Abstract
The c-myb gene is implicated in the differentiation and proliferation of hematopoietic cells. Truncations of the N and/or C terminus of c-Myb, found in v-Myb, can potentiate its transforming ability. Two negative regulatory subregions, located in the C terminus, were mapped previously by using GAL4–c-Myb fusion proteins in transient transfection assays for the transcriptional activation of a GAL4-responsive reporter gene. To dissect the C terminus of c-Myb in terms of its involvement in transcriptional activation and oncogenic transformation, a series of C-terminal deletion mutants of c-Myb were analyzed. In addition, linker insertion mutants within the transactivation domain and/or heptad leucine repeat of c-Myb were examined along with those deletion mutants. In this study, we demonstrated that the removal of both of the two previously mapped negative regulatory subregions from the native form of c-Myb not only supertransactivates a Myb-responsive reporter gene but also potentiates its transforming ability in culture. However, in contrast to previous results, cells transformed by all of the mutants analyzed here except v-Myb itself exhibited the same phenotype as those transformed by c-Myb. The proliferating cells were bipotenial and differentiated into both the granulocytic and monocytic lineages. This result implies that the C terminus of c-Myb alone has no effect on the lineage determination. Finally, the transactivation activities of these mutants correlated with their transforming activities when a mim-1 reporter gene was used but not when a model promoter containing five tandem Myb-binding sites was used. In particular, a very weakly transforming mutant with a linker insertion in the heptad leucine repeat superactivated the model promoter but not the mim-1 reporter gene.
The nuclear proto-oncogene c-myb is implicated in the differentiation and proliferation of hematopoietic cells (34). The expression of c-Myb is high in immature hematopoietic cells and decreases as differentiation progresses (18, 63). Two naturally arising avian retroviruses, avian myeloblastosis virus (AMV) and E26, carry a transduced myb gene and cause acute monoblastic leukemia and erythroblastosis in chickens, respectively (58). The transduced v-myb oncogenes of AMV and E26 encode N- and C-terminally truncated forms of c-Myb. In the case of AMV, v-Myb contains 10 amino acid substitutions throughout the protein, whereas E26 v-Myb contains a single unrelated substitution. A proviral insertion within the c-myb locus can lead to B-cell lymphomas and myeloid leukemias in chickens and mice, respectively. In both cases, the insertional activations cause the expression of truncated forms of c-Myb (35, 58). Recently, a truncation of the C terminus of c-Myb was observed during the progression of a case of human chronic myelogenous leukemia (62). Further analyses of in vitro transformation by mutants of c-Myb imply that the N- and/or C-terminal truncation is important for tumorigenesis (15, 27, 29).
c-Myb and its transduced version, v-Myb, can bind to the same DNA sequence (YAAC[G/T]G) and mediate transcriptional activation (5, 25, 56, 64). The DNA-binding domain of c-Myb, located at the N terminus, is composed of three imperfect repeats, each with a helix-turn-helix structure (55). This domain is the most conserved region among Myb-related proteins. Part of the first repeat is deleted in v-Myb. Next to the DNA-binding domain is the centrally localized transcriptional activation (TA) domain. The minimal transactivation domain, mapped by fusion with the GAL4 DNA-binding domain, is 50 amino acids and contains 15 charged residues with a predicted amphipathic helix (64). However, a much larger region encompassing the minimal TA domain was found to be essential for a native v-Myb protein to exert its transactivational function (10, 32). Interestingly, it was shown that the transcriptional coactivator CREB binding protein can bind to c-Myb through a region containing the TA domain (12, 54). The part of c-Myb C-terminal to the TA domain has a negative regulatory role in terms of transactivation and transformation (17, 28, 29, 56). The N-terminal region of the negative regulatory domain contains a heptad leucine repeat (HLR). The disruption of the HLR can potentiate the transcriptional and transforming activities in cell culture with a murine c-Myb from which the first 17 amino acids at its N-terminal end have been deleted (27, 36). However, the HLR region has a positive role in transcriptional activation and transformation by AMV v-Myb (4, 22). Also, E26 virus, which lacks this region, does not transform in the absence of the v-ets oncogene (48). The deletion of the FAETL motif, which is in the region of the HLR, from AMV v-Myb renders the protein incapable of transactivation and transformation in cell culture (22). The substitution of alanine for leucine in this region causes v-Myb to lose its leukemogenicity in vivo but not its transformation ability in cell culture, presumably due to temperature sensitivity (4, 22).
The rest of the C terminus, following the HLR of c-Myb, is absent in both the AMV and E26 Myb proteins. The truncation of this region has been linked to the activation of the proto-oncogene, and analyses of various deletion mutants imply a negative regulatory function. Finer deletions of the C-terminal domain of c-Myb fused to the GAL4 DNA-binding domain were previously tested for their abilities to transactivate a GAL4-responsive promoter (17). Two subregions (positions corresponding to residues 425 to 464 and 499 to 558 of c-Myb) in the C terminus were important in negatively regulating the transcriptional activity of GAL4-Myb fusion proteins. These two noncontiguous regions flank a domain that is highly conserved among vertebrate A-, B-, and c-Myb as well as Drosophila Myb proteins (43). The same study also pointed out that the negative regulatory effect mediated by the C terminus is c-Myb DNA-binding domain independent and can be exerted in trans. In addition, it was previously demonstrated that either the N- or C-terminal truncation (dCC or CCd, respectively) can promote the transforming activity of c-Myb, particularly the former (15, 29). Both the dCC- and CCd-transformed cells exhibited a promyelocytic or granulocytic phenotype rather than the monocytic phenotype displayed by v-Myb-transformed cells. This result raised the interesting question of whether the C terminus of c-Myb controls lineage determination.
Recently it was shown that the constitutive expression of the full-length chicken c-Myb from an internal ribosomal binding site (IRES)-containing retroviral vector can transform myelomonocytic cells in cultures (21). Similar results were observed with full-length murine c-Myb in fetal liver cell cultures (20). Because of these results, we sought to further investigate the subregions of the C-terminal part of c-Myb with regard to their transactivating and transforming abilities in the context of full-length chicken c-Myb. Furthermore, two linker insertion mutations that disrupt the central transactivation domain (Gly-Pro insertion at residue 304) and the HLR (Gly-Pro-Asp at residue 389) were also tested for oncogenic transformation.
MATERIALS AND METHODS
Plasmid constructions.
The avian retroviral constructs and a series of C-terminal deletion and linker insertion mutants of c-Myb have been previously described or were constructed from GAL4–c-Myb fusions (17). An additional deletion mutant, CCC-dPSBB, was made by digesting the double c-Myb deletion CCC-dPSBN in the pSP73 vector with NsiI and BamHI (3′ of the c-Myb coding sequence), treating it with Klenow fragment and deoxynucleoside triphosphates, and reclosing the plasmid by ligation to an SpeI linker with stop codons in all three reading frames. To construct the other undescribed mutant, CCC-dMN, a partial MscI digestion was performed, followed by heat inactivation of the MscI enzyme and digestion with Ppu10I. The latter enzyme is an isoschizomer of NsiI that maintains the c-Myb reading frame when it is filled in with Klenow fragment and deoxynucleoside triphosphates and fused to the MscI blunt end. To convert the previously described splice acceptor-driven C-terminal deletion or insertion mutants into IRES-driven ones, the BstEII fragments containing the deletion(s) or insertion(s) were swapped with the BstEII fragment from N-I-CCC (21).
Cells and media.
QT6 cells, a quail fibroblast cell line, were used for transient transfection assays and virus production. The cells were cultured in Dulbecco’s modified essential medium with 5% fetal calf serum, 4.5 g of glucose per liter, 1× MEM nonessential amino acids, 1 mM sodium pyruvate, 2 mM glutamine, 100 μg of streptomycin per ml, and 100 U of penicillin per ml at 37°C in a humidified 10% CO2–90% air incubator. Yolk sac and bone marrow cells were obtained from 10- to 13- and 19-day-old White Leghorn chicken embryos, respectively. Both types of the primary cells were grown in Iscove’s medium containing 10% fetal calf serum, 5% heat-inactivated chicken serum, 1× MEM vitamins, and other medium supplements as described above for QT6 cells at 37°C in a humidified 5% CO2–90% air incubator.
Transfections, immunoblotting, and assays for transactivation.
