Abstract
High-risk human papillomaviruses (HPVs) are the causative agents of certain human cancers. HPV type 16 (HPV16) is the papillomavirus most frequently associated with cervical cancer in women. The E6 and E7 genes of HPV are expressed in cells derived from these cancers and can transform cells in tissue culture. Animal experiments have demonstrated that E6 and E7 together cause tumors. We showed previously that E6 and E7 together or E7 alone could induce skin tumors in mice when these genes were expressed in the basal epithelia of the skin. In this study, we investigated the role that the E6 gene plays in carcinogenesis. We generated K14E6 transgenic mice, in which the HPV16 E6 gene was directed in its expression by the human keratin 14 promoter (hK14) to the basal layer of the epidermis. We found that E6 induced cellular hyperproliferation and epidermal hyperplasia and caused skin tumors in adult mice. Interestingly, the tumors derived from E6 were mostly malignant, as opposed to the tumors from E7 mice, which were mostly benign. This result leads us to hypothesize that E6 may contribute differently than E7 to HPV-associated carcinogenesis; whereas E7 primarily contributes to the early stages of carcinogenesis that lead to the formation of benign tumors, E6 primarily contributes to the late stages of carcinogenesis that lead to malignancy.
Human papillomaviruses (HPVs) are small DNA tumor viruses that cause papillomas in human skin, genitalia, and upper respiratory tract. Certain types of HPVs, referred to as high-risk HPVs, are associated with malignant tumors (55). More than 90% of cervical carcinomas, for example, are related to HPV infections (11). In these cancer cells, HPV genomes commonly are found integrated into the cellular genome (11, 41). The integration events leave intact the early region of the HPV genome containing the E6 and E7 genes (41, 53). Transcripts of the E6 and E7 genes are detected in the cervical cancer-derived cell lines (47, 52), indicating these two genes potentially play a role in HPV-associated carcinogenesis. Cell culture experiments have demonstrated that E6 and E7 possess transforming activities; E6 or E7 each can transform established cells; E6 is also found to immortalize primary human mammary epithelial cells (3), whereas E7 is sufficient to immortalize primary human keratinocytes (22).
The discovery of cellular targets of E6 and E7 provided insight into the potential molecular mechanisms by which E6 and E7 cause cellular transformation. E6 can associate with p53 through the E6 associate protein, leading to degradation of p53 by the ubiquitin proteasome pathway (39, 40, 49). Recently, other cellular factors, such as E6 binding protein (9), paxillin (46), the human homologue of Drosophila large disc tumor suppressor gene (29, 31), and E6TP1 (18), have been reported to associate with E6. Whether and how these interactions contribute to carcinogenesis is not yet clear. E7 can also associate with multiple cellular factors, one of which is the retinoblastoma tumor susceptibility gene product, pRb (8, 16). Binding of E7 to pRb promotes degradation of pRb and leads to the inhibition of pRb functions (5, 26). Other proteins that associate with E7 are prominently related to the regulation of the cell cycle, such as cyclins/cyclin-dependent kinase inhibitors (17, 25, 54) and other members of the Rb gene family (12, 15). As many of the cellular factors that E6 and E7 interact with are either tumor suppressors or cell cycle regulators, it is not surprising that E6 and E7 can cause fundamental changes in cell growth behavior.
Cells transformed by E6 or E7 rarely grow into tumors when transplanted into nude mice, indicating that other molecular events must be required for the transformed cells to become tumorigenic (27, 35, 51). This observation is consistent with the epidemiological data from human cancers in that high-risk HPV infections require long latency periods before their associated cervical cancers develop. To test whether E6 and E7 are oncogenic in vivo, experiments using transgenic mice have been conducted. E6 and E7 together cause tumors in mice (2, 10, 20). To dissect the roles of E6 and E7 in HPV carcinogenesis, we generated transgenic mice in which the HPV type 16 (HPV16) E7 gene was expressed in the epidermis by the human keratin 14 (hK14) promoter. These mice had multiple phenotypes, including epidermal hyperplasia and skin tumors (23). Thus, HPV16 E7 alone causes tumorigenesis in animals. In the present study, we have addressed the role of E6 in HPV-induced carcinogenesis. We generated K14E6 transgenic mice in which the HPV16 E6 gene was expressed in the basal layer of epithelia, using the hK14 promoter. Expression of E6 increased cell proliferation and induced epidermal hyperplasia. Skin tumors developed in adult K14E6 mice with an incidence of about 7% at 1 year of age. In contrast to the tumors derived from K14E7 transgenic mice, which were primarily benign, tumors derived from K14E6 transgenic mice were mostly malignant, indicating that E6 alone not only is sufficient to induce tumors but may contribute to the development of malignancy in animals.
MATERIALS AND METHODS
Construction of transgene and generation of transgenic mouse.
The K14E6 transgene was constructed similarly to the K14E7 transgene (23). Briefly, a DNA fragment from the HPV genome spanning nucleotides 79 to 883 and encompassing the E6 and E7 open reading frames (ORFs) was PCR amplified with primers containing BamHI sites. In this amplified DNA fragment, a translational termination linker (TTL) is present in the early region of the E7 ORF, precluding expression of E7. The BamHI fragment was inserted into the unique BamHI site between the hK14 promoter and hK14 polyadenylation sequences in plasmid pG1Z-K14 to generate plasmid pK14HPV16E6. The construct was sequenced to verify the intact state of the E6 ORF. The presence of TTL in the E7 ORF was verified by presence of an engineered HpaI site in the linker. This recombinant plasmid was digested with HindIII and EcoRI to release a 3.2-kbp fragment that contains hK14 promoter, HPV16 sequences, and the K14 polyadenylation sequences. The fragment was purified by gel electrophoresis and microinjected into fertilized mouse FVB/N eggs as described previously (20, 24). Mice born from these eggs were screened for the presence of transgene in their genome by Southern analysis of genomic DNA prepared from tail biopsies. The hybridization probe was the approximately 800-bp HPV16-specific DNA fragment released from plasmid pK14HPV16E6 by digestion with BamHI; it was 32P labeled by random primer extension. To estimate the copy number of each of the transgenic lineages, standard DNA of plasmid pK14HPV16E6 equal in amount to 1, 10, or 20 copies per mouse genome was included in the Southern analysis. The blot was quantified with a Molecular Dynamics PhosphorImager. Multiple lineages of K14E6 mice were bred in the Association for Assessment and Accreditation of Laboratory Animal Care-approved animal facilities in the McArdle Laboratory for Cancer Research. All offspring were screened by Southern analysis or PCR.
