occupancy. We observed that 80% of the villus-enriched transcripts and 72% of the crypt-enriched transcripts within the main set of 670 genes exhibited a compartment-specific increase in mRNA stability (Figure S3B). Gene Ontology (GO) terms associated with transcripts that are stable and differentially expressed in crypts predominantly pertain to proliferation, whereas those linked with transcripts stable and enriched in villi are primarily related to immunity and lipid metabolism (Figure S3C; Table S2). To further explore the influence of stability on the expression of genes specific to differentiation, we examined two genes responsible for promoting barrier health and providing protection against colitis (Muc1332,33 and Sdc434) (Figures 2F and 2G). Muc13 and Sdc4 each exhibit strikingly similar Pol II distributions in both the villus and crypt compartments, despite their mRNA transcript enrichment in only one of these compartments (Muc13 in villus [Figure 2F] and Sdc4 in crypts [Figure 2G]). Interestingly, there are minimal variations in intronic reads for both genes relative to the changes in exonic reads, indicating similar transcription rates but more substantial changes in the abundance of spliced mRNA. This finding suggests that the changes in crypt-villus expression levels of Muc13 and Sdc4 can primarily be attributed to post-transcriptional effects. In summary, our study indicates that mRNA transcript stability serves as a mechanism regulating dynamic expression during intestinal differentiation in a subset of genes.
Figure 2. A subset of genes with similar Pol II occupancy yet differential expression is regulated post-transcriptionally.
(A) Quadrant plots comparing 6,429 genes (from Figure S1)with gene expression from crypt versus villus RNA-seq (GEO: GSE133949). The red and blue points indicate crypt- and villus-enriched Pol II gene sets respectively (differential Pol II; DESeq2, l2FC > 0.58 or < −0.58; FDR < 0.05). Black points indicate genes that have similar Pol II signal (differential Pol II; DESeq2, l2FC < 0.58 or > −0.58). Black points highlighted with red boxes indicate genes with similar Pol II occupancy yet are differentially expressed (crypt vs. villus RNA-seq; DESeq2 l2FC > 1 or < −1, FDR < 0.05, FPKM > 1; GEO: GSE133949). The dotted purple lines show a FC cut-off of 2.
(B) Venn diagram shows there are 700 genes with similar Pol II occupancy but differential gene expression between crypt and villus cells (see Table S2).
(C) Metagene profile shows similarity in Pol II occupancy patterns between crypt and villus in the 700 genes. Confidence bands were generated at 95%.
(D) Schematic showing principle behind DiffRAC,31 a computational framework used to assess differential mRNA stability based on differential exonic and intronic read counts.
(E) Plot shows 670 genes (genes without introns were excluded) with similar Pol II occupancy yet differential expression are more likely to be regulated by differential mRNA stability than any differences in Pol II, as evidenced by comparison with differential crypt-villus gene expression (differential mRNA stability: crypt vs. villus RNA-seq, DiffRAC DESeq2, l2FC > 0 or < 0; differential gene expression: crypt vs. villus RNA-seq, DESeq2 l2FC > 1 or < − 1, FDR < 0.05, FPKM > 1; GEO: GSE133949) (see Table S3).
(F and G) Representative examples of genes showing similar Pol II occupancies (genomic tracks) yet significantly different exonic counts (bar plots). Villus-enriched is shown in (F) and crypt-enriched is shown in (G). Loci are indicated above, data visualized using IGV. Villus tracks are depicted in blue and crypt tracks are depicted in red (demonstrated through merged Pol II ChIP-seq replicate data, n=3 per group). Bar plots depict FPKM values, exonic counts and intronic counts for each gene in crypt and villus derived from RNA-seq (GEO: GSE133949). The data are presented as mean ± SEM (n = 13 villi and 12 crypts, two-sided Student’s t test).
Dynamic pause-release of Pol II governs regulation of genes during crypt-villus transitions
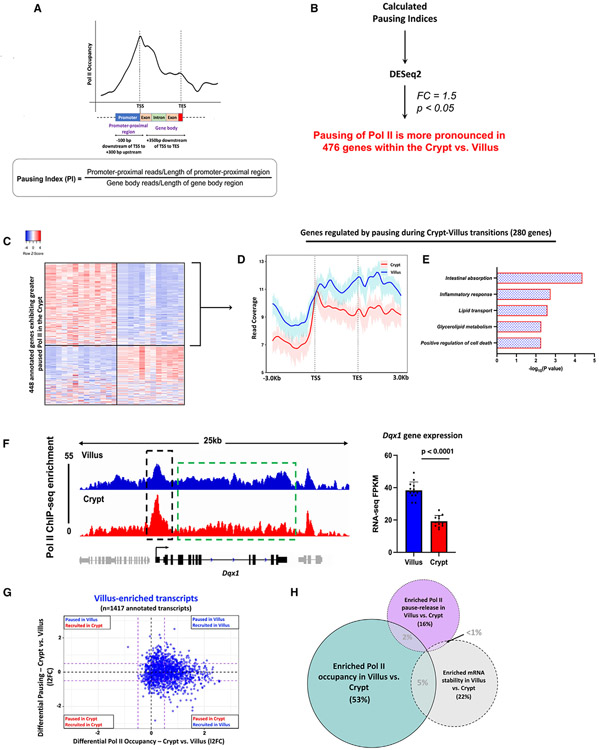
The dynamic pause-release mechanism of Pol II serves as a rapid tool for finely adjusting gene expression in response to evolving cellular conditions.23,26 We contemplated the possibility that there might be genes that achieve higher expression levels in the villus by transitioning from a paused and unexpressed state in the crypts. Leveraging Pol II ChIP-seq data, we binned Pol II reads into two zones: the promoter-proximal region (−100 bp to +300 bp from TSS) and the gene body (+350 bp from TSS to TES). This enabled us to compute the pausing index (PI),35-38 which gauges the ratio of reads in the promoter-proximal region to those in the gene body, normalized by region length (Figure 3A). The PI provides insights into Pol II pausing at gene promoters. We initially validated the utility of the PI as an analytical tool for assessing promoter-proximal pausing of Pol II (Figure S4A). Genes were classified based on their PI in crypt and villus cells. We observed that genes with a PI > 2 displayed a distinct peak in Pol II signal at the TSS, followed by a decline in signal toward the TES. The sharpness of this TSS peak became more pronounced for genes with PI > 3 and PI > 4. Conversely, genes with a PI < 2 lacked a distinct TSS peak and tended to exhibit a higher signal in the gene body, with a pile-up of Pol II toward the TES, likely due to a slowdown of elongation39,40 (Figure S4A). We applied DESeq2 to the calculated PIs and identified 476 genes with more pronounced pausing of Pol II in the crypts (l2FC < 0.58, p < 0.05) (Figure 3B; Tables S2, and S3). Of these crypt-paused/villus elongating genes, 280 genes displayed higher mRNA expression in the villus, suggesting that they could be regulated by changes in pause-release between crypts and villi (Figure 3C; Table S2). A metagene analysis of Pol II distribution across these 280 genes revealed an elevated Pol II signal along the gene body in villus cells compared to the crypts (Figure 3D). However, this analysis also demonstrated a distinct and prominent peak near the TSS in the crypts, suggesting the presence of Pol II pausing in the crypts, but not in the villi. This observation is consistent with pause-release as the regulatory mechanism governing this subset of villus-enriched mRNAs. Functional annotation shows this set primarily encompassed genes with established villus-specific roles, including functions such as nutrient absorption, lipid transport, immune-related activities, and apoptosis. (Figure 3E; Table S2). An illustrative example showing a gene from this set, Dqx1, provides additional confirmation of the heightened Pol II signal near the promoter-proximal region within the crypts. Moreover, there is a general enhancement in the Pol II signal across the Dqx1 gene body in the villus. Crucially, there is also clear evidence of significantly higher steady-state gene expression in the villus, as evident by the differences in RNA-seq FPKMs (Figure 3F).
Figure 3. Pause-release of Pol II during crypt-villus transitions is an additional gene regulatory mechanism.
(A) Diagram illustrating the procedure for calculating the PI as a measure of promoter-proximal pausing of Pol II (see Table S1).
(B) Schematic illustrating the approach used to identify 476 genes demonstrating significantly elevated promoter-proximal pausing of Pol II within the crypt (see Table S2).
(C) Heatmap shows expression patterns of 448 annotated genes which exhibit higher Pol II pausing in the crypt versus the villus (Pol II ChIP-seq; differential PI: DESeq2, l2FC < −0.58, p < 0.05) (see Table S2).
(D) Metagene plot of 280 genes that exhibit greater Pol II pausing in the crypt yet higher expression in the villus, pointing to Pol II pause-release being the major regulatory mechanism. Confidence bands were generated at 95%.
(E) Functional annotation of 280 crypt-paused, villus-expressed genes shows gene classes which preferentially undergo pause-release in the villus versus the crypt (see full table in Table S2).
(F) An illustration of Pol II occupancy patterns at the Dqx1 gene. Genomic tracks show elevated Pol II presence at promoters in crypt cells contrasted with heightened Pol II occupancy along gene bodies in villus cells (demonstrated through merged Pol II ChIP-seq replicate data, n=3 per group). Villus tracks are depicted in blue and crypt tracks are depicted in red, data visualized using IGV. Bar plots show FPKM values for each gene in crypt and villus derived from crypt versus villus RNA-seq (GEO: GSE133949). FPKM data are presented as mean ± SEM (n = 13 villi and 12 crypts, two-sided Student’s t test).