A calcium phosphate-mediated procedure described previously was used for the transient transfection of QT6 cells (9, 32). To assay the transactivation activity of c-Myb mutants in QT6 cells, each reaction included 3 μg of activator plasmid, 1 μg of reporter plasmid, and 0.5 μg of pCMV-β-Gal as an internal control for transfection efficiency. Yeast tRNA was added to bring the total nucleic acid level to 10 μg. Cells from a 10-cm-diameter plate were harvested 48 h after transfection. Half of the cells from each transfection were extracted and subjected to β-galactosidase (β-Gal) and luciferase assays as described previously (1, 57). The other half of the cells were lysed and boiled in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading dye for immunoblotting. Cell lysates from transfected QT6 cells were normalized to equal β-Gal activities to account for differences in transfection efficiency and then subjected to SDS-PAGE on 10% polyacrylamide gels. The resolved proteins were transferred to nitrocellulose (BA85; Schleicher and Schuell, Keene, N.H.) and probed with mouse monoclonal anti-Myb antibodies (2.7 and 5E) (19, 59) or a rabbit polyclonal anti-Mim-1 antibody (51). Proteins were detected by alkaline phosphatase-conjugated anti-mouse or anti-rabbit immunoglobulin G with the use of 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium as substrates (Promega). Alternatively, the ECL Western blotting detection system was employed according to the manufacturer’s instructions (Amersham Life Science).
Transformation assays.
The transformation assays were performed by the cocultivation of chicken hematopoietic cells from either yolk sac or bone marrow with mitomycin C-treated, virus-producing QT6 cells. To produce infectious viruses, QT6 cells were cotransfected with 1 μg of helper virus (pMAV-dX) and 9 μg of a replication-defective retroviral construct expressing both the neomycin resistance protein and c-Myb or a c-Myb variant. The supernatant of the 2-day-old transfected culture was collected and used to infect fresh QT6 cells (45). Either the transfected or the virally infected QT6 cells were selected with G418 at a concentration of 200 μg/ml for two weeks. The G418-resistant QT6 cells were treated with 10 μg of mitomycin C per ml for 2 h, washed, and then used for overnight cocultivation with primary hematopoietic cells as previously described (31, 44). The yolk sac or bone marrow cells were fed by partially changing the medium every 2 to 3 days. The growth of the transformed cells was monitored by cell counting. Rapidly growing cells from liquid culture were harvested for immunoblotting and fluorescence-activated cell sorter (FACS) analysis. To determine the colony-forming ability, a methylcellulose assay was carried out. After 8 days of cocultivation, 105 cells from each plate were seeded into 4 ml of a medium with 0.8% methylcellulose (MethoCult H4100; Stem Cell Technologies, Inc.) per 5-cm-diameter dish. The colonies were counted after 2 weeks at 37°C.
Cytocentrifugation and FACS analyses.
The morphology of the transformed cells was observed by performing a cytocentrifugation (Cytospin 2; Shandon) of 5 × 104 cells onto a glass slide at 500 × g for 5 min followed by modified Wright-Giemsa staining (Diff-Quick; Baxter). To determine the cell surface markers, FACS analyses were carried out with a Coulter EPICS XL-MCL instrument. The preparation and staining of cells with 1C3 and HLO72 antibodies were described previously (21). A fluorescein-conjugated goat anti-mouse immunoglobulin G (Cappel) at a 250× dilution was used for the detection of primary antibodies. The final cell pellet was resuspended in Hanks’ balanced salt solution containing 1 μg of propidium iodide per ml for gating out dead cells.
RESULTS
Construction, expression, and transcriptional activity of c-Myb mutants with linker insertions or C-terminal deletions.
To use retroviral vectors for gene expression, one commonly places more than one gene onto a vector. For our in vitro transformation assay, a retroviral vector derived from myeloblastosis-associated virus type 1 was used (45). This retroviral vector contains a neomycin resistance selectable marker gene and a tester gene. It has been demonstrated that an IRES element can mediate the expression of a second gene from a retroviral vector more efficiently than an internal promoter or splice acceptor (26). Previous data showed that the constitutive expression of c-Myb through the encephalomyocarditis virus IRES renders the protein weakly transforming (21). Because IRES-driven c-Myb is a weak transformer, this provided us a means to map the C-terminal negative regulatory subregions of c-Myb with respect to transformation. We therefore constructed a series of encephalomyocarditis virus IRES-driven c-Myb mutants encoded by the myeloblastosis-associated virus-derived vector as described in Materials and Methods.
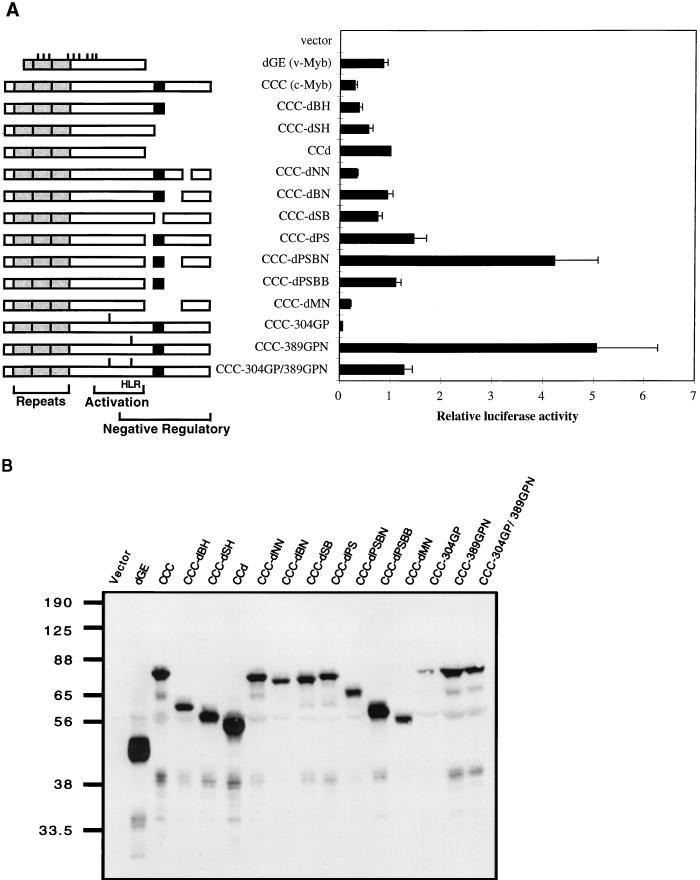
Many of the c-Myb mutants used here were previously tested for transcriptional activation in the context of GAL4-Myb fusion proteins (17). Two negatively regulating subregions were mapped, and the deletion of both of them was required for the activation of the GAL4–c-Myb fusion protein. These two subregions, corresponding to the residues 425 to 464 and 499 to 558 (PS and BN), flank a very highly conserved domain found in c-Myb, A-Myb, B-Myb, and Drosophila Myb. A series of C-terminal deletions used previously as GAL4-Myb fusions were reconstructed back into the native form of protein and expressed from the IRES-containing retroviral vector. In addition, a small set of linker insertion mutants of c-Myb was examined. The linkers were introduced into the acidic region and/or HLR of c-Myb. Schematic diagrams for all the c-Myb mutants are shown in Fig. 1A.
FIG. 1.
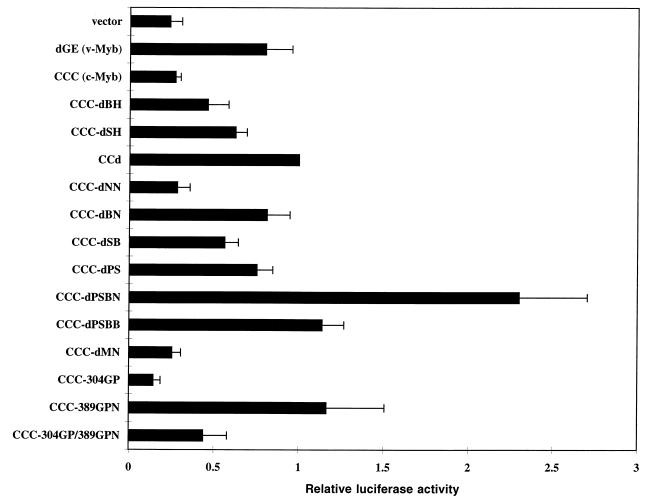
Transactivation of the EW5 reporter gene by c-Myb mutants. (A) A series of deletion or insertion mutants of c-Myb expressed from a retroviral vector were assayed for transcriptional activity in QT6 cells by transient transfection. For each transfection, 3 μg of activator and 1 μg of EW5 reporter were cotransfected into QT6 cells, along with 0.5 μg of a β-galactosidase-expressing vector (CMV-β-Gal) as an internal control. Half of the cells from each transfection were used to determine transactivation activity, as described in Materials and Methods. The other half of the cells were reserved for immunoblotting. Relative luciferase activities were obtained by assigning the luciferase activity of CCd a value of 1. Shown are the mean values of relative luciferase activities from at least three transfections and average deviations (error bars) from the mean. Schematic diagrams of c-Myb mutants are shown at the left. The point mutations in v-Myb are indicated as short bars above the v-Myb diagram. The gray boxes represent the DNA-binding domain and are also marked by repeats. The C-terminal most highly conserved domain is shown as a black box. The linker insertions are indicated as bars above the diagrams for CCC-304GP, CCC-389GPN, and CCC-304GP/389GPN. (B) Representative immunoblot of transiently transfected QT6 cells from the experiments whose results are shown in panel A. Cell lysates with equal amounts of β-Gal activity were resolved on an SDS–10% PAGE gel and immunoblotted with anti-Myb antibodies (5E and 2.7). The relative mobilities of protein markers are indicated in kilodaltons.
