Abstract
Some viruses including herpesviruses have undergone evolution to benefit viral infection and propagation by pirating and modifying host genes such as chemokine genes. Human herpesvirus 6 (HHV-6), acutely or persistently infects mononuclear cells in vitro. DNA sequence analysis of HHV-6 has revealed that the putative protein encoded by an open reading frame (ORF) of the U83 gene in HHV-6 variant B resembled a human chemokine. We have cloned the U83 gene and analyzed the biological function of this gene. The U83 gene contained an ORF encoding a 113-amino-acid peptide, starting at the first methionine and containing a possible signal peptide and the typical cysteine residues characteristic of the chemokines. Reverse transcription-PCR analysis of mRNA and immunofluorescent-antibody testing of infected cells both indicated that the encoded protein was a late protein. The ORF U83 gene fused to the Fc gene was expressed as a fusion protein in COS-7 cells by transfection, and the fusion protein was purified from the supernatant of transfected cells to test its biological function. The purified protein was capable of inducing transient calcium mobilization in THP-1 cells and of chemotactically activating THP-1 cells. These findings suggested that the U83 protein might play an important role in HHV-6 propagation in vivo by activating and trafficking mononuclear cells to sites of viral replication, thus aiding the development of superbly efficient virus production mechanisms.
Human herpesvirus 6 (HHV-6) was first isolated in 1986 from the peripheral blood of patients with lymphoproliferative disorders (41). The distinct nature of HHV-6 with respect to other human herpesviruses was confirmed by molecular and immunological analyses (24). The virus replicates predominantly in CD4-positive lymphocytes in vivo and in vitro (31, 50) and may establish latent infection in the monocyte/macrophage-lineage cells (28). Nucleotide sequence analysis of the genome has demonstrated that HHV-6 is closely related to human cytomegalovirus and human herpesvirus 7 (HHV-7) and that these three viruses belong to the betaherpesvirus subfamily (30, 36). Two variants of HHV-6, i.e., HHV-6A and HHV-6B, have been identified based on differences in epidemiology, growth in vitro, antigenic properties, restriction endonuclease profile, and nucleotide sequence (1–3, 9, 18, 44, 57, 59). HHV-6B appears to be isolated more frequently than HHV-6A from the blood except in patients with AIDS (1, 26). In addition, it has been proved that HHV-6B is the etiologic agent of exanthem subitum (60), which is a common childhood illness characterized by a high fever and skin rash. HHV-6B has also been reported to cause bone marrow suppression (16), interstitial pneumonia (8, 13), and encephalitis (17), as well as being associated with an infectious mononucleosis-like illness in adults (48), whereas HHV-6A has not yet been associated with any human diseases.
The entire genome of HHV-6A has been sequenced by Gompels et al. (19), and we have also performed DNA sequencing of the entire HHV-6B strain HST genome (unpublished data). The homology between HHV-6A U1102 and HHV-6B HST was approximately 95% for the DNA sequence and for the amino acid sequence (unpublished data). Analyses of the sequence comparisons have led to the identification of a candidate for a chemokine open reading frame (ORF), ORF U83, within the HHV-6 genome (19). Recently, it has been reported that certain viruses have evolved molecular piracy and mimicry mechanisms so that acquired host genes within virus genomes are able to produce proteins capable of interfering with the normal host defense response (56). Kaposi’s sarcoma-associated herpesvirus, belonging to the gammaherpesvirus subfamily, encodes three chemokine homologs, i.e., viral macrophage inflammatory protein I (vMIP-I), vMIP-II, and vMIP-III (14). Although the functions of vMIP-III have not yet been characterized, vMIP-I and vMIP-II were shown to inhibit human immunodeficiency virus entry into cells through CCR3, CCR5, and CXCR4, which are specific receptors for chemokines (25). In addition, in the chicken chorioallantoic membrane assay, vMIP-I was found to have strong angiogenic properties (6). Furthermore, the vMIP-II protein has in vitro antagonistic activity against CCR1, CCR2, CCR5, CXCR4, and CXCR3 but not against CXCR1 and CXCR2. In vivo, vMIP-II potently inhibits MIP-1α-, MIP-1β-, and RANTES-induced leukocyte infiltration and markedly attenuates proteinuria (10). Additionally, vMIP-II induces a significant cytoplasmic calcium flux in human eosinophils and can induce angiogenesis (6). Molluscum contagiosum virus, a member of the poxvirus family, encodes a secreted CC chemokine homolog, MC148, that potently interferes with the chemotaxis of human monocytes, lymphocytes, and neutrophils that occurs in response to a large number of CC and CXC chemokines with diverse receptor specificity. These findings provide a possible explanation for the absent or delayed inflammatory response in molluscum contagiosum virus lesions (15). In murine cytomegalovirus, belonging to the betaherpesvirus family, a chemokine homolog was also found (32).