(G and H) Comparison of differential Pol II pausing and differential Pol II occupancy at 1,417 genes with villus-enriched transcripts (crypt vs. villus RNA-seq: DESeq2 l2FC > 1, FDR < 0.05, FPKM > 1; GEO: GSE133949) (see Table S3) reveals differential Pol II occupancy to be the major gene regulatory mechanism driving differentiation-specific gene expression compared with changes in Pol II pause-release or mRNA stability.
In our exploration of differentially expressed genes within the intestinal epithelium, it became apparent that a combination of different regulatory mechanisms was involved in achieving the distinct expression patterns seen during differentiation. To assess the relative impact of various mechanisms contributing to differentiation-induced gene expression, we first plotted the measured differential Pol II occupancy of villus-enriched genes against their corresponding measurement of differential Pol II pausing. We observed that among transcripts enriched in the villus, most of them exhibited increased Pol II occupancy in the villus compared with the crypts. A smaller proportion of villus-enriched transcripts underwent pause-release upon differentiation from crypts onto villi (Figure 3G). Our analysis showed that 53% of villus-enriched transcripts were primarily governed by differential Pol II occupancy (Pol II ChIP-seq; differential Pol II occupancy: l2FC > 0.58), whereas only 16% were regulated by changes in pausing and pause-release from crypts to villi (Pol II ChIP-seq; differential PI: l2FC < 0.58) (Figure 3H). Interestingly, within the genes corresponding to villus-enriched transcripts, while there was a noticeable increase in Pol II occupancy in villus cells, Pol II pausing seemed to be relatively similar in both crypt and villus cells. These observations imply that in addition to genes undergoing pause-release during the crypt-villus transition, other genes might still experience pausing in the villus and engage in pause-release when required. This aligns with previous research suggesting that Pol II pausing is a mechanism that readies genes for prompt initiation of transcriptional elongation.23,36,41,42 Finally, 22% of villus-enriched transcripts showed enriched mRNA stability in the villus compared with the crypts (Figure 3H), providing another mechanism that fine-tunes differentiation-specific gene expression. In summary, it appears that the primary mechanism driving differential gene expression during differentiation across the crypt-villus axis is the differential occupancy of RNA polymerase II.
Dynamic transcriptional enhancer activities are associated with differential Pol II occupancy during intestinal differentiation
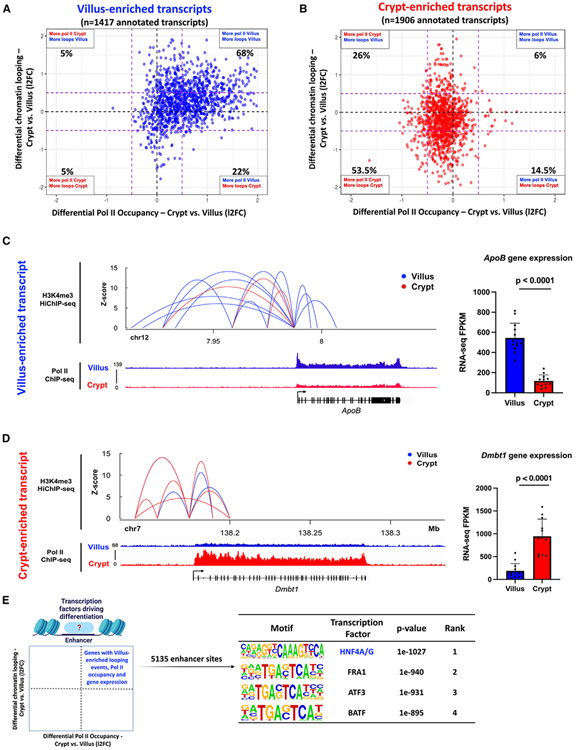
Given that the primary regulatory mechanism in the intestinal epithelium involves the dynamic recruitment and occupancy of Pol II, we recognized the necessity of incorporating the impact of distal regulatory elements, especially enhancers, to gain a comprehensive grasp of the alterations in gene expression linked to cellular differentiation. We postulated that alterations in Pol II occupancy would be concomitant with shifts in chromatin looping patterns between enhancers and promoters. To explore this, we compared associations between dynamic Pol II occupancy and the frequency of compartment-specific enhancer-promoter looping interactions, as measured by H3K4me3 HiChIP-seq in intestinal villus and crypt cells.17 Ligation events captured between distal regulatory elements and each gene promoter were measured in both crypts and villus. Our analysis revealed that 68% of the villus-enriched transcripts (Figure 4A) and 53.5% of crypt-enriched transcripts (Figure 4B) displayed a higher frequency of chromatin loops (H3K4me3 Hi-ChIP-seq; l2FC > 0.58 or < −0.58) and Pol II occupancy (Pol II ChIP-seq; l2FC >0.58 or < −0.58) in the corresponding compartment. An illustrative example at the ApoB gene promoter shows an increase in villus enhancer-promoter interactions (as measured by H3K4Me3 HiChIP-seq) had a corresponding increase in its Pol II occupancy (as measured by Pol II ChIP-seq) and gene transcript levels (as measured by FPKM values from crypt vs. villus RNA-seq) (Figure 4C). A similar relationship was observed at a crypt-enriched gene promoter, Dmbt1 (Figure 4D). To confirm that the observed differential chromatin looping at villus- and crypt-enriched Pol II promoters is indeed attributable to differences in looping patterns rather than variations in the H3K4me3 promoter mark, we examined the enrichment of H3K4me3 at promoters associated with both villus- and crypt-enriched chromatin loops, Pol II occupancy, and gene expression patterns.43 Our analysis did not reveal any remarkable differences in H3K4me3 binding between the crypt and villus regions at these promoters (Figure S4B).
Figure 4. Dynamic transcriptional enhancer activities are associated with differential Pol II occupancy during intestinal differentiation.
(A and B) Quadrant plots showing distribution of genes with villus-enriched (A) and crypt-enriched (B) transcripts from RNA-seq data (crypt vs. villus RNA-seq: DESeq2 l2FC > 1 or < −1, FDR < 0.05, FPKM > 1; GEO: GSE133949) with respect to their associated differential enhancer-promoter loops (H3K4Me3 HiChIP-seq: GEO: GSE148691) and differential Pol II occupancy (Pol II ChIP-seq). The dotted purple lines show a FC cut-off of 1.5.
(C and D) Gene loci of ApoB (C) and Dmbt1 (D) with corresponding enhancer-promoter loops and Pol II occupancy. Villus tracks are depicted in blue and crypt tracks are depicted in red (demonstrated through merged Pol II ChIP-seq replicate data, n=3 per group). All loops shown have q < 0.0001 and counts of >4. Bar plots show FPKM values for each gene in crypt and villus derived from RNA-seq (GEO: GSE133949). FPKM data is presented as mean ± SEM (n = 13 villi and 12 crypts, two-sided Student’s t test).
(E) Schematic showing the gene set selected for motif analysis using HOMER (genes with higher chromatin looping events, increased Pol II occupancy, and elevated gene expression in the villus) to identify enhancer-bound transcription factors driving differentiation. The highest-scoring motif identified in the analysis corresponds with the transcription factor HNF4 (see full table in Table S4).
Since there was a correlation between looping events, dynamic Pol II activity, and variations in gene expression, our subsequent focus was on pinpointing the potential transcription factors responsible for driving these dynamic occurrences within the differentiated intestinal epithelium. A total of 5,135 enhancer regions linked with promoters displaying villus-enriched looping events, Pol II occupancy, and gene expression were identified. To discern the transcription factors operating within these 5,135 villus-enriched enhancer regions, we conducted a DNA-binding motif analysis. Notably, the motif corresponding with hepatocyte nuclear factor 4 alpha/gamma (HNF4A/G) emerged as the highest-ranking factor, implicating HNF4 as a key regulator of Pol II dynamics and gene expression in differentiated villi (Figure 4E; Table S4). Collectively, our findings lead us to the conclusion that the dynamic occupancy of Pol II during intestinal differentiation correlates with dynamic chromatin looping events. This interplay ultimately gives rise to unique gene expression patterns, with this process seemingly under the influence of HNF4 transcription factors.