To test for transcriptional activation by the c-Myb mutants, transient transfection assays were carried out with QT6 quail fibroblasts with a Myb-responsive reporter gene (EW5 reporter gene). The EW5 reporter gene uses a recombinant promoter with five concatamerized mim-1A binding sites (the strongest Myb-binding site from the mim-1 promoter) and an E1B TATA box to drive a luciferase gene (11). A retroviral vector (N-Cla) (15) bearing the gene encoding only the neomycin resistance gene product was used as a negative control in all transfection and transformation assays. All transfection assays were performed at least three times, and an internal pCMV–β-Gal control was used to normalize for transfection efficiency. Relative luciferase activities were obtained by assigning the luciferase activity of CCd a value of 1. CCd is a truncated c-Myb with a C-terminal deletion similar to that of AMV v-Myb. As we observed before, the level of transactivation by full-length c-Myb (CCC) was fairly low (Fig. 1A). The removal of the C terminus in CCd increased the transactivation by two- to threefold. The removal of the two negative regulatory subregions previously deleted in the GAL4-Myb fusion, PS and BN, superactivated c-Myb. A single deletion in either of these two subregions could activate c-Myb, but only to an extent similar to that of the activation by C-terminal truncation (CCd). The double deletion had a much greater effect than the single deletions. The transactivation by CCC-dPSBN was four times greater than that of CCd and up to 15 times that of CCC. All of the other deletion mutants had activities in the range of c-Myb and CCd. The other superactivating c-Myb mutant was CCC-389GPN, which has a linker insertion (Gly-Pro-Asn) within the HLR. The linker insertion (Gly-Pro) mutant at position 304 inactivated c-Myb as previously observed in v-Myb (40). The double linker insertion mutant (CCC-304GP/389GPN) appeared to have competing effects from the inactivation by the 304 insertion and superactivation by the 389 insertion and as a result had an activity similar to that of CCd.
To confirm the proper expression of all these mutants, immunoblotting was performed on the transiently transfected QT6 cells. After harvesting the transfected QT6 cells, half of each sample was used for the transactivation assay and the other half was reserved for the protein assay. Figure 1B shows an immunoblot probed with anti-Myb antibodies on a representative set of transient transfections with a loading of equal β-Gal activity to normalize for variations in transfection efficiency. From the immunoblot, the protein level of each mutant varied somewhat. This may be due to the variation of protein stability from mutant to mutant. No other assays of protein stability were performed, but the patterns of the mutant protein levels were consistent among different sets of transfection experiments. Most importantly, each mutant migrated per its predicted relative molecular weight and at a correct position relative to the others. Also, the superactivation by CCC-dPSBN and CCC-389GPN was clearly not due to increased protein levels relative to that of wild-type c-Myb (CCC).
In vitro transformation assay of c-Myb mutants.
To determine the transforming activity of the c-Myb mutants, a yolk sac assay was carried out. The yolk sac cells obtained from 13-day-old chicken embryos are rich in myelomonocytic progenitor cells. It is known that v-Myb efficiently transforms primary yolk sac cells and blocks the differentiation process at the monoblast stage. The infection of primary yolk sac cells was performed by cocultivation with virus-producing QT6 cells. Then, the infected yolk sac cells were cultured by a partial change of media every 2 to 3 days, and no exogenous growth factor was included in the media. Cell proliferation was monitored directly by cell counting. However, cells transformed by c-Myb and some of the weak mutants exhibited a tendency to rely on feeder cells to grow. This made the growth of transformed cells somewhat variable with weaker mutants when assayed in different experiments. The only c-Myb mutant that consistently displayed a robust growth was CCC-dPSBN (determined by cell counting, as shown in Table 1). In order to further assess the relative transforming potential of CCC-dPSBN, a methylcellulose colony-forming assay was used. This assay is more stringent and quantitative. The averages of normalized colony numbers from three independent assays of a 5-cm-diameter tissue culture dish seeded with 105 cocultivated yolk sac cells are listed in Table 1. The viral titers of the cultures used for cocultivation in the methylcellulose assay were determined by the formation of G418-resistant colonies in QT6 cells. Two independent sets were examined and exhibited similar levels of viral titers for all the tested viruses (data not shown). The overall transforming activity was determined by cell number at day 22 or 23, the frequency of liquid culture outgrowth, and the colony count on methylcellulose assay.
TABLE 1.
Transforming activities of c-Myb mutants
| Construct | No. of cells/ml in expta
|
Frequency of liquid culture outgrowthb | Relative colony no.c
|
Transformation leveld | ||
|---|---|---|---|---|---|---|
| 1 (day 23) | 2 (day 22) | Avg | ADe | |||
| N-Cla (vector) | <103 | <103 | 0/4 | 0 | 0 | − |
| dGE (v-Myb) | 1.4 × 107 | 9 × 106 | 4/4 | 100 | 0 | +++++ |
| CCC (c-Myb) | <103 | 4.1 × 104 | 2/4 | 1.8 | 1.2 | + |
| CCd | 2 × 104 | 1 × 104 | 4/4 | 2.5 | 1.1 | + |
| CCC-dMN | <103 | <103 | 0/4 | 0.5 | 0.4 | − |
| CCC-dNN | <103 | <103 | 0/4 | 3.1 | 2.5 | −/+ |
| CCC-dBN | 1.5 × 104 | 1 × 104 | 3/4 | 13.3 | 3.6 | ++ |
| CCC-dPSBB | 6 × 104 | 1.6 × 104 | 4/4 | 11.6 | 5.5 | ++ |
| CCC-dPSBN | 1.6 × 105 | 1.1 × 105 | 4/4 | 37.4 | 17.1 | +++ |
| CCC-dSB | 3.5 × 104 | 2 × 104 | 2/4 | 4.7 | 1.0 | + |
| CCC-dSH | 0.7 × 104 | 1.4 × 104 | 2/4 | 15.2 | 5.8 | ++ |
| CCC-dPS | 3 × 104 | 4.8 × 104 | 2/4 | 8.8 | 8.0 | ++ |
| CCC-dBH | 2 × 104 | 3.8 × 104 | 4/4 | 16.0 | 7.0 | ++ |
| CCC-304GP | <103 | <103 | 0/4 | 0 | 0 | − |
| CCC-389GPN | 2 × 104 | <103 | 1/4 | 2.9 | 0.9 | + |
| CCC-304GP/389GPN | <103 | <103 | 0/4 | NDf | ND | − |
The cell numbers were determined on the day indicated after cocultivation.
The frequency of liquid culture outgrowth is from three yolk sac assays and one bone marrow assay.
In methyl cellulose assays, relative colony numbers were obtained by assigning the colony number of N-dGE (v-Myb) a value of 100. Averages of relative colony numbers were from three independent methylcellulose assays.
The nontransforming viruses and v-Myb are designated arbitrarily − and +++++, respectively. The transforming activities of c-Myb and c-Myb mutants were determined relative to v-Myb activity after considering cell number count, frequency of liquid culture outgrowth, and colony number count in the methylcellulose assay.
AD, average deviation.
ND, not done.
v-Myb, our positive control in the yolk sac assay, transformed every time and gave the highest colony count in each set of methylcellulose assays. c-Myb and CCd were very weak transformers as previously reported (15, 21, 29). Although the C terminus of c-Myb was assigned a negative role in Myb-mediated transformation, the removal of the C terminus from c-Myb only slightly improved the transforming activity, as seen in CCd. The nontransforming mutants, including CCC-dMN, CCC-dNN, CCC-304GP, and CCC-304GP/389GPN, generally had lower transactivation activities than that of c-Myb on our model Myb promoter assay (with the exception of CCC-304GP/389GPN). The superactivating double deletion mutant, CCC-dPSBN, was consistently demonstrated to be a better transformer than c-Myb and the other c-Myb deletion mutants in terms of cell count, frequency of liquid culture outgrowth, and colony count by a methylcellulose assay. Surprisingly, the other superactivating mutant, CCC-389GPN, had only a weak transforming activity similar to that of c-Myb. The double linker insertion mutant, CCC-304GP/389GPN, exhibited a transforming ability similar to that of CCC-389GPN.
To test whether the transcriptional activity of Myb is essential for the transformation process, two Myb mutants that lacked transactivation activity were specifically analyzed for transformation. c-Myb mutants with a disrupted transactivation domain (CCC-304GP) or a missing transactivation domain (CCC-dSM) with a deletion from the SmaI site to the MscI site of c-myb failed to transactivate the EW5 reporter gene and transform primary yolk sac cells (Table 1 and data not shown). The linker insertion at position 389, which was predicted to disrupt the HLR and abolish a negative regulatory activity of c-Myb, affected only transcriptional activation but not oncogenic transformation.