Our purpose in the present experiments was to characterize the chemokine homolog U83 of HHV-6B and test whether this gene product has the chemokine properties. We demonstrated that the U83 protein induces transient calcium mobilization in THP-1 cells and efficient chemotactic activity to the cells. Thus, these results suggest that the U83 protein plays a role in HHV-6B pathogenesis by activating mononuclear cells and recruiting them to sites of viral replication in vivo, thus aiding the spread of HHV-6B.
MATERIALS AND METHODS
Cells and virus.
Umbilical cord blood mononuclear cells (CBMCs) were separated on a Ficoll-Conray gradient and cultured for 2 or 3 days in RPMI 1640 medium containing 10% fetal calf serum (FCS) and 5 μg of phytohemagglutinin per ml. HHV-6 HST, which had been isolated from a patient with exanthem subitum (60) and belongs to the HHV-6B variant (59), was grown in CBMCs stimulated with phytohemagglutinin. To prepare a virus stock, the stimulated cells were infected with virus. When more than 80% of the cells showed cytopathic effects, the infected cells were frozen and thawed twice, and after centrifugation at 1,500 × g for 10 min at 4°C, the supernatant was stored at −80°C as a cell-free virus stock. Nonadherent THP-1 cells (53) and MT-4 cells (33) were grown in RPMI 1640 medium supplemented with 10% FCS. COS-7 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% FCS.
Preparation of RNA.
Stimulated CBMCs (approximately 107 cells) were infected with strain HST at a multiplicity of infection of 0.1 50% tissue culture infective dose per cell and centrifuged at 1,500 × g for 40 min at 37°C for adsorption. After being washed twice with phosphate-buffered saline (PBS), the cells were cultured for 3 days in RPMI 1640 medium supplemented with 10% FCS and harvested. Virus-infected cells were pelleted by centrifugation and suspended in 4 M guanidium isothiocyanate containing 0.5% sodium N-lauroylsarcosine and 0.1 M 2-mercaptoethanol. Cycloheximide (CHX) and phosphonoformic acid (PFA) were used as inhibitors of protein and DNA synthesis at 50 and 200 μg/ml, respectively. CHX or PFA was added to cultures from the initiation of infection for 24 h. Total RNA was extracted by the guanidium isothiocyanate method (11).
Preparation of cDNA and 5′ and 3′ RACE.
A primer, U83-5a (5′-ACTAGTACTTACTTGATTCTTTGTCTAATTTCGACA), was used to synthesize first-strand cDNA from 1 μg of total RNA with Superscript II reverse transcriptase (Gibco BRL). After hydrolysis of RNA with RNase H, the first-strand cDNA was purified with a GlassMax DNA isolation spin cartridge (Gibco BRL) and subjected to the oligo(dC) tailing reaction with terminal deoxynucleotidyl transferase. The 5′ ends of the U83 cDNA were obtained by the 5′ rapid amplification of cDNA ends (RACE) method by using the 5′ RACE system of the Rapid Amplification of cDNA Ends kit (Gibco BRL). The 5′ RACE procedure was carried out as specified by the manufacturer. In brief, PCR of dC-tailed cDNA was carried out with a 5′ RACE-abridged anchor primer (5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′) and U83-5b (5′-GATGCGTTTGCCATATCACACATCG-3′) under the following conditions: a total of 35 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, and one final extension at 72°C for 5 min. One-fiftieth of the resulting PCR product was amplified with an abridged universal amplification primer (AUAP) (5′-GGCCACGCGTCGACTAGTAC-3′) and a nested primer, U83-5c (5′-TGCAACACAACAAACATCCTAA-3′). The PCR conditions of the nested PCR were as follows: 30 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, and one final extension at 72°C for 5 min. The PCR product of the nested PCR was cloned into pCR2.1 vector and subjected to DNA sequence analysis. The 3′ ends of the U83 cDNA were obtained by the 3′ RACE method by using the 3′ RACE system of the Rapid Amplification of cDNA Ends kit. First-strand cDNA was synthesized from 5 μg of the total RNA with an adapter primer (5′-GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT-3′) and SuperScript II reverse transcriptase (Gibco BRL). After hydrolysis of the RNA with RNase H, PCR of cDNA was performed with AUAP and U83-3a primer (5′-GTCGACCATGTTCATTTGGCTTTTTATTGTT-3′) under the following conditions: 30 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, and one final extension at 72°C for 5 min. Nested amplification was performed by PCR with AUAP and U83-3b (5′-TGTCGAAATTAGACAAAGAATCATG-3′) under the following conditions: 25 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, and one final extension at 72°C for 5 min. The PCR product of the nested PCR was cloned into pCR2.1 vector and sequenced.
RT-PCR.