Analysis of transgene expression by in situ hybridization.
Eight-day-old and six-week-old mice were sacrificed, and skin and ear samples were collected. The samples were fixed in buffered formalin, embedded in paraffin, and cut into 5-μm-thick sections. In situ hybridization was performed as described previously (21). Sense and antisense E6 cRNA probes were transcribed in vitro from a plasmid containing the HPV16 E6 and E7 ORFs downstream of SP6, and both [35S]UTP and [35S]CTP were incorporated. Photographic emulsion-coated slides were kept at −20°C for 2 weeks before development. The hybridization signals were examined by dark- or bright-field microscopy.
Immunohistochemistry for BrdU, PCNA, and K14.
Skin sections from torso skin of 8-day-old mice or from the ears of 6-week-old mice were deparaffinized in xylenes and rehydrated in graded alcohol and phosphate-buffered saline (PBS). Endogenous peroxidase was quenched by treatment of skin sections with 3% hydrogen peroxide for 15 min. For detection of 5′-bromo-2′-deoxyuridine (BrdU), the mice were pulse-labeled with BrdU (100 μg/g of body weight; Sigma catalog no. B-5002) 1 h before sacrifice, and tissue sections were stained by the protocol provided with the BrdU staining kit (catalog no. HCS24; Oncogene Research Products, Calbiochem). Briefly, tissue sections were digested with trypsin and treated with a denaturing solution. After incubation with biotinylated mouse anti-BrdU antibody and then streptavidin-peroxidase, the slides were exposed to the peroxidase substrate (diaminobenzidine) mixture for 5 min and counterstained with hematoxylin. To compare the proliferation index of 8-day-old skin among lineages, the total numbers of cells and the number of BrdU-positive cells in the epidermis were counted in 30 randomly selected microscopic fields (magnification of ×400) of skin sections from three mice.
For proliferating cell nuclear antigen (PCNA) detection, tissue sections were blocked with 5% nonfat dry milk–PBS and/or 5% normal goat serum for 30 min, mouse monoclonal anti-mouse PCNA antibody (Boehringer Mannheim), diluted 1:200 in 5% milk–PBS, was added, and the the mixture was incubated for 3 h at room temperature. After incubation with peroxidase-labeled anti-mouse secondary antibody (30 min) and then with Vectastain ABC reagents (30 min), the slides were exposed to diaminobenzidine substrate. The slides were counterstained with fast green and examined for nuclear staining of PCNA. Mouse K14 staining was performed similarly except that the slides were incubated with 1:500-diluted rabbit polyclonal antibody against mouse K14 (catalog no. PRB-155P-100; BAbco, Richmond, Calif.) for 1 h, and the slides were counterstained with hematoxylin.
Monitoring and statistical analysis of skin tumors.
Mice were checked every 2 weeks for 15 months to monitor the development of skin tumors. At time of animal sacrifice, part of tumor tissue was fixed in buffered formalin for histological analysis; another part was frozen and kept in −20°C for preparation of genomic DNA. Fixed tissue was embedded in paraffin and cut into 5-μm sections for staining with hematoxylin and eosin. Tumor type was determined by histological analysis. The incidence of tumors from different lineages was analyzed for statistical difference by the chi square test.
Analysis of H-ras mutations with PCR and RFLP.
H-ras mutations were analyzed by PCR and restriction fragment length polymorphism RFLP. The DNA fragments encompassing codons 12 and 13 (collectively referred to as codon 12/13) (GGAGGC) or codon 61 (CAA) were amplified via PCR from tumor genomic DNA with two pairs of primers. The primers for codon 12/13 were 5′-GGGTCAGGCATCTATTAGCCG and 5′-CCAGCCTACACCCTTGCACCTC; primers for codon 61 were CTCCTACCGGAAACAGGTGGTC and 5′-GCTAGCCATAGGTGGCTCACC. Activated H-ras mutations commonly are G-to-A transitions or G-to-T transversions at the second position of codon 12 or 13 (6, 33). Transition mutations at codon 61, from CAA to CTA or CAT or GAA, are also frequently involved in H-ras activation, especially in some chemically induced tumors (7). These mutations create new restriction sites. At codon 12, GGA-to-GTA and GGA-to-GAA changes can be detected by digestion with SfcI and Eco57I, respectively; at codon 13, GGC-to-GTA and GGC-to-GAC changes can be detected by digestion with MaeII and HinfI, respectively; at codon 61, CAA-to-CTA and CAT-or-GAA changes can be detected by digestion with XbaI, BspHI or TaqI. PCR products digested with these enzymes were run on 3% Metaphor agarose gels and examined visually by ethidium bromide staining.
Analysis of DNA damage responses.
Eight-day-old mice from line 5718 and line 5737 were irradiated with γ rays from a (137Cs) source at a dose rate of 3.1 Gy/min. A total dose of 5 Gy was delivered to the whole body of each mouse individually. Three mice from each lineage were sacrificed at each time point of 4, 24, or 48 h following irradiation. Unirradiated mice were used as controls. Mice were injected with BrdU 1 h before sacrifice, similarly as described above. Skin samples were obtained from the dorsal area, fixed in 10% buffered formalin. Paraffin-embedded tissue sections were stained for BrdU. Total epidermal cells and BrdU-positive epidermal cells in 10 microscopic fields of skin section from each mouse were counted. The average percentage of BrdU-positive cells and standard deviation were calculated from the data generated from three mice for each time point.