The transforming activities of all other c-Myb deletion mutants were lower than that of CCC-dPSBN. This demonstrated that the two previously mapped subregions, with a negative regulatory activity in transcription, are also involved in transformation. The deletion of both negative regulatory subregions not only superactivates transcription but also greatly potentiates its transforming activity. To look into the possibility of an effect by c-Myb mutants on other hematopoietic lineages, a bone marrow transformation assay was also carried out. The results of outgrowth from the bone marrow assay were similar to those observed with yolk sac cells, and the data are shown in Table 1.
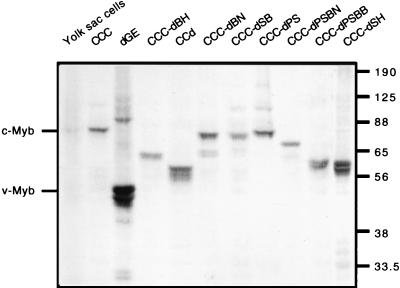
To confirm that the outgrowth of primary yolk sac cells arises from the expression of the predicted forms of c-Myb, the transformed cells were subjected to an immunoblotting analysis with anti-Myb antibodies. A representative immunoblot for all the yolk sac cells transformed by c-Myb and its mutants is shown in Fig. 2. The transformed yolk sac cells expressed mutant Myb proteins with the predicted molecular sizes and mobilities relative to each other. Primary yolk sac cells freshly isolated from 13-day-old chicken embryos were used as an endogenous source of c-Myb. Furthermore, the mobility of each Myb mutant from transformed cells was verified by coelectrophoresis on an SDS-PAGE gel with transiently transfected QT6 cells expressing the corresponding form of Myb (data not shown).
FIG. 2.
Immunoblotting analysis of c-Myb mutant-transformed chicken yolk sac cells. From the above yolk sac assays, v-Myb, c-Myb, and c-Myb mutant-transformed yolk sac cells were collected and subjected to immunoblot analysis. All yolk sac cell lysates were derived from 5 × 105 cells with the exception of primary yolk sac cells obtained from a 13-day-old chicken embryo (6 × 106 cells/lane) and CCC-dPS (3 × 105 cells/lane). The cell lysates were resolved on an SDS–10% PAGE gel and immunoblotted with Myb antibodies (5E and 2.7). The relative mobilities of protein markers are indicated in kilodaltons.
Characterization of the yolk sac cells transformed by c-Myb mutants.
To characterize the phenotypes of the transformed cells, three different assays were used. First, the cell morphology was examined by cytocentrifugation followed by a modified Wright-Giemsa staining. Second, the expression of two cell surface markers was determined by FACS analysis with the use of 1C3 (granulocyte-specific marker [47]) and HLO72 (monoblast-specific marker [46]) antibodies. Third, the expression of Mim-1, a promyelocyte- or granulocyte-specific marker, was examined by immunoblotting with an anti-Mim-1 antibody.
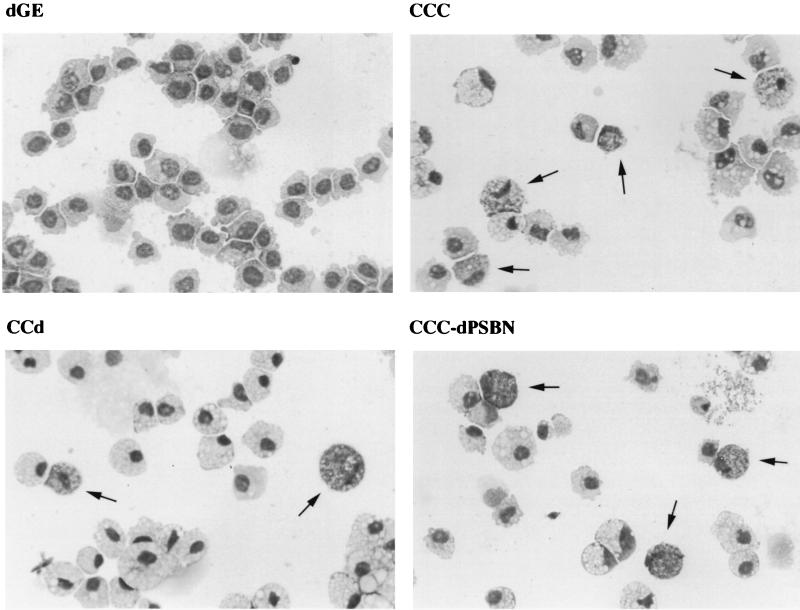
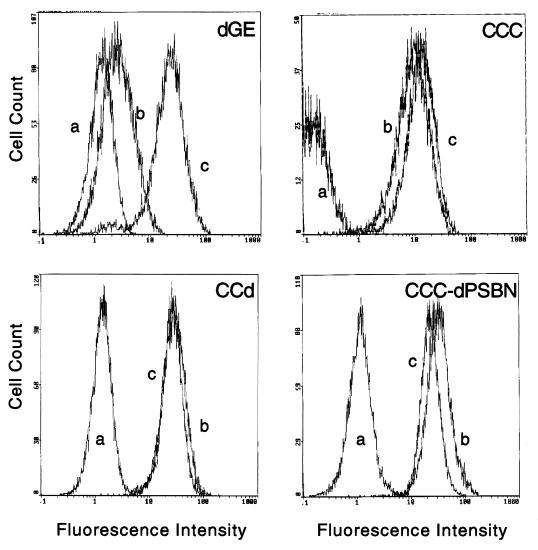
As previously reported, v-Myb-transformed cells are exclusively monoblasts, whereas c-Myb-transformed cells are heterogeneous in nature and exhibit cell types with characteristics of either granulocytic or monocytic lineage (14, 21). Cells transformed by all of the c-Myb mutants displayed a phenotype similar to that of c-Myb, with the one exception of CCC-389GPN. However, because CCC-389GPN had a very weak transforming activity and grew out only once in four assays, the exclusively granulocytic phenotype seen here may be dependent upon culture conditions. Two representative c-Myb mutants, CCd and CCC-dPSBN, transformed yolk sac cells similar to c-Myb, with heterogeneous populations resembling both the granulocytic and monocytic lineages, as shown in Fig. 3. The FACS profiles of the same representative set shown in Fig. 3 are shown in Fig. 4. v-Myb-transformed cells were stained positively only for a monocytic cell surface marker (HLO72) but not a granulocytic marker (1C3). In contrast, cells transformed by c-Myb, CCd, and CCC-dPSBN were stained positively for both the granulocytic and monocytic cell surface markers. Furthermore, since the entire populations were shifted with both 1C3 and HLO72 stainings, it followed that all cells transformed by c-Myb, CCd, and CCC-dPSBN are double positive. In addition, the expression of Mim-1 was examined by immunoblotting. v-Myb-transformed cells did not express Mim-1, whereas cells transformed by c-Myb and its mutants were Mim-1 positive (Table 2). As mentioned previously, one bone marrow assay was carried out and the cells transformed by c-Myb mutants also displayed a heterogeneous granulocytic and monocytic phenotype (data not shown). In addition, the proliferating cells transformed by CCC-dBN and CCC-dPSBN were double positive for granulocytic and monocytic cell surface markers (data not shown). Together, there was no significant difference between the results from bone marrow and yolk sac assays in terms of the transforming ability, cell morphology, and expression of cell surface markers. Table 2 presents all the results obtained from the above assays with v-Myb, c-Myb, and c-Myb mutants. Occasionally, we also saw the exclusive outgrowth of granulocytes from the yolk sac assays in mutants CCd and CCC-dPS. This may be due to feeder cells, which may provide cytokines that promote the growth and/or differentiation of one particular type of cell. In summary, cells transformed by c-Myb and most of its C-terminal deletion mutants, including CCd, have a bilineage phenotype as determined by cell morphology and cell surface markers.
FIG. 3.
Morphology of yolk sac cells transformed by dGE, CCC, CCd, and CCC-dPSBN. The morphologies of a representative set of transformed yolk sac cells are shown. The yolk sac cells from liquid culture outgrowth were cytocentrifuged onto glass slides and stained with Diff-Quick (DADE). The v-Myb-transformed cells (dGE) are a homogeneous population of monoblasts. The cells transformed by c-Myb (CCC), a double deletion mutant of c-Myb (CCC-dPSBN), and a C-terminal truncation of c-Myb (CCd) exhibited heterogeneous phenotypes. Cells of one type (marked with arrowheads) resemble granulocytes with distinct cytoplasmic granules. The rest of the cells are macrophage-like and have an eccentric nucleus with many vacuoles in the cytoplasm.
FIG. 4.
