ABSTRACT
Acinetobacter baumannii, an opportunistic pathogen, poses a significant threat in intensive care units, leading to severe nosocomial infections. The rise of multi-drug-resistant strains, particularly carbapenem-resistant A. baumannii, has created formidable challenges for effective treatment. Given the prolonged development cycle and high costs associated with antibiotics, phages have garnered clinical attention as an alternative for combating infections caused by drug-resistant bacteria. However, the utilization of phage therapy encounters notable challenges, including the narrow host spectrum, where each phage targets a limited subset of bacteria, increasing the risk of phage resistance development. Additionally, uncertainties in immune system dynamics during treatment hinder tailoring symptomatic interventions based on patient-specific states. In this study, we isolated two A. baumannii phages from wastewater and conducted a comprehensive assessment of their potential applications. This evaluation included sequencing analysis, genome classification, pH and temperature stability assessments, and in vitro bacterial inhibition assays. Further investigations involved analyzing histological and cytokine alterations in rats undergoing phage cocktail treatment for pneumonia. The therapeutic efficacy of the phages was validated, and transcriptomic studies of rat lung tissue during phage treatment revealed crucial changes in the immune system. The findings from our study underscore the potential of phages for future development as a treatment strategy and offer compelling evidence regarding immune system dynamics throughout the treatment process.
IMPORTANCE
Due to the growing problem of multi-drug-resistant bacteria, the use of phages is being considered as an alternative to antibiotics, and the genetic safety and application stability of phages determine the potential of phage application. The absence of drug resistance genes and virulence genes in the phage genome can ensure the safety of phage application, and the fact that phage can remain active in a wide range of temperatures and pH is also necessary for application. In addition, the effect evaluation of preclinical studies is especially important for clinical application. By simulating the immune response situation during the treatment process through mammalian models, the changes in animal immunity can be observed, and the effect of phage therapy can be further evaluated. Our study provides compelling evidence that phages hold promise for further development as therapeutic agents for Acinetobacter baumannii infections.
KEYWORDS: bacteriophage, Acinetobacter baumannii, multi-drug resistant, pneumonia, immune processes
INTRODUCTION
Acinetobacter species are recognized as prominent hospital pathogens, with Acinetobacter baumannii emerging as a paramount concern in nosocomial infections (1). The World Health Organization underscored the severity of antibiotic-resistant pathogens, listing A. baumannii as the most menacing in 2017 (2). Multidrug-resistant (MDR) A. baumannii, an opportunistic pathogen, poses a substantial threat to human health, giving rise to various untreatable infectious diseases, including pneumonia, bacteremia, and meningitis (3–5). The China Antimicrobial Resistance Surveillance System (CHINET) reported that in 2023, A. baumannii ranked as the fourth most frequently isolated pathogen in clinical settings and the second most common pathogen isolated from respiratory tract specimens (http://www.chinets.com). This highlights the critical role of A. baumannii in clinical infections and the urgent need for effective intervention strategies to mitigate its impact on public health.
Carbapenem antibiotics, such as imipenem and meropenem, serve as the last line of defense against severe Gram-negative bacterial infections (6). However, 2023 CHINET reported the alarming finding that clinical isolates of A. baumannii from tertiary hospitals in China had significantly higher rates of resistance to imipenem (78.6%) and meropenem (79.5%) compared to other multidrug-resistant bacteria (http://www.chinets.com). This heightened resistance underscores the critical and urgent necessity for effective therapeutic agents to combat infections caused by multidrug-resistant A. baumannii. In response to the growing antibiotic resistance crisis, bacteriophages (phages) are re-emerging as a promising alternative to antibiotics, leveraging their capacity to effectively eliminate MDR bacteria (7, 8). There have been many studies reported on the use of phages for the control of A. baumannii infections (9–12).
Currently, a primary approach to assess the potential of phage application involves analyzing novel phage genomes. The evaluation of lytic phages includes considerations of pH and temperature stability (13, 14) and in vitro bacterial inhibition. These assessments contribute to determining the potential for phage applications, and their characteristics extend the possibility of in vivo applications. Although numerous preclinical animal studies have demonstrated the feasibility of phage therapies for various diseases, including MDR bacterial lung infections (15–17), current research on phage therapy for lung infections caused by A. baumannii has predominantly focused on its efficacy as a treatment. Rigorous in vivo studies, especially those addressing critical questions about the impact on the immune system, are limited. Therefore, additional comprehensive studies are imperative before considering the application of phages for the treatment of A. baumannii pneumonia.
In this investigation, we isolated two lytic phages specific to A. baumannii from wastewater. Subsequently, the safety of phage application was assessed by excluding the presence of antibiotic-resistance genes and virulence genes in their genomes. We determined phage pH, temperature stability, and in vitro capabilities to lay the foundation for phage applications. A rat model of A. baumannii pneumonia was established, and phage cocktails were administered to evaluate the efficacy of phage therapy. Significantly, our study employed a combined approach of RNA-seq and qPCR to monitor alterations in the immune system of rats throughout the treatment. As anticipated, the phage cocktail exhibited efficient bacterial eradication and contributed to the alleviation of stress on the immune system. The outcomes of our study furnish a more comprehensive basis for advocating the application of phages in the treatment of A. baumannii lung infections.
RESULTS
Description of the phage genome
The whole genome sequence of vB_AbaM_P1 contains 107,537 bp with 37.68% G + C content. A total of 197 gene regions were predicted [183 open reading frames (ORFs) plus 14 tRNAs]; 36 of them were leftward-directed, and 162 were rightward-directed. Among them, 58 of the predicted ORFs have designated functions (Table S1). The whole genome of phage vB_AbaM_DP45 contains 164,031 bp with a G + C content of 36.77%. A total of 256 gene regions were predicted (248 ORFs plus 8 tRNAs), of which 216 were leftward-directed and 40 were rightward-directed (Table S2). Of these, 123 putative ORFs predict function. Both PHaTYP and Bacphlip predictions showed vB_AbaM_P1 and vB_AbaM_DP45 as lytic phages. As predicted by Pharokka’s results, the genomes of both phages did not contain integrase genes, virulence factors, or antibiotic resistance genes, suggesting that the two phages were lytic phages eligible for application.