RESULTS
Generation of K14E6 transgenic mice.
To direct expression of HPV-16 E6 to the basal layer of squamous epithelia, we cloned a fragment of HPV16 DNA sequence from HPV16 nucleotides 79 to 883 that contains both the E6 and E7 ORFs behind the hK14 promoter. A TTL was inserted into the 5′ region of E7 gene so that translation of E7 is disrupted. The 3.2-kbp recombinant DNA fragment containing the intact HPV16 E6 gene flanked by hK14 promoter and hK14 polyadenylation sequences was injected into fertilized eggs from inbred FVB mice. Of the 18 mice born, 5 were positive for the K14E6 transgene, based on Southern analysis. The F0 mice were bred to establish five lines of E6 transgenic mice (Table 1).
TABLE 1.
Phenotypes of K14E6 transgenic lineages
| Line | Transgene copy no. | Skin thickening | Cataract | Testicular degenerative syndrome | Skin tumor incidence by 15 mo | % Malignancya |
|---|---|---|---|---|---|---|
| 5718 | 2 | − | − | − | 0/129 | |
| 5719 | 1 | − | − | − | NDb | |
| 5737 | 15 | + | + | − | 26/180 | 75 |
| 5738 | 2 | − | − | − | 0/93 | |
| 5743 | 20 | + | + | + | 7/73 | 57 |
Percentage of malignant tumors in skin tumors derived from lines 5737 and 5743, based on histopathological screening of tumors.
ND, not done.
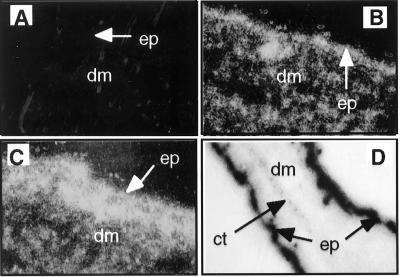
To determine whether E6 was expressed specifically in the epidermis, we examined the pattern of E6 mRNA expression by in situ hybridization with a probe specific for E6 mRNA. E6 mRNA was detected in the epidermis in both low- and high-copy-number transgenic mice (Fig. 1B and C). Lower levels of E6 expression were also detected in the underlying dermis, but restricted primarily to the hair follicles. Epidermis from the ear demonstrated high levels of mRNA of E6. There was an absence of signal in the dermis, which in the ear has a paucity of hair follicles (Fig. 1D).
FIG. 1.
Examples of skin sections analyzed for E6 mRNA by in situ hybridization with a specific antisense RNA probe. (A to C) Dark-field images (magnification, ×400) of skin sections from nontransgenic (A), K14E6 transgenic line 5718 (B) and K14E6 transgenic line 5737 (C) mice. Bright areas indicate strong hybridization to an E6-specific probe. Note that signal is brightest in the epidermis (ep) but also present in underlying epithelial structures (hair follicles) in the dermis (dm). (D) Bright-field image (×100) of an ear section from K14E6 line 5737. Dark areas in the epidermis of both sides of the ear indicate strong hybridization to the E6-specific probe. Note absence of signal in the underlying dermis and cartilage (ct).
Phenotypes of K14E6 transgenic mice and hyperproliferation of E6-expressing cells.
All founder mice and their offspring were carefully examined for overt and microscopic phenotypes. Of the five K14E6 transgenic lineages, lines 5737 and 5743, which have high copy numbers of the transgene per cells, developed overt phenotypes, including thickened ears and cataracts in the eye (Table 1). All mice in these lines, including founders, developed these overt phenotypes.
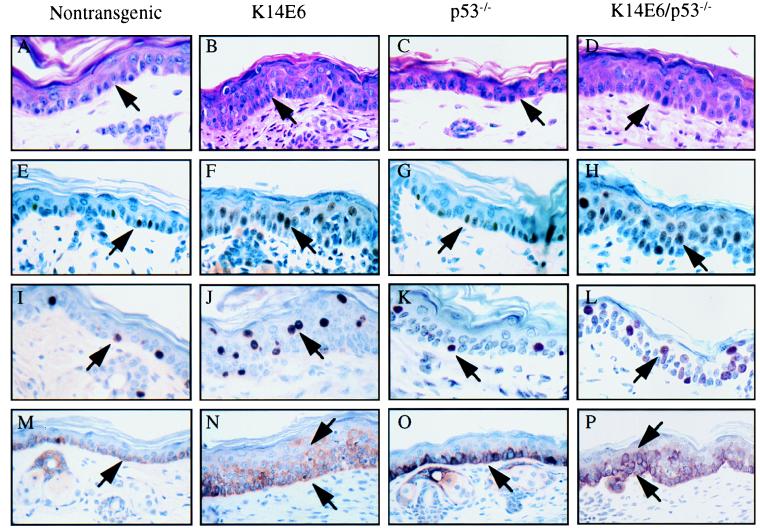
Histologically, the thickening of the ear was due to the thickening of epidermis (Fig. 2A and B). The epidermis of the normal (i.e., nontransgenic) adult mouse contains no more than two layers of nucleated cells beneath a keratinized layer. The basal cells in the stratum basale are normally well organized and are the only cells positively stained for keratin 14 (Fig. 2M). In the K14E6 transgenic mice, there was an expansion in the number of layers positively stained for K14 (Fig. 2N). To determine whether E6 induces hyperproliferation in the epidermis, we monitored nuclear staining for PCNA, a marker for cell proliferation. Compared with nontransgenic mice, the number of PCNA-positive cells in the epidermis from the ear of the K14E6 transgenic mice was significantly increased, and PCNA-positive cells were not restricted to the basal layer but included suprabasal cells (Fig. 2E and F). To test whether the suprabasal cells were actively synthesizing DNA, we pulse-labeled the mice with BrdU before sacrifice and measured BrdU incorporation immunohistochemically. As expected, BrdU-positive cells were seen only in the stratum basale of the epidermis in the nontransgenic mice. In the K14E6 transgenic epidermis of the ear, however, many BrdU-positive cells were also seen in the suprabasal compartment. All nucleated cell layers in the K14E6 epidermis had BrdU-positive cells, indicating that these cells were aberrantly passing through S phase (Fig. 2I and J). The proliferation index of the epidermis from skin taken from the torso was also measured by using the same BrdU incorporation assay. The percentage of BrdU-positive cells in the torso epidermis was significantly higher in K14E6 transgenic mice than in the nontransgenic mice (Table 2). An increase in the proliferation index was observed in the skin from both low-copy-number (line 5718) and high-copy-number (line 5737) mice, but it was statistically significant only in the latter mice. The ability of E6 to promote cell proliferation seemed to be p53 independent, as the epidermis from p53-knockout mice did not display an increase in the BrdU labeling index in body skin (Table 2).