FACS analysis of cell surface markers on yolk sac cells transformed by dGE, CCC, CCd, and CCC-dPSBN. The same representative set of Myb-transformed yolk sac cells shown in Fig. 3 were stained with antibodies specific for cell surface markers. The antibodies 1C3 and HLO72 were used to detect granulocytic and monocytic lineages, respectively. Fluorescence profiles are shown after the cells were stained with wash medium control (a), 1C3 (b), or HLO72 (c).
TABLE 2.
Phenotypes of cells transformed by c-Myb mutants
| Construct | Morphologya | Expression of cell surface markerb
|
||
|---|---|---|---|---|
| 1C3 | HLO72 | Mim-1 | ||
| N-Cla (vector) | ND | ND | ND | ND |
| dGE (v-Myb) | Monocytic | − | + | − |
| CCC (c-Myb) | Bilineage | + | + | + |
| CCd | Bilineage | + | + | + |
| CCC-dMN | ND | ND | ND | ND |
| CCC-dNN | ND | ND | ND | ND |
| CCC-dBN | Bilineage | + | + | + |
| CCC-dPSBB | Bilineage | + | + | + |
| CCC-dPSBN | Bilineage | + | + | + |
| CCC-dSB | Bilineage | + | + | + |
| CCC-dSH | Bilineage | ND | ND | ND |
| CCC-dPS | Bilineage | + | + | + |
| CCC-dBH | Bilineage | + | + | + |
| CCC-304GP | ND | ND | ND | ND |
| CCC-389GPN | Granulocytic | ND | ND | ND |
| CCC-304GP/389GPN | ND | ND | ND | ND |
Cell morphology was determined by staining cytospins with Diff-Quick. The bilineage phenotype is based on the fact that the transformed cells display heterogeneous populations with both monocytic and granulocytic features. ND, not determined.
The expression of cell type-specific markers was determined by FACS analysis with 1C3 (granulocytic lineage) and HLO72 (monocytic lineage) antibodies and by immunoblotting with an anti-Mim-1 antibody (promyelocytic or granulocytic lineage).
Transactivation by c-Myb mutants with a mim-1 reporter gene.
The transactivation of the EW5 reporter gene did not correlate well with the transforming activity of c-Myb mutants. For example, CCC-389GPN superactivated the EW5 reporter but failed to consistently transform primary yolk sac cells. Since all the c-Myb mutant-transformed cells expressed Mim-1, we wanted to see whether the transactivation of the mim-1 reporter gene correlated better with oncogenic transformation. We therefore conducted transient transfections in the same way as shown in Fig. 1, except that we used a mim-1 reporter gene to score for transactivation. This mim-1 reporter gene comprises a sequence from nucleotide −242 to +102 of mim-1 and a luciferase gene (51). Figure 5 displays the results from three different independent transfections. It shows that the mim-1 promoter had a much higher basal activity than the EW5 reporter in QT6 cells with our control retroviral vector. For most of the c-Myb mutants, the transactivating profiles were very similar between EW5 and mim-1 reporters. c-Myb, CCC-dMN, CCC-dNN, and CCC-304GP were still fairly inactive. Most of the c-Myb mutants, having a transactivational activity between those of c-Myb and CCd on the EW5 reporter, remained at similar levels of activity on the mim-1 reporter. The double deletion mutant, CCC-dPSBN, had the highest level of activity among the constructs tested on the mim-1 reporter. Its level of activity was about twofold higher than the activity of CCd, which was less of an increase than we had observed with the EW5 reporter. The decrease in the degree of superactivation for CCC-dPSBN may relate to the high basal activity of the mim-1 promoter. Interestingly, CCC-389GPN was no longer superactivating and exhibited an activity similar to that of CCd.
FIG. 5.
Transactivation by c-Myb mutants with the mim-1 reporter gene. The same series of c-Myb mutants shown in Fig. 1 was analyzed in QT6 cells for transactivation with a reporter promoter derived from mim-1. The transfections were performed in the way mentioned previously, except for the use of a mim-1 reporter gene. Relative luciferase activities were obtained by assigning the luciferase activity of CCd a value of 1. Shown are the mean values of relative luciferase activities from at least three transfections and average deviations (error bars) from the mean.
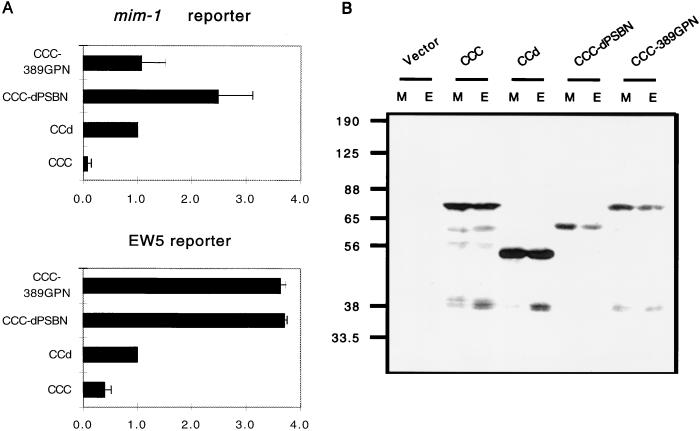
In order to compare the activation of these two different reporters despite the high-level basal activity of the mim-1 promoter, the transcriptional activity obtained from the control retroviral vector (N-Cla) was subtracted from the transcriptional activities of c-Myb, CCd, CCC-dPSBN, and CCC-389GPN. All the subtracted activities are shown relative to the subtracted activity of CCd and plotted in Fig. 6A. The EW5 reporter gene was superactivated equally well by CCC-dPSBN and CCC-389GPN, whereas the mim-1 reporter gene was superactivated only by CCC-dPSBN. To determine whether the change in the transactivation was due to differences in protein levels, cell lysates from the transfected cells normalized for equal β-Gal activities were resolved on an SDS-PAGE gel and examined by immunoblotting with anti-Myb antibodies (Fig. 6B). Regardless of the reporters used in the transfection, the intensities of the Myb-specific bands were similar. It is clear that CCC-389GPN failed to superactivate the mim-1 promoter, despite an equivalent protein level. This result implies that the superactivating activity of CCC-389GPN on the EW5 reporter gene does not correlate with transformation, whereas the transactivational activities of c-Myb mutants on the mim-1 promoter correlate better with their transforming abilities. However, this correlation did not hold for v-Myb, which transformed primary hematopoietic cells with a high level of efficiency but transactivated both of the reporter genes only moderately.
FIG. 6.
Differential activation of promoters by CCC-dPSBN and CCC-389GPN. (A) Transfections were repeated with both reporters in the same experiment to show the differential effect of CCC-389GPN on EW5 and mim-1 reporter genes. Because the mim-1 reporter gene has a high-level basal activity, we subtracted the activity obtained from the vector (N-Cla). Relative luciferase activities were then calculated by assigning the subtracted luciferase activity of CCd a value of 1. Shown are the mean values of relative luciferase activities from at least three transfections and average deviations (error bars) from the mean. (B) Cell lysates prepared from the same transfection assays were resolved by SDS–10% PAGE and immunoblotted with anti-Myb antibodies. The relative mobilities of protein markers are indicated in kilodaltons. M, mim-1; E, EW5.
DISCUSSION
c-Myb is a nuclear protein and mediates transcriptional activation through a specific DNA sequence. The truncation of either terminus of c-Myb as found in AMV v-Myb activates its transcriptional and transforming activities in culture. Therefore, a negative regulatory function has been ascribed to the C terminus of c-Myb. We have now studied two linker insertions and a series of C-terminal deletion mutants within the context of full-length c-Myb with the use of an IRES-containing retroviral vector. Both the transcriptional activities and oncogenic transformation abilities of these mutants were analyzed in cell culture.
We previously showed that a linker insertion mutant (CCC-304GP) at position 304 in the TA domain abolishes the transactivation by c-Myb, whereas a linker insertion mutant (CCC-389GPN) at position 389 within the HLR can superactivate the transcriptional activity of c-Myb on a model reporter gene. The double insertion mutant (CCC-304GP/389GPN) restores part of the transactivating activity but not to the full extent of CCC-389GPN (16). This implies that c-Myb can regulate transcription through at least two different domains. One domain, involved in the activation of transcription, contains many charged residues and a predicted helical structure. The other domain having a negative regulatory activity comprises a HLR also referred to as the leucine zipper (36, 52). Because of the increased transcriptional activation by these linker insertion mutants, we further looked into their transforming activities in this study. It was first reported by Kanei-Ishii et al. that the disruption of the HLR with substitutions of proline for leucine led to an increase in both transcriptional and transforming activities of murine c-Myb (36). However, our CCC-389GPN mutant displayed an increase of activity only in transactivation and not in oncogenic transformation. This is similar to our recent studies of an alternatively spliced form of c-Myb, which contains an insertion of an additional 120 amino acids (encoded by exon 9A) within the C-terminal end of the HLR (65). The expression of exon 9A in the context of c-Myb results in an elevated level of transcriptional activity without an effect on transformation.