Reverse transcription-PCR (RT-PCR) for cDNA synthesis of the regions encoding HHV-6B, immediate-early 1 (IE-1), DNA polymerase (Pol), glycoprotein H (gH), U83, and elongation factor 1a (EF) was performed in a 20-μl solution containing 50 mM Tris-HCl (pH 8.3), 50 mM KCl, 10 mM MgCl2, and 3 mM dithiothreitol and carried out at 42°C for 30 min with 20 U of RAV-2 reverse transcriptase (Takara Shuzo, Kyoto, Japan), 1 μg of cellular RNA, and 0.4 μM oligo(dT). PCR was then performed as described above with EX Taq DNA polymerase (Takara Shuzo); the appropriate pairs of primers for IE-1, Pol, gH, U83, and EF were IE03-F340 (5′-CAGAATTCATGGAAGTACAATCTCCTACTG-3′) and IE03 MR (5′-CACTGCAGTTAATGACTTTTGACAGGAGTTGC-3′), O6-PolR1 (5′-CGAACAGTTTTGCATCTCCGC-3′) and O6-PolC3 (5′-GTTTGTATCCGAGCATTATG-3′), gHF4 (5′-CCAGTCCAAGTCAGATGCGC-3′) and gHR5 (5′-AATAGGGTTTGGATTCCTAGG-3′), U83-F (5′-GTCGACCATGTTCATTTGGCTTTTTATTGTT-3′) and U83-R (5′-ATGAATTCTCATGATTCTTTGTCTAATTTC-3′), and CEF1A (5′-GCTCCAGCATGTTGTCACCATTC-3′) and EF1A (5′-GGTGAATTTGAAGCTGGTATCTC-3′), respectively.
Expression of U83 in E. coli and generation of anti-U83 sera.
To express the U83 protein, an expression plasmid was constructed by using Escherichia coli expression vector pGEX-3X (Pharmacia, Uppsala, Sweden). In this plasmid, the U83 gene was fused to the glutathione S-transferase (GST) gene. Virus DNA from HHV-6B HST was used as a template for PCR. PCR was performed with EX Taq DNA polymerase by using the primers U83–GST-1 (5′-TGGGATCCCCGACGATGACGACAAGTTTA-3′) and U83–GST-2 (5′-ATGAATTCTCATGATTCTTTGTCTAATTTC-3′), which were a sense strand with a BamHI recognition sequence appended at the 5′ end and an antisense strand with an EcoRI recognition sequence at the 5′ end, respectively. The PCR conditions were as follows: 25 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, and one final extension at 72°C for 10 min, in a TP480 PCR thermal cycler (Takara Shuzo). The PCR product was cloned into pCR2.1 (Invitrogen, San Diego, Calif.), and the entire insert was sequenced by using a SequiTher Long-Read cycle-sequencing kit and a 4000L DNA sequencer (Li-Cor, Lincoln, Nebr.). After digestion with BamHI and EcoRI, the product was inserted into the BamHI-EcoRI sites of pGEX-3X to create plasmid pGEXU83. A polypeptide containing amino acid residues 18 to 113 of the U83 protein was produced as a GST fusion protein in E. coli and purified. This fusion protein was used for immunizing rabbits to obtain monospecific antisera. Antibodies were purified with the ImmunoPure (A) immunoglobulin G (IgG) purification kit (Pierce).
Preparation of rU83 virus.
Recombinant U83 (rU83) viruses were prepared by using the Bac-to-Bac baculovirus expression system (Gibco BRL) as recommended by the manufacturer. Hi-5 insect cells (Gibco BRL) were infected with the recombinant viruses and were cultured in Express Five SFM (Gibco BRL). The culture supernatants were collected 2 days after infection. rU83 protein was partially purified with a 25-ml cation-exchange Hiload column (Pharmacia) in a fast protein liquid chromatography system (Pharmacia).
Western blotting.
Samples for Western blot analysis were prepared from recombinant U83 virus-infected insect cells. The samples were dissolved in sample buffer (0.1 M Tris-HCl [pH 6.8], 15% glycerol, 4% sodium dodecyl sulfate [SDS], 0.1% Coomassie brilliant blue G-250) with 5% β-mercaptoethanol, and separated by Tricine-SDS-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (42). Western blot analysis was done as described by Towbin et al. (52). A semidry transfer unit with a polyvinylidene difluoride membrane (Bio-Rad) was used. The membrane was soaked in 5% skim milk–PBS and reacted sequentially with the purified anti-U83 (1:1,000) and alkaline phosphatase-conjugated anti-rabbit IgG (1:5,000) (Promega), and the signals were detected with the ProtBlot NBT and BCIP Color Development System (Promega).
Indirect immunofluorescence assay (IFA).
HST- or mock-infected MT-4 cells were spotted on glass slides, air dried, and fixed with cold acetone. Antibodies diluted (1:100) with a dilution buffer (1× PBS containing 2% bovine serum albumin, 0.2% Tween 20, and 0.05% NaN3) were spotted onto the slides and incubated at 37°C for 45 min. After the slides were washed with PBS for 10 min twice, fluorescein-conjugated goat antibodies against rabbit IgG were spotted onto the slides and incubated at 37°C for another 45 min. Then, after the slides were washed as above, signals were detected by immunofluorescence microscopy.