FIG. 2.
Histological and immunohistochemical analysis of epidermis from the ears of 6-week-old nontransgenic, K14E6 transgenic line 5737, p53-null, and E6/p53-null mice. All tissue sections were paraffin embedded. (A to D) Hematoxylin-and-eosin-stained sections of the ear at high magnification (×200). The epidermis is indicated by the arrow. Note thickening of the epidermis in the ears of K14E6 and K14E6/p53-null mice. Ear sections were stained immunohistochemically for PCNA (E to H) and BrdU (I to L). Positive cells are indicated by arrows. Note increased numbers of PCNA-positive and BrdU-positive cells and their appearance in the suprabasal portion of the epidermis in both K14E6 mice and K14E6/p53-null mice. In panels M to P, ear sections were stained for K14. Note thickening of the K14-positive portion of the epidermis in the K14E6 and K14E6/p53-null ear sections.
TABLE 2.
Proliferation index of epidermis from different mouse strains
| Mice strain | Avg % BrdU-positive cells ± SDa |
|---|---|
| FVB (nontransgenic) | 4.51 ± 0.26 |
| K14E6 line 5718 | 5.39 ± 1.19 |
| K14E6 line 5737 | 7.93 ± 1.54 |
| FVB p53 null | 4.48 ± 0.72 |
Data were generated from 30 microscopic fields on tissue sections of three mice from each mouse strain.
To determine further whether E6-induced thickening of the epidermis is due to E6’s p53-independent activities, we generated K14E6 transgenic/p53-null mice and compared their phenotypes with that of p53-null mice. The epidermis from p53-null mice was not different from that of nontransgenic mice (Fig. 2C, G, K, and O). In contrast, the epidermis from K14E6/p53-null mice was thickened, the K14-positive compartment was expanded, and there was an increase in the number of PCNA- and BrdU-positive cells, including their abundance in the suprabasal compartment (Fig. 2D, H, L, and P). These data indicate that E6 activities other than its inactivation of p53 contribute to its induction of epithelial hyperplasia.
Histological analysis of the eyes from K14E6 mice indicated that the cataracts in their eyes resulted from the existence of nucleated cells in the interior of the lens (data not shown). Preliminary evidence indicates that the K14E6 transgene was expressed in the transitional zone of the lens epithelium (33a). BrdU incorporation assays demonstrated that some of the nucleated cells in the interior of the lens were able to synthesize DNA. Similar phenotypes were also seen in the lens of the K14E6/p53-null mice but not in the lens of p53-null mice.
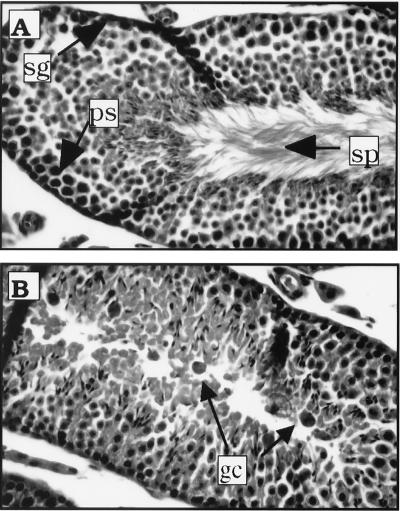
One line of K14E6 mice (line 5743) showed a testicular degenerative syndrome. Mice from this lineage were initially found difficult to breed. Their litter sizes were small, and only some of the male mice were fertile. We examined the tissue section of testes from these mice and found that the seminiferous tubules in these sections were structurally normal but lacked mature sperm cells in the seminiferous lumen. Instead, many giant cells with dark nuclei were observed inside the seminiferous tubules (Fig. 3). These cells are hallmarks of a degenerative testicular syndrome. Interestingly, this syndrome occurs in some p53-knockout mice and mice with reduced levels of p53 protein (37). Loss of p53 function in the mice from the K14E6 lineage may be responsible, therefore, for the development of this syndrome.
FIG. 3.
Histological analysis of mouse testes. Normal mature sperm cells (sp), diploid spermatogonia stem cells (sg), and 4N spermatocytes in the seminiferous tubules from nontransgenic mice are indicated by arrows (A). Many giant cells (gc), not mature sperm cells, are seen in seminiferous tubules from K14E6 transgenic line 5743 mice (B).
HPV16 E6 induces primarily malignant skin tumors.