HNF4 transcription factors interact with chromatin remodeling and chromatin looping proteins and are required for Pol II occupancy to differentiation genes
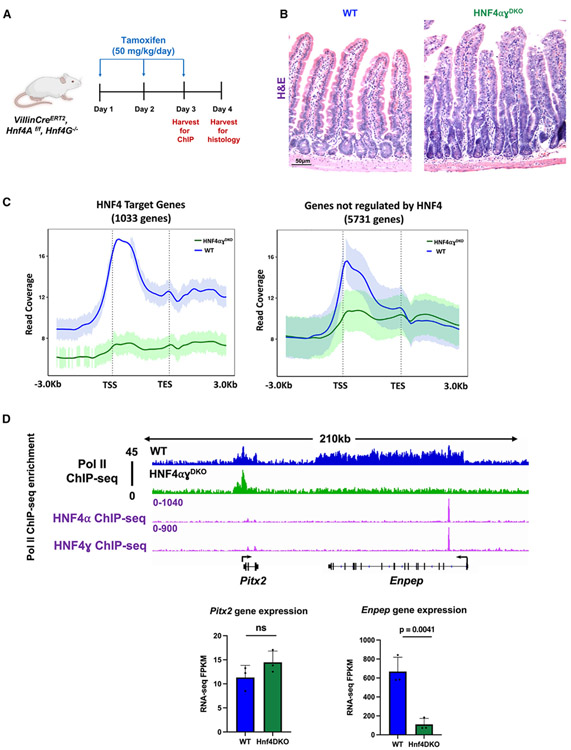
HNF4 transcription factors have roles in activating enhancer chromatin and are essential for the expression of vital villus-enriched genes.44-46 Nevertheless, the influence of HNF4 on the dynamics of Pol II remains uncertain, specifically regarding its influence on Pol II occupancy and the subsequent implications for its role as a pro-differentiation factor. To study this, we leveraged a previously generated mouse model that integrates the Villin-CreERT2 transgenic allele47 with Hnf4aflox/flox48 on an Hnf4g−/− background44 (hereafter referred to as HNF4αγDKO) (Figure 5A). Loss of both HNF4 paralogs in the intestinal epithelium upon tamoxifen treatment triggered a surplus of proliferative cells, loss of villus integrity, and differentiation failure at 4 days after the initial tamoxifen-induced gene knockout (Figure 5B), consistent with previous reports.44 We sought to determine the impact of HNF4 loss on the occupancy of Pol II at the genes analyzed above (6,429 genes identified in Figure S1) using ChIP-seq in control and HNF4αγDKO duodenal villus cells 3 days after tamoxifen treatment, when HNF4 factors are depleted, but the tissue lacks an overt phenotype.44 Upon loss of HNF4 factors, we observed a notable decrease in Pol II occupancy that was particularly pronounced at HNF4-dependent target genes (defined as bound and regulated by HNF4 as measured by HNF4A/G ChIP-seq44; within 30 kb of HNF4-binding sites; and differentially regulated in wild-type [WT] vs. HNF4αγDKO RNA-seq44) (Figures 5C, S5A, and S5B; Table S2). For example, within a 210-kb window on chromosome 3, Pitx2, a gene not previously identified as being influenced by HNF4, displayed minimal differences in Pol II occupancy between WT and HNF4αγDKO villus cells. In stark contrast, Enpep, a known HNF4 target, exhibited a significant decrease in Pol II occupancy upon the absence of HNF4 factors (Figure 5D). Additionally, our research revealed that the influence of HNF4 was predominantly observed in villus-enriched Pol II genes, with approximately 34% of them demonstrating dependence on HNF4. Conversely, crypt-enriched Pol II genes, genes undergoing pause-release upon differentiation onto villi, and genes displaying post-transcriptional stability changes exhibited lower dependence on HNF4, at rates of 4%, 17.5%, and 18%, respectively (Figure S5C). Additionally, we observed a dependence on HNF4 for the chromatin looping events acquired during differentiation. Among 618 genes identified with both villus-enriched Pol II occupancy (Pol II ChIP-seq: l2FC > 0.58, FDR < 0.05) and villus-enriched steady-state gene expression (crypt vs. villus RNA-seq: l2FC > 1, FDR < 0.05, FPKM > 1), 512 genes gained interactions with their corresponding enhancers upon differentiation (identified by changes in crypt vs. villus H3K4me3 HiChIP-seq: l2FC > 0). Approximately 30% of these enhancers exhibited HNF4 occupancy (Figure S5D).
Figure 5. Loss of the transcription factor HNF4 compromises Pol II recruitment and occupancy at its target genes.
(A) Schematic illustrating the timepoint at which cell collection was performed in HNF4αγDKO mice. For ChIP studies, HNF4αγDKO mice were injected with tamoxifen for 2 consecutive days and harvested the following day. For histology, HNF4αγDKO mice were injected with tamoxifen for 3 consecutive days and harvested the following day.
(B) Hematoxylin and eosin (H&E) staining shows loss of HNF4 paralogs leads to loss of villus integrity and prevalence of elongating crypts (representative of two biological replicates per group).
(C) Metagene plots show a reduction in Pol II occupancy in HNF4αγDKO villus cells (n = 3 biological replicates). This reduction is particularly pronounced at the genes dependent on HNF4, compared with those which are not (HNF4 targets defined as bound and regulated by HNF4 as measured by HNF4A/G ChIP-seq: within 30 kb of HNF4-binding sites; and differentially regulated in WT versus HNF4αγDKO RNA-seq; GEO: GSE112946). Confidence bands were generated at 95%.
(D) Examples of differential Pol II binding to gene loci in WT and HNF4αγDKO villus within a 210-kb window on chromosome 3 (as illustrated using merged Pol II ChIP-seq replicate data, n=3 per group). WT tracks are depicted in blue and HNF4αγDKO tracks are depicted in green. Loci are indicated above, data visualized using IGV. Bar plots show FPKM values for each gene in WT and HNF4αγDKO cells derived from RNA-seq (GEO: GSE112946). FPKM data are presented as mean ± SEM (n = 3 biological replicates per group, two-sided Student’s t test).
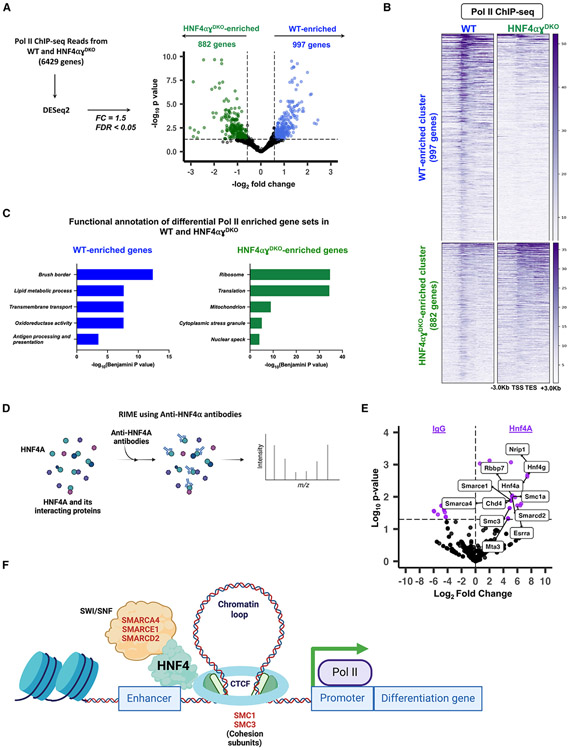
Building on these findings, we sought to identify all genes exhibiting distinct Pol II patterns in WT mice compared with HNF4αγDKO mice. By applying DESeq2 on Pol II ChIP-seq read counts (l2FC > 0.58 or < −0.58, FDR < 0.05), we identified nearly 2,000 genes that had differential Pol II occupancy between control and HNF4αγDKO villus epithelia (997 WT-enriched Pol II genes; 882 HNF4αγDKO -enriched Pol II genes) (Figure 6A; Tables S2 and S3). Clear differential Pol II signals at both genes gaining and losing HNF4 were demonstrated in the heatmap (Figure 6B). Moreover, at the individual gene level, the contrasting Pol II occupancy is clearly visible in the representative examples (Figure S6A). Functional annotation of genes showing loss of Pol II occupancy in the HNF4αγDKO reveals functions associated with differentiation, whereas those genes gaining Pol II occupancy in the HNF4αγDKO are associated with cellular proliferation and stress responses (Figure 6C; Table S2). We observed a strong correlation between dynamic Pol II occupancy and gene expression changes in the HNF4αγDKO (Figures S6B and S6C). We also discovered that approximately 40% of the genes losing Pol II occupancy in the HNF4αγDKO were also bound by HNF4 in ChIP-seq and show decreased RNA levels in the HNF4αγDKO, underscoring the critical role of HNF4 in regulating Pol II occupancy and transcriptional activity during intestinal differentiation (Figure S6D).
Figure 6. HNF4 fosters open chromatin and enhances Pol II recruitment and occupancy at genes specific to differentiation.
(A) Volcano plot of differential Pol II occupancy between WT and HNF4αγDKO villus cells (n = 3 biological replicates). Significant Pol II occupancy was called with DESeq2 (Pol II ChIP-seq: l2FC > 0.58 or < −0.58, FDR < 0.05) (see Table S3). Genes with significant Pol II enrichment in WT and HNF4αγDKO were identified as blue and green points, respectively. Points in black represent genes with similar Pol II binding patterns in both cell types. 997 genes in the WT and 882 genes in the HNF4αγDKO exhibit differential Pol II occupancy (see Table S2).
(B) Heatmap shows the distribution of Pol II signal across the gene loci for 997 and 882 WT- and HNF4αγDKO-enriched Pol II gene sets, respectively (Pol II ChIP-seq: DESeq2, l2FC > 0.58 or < −0.58; FDR < 0.05).
(C) Functional annotation of differential Pol II gene sets shows an enrichment in HNF4-dependent, villus-specific functions in WT and a reversion to a crypt-like state in the HNF4αγDKO. p values were calculated using DAVID (see full table in Table S2).
