ABSTRACT
Rotavirus causes severe diarrhea in infants. Although live attenuated rotavirus vaccines are available, vaccine-derived infections have been reported, which warrants development of next-generation rotavirus vaccines. A single-round infectious virus is a promising vaccine platform; however, this platform has not been studied extensively in the context of rotavirus. Here, we aimed to develop a single-round infectious rotavirus by impairing the function of the viral intermediate capsid protein VP6. Recombinant rotaviruses harboring mutations in VP6 were rescued using a reverse genetics system. Mutations were targeted at VP6 residues involved in virion assembly. Although the VP6-mutated rotavirus expressed viral proteins, it did not produce progeny virions in wild-type cells; however, the virus did produce progeny virions in VP6-expressing cells. This indicates that the VP6-mutated rotavirus is a single-round infectious rotavirus. Insertion of a foreign gene, and replacement of the VP7 gene segment with that of human rotavirus clinical isolates, was successful. No infectious virions were detected in mice infected with the single-round infectious rotavirus. Immunizing mice with the single-round infectious rotavirus induced neutralizing antibody titers as high as those induced by wild-type rotavirus. Taken together, the data suggest that this single-round infectious rotavirus has potential as a safe and effective rotavirus vaccine. This system is also applicable for generation of safe and orally administrable viral vectors.
IMPORTANCE
Rotavirus, a leading cause of acute gastroenteritis in infants, causes an annual estimated 128,500 infant deaths worldwide. Although live attenuated rotavirus vaccines are available, they are replicable and may cause vaccine-derived infections. Thus, development of safe and effective rotavirus vaccine is important. In this study, we report the development of a single-round infectious rotavirus that can replicate only in cells expressing viral VP6 protein. We demonstrated that (1) the single-round infectious rotavirus did not replicate in wild-type cells or in mice; (2) insertion of foreign genes and replacement of the outer capsid gene were possible; and (3) it was as immunogenic as the wild-type virus. Thus, the mutated virus shows promise as a next-generation rotavirus vaccine. The system is also applicable to orally administrable viral vectors, facilitating development of vaccines against other enteric pathogens.
KEYWORDS: rotavirus, VP6, single-round infection, vaccine
INTRODUCTION
Rotavirus infection is a serious public health concern because it causes severe diarrhea, vomiting, and abdominal pain in infants (1). It is estimated that about 128,500 infants worldwide die every year due to rotavirus infection (2). Live attenuated vaccines against rotavirus infection are available (3, 4); however, dissemination of vaccine strains into the environment and vaccine-derived rotavirus infections have been reported (5–7). Therefore, development of a next-generation rotavirus vaccine is needed urgently.
A single-round infectious virus, which lacks part of the gene essential for virion assembly, could serve as a safe and effective vaccine (8). Such a single-round infectious virus would express intracellular viral proteins but would be unable to produce virus progeny in wild-type cells. The virus would be capable of replicating only in cells expressing the viral protein that was deleted from the viral genome. Due to these characteristics, a single-round infectious virus will be highly immunogenic and safer than conventional live attenuated vaccines. In addition, a single-round infectious virus will be amenable to insertion of foreign genes, making it a useful viral vector for many purposes. For example, it could be used as a viral-vectored vaccine and as a viral antigen for safe and easy-to-use antiviral evaluation and neutralization tests. The single-round infectious virus system has been reported for various viruses (9–11). In the field of rotavirus research, a trans-complementation system using recombinant rotaviruses lacking functional nonstructural protein (NSP) 5 has been reported (12); however, its utility as a vaccine or viral vector has not been reported. Thus, development of a single-round infectious rotavirus and its application as a vaccine or viral vector is warranted.
Rotavirus belongs to the genus Rotavirus within the family Reoviridae (13). It has an 11-segmented double-stranded RNA (dsRNA) genome encoding six structural proteins (VP1–4, VP6, and VP7) and six NSPs (NSP1–6). The infectious virions comprise three protein layers (14). The VP2, VP6, and VP7 proteins comprise the inner capsid, intermediate capsid, and outer capsid, respectively. VP4 functions as a spike protein that mediates viral attachment to susceptible cells. Since VP4 and VP7 are located on the surface of the virion, they are considered to be the main target of neutralizing antibodies (15). Upon viral infection, viral mRNA is transcribed, and viral proteins are translated. VP1 and VP3 serve as an RNA-dependent RNA polymerase and a capping enzyme, respectively (16, 17). These proteins, together with VP2, are indispensable for transcription and replication of viral RNA (18). The NSP2 and NSP5 proteins form inclusion bodies, termed viroplasm, in which viral dsRNA synthesis and particle formation take place (19).
VP6, an intermediate capsid protein, plays important roles in viral particle formation by interacting with VP2 (inner capsid) and VP7 (outer capsid). Cryo-EM studies show that VP6 has two domains: domain H and domain B (20, 21). Domain H interacts with VP7, while domain B interacts with VP2. Domain H contains loop structures that, presumably, are involved in the interaction with VP7 (21). In addition to viral particle formation, VP6 is also involved in viral RNA transcription through the VP6–VP2 interaction (22). Because VP6 is not located on the surface of the virion, it is not a main target or neutralizing antibodies. Nevertheless, humoral and cellular immunity against VP6 still plays a role in protection from rotavirus infection (23).
In the present study, we aimed to generate a single-round infectious rotavirus by impairing the functions of the viral VP6 gene. The VP6 gene was chosen as a target gene for development of a single-round rotavirus infection system because (1) it is indispensable for virion assembly; (2) it is not a main target for the neutralizing antibodies; (3) it is not a major player in RNA replication; and (4) replacement of the outer capsid gene (VP7), which alters both antigenicity and immunogenicity, is possible. We evaluated the potency of the single-round infectious rotavirus as a vaccine and as a viral vector.
RESULTS
Generation of VP6-knockout rotavirus
We aimed to develop a single-round infectious rotavirus by introducing mutations into the viral VP6 gene. A prototype simian rotavirus SA11 strain was used, unless stated otherwise (24). Recombinant rotaviruses harboring mutations in VP6 were generated using a reverse genetics system, as described previously, but with some modifications (see Materials and Methods) (24–26). The recombinant SA11 strain was referred as rSA11 and used as the wild-type virus. MA104 cells stably expressing functional VP6 (MA104-VP6) were established using a lentivirus vector and used for propagation and titration of VP6-mutated rotaviruses (Fig. 1A).
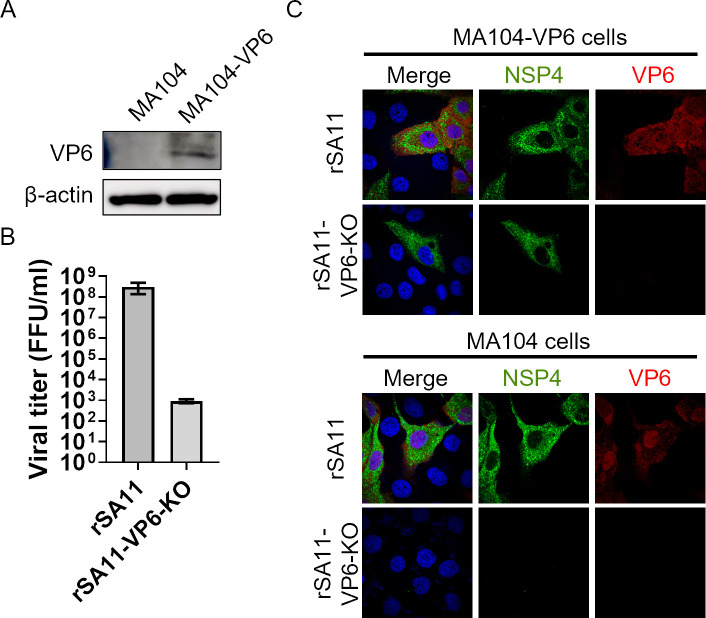
Fig 1.
Generation of a VP6-knockout rotavirus. (A) Generation of MA104 cells stably expressing VP6. VP6 expression was confirmed by Western blotting. (B) Growth of rSA11-VP6-KO. MA104-VP6 cells were infected with viruses at an MOI of 0.001 and then harvested at 72 hours post-infection. (C) Expression of viral proteins by rSA11-VP6-KO. Cells were infected with the virus at an MOI of 1.0. At 8 hours post-infection, cells were fixed and examined in an immunofluorescence assay.