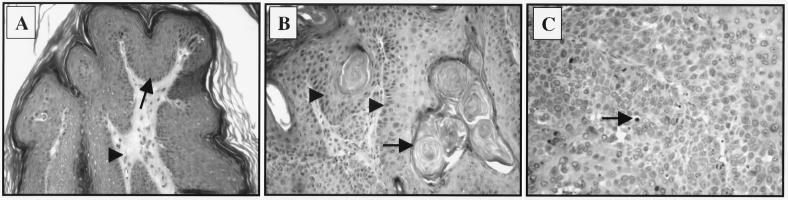
To determine whether E6 alone is sufficient to induce tumors in vivo, mice from each K14E6 transgenic lineage were monitored closely for tumorigenesis over their life span. The two lines with high copy numbers of the E6 transgene, line 5737 and 5743, developed skin tumors with incidences about 14 and 10% at 15 months of age, respectively (Table 1). All tumors arose on the torso and limbs of the mice. We did not observe any skin tumors in the nontransgenic mice in the same period of time. The first onset of tumor was at 6 months of age; most tumors, however, developed in late adult life, indicating that a long latency is required. Interestingly, majority of these tumors showed signs of being malignant, as evidenced by their open and invasive characteristics. Upon histopathological examination, these malignant tumors were diagnosed as grade I, II, or III epidermoid carcinomas (Fig. 4). Among 24 tumors examined, 6 were papillomas and 5 were grade I, 11 were grade II, and 2 were grade III epidermoid carcinomas.
FIG. 4.
Histology of skin tumors derived from K14E6 transgenic mice. The K14E6 mice developed both benign and malignant skin tumors. (A) Section of papilloma, a benign outgrowth of skin with epidermal hyperplasia. The morphology of cells are relatively normal and the basement membrane in this lesion is intact (arrow). The epidermis and underlying dermis (arrowhead) are clearly demarcated. (B) Epidermoid carcinoma grade I. The cells form a concentric pattern that is poorly demarcated (arrowheads). In the center of these structures are keratinized and differentiated cells. Note the presence of “pearls” of keratin (arrow), characteristic of well-differentiated epidermoid carcinoma. (C) Example of grade III epidermoid carcinoma. The cells are more anaplastic than in panel B and do not form keratin pearls. These cells are still able to be keratinized individually (arrow).
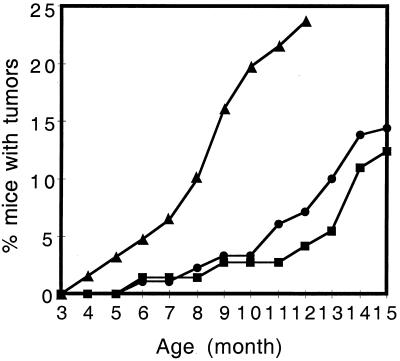
The overt phenotype and tumor incidence appeared to be dependent on gene dosage of E6 transgene, because only the two lines with higher copy number spontaneously developed tumors. To examine the gene dosage effect further, transgenic mice homozygous for the E6 transgene were generated from line 5737 and monitored for tumor development. The mice homozygous for the K14E6 transgene (5737-H) developed skin tumors earlier and with a higher incidence than did mice from the same line hemizygous for the K14E6 transgene (Fig. 5). Tumors in the homozygous line 5737 were seen as early as 4 months of age, and the incidence of skin tumors increased to more than 20% by 1 year of age, compared to 7% in the line 5737 mice hemizygous for the K14E6 transgene. The difference in the incidence of skin tumors in line 5737 hemizygotes and homozygotes was statistically significant (P < 0.01). This result demonstrated that E6 gene dosage strongly influences tumor development in these animals.
FIG. 5.
Skin tumor development over 15 months of life in K14E6 line 5737 (●), line 5743 (■), and line 5737 homozygous (▴) mice. The numbers of mice monitored up to at least 12 months in the three groups were 181, 73, and 80, respectively.
Abrogation of DNA damage response is not sufficient for tumor induction.
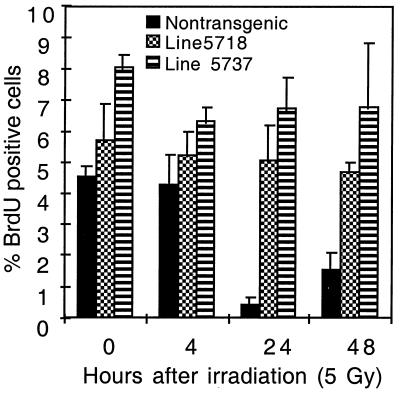
We have reported earlier that E6 can abrogate responses to DNA damage in vivo (45). Abrogation of DNA damage responses may contribute to carcinogenesis. Because tumors developed in the K14E6 mice with high copy numbers of transgene but not in low-copy-number mice, we tested whether there was a similar difference in the abrogation of responses to DNA damage in these different transgenic lineages. Growth arrest was abrogated in the mice of the line with low copy number as efficiently as in the mice with high copy numbers (Fig. 6). Furthermore, induction of levels of p53 protein by ionizing radiation was not detectable in either line (reference 45 and unpublished data). This observation indicated that the responses to DNA damage in the skin must be sensitive to the presence of E6 and therefore were equally abrogated in both lines with high and low copy numbers of the transgene.
FIG. 6.
Abrogation of radiation-induced growth arrest in the epidermis from K14E6 mice with low or high transgene copy number. Shown are percentages of BrdU-positive cells in the epidermis of K14E6 line 5737 mice carrying a high copy number of the transgene, K14E6 line 5718 mice carrying a low copy number of the transgene, and nontransgenic FVB/N mice that were treated or not treated with 5 Gy of ionizing radiation. Provided are the average values for three mice in each group ± 1 standard deviation. The 10-fold reduction in percentage of BrdU-positive cells in nontransgenic FVB/N mice is an indicator of the radiation-induced growth arrest. Note absence of reduction in the percentage of BrdU-positive cells in the epidermis from mice of both lines 5737 and 5718.
H-ras gene mutation does not play a role in E6-induced tumorigenesis.