Taxonomic relationships of the two phages
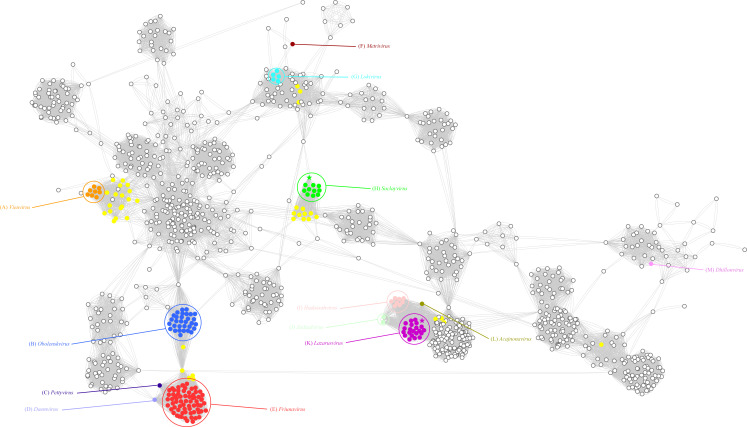
The taxonomic relationships of the two A. baumannii phage strains were evaluated within a broader context of phages using vContact2 (Fig. 1). Within the network, the Acinetobacter phages were classified into distinct groups, and the relationships between these groups reflected their nucleic acid similarity and shared protein content. Specifically, Phage vB_AbaM_P1 was assigned to the Saclayvirus category. Phages in this group typically have genomes ranging from 99.7 to 113 kb and contain 153–184 ORFs. According to the classification by the International Committee on Taxonomy of Viruses (ICTV), Saclayvirus phages have three representative phages, and the prediction of their lifestyles showed that all of them are lytic phages. vB_AbaM_DP45 belongs to the Lazarusvirus. Lazarusvirus phages have genomes ranging from 164 to 168.2 kb and contain 244–253 ORFs. All eight Lazarusvirus representative phages in ICTV were predicted to be lytic phages.
Fig 1.
The network analysis conducted using vContact2 focuses on the double-stranded DNA (dsDNA) prokaryotic virus genome. In the visual representation of the network, phage genomes are depicted as circles, and the connections between them indicate shared protein clusters. Each cluster is assigned a different color, and yellow circles specifically denote unclassified Acinetobacter phages. Notably, phages vB_AbaM_P1 and vB_AbaM_DP45 are marked with an asterisk in the network, drawing attention to their significance within the analyzed data set.
Temperature and pH stability of phages
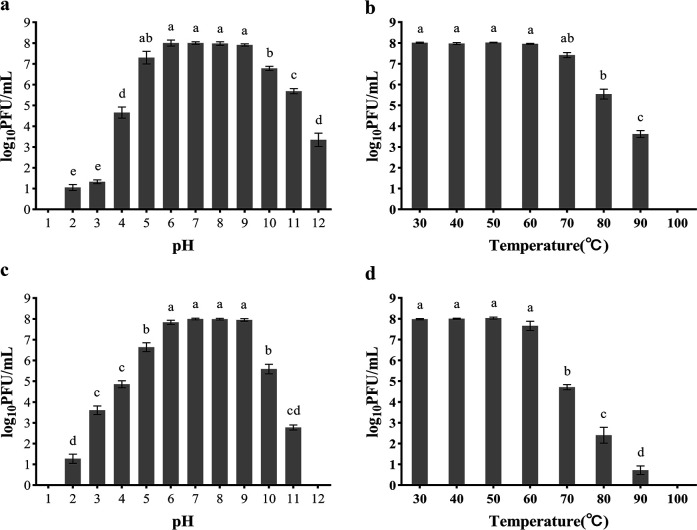
The phage could be considered stable when the free phage titer after 1 h of incubation decreases within 1 log10PFU/mL (plaque forming unit) compared to the initial phage titer. Phage P1 was stable in the range of pH 5–9 (Fig. 2a), beyond which the titer decreased significantly to 4.7 log10 PFU/mL and 6.8 log10 PFU/mL at pH 4 and 10, respectively. DP45 was stable in the pH 6–9 range (Fig. 2c) with a drop in titer at pH 5 and 10 to 6.6 log10 PFU/mL and 5.6 log10 PFU/mL, respectively, and inactivated at pH 1 and 12. P1 was stable within the range of 30°C–70°C (Fig. 2b), with a drop in titer of more than 2 log10 from 8 log10 PFU/mL to 5.5 log10 PFU/mL when the temperature was higher than 80°C and complete inactivation at 100°C. DP45 was stable in the range of 30°C–60°C (Fig. 2d), with a drop in titer from 8 log10 PFU/mL to 4.7 log10 PFU/mL when the temperature was higher than 70°C and complete inactivation at 100°C.
Fig 2.
pH and thermal stability of phage P1 and DP45. P1 was incubated at different pH (a) and temperatures (b), while DP45 was incubated at different pH (c) and temperatures (d) for 1 h. The surviving phage titers were determined by the double-layer plate method. Error bars represent the SD of three independent replicate experiments, and different letters represent significant differences.
Bacteriostatic effect of phage
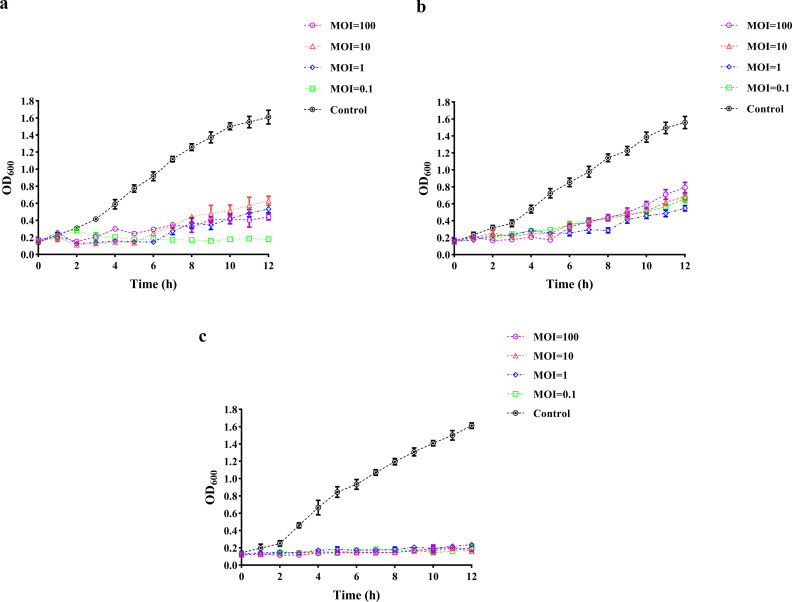
Both phage P1 and DP45 were able to inhibit bacterial growth for more than 6 h at all multiplicity of infections (MOIs; 100, 10, 1, and 0.1). This was a significant difference from what occurred when phosphate-buffered saline (PBS) was added to the bacteria. P1 was able to inhibit bacterial growth within 12 h of the assay when the MOI was 0.1 (Fig. 3a), and DP45 could also inhibit bacterial growth for 8 h when the MOI was 1 (Fig. 3b). After combining the two phages, both could inhibit bacterial growth for more than 12 h at all MOIs (Fig. 3c).
Fig 3.