(D) Schematic for RIME using anti-HNF4α antibodies.
(E) Differential analysis of peptide counts from RIME directed against HNF4α and IgG in primary mouse epithelium shows that cohesin subunits and peptides of the SWI/SNF complex co-IP with anti-HNF4α. Peptide fragments with more than one spectral count and more than two unique peptide fragments were used for analysis (DESeq2 of peptide counts, FDR < 0.05) (see Table S5).
(F) Proposed model suggesting a mechanism by which HNF4 facilitates enhancer-promoter looping and chromatin remodeling, thus providing a conducive environment for Pol II recruitment and occupancy at differentiation-specific promoters.
To gain insights into the mechanisms underlying HNF4-mediated Pol II occupancy, we performed rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME)49 using anti-HNF4α antibodies in primary mouse epithelium (Figure 6D; Table S5). While analysis of the HNF4 interactome has been performed in cell lines50,51 and murine colon,52 we provide here an analysis focused on the differentiated small intestine. Differential analysis of raw peptide counts from IgG and HNF4α co-immuno-precipitated samples led to the identification of 29 proteins that exhibit significant interaction with HNF4α (l2FC > 0.58, FDR < 0.05) (Table S5). Our analysis showed that 48% of the HNF4α interactome was associated with transcription, functioning as transcription factors (10%), transcriptional co-regulators (7%), members of chromatin remodeling complexes (21%), or proteins involved in chromatin looping (10%) (Figures 6E and S6E; Table S5). Of note was the substantial presence of chromatin remodeling proteins, particularly those within the SWI/SNF complex, such as SMARCA4, SMARCE1, and SMARCD2, among others. Additionally, we detected interactions with SMC1A and SMC3, key constituents of the cohesin protein complex, pivotal for enabling DNA looping53,54 (Figures 6E and S6E). Based on these findings, we propose a model that suggests the following mechanism: HNF4 binding to an enhancer region initiates chromatin remodeling facilitated by the SWI/SNF complex. This remodeling leads to the formation of a chromatin loop between the enhancer and gene promoter, thus, creating a favorable environment for the recruitment and occupancy of Pol II to a target gene regulated by HNF4 (Figure 6F). By influencing chromatin accessibility, interacting with transcriptional regulators, and facilitating the assembly of transcriptional complexes, HNF4 ensures proper Pol II occupancy and subsequent gene expression driving intestinal differentiation.
DISCUSSION
The regulation of gene expression during tissue differentiation is a multifaceted process, and the relative significance of Pol II recruitment and occupancy, pause-release, and mRNA stability can vary across different tissue types and developmental contexts. While de novo Pol II recruitment is a fundamental step in transcription and often contributes to differential gene expression during differentiation, other mechanisms may take precedence in different tissues. For instance, in embryonic stem cells, the dynamics of Pol II pause-release and elongation rates are prominent as cells differentiate into various lineages.42 In contrast, during skeletal muscle regeneration, both Pol II pause-release and mRNA stability have been implicated in sculpting the muscle-specific transcriptome.55,56 In immune cells, such as T cells, mRNA stability and post-transcriptional regulation are prominent mechanisms for orchestrating rapid and precise gene expression changes in response to immune challenges.57 These diverse examples underscore the tissue-specific and context-dependent nature of gene expression regulation for maintaining cellular homeostasis.
The intestinal epithelium, characterized by its constant renewal, presents a valuable yet underexplored model system for studying the mechanisms of transcriptional differentiation. Maintaining the equilibrium between epithelial cell proliferation and differentiation constitutes a critical physiological response during viral infections,58 periods of fasting,59 or during the consumption of fat-rich diets.60 Deviations from the normal levels of differentiation can render individuals more susceptible to diseases pathologies, including metabolic syndrome60 and cancer.60-62 Understanding the mechanisms governing differentiation is also of paramount importance in the field of regenerative medicine, where the delicate balance between cell expansion and the generation of fully functional, differentiated tissue is pivotal to success. The intestinal crypt-villus unit plays a vital role in upholding intestinal equilibrium and overall health. Nonetheless, the precise gene regulatory mechanisms during the transition from crypts onto villi remain unclear. Our research has extensively examined the impact of diverse Pol II patterns driving alterations in gene expression and cell-specific functions along this axis. A key result from our study was that the driver of gene expression changes across the crypt-villus axis stemmed from the dynamic recruitment and occupancy of Pol II to gene promoters. Additionally, smaller subsets of genes were regulated through changes in Pol II pausing and post-transcriptional mechanisms. Moreover, we demonstrated that dynamic chromatin looping also plays a role in shaping the observed patterns of Pol II occupancy in intestinal genes. This phenomenon is greatly influenced by the transcription factor HNF4 in differentiated villus cells.
Current research in intestinal biology frequently highlights the bone morphogenetic protein (BMP) signaling pathway as the central force behind intestinal differentiation.63 This assertion is grounded in sophisticated studies employing transgenic expression of BMP antagonists leading to ectopic crypt formation in the villus.64,65 A focal point for future investigations could include the molecular mechanisms of how the BMP signaling pathway activates differentiation-specific gene expression. SMAD4 acts as the transcriptional effector of BMP, and mutations within it are among the most prevalent in colon cancers.62,66-69 SMAD4 and HNF4 mutually activate each other’s expression and subsequently bind to and activate genes associated with enterocyte differentiation.44 Yet the impact of this interaction on cis- and trans-regulatory factors within the differentiated intestinal genome remains undocumented. It raises intriguing questions about whether HNF4’s role in mediating open chromatin might facilitate the access of other pro-differentiation factors like SMAD4, to DNA, thereby modulating the expression of differentiation genes. Or if they collaborate in processes such as chromatin opening, recruitment of Pol II, or the release of transcriptional pauses. These findings have the potential to uncover crucial transcriptional mechanisms of pro-differentiation factors in normal intestinal physiology, shedding light on their relevance to conditions such as inflammatory bowel disease and cancer.
In conclusion, this comprehensive understanding of the regulatory network underlying dynamic crypt-villus gene expression patterns not only advances our knowledge of intestinal biology, but also provides insights into the broader mechanisms of tissue specialization and homeostasis. Further investigations into intestinal regulatory mechanisms promise to uncover additional layers of complexity and deepen our understanding of the remarkable adaptability and functionality of the intestinal epithelium.
Limitations of the study
This study primarily utilized Pol II ChIP-seq to track the dynamics of Pol II occupancy along the intestinal crypt-villus axis. However, this approach has limitations as its resolution does not facilitate a direct distinction of the various states of engaged Pol II; rather, it infers them based upon the total ChIP-seq signal distribution. Another limitation of our research is its primary focus on bulk crypt-villus populations, which could mask differences in Pol II function between the heterogeneous cell types of the epithelium. A logical next step would involve validating the outcomes of this study using more refined cell populations along the duodenal crypt-villus axis, as well as in the jejunum, ileum, and colon, to account for regional variations in gene expression. Recent advances in single-cell sequencing methodologies offer promising avenues for such validation. Finally, our study primarily focused on the small intestine; understanding colonic regulatory mechanisms concerning Pol II represents important future research.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Michael P. Verzi (verzi@biology.rutgers.edu).
Materials availability
This study did not generate any unique reagents and the mouse lines generated in this study are available upon execution of a suitable Materials Transfer Agreement.
Data and code availability
ChIP-seq data from this publication have been deposited to GEO accession number GEO: GSE244918. Mass spectrometry data from the RIME analysis have been deposited into the MassIVE database with the dataset identifier MassIVE: MSV000093071. This paper analyzes existing, publicly available data. The following datasets from GEO were reanalyzed with our sequencing data: GEO: GSE13394918 was used to perform RNA-seq analysis of villus-enriched genes and crypt-enriched genes, and to identify regions of open chromatin in the villus; GEO: GSE14869117 was used to analyze enhancer-promoter looping across the crypt-villus axis; GEO: GSE11294644 was used to identify HNF4 binding patterns in intestinal epithelial cells; GEO: GSE16077643 was used to evaluate distribution of H3K4me3 binding at villus- and crypt-enriched gene promoters.
All original code used in this study as well as data files and additional documentation can be found in our GitHub repository at https://github.com/VerziLab/Intestinal-RNA-Pol-II-Dynamics (https://doi.org/10.5281/zenodo.11051235).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Mice
Villin-CreERT2 transgene,47 Hnf4αf/f48 and Hnf4γCrispr/Crispr44 alleles were integrated to generate conditional compound mutants and controls. Experimental Villin-CreERT2 Hnf4 mutant mice (8–12 weeks old) were intraperitoneally administered tamoxifen at 50 mg/kg/day (Sigma, T5648). Wild-type, cage mate controls were administered corn oil vehicle (Sigma, C8267). For Pol II ChIP-seq experiments, duodenal villi were collected after 2 consecutive days of tamoxifen or vehicle treatment. For histology experiments, tissue was collected after 3 consecutive days of tamoxifen or vehicle treatment. For RIME, villi from the proximal half of the small intestine of C57BL/6 mice was collected. Mice of both sexes were used in all experiments. Mouse protocols and experiments were approved by the Rutgers Institutional Animal Care and Use Committee and all relevant ethical regulations were followed. All samples were collected between 9:00a.m. to 12:00p.m. to avoid circadian variability.