Production of U83-Fc fusion protein.
Virus DNA prepared from cells infected with strain HST was used as a template. The U83 gene was amplified by PCR. PCR was performed with EX Taq DNA polymerase and primers U83-SalI (5′-GTCGACCATGTTCATTTGGCTTTTTATTGTT-3′) and U83-SpeI (5′-ACTAGTACTTACTTGATTCTTTGTCTAATTTCGACA-3′), which were a sense strand with a SalI recognition sequence appended at the 5′ end and an antisense strand with an SpeI recognition sequence appended at the 5′ end, respectively. The PCR product was cloned into pCR2.1 (Invitrogen), and the entire insert was sequenced. After digestion with SalI and SpeI, the DNA fragment was inserted into the SalI-SpeI sites of the pEF-FC mammalian expression vector (49), which was derived from pEF-Bos (34). To produce a U83-Fc fusion protein, COS-7 cells were transfected with the plasmid together with SuperFectant transfection reagent (Qiagen). The transfected cells were incubated for 24 h in medium containing 10% FCS and then for 48 h in serum-free medium. The supernatants were combined, centrifuged, and passed through a 0.45-μm-pore-size filter to remove cell debris. Then the U83-Fc proteins were purified with the ImmunoPure(A) IgG purification kit (Pierce). Purified fusion proteins were equilibrated with HEPES-buffered Krebs solution (1.24 mM NaCl, 5 mM KCl, 1.24 mM KH2PO4, 1.3 mM MgSO4, 2.4 mM CaCl2, 10 mM glucose, 25 mM HEPES [pH 7.4] (HBKS) and concentrated with a Centricon 10 apparatus (Amicon, Inc.), and the protein concentration was estimated by measuring the optical density at 280 nm.
Intracellular [Ca2+] measurements.
Cells were washed twice in HBKS. Then 107 cells were incubated for 30 min at 37°C in the dark in 1 ml of HBKS containing 5 μM Indo-1AM (Dojin Chemical Co.). The cells were subsequently washed twice with HBKS and resuspended at 2.5 × 106 cells/ml. A 1-ml volume of the cell suspension was placed in a continuously stirred cuvette at 37°C in a CAF-110 fluorometer (Jasco). Fluorescein was monitored at an excitation wavelength of 355 nm and emission wavelengths of 405 and 485 nm, and the data were presented as relative ratios of fluorescein levels excited at 405 and 485 nm. Data were collected every 10 ms.
Chemotaxis assay.
Cell migration was assessed by using Transwell chambers with 3-μm pores (Costar) as described elsewhere (38). A THP-1 cell suspension in 100 μl of assay medium (RPMI 1640 medium supplemented with 20 mM HEPES [pH 7.4] and 0.5% bovine serum albumin) was placed in the upper compartment (106 cells/well), and 0.6 ml of assay medium without or with a chemokine was placed in the lower compartment. After incubation at 37°C for 4 h in 5% CO2, the plates were centrifuged at 300 × g for 5 min. The cells from the lower compartment were counted on a FACSCalibur apparatus (Becton Dickinson). Each experiment was carried out three times.
RESULTS
DNA cloning and cDNA analysis.
Inspection of the nucleotide sequence of HHV-6B HST (29) revealed that ORF U83 is located approximately 4 kb from the right end. We have been particularly interested in this ORF because its deduced amino acid sequence was similar to a chemokine. We have obtained an ORF U83-containing cDNA from infected cells as described in Materials and Methods. Then the 5′ and 3′ ends of the U83 cDNA were obtained by RACE methods. The U83 cDNA was approximately 1.0 kb in size, with the ORF of 345 bp starting with the first methionine codon and encoding a polypeptide of 113 amino acid residues (Fig. 1). The 3′-noncoding region contained the putative polyadenylation signal (AATAAA) and also contained the mRNA destabilization signal (ATTTA) that is often found in cytokine and chemokine sequences (45). The NH2-terminal end of the deduced amino acid sequence was highly hydrophobic and was consistent with the typical signal peptide sequence (54). The cleavage site was predicted to be between amino acid positions 20 (Glu) and 21(Phe) (Fig. 1). The primary sequence of the U83 polypeptide contained five properly placed cysteine residues as well as a putative N-glycosylation site (Fig. 1). The putative molecular mass of the U83 protein without the signal sequence is approximately 10 kDa. However, compared with other chemokines, the polypeptide had an extra NH2-terminal extension of about 14 amino acids, which had relatively low homology to human chemokines. There was no splicing site within this gene.
FIG. 1.