Phage inhibition time at different MOIs. Inoculated with different MOIs, the OD600 values were measured every hour to observe the production of resistant bacteria. Equal amounts of PBS-inoculated bacteria were used as controls. (a) Inhibition curve of P1. (b) Inhibition curve of DP45. (c) Inhibition curve after combination of two phages. Error bars indicate the SD of three replicate counts.
Endotoxin content
Endotoxin, also known as lipopolysaccharide (LPS), constitutes a component of the cell wall in Gram-negative bacteria and is commonly found in phage lysates. In our investigation, we quantified the endotoxin content in the purified phage lysate. The determined amount of LPS was 62.957 EU/109 PFU, significantly lower than the pre-purification level of 4132.655 EU/109 PFU. Consequently, the presence of LPS carried by the phages did not result in any notable adverse effects.
Phage therapy improves survival of A. baumannii pneumonia
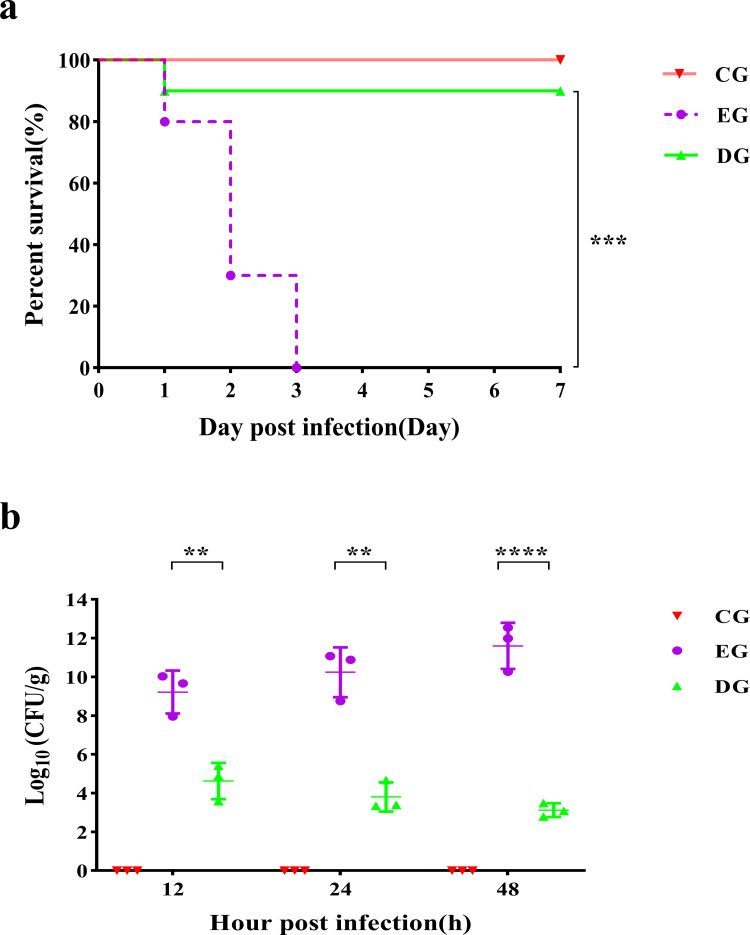
For the EG group, A. baumannii pneumonia was fatal, and all rats died within 72 h after infection (Fig. 4a). Phage treatments significantly increased the survival rate of the rats infected with A. baumannii (90%, P < 0.001).
Fig 4.
Survival and lung bacterial load in rats after phage treatment. (a) The survival rates of rats in different groups were observed over 7 days. (b) Lung bacterial load in different groups after infection. ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
Bacterial load
The bacteria grew rapidly in the lungs after being infected with A. baumannii, reaching 9.22 ± 1.11 log10 CFU/g at 12 h post-infection and growing to 11.60 ± 1.19 log10 CFU/g at 48 h (Fig. 4b). When compared to the infection group, the bacteria in the lungs were effectively eradicated after the administration of the phage treatment. Bacterial counts after administering the phage therapy decreased by 4.59, 6.43, and 8.48 log10 at 12 h, 24 h, and 48 h after infection, respectively, and the mean colony count was 4.63 ± 0.94 log10, 3.81 ± 0.75 log10, and 3.12 ± 0.35 log10 CFU/g, respectively.
Histology
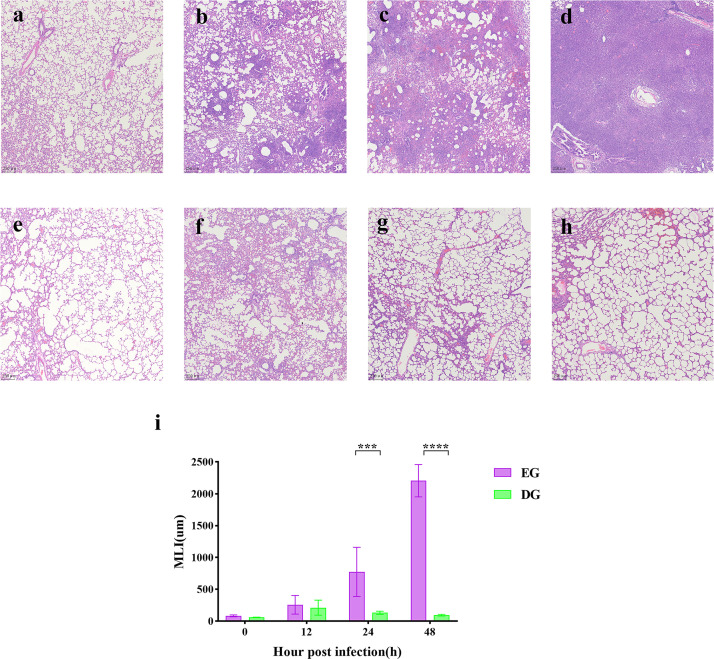
The results of the hematoxylin-eosin (HE) staining of lung tissue collected at each time point are shown in Fig. 5a through h. At 0 h of infection, HE staining results of lung tissues were similar in the EG (Fig. 5a) and DG (Fig. 5e) groups. At 12 h post-infection (Fig. 5b), the lung tissue showed obvious alveolar wall thickening and inflammatory cell infiltration. Large numbers of alveolar structures disappeared, and a small amount of light pink edema fluid was visible in the alveolar cavity. As the infection time increased, the alveolar structure continued to gradually decrease. The inflammatory cell infiltration was aggravating, and the edema fluid in the alveolar cavity increased (Fig. 5c). At 48 h post-infection (Fig. 5d), the alveolar structures were largely lost, and the lung tissue showed a large number of substantial lesions of inflammatory cell infiltration. Compared with the infected group, the lung tissue from rats treated with the phage cocktail showed a lower degree of pathological changes and alveolar fusion. At 12 h post-infection (Fig. 5f), the inflammatory cell infiltration and the general inflammatory conditions showed a decrease with time (Fig. 5g and h).