At first, we aimed to generate a VP6-knockout (KO) rotavirus containing a stop codon at the 8th amino acid of VP6 (named rSA11-VP6-KO). Although rSA11-VP6-KO was rescued successfully, viral replication was 100,000-fold lower than that of rSA11 (Fig. 1B). In addition, rSA11-VP6-KO expressed viral protein (NSP4) in MA104-VP6 cells, but not in MA104 cells (Fig. 1C). This indicates that the VP6 gene is essential for viral protein expression. However, when considering use of a single-round infectious virus as a vaccine, such a low yield is a major obstacle. Additionally, a single-round infectious virus should express viral proteins in wild-type cells to induce robust immune responses. Therefore, these data suggest that the VP6-KO rotavirus will not be an effective vaccine or viral vector.
Generation of VP6-mutated rotaviruses in which virion assembly is selectively defective
Since VP6 is involved in both virion assembly and viral RNA transcription, knocking out VP6 impairs those functions. We speculated therefore that impaired transcription of rSA11-VP6-KO viral mRNA might have caused the low viral titer, as well as the lack of viral protein expression in wild-type cells. In general, single-round infectious viruses are selectively defective for virion assembly. To maintain viral protein expression, the function of VP6 in viral RNA transcription should be intact. Therefore, we aimed to generate VP6-mutated rotaviruses in which only virion assembly is defective.
Domain H and domain B of VP6 interact with VP7 and VP2, respectively (Fig. 2A and B). Since the VP6–VP2 interaction plays a role in viral RNA transcription and subsequent protein translation (22), we hypothesized that domain B is essential for viral RNA transcription, while domain H is involved only in virion assembly. Therefore, we selectively deleted domain H to generate a virion assembly-defective rotavirus. However, deletion of domain H led to a marked reduction in viral replication of the rescued viruses (rSA11-Δ150–397, rSA11-Δ150–331, rSA11-151-stop, and rSA11-332-stop), as evidenced by the low number (~103) of focus-forming units (FFU)/mL even at 72 hours post-infection (Fig. 2C). This suggests that deleting domain H might still affect RNA transcription and that more targeted deletions/mutations are needed.
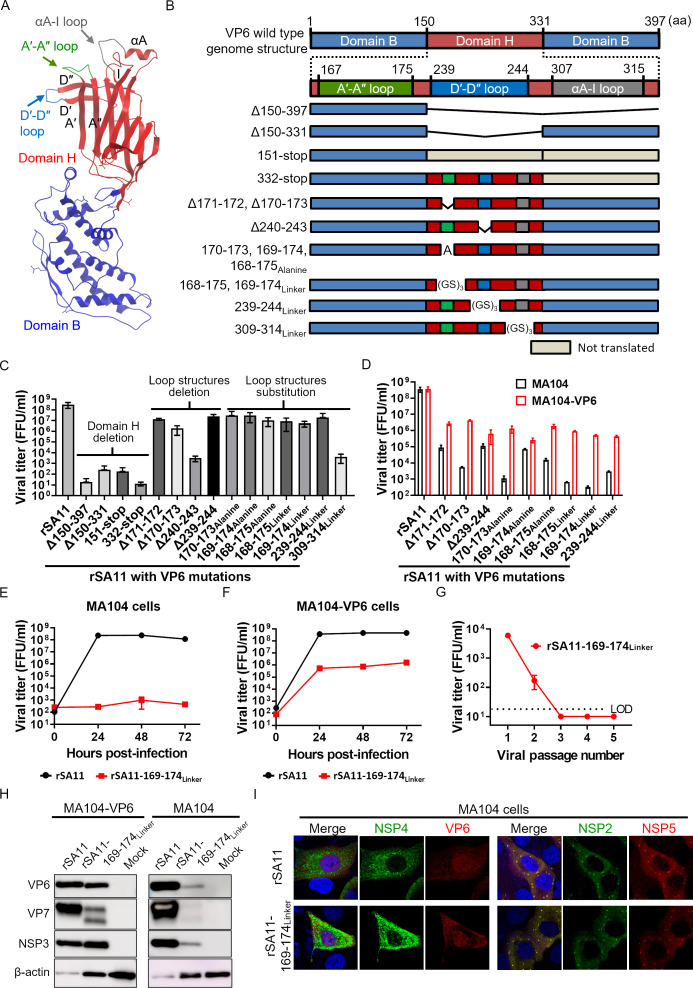
Fig 2.
Generation of VP6-mutated rotaviruses. (A) Structure of VP6. The ribbon diagram of the VP6 protein is based on Protein Data Bank (PDB) accession no. 1QHD. Domain B and domain H are indicated in blue and red, respectively. The A′-A″, D′-D″, and αA-I loop structures are highlighted in green, blue, and gray, respectively. (B) Construction of the VP6 gene. The number of amino acid residues is shown above. (C) Growth of the rescued recombinant viruses. MA104-VP6 cells were infected with viruses at an MOI of 0.001 and then harvested at 72 hours post-infection. (D) Growth of the recombinant viruses in MA104 and MA104-VP6 cells. Cells were infected with viruses at an MOI of 0.01 and then harvested at 24 hours post-infection. (E and F) Growth kinetics of rSA11-169-174Linker. Cells were infected with viruses at an MOI of 0.01. (G) Passage of the rSA11-169-174Linker in MA104 cells. Cells were infected at an MOI of 1.0, cultured for 7 days, and then freeze–thawed. Then, 10% of the cell lysate was inoculated onto new MA104 cells. This was repeated five times. The lysates were then titrated. The limit of detection (LOD) was 200 FFU/mL. Viral titers below the detection limit are plotted as half of the detection limit (100 FFU/mL). (H and I) Expression of viral proteins by rSA11-169-174Linker. For Western blotting, cells were infected with viruses at an MOI of 10 and then harvested at 8 hours post-infection. For the immunofluorescence assay, cells were infected with viruses at an MOI of 1.0 and then fixed at 8 hours post-infection.
Domain H contains loop structures that are, presumably, involved in the interaction with VP7 (Fig. 2A and B) (21). Therefore, to specifically abolish viral particle formation, we generated viruses in which amino acids in the loop structures were deleted (rSA11-Δ171–172, rSA11-Δ170–173, rSA11-Δ240–243, and rSA11-Δ239–244), replaced with alanine (rSA11-170-173Alanine, rSA11-169-174Alanine, and rSA11-168-175Alanine), or replaced with a linker sequence (GSGSGS) (rSA11-168-175Linker, rSA11-169-174Linker, rSA11-239-244Linker, and rSA11-309-314Linker). After these changes, some of the recombinant viruses exhibited relatively high viral replication, as evidenced by more than 105 FFU/mL at 72 hours post-infection (Fig. 2C). These viruses were used for subsequent analysis.
Because the single-round infectious rotavirus must replicate in MA104-VP6, but not in MA104 cells, we next assessed virus growth in MA104 and MA104-VP6 cells. There was no difference in the growth of rSA11 in either cell line (Fig. 2D); however, there was a tenfold to 1,000-fold difference in the growth of VP6-mutated viruses in the two cell lines, suggesting that VP6 is impaired in these recombinant viruses and is trans-complemented by functional VP6 present in MA104-VP6 cells. Of the VP6-mutated viruses tested, the recombinant rotavirus harboring the 169–174Linker mutation, in which amino acids 169–174 (SQPAHD) were replaced by the linker sequence (GSGSGS), showed the largest growth difference in the two cell lines. Thus, this virus (rSA11-169–174Linker) was used for all subsequent analyses.
The rSA11-169–174Linker is a practical single-round infectious rotavirus
To examine the potency of the rSA11-169–174Linker as a single-round infectious virus, we conducted a series of experiments. First, a viral growth curve assay was performed in MA104 and MA104-VP6 cells. The data showed that rSA11-169–174Linker did not propagate in MA104 cells for at least 72 hours post-infection (Fig. 2E). Moreover, it replicated well (peak titer = 106 FFU/mL) in MA104-VP6 cells (Fig. 2F). However, this titer was still 100-fold lower than that of the rSA11.
To further confirm that the rSA11-169–174Linker did not replicate in wild-type cells, the virus was passaged five times in MA104 cells. The viral titer decreased with increase in passage number (Fig. 2G). Infectious virions detected at passages 1 and 2 were considered to be virions carried over from the initial infection. No infectious virus was detected after the third passage.