METHOD DETAILS
Epithelial cell Isolation
The initial third of the small intestine (duodenum) was dissected and opened longitudinally to wash the inter-lumen space and expose epithelial cells. Tissue was cut into 1-inch pieces and rinsed with ice-cold PBS. Tissue was treated with 3mM EDTA/PBS for 25 min for mutant mice and 35 min for wild-type, with changes in EDTA solution at the initial 5- and 10-min time points. Mechanical force was applied to release the epithelial cell layer from the underlying mesenchyme. Crypts and villi were separated with a 70uM filter. The smaller crypts pass through the pores of the filter while the larger villi stay on top. Both crypts and villi were collected from wild-type mice, while only villi were collected from Hnf4 mutant mice. Cells were washed twice with ice-cold PBS and pelleted by centrifugation at 300 rcf at 4°C.
Histology and immunostaining
Intestinal tissues were fixed overnight in 4% paraformaldehyde at 4°C, washed with PBS, and dehydrated through ascending alcohols before paraffin embedding. Five-micrometer paraffin sections were used for Hematoxylin & Eosin staining (H&E). The staining was imaged with a Lumenera INFINITY3 camera and Infinity Analyze imaging software (v6.5.6).
ChIP-seq
For Pol II ChIP-seq, crypt and villus cell pellets were cross-linked with 1% formaldehyde (Sigma, F8775) at 4°C for 10 min and then at 25°C for 50 min. Cells were pelleted and washed with ice-cold PBS twice by centrifugation at 300 rcf at 4°C. Fixed cells were lysed in 3X volume of lysis buffer consisting of 1% SDS, 10mM EDTA, 50mM Tris-HCl pH 8.0, 1X protease inhibitor cocktail (G-Biosciences, 786-433). Cells were sonicated for 35 min using a Diagenode Bioruptor to shear chromatin to 200-500bp.
Protein A/G beads (Invitrogen, 10001D and 10004D) were washed with 1% BSA in PBS and pre-loaded with Anti-RNA polymerase II antibody (Millipore, 05–623, Lot 3506610). Antibody-conjugated beads were washed twice with 1% BSA and incubated with sheared chromatin in dilution buffer (1% Triton X–100, 2mM EDTA, 150mM NaCl, 20mM Tris-HCl, pH 8.0). The final concentration of SDS in the sonicate was 0.26%. Chromatin and beads were incubated on a rotator at 4°C overnight. Chromatin-bound, antibody-conjugated beads were collected by magnetic separation. Unbound chromatin was discarded. Beads were washed five times with RIPA buffer (50mM HEPES pH 7.6, 1mM EDTA, 0.7% Sodium deoxycholate, 1% NP-40, 0.5M LiCl), and once with TE buffer (10mM Tris, 0.1mM EDTA). Cross-links were reversed in buffer containing 0.1M NaHCO3 and 1% SDS overnight at 65°C to release ChIP DNA. DNA was column purified using the MinElute PCR purification kit (Qiagen, 28004) and quantified using Picogreen (Invitrogen, P7581). Libraries were prepared using the ThruPLEX DNA-Seq kit (Takara, R400675) and DNA Unique Dual Index Kit (Takara, R400666). Paired-end sequencing of ChIP-seq libraries was performed (150 bp) to a depth of at least 30 million reads on the Illumina NovaSeq 6000 platform, demultiplexed and converted to fastq format.
Rapid Immunoprecipitation Mass Spectrometry of Endogenous Proteins (RIME)
Villus cells from the proximal small intestine of C57BL/6 mice were collected in ice-cold PBS by scraping the everted tissue with a coverslip. Care was taken to avoid collecting the muscle layer. Cell suspensions were pooled, snap-frozen in liquid nitrogen and submitted to Active Motif for RIME analysis according to published protocols.49 RIME was carried out using an antibody against HNF4α (Santa Cruz Biotechnology, sc-6556X, Lot B1015) to identify peptides which interact with HNF4α using mass spectrometry. An isotype-matched IgG antibody was used as control.
QUANTIFICATION AND STATISTICAL ANALYSIS
ChIP-seq data processing
FastQC (v0.11.3)72 was used to check the quality of raw sequencing reads. Alignment to the mouse reference genome (MGSCv37/mm9) was done using bowtie2 (v2.2.6)74 and alignments were sorted using Samtools (v0.1.19).91 When required, MergeSamFiles from Picard Tools (v1.95) was used to merge replicate bam files (http://broadinstitute.github.io/picard). deepTools2 bamCoverage (v2.4.2)75 was used to generate RPKM-normalized bigwig files from individual and merged bam files. The Integrative Genomic Viewer (v2.8.13)82 was used to visualize normalized bigwig tracks. The BEDTools utility (v2.17.0)73 was used to merge, intersect, or subtract the intervals of bed files. Enriched genes and/or ontologies were identified with GREAT (v4.0.4)80 or DAVID (v2021).81,92 For generation of heatmaps and metagene profiles, bigwigs were first quantile-normalized using Haystack (v0.4.0).77 deepTools2 (v2.4.2) computeMatrix, plotHeatmap and plotProfile were used to generate plots at defined genomic regions. multiBamSummary and plotCorrelation from deepTools2 (v2.4.2) were used to compute Pearson correlations showing similarity between replicates.
RNA-seq and HiChIP-seq data processing
RNA-seq data (GEO: GSE133949) was re-analyzed using Kallisto (v0.44.0)70 and DESeq2 (v1.36.0)71 for the R programming language (R v4.2/RStudio v2022.02.3/Bioconductor v3.15). GSEA (v4.2.2)83 was performed on pre-ranked gene lists. Genes with FPKM > 1 were used for further analysis. Heatmapper84 and Morpheus (https://software.broadinstitute.org/morpheus) were used for k-means clustering and/or visualization of normalized transcript levels of genes of interest.
For identification of differential loops from H3K4me3 HiChIP-seq data, we considered all loops with q ≤ 0.0001 in at least one replicate and raw counts ≥ 4 in both replicates. DESeq2 (v1.36.0) was applied to identify differential looping events using sequencing counts. Promoters involved in these differential looping events were extracted and annotated using BEDTools (v2.17.0) by intersection with a whole genome promoter bed file (UCSC transcription start site ±2 kb). The number of loops associated with each gene promoter was summed and averaged between replicates, and DESeq2 (v1.36.0) was run again to identify differential looping between each gene promoter. The R package, Sushi (v1.20.0)76 was used to visualize HiChIP-seq looping data. For simplicity, we combined biological replicates for visualizing loops in Sushi.
Pol II ChIP-seq bioinformatics data analysis
For analysis of Pol II ChIP-seq data, each gene was partitioned into the promoter-proximal region and the gene body. Raw gene-level counts data for each region was obtained using featureCounts from the Subread package (v2.03).78 Modified whole genome annotation files specific to the promoter-proximal region and gene body regions were generated as input for featureCounts. Custom annotation files were generated as follows: 1) From the mm9 transcript-level annotation (RefSeq), TSS coordinates were extracted from forward and reverse strands for all isoforms associated with a gene; 2) SAF annotation format files were generated with only promoter-proximal region (TSS, −100bp to +300bp) and gene body regions (TSS+350bp to 500bp before TES); 3) Finally, transcripts shorter than 1000bp were removed. The isoform with the strongest signal at the promoter-proximal region was retained for further analyses. For analysis of Pol II occupancy and pausing, transcripts from non-standard chromosomes, non-coding transcripts, and those with less than 18 counts/kb gene length were excluded. For the remaining transcripts, promoter-proximal and gene body counts were normalized by region length. Pausing Index (PI) was calculated as the ratio of normalized reads at the promoter proximal region to the normalized gene body reads for each gene.
Differential expression analyses were performed using the DESeq2 package (v1.36.0). For analysis of differential Pol II occupancy, total reads mapped to the full gene were used. For differential pausing analyses, the PI multiplied by a factor of 1000 was used as DESeq2 input.
Data visualization was performed using GraphPad Prism (v9.50) and the R packages ggplot2 (v3.3.6),85 ggpubr (v0.6.0),86 metagene (v2.8.1)87 and eulerr (v7.0.0).88
Differential mRNA stability analysis
Differential mRNA stability analysis was performed as previously described.31 Briefly, crypt vs. villus RNA-seq data was aligned to Ensembl GRCm39 version 109 using HISAT2 (v 2.2.0).93 Using the CRIES workflow (https://github.com/csglab/CRIES),89 reads were mapped to constitutive exonic and intronic regions for transcripts supported by both Ensembl and Havana consortia and quantified with featureCounts (v2.03). Genes without introns or those with zero intron counts in all replicates across all conditions were removed from subsequent analyses. DiffRAC was used to infer changes in mRNA stability as described previously.31
Transcription factor motif analysis
Enhancers associated with villus-enriched gene sets were identified by employing BEDtools intersect and pairToBed to overlay their promoters with H3K4me3 HiChIP-seq regions. Enhancer locations were further refined by use of a subsequent BEDtools intersect step with villus ATAC-seq peaks18 to center enhancer regions within areas of open chromatin. HOMER findMotifsGenome.pl (v.4.8.3) was used to call transcription factor motifs enriched at enhancers.79
RIME data analysis
Analysis of interacting proteins was performed in duplicate for each immunoprecipitation. Bioinformatic analysis was performed with the software Scaffold (v5.2.2)90 and enriched peptide fragments which showed 2-fold enrichment over IgG controls, > 1 spectral count and > 2 unique peptide fragments were listed as interacting partners of HNF4α.