Fig 5.
Histological analysis of lung tissue. Histopathological changes in lung tissues with HE staining. Lung tissue from the EG group was collected at 0, 12, 24, and 48 h (a to d). Lung tissues from the DG group were collected at 0, 12, 24, and 48 h (e to h). (i) Results of mean linear intercept (MLI) analysis of the EG and DG groups. ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
As shown in (Fig. 5i), phage treatment can significantly reduce pulmonary mean linear intercept (MLI), with an increasing disparity between the two groups as time progresses.
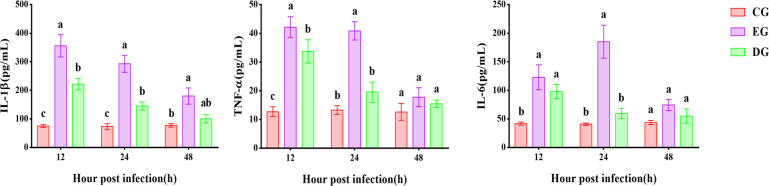
Cytokine levels
The expression of all three inflammatory factors increased to varying degrees. As shown in Fig. 6, for the EG group, TNF-α and IL-1β had the highest expression at 12 h, followed by a gradual decrease. The expression of IL-6 was highest at 24 h, followed by a decrease in expression. Unlike the EG group, the DG group showed a lower degree of change in cytokines. The expression of TNF-α showed a significant increase at 12 h, followed by a gradual decrease. The expression of IL-1β was significantly different from both the CG and EG groups at 12 h and then gradually decreased. IL-6 expression rose at 12 h and then fell back rapidly. All three cytokines were similar in the CG group at 48 h.
Fig 6.
Cytokine expression in the serum of rats in CG, EG, and DG groups at 12 h, 24 h, and 48 h after A. baumannii infection. Expressed as mean ± SD, with different letters representing significant differences.
Transcriptome changes
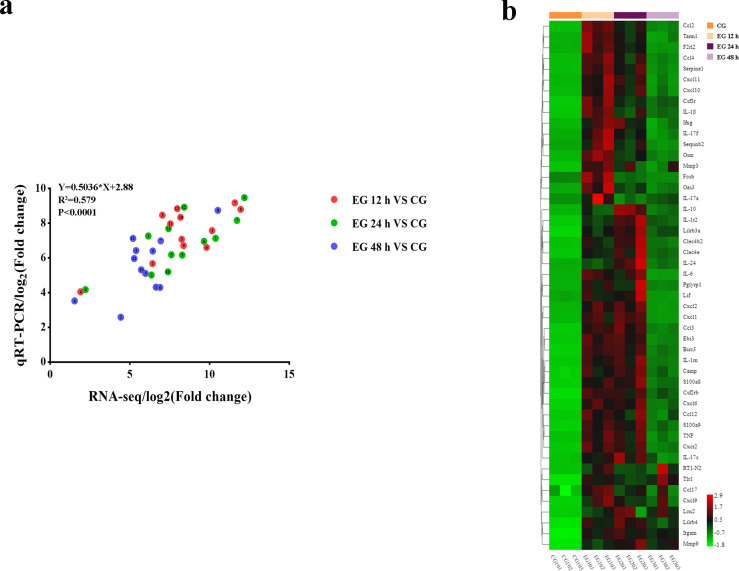
Immunological alterations in the processes of A. baumannii pneumonia were observed using transcriptome analysis. A large number of differentially expressed genes (DEGs) were observed after infection. To validate the results of our RNA-seq analysis, several highly expressed DEGs were selected for validation using the qRT-PCR method (primer sequences for qRT-PCR are listed in Table S3). The results showed that the expression trends of the selected DEGs during A. baumannii pneumonia were consistent with RNA-seq (Fig. 7a), indicating that the RNA-seq results reliably reflected the gene expression trends.
Fig 7.
Analysis of RNA-seq results. (a) Expression relationships between gene expression measured by RNA-seq and qRT-PCR. The numbers indicate genes: 1. Il-6; 2. TNF; 3. Il-1β; 4. Myd88; 5. Ccl2; 6. Nos2; 7. Csf3; 8. Acod1; 9. Ebi3; 10. Ptges; 11. Cxcl10; 12. Cxcl11. (b) Heat map showing the expression levels of 50 immune-related DEGs. Colors represent the expression level of each gene. Red indicates high expression, while green indicates low expression.
The results of the clustering analysis of the expression of 50 immune-related genes are shown in Fig. 7b. Compared with the CG group, the expression of immune genes was significantly upregulated in the EG-12 h and EG-24 h groups and then decreased in the EG-48 h groups.
Effect of phage treatment on immune gene expression
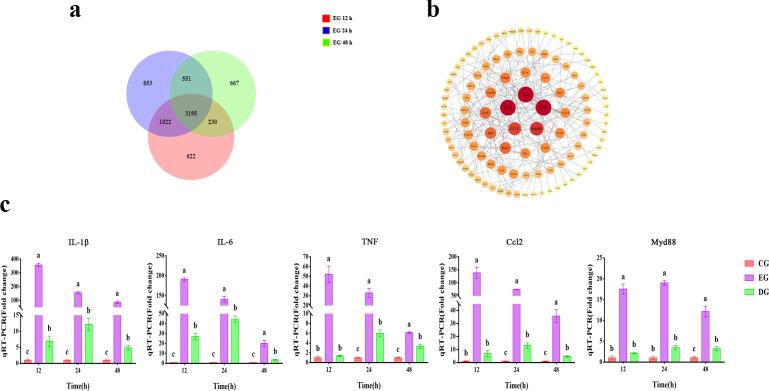
DEGs in all groups after infection were analyzed, and a total of 3,195 DEGs were found (Fig. 8a). To investigate the gene functions of the DEGs, we performed KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway annotation and enrichment analysis on 3,195 DEGs. The results of the KEGG analysis showed that there were 41 pathways with q < 0.01 (Table S4). Of these, seven pathways are immune-related: cytokine-cytokine receptor interactions, chemokine signaling pathways, tumor necrosis factor (TNF) signaling pathway, NOD-like receptor signaling pathway, Toll-like receptor signaling pathway, C-type lectin receptor signaling pathway, and Platelet activation. Based on the enrichment results of KEGG, the protein-protein interaction (PPI) analysis of the selected DEGs in immune-related pathways is shown in Fig. 8b, in which the genes with the highest association and expression were TNF-α, IL-6, IL-1β, Myd88, and Ccl2. For the expression results of these hub genes after being treated with phages, our qRT-PCR result is shown in Fig. 8c, which showed a continuous decrease in the expression of the highly associated genes.
Fig 8.