Alignment of the HHV-6BU83 cDNA sequence and its deduced amino acid sequence. The 5′ and 3′ ends of the U83 cDNA were obtained by RACE. The U83 cDNA contained an ORF encoding 113 amino acids, starting at the first methionine codon, which contains the five cysteine residues (asterisks). The vertical arrow indicates the experimentally determined cleavage site of the signal sequence. The putative N-glycosylation site NAS is indicated by a dotted underline. The AATAAA sequence in the 3′ noncoding region indicates the putative polyadenylation signal, and the ATTTA is the mRNA destabilization signal that is often found among cytokine and chemokine cDNAs.
Analysis of U83 expression in HHV-6B-infected cells by RT-PCR.
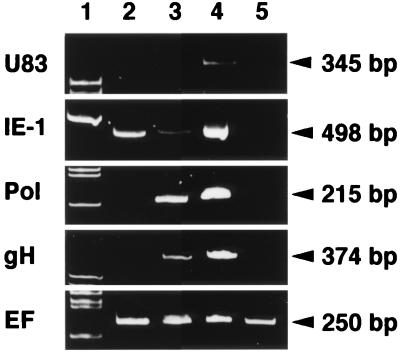
We have examined the transcription of the U83 gene in the presence of an inhibitor of protein or DNA synthesis. RNA was purified from CBMCs infected with strain HST in the presence of CHX or PFA as described in Materials and Methods. Specific RNAs were amplified by RT-PCR with primers for the U83, IE-1, Pol, gH, and EF genes. The PCR products were loaded onto a 10% polyacrylamide gel (Fig. 2). The EF bands, which were amplified from cellular RNA as a control, were displayed at approximately equal intensities in all lanes. No products were amplified from mRNA of mock-infected cells with the primers related to the HHV-6 genome (Fig. 2, lane 5). In the presence of CHX, only the IE-1 band (498 bp) was detected by RT-PCR (lane 2). In the presence of PFA, IE-1, Pol (215-bp), and gH (374-bp) bands appeared strongly but the U83 band was not detected (lane 3). However, in virus-infected but untreated cells, a clear 345-bp band of U83 was detected (lane 4). These results indicated that the U83 gene was expressed as a late gene.
FIG. 2.
RT-PCR assay of the transcripts in the HST-infected cells treated with CHX and PFA. Total RNAs purified from mock-infected (lane 5) and HST-infected (lanes 2 to 4) CBMCs, which had been treated with CHX for 24 h (lane 2) or PFA for 24 h (lane 3) or left untreated (lane 4), were used for the RT-PCR assay of mRNA expression of HHV-6 IE-1, Pol, gH, U83, and cellular EF genes. The EF band, which was amplified from the endogeneous cellular control RNA, was expressed at approximately equal intensities in all lanes. In the presence of CHX or PFA, the U83 band was not detected. In the untreated sample in lane 4, the U83 band was detected. The results indicate that U83 is expressed as a late gene. Lane 1 shows molecular size markers.
Expression of the U83 gene in HHV-6B-infected cells.
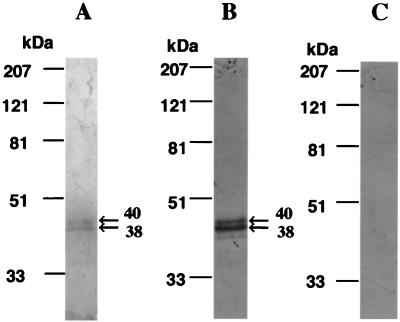
To characterize the protein encoded by the U83 gene, the GST-U83 fusion protein was expressed in E. coli as described in Materials and Methods and later used for raising monospecific antibody against U83 protein. Using the antibody, we examined its specificity to the U83 protein. Western blot analysis revealed that the antibody specifically recognized the purified U83-Fc fusion protein of 38 and 40 kDa (see Fig. 5B) and also recognized purified recombinant U83 protein of 10 and 12.5 kDa in supernatants from the rU83-virus infected Hi-5 cell cultures (see Materials and Methods) (Fig. 3A); however, no specific band was observed by using preimmune serum (Fig. 3B). Thus, our results showed that antibodies specifically recognized the U83 protein.
FIG. 5.
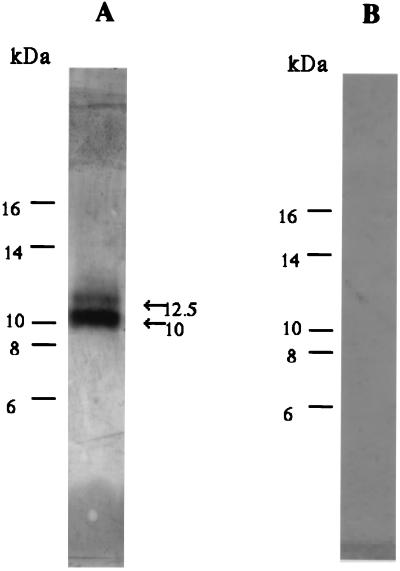
(A) U83-Fc proteins were purified from culture supernatants of transfected COS-7 cells by protein A affinity chromatography, subjected to electrophoresis on a 10% polyacrylamide gel, and stained with Coomassie brilliant blue. (B and C) Western blot analysis showed that purified U83-Fc proteins were detected with anti-U83 serum (B) but not with preimmune serum (C). Amino acid sequencing demonstrated that the NH2 termini of both the 38- and 40-kDa U83-Fc proteins started at Phe-21 of the predicted sequence. These results agreed with the putative signal cleavage site and molecular mass of the cleaved mature protein. Positions of size markers (in kilodaltons) are shown on the left.