Statistical analysis
Data is presented as mean ± s.e.m., and statistical comparisons were performed using two-way analysis of variance (ANOVA) or two-sided Student’s t-test at p < 0.05 with GraphPad Prism (v.9.50) or ggplot2 (v3.3.6). Mann-Whitney U-test and Kruskal–Wallis test (followed by post hoc Dunn’s test) were used as part of RNA-seq analysis. Bioinformatics-related statistical analysis was performed with the embedded statistics in each package, including DESeq2,71 GSEA,83,94 and DAVID.81,92
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-RNA polymerase II antibody | Millipore | Cat # 05–623, RRID:AB_309852 |
| Anti-HNF4α antibody | Santa Cruz Biotechnology | Cat # sc-6556, RRID:AB_2117025 |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma | T5648 |
| Corn Oil | Sigma | C8267 |
| Formaldehyde | Sigma | F8775 |
| Paraformaldehyde | Electron Microscopy Sciences | 15714-S |
| Hematoxylin solution (Harris) | VWR | 95057–858 |
| Eosin Y solution, alcoholic | Sigma | HT110180 |
| Critical commercial assays | ||
| MinElute PCR Purification Kit | Qiagen | 28004 |
| Thruplex DNA-seq kit | Takara | R400675 |
| DNA Unique Dual Index Kit | Takara | R400666 |
| Quant-iT™ PicoGreen™ dsDNA Reagent | Invitrogen | P7581 |
| Dynabeads™ Protein A for Immunoprecipitation | Invitrogen | 10001D |
| Dynabeads™ Protein G for Immunoprecipitation | Invitrogen | 10004D |
| EvaGreen Dye | Biotium | 31000 |
| Deposited data | ||
| Pol II ChIP-seq | This study | GEO: GSE244918 |
| HNF4α RIME mass spectrometry | This study | MassIVE: MSV000093071 |
| Experimental models: Organisms/strains | ||
| Mouse: Villin-CreERT2 | el Marjou et al., 200447 | MGI:3053826 |
| Mouse: Hnf4αf/f; Villin-CreERT2 | Hayhurst et al., 200148 | MGI:2183520 |
| Mouse: Hnf4αf/f; Hnf4γcrispr/crispr; Villin-CreERT2 | Chen et al., 2019b44 | N/A |
| Software and algorithms | ||
| Kallisto v0.44.0 | Bray et al., 201670 | https://github.com/pachterlab/kallisto |
| DESeq2 v1.36.0 | Love et al., 201471 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| FastQC v0.11.3 | Andrews, 201072 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| BEDTools v2.17.0 | Quinlan and Hall, 201073 | https://bedtools.readthedocs.io/en/latest/ |
| Bowtie2 v2.2.6 | Langmead and Salzberg, 201274 | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| deepTools2 v2.4.2 | Ramirez et al., 201675 | https://deeptools.readthedocs.io/en/develop |
| Sushi v1.20.0 | Phanstiel et al., 201476 | https://github.com/PhanstielLab/Sushi |
| Picard Tools v1.95 | Broad Institute | http://broadinstitute.github.io/picard/ |
| Haystack v0.4.0 | Pinello et al., 201877 | https://github.com/lucapinello/Haystack |
| featureCounts v2.03 | Liao et al., 201478 | https://subread.sourceforge.net/featureCounts.html |
| HOMER v4.8.3 | Heinz et al., 201079 | http://homer.ucsd.edu/homer/motif/ |
| GREAT v4.0.4 | McLean et al., 201080 | http://great.stanford.edu/public/html/ |
| DAVID v2021 | Huang da et al., 200981 | https://david.ncifcrf.gov |
| IGV v2.8.13 | Robinson et al., 201182 | https://igv.org |
| GSEA v4.2.2 | Subramanian et al., 200583 | https://www.gsea-msigdb.org/gsea/index.jsp |
| Heatmapper | Babicki et al., 201684 | http://www.heatmapper.ca |
| Morpheus | Broad Institute | https://software.broadinstitute.org/morpheus/ |
| ggplot2 v3.3.6 | Wickham, 201685 | https://ggplot2.tidyverse.org |
| ggpubr v0.6.0 | Kassambara, 202386 | https://rpkgs.datanovia.com/ggpubr/ |
| Metagene v2.8.1 | Beauparlant et al., 202287 | https://bioconductor.org/packages/release/bioc/html/metagene.html |
| Eulerr v7.0.0 | Larsson and Gustafsson, 201888 | https://cran.r-project.org/web/packages/eulerr/vignettes/introduction.html |
| CRIES | Alkallas et al., 201789 | https://github.com/csglab/CRIES |
| DiffRAC | Perron et al., 202231 | https://github.com/csglab/DiffRAC |
| Scaffold v5.2.2 | Searle, 201090 | https://www.proteomesoftware.com/products/scaffold-5 |
| Prism v9.50 | GraphPad | https://www.graphpad.com/features |
| Other | ||
| Custom scripts and code | This study | https://github.com/VerziLab/Intestinal-RNA-Pol-II-Dynamics/ (DOI: https://doi.org/10.5281/zenodo.11051235) |
| RNA-seq analysis of villus-enriched genes and crypt-enriched genes; ATAC-seq analysis of open chromatin in the villus | Aita et al., 202218 | GEO: GSE133949 |
| HiChIP-seq analysis of villus-enriched and crypt-enriched enhancer-promoter looping events | Chen et al., 202117 | GEO: GSE148691 |
| HNF4 ChIP-seq | Chen et al., 201944 | GEO: GSE112946 |
| H3K4me3 ChIP-seq analysis of villus-enriched and crypt-enriched gene promoters | Ferrari et al., 202143 | GEO: GSE160776 |
Highlights.
Dynamic Pol II occupancy shapes transcriptomic alterations during intestinal differentiation
Fewer genes are regulated by shifts in Pol II pause-release and mRNA stability
Chromatin remodelers are a major component of the HNF4 interactome in the mature intestine
HNF4 is crucial for recruiting Pol II to intestinal differentiation gene promoters
ACKNOWLEDGMENTS
This research was funded by grants from the National Institutes of Health (NIH) to M.P.V. (R01DK121915 and R01DK126446). K.V. was supported by an American Heart Association pre-doctoral fellowship (906006). S.K. was supported by a Rutgers DLS Summer Undergraduate Research Fellowship. L.C. was supported by grants from the National Natural Science Foundation of China (32270830), the Nature Science Foundation of Jiangsu Province (BK20230026), the National Key R&D Program of China (2023YFA1801500), and the Start-up Research Fund of Southeast University (RF1028623015). Figures 1A, 1B, 2D, 4E, 5A, 6D, 6F, S1C, and the graphical abstract were created with the aid of Biorender.com. The authors acknowledge the Office of Advanced Research Computing (OARC) at Rutgers University for providing access to the Amarel cluster and associated research computing resources, which have contributed to the results reported here.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114242.