Effect of phage treatment on the expression of hub immune-related DEGs. (a) Venn diagram of DEGs at different time points in the EG group. (b) PPI analysis of DEGs in the EG group. (c) Expression of hub immune-related genes in the CG, EG, and DG groups at different time points. Expressed as mean ± SD, with different letters representing significant differences.
DISCUSSION
In this study, both phages composing the phage cocktail showed reliable performance for application. The two phages remained stably active after 1 h of incubation at pH 6–9 (Fig. 2a and c), and they also remained stable when the temperature was lower than 60°C (Fig. 2b and d). Interestingly, both phages we used could survive at 70°C, but the stability decreased as the temperature increased, similar to the results of previous studies (18). The optimal MOI for phage vB_AbaM_P1 was 0.1, and the host did not develop phage resistance within 12 h (Fig. 3a). Phage vB_AbaM_DP45 also inhibited the growth of host bacteria for 8 h at an MOI of 1 (Fig. 3b). The phage cocktail they formed showed a cooperative effect (Fig. 3c), which could better kill the pathogenic host bacteria and reduced the possibility of the generation of resistant bacteria. Therefore, the cocktail formed by the combination of two phages can be more effective in killing pathogenic bacteria.
In clinical studies of lethal pneumonia caused by MDR A. baumannii, the main concerns include bacterial infection load, tissue damage, and immune system status. However, these factors are almost impossible to rapidly and precisely determine in actual clinical situations. Therefore, studying changes in various factors in mammals after phage application is important for the wide application of phages and the pharmacodynamic study of phages. In the actual clinical setting, it is almost impossible to accurately determine the bacterial load. Therefore, we used a high dose of A. baumannii for modeling to ensure the stability of the pneumonia model. In addition, since phage concentration in vivo is dose-dependent and high phage/bacteria ratio conditions have better therapeutic efficacy (19–21), we used a high MOI for treatment. When phages enter the animal body, they are phagocytosed by mononuclear macrophages, filtered and cleared by the immune organs, and gradually reduced over time. In previous studies, phages have been shown to achieve therapeutic efficacy by killing the target bacteria before they are cleared (22).
As bacteria are more likely to develop resistance mutations against phages when treated with a single phage (23–25), we used a cocktail of two phages. Our results showed that treatment using two phages was effective in improving the survival rates in rats infected with lethal A. baumannii pneumonia (Fig. 4a), indicating the potential use of phages to treat lethal pneumonia. After treatment, the number of bacteria in the lung tissue rapidly decreased to low levels. Interestingly, the bacteria were not completely cleared within 48 h (Fig. 4b). Since the bacteria were not uniformly distributed throughout the lungs, some of the bacteria were hidden in the crevices of the tissue and, therefore, were not infected by phages.
In the immune system, immune cells are the first line of defense against pathogens, and the recruitment of neutrophils and monocytes/macrophages plays a crucial role in the response to bacterial infections. The depletion of neutrophils, monocytes, and macrophages increases the bacterial burden of the organism, and the recruitment of a large number of immune cells can cause unrecoverable damage to infected tissues. As shown in the histological results (Fig. 5a and e), the lung tissue in the DG and EG groups was similar in the initial phases of infection (12 h). Unsurprisingly, recruitment of inflammatory cells also occurred in the lung tissue after phage treatment. This is a normal immune response caused by the entry of bacteria and the lipopolysaccharide released by phage lysis of bacteria, which can lead to the aggregation of immune cells (26–28). After 24 h, the DG group normalized due to a significant reduction in bacterial numbers and diminished immune cell recruitment. This indicates that a small number of bacteria cannot induce a strong immune response. In contrast, the increasing number of bacteria in the EG group caused the immune system to overreact, large numbers of inflammatory cells infiltrated, and the alveolar structures gradually disappeared, which is similar to the results of previous studies (19, 22).
Immunity is a complex process in which cytokines, as signaling molecules in the immune system, play an important role. Most immune responses caused by severe bacterial infections are systemic, and the levels of cytokines in the serum better reflect the immune status. In the EG group, measurements of serum levels for three important cytokines showed a rapid increase in cytokine release after infection (Fig. 6), and the results were consistent with our expectations. After phage treatment, the release of cytokines decreased rapidly after the initial upregulation of release. Similar to immune cell recruitment in the lungs, cytokines in the blood vary with the degree of the infection. Notably, cytokine release in the EG group suddenly dropped to lower values at 48 h, similar to the DG and CG groups, which we analyzed as a result of infected rats reaching a near-death state and the organism becoming dysfunctional and unable to respond normally to the infection.
A sustained state of high immune activation may cause immune exhaustion (29–31). In our transcriptomic studies, we focused on immune-related genes and investigated immune system changes at the genetic level during A. baumannii pneumonia. Clustering analysis of immune-related genes showed (Fig. 7b) that the expression profile of immune-related genes showed a rapid increase followed by a rapid decrease, suggesting that the expression of immune-related genes was impaired after severe infection. Interestingly, several genes related to signal transduction and metabolism were expressed abnormally in the late stages of infection, indicating a severe dysregulation of the gene expression system in infected animals, which further led to a reduction in cytokine release in the late stages of infection.
Throughout the immunization process, different genes contribute to immunity to different degrees. The genes that have been differentially expressed across the different samples (Fig. 8a) were enriched by KEGG. We conducted a PPI analysis of genes enriched for immune-related pathways and screened five key genes that play important roles in the immune pathways involved in A. baumannii pneumonia (Fig. 8b). The effect of phages on the expression profile of the immune system can be derived by measuring the expression of these key genes after phage treatment. Based on previous studies, the levels of IL-6, TNF-α, and IL-1β were strongly correlated with the activity of macrophages (32, 33). Ccl2 is an important chemokine that regulates the migration and infiltration of monocytes/macrophages (34). Myd88 is a key regulator that plays an important role in immunity (35). The qPCR results (Fig. 8c) showed that the expression of these genes was reduced to different degrees after phage treatment, indicating that phage treatment reduced the burden on the immune system and avoided high loads and exhaustion of the immune system.
In summary, our study demonstrates that severe and uncontrolled inflammatory responses lead to lung injury and immune system exhaustion. In comparison, the use of phages killed A. baumannii and prevented lethal immune changes. Up until now, no mammalian immune changes have been reported during phage treatment of A. baumannii pneumoniae. These data provide a deeper understanding of immune changes in mammals during phage treatments.
MATERIALS AND METHODS
Animal maintenance
Male Wistar rats, aged 6 weeks and weighing 180–200 g, were acquired from Liaoning Changsheng Biotechnology Co., Ltd. Throughout the entire experiment, rats were placed in specific pathogen-free, light-controlled, and temperature-controlled conditions and supplemented with standard feed and water.