Finally, we examined viral protein expression by Western blotting and immunofluorescence assay (IFA). Unlike rSA11-VP6-KO, rSA11-169–174Linker expressed detectable viral proteins in MA104 cells (Fig. 2H and I). However, it showed lower viral protein expression than rSA11, especially in MA104 cells (Fig. 2H). This indicates that the A′–A″ loop (amino acids 169–174) might still affect viral RNA transcription and protein translation. Nevertheless, viroplasm, in which viral replication takes place, was detected (Fig. 2I). These results indicate that rSA11-169–174Linker can express detectable levels of viral proteins in wild-type cells, indicating that it has potential as a vaccine. Taken together, the results suggest that rSA11-169–174Linker could have a practical application as a single-round infectious rotavirus.
Generation of a single-round infectious rotavirus carrying foreign genes
To utilize this single-round infectious rotavirus as a viral vector, we attempted to insert a foreign gene (ZsGreen: ZsG or NanoLuc luciferase: NLuc) in its genome. The foreign gene was inserted into the NSP1 segment, as described previously (Fig. 3A) (26). The single-round infectious rotaviruses harboring ZsG or NLuc (named rSA11-169–174Linker-ZsG or rSA11-169–174Linker-NLuc, respectively) were rescued successfully, although viral replication was 100- to 1000-fold lower than that of rSA11-169–174Linker (Fig. 3B). rSA11-169–174Linker-ZsG expressed ZsG in infected cells (Fig. 3C), and rSA11-169–174Linker-NLuc induced expression of NLuc in a time-dependent manner (Fig. 3D). These results indicate that it is possible to insert foreign genes into this single-round infectious rotavirus. Of note, the NLuc activity shown by rSA11-169–174Linker-NLuc was 10–100 fold lower than that of a replication-competent NLuc-expressing rotavirus (named rSA11-NLuc) (26). This suggests that protein expression of rSA11-169–174Linker is compromised, consistent with the data shown in Fig. 2H.
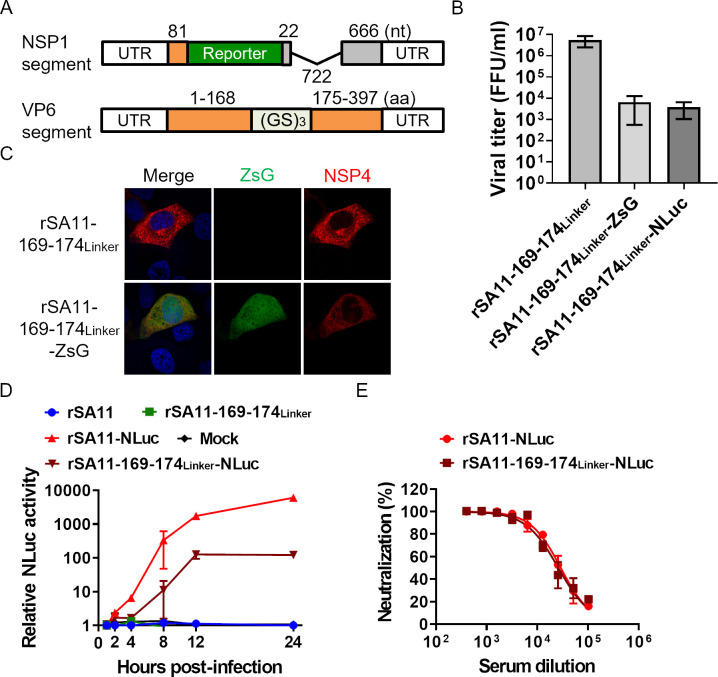
Fig 3.
Generation of single-round infectious rotaviruses expressing foreign genes. (A) Construction of the VP6 and NSP1 gene segments used to generate foreign gene-expressing rotaviruses. The number of nucleotides (nt) or amino acid residues (aa) is shown above. (B) Growth of the rescued recombinant rotavirus. MA104-VP6 cells were infected with viruses at an MOI of 0.001 and then harvested at 72 hours post-infection. (C) Fluorescence image of ZsG expression by rSA11-169–174Linker-ZsG. Cells were infected with viruses at an MOI of 1.0. At 8 hours post-infection, cells were fixed and examined in an immunofluorescence assay. (D) Kinetics of NLuc expression by rSA11-169–174Linker-NLuc. Cells were infected with viruses at an MOI of 0.01. The cells were harvested at the designated times. (E) NLuc-based neutralization test using serum from a mouse immunized with rSA11.
Single-round infectious reporter viruses can form the basis of safe and easy-to-use antiviral evaluation and neutralization tests (11). To examine whether rSA11-169–174Linker-NLuc can be used for this purpose, we performed an NLuc-based neutralization test. rSA11-NLuc was used as a control (26). Serum from a mouse immunized with the rSA11 strain was used as an antibody for the neutralization test (27). We found no apparent differences in the neutralization curves of rSA11-169–174Linker-NLuc and rSA11-NLuc (Fig. 3E), suggesting that both viruses have similar antigenicity and that the 169–174Linker mutation does not affect neutralization. Thus, the rSA11-169–174Linker-NLuc has potential for use in a neutralization test.
Generation of single-round infectious rotavirus harboring the VP7 segment of human rotavirus
The single-round infectious rotavirus generated in the present study is based on the simian rotavirus SA11 strain. If this system is to be used as a human rotavirus vaccine, then replacement of the VP7 gene, a major target of neutralizing antibodies, with that from a human rotavirus is desirable. Thus, we aimed to generate single-round infectious rotaviruses carrying the human rotavirus VP7 segment (named hereinafter as the single-round VP7 monoreassortant). Human rotavirus VP7 genes derived from clinical isolates obtained in Japan were used (27). The genotypes of the VP7 genes were G1, G2, G3, G8, and G9, all of which are commonly prevalent in Japan. The single-round VP7 monoreassorant viruses were rescued successfully, although their replication was lower than that of rSA11-169–174Linker (Fig. 4A).
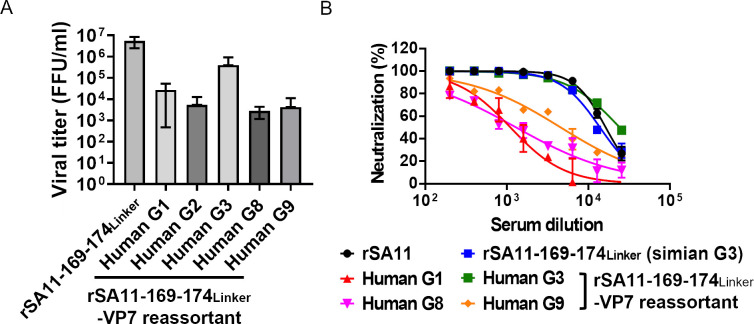
Fig 4.
Generation of single-round infectious rotaviruses harboring the VP7 segment of human rotavirus. (A) Growth of the rescued recombinant rotavirus. MA104-VP6 cells were infected with viruses at an MOI of 0.001 and then harvested at 72 hours post-infection. (B) Neutralization test using single-round VP7 monoreassortant viruses; serum from a mouse immunized with rSA11 (simian G3 genotype) was used.
To examine the antigenicity of the single-round VP7 monoreassortant viruses, we performed a focus number-based neutralization test. As mentioned previously, antiserum from mice immunized with the rSA11 was used as a source of antibodies. As observed for the NLuc-based neutralization tests, the neutralization curves for rSA11 and rSA11-169–174Linker were similar (Fig. 3E and 4B). The single-round VP7 monoreassortant harboring human G3 exhibited a neutralization curve similar to that of rSA11, possibly because the antiserum was generated by immunization with SA11 (simian G3 genotype) (Fig. 4B). Importantly, single-round VP7 monoreassortants harboring G1, G8, or G9 were less susceptible to neutralizing antibodies, suggesting that the antigenicity of these single-round VP7 monoreassortants was altered. This is consistent with the finding of a previous study in which we demonstrated that replacing VP7 of the SA11 strain with that of Japanese clinical isolates alters the antigenicity and immunogenicity (27). Thus, these single-round VP7 monoreassortants have potential for use as human rotavirus vaccines.