The differences between the results of Kanei-Ishii et al. and the lack of activation of oncogenic transformation by CCC-389GPN may be ascribed to the nature of mutations (substitutions of Pro for Leu versus Gly-Pro-Asn linker insertion) in the HLR and the assay systems (mouse versus chicken). However, we have previously shown that the same linker insertion and similar leucine substitutions in v-Myb are both compatible with oncogenic transformation (22, 40). In addition, the murine c-Myb mutants used in the assays done by Kanei-Ishii et al. were generated from a c-Myb background (Red [FLmyb]), lacking the N-terminal 17 amino acids (28). A recent report showed that a truncation as small as 20 amino acids at the N-terminal end of c-Myb can cause rapid-onset B-cell lymphomas at a high frequency in chickens (35). Thus, it is possible that the 17-amino-acid truncation in Red(FLmyb) may potentiate its transforming activity in conjunction with a leucine repeat disruption. The HLR has been implicated in mediating the interaction with a negative regulatory protein. Thus far, the only protein that has been shown to interact directly with Myb in this region is p160 (61). However, p160 localizes mainly in nucleolus, whereas Myb localizes diffusely in the nucleus while sparing the nucleolus (7, 39). Therefore, the biological significance of the interaction of c-Myb with p160 remains to be determined.
As mentioned previously, the deletion of both the PS and BN subregions potentiates the transcriptional activation by GAL4-Myb fusions. We now report that CCC-dPSBN, a double deletion in both the PS and BN subregions of c-Myb, contains an increased activity in both transactivation and oncogenic transformation. We have previously shown that at least the BN subregion can block the transactivation by v-Myb or LexA-Myb in trans (17). These results imply that the negative regulatory effect of the C terminus of c-Myb is Myb DNA-binding domain independent and may be involved in protein-protein interactions. Additionally, c-Myb is a very good transactivator on a Myb-responsive reporter gene in yeast but behaves in an opposite manner in animal cells. Indeed, the deletion of the C terminus of c-Myb decreased transcription in yeast (11). These results suggest the presence of an animal cell protein that specifically interacts with the C terminus of c-Myb and mediates the negative regulatory function. Together with our observations here, this hypothetical interaction of the C terminus of c-Myb with an animal repressor protein may most likely reside in the PS and/or BN subregion.
A number of the functions of the C terminus have been mapped near the PS and/or BN subregion. First, the effect of the C terminus on the DNA-binding activity was analyzed by Ishii’s group with an electrophoretic mobility shift assay (60). Three C-terminal truncation mutants (CT1, CTV, and CT2) used in that assay elevated relative DNA-binding activity levels by progressive deletion. Interestingly, the negative regulatory domain NRD2 (equivalent to chicken c-Myb residues 477 to 505) appears to lie largely between PS (residues 425 to 464) and BN (residues 499 to 558). Furthermore, the deletion of PS and BN greatly increases the transcriptional activation of GAL4-Myb fusion proteins that do not contain the Myb DNA-binding domain (17).
Second, an EVES domain (residues 513 to 563) mapped by Dash et al. coincides closely with the BN subregion and can interact with N-terminal Myb DNA-binding repeats in a yeast two-hybrid assay (13). Dash et al. further implicated another EVES-containing protein, p100, in the process of negatively regulating c-Myb activity. Therefore, they have proposed that the EVES motif itself is involved in regulating c-Myb activity intramolecularly and intermolecularly.
Third, it has also been reported that serine-533 residing in the EVES motif of the BN subregion of chicken c-Myb can be phosphorylated in vitro and in vivo by the 42-kDa mitogen-activated protein kinase (p42mapk) (3). A mutation to alanine at the homologous residue results in a promoter-specific superactivation of c-Myb (2). This phosphorylation-dependent regulation of Myb transactivation in vitro suggests a mechanism for signal integration by phosphorylation of the C-terminal portion of c-Myb. However, no increase in oncogenic transformation by an in vitro assay was observed with this mutation (22a).
Fourth, the BS69 protein was found to interact with the C terminus of c-Myb by a yeast two-hybrid assay (39a). The interaction of c-Myb with BS69 was mapped to the PS and BN subregions with a transient transfection assay. The deletion of the PS and/or BN subregion confers the ability to evade the transcriptionally inhibitory effect of BS69 on Myb proteins. The biological function of BS69 is not clear at this point, but the protein was first identified as a transcriptional repressor that interacts with the adenoviral E1A protein and contains a number of conserved protein domains found in other transcriptional regulators.
Finally, although the BN subregion also contains a predicted PEST domain, this region of c-Myb has not been implicated in the regulation of c-Myb protein stability (6). Consistent with this, our immunoblotting analysis shows no evidence of increased protein levels of CCC-dBN and CCC-dPSBN. Therefore, the regulation of the c-Myb activity by the BN subregion does not appear to depend upon protein stability. In summary, the negative regulatory activity that is disrupted by the removal of the PS and BN subregions may be due to protein-protein interaction and phosphorylation.
The c-Myb mutant CCC-dPSBB not only contains deletions in the PS and BN subregions but also extends further from BN to the C-terminal end of the protein. This extra deletion of the C-terminal distal part of c-Myb also contains a small conserved domain shared by vertebrate A-, B-, and c-Myb and Drosophila Myb (24). It is interesting that CCC-dPSBB did not exhibit transcription superactivation or a strong transforming phenotype as seen for CCC-dPSBN. These results suggest that an additional regulatory activity resides in the extreme C terminus of c-Myb. Two other lines of evidence point to the importance of this region. One comes from a report by Nazarov and Wolff that a minimal 38-amino-acid truncation at the C-terminal end of c-Myb correlates with promonocytic leukemias by retroviral insertions (Friend MuLV) in immunocompromised mice (49). In addition, one of the two temperature-sensitive, recessive lethal mutants in Drosophila Myb falls in this extreme C-terminal subregion (38).
The yolk sac cells transformed by c-Myb (21) and various C-terminal deletion mutants of c-Myb all displayed the same phenotype. They all expressed Mim-1 and cell surface markers of both the granulocytic and monocytic lineages. In contrast, v-Myb, which has sustained point mutations and both N- and C-terminal truncations, exclusively transforms cells of the monocytic lineage (14, 33). Thus, it is clear that the C terminus of c-Myb alone does not have a functional impact on the lineage determination. Other alterations of the protein are needed in order to confer the phenotype of v-Myb. This is consistent with other reports that the amino acid substitutions in v-Myb are responsible for the distinct monocytic phenotype and autocrine growth of transformed cells (14, 33). It is worth noting that CCd-transformed cells in our assays were of dual lineage. This is different from our previous report that CCd, CCC-dSH, and CCC-dBH transformed only cells of the granulocytic lineage (29). The difference in phenotype in our previous study may have been due to the inclusion of crude chicken myelomonocytic growth factor in the culture media. Molecular cloning and sequence analysis have shown that chicken myelomonocytic growth factor is most closely related to the mammalian granulocyte colony-stimulating factor (41). The cells transformed by c-Myb and c-Myb deletion mutants generally have a growth dependency on feeder cells which secrete cytokines. Depending on the type and number of feeder cells present during the culturing, we occasionally observed an exclusively granulocytic phenotype with CCd-transformed cells. In both cases, the cytokine(s) may drive the growth of the transformed cells to one particular lineage. However, our results with c-Myb and CCd agree with the observation that four murine myeloid cell lines generated from c-Myb and C-terminally truncated c-Myb express cell surface markers of both the monocytic and granulocytic lineages (28).
The transcriptional activation by the c-Myb mutants generally correlated well with their transforming activities. The only exception was the linker insertion mutant CCC-389GPN. This disruption of the HLR strongly activated transcription when we used a recombinant Myb-responsive reporter gene (EW5) containing only Myb-binding sites but not a natural mim-1 reporter gene. However, CCC-389GPN was a very weak transformer. It seems that the transforming ability of these c-Myb mutants correlates better with the transactivating ability on the mim-1 reporter gene. The mim-1 promoter contains three Myb-binding sites, with one of them juxtaposed to a CAAT/enhancer-binding protein β (C/EBPβ) binding site. Two other Myb target genes, tom-1 and the elastase gene, have a similar composition in their promoters (8, 53). C/EBPβ was shown to play an important role in the commitment to the myelomonocytic lineage and to synergize with Myb on the mim-1 promoter in vitro (37, 50). The loss of the cooperativity with C/EBPβ may in part account for the decreased transcriptional activity of CCC-389GPN on the mim-1 promoter. However, the correlation between transcription and transformation does not extend to v-Myb. In this study, AMV v-Myb efficiently transformed monoblasts but only moderately activated both the EW5 and mim-1 reporter genes. Furthermore, cells transformed by v-Myb lacked the expression of Mim-1. This suggests that the transformation by v-Myb involves mechanisms other than cooperation with C/EBPβ in transcription. Overall, c-Myb mutant-transformed cells are bipotential in myelocytic and monocytic lineages and express Mim-1. This phenotype agrees with our findings in transactivation assays with the mim-1 reporter gene, in which the activity correlates with transformation.