Bacteria and bacteriophages
The strains used in this experiment are listed in Table S5, and the strains were grown at 37°C in Luria-Bertani (LB) with shaking or on LB plates containing 1.5% agar (wt/vol). The multidrug-resistant strain AB-5 was isolated from the sputum of patients with A. baumannii pneumonia. Two phages used for phage therapies, named vB_AbaM_P1 (OL960030) and vB_AbaM_DP45 (OP585103), were isolated from medical sewage. The host spectrum of two phages is listed in Table S5.
Phage identification and analysis
The genomes of two phage strains were sequenced using the Illumina Hiseq 2500 platform, and phage lifestyles were predicted using PhaTYP (36) and Bacphlip (37). Gene function prediction of phages was performed using Pharokka (38), a phage rapid annotation tool, and ORF function analysis of phages was performed combined with RASTtk (https://rast.nmpdr.org/) (39). Functional validation of the annotated ORFs was performed using BLASTP, HHpred (40) (https://toolkit.tuebingen.mpg.de/tools/hhpred), and InterPro (41) (https://www.ebi.ac.uk/interpro/). The presence of transfer RNA (tRNA) was detected using tRNAscan-SE (42) (http://lowelab.ucsc.edu/tRNAscan-SE/). Based on Pharokka predictions, the Comprehensive Antibiotic Resistance Database (CARD) (43) and the Virulence Factor Database for Pathogenic Bacteria (VFDB) (44) were used to determine that the genome did not contain antibiotic resistance genes and virulence factors.
Taxonomic relationships of phages
To explore the association between Acinetobacter phages and the broader phage population, a gene-sharing network comprising 199 Acinetobacter phage genomes from GenBank was established using vContact2 (45). Subsequently, the generated network was visualized and examined utilizing Cytoscape (46).
Effects of temperature and pH on phage activity
The temperature and pH stability of phages were determined as described previously (47). Briefly, 108 PFU/mL of phages were incubated at temperatures ranging from 30°C to 100°C for 1 h, and the number of surviving phages was then counted by the double-layer plate method. Using 1 M HCl and 1 M NaOH to prepare SM buffers with different pH values (1–12), 108 PFU of phages were added to each pH solution, and the surviving phages were counted after 1 h of incubation.
Bactericidal assays
To assess the inhibitory potential of the phages, a 200 µL suspension of mid-log phase A. baumannii strain CGMCC 1.90331 was introduced into 96-well plates along with 20 µL of phage at various MOI levels. A blank control was included with an equivalent volume of PBS. All cultures were incubated at 37°C and 160 rpm, and OD600 was measured at 1 h intervals using a microtiter plate reader. Based on the results of the single phage assay, a 1:1 mixture of the two phages was used to determine the bacteriostatic effect of the phage cocktail under the same conditions. The experiments lasted for 12 h, and all experiments were performed in triplicate.
Endotoxin assay
Endotoxin removal from the phage lysate was achieved through the dialysis method. The endotoxin level in the phage lysates was measured using a dynamic colorimetric assay in accordance with the manufacturer’s guidelines (abs50045, absin Biotechnology Co. LTD, Shanghai, China). The result was expressed as endotoxin units per phage titer (EU/PFU).
Phage treatment of pneumonia
Rats were assigned to A. baumannii infection and phage therapy groups (n = 10) and were anesthetized with sodium pentobarbital (50 mg/kg). MDR A. baumannii was administered by non-invasive intratracheal drops of 2 × 109 CFU under direct vision, while the phage therapy group was treated 1 h after infection in the same manner but using the phage cocktail (2 × 1010 PFU). The survival rate of the rats was observed 7 days post-infection.
Based on 7-day survival, rats were partitioned into three groups (n = 15): the control group (CG), the A. baumannii infection group (EG), and the phage therapy group (DG). At different time points after infection (12 h, 24 h, and 48 h), rats (n = 3) were euthanized, and samples of lung tissue and serum were collected. After sampling, the lung tissue was immediately homogenized and prepared for bacterial enumeration.
Histological tests
Lung tissues from rats were collected at different time points and fixed with 4% paraformaldehyde for 48 h. The fixed tissues were embedded using paraffin, sectioned into 5 µm slices, and then stained using HE to observe any histomorphological changes. The extent of lung injury was assessed using Image Pro Plus 6.0 software to quantify the MLI of the lung.
Cytokines quantification
The concentrations of inflammatory factors were measured from the collected rat serum at different time points. Factors including tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), and interleukin-6 (IL-6) from each group were determined by ELISA.
Transcriptome analysis
For the lung tissues of the CG and EG groups collected at different times, total RNA was extracted, followed by isolating and purifying the mRNA to create cDNA libraries. Transcriptome sequencing was performed using an Illumina NovaSeq 6000 sequencing system. Gene expression with paired-end clean reads was aligned to the reference genome of Rattus norvegicus (Rnor_6.0_release95). Genes with significantly different expression (|FC|≥ 2, FDR < 0.01) were selected for analysis, and those DEGs with high expression were selected for qPCR to verify the quality of the transcriptome results.
Variation in immune gene expression
DEGs at all three time points were enriched in the KEGG pathway. Immune-related pathways were selected from the enriched pathways, and PPI were performed on DEGs using the Search Tool for Retrieval of Interacting Genes and Proteins (STRING) database, with the highest confidence interaction score of 0.900, and Cytoscape software was used to determine the ranking of gene associations in these immune-related pathways.
The expression of the most strongly associated genes in the EG group and DG group was quantified using the ABI7500 system. We used β-actin as a housekeeping gene, and we quantified gene expression using the 2-ΔΔCT method. Gene expressions were compared between the groups. Primer sequences are detailed in Table S3.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software. The survival rate was calculated by Log-rank tests, and all experimental data were expressed as mean ± SD. A t test and one-way ANOVA were used for comparison between groups, followed by Tukey’s multiple comparison test. Differences were considered significant when P-values were less than 0.05 (*), 0.01 (**), 0.001 (***), or 0.0001 (****).
ACKNOWLEDGMENTS
This study was funded by the third batch of “the open competition mechanism to select the best candidate” science and technology research project of Liaoning Province (No.2022JH1/10900013); A project admitted in the Dalian Deng Feng Program: key medical specialties in construction funded by the People’s Government of Dalian Municipality (No. 243, 2021); Dalian Key R&D Program (2021YF16SN012).
S.L. designed and coordinated the trial, as well as wrote and revised the manuscript. B.W. contributed to the trial’s design and organization and revised the manuscript. L.X. participated in writing and revising the manuscript. C.C. directed the trial. B.M. contributed to writing and revising the manuscript. L.W. designed and organized the trial. X.L. designed and organized the trial. Y.X., J.L., M.X., and J.Y. designed, organized, and directed the manuscript. All authors have reviewed and approved the final manuscript.
Contributor Information
Jiajun Yin, Email: yinjiajun@dlu.edu.cn.
Yongping Xu, Email: xyping@dlut.edu.cn.