Infection of single-round infectious rotavirus into mouse models
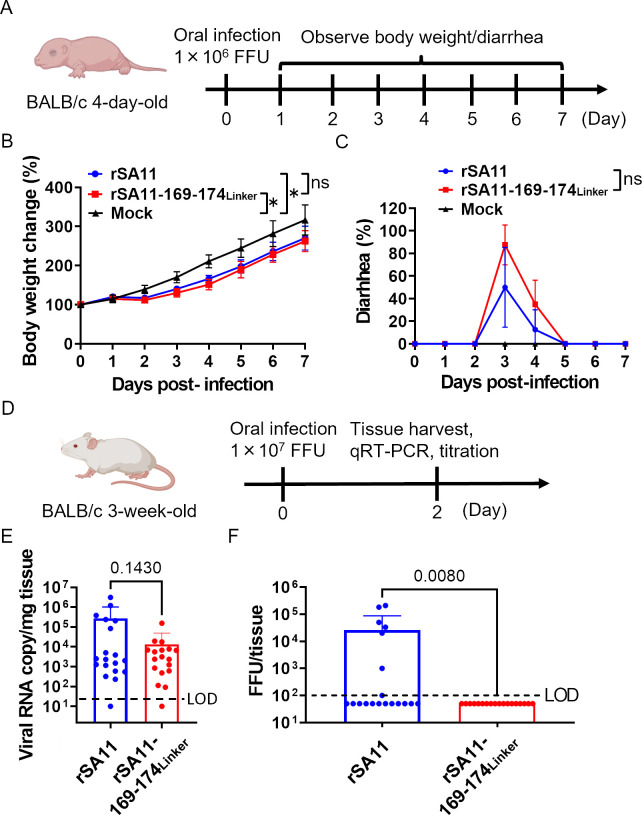
To further examine the potency of this system, we performed experiments in mice. To examine whether the pathogenicity of the single-round infectious rotavirus is attenuated, we orally infected newborn 4-day-old BALB/c mice with 1 × 106 FFU of the virus (Fig. 5A) and observed changes in body weight and diarrhea symptoms daily. Neonatal mice were used because adult mice do not develop symptoms despite rotavirus infection. Weight gain of infected mice in both groups was significantly slower than that of mice in the mock group (Fig. 5B). Unexpectedly, there was no significant difference in body weight or diarrhea symptoms between mice infected with rSA11 or rSA11-169–174Linker (Fig. 5B and C). These results suggest that the single-round virus can infect newborn mice and can still cause body weight loss and diarrhea.
Fig 5.
Viral challenge experiments using mouse models. (A) Schematic showing animal challenge experiments in newborn mice. Three groups of 4-day-old BALB/c mice (n = 8–9/group) were orally infected with 1 × 106 FFU of the virus. (B) Changes in body weight. Statistical significance was analyzed using two-way ANOVA. (C) Diarrhea symptoms. The graph shows the percentage of mice (per group) that had diarrhea. Statistical significance was analyzed using two-way ANOVA. P values < 0.05 were considered statistically significant (*P < 0.05). ns = not significant. (D) Schematic showing the animal challenge experiments using adult mice. Two groups of 3-week-old BALB/c mice (n = 18–19/group) were orally infected with 1 × 107 FFU of viruses. (E) Viral RNA copy number in intestinal samples. Intestinal samples were homogenized and subjected to RNA extraction and qRT-PCR. Statistical significance was analyzed using a t-test. The P value is shown. The limit of detection (LOD) was 20 copy/mg tissue. Viral titers below the detection limit are plotted as half of the detection limit (10 copy/mg tissue). (F) Titer of infectious virions in intestinal samples. The tissue homogenate was titrated in MA104-VP6 cells. The LOD was 100 FFU/tissue. Viral titers below the detection limit are plotted as half of the detection limit (50 FFU/tissue). Statistical significance was analyzed using the Mann–Whitney test. The P value is shown.
One of the advantages of a single-round infectious virus is that it cannot disseminate into the environment. To check this, we orally infected 3-week-old adult BALB/c mice with 107 FFU of the virus (Fig. 5D). We used adult mice for this because administration of a high dose of the SA11 strain results in production of progeny virions. Intestinal samples were harvested at 2 days post-infection. We found that the viral copy number in the intestine of mice infected with rSA11 was not significantly different from that in mice infected with rSA11-169–174Linker, although the latter tended to be lower (Fig. 5E); however, there was a significant difference in the amount of infectious virions (Fig. 5F). Virions were detected in 7/19 mice infected with rSA11, but not in any of the mice infected with rSA11-169–174Linker (0/18 mice). These data confirm that rSA11-169–174Linker does not disseminate into the environment.
Immunization of mice with a single-round infectious rotavirus
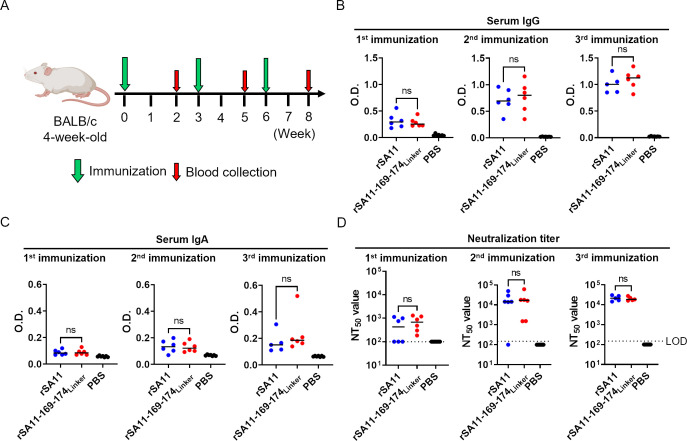
Finally, mice were immunized with the single-round infectious rotavirus to evaluate its potential as a vaccine. Adult 4-week-old BALB/c mice were orally immunized with 107 FFU of the virus (three vaccinations, with an interval of 3 weeks between each; Fig. 6A). Blood samples were taken 2 weeks after each vaccination and examined for the induction of serum IgG, serum IgA, and neutralizing antibodies, which are surrogate markers for protection against rotavirus infection (28, 29). There was no significant difference in the serum IgG, serum IgA, and neutralizing antibody titers between mice immunized with rSA11 and those immunized with rSA11-169–174Linker (Fig. 6B through D). This indicates that the single-round infectious rotavirus is as immunogenic as the wild-type virus, suggesting that it has potential as a rotavirus vaccine and a viral vector.
Fig 6.
Immunization of mice with the single-round infectious rotavirus. (A) Schematic showing the immunization protocol. Three groups of 4-week-old BALB/c mice (n = 5–7/group) were orally immunized with 1 × 107 FFU of viruses. (B and C) Detection of serum IgG and IgA against rotavirus. Statistical significance was analyzed using a t-test. P values < 0.05 were considered statistically significant. ns = not significant. (D) Neutralization titer in serum from immunized mice. The neutralization test was performed using rSA11-NLuc. The neutralizing titer is calculated as the maximum serum dilution yielding more than 50% neutralization. The limit of detection (LOD) was 1:200. Neutralization titers below the detection limit are plotted as half of the detection limit (1:100). Statistical significance was analyzed using the Mann–Whitney test. P values < 0.05 were considered statistically significant. ns = not significant.
DISCUSSION
A single-round infectious virus is a promising platform for a vaccine and viral vector. In the present study, we developed a single-round infectious rotavirus by impairing the function of VP6. This single-round infectious rotavirus did not replicate in wild-type cells in vitro (Fig. 2E and G), and no infectious virions were detected in infected mice (Fig. 5F). Considering the detection of rotavirus vaccine strains in the environment, this feature makes the current virus a promising next-generation rotavirus vaccine. Single-round infectious viruses have been studied extensively, including SARS-CoV-2, influenza virus, and flaviviruses (9–11), and replication-defective viral vectors are currently approved for clinical use (30). These studies demonstrate the safety of the single-round infection system.
Because foreign genes can be inserted into the rSA11-169–174Linker (Fig. 3), and rotavirus is a non-enveloped enteric virus that can tolerate low-pH conditions in the stomach and replicate in the intestine, this system could be used as a safe and orally administrable viral vector. Insertion of antigens derived from other enteric pathogens such as norovirus would facilitate development of a bivalent vaccine (31). We have recently developed rotavirus vectors stably expressing foreign genes (32). Further application of this single-round infection system to a viral vector is warranted.
Replacement of the outer capsid (VP7) with that of human rotavirus was successful (Fig. 4). This is beneficial because the VP7 genotype has a marked effect on susceptibility to neutralizing antibodies (33). Indeed, the currently approved live attenuated rotavirus vaccine, Rotateq, contains bovine rotaviruses harboring various genotypes of the human rotavirus VP7 segment (3). Since the single-round infectious rotavirus is as immunogenic as the wild-type virus (Fig. 6), and replacement of VP7 alters the antigenicity and immunogenicity (27), single-round VP7 monoreassortants could have potential as human rotavirus vaccines. However, viral replication of the single-round VP7 monoreassortant was lower than that of rSA11-169–174Linker, which would hamper vaccine production (Fig. 4A). Further improvement, such as serial passage of the viruses to introduce adaptive mutations, is warranted. Replacement of the VP4 gene, another target of neutralizing antibodies, has not been achieved yet (data not shown), probably because replacing the VP4 gene reduces the rescue efficiency of recombinant rotaviruses (27).