FIG. 3.
The monospecific antiserum specifically recognized HHV-6B U83 protein by Western blotting. Partially purified U83 protein from supernatants of rU83 virus-infected Hi-5 cell cultures was reacted with antiserum against GST-U83 fusion protein (A) or with preimmune serum (B).
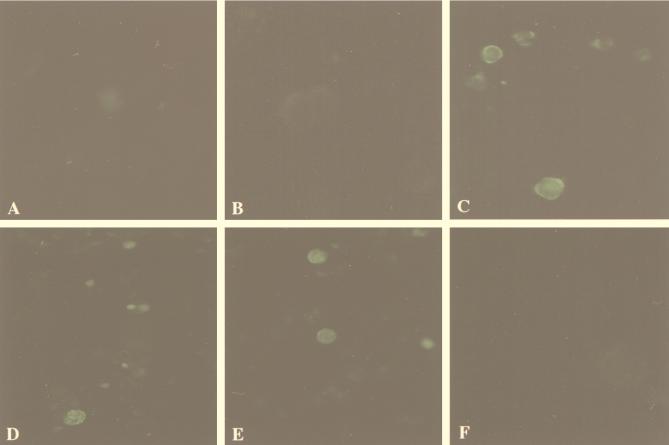
To characterize the viral protein in HHV-6-infected cells, MT-4 cells were mock infected or infected with HHV-6B HST, harvested 48 h after infection, and analyzed by IFA with antibody. As shown in Fig. 4, the antibody reacted with the U83 viral antigen in cytoplasm and on the membrane of HST-infected cells (Fig. 4C). No staining was observed in uninfected cells (Fig. 4A), and no specific staining was observed with preimmune sera in HST-infected cells (Fig. 4B). In the presence of an inhibitor of DNA polymerase, PFA, at 200 μg/ml, U83 antigen in MT4 cells infected with HST was not detected at 48 h by IFA (Fig. 4F) whereas positive control U89 antigen (an immediate-early gene product) and U41 protein (an early gene product) were detected in the nuclear region with anti-U89 rabbit sera (51) and monoclonal antibody OHV-2 against U41 protein (unpublished data), respectively (Fig. 4D and E). These data further demonstrated that U83 antigen was a late protein.
FIG. 4.
Immunofluorescence micrographs. (A) Mock-infected MT4 cells. (B) HST-infected MT4 cells stained with preimmune rabbit sera. (C) HST-infected MT4 cells stained with rabbit anti-U83 sera. (D to F) HST-infected MT4 cells cultured for 48 h in the presence of PFA (200 μg/ml) and stained with rabbit anti-U89 sera (D), monoclonal antibody OHV-2 for U41 (E), and rabbit anti-U83 sera (F).
Expression of U83-Fc fusion protein.
To obtain the U83 protein, the COOH terminus of ORF U83 was fused with the Fc portion of human IgG1 and the U83-Fc fusion protein was produced in COS-7 cells by transfection with the DNA. The U83-Fc fusion protein in the culture supernatants was then purified by protein A affinity chromatography. When the protein was analyzed by SDS-PAGE and stained with Coomassie brilliant blue, the purified fusion protein migrated as double bands of 38 and 40 kDa (Fig. 5A). Sequence analysis of 8 amino acids of the NH2 terminus of 38- and 40-kDa polypeptides was performed, and the NH2 termini of both the U83-Fc proteins were found to start at position 21(Phe). Western blot analysis showed that purified U83-Fc proteins were detected with anti-U83 serum (Fig. 5B) but not with preimmune serum (Fig. 5C). These results showed the polypeptide was cleaved between positions 20 and 21, which corresponded to the putative signal cleavage site. The difference in molecular masses between the two polypeptides was probably due to glycosylation.
Induction of calcium flux by U83 protein.
It has been reported that RANTES was chemotactic for human peripheral blood monocytes and T lymphocytes (43). RANTES also has been shown to induce chemotaxis and calcium mobilization in THP-1 cells (55). THP-1 cells, derived from an acute monocytic leukemia patient, exhibit a rare chromosomal abnormality (53) and express at least CCR1, CCR2B, and V28 (40, 58). We have examined whether U83-Fc was capable of inducing calcium flux in THP-1 cells. THP-1 cells were loaded with Indo-1, a calcium indicator. As shown in Fig. 6, RANTES (R & D System), used as the positive control, showed clear induction. Similarly, U83-Fc protein induced calcium flux in THP-1 whereas a control protein, Fas-Fc protein, as well as a negative control buffer alone, did not induced any calcium flux in THP-1 cells. These data clearly indicated that the U83 had the ability to induce calcium flux in THP-1 cells.
FIG. 6.
THP-1 cells were loaded with Indo-1 AM and stimulated with U83-Fc (100 nM), Fas-Fc (100 nM), or RANTES (100 nM). The arrows indicate the time of application. Intercellular concentrations of calcium were monitored by measuring the fluorescence ratio. U83 protein induced a calcium flux in THP-1 cells.
Chemotaxis activity of U83 protein.
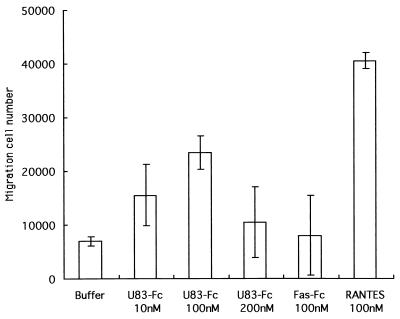
We next examined the chemotactic ability of the U83 protein to induce the migration of THP-1 cells. By transwell migration assays, we analyzed its ability to stimulate chemotaxis of THP-1 cells. RANTES as the positive control induced significant migration compared with that induced by the negative control buffer. The U83-Fc protein also induced efficient migration of THP-1 cells (Fig. 7). U83-Fc, which was capable of attracting THP-1 cells, showed the typical dose-response curve with the maximal effect at 100 nM, whereas the Fas-Fc protein, a negative control protein, was unable to induce chemotaxis in THP-1 cells. These data clearly indicate that the U83 protein has the ability to induce migration in THP-1 cells, as does RANTES. Thus, taken together, our observations clearly demonstrated that U83 is a functional chemokine.
FIG. 7.
Chemotaxis of THP-1 cells by U83-Fc protein. THP-1 cell migration was measured in response to increased concentrations of U83-Fc protein (10, 100, and 200 nM) by using the transwell migration assay system (see Materials and Methods). RANTES at 100 nM served as a positive control. Each experiment was performed at least three times. Representative data from the three experiments are shown. The data are expressed as means and standard deviations.
DISCUSSION
The HHV-6 genome is composed of linear, double-stranded DNA, which has two GC-rich terminal direct repeats, direct repeat left and direct repeat right, and a long AT-rich unique long sequence (UL). In the present study, we have identified and characterized a novel viral gene of chemokine, designated the U83 gene, which encoded a product of 113 amino acids. The chemokine homolog is encoded by HHV-6 ORF U83, and its genomic location is similar between HHV-6A and HHV-6B. It has been speculated from the sequencing results that the U83 gene of HHV-6B would consist of 345 bp encoding 113 amino acids whereas that of HHV-6A would consist of 294 bp encoding 97 amino acids (19). The predicted mature protein of U83 had 93 amino acids, and the U83 shows relatively low sequence similarity to human chemokines: 17.6% to MIP-3β and 11.8% to RANTES over a stretch of 34 amino acids (Mac DNAsis; Hitachi Software Engineering Co., Ltd., Yokohama, Japan). The ORF U83 product of HHV-6B HST had 85.6% amino acid identity to that of HHV-6A U. The major difference between the HHV-6A ORF U83 product and the HHV-6B ORF U83 product was present at their NH2 termini. The HHV-6B ORF U83 product was 16 amino acids longer than that of HHV-6A. The NH2-terminal end of the HHV-6B ORF U83 product was hydrophobic and roughly corresponded to the region containing a typical signal peptide sequence. The cleavage site was present between positions 20 and 21, whereas no clear cleavage site of the leader peptide has been identified near the CC motif of the U83 protein of HHV-6A (15), suggesting that the U83 protein of HHV-6A may not be secreted from infected cells. We next found that the typical poly(A)+ signal sequence AATAAAT was located at the 3′ noncoding region. The 3′ noncoding region also contained the consensus sequence ATTTA, which has been commonly found in mRNAs encoding a variety of cytokines and chemokines (45) and in those encoding proteins related to the inflammatory response (7) but not in mammalian mRNAs in general. The sequence ATTTA appear to correlate with relative instability of mRNA. Thus, its presence in the U83 gene may support the idea that this gene represents a novel viral chemokine.
The chemokine is a small protein with four conserved cysteines capable of forming two essential disulfide bonds (Cys1-Cys3 and Cys2-Cys4). The family of chemokines comprises four subfamilies defined by the distribution profiles of the cystein residues in the NH2 terminal: the CC, CXC, C, and CX3C subfamilies. CC, CXC, and CX3C chemokines are distinguished according to the position of the first two cysteines, which are adjacent (CC), separated by one amino acid (CXC), and separated by three amino acids (CX3C). The ORF U83 protein has five cysteines, the first three of which are found in the sequence of CXXCC, which may correspond to the above CC or/and CX3C profiles. It remains to be determined which profile is functional in the U83 protein.
It is supposed that the gene expression of HHV-6 follows a sequential and regulated pattern. The transcripts can be divided into three broad categories, immediate-early (IE), early (E), and late (L), as is the case with other herpesviruses (22). IE transcriptions require only preformed host factors, and thus the transcripts are expressed despite inhibition of host cell translation. E transcripts require the IE gene products for their expression, whereas L transcripts are expressed after the initiation of viral DNA replication. Our RT-PCR analysis showed that the U83 mRNA belongs to the late kinetic class in HHV-6B-infected CBMCs (Fig. 2). Moreover, our IFA revealed that the expression of U83 protein was not detected in HST-infected cells in the presence of PFA by staining with a specific antibody (Fig. 4). When the recombinant U83-Fc fusion protein was expressed in mammalian cells, purified from culture supernatants, subjected to affinity chromatography, and analyzed by SDS-PAGE, it migrated as double bands of 38 and 40 kDa (Fig. 5). In addition, recombinant U83 partially purified from supernatants of Hi-5 insect cells was detected as double bands by Western blotting analysis. Since amino acid sequencing demonstrated that the NH2 termini of the two polypeptides were the same (data not shown), the differences may be due to some other modification such as glycosylation on the U83 protein. In contrast, only one band was seen in the purified Fas-Fc fusion protein, which was used as a control (data not shown). Computer-aided analysis of the U83 protein sequence showed that the NH2-terminal region was highly hydrophobic, corresponding to the signal peptide. When the recombinant U83-Fc fusion protein was expressed in COS-7 cells, U83 protein was secreted into the medium. Moreover, NH2-terminal sequencing of the U83 protein showed that the signal peptide was cleaved as predicted. Even though the 93-amino-acid sequence of this viral chemokine showed relatively low homology to human chemokines, the ORF U83 retained four properly spaced cysteine residues, which is a hallmark for the chemokines.
Our data showed U83 was able to transduce signals involving the Ca2+ flux in THP cells. However, its inducing level was apparently lower than that of RANTES. Chemokines have two main sites interacting with their receptors, one in the NH2-terminal region and the other within an exposed loop of the backbone that extends between the second and third cysteines (5). The NH2-terminal binding site is essential for triggering the receptor. It is believed that the receptor first recognizes and interacts with the chemokine loop region to correctly present its triggering domain at the NH2-terminus. The mature NH2-terminal region of the chemokines is thought to be involved in biological activity and leukocyte selectivity as described for MCP-1 and interleukin-8 (5, 20). Thus, truncation or elongation of the NH2-terminal sequence leads to considerable loss of those activity (4, 5, 12). In comparison with other chemokines, the U83 protein had an extra NH2-terminal region of 14 amino acids. Thus, the extra region is probably involved in a particular function upon triggering the receptor. So far, there are at least three distinct classes of receptors for chemokines on monocytic THP-1 cell (40, 58). Whether U83 protein belongs to one of these classes or to some other, unknown class remains to be determined.
Molecular piracy of host cellular genes is a newly recognized feature of some herpesviruses. It is noteworthy that in addition to a gene encoding a viral chemokine, there is a gene encoding a viral chemokine receptor within the same viral genome. In HHV-6, there are genes that encode proteins homologous genes to G-protein-coupled receptors, U12 and U51 (19). U12 encodes a functional chemokine receptor for RANTES, MIP-1α, MIP-1β, and MCP-1 (23). These chemokines exert their effects through binding to target cell surface chemokine receptors that belong to the family of G-protein-coupled seven-transmembrane-domain receptors. To date, five CXC chemokine receptors (CXCR1 to CXCR5), at least eight CC chemokine receptors (CCR1 to CCR8), one CX3C chemokine receptor (V28), and one C chemokine receptor (XCR1) have been cloned and characterized (38). The various chemokine receptors are known to exhibit overlapping ligand specificities. In addition, there are numerous orphan chemokine receptors whose ligands have not been identified. The receptor(s) for the U83 protein remains to be determined. At present, it is not known whether the U83 protein binds to the same receptor(s) as that for RANTES. The structure-function studies of the viral chemokine could lead to a greater understanding of the viral chemokine and provide an insight into useful therapeutic strategies that broadly target chemokine-signaling systems. At present, it is also not known whether the U83 viral chemokine affects the signaling of U12. Identification of the viral chemokine and its corresponding receptor will probably lead to a further understanding of their roles in the survival of viruses in the hostile environment.
Chemokines recruit specific leukocyte subsets. The CXC chemokines are chemotactic for neutrophils, whereas the CC chemokines generally attract monocytes and other leukocytes, including lymphocytes, eosonophils, and basophils; the C chemokine is specific for T and NK lymphocytes (21); and the CX3C chemokine appears to be a potent chemoattractant for monocytes and lymphocytes. In addition, chemokines activate leukocytes, promote leukocyte adhesion, and