REFERENCES
- 1.Barker N, Van De Wetering M, and Clevers H (2008). The intestinal stem cell. Genes Dev. 22, 1856–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potten CS (1998). Stem cells in gastrointestinal epithelium: numbers, characteristics and death. Philos. Trans. R. Soc. Lond. B Biol. Sci 353, 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Creamer B, Shorter RG, and Bamforth J (1961). The turnover and shedding of epithelial cells: Part I The turnover in the gastro-intestinal tract. Gut 2, 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiela PR, and Ghishan FK (2016). Physiology of Intestinal Absorption and Secretion. Best Pract. Res. Clin. Gastroenterol 30, 145–159. 10.1016/j.bpg.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santaolalla R, Fukata M, and Abreu MT (2011). Innate immunity in the small intestine. Curr. Opin. Gastroenterol 27, 125–131. 10.1097/MOG.0b013e3283438dea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eshleman EM, Rice T, Potter C, Woo V, Field S, Hashimoto-Hill S, Waddell A, Engelman L, Finkelman FD, and Alenghat T (2023). Microbiota dampen type 2 immunity by epigenetically restricting tuft cell differentiation. J. Immunol 210, 150.08–150.108. 10.4049/jimmunol.210.Supp.150.08. [DOI] [Google Scholar]
- 7.Kazakevych J, Sayols S, Messner B, Krienke C, and Soshnikova N (2017). Dynamic changes in chromatin states during specification and differentiation of adult intestinal stem cells. Nucleic Acids Res. 45, 5770–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jadhav U, Saxena M, O’Neill NK, Saadatpour A, Yuan G-C, Herbert Z, Murata K, and Shivdasani RA (2017). Dynamic reorganization of chromatin accessibility signatures during dedifferentiation of secretory precursors into Lgr5+ intestinal stem cells. Cell Stem Cell 21, 65–77.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raab JR, Tulasi DY, Wager KE, Morowitz JM, Magness ST, and Gracz AD (2020). Quantitative classification of chromatin dynamics reveals regulators of intestinal stem cell differentiation. Development 147, dev181966. 10.1242/dev.181966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh PNP, Madha S, Leiter AB, and Shivdasani RA (2022). Cell and chromatin transitions in intestinal stem cell regeneration. Genes Dev. 36, 684–698. 10.1101/gad.349412.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindeboom RG, van Voorthuijsen L, Oost KC, Rodríguez-Colman MJ, Luna-Velez MV, Furlan C, Baraille F, Jansen PW, Ribeiro A, Burgering BM, et al. (2018). Integrative multi-omics analysis of intestinal organoid differentiation. Mol. Syst. Biol 14, e8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, Montgomery RK, Fleet JC, Brown M, Liu XS, and Shivdasani RA (2010). Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell 19, 713–726. 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim T-H, Li F, Ferreiro-Neira I, Ho L-L, Luyten A, Nalapareddy K, Long H, Verzi M, and Shivdasani RA (2014). Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature 506, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaaij LTJ, van de Wetering M, Fang F, Decato B, Molaro A, van de Werken HJG, van Es JH, Schuijers J, de Wit E, de Laat W, et al. (2013). DNA methylation dynamics during intestinal stem cell differentiation reveals enhancers driving gene expression in the villus. Genome Biol. 14, R50. 10.1186/gb-2013-14-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheaffer KL, Kim R, Aoki R, Elliott EN, Schug J, Burger L, Schübeler D, and Kaestner KH (2014). DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 28, 652–664. 10.1101/gad.230318.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tremblay E, Auclair J, Delvin E, Levy E, Ménard D, Pshezhetsky AV, Rivard N, Seidman EG, Sinnett D, Vachon PH, and Beaulieu JF (2006). Gene expression profiles of normal proliferating and differentiating human intestinal epithelial cells: A comparison with the Caco-2 cell model. J. Cell. Biochem 99, 1175–1186. [DOI] [PubMed] [Google Scholar]
- 17.Chen L, Cao W, Aita R, Aldea D, Flores J, Gao N, Bonder EM, Ellison CE, and Verzi MP (2021). Three-dimensional interactions between enhancers and promoters during intestinal differentiation depend upon HNF4. Cell Rep. 34, 108679. 10.1016/j.celrep.2020.108679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aita R, Aldea D, Hassan S, Hur J, Pellon-Cardenas O, Cohen E, Chen L, Shroyer N, Christakos S, Verzi MP, and Fleet JC (2022). Genomic analysis of 1,25-dihydroxyvitamin D(3) action in mouse intestine reveals compartment and segment-specific gene regulatory effects. J. Biol. Chem 298, 102213. 10.1016/j.jbc.2022.102213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hnisz D, Day DS, and Young RA (2016). Insulated Neighborhoods: Structural and Functional Units of Mammalian Gene Control. Cell 167, 1188–1200. 10.1016/j.cell.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dekker J, and Mirny L (2016). The 3D Genome as Moderator of Chromosomal Communication. Cell 164, 1110–1121. 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smallwood A, and Ren B (2013). Genome organization and long-range regulation of gene expression by enhancers. Curr. Opin. Cell Biol 25, 387–394. 10.1016/j.ceb.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cramer P. (2019). Organization and regulation of gene transcription. Nature 573, 45–54. 10.1038/s41586-019-1517-4. [DOI] [PubMed] [Google Scholar]
- 23.Adelman K, and Lis JT (2012). Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet 13, 720–731. 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, and Ren B (2005). A high-resolution map of active promoters in the human genome. Nature 436, 876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Core L, and Adelman K (2019). Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 33, 960–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abuhashem A, Garg V, and Hadjantonakis AK (2022). RNA polymerase II pausing in development: orchestrating transcription. Open Biol. 12, 210220. 10.1098/rsob.210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.’t Hoen PAC, Hirsch M, de Meijer EJ, de Menezes RX, van Ommen GJ, and den Dunnen JT (2011). mRNA degradation controls differentiation state-dependent differences in transcript and splice variant abundance. Nucleic Acids Res. 39, 556–566. 10.1093/nar/gkq790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hung Y-H, Capeling M, Villanueva JW, Kanke M, Shanahan MT, Huang S, Cubitt R, Rinaldi VD, Schimenti JC, Spence JR, and Sethupathy P (2023). Integrative genome-scale analyses reveal post-transcriptional signatures of early human small intestinal development in a directed differentiation organoid model. BMC Genom. 24, 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe K, Schauer T, and Torres-Padilla ME (2022). Distinct patterns of RNA polymerase II and transcriptional elongation characterize mammalian genome activation. Cell Rep. 41, 111865. 10.1016/j.celrep.2022.111865. [DOI] [PubMed] [Google Scholar]
- 30.Mikula M, Skrzypczak M, Goryca K, Paczkowska K, Ledwon JK, Statkiewicz M, Kulecka M, Grzelak M, Dabrowska M, Kuklinska U, et al. (2016). Genome-wide co-localization of active EGFR and downstream ERK pathway kinases mirrors mitogen-inducible RNA polymerase 2 genomic occupancy. Nucleic Acids Res. 44, 10150–10164. 10.1093/nar/gkw763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perron G, Jandaghi P, Moslemi E, Nishimura T, Rajaee M, Alkallas R, Lu T, Riazalhosseini Y, and Najafabadi HS (2022). Pan-cancer analysis of mRNA stability for decoding tumour post-transcriptional programs. Commun. Biol 5, 851. 10.1038/s42003-022-03796-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams SJ, Wreschner DH, Tran M, Eyre HJ, Sutherland GR, and McGuckin MA (2001). Muc13, a novel human cell surface mucin expressed by epithelial and hemopoietic cells. J. Biol. Chem 276, 18327–18336. [DOI] [PubMed] [Google Scholar]
- 33.Sheng YH, Lourie R, Lindén SK, Jeffery PL, Roche D, Tran TV, Png CW, Waterhouse N, Sutton P, Florin THJ, and McGuckin MA (2011). The MUC13 cell-surface mucin protects against intestinal inflammation by inhibiting epithelial cell apoptosis. Gut 60, 1661–1670. 10.1136/gut.2011.239194. [DOI] [PubMed] [Google Scholar]
- 34.Fröhling M, Tepasse P, Intemann J, Sambale M, Sherwood J, Paruzel P, Tiemeyer NM, Nowacki TM, Brückner M, Mennigen R, et al. (2018). Syndecan-4 Modulates Epithelial Gut Barrier Function and Epithelial Regeneration in Experimental Colitis. Inflamm. Bowel Dis 24, 2579–2589. 10.1093/ibd/izy248. [DOI] [PubMed] [Google Scholar]
- 35.Day DS, Zhang B, Stevens SM, Ferrari F, Larschan EN, Park PJ, and Pu WT (2016). Comprehensive analysis of promoter-proximal RNA polymerase II pausing across mammalian cell types. Genome Biol. 17, 120. 10.1186/s13059-016-0984-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Core LJ, Waterfall JJ, and Lis JT (2008). Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, and Young RA (2012). Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151, 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeitlinger J, Stark A, Kellis M, Hong J-W, Nechaev S, Adelman K, Levine M, and Young RA (2007). RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet 39, 1512–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cortazar MA, Sheridan RM, Erickson B, Fong N, Glover-Cutter K, Brannan K, and Bentley DL (2019). Control of RNA Pol II Speed by PNUTS-PP1 and Spt5 Dephosphorylation Facilitates Termination by a "Sitting Duck Torpedo" Mechanism. Mol. Cell 76, 896–908.e4. 10.1016/j.molcel.2019.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ehrensberger AH, Kelly GP, and Svejstrup JQ (2013). Mechanistic interpretation of promoter-proximal peaks and RNAPII density maps. Cell 154, 713–715. [DOI] [PubMed] [Google Scholar]
- 41.Henriques T, Scruggs BS, Inouye MO, Muse GW, Williams LH, Burkholder AB, Lavender CA, Fargo DC, and Adelman K (2018). Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 32, 26–41. 10.1101/gad.309351.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, and Young RA (2010). c-Myc regulates transcriptional pause release. Cell 141, 432–445. 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrari KJ, Amato S, Noberini R, Toscani C, Fernández-Pérez D, Rossi A, Conforti P, Zanotti M, Bonaldi T, Tamburri S, and Pasini D (2021). Intestinal differentiation involves cleavage of histone H3 N-terminal tails by multiple proteases. Nucleic Acids Res. 49, 791–804. 10.1093/nar/gkaa1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen L, Toke NH, Luo S, Vasoya RP, Fullem RL, Parthasarathy A, Perekatt AO, and Verzi MP (2019). A reinforcing HNF4-SMAD4 feed-forward module stabilizes enterocyte identity. Nat. Genet 51, 777–785. 10.1038/s41588-019-0384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vemuri K, Radi SH, Sladek FM, and Verzi MP (2023). Multiple roles and regulatory mechanisms of the transcription factor HNF4 in the intestine. Front. Endocrinol 14, 1232569. 10.3389/fendo.2023.1232569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herrera-Pulido JA, Boisvert F-M, and Boudreau F (2023). Hepatocyte nuclear factor 4α multiple isoforms, their functions, and their interactomes. Proteomics 23, 2200372. 10.1002/pmic.202200372. [DOI] [PubMed] [Google Scholar]