Generation of VP6-defective virus is also useful for functional analysis of VP6. Previous functional analyses of VP6 relied on generation of virus-like particles and siRNA-mediated knockdown of viral genes (22, 34). In this study, we used a reverse genetics system to demonstrate that VP6 plays an important role in viral protein expression and viral replication (Fig. 1). In addition, both domain B and domain H are important for viral replication (Fig. 2C). Furthermore, deletions or mutations in the loop structures of domain H led to a marked reduction in protein expression and viral replication (Fig. 2C, D, H and 3D). Further functional analysis of VP6 using this system will be of interest.
A previous study developed a single-round infectious rotavirus by knocking out the viral NSP5 gene (12). NSP5 is a main component of viroplasm (19); thus, it is essential for viral genome assembly, viral particle formation, and dsRNA synthesis, which in turn facilitate secondary mRNA transcription and subsequent protein translation (35). Nevertheless, the NSP5-knockout rotavirus expressed detectable viral proteins in MA104-wild type cells and reached a titer of 107 FFU/mL in MA104-NSP5 cells. These characteristics suggest its potential as a single-round infectious rotavirus. Of note, the NSP5-knockout rotavirus cannot induce formation of viroplasm in infected cells, whereas our rSA11-169–174Linker can induce formation of viroplasm in MA104 cells (Fig. 2I) (35). It would be intriguing to compare the protein expression levels and immunogenicity of these two single-round infectious rotaviruses.
Regarding the viral challenge experiment, the results using newborn mice were unexpected. There was no significant difference between the rSA11 and rSA11-169–174Linker with respect to body weight loss and diarrhea symptoms in infected mice (Fig. 5B and C). This suggests that even expression of single-round viral protein, especially viral enterotoxin NSP4, is sufficient to cause symptoms of rotavirus infection in newborn mice (36). Conversely, the result implies that rSA11 does not replicate well in newborn mice; symptoms induced by rSA11 infection could also be caused by expression of single-round proteins. Since the SA11 strain is a simian rotavirus, its replication would be low in a mouse model, which would result in no difference in pathogenicity between rSA11 and rSA11-169–174Linker. The low replication of rSA11 could also be a reason for no statistical differences in viral RNA levels in the infected mouse intestine (Fig. 5E) and immunogenicity (Fig. 6) between rSA11 and rSA11-169–174Linker. Use of a highly virulent rotavirus strain as a backbone for the single-round infectious rotavirus, as well as an appropriate animal model, could improve the determination of immunogenicity and attenuated pathogenicity. Specifically, the combination of a murine-like rotavirus and a mouse model, or a simian rotavirus and a non-human primate model, will be of interest (37).
Even though the current single-round infectious rotavirus shows promise, it has some limitations. For example, replication of the rSA11-169–174Linker was still 100-fold lower than that of rSA11 (Fig. 2F). In addition, viral protein expression was attenuated (Fig. 2H and 3D). In general, the titer of the single-round infectious virus was lower than that of the wild-type virus (11, 38). Passage of the single-round virus in MA104-VP6 cells may introduce adaptive mutations that increase the viral titer (11, 38).
Another limitation is the possibility that a viral revertant may emerge. Since VP6 is involved in transcription of viral RNA, knocking out VP6 was not practical in terms of application to a single-round infectious rotavirus (Fig. 1). For the single-round infectious virus to maintain the expression of viral proteins in wild-type cells, only six amino acid substitutions were introduced into the rSA11-169–174Linker. Even though no viral revertant emerged after passage of the virus in MA104 cells (Fig. 2G), the possibility of viral reversion remains.
However, generation of single-round virus with minimal amino acid substitutions has positive aspects. IgA and IgG antibodies targeting VP6 inhibit infection by rotavirus (39, 40). In addition, VP6 has adjuvant activity, which enhances generation of antibodies specific for VP4 and VP7 (41). Minimal mutations in VP6 have less effect on these functions. Of note, the A′–A″ loop region (169–174th amino acids) that is mutated in our single-round infectious virus is a part of the epitope region (42); thus, the mutation in the A′–A″ loop may compromise immune responses to VP6. Taken together, the data suggest that introduction of other mutations into VP6, or deletion of other viral genes that are not involved in viral RNA transcription/protein translation, is warranted to develop a second-generation single-round infectious rotavirus.
In conclusion, we developed a single-round infectious rotavirus by introducing mutations into the VP6 gene. This virus has potential for use as a single-round infectious virus because it does not replicate in wild-type cells/mice; insertion of foreign genes and replacement of the outer capsid gene were possible, and it was as immunogenic as the wild-type virus. Further improvement of this system and its application to vaccine and viral vector development are warranted.
MATERIALS AND METHODS
Cells
Monkey kidney MA104 cells and human embryonic kidney HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Nacalai Tesque) supplemented with 5% fetal bovine serum (FBS) (Gibco). MA104-VP6 cells were cultured in DMEM supplemented with 5% FBS and 800 µg/mL G418 (Nacalai Tesque). Baby hamster kidney cells stably expressing T7 RNA polymerase (BHK-T7) were cultured in DMEM supplemented with 5% FBS and 1 µg/mL puromycin (Sigma-Aldrich). All cells were cultured at 37°C/5% CO2.
Viruses
The simian rotavirus SA11 strain was used in this study, unless otherwise stated. Recombinant rotavirus SA11 strain was generated as described previously and designated as rSA11 (24–26). The recombinant VP6-mutated rotaviruses were generated as described below. Single-round infectious rotaviruses carrying a foreign gene or harboring the VP7 segment of human rotavirus were also generated (26, 27). Replication-competent rotavirus expressing NLuc was used (rSA11-NLuc; previously named rsSA11-NLuc-Δ722bp) (26). The rSA11 and VP6-mutated rotaviruses were propagated in MA104 and MA104-VP6, respectively. DMEM supplemented with 0.5 µg/mL trypsin (Sigma-Aldrich) was used as the culture medium for virus propagation. The recombinant viruses of passage number 2 were used. The sequences of recombinant viruses were confirmed by Sanger DNA sequencing.
Antibodies
Rabbit anti-NSP4 antiserum was prepared by immunization with a synthetic peptide spanning amino acid residues 158–171 of rotavirus NSP4 (Eurofins Genomics) (43). A mouse monoclonal antibody specific for rotavirus VP6 was generated as previously described (43). Rabbit anti-NSP3 antiserum was raised against a synthetic SA11 NSP3 peptide spanning amino acid residues 143–156 (Eurofins Genomics). A rabbit anti-VP7 antibody was purchased (MyBioSource). A mouse monoclonal antibody against β-actin was purchased (Sigma-Aldrich). Rabbit anti-NSP2 antiserum was raised against a synthetic SA11 NSP2 peptide spanning amino acid residues 299–312 (Eurofins Genomics). Guinea pig anti-NSP5 antiserum was raised against a synthetic SA11 NSP5 peptide spanning amino acid residues 48–66 (Eurofins Genomics).
Plasmids
To generate lentiviral vectors, the rotavirus VP6 gene was cloned into a pLVSIN-CMV-Neo vector (Takara). The resulting plasmid was named pLVSIN-VP6. The psPAX2 (#12260) and pCMV-VSV-G (#8454) plasmids were purchased from Addgene.
To generate recombinant rotaviruses, viral rescue plasmids that contained each rotavirus genome segment flanked by the T7 promoter and the HDV ribozyme were used (named pT7-VP1SA11, pT7-VP2SA11, pT7-VP3SA11, pT7-VP4SA11, pT7-VP6SA11, pT7-VP7SA11, pT7-NSP1SA11, pT7- NSP2SA11, pT7- NSP3SA11, pT7- NSP4SA11, and pT7-NSP5SA11) (24). Additionally, pCAG vector plasmids that contain rotavirus NSP2 or NSP5 or vaccinia virus D1R or D12L were used (named pCAG-NSP2SA11, pCAG-NSP5SA11, pCAG-D1R, and pCAG-D12L, respectively) (24–26). The rotavirus VP6 gene was cloned into the pcDNA3.1 vector to yield pcDNA-VP6SA11. Designated mutations were introduced into pT7-VP6SA11 by PCR-directed mutagenesis. Plasmid sequences were confirmed by Sanger DNA sequencing. To generate single-round infectious rotaviruses harboring a reporter gene, pT7-NSP1SA11 containing ZsG or NLuc with a 722-bp deletion of NSP1 (pT7-NSP1-ZsG or pT7-NSP1-NLuc, respectively) were used (26). Additionally, to generate single-round infectious rotaviruses carrying the human rotavirus VP7 gene, plasmids encoding the VP7 segment of Japanese clinical isolates were used (27), specifically pT7-VP7U14 (G1), pT7-VP7U6 (G2), pT7-VP7U4 (G3), pT7-VP7U2 (G8), and pT7-VP7U8 (G9).