In summary, the transforming activities of the C-terminal mutants of c-Myb correlate well with their transactivational activities on the mim-1 reporter genes. A linker insertion mutant that is disrupted in the HLR superactivates transcription on the EW5 reporter gene but fails to strongly activate the mim-1 promoter and potentiate its transforming ability. All the C-terminal deletion mutants of c-Myb tested here induce a bilineage phenotype similar to that of full-length c-Myb. This indicates that the C terminus alone does not have a functional impact on lineage determination. However, the removal of the two negative regulatory subregions PS and BN can markedly improve the transactivational and transforming activities of c-Myb in culture. By demonstrating the importance of these two subregions in a biological assay, this paper provides a framework for future studies on the negative regulatory mechanism of c-Myb. It will now be interesting to determine how these regions affect the interaction of c-Myb with putative Myb-interacting proteins, including p100, p160, cyclin D, Maf (23, 30, 42, 61), and BS69 (39a).
ACKNOWLEDGMENTS
We thank members of the Lipsick laboratory for helpful discussions and Sara Roberts for expert technical assistance. We also thank Scott Ness for providing the mim-1 reporter and antiserum.
This work was supported by U.S. Public Health Service research grant R01 CA56509, training grants T32 CA09151 (D.-M.W) and T32 CA09176 (J.W.D.), and a Markey Fellowship in Molecular Mechanisms of Disease (C.H.W.).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 2.Aziz N, Miglarese M R, Hendrickson R C, Shabanowitz J, Sturgill T W, Hunt D F, Bender T P. Modulation of c-Myb-induced transcription activation by a phosphorylation site near the negative regulatory domain. Proc Natl Acad Sci USA. 1995;92:6429–6433. doi: 10.1073/pnas.92.14.6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aziz N, Wu J, Dubendorff J W, Lipsick J S, Sturgill T W, Bender T P. c-Myb and v-Myb are differentially phosphorylated by p42mapk in vitro. Oncogene. 1993;8:2259–2265. [PubMed] [Google Scholar]
- 4.Bartunek P, Karafiat V, Dvorakova M, Zahorova V, Mandikova S, Zenke M, Dvorak M. The Myb leucine zipper is essential for leukemogenicity of the v-Myb protein. Oncogene. 1997;15:2939–2949. doi: 10.1038/sj.onc.1201457. [DOI] [PubMed] [Google Scholar]
- 5.Biedenkapp H, Borgmeyer U, Sippel A E, Klempnauer K H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988;335:835–837. doi: 10.1038/335835a0. [DOI] [PubMed] [Google Scholar]
- 6.Bies J, Nazarov V, Wolff L. Identification of protein instability determinants in the carboxy-terminal region of c-Myb removed as a result of retroviral integration in murine monocytic leukemias. J Virol. 1999;73:2038–2044. doi: 10.1128/jvi.73.3.2038-2044.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle W J, Lampert M A, Lipsick J S, Baluda M A. Avian myeloblastosis virus and E26 virus oncogene products are nuclear proteins. Proc Natl Acad Sci USA. 1984;81:4265–4269. doi: 10.1073/pnas.81.14.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burk O, Worpenberg S, Haenig B, Klempnauer K H. Tom-1, a novel v-Myb target gene expressed in Amv- and E26-transformed myelomonocytic cells. EMBO J. 1997;16:1371–1380. doi: 10.1093/emboj/16.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C H, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen R H, Fields S, Lipsick J S. Dissociation of transcriptional activation and oncogenic transformation by v-Myb. Oncogene. 1995;11:1771–1779. [PubMed] [Google Scholar]
- 11.Chen R-H, Lipsick J S. Differential transcriptional activation by v-Myb and c-Myb in animal cells and Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:4423–4431. doi: 10.1128/mcb.13.7.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai P, Akimaru H, Tanaka Y, Hou D X, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 13.Dash A B, Orrico F C, Ness S A. The EVES motif mediates both intermolecular and intramolecular regulation of c-Myb. Genes Dev. 1996;10:1858–1869. doi: 10.1101/gad.10.15.1858. [DOI] [PubMed] [Google Scholar]
- 14.Dini P W, Eltman J T, Lipsick J S. Mutations in the DNA-binding and transcriptional activation domains of v-Myb cooperate in transformation. J Virol. 1995;69:2515–2524. doi: 10.1128/jvi.69.4.2515-2524.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dini P W, Lipsick J S. Oncogenic truncation of the first repeat of c-Myb decreases DNA binding in vitro and in vivo. Mol Cell Biol. 1993;13:7334–7348. doi: 10.1128/mcb.13.12.7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubendorff, J., and J. Lipsick. Transcriptional regulation by the carboxyl terminus of c-Myb depends upon both the Myb DNA-binding domain and the DNA recognition site. Oncogene, in press. [DOI] [PubMed]
- 17.Dubendorff J W, Whittaker L J, Eltman J T, Lipsick J S. Carboxy-terminal elements of c-Myb negatively regulate transcriptional activation in cis and in trans. Genes Dev. 1992;6:2524–2535. doi: 10.1101/gad.6.12b.2524. [DOI] [PubMed] [Google Scholar]
- 18.Duprey S P, Boettiger D. Developmental regulation of c-myb in normal myeloid progenitor cells. Proc Natl Acad Sci USA. 1985;82:6937–6941. doi: 10.1073/pnas.82.20.6937. . (Erratum, 83:2281, 1986.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evan G I, Lewis G K, Bishop J M. Isolation of monoclonal antibodies specific for products of avian oncogene myb. Mol Cell Biol. 1984;4:2843–2850. doi: 10.1128/mcb.4.12.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrao P, Macmillan E M, Ashman L K, Gonda T J. Enforced expression of full-length c-Myb leads to density-dependent transformation of murine haemopoietic cells. Oncogene. 1995;11:1631–1638. [PubMed] [Google Scholar]
- 21.Fu S L, Lipsick J S. Constitutive expression of full-length c-Myb transforms avian cells characteristic of both the monocytic and granulocytic lineages. Cell Growth Differ. 1997;8:35–45. [PubMed] [Google Scholar]
- 22.Fu S-L, Lipsick J S. FAETL motif required for leukemic transformation by v-Myb. J Virol. 1996;70:5600–5610. doi: 10.1128/jvi.70.8.5600-5610.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Fu, S.-L., and J. S. Lipsick. Unpublished data.