H-ras mutations are considered to be an initiation event in development of skin tumor, especially in chemically induced tumorigenesis (13, 32). H-ras mutations at codon 61 are predominant events in the dimethylbenzanthracene-induced tumors; most papillomas induced by chemical carcinogens contain A-to-T transversions at this codon (7, 38). Mutations at codon 12/13, especially G-to-T or G-to-A mutations at the second nucleotide of codon 12 or 13, also can activate H-ras and are found in murine experimental skin tumors (33). Earlier investigations have implicated the involvement of ras mutations in the carcinogenesis by HPV in humans (1) or in mice (19); in vitro experiments demonstrated that activated ras cooperates with E6 in cell transformation (4, 34). To learn whether activation of H-ras plays a role in the E6-induced tumor development, we amplified portions of the H-ras gene encompassing either codon 12/13 or codon 61 by PCR. The PCR product was digested with different restriction enzymes to identify activating mutations at these codons which generate new restriction sites. Twenty-four tumors derived from E6 transgenic mice were screened. No mutations at either codon 12/13 or codon 61 were detected.
DISCUSSION
E6 induces cellular hyperproliferation and epidermal hyperplasia.
In the K14E6 mice, the epidermis was thickened. There was an increase in the number of PCNA-positive and BrdU-positive cells in the K14E6 epidermis, and these cells were found not only in the basal layer but also in the suprabasal compartment. A similar observation was made in the lens from these mice, in which the transgene was also found to be expressed. Nucleated cells were present in the center of the lens from these mice, whereas nontransgenic lens lack nucleated cells. PCNA and BrdU staining experiments indicated that some of these lens cells were supporting DNA synthesis (data not shown). The observations made in the skin and lens of K14E6 mice demonstrate that E6 induces cell proliferation. This induction of cell proliferation also was associated with an expansion in the K14-positive compartment in the K14E6 epidermis, indicating that the normal differentiation program is delayed. This delay may explain why other investigations have observed E6-positive human foreskin keratinocytes in tissue culture to be at least partially resistant to signals that normally induce their differentiation (42, 43).
E6 appears to induce hyperproliferation independently of p53 inactivation, because the cellular proliferation index was not increased in the epidermis of p53-null mice. Furthermore, the epidermis of p53-null mice appeared macroscopically and microscopically normal. That the p53-null epidermis is normal conforms with the observation made previously that p53 does not interfere with cell division and differentiation under normal conditions (48). Therefore, the hyperproliferation and the related delay in differentiation in the epidermis of K14E6 mice must be caused, at least in part, by activities of E6 other than its inactivation of p53. This prediction was supported by the data obtained with K14E6/p53-null mice (Fig. 2) in which E6 retained its capacity to induce epithelial hyperplasia. A requirement for p53-independent activities of E6 also has been posited to contribute to E6’s ability to bestow on human foreskin keratinocytes resistance to differentiation in tissue culture.
E6 primarily induces malignant tumors.
Our animal studies demonstrate that E6 performs a different role than E7 in carcinogenesis. Earlier, we found that E7 alone usually induces benign tumors in adult K14E7 mice (23). By contrast, we found E6 alone usually induces malignant tumors in the K14E6 mice (this study). Carcinogenesis is a multistaged process, with multiple genetic events being required for induction of benign tumors and additional genetic events being required for progression of these benign tumors to malignancies. The simplest interpretation of our tumor data is that E7 contributes primarily to the early stages in carcinogenesis required for formation of benign tumors whereas E6 contributes to late stages in carcinogenesis involved in the evolution to malignancy. Interestingly, the activation of telomerase is preferentially detected in cervical carcinomas and a subset of cervical intraepithelial neoplasia grade III lesions (44). E6, but not E7, can activate telomerase (30). Thus, E6’s activation of telomerase may account at least in part for its capacity to induce malignant tumors.
Multiple factors may be involved in E6 carcinogenesis.
A well-recognized cellular target of E6 is p53. Loss of p53 in mice leads to early spontaneous development of tumors (14) and enhances the malignant progression of chemically induced skin tumors (28). Therefore, inactivation of p53 by E6 likely also contributes to E6’s capacity to induce malignant skin tumors.
Inactivation of p53, however, may not be the only mechanism for E6-induced carcinogenesis. The K14E6 mice that developed tumors also displayed hyperplastic changes in the epidermis, a property that, as discussed above, cannot be ascribed to E6’s inactivation of p53. It is likely that the hyperproliferation induced by E6 is important for its carcinogenesis. Another activity that may contribute to E6-associated carcinogenesis is its apparent inhibition of cellular differentiation. Cancer cells usually lack the ability to terminally differentiate. In this study, we found that the suprabasal compartment of the K14E6 transgenic epidermis stained for K14, indicating that E6 may also perturb cellular differentiation; this perturbation was caused by p53-independent activities of E6.
The fact that E6 needs such a long latency to induce tumors indicates that although E6 inhibits functions of p53 and causes hyperplasia, other genetic events must be required for it to cause cancer. Mutations of H-ras have been implicated casually in the development of spontaneous and chemically induced skin tumor. We therefore assayed for mutations at H-ras codons 12, 13, and 61. No mutations were detected, indicating that H-ras mutations are not involved in E6-induced carcinogenesis. Genomic instability is thought to be one of the important events in malignant progression of tumors. In cell culture studies, E6 was demonstrated to induce genomic instability, including aneuploidy and gene amplification, and this induction was ascribed to E6’s ability to inactivate p53 (36, 50). E6’s ability to induce malignant tumors may be related to its capacity to induce genomic instability. We now are in the process of investigating whether there are genomic changes in tumors derived from E6 transgenic mice.
ACKNOWLEDGMENTS
We thank Renee Herber for providing plasmid RLH66.2 containing HPVE6E7ttl, Kathleen Helmuth for technical assistance with microinjection, Amy Liem for assistance with breeding and maintenance of the transgenic lineages, and Jane Weeks, Angie Buehl, and Harlene Edwards for processing tissue sections. We thank Anne Griep for communication of unpublished results and Bill Sugden for critical review of the manuscript.
This study was supported by a grant from the American Cancer Society (VM164) and by grants from the National Institutes of Health (CA22443 and CA07175).