Generation of MA104 cells stably expressing VP6
The VP6 gene was transduced into MA104 cells using a lentiviral vector. To generate the lentiviral vector, HEK293T cells were transfected with pLVSIN-VP6, psPAX2, and pCMV-VSV-G at a ratio of 3:4:1. At 48 hours post-transfection, the culture supernatant was inoculated onto MA104 cells for lentivirus infection. At 48 hours post-infection, the culture medium was replaced with DMEM containing 5% FBS and 800 µg/mL G418. The cells were then subjected to cell cloning by limiting dilution. Expression of VP6 was confirmed by IFA using an anti-rotavirus VP6 antibody as a primary antibody.
Generation of recombinant rotaviruses
rSA11 was generated as described previously (24). Recombinant rotaviruses carrying a mutation in VP6 were generated as described previously, with some modifications (24, 25). Briefly, BHK-T7 cells were seeded into a 12-well plate (1 × 105 cells/well). On the next day, the BHK-T7 cells were co-transfected with 11 viral rescue plasmids (pT7-VP1SA11, pT7-VP2SA11, pT7-VP3SA11, pT7-VP4SA11, pT7-VP6SA11 with mutation, pT7-VP7SA11, pT7-NSP1SA11, pT7- NSP2SA11, pT7- NSP3SA11, pT7-NSP4SA11, and pT7-NSP5SA11) and five plasmids (pCAG-NSP2SA11, pCAG-NSP5SA11, pCAG-D1R, pCAG-D12L, and pcDNA-VP6SA11). pcDNA-VP6SA11 were transfected to trans-complement the function of VP6 in BHK-T7 cells. Thus, a total of 16 plasmids (0.125 µg each, 2 µg in total) were transfected into the cells using TransIT-LT1 (Mirus). At 24 hours post-transfection, MA104-VP6 cells (1.5 × 105 cells/well) suspended in DMEM containing 0.5 µg/mL trypsin were added to the BHK-T7 cells and co-cultured for 5 days, followed by lysis via three cycles of freeze–thawing. Then, 10% of the cell lysate was inoculated onto a new MA104-VP6 monolayer in a 12-well plate (1.5 × 105 cells/well), followed by incubation for 5 days. The cells were then lysed by three cycles of freeze–thawing and inoculated onto MA104-VP6 cells followed by IFA using an anti-NSP4 antibody (see below). The presence of the NSP4 antigen indicated successful rescue of recombinant rotavirus. The rescued virus was inoculated onto a new MA104-VP6 monolayer in a 10-cm dish (2 × 106 cells) and incubated for 7 days, or until apparent cytopathic effects were observed. The titer of the virus was determined as described in the following section.
To generate single-round infectious rotaviruses expressing a foreign gene, pT7-VP6SA11 was replaced with pT7-VP6SA11-169–174Linker and pT7-NSP1SA11 was replaced with pT7-NSP1-ZsG or pT7-NSP1-NLuc. To generate single-round monoreassortants harboring the human rotavirus VP7 segment, pT7-VP6SA11 was replaced with pT7-VP6SA11-169–174Linker and pT7-VP7SA11 was replaced with one of the plasmids encoding the human rotavirusVP7 segment.
Virus titration
MA104 or MA104-VP6 cells were seeded in 96-well plates (1 × 104 cells/well). On the next day, serially diluted rotavirus samples were inoculated onto the cell monolayer, followed by overnight incubation at 37°C. The cells were then fixed for 30 minutes with 4% formaldehyde, followed by permeabilization for 15 minutes with 0.5% Triton-X. Then, the cells were blocked for 1 hour in PBS containing 2% FBS. The cells were then incubated with the anti-rotavirus NSP4 antibody, followed by a secondary antibody conjugated to a fluorescent dye. Each antibody was incubated for 1 hour at room temperature. The cells were washed three times with PBS at the end of each incubation with the antibodies. The nuclei were stained with Hoechst (Thermofisher). Fluorescence images were acquired under an Axio Observer 7 fluorescence microscope (Zeiss). Infected cells were counted manually under the fluorescence microscope, and the viral titer was expressed as FFU/mL.
Viral growth kinetics
The viral growth curve assay was conducted as described previously (43). Briefly, MA104 or MA104-VP6 cells were seeded into 24-well plates (7.5 × 104 cells/well). The cells were then infected with viruses at a multiplicity of infection (MOI) of 0.01 or 0.001 and incubated at 37°C for 1 hour. The infected cells were washed three times with PBS, followed by addition of DMEM containing 0.5 µg/mL trypsin. The infected cells were then incubated for the designated times and freeze–thawed three times. The cell lysates were then subjected to viral titration.
Virus passage
MA104 cells were seeded in a 12-well plate (1.5 × 105 cells/well). On the next day, the cells were infected with rSA11-169–174Linker at an MOI of 1.0. The infected cells were washed three times with PBS, followed by addition of DMEM containing 0.5 µg/mL trypsin. The cells were incubated for a further 7 days and freeze–thawed three times. Then, 10% of the cell lysate was inoculated onto a new MA104 cell monolayer in DMEM/0.5 µg/mL trypsin. These procedures were repeated five times. The virus titer in the lysate was measured at each passage.
Western blotting
MA104 or MA104-VP6 cells were seeded into 24-well plates (7.5 × 104 cells /well). The cells were infected with the virus at an MOI of 10. At 8 hours post-infection, cells were washed three times with PBS and lysed with RIPA buffer (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Nonidet P-40, 1% sodium-deoxycholate, 0.1% SDS). The lysate samples were then mixed with 2 × Laemmli sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 6% 2-mercaptoethanol, 10% glycerol, and 0.01% bromophenol blue) and boiled at 95°C for 5 minutes. The samples were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using a precast 10% polyacrylamide gel (ATTO), followed by transfer to an Immobilon-P PVDF membrane (Merck). Viral proteins and β-actin were detected through serial incubation with primary antibodies anti-VP6, anti-VP7, anti-NSP3, or anti-β-actin; secondary antibodies HRP-conjugated anti-mouse or anti-rabbit IgG; and Chemi-Lumi One Ultra (Nacalai Tesque). Images were acquired by an Amersham ImageQuant 800 (Cytiva).
IFA analysis of viral protein localization
For confocal imaging, a monolayer of MA104 or MA104-VP6 cells was prepared on 24-well plates (7.5 × 104 cells /well) containing glass cover slips. Cells were infected with virus at an MOI of 1.0. At 8 hours post-infection, cells were fixed and permeabilized as described previously. The cells were serially incubated with the designated primary antibodies, followed by a secondary antibody conjugated to a fluorescent dye. Fluorescence images were acquired under a C2 +Eclipse Ti2 confocal microscope (Nikon).
Detection of NLuc activity
MA104-VP6 cells were seeded into 96-well plates (1 × 104 cells/well). The cells were then infected with viruses at an MOI of 0.01. After incubation for 1 hour at 37°C, the cells were washed three times with PBS, followed by addition of DMEM containing 5% FBS. The infected cells were then incubated for the designated times and freeze–thawed twice. NLuc activity in the cell lysates was measured in a NanoGlo Luciferase Assay system (Promega).
NLuc-based neutralization assay
MA104-VP6 cells were seeded in a 96-well plate (1 × 104 cells/well). On the next day, serially diluted antibody was mixed with 100 FFU of NLuc-expressing rotavirus and incubated for 1 hour at 37°C, followed by inoculation onto the seeded cells. On the following day, the cells were freeze–thawed twice, and NLuc activity was measured as described above. Percent neutralization was calculated based on NLuc activity. Serum from a mouse immunized seven times with SA11 was used as an antibody (27).
Focus number-based neutralization assay
The focus number-based neutralization test was performed in essentially the same manner as the NLuc-based neutralization test. After overnight incubation of the cells infected with the virus–antibody mixture, the cells were fixed and subjected to IFA to visualize the infected cells. The infected cells were counted manually, and percent neutralization was calculated based on the focus number.
Virus challenge experiment
The virus challenge experiment using newborn mice was conducted as described previously (43). Pregnant BALB/cAJcl mice were purchased (CLEA Japan). Eight or nine 4-day-old BALB/c mice per group were orally infected with 1 × 106 FFU of the virus. Body weight changes and diarrhea symptoms were monitored daily. Diarrhea was defined as described previously (44).