Kristin N. Parent, Michigan State University, East Lansing, Michigan, USA
DATA AVAILABILITY
The transcriptome sequencing results have been uploaded to the NCBI database, and the BioProject ID is PRJNA1009251. Phages used in this study can be found in GenBank with accession numbers OL960030 and OP585103; pathogenic bacteria used here were deposited in the China General Microbiological Culture Collection Center (CGMCC) with accession number CGMCC 1.90331.
ETHICS APPROVAL
All animal experiments followed the ARRIVE guidelines. All procedures were approved by the Ethics Committee of Dalian University of Technology. The ethical approval number is DUTSBE220624_01.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.00467-24.
Tables S1 to S5.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Almasaudi SB. 2018. Acinetobacter spp. as nosocomial pathogens: epidemiology and resistance features. Saudi J Biol Sci 25:586–596. doi: 10.1016/j.sjbs.2016.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Willyard C. 2017. Drug-resistant bacteria ranked. Nature 543:15–15. doi: 10.1038/nature.2017.21550 [DOI] [PubMed] [Google Scholar]
- 3. Alquethamy SF, Khorvash M, Pederick VG, Whittall JJ, Paton JC, Paulsen IT, Hassan KA, McDevitt CA, Eijkelkamp BA. 2019. The role of the CopA copper efflux system in Acinetobacter baumannii virulence. Int J Mol Sci 20:575. doi: 10.3390/ijms20030575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nie D, Hu Y, Chen Z, Li M, Hou Z, Luo X, Mao X, Xue X. 2020. Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection. J Biomed Sci 27:8. doi: 10.1186/s12929-020-0617-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sato Y, Tansho-Nagakawa S, Ubagai T, Ono Y. 2020. Analysis of immune responses in Acinetobacter baumannii-infected klotho knockout mice: a mouse model of Acinetobacter baumannii infection in aged hosts. Front Immunol 11:601614. doi: 10.3389/fimmu.2020.601614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen XL, Sun SB, Huang S, Yang H, Ye Q, Lv L, Liang YS, Shan JJ, Xu JQ, Liu WK, Ma TH. 2023. Gold(I) selenium N-heterocyclic carbene complexes as potent antibacterial agents against multidrug-resistant gram-negative bacteria via inhibiting thioredoxin reductase. Redox Biol 60:102621. doi: 10.1016/j.redox.2023.102621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kortright KE, Doss-Gollin S, Chan BNK, Turner PE. 2021. Evolution of bacterial cross-resistance to lytic phages and albicidin antibiotic. Front Microbiol 12:658374. doi: 10.3389/fmicb.2021.658374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hietala V, Horsma-Heikkinen J, Carron A, Skurnik M, Kiljunen S. 2019. The removal of endo- and enterotoxins from bacteriophage preparations. Front Microbiol 10:1674. doi: 10.3389/fmicb.2019.01674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Altamirano FLG, Kostoulias X, Subedi D, Korneev D, Peleg AY, Barr JJ. 2022. Phage-antibiotic combination is a superior treatment against Acinetobacter baumannii in a preclinical study. EBioMedicine 80:104045. doi: 10.1016/j.ebiom.2022.104045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, Barr JJ, Reed SL, Rohwer F, Benler S, et al. 2018. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother 62:e00954-17. doi: 10.1128/AAC.02221-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tan X, Chen HS, Zhang M, Zhao Y, Jiang YC, Liu XY, Huang W, Ma YF. 2021. Clinical experience of personalized phage therapy against carbapenem-resistant Acinetobacter baumannii lung infection in a patient with chronic obstructive pulmonary disease. Front Cell Infect Microbiol 11:631585. doi: 10.3389/fcimb.2021.631585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu NN, Dai J, Guo MQ, Li JH, Zhou X, Li F, Gao Y, Qu HP, Lu HZ, Jin J, et al. 2021. Pre-optimized phage therapy on secondary Acinetobacter baumannii infection in four critical COVID-19 patients. Emerg Microbes Infect 10:612–618. doi: 10.1080/22221751.2021.1902754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee D, Im J, Na H, Ryu S, Yun CH, Han SH. 2019. The novel enterococcus phage VB_EfaS_HEf13 has broad lytic activity against clinical isolates of enterococcus faecalis. Front Microbiol 10:2877. doi: 10.3389/fmicb.2019.02877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Born Y, Fieseler L, Marazzi J, Lurz R, Duffy B, Loessner MJ. 2011. Novel virulent and broad-host-range erwinia amylovora bacteriophages reveal a high degree of mosaicism and a relationship to enterobacteriaceae phages. Appl Environ Microbiol 77:5945–5954. doi: 10.1128/AEM.03022-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen F, Cheng XJ, Li JB, Yuan XF, Huang XH, Lian M, Li WF, Huang TF, Xie YL, Liu J, Gao P, Wei XW, Wang ZL, Wu M. 2021. Novel lytic phages protect cells and mice against Pseudomonas aeruginosa infection. J Virol 95:e01832-20. doi: 10.1128/JVI.01832-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jeon J, Ryu CM, Lee JY, Park JH, Yong D, Lee K. 2016. In vivo application of bacteriophage as a potential therapeutic agent to control OXA-66-like carbapenemase-producing Acinetobacter baumannii strains belonging to sequence type 357. Appl Environ Microbiol 82:4200–4208. doi: 10.1128/AEM.00526-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prazak J, Iten M, Cameron DR, Save J, Grandgirard D, Resch G, Goepfert C, Leib SL, Takala J, Jakob SM, Que YA, Haenggi M. 2019. Bacteriophages improve outcomes in experimental Staphylococcus aureus ventilator-associated pneumonia. Am J Respir Crit Care Med 200:1126–1133. doi: 10.1164/rccm.201812-2372OC [DOI] [PubMed] [Google Scholar]
- 18. Tajuddin S, Khan AM, Chong LC, Wong CL, Tan JS, Ina-Salwany MY, Lau HY, Ho KL, Mariatulqabtiah AR, Tan WS. 2023. Genomic analysis and biological characterization of a novel schitoviridae phage infecting vibrio alginolyticus. Appl Microbiol Biotechnol 107:749–768. doi: 10.1007/s00253-022-12312-3 [DOI] [PubMed] [Google Scholar]
- 19. Hua YF, Luo TT, Yang YQ, Dong D, Wang R, Wang YJ, Xu MS, Guo XK, Hu FP, He P. 2017. Phage therapy as a promising new treatment for lung infection caused by carbapenem-resistant Acinetobacter baumannii in mice. Front Microbiol 8:2659. doi: 10.3389/fmicb.2017.02659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chang RYK, Chow MYT, Wang Y, Liu C, Hong Q, Morales S, McLachlan AJ, Kutter E, Li J, Chan HK. 2022. The effects of different doses of inhaled bacteriophage therapy for Pseudomonas aeruginosa pulmonary infections in mice. Clin Microbiol Infect 28:983–989. doi: 10.1016/j.cmi.2022.01.006 [DOI] [PubMed] [Google Scholar]