REFERENCES
- 1.Anwar K, Nakakuki K, Naiki H, Inuzuka M. ras gene mutations and HPV infection are common in human laryngeal carcinoma. Int J Cancer. 1993;53:22–28. doi: 10.1002/ijc.2910530106. [DOI] [PubMed] [Google Scholar]
- 2.Arbeit J M, Munger K, Howley P M, Hanahan D. Neuroepithelial carcinomas in mice transgenic with human papillomavirus type 16 E6/E7 ORFs. Am J Pathol. 1993;142:1187–1197. [PMC free article] [PubMed] [Google Scholar]
- 3.Band V, De C J, Delmolino L, Kulesa V, Sager R. Loss of p53 protein in human papillomavirus type 16 E6-immortalized human mammary epithelial cells. J Virol. 1991;65:6671–6676. doi: 10.1128/jvi.65.12.6671-6676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedell M A, Jones K H, Grossman S R, Laimins L A. Identification of human papillomavirus type 18 transforming genes in immortalized and primary cells. J Virol. 1989;63:1247–1255. doi: 10.1128/jvi.63.3.1247-1255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer S N, Wazer D E, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 6.Brown K, Buchmann A, Balmain A. Carcinogen-induced mutations in the mouse c-Ha-ras gene provide evidence of multiple pathways for tumor progression. Proc Natl Acad Sci USA. 1990;87:538–542. doi: 10.1073/pnas.87.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarti D, Pelling J C, Cavalieri E L, Rogan E G. Relating aromatic hydrocarbon-induced DNA adducts and c-H-ras mutations in mouse skin papillomas: the role of apurinic sites. Proc Natl Acad Sci USA. 1995;92:10422–10422. doi: 10.1073/pnas.92.22.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chellappan S, Kraus V B, Kroger B, Munger K, Howley P M, Phelps W C, Nevins J R. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc Natl Acad Sci USA. 1992;89:4549–4553. doi: 10.1073/pnas.89.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J J, Reid C E, Band V, Androphy E J. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science. 1995;269:529–531. doi: 10.1126/science.7624774. [DOI] [PubMed] [Google Scholar]
- 10.Comerford S A, Maika S D, Laimins L A, Messing A, Elsasser H P, Hammer R E. E6 and E7 expression from the HPV 18 LCR: development of genital hyperplasia and neoplasia in transgenic mice. Oncogene. 1995;10:587–597. [PubMed] [Google Scholar]
- 11.Das B C, Sharma J K, Gopalkrishna V, Das D K, Singh V, Gissmann L, zur Hausen H, Luthra U K. A high frequency of human papillomavirus DNA sequences in cervical carcinomas of Indian women as revealed by Southern blot hybridization and polymerase chain reaction. J Med Virol. 1992;36:239–245. doi: 10.1002/jmv.1890360402. [DOI] [PubMed] [Google Scholar]
- 12.Davies R, Hicks R, Crook T, Morris J, Vousden K. Human papillomavirus type 16 E7 associates with a histone H1 kinase and with p107 through sequences necessary for transformation. J Virol. 1993;67:2521–2528. doi: 10.1128/jvi.67.5.2521-2528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiGiovanni J. Multistage carcinogenesis in mouse skin. Pharmacol Ther. 1992;54:63–128. doi: 10.1016/0163-7258(92)90051-z. [DOI] [PubMed] [Google Scholar]
- 14.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 15.Dyson N, Guida P, Munger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyson N, Howley P M, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 17.Funk J O, Waga S, Harry J B, Espling E, Stillman B, Galloway D A. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Q, Srinivasan S, Boyer S N, Wazer D E, Band V. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative GAP protein, E6TP1, and target it for degradation. Mol Cell Biol. 1999;19:733–744. doi: 10.1128/mcb.19.1.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenhalgh D A, Wang X J, Rothnagel J A, Eckhardt J N, Quintanilla M I, Barber J L, Bundman D S, Longley M A, Schlegel R, Roop D R. Transgenic mice expressing targeted HPV-18 E6 and E7 oncogenes in the epidermis develop verrucous lesions and spontaneous, rasHa-activated papillomas. Cell Growth Differ. 1994;5:667–675. [PubMed] [Google Scholar]
- 20.Griep A E, Herber R, Jeon S, Lohse J K, Dubielzig R R, Lambert P F. Tumorigenicity by human papillomavirus type 16 E6 and E7 in transgenic mice correlates with alterations in epithelial cell growth and differentiation. J Virol. 1993;67:1373–1384. doi: 10.1128/jvi.67.3.1373-1384.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gulliver G, Herber R, Liem A, Lambert P F. Both the CR1 and CR2 domains of human papillomavirus type 16 E7 are required for the induction of epidermal hyperplasia and tumor formation in transgenic mice. J Virol. 1997;71:5905–5914. doi: 10.1128/jvi.71.8.5905-5914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halbert C L, Demers G W, Galloway D A. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herber R, Liem A, Pitot H, Lambert P F. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J Virol. 1996;70:1873–1881. doi: 10.1128/jvi.70.3.1873-1881.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogan B, Costantini F, Lacy E. Manipulating the mouse embro: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor; 1986. [Google Scholar]
- 25.Jones D L, Alani R M, Munger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones D L, Thompson D A, Munger K. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology. 1997;239:97–107. doi: 10.1006/viro.1997.8851. [DOI] [PubMed] [Google Scholar]
- 27.Kaur P, McDougall J K. Characterization of primary human keratinocytes transformed by human papillomavirus type 18. J Virol. 1988;62:1917–1924. doi: 10.1128/jvi.62.6.1917-1924.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kemp C J, Donehower L A, Bradley A, Balmain A. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell. 1993;74:813–822. doi: 10.1016/0092-8674(93)90461-x. [DOI] [PubMed] [Google Scholar]
- 29.Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klingelhutz A J, Foster S A, McDougall J K. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 31.Lee S S, Weiss R S, Javier R T. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangues R, Pellicer A. ras activation in experimental carcinogenesis. Semin Cancer Biol. 1992;3:229–239. [PubMed] [Google Scholar]
- 33.Munoz E F, Diwan B A, Calvert R J, Weghorst C M, Anderson J, Rice J M, Buzard G S. Transplacental mutagenicity of cisplatin: H-ras codon 12 and 13 mutations in skin tumors of SENCAR mice. Carcinogenesis. 1996;17:2741–2745. doi: 10.1093/carcin/17.12.2741. [DOI] [PubMed] [Google Scholar]
- 33a.Nguyen, M., and A. Griep. Unpublished observations.