The virus challenge experiment using adult mice was conducted essentially as described previously (45). A total of 18 or 19 3-week-old BALB/c mice per group were orally infected with 1 × 107 FFU of viruses. At 2 days post-infection, cecum samples were harvested and homogenized in a beads shocker. RNA was extracted from the homogenate using TRIzol reagent (Invitrogen) followed by qRT-PCR using a Thunderbird One-step qRT-PCR kit (Toyobo) to quantify the viral copy in the intestine (43). Additionally, the homogenate was subjected to viral titration to quantify the amount of the infectious virus.
Immunization of mice
Six or seven 4-week-old BALB/c mice per group were orally administered 1 × 107 FFU of the virus. Oral immunization was repeated three times, with a 3-week interval. Blood samples were collected 2 weeks after each immunization. Sera were isolated from blood samples by centrifugation at 3,000 rpm for 10 min. The sera were subjected to NLuc-based neutralization tests using rSA11-NLuc (26). The neutralizing antibody titer was expressed as the highest serum dilution factor that yielded 50% neutralization of NLuc activity.
Enzyme-linked immunosorbent assay (ELISA)
The infectious virion (triple-layered particle) of rSA11 was purified by CsCl density-gradient centrifugation (43). A 96-well flat-bottomed MaxiSorp plate (Thermo Fisher Scientific) was coated with the purified virus (1 × 106 FFU/well) overnight at 4°C in carbonate coating buffer (15 mM Na2CO3, 7 mM NaHCO3, pH 9.6). The plate was washed with ELISA wash buffer (PBS containing 0.05% Tween) three times at the end of each incubation. All the incubation steps were performed for 1 hour at 37°C. The coated plate was blocked with ELISA diluent buffer (PBS containing 1% bovine serum albumin and 0.05% Tween) followed by incubation with immunized mice sera (1:1000) and HRP-conjugated anti-mouse IgG or IgA (1:1000) (Abcam). Then, the 3,3',5,5′-tetramethylbenzidine substrate (Sigma Aldrich) was added for color development. The absorbance was measured at 450 nm using a Cytation 5 Cell Imaging Multimode Reader (Agilent).
Statistical analysis
Data analysis was performed using GraphPad Prism 9 (GraphPad Software, Inc.). Data are expressed as mean ± standard deviation of at least two independent experiments (each with three biological replicates, unless stated otherwise). P values < 0.05 were considered statistically significant.
ACKNOWLEDGMENTS
We thank M. Yoshida and Y. Hashimoto for secretarial work.
This work was supported in part by AMED grant numbers JP21fk0108122, JP22gm1610008, and JP23fk0108668; by KAKENHI grant numbers JP18H02663, JP21K19379, JP21H02739, and JP22K07100; by JST Moonshot R&D-MILLENNIA Program grant number JPMJMS2025; by The Research Foundation for Microbial Diseases of Osaka University (BIKEN); and by the Center for Advanced Modalities and DDS, Osaka University, Osaka, Japan.
Contributor Information
Takeshi Kobayashi, Email: tkobayashi@biken.osaka-u.ac.jp.
Christiane E. Wobus, University of Michigan Medical School, Ann Arbor, Michigan, USA
ETHICS APPROVAL
All animal experiments were approved by the Osaka University institutional animal experiment committee (approval number BidouR03-10-0).
REFERENCES
- 1. Kotloff KL, Platts-Mills JA, Nasrin D, Roose A, Blackwelder WC, Levine MM. 2017. Global burden of diarrheal diseases among children in developing countries: incidence, etiology, and insights from new molecular diagnostic techniques. Vaccine 35:6783–6789. doi: 10.1016/j.vaccine.2017.07.036 [DOI] [PubMed] [Google Scholar]
- 2. Troeger C, Khalil IA, Rao PC, Cao S, Blacker BF, Ahmed T, Armah G, Bines JE, Brewer TG, Colombara DV, Kang G, Kirkpatrick BD, Kirkwood CD, Mwenda JM, Parashar UD, Petri WA, Riddle MS, Steele AD, Thompson RL, Walson JL, Sanders JW, Mokdad AH, Murray CJL, Hay SI, Reiner RC. 2018. Rotavirus vaccination and the global burden of rotavirus diarrhea among children younger than 5 years. JAMA Pediatr 172:958–965. doi: 10.1001/jamapediatrics.2018.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, Rodriguez Z, Dallas MJ, Heyse JF, Goveia MG, Black SB, et al. 2006. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N Engl J Med 354:23–33. doi: 10.1056/NEJMoa052664 [DOI] [PubMed] [Google Scholar]
- 4. Ruiz-Palacios GM, Pérez-Schael I, Velázquez FR, Abate H, Breuer T, Clemens SC, Cheuvart B, Espinoza F, Gillard P, Innis BL, et al. 2006. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med 354:11–22. doi: 10.1056/NEJMoa052434 [DOI] [PubMed] [Google Scholar]
- 5. Anderson EJ. 2008. Rotavirus vaccines: viral shedding and risk of transmission. Lancet Infect Dis 8:642–649. doi: 10.1016/S1473-3099(08)70231-7 [DOI] [PubMed] [Google Scholar]
- 6. Simsek C, Bloemen M, Jansen D, Descheemaeker P, Reynders M, Van Ranst M, Matthijnssens J. 2022. Rotavirus vaccine-derived cases in Belgium: evidence for reversion of attenuating mutations and alternative causes of gastroenteritis. Vaccine 40:5114–5125. doi: 10.1016/j.vaccine.2022.06.082 [DOI] [PubMed] [Google Scholar]
- 7. Ito E, Pu J, Miura T, Kazama S, Nishiyama M, Ito H, Konta Y, Omura T, Watanabe T. 2021. Detection of rotavirus vaccine strains in oysters and sewage and their relationship with the gastroenteritis epidemic. Appl Environ Microbiol 87:e02547-20. doi: 10.1128/AEM.02547-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dudek T, Knipe DM. 2006. Replication-defective viruses as vaccines and vaccine vectors. Virology 344:230–239. doi: 10.1016/j.virol.2005.09.020 [DOI] [PubMed] [Google Scholar]
- 9. Nogales A, Baker SF, Domm W, Martínez-Sobrido L. 2016. Development and applications of single-cycle infectious influenza A virus (sciIAV). Virus Res 216:26–40. doi: 10.1016/j.virusres.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mason PW, Shustov AV, Frolov I. 2006. Production and characterization of vaccines based on flaviviruses defective in replication. Virology 351:432–443. doi: 10.1016/j.virol.2006.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang X, Liu Y, Liu J, Bailey AL, Plante KS, Plante JA, Zou J, Xia H, Bopp NE, Aguilar PV, Ren P, Menachery VD, Diamond MS, Weaver SC, Xie X, Shi PY. 2021. A trans-complementation system for SARS-CoV-2 recapitulates authentic viral replication without virulence. Cell 184:2229–2238. doi: 10.1016/j.cell.2021.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Papa G, Venditti L, Arnoldi F, Schraner EM, Potgieter C, Borodavka A, Eichwald C, Burrone OR. 2019. Recombinant rotaviruses rescued by reverse genetics reveal the role of NSP5 hyperphosphorylation in the assembly of viral factories. J Virol 94:e01110-19. doi: 10.1128/JVI.01110-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Estes MK, Greenberg HB. 2013. Rotaviruses, p 1347–1401. In Knipe DM, Howley PM (ed), Fields virology, 6th ed. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- 14. Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. 2011. Atomic model of an infectious rotavirus particle. EMBO J 30:408–416. doi: 10.1038/emboj.2010.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Desselberger U, Huppertz HI. 2011. Immune responses to rotavirus infection and vaccination and associated correlates of protection. J Infect Dis 203:188–195. doi: 10.1093/infdis/jiq031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Estrozi LF, Settembre EC, Goret G, McClain B, Zhang X, Chen JZ, Grigorieff N, Harrison SC. 2013. Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles. J Mol Biol 425:124–132. doi: 10.1016/j.jmb.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumar D, Yu X, Crawford SE, Moreno R, Jakana J, Sankaran B, Anish R, Kaundal S, Hu L, Estes MK, Wang Z, Prasad BVV. 2020. 2.7Å cryo-EM structure of rotavirus core protein VP3, a unique capping machine with a helicase activity. Sci Adv 6:eaay6410. doi: 10.1126/sciadv.aay6410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McDonald SM, Patton JT. 2011. Rotavirus VP2 core shell regions critical for viral polymerase activation. J Virol 85:3095–3105. doi: 10.1128/JVI.02360-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fabbretti E, Afrikanova I, Vascotto F, Burrone OR. 1999. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J Gen Virol 80 (Pt 2):333–339. doi: 10.1099/0022-1317-80-2-333 [DOI] [PubMed] [Google Scholar]