- 23.Ganter B, Fu S, Lipsick J. D-type cyclins repress transcriptional activation by the v-Myb but not the c-Myb DNA-binding domain. EMBO J. 1998;17:255. doi: 10.1093/emboj/17.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganter, B., and J. Lipsick. Myb and oncogenesis. Adv. Cancer Res., in press. [DOI] [PubMed]
- 25.Garcia A, LaMontagne K, Reavis D, Stober-Grasser U, Lipsick J S. Determinants of sequence-specific DNA-binding by p48v-myb. Oncogene. 1991;6:265–273. [PubMed] [Google Scholar]
- 26.Ghattas I R, Sanes J R, Majors J E. The encephalomyocarditis virus internal ribosome entry site allows efficient coexpression of two genes from a recombinant provirus in cultured cells and in embryos. Mol Cell Biol. 1991;11:5848–5859. doi: 10.1128/mcb.11.12.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonda T J, Buckmaster C, Ramsay R G. Activation of c-myb by carboxy-terminal truncation: relationship to transformation of murine haemopoietic cells in vitro. EMBO J. 1989;8:1777–1783. doi: 10.1002/j.1460-2075.1989.tb03571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonda T J, Ramsay R G, Johnson G R. Murine myeloid cell lines derived by in vitro infection with recombinant c-myb retroviruses express myb from rearranged vector proviruses. EMBO J. 1989;8:1767–1775. doi: 10.1002/j.1460-2075.1989.tb03570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grässer F A, Graf T, Lipsick J S. Protein truncation is required for the activation of the c-myb proto-oncogene. Mol Cell Biol. 1991;11:3987–3996. doi: 10.1128/mcb.11.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hegde S P, Kumar A, Kurschner C, Shapiro L H. c-Maf interacts with c-Myb to regulate transcription of an early myeloid gene during differentiation. Mol Cell Biol. 1998;18:2729–2737. doi: 10.1128/mcb.18.5.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ibanez C E, Garcia A, Stober-Grässer U, Lipsick J S. DNA-binding activity associated with the v-myb oncogene product is not sufficient for transformation. J Virol. 1988;62:4398–4402. doi: 10.1128/jvi.62.11.4398-4402.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibanez C E, Lipsick J S. trans activation of gene expression by v-myb. Mol Cell Biol. 1990;10:2285–2293. doi: 10.1128/mcb.10.5.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Introna M, Golay J, Frampton J, Nakano T, Ness S A, Graf T. Mutations in v-myb alter the differentiation of myelomonocytic cells transformed by the oncogene. Cell. 1990;63:1289–1297. doi: 10.1016/0092-8674(90)90424-d. [DOI] [PubMed] [Google Scholar]
- 34.Introna M, Luchetti M, Castellano M, Arsura M, Golay J. The myb oncogene family of transcription factors: potent regulators of hematopoietic cell proliferation and differentiation. Semin Cancer Biol. 1994;5:113–124. [PubMed] [Google Scholar]
- 35.Jiang W, Kanter M R, Dunkel I, Ramsay R G, Beemon K L, Hayward W S. Minimal truncation of the c-myb gene product in rapid-onset B-cell lymphoma. J Virol. 1997;71:6526–6533. doi: 10.1128/jvi.71.9.6526-6533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanei-Ishii C, MacMillan E M, Nomura T, Sarai A, Ramsay R G, Aimoto S, Ishii S, Gonda T J. Transactivation and transformation by Myb are negatively regulated by a leucine-zipper structure. Proc Natl Acad Sci USA. 1992;89:3088–3092. doi: 10.1073/pnas.89.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katz S, Kowenz-Leutz E, Muller C, Meese K, Ness S A, Leutz A. The NF-M transcription factor is related to C/EBP beta and plays a role in signal transduction, differentiation and leukemogenesis of avian myelomonocytic cells. EMBO J. 1993;12:1321–1332. doi: 10.1002/j.1460-2075.1993.tb05777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katzen A L, Jackson J, Harmon B P, Fung S, Ramsay G, Bishop J M. Drosophila myb is required for the G2/M transition and maintenance of diploidy. Genes Dev. 1998;12:831–843. doi: 10.1101/gad.12.6.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klempnauer K H, Symonds G, Evan G I, Bishop J M. Subcellular localization of proteins encoded by oncogenes of avian myeloblastosis virus and avian leukemia virus E26 and by chicken c-myb gene. Cell. 1984;37:537–547. doi: 10.1016/0092-8674(84)90384-2. [DOI] [PubMed] [Google Scholar]
- 39a.Ladendorff, N., and J. S. Lipsick. Submitted for publication.
- 40.Lane T, Ibanez C, Garcia A, Graf T, Lipsick J. Transformation by v-myb correlates with trans-activation of gene expression. Mol Cell Biol. 1990;10:2591–2598. doi: 10.1128/mcb.10.6.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leutz A, Damm K, Sterneck E, Kowenz E, Ness S, Frank R, Gausepohl H, Pan Y C, Smart J, Hayman M, et al. Molecular cloning of the chicken myelomonocytic growth factor (cMGF) reveals relationship to interleukin 6 and granulocyte colony stimulating factor. EMBO J. 1989;8:175–181. doi: 10.1002/j.1460-2075.1989.tb03362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leverson J, Koskinen P, Orrico F, Rainio E, Jalkanen K, Dash A, Eisenman R, Ness S. Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol Cell. 1998;2:417–425. doi: 10.1016/s1097-2765(00)80141-0. [DOI] [PubMed] [Google Scholar]
- 43.Lipsick J S. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- 44.Lipsick J S, Ibanez C E. env-encoded residues are not required for transformation by p48v-myb. J Virol. 1987;61:933–936. doi: 10.1128/jvi.61.3.933-936.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lipsick J S, Ibanez C E, Baluda M A. Expression of molecular clones of v-myb in avian and mammalian cells independently of transformation. J Virol. 1986;59:267–275. doi: 10.1128/jvi.59.2.267-275.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J L, Klein P A, Moscovici M G, Moscovici C. Monoclonal antibodies recognizing normal and retrovirus-transformed chicken hematopoietic cells. Virology. 1992;189:583–591. doi: 10.1016/0042-6822(92)90581-9. [DOI] [PubMed] [Google Scholar]
- 47.Mandi Y, Veromaa T, Baranji K, Miczak A, Beladi I. Granulocyte-specific monoclonal antibody inhibiting cytotoxicity reactions in the chicken. Immunobiology. 1987;174:292–299. doi: 10.1016/s0171-2985(87)80004-9. [DOI] [PubMed] [Google Scholar]
- 48.Metz T, Graf T. Fusion of the nuclear oncoproteins v-Myb and v-Ets is required for the leukemogenicity of E26 virus. Cell. 1991;66:95–105. doi: 10.1016/0092-8674(91)90142-l. [DOI] [PubMed] [Google Scholar]
- 49.Nazarov V, Wolff L. Novel integration sites at the distal 3′ end of the c-myb locus in retrovirus-induced promonocytic leukemias. J Virol. 1995;69:3885–3888. doi: 10.1128/jvi.69.6.3885-3888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ness S A, Kowenz-Leutz E, Casini T, Graf T, Leutz A. Myb and NF-M: combinatorial activators of myeloid genes in heterologous cell types. Genes Dev. 1993;7:749–759. doi: 10.1101/gad.7.5.749. [DOI] [PubMed] [Google Scholar]
- 51.Ness S A, Marknell A, Graf T. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell. 1989;59:1115–1125. doi: 10.1016/0092-8674(89)90767-8. [DOI] [PubMed] [Google Scholar]
- 52.Nomura T, Sakai N, Sarai A, Sudo T, Kanei-Ishii C, Ramsay R G, Favier D, Gonda T J, Ishii S. Negative autoregulation of c-Myb activity by homodimer formation through the leucine zipper. J Biol Chem. 1993;268:21914–21923. [PubMed] [Google Scholar]
- 53.Nuchprayoon I, Simkevich C P, Luo M, Friedman A D, Rosmarin A G. GABP cooperates with c-Myb and C/EBP to activate the neutrophil elastase promoter. Blood. 1997;89:4546–4554. [PubMed] [Google Scholar]
- 54.Oelgeschlager M, Janknecht R, Krieg J, Schreek S, Luscher B. Interaction of the co-activator CBP with Myb proteins: effects on Myb-specific transactivation and on the cooperativity with NF-M. EMBO J. 1996;15:2771–2780. [PMC free article] [PubMed] [Google Scholar]
- 55.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 56.Sakura H, Kanei-Ishii C, Nagase T, Nakagoshi H, Gonda T J, Ishii S. Delineation of three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc Natl Acad Sci USA. 1989;86:5758–5762. doi: 10.1073/pnas.86.15.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 58.Shen-Ong G L. The myb oncogene. Biochim Biophys Acta. 1990;1032:39–52. doi: 10.1016/0304-419x(90)90011-o. [DOI] [PubMed] [Google Scholar]
- 59.Sleeman J P. Xenopus A-myb is expressed during early spermatogenesis. Oncogene. 1993;8:1931–1941. [PubMed] [Google Scholar]
- 60.Tanaka Y, Nomura T, Ishii S. Two regions in c-myb proto-oncogene product negatively regulating its DNA-binding activity. FEBS Lett. 1997;413:162–168. doi: 10.1016/s0014-5793(97)00900-9. [DOI] [PubMed] [Google Scholar]
- 61.Tavner F J, Simpson R, Tashiro S, Favier D, Jenkins N A, Gilbert D J, Copeland N G, Macmillan E M, Lutwyche J, Keough R A, Ishii S, Gonda T J. Molecular cloning reveals that the p160 Myb-binding protein is a novel, predominantly nucleolar protein which may play a role in transactivation by Myb. Mol Cell Biol. 1998;18:989–1002. doi: 10.1128/mcb.18.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomita A, Watanabe T, Kosugi H, Ohashi H, Uchida T, Kinoshita T, Mizutani S, Hotta T, Murate T, Seto M, Saito H. Truncated c-Myb expression in the human leukemia cell line TK-6. Leukemia. 1998;12:1422–1429. doi: 10.1038/sj.leu.2401113. [DOI] [PubMed] [Google Scholar]
- 63.Westin E H, Gallo R C, Arya S K, Eva A, Souza L M, Baluda M A, Aaronson S A, Wong-Staal F. Differential expression of the amv gene in human hematopoietic cells. Proc Natl Acad Sci USA. 1982;79:2194–2198. doi: 10.1073/pnas.79.7.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weston K, Bishop J M. Transcriptional activation by the v-myb oncogene and its cellular progenitor, c-myb. Cell. 1989;58:85–93. doi: 10.1016/0092-8674(89)90405-4. [DOI] [PubMed] [Google Scholar]
- 65.Woo C H, Sopchak L, Lipsick J S. Overexpression of an alternatively spliced form of c-Myb results in increases in transactivation and transforms avian myelomonoblasts. J Virol. 1998;72:6813–6821. doi: 10.1128/jvi.72.8.6813-6821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]