- 34.Phelps W C, Yee C L, Munger K, Howley P M. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell. 1988;53:539–547. doi: 10.1016/0092-8674(88)90570-3. [DOI] [PubMed] [Google Scholar]
- 35.Pirisi L, Creek K E, Doniger J, DiPaolo J A. Continuous cell lines with altered growth and differentiation properties originate after transfection of human keratinocytes with human papillomavirus type 16 DNA. Carcinogenesis. 1988;9:1573–1579. doi: 10.1093/carcin/9.9.1573. [DOI] [PubMed] [Google Scholar]
- 36.Reznikoff C A, Belair C, Savelieva E, Zhai Y, Pfeifer K, Yeager T, Thompson K J, De Vries S, Bindley C, Newton M A. Long-term genome stability and minimal genotypic and phenotypic alterations in HPV16 E7-, but not E6-, immortalized human uroepithelial cells. Genes Dev. 1994;8:2227–2240. doi: 10.1101/gad.8.18.2227. [DOI] [PubMed] [Google Scholar]
- 37.Rotter V, Schwartz D, Almon E, Goldfinger N, Kapon A, Meshorer A, Donehower L A, Levine A J. Mice with reduced levels of p53 protein exhibit the testicular giant-cell degenerative syndrome. Proc Natl Acad Sci USA. 1993;90:9075–9079. doi: 10.1073/pnas.90.19.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaki K, Bertrand O, Nakazawa H, Fitzgerald D J, Mironov N, Yamasaki H. Cell-type-specific ras mutations but no microsatellite instability in chemically induced mouse skin tumors and transformed 3T3 cells. Cancer Res. 1995;55:3513–3516. [PubMed] [Google Scholar]
- 39.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 40.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 41.Schwarz E, Freese U K, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature. 1985;314:111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 42.Sherman L, Jackman A, Itzhaki H, Stoppler M C, Koval D, Schlegel R. Inhibition of serum- and calcium-induced differentiation of human keratinocytes by HPV16 E6 oncoprotein: role of p53 inactivation. Virology. 1997;237:296–306. doi: 10.1006/viro.1997.8778. [DOI] [PubMed] [Google Scholar]
- 43.Sherman L, Schlegel R. Serum- and calcium-induced differentiation of human keratinocytes is inhibited by the E6 oncoprotein of human papillomavirus type 16. J Virol. 1996;70:3269–3279. doi: 10.1128/jvi.70.5.3269-3279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snijders P J, van Duin M, Walboomers J M, Steenbergen R D, Risse E K, Helmerhorst T J, Verheijen R H, Meijer C J. Telomerase activity exclusively in cervical carcinomas and a subset of cervical intraepithelial neoplasia grade III lesions: strong association with elevated messenger RNA levels of its catalytic subunit and high-risk human papillomavirus DNA. Cancer Res. 1998;58:3812–3818. [PubMed] [Google Scholar]
- 45.Song S, Gulliver G A, Lambert P F. Human papillomavirus type 16 E6 and E7 oncogenes abrogate radiation-induced DNA damage responses in vivo through p53-dependent and p53-independent pathways. Proc Natl Acad Sci USA. 1998;95:2290–2295. doi: 10.1073/pnas.95.5.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tong X, Howley P M. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc Natl Acad Sci USA. 1997;94:4412–4417. doi: 10.1073/pnas.94.9.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Brule A J, Cromme F V, Snijders P J, Smit L, Oudejans C B, Baak J P, Meijer C J, Walboomers J M. Nonradioactive RNA in situ hybridization detection of human papillomavirus 16-E7 transcripts in squamous cell carcinomas of the uterine cervix using confocal laser scan microscopy. Am J Pathol. 1991;139:1037–1045. [PMC free article] [PubMed] [Google Scholar]
- 48.Weinberg W C, Azzoli C G, Chapman K, Levine A J, Yuspa S H. p53-mediated transcriptional activity increases in differentiating epidermal keratinocytes in association with decreased p53 protein. Oncogene. 1995;10:2271–2279. [PubMed] [Google Scholar]
- 49.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 50.White A E, Livanos E M, Tlsty T D. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 51.Woodworth C D, Bowden P E, Doniger J, Pirisi L, Barnes W, Lancaster W D, DiPaolo J A. Characterization of normal human exocervical epithelial cells immortalized in vitro by papillomavirus types 16 and 18 DNA. Cancer Res. 1988;48:4620–4628. [PubMed] [Google Scholar]
- 52.Woodworth C D, Doniger J, DiPaolo J A. Immortalization of human foreskin keratinocytes by various human papillomavirus DNAs corresponds to their association with cervical carcinoma. J Virol. 1989;63:159–164. doi: 10.1128/jvi.63.1.159-164.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yee C, Krishnan H I, Baker C C, Schlegel R, Howley P M. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am J Pathol. 1985;119:361–366. [PMC free article] [PubMed] [Google Scholar]
- 54.Zerfass-Thome K, Zwerschke W, Mannhardt B, Tindle R, Botz J W, Jansen-Durr P. Inactivation of the cdk inhibitor p27KIP1 by the human papillomavirus type 16 E7 oncoprotein. Oncogene. 1996;13:2323–2330. [PubMed] [Google Scholar]
- 55.zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]