- 20. Mathieu M, Petitpas I, Navaza J, Lepault J, Kohli E, Pothier P, Prasad BV, Cohen J, Rey FA. 2001. Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the virion. EMBO J 20:1485–1497. doi: 10.1093/emboj/20.7.1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen JZ, Settembre EC, Aoki ST, Zhang X, Bellamy AR, Dormitzer PR, Harrison SC, Grigorieff N. 2009. Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc Natl Acad Sci U S A 106:10644–10648. doi: 10.1073/pnas.0904024106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Charpilienne A, Lepault J, Rey F, Cohen J. 2002. Identification of rotavirus VP6 residues located at the interface with VP2 that are essential for capsid assembly and transcriptase activity. J Virol 76:7822–7831. doi: 10.1128/jvi.76.15.7822-7831.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Choi AH, Basu M, McNeal MM, Clements JD, Ward RL. 1999. Antibody-independent protection against rotavirus infection of mice stimulated by intranasal immunization with chimeric VP4 or VP6 protein. J Virol 73:7574–7581. doi: 10.1128/JVI.73.9.7574-7581.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanai Y, Komoto S, Kawagishi T, Nouda R, Nagasawa N, Onishi M, Matsuura Y, Taniguchi K, Kobayashi T. 2017. Entirely plasmid-based reverse genetics system for rotaviruses. Proc Natl Acad Sci U S A 114:2349–2354. doi: 10.1073/pnas.1618424114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kawagishi T, Nurdin JA, Onishi M, Nouda R, Kanai Y, Tajima T, Ushijima H, Kobayashi T. 2020. Reverse genetics system for a human group A rotavirus. J Virol 94:e00963-19. doi: 10.1128/JVI.00963-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanai Y, Kawagishi T, Nouda R, Onishi M, Pannacha P, Nurdin JA, Nomura K, Matsuura Y, Kobayashi T. 2019. Development of stable rotavirus reporter expression systems. J Virol 93:e01774-18. doi: 10.1128/JVI.01774-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanai Y, Onishi M, Kawagishi T, Pannacha P, Nurdin JA, Nouda R, Yamasaki M, Lusiany T, Khamrin P, Okitsu S, Hayakawa S, Ebina H, Ushijima H, Kobayashi T. 2020. Reverse genetics approach for developing rotavirus vaccine candidates carrying VP4 and VP7 genes cloned from clinical isolates of human rotavirus. J Virol 95:e01374-20. doi: 10.1128/JVI.01374-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Velázquez FR, Matson DO, Guerrero ML, Shults J, Calva JJ, Morrow AL, Glass RI, Pickering LK, Ruiz-Palacios GM. 2000. Serum antibody as a marker of protection against natural rotavirus infection and disease. J Infect Dis 182:1602–1609. doi: 10.1086/317619 [DOI] [PubMed] [Google Scholar]
- 29. Chiba S, Yokoyama T, Nakata S, Morita Y, Urasawa T, Taniguchi K, Urasawa S, Nakao T. 1986. Protective effect of naturally acquired homotypic and heterotypic rotavirus antibodies. Lancet 2:417–421. doi: 10.1016/s0140-6736(86)92133-1 [DOI] [PubMed] [Google Scholar]
- 30. Li X, Le Y, Zhang Z, Nian X, Liu B, Yang X. 2023. Viral vector-based gene therapy. Int J Mol Sci 24:7736. doi: 10.3390/ijms24097736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kawagishi T, Sánchez-Tacuba L, Feng N, Costantini VP, Tan M, Jiang X, Green KY, Vinjé J, Ding S, Greenberg HB. 2023. Mucosal and systemic neutralizing antibodies to norovirus induced in infant mice orally Inoculated with recombinant rotaviruses. Proc Natl Acad Sci U S A 120:e2214421120. doi: 10.1073/pnas.2214421120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kanai Y, Onishi M, Yoshida Y, Kotaki T, Minami S, Nouda R, Yamasaki M, Enoki Y, Kobayashi T. 2023. Genetic engineering strategy for generating a stable dsRNA virus vector using a virus-like codon-modified transgene. J Virol 97:e0049223. doi: 10.1128/jvi.00492-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Diller JR, Carter MH, Kanai Y, Sanchez SV, Kobayashi T, Ogden KM. 2021. Monoreassortant rotaviruses of multiple G types are differentially neutralized by sera from infants vaccinated with ROTARIX and RotaTeq. J Infect Dis 224:1720–1729. doi: 10.1093/infdis/jiab479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ayala-Breton C, Arias M, Espinosa R, Romero P, Arias CF, López S. 2009. Analysis of the kinetics of transcription and replication of the rotavirus genome by RNA interference. J Virol 83:8819–8831. doi: 10.1128/JVI.02308-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Papa G, Venditti L, Braga L, Schneider E, Giacca M, Petris G, Burrone OR. 2020. CRISPR-Csy4-mediated editing of rotavirus double-stranded RNA genome. Cell Rep 32:108205. doi: 10.1016/j.celrep.2020.108205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ball JM, Tian P, Zeng CQ, Morris AP, Estes MK. 1996. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 272:101–104. doi: 10.1126/science.272.5258.101 [DOI] [PubMed] [Google Scholar]
- 37. Sánchez-Tacuba L, Feng N, Meade NJ, Mellits KH, Jaïs PH, Yasukawa LL, Resch TK, Jiang B, López S, Ding S, Greenberg HB. 2020. An optimized reverse genetics system suitable for efficient recovery of simian, human, and murine-like rotaviruses. J Virol 94:e01294-20. doi: 10.1128/JVI.01294-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shan C, Xie X, Zou J, Züst R, Zhang B, Ambrose R, Mackenzie J, Fink K, Shi PY. 2018. Using a virion assembly-defective dengue virus as a vaccine approach. J Virol 92:e01002-18. doi: 10.1128/JVI.01002-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burns JW, Siadat-Pajouh M, Krishnaney AA, Greenberg HB. 1996. Protective effect of rotavirus VP6-specific IgA monoclonal antibodies that lack neutralizing activity. Science 272:104–107. doi: 10.1126/science.272.5258.104 [DOI] [PubMed] [Google Scholar]
- 40. Caddy SL, Vaysburd M, Wing M, Foss S, Andersen JT, O‘Connell K, Mayes K, Higginson K, Iturriza-Gómara M, Desselberger U, James LC. 2020. Intracellular neutralization of rotavirus by VP6-specific IgG. PLoS Pathog 16:e1008732. doi: 10.1371/journal.ppat.1008732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Esquivel FR, Lopez S, Guitierrez-X L, Arias C. 2000. The internal rotavirus protein VP6 primes for an enhanced neutralizing antibody response. Arch Virol 145:813–825. doi: 10.1007/s007050050674 [DOI] [PubMed] [Google Scholar]
- 42. López S, Espinosa R, Greenberg HB, Arias CF. 1994. Mapping the subgroup epitopes of rotavirus protein VP6. Virology 204:153–162. doi: 10.1006/viro.1994.1519 [DOI] [PubMed] [Google Scholar]
- 43. Nurdin JA, Kotaki T, Kawagishi T, Sato S, Yamasaki M, Nouda R, Minami S, Kanai Y, Kobayashi T. 2023. N-glycosylation of rotavirus NSP4 protein affects viral replication and pathogenesis. J Virol 97:e0186122. doi: 10.1128/jvi.01861-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li B, Ding S, Feng N, Mooney N, Ooi YS, Ren L, Diep J, Kelly MR, Yasukawa LL, Patton JT, Yamazaki H, Shirao T, Jackson PK, Greenberg HB. 2017. Drebrin restricts rotavirus entry by inhibiting dynamin-mediated endocytosis. Proc Natl Acad Sci U S A 114:E3642–E3651. doi: 10.1073/pnas.1619266114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yamasaki M, Kanai Y, Wakamura Y, Kotaki T, Minami S, Nouda R, Nurdin JA, Kobayashi T. 2023. Characterization of sialic acid-independent simian rotavirus mutants in viral infection and pathogenesis. J Virol 97:e0139722. doi: 10.1128/jvi.01397-22 [DOI] [PMC free article] [PubMed] [Google Scholar]