- 47.El Marjou F, Janssen K-P, Chang BHJ, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, and Robine S (2004). Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193. 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 48.Hayhurst GP, Lee YH, Lambert G, Ward JM, and Gonzalez FJ (2001). Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell Biol 21, 1393–1403. 10.1128/mcb.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohammed H, Taylor C, Brown GD, Papachristou EK, Carroll JS, and D’santos CS (2016). Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc 11, 316–326. [DOI] [PubMed] [Google Scholar]
- 50.Daigo K, Kawamura T, Ohta Y, Ohashi R, Katayose S, Tanaka T, Aburatani H, Naito M, Kodama T, Ihara S, and Hamakubo T (2011). Proteomic analysis of native hepatocyte nuclear factor-4α (HNF4α) isoforms, phosphorylation status, and interactive cofactors. J. Biol. Chem 286, 674–686. 10.1074/jbc.M110.154732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lambert É, Babeu JP, Simoneau J, Raisch J, Lavergne L, Lévesque D, Jolibois É, Avino M, Scott MS, Boudreau F, and Boisvert FM (2020). Human Hepatocyte Nuclear Factor 4-α Encodes Isoforms with Distinct Transcriptional Functions. Mol. Cell. Proteomics 19, 808–827. 10.1074/mcp.RA119.001909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chellappa K, Deol P, Evans JR, Vuong LM, Chen G, Briançon N, Bolotin E, Lytle C, Nair MG, and Sladek FM (2016). Opposing roles of nuclear receptor HNF4α isoforms in colitis and colitis-associated colon cancer. Elife 5, e10903. 10.7554/eLife.10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun M, Nishino T, and Marko JF (2013). The SMC1-SMC3 cohesin heterodimer structures DNA through supercoiling-dependent loop formation. Nucleic Acids Res. 41, 6149–6160. 10.1093/nar/gkt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oldenkamp R, and Rowland BD (2022). A walk through the SMC cycle: From catching DNAs to shaping the genome. Mol. Cell 82, 1616–1630. 10.1016/j.molcel.2022.04.006. [DOI] [PubMed] [Google Scholar]
- 55.Dumont NA, Wang YX, and Rudnicki MA (2015). Intrinsic and extrinsic mechanisms regulating satellite cell function. Development 142, 1572–1581. 10.1242/dev.114223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson DCL, Ritso M, Nelson GM, Mokhtari Z, Nakka K, Bandukwala H, Goldman SR, Park PJ, Mounier R, Chazaud B, et al. (2021). Negative elongation factor regulates muscle progenitor expansion for efficient myofiber repair and stem cell pool repopulation. Dev. Cell 56, 1014–1029.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turner M, Galloway A, and Vigorito E (2014). Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat. Immunol 15, 484–491. 10.1038/ni.2887. [DOI] [PubMed] [Google Scholar]
- 58.Zou WY, Blutt SE, Zeng XL, Chen MS, Lo YH, Castillo-Azofeifa D, Klein OD, Shroyer NF, Donowitz M, and Estes MK (2018). Epithelial WNT Ligands Are Essential Drivers of Intestinal Stem Cell Activation. Cell Rep. 22, 1003–1015. 10.1016/j.celrep.2017.12.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mihaylova MM, Cheng CW, Cao AQ, Tripathi S, Mana MD, Bauer-Rowe KE, Abu-Remaileh M, Clavain L, Erdemir A, Lewis CA, et al. (2018). Fasting Activates Fatty Acid Oxidation to Enhance Intestinal Stem Cell Function during Homeostasis and Aging. Cell Stem Cell 22, 769–778.e4. 10.1016/j.stem.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aliluev A, Tritschler S, Sterr M, Oppenländer L, Hinterdobler J, Greisle T, Irmler M, Beckers J, Sun N, Walch A, et al. (2021). Diet-induced alteration of intestinal stem cell function underlies obesity and prediabetes in mice. Nat. Metab 3, 1202–1216. 10.1038/s42255-021-00458-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tong K, Pellón-Cárdenas O, Sirihorachai VR, Warder BN, Kothari OA, Perekatt AO, Fokas EE, Fullem RL, Zhou A, Thackray JK, et al. (2017). Degree of Tissue Differentiation Dictates Susceptibility to BRAF-Driven Colorectal Cancer. Cell Rep. 21, 3833–3845. 10.1016/j.celrep.2017.11.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perekatt AO, Shah PP, Cheung S, Jariwala N, Wu A, Gandhi V, Kumar N, Feng Q, Patel N, Chen L, et al. (2018). SMAD4 Suppresses WNT-Driven Dedifferentiation and Oncogenesis in the Differentiated Gut Epithelium. Cancer Res. 78, 4878–4890. 10.1158/0008-5472.Can-18-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beumer J, and Clevers H (2021). Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol 22, 39–53. 10.1038/s41580-020-0278-0. [DOI] [PubMed] [Google Scholar]
- 64.Davis H, Irshad S, Bansal M, Rafferty H, Boitsova T, Bardella C, Jaeger E, Lewis A, Freeman-Mills L, Giner FC, et al. (2015). Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nat. Med 21, 62–70. 10.1038/nm.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haramis APG, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJA, and Clevers H (2004). De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 303, 1684–1686. 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- 66.Tong K, Kothari OA, Haro KS, Panda A, Bandari MM, Carrick JN, Hur JJ, Zhang L, Chan CS, Xing J, et al. (2021). SMAD4 is critical in suppression of BRAF-V600E serrated tumorigenesis. Oncogene 40, 6034–6048. 10.1038/s41388-021-01997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang X, Duronio GN, Yang Y, Bala P, Hebbar P, Spisak S, Sahgal P, Singh H, Zhang Y, Xie Y, et al. (2022). An Enhancer-Driven Stem Cell-Like Program Mediated by SOX9 Blocks Intestinal Differentiation in Colorectal Cancer. Gastroenterology 162, 209–222. 10.1053/j.gastro.2021.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bala P, Rennhack JP, Aitymbayev D, Morris C, Moyer SM, Duronio GN, Doan P, Li Z, Liang X, Hornick JL, et al. (2023). Aberrant cell state plasticity mediated by developmental reprogramming precedes colorectal cancer initiation. Sci. Adv 9, eadf0927. 10.1126/sciadv.adf0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.(2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337. 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527. 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 71.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinformatics (Babraham Institute). [Google Scholar]
- 73.Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165. 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Phanstiel DH, Boyle AP, Araya CL, and Snyder MP (2014). Sushi. R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics 30, 2808–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pinello L, Farouni R, and Yuan G-C (2018). Haystack: systematic analysis of the variation of epigenetic states and cell-type specific regulatory elements. Bioinformatics 34, 1930–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 79.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, and Bejerano G (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol 28, 495–501. 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang DW, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 82.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, and Wishart DS (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44, W147–W153. 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wickham H. (2016). ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag; ). [Google Scholar]
- 86.Kassambara A. (2023). ggpubr: ‘ggplot2’ Based Publication Ready Plots. https://rpkgs.datanovia.com/ggpubr/. [Google Scholar]
- 87.Fournier E, Joly Beauparlant C, Lippens C, and Droit A (2024). Metagene: A Package to Produce Metagene Plots. R package version 1.21.0. https://github.com/ArnaudDroitLab/metagene2. [Google Scholar]
- 88.Larsson J, and Gustafsson P (2018). A case study in fitting area-proportional euler diagrams with ellipses using eulerr. SetVR@ Diagrams, 84–91. [Google Scholar]
- 89.Alkallas R, Fish L, Goodarzi H, and Najafabadi HS (2017). Inference of RNA decay rate from transcriptional profiling highlights the regulatory programs of Alzheimer’s disease. Nat. Commun 8, 909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Searle BC (2010). Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 10, 1265–1269. 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 91.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, and Chang W (2022). DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221. 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim D, Paggi JM, Park C, Bennett C, and Salzberg SL (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol 37, 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tamayo P, Steinhardt G, Liberzon A, and Mesirov JP (2016). The limitations of simple gene set enrichment analysis assuming gene independence. Stat. Methods Med. Res 25, 472–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq data from this publication have been deposited to GEO accession number GEO: GSE244918. Mass spectrometry data from the RIME analysis have been deposited into the MassIVE database with the dataset identifier MassIVE: MSV000093071. This paper analyzes existing, publicly available data. The following datasets from GEO were reanalyzed with our sequencing data: GEO: GSE13394918 was used to perform RNA-seq analysis of villus-enriched genes and crypt-enriched genes, and to identify regions of open chromatin in the villus; GEO: GSE14869117 was used to analyze enhancer-promoter looping across the crypt-villus axis; GEO: GSE11294644 was used to identify HNF4 binding patterns in intestinal epithelial cells; GEO: GSE16077643 was used to evaluate distribution of H3K4me3 binding at villus- and crypt-enriched gene promoters.
All original code used in this study as well as data files and additional documentation can be found in our GitHub repository at https://github.com/VerziLab/Intestinal-RNA-Pol-II-Dynamics (https://doi.org/10.5281/zenodo.11051235).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.