- 21. Dąbrowska K. 2019. Phage therapy: what factors shape phage pharmacokinetics and bioavailability? Systematic and critical review. Med Res Rev 39:2000–2025. doi: 10.1002/med.21572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dhungana G, Nepal R, Regmi M, Malla R. 2021. Pharmacokinetics and pharmacodynamics of a novel virulent Klebsiella phage Kp_pokalde_002 in a mouse model. Front Cell Infect Microbiol 11:684704. doi: 10.3389/fcimb.2021.684704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kong M, Ryu S. 2015. Bacteriophage PBC1 and its endolysin as an antimicrobial agent against Bacillus cereus. Appl Environ Microbiol 81:2274–2283. doi: 10.1128/AEM.03485-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ha E, Chun J, Kim M, Ryu S. 2019. Capsular polysaccharide is a receptor of a clostridium perfringens bacteriophage CPS1. Viruses 11:1002. doi: 10.3390/v11111002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petsong K, Benjakul S, Chaturongakul S, Switt AIM, Vongkamjan K. 2019. Lysis profiles of Salmonella phages on salmonella isolates from various sources and efficiency of a phage cocktail against S. enteritidis and S. typhimurium. Microorganisms 7:100. doi: 10.3390/microorganisms7040100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okamoto H, Muraki I, Okada H, Tomita H, Suzuki K, Takada C, Wakayama Y, Kuroda A, Fukuda H, Kawasaki Y, et al. 2021. Recombinant antithrombin attenuates acute respiratory distress syndrome in experimental endotoxemia. Am J Pathol 191:1526–1536. doi: 10.1016/j.ajpath.2021.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Su VYF, Lin CS, Hung SC, Yang KY. 2019. Mesenchymal stem cell-conditioned medium induces neutrophil apoptosis associated with inhibition of the NF-B pathway in endotoxin-induced acute lung injury. Int J Mol Sci 20:2208. doi: 10.3390/ijms20092208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang YL, Liang DD, Dong LL, Ge XT, Xu FL, Chen WB, Dai YR, Li HM, Zou P, Yang SL, Liang G. 2015. Anti-inflammatory effects of novel curcumin analogs in experimental acute lung injury. Respir Res 16:13. doi: 10.1186/s12931-015-0199-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li SY, Zhang ZN, Jiang YJ, Fu YJ, Shang H. 2020. Transcriptional insights into the CD8(+) T cell response in mono-HIV and HCV infection. J Transl Med 18:96. doi: 10.1186/s12967-020-02252-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nikolich-Zugich J. 2008. Ageing and life-long maintenance of T-cell subsets in the face of latent persistent infections. Nat Rev Immunol 8:512–522. doi: 10.1038/nri2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zeng X, Gu H, Peng LS, Yang Y, Wang N, Shi Y, Zou Q. 2020. Transcriptome profiling of lung innate immune responses potentially associated with the pathogenesis of Acinetobacter baumannii acute lethal pneumonia. Front Immunol 11:708. doi: 10.3389/fimmu.2020.00708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lv J, Chen J, Wang MJ, Yan F. 2020. Klotho alleviates indoxyl sulfate-induced heart failure and kidney damage by promoting M2 macrophage polarization. Aging (Albany NY) 12:9139–9150. doi: 10.18632/aging.103183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gou XM, Xu WC, Liu YS, Peng Y, Xu WL, Yin YB, Zhang XM. 2022. IL-6 prevents lung macrophage death and lung inflammation injury by inhibiting GSDME- and GSDMD-mediated pyroptosis during pneumococcal pneumosepsis. Microbiol Spectr 10:e0204921. doi: 10.1128/spectrum.02049-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aikawa S, Deng WB, Liang XH, Yuan J, Bartos A, Sun XF, Dey SK. 2020. Uterine deficiency of high-mobility group box-1 (HMGB1) protein causes implantation defects and adverse pregnancy outcomes. Cell Death Differ 27:1489–1504. doi: 10.1038/s41418-019-0429-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wirsdörfer F, Jendrossek V. 2017. Modeling DNA damage-induced pneumopathy in mice: insight from danger signaling cascades. Radiat Oncol 12:142. doi: 10.1186/s13014-017-0865-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shang JY, Tang XB, Sun YN. 2022. PhaTYB: predicting the lifestyle for bacteriophages using BERT. Brief Bioinformatics 24. doi: 10.1093/bib/bbac487:11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hockenberry AJ, Wilke CO. 2021. BACPHLIP: predicting bacteriophage lifestyle from conserved protein domains. PeerJ 9:e11396. doi: 10.7717/peerj.11396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bouras G, Nepal R, Houtak G, Psaltis AJ, Wormald PJ, Vreugde S. 2023. Pharokka: a fast scalable bacteriophage annotation tool. Bioinformatics 39:4. doi: 10.1093/bioinformatics/btac776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Söding J, Biegert A, Lupas AN. 2005. The hhpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Paysan-Lafosse T, Blum M, Chuguransky S, Grego T, Pinto BL, Salazar GA, Bileschi ML, Bork P, Bridge A, Colwell L, et al. 2023. InterPro in 2022. Nucleic Acids Res 51:D418–D427. doi: 10.1093/nar/gkac993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chan PP, Lin BY, Mak AJ, Lowe TM. 2021. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res 49:9077–9096. doi: 10.1093/nar/gkab688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alcock BP, Huynh W, Chalil R, Smith KW, Raphenya AR, Wlodarski MA, Edalatmand A, Petkau A, Syed SA, Tsang KK, et al. 2023. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res 51:D690–D699. doi: 10.1093/nar/gkac920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–D328. doi: 10.1093/nar/gki008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oliveira H, Domingues R, Evans B, Sutton JM, Adriaenssens EM, Turner D. 2022. Genomic diversity of bacteriophages infecting the genus Acinetobacter. Viruses 14:181. doi: 10.3390/v14020181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Imam M, Alrashid B, Patel F, Dowah ASA, Brown N, Millard A, Clokie MRJ, Galyov EE. 2019. VB_PaeM_Mij3, a novel jumbo phage infecting Pseudomonas aeruginosa, possesses unusual genomic features. Front Microbiol 10:2772. doi: 10.3389/fmicb.2019.02772 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S5.
Data Availability Statement
The transcriptome sequencing results have been uploaded to the NCBI database, and the BioProject ID is PRJNA1009251. Phages used in this study can be found in GenBank with accession numbers OL960030 and OP585103; pathogenic bacteria used here were deposited in the China General Microbiological Culture Collection Center (CGMCC) with accession number CGMCC 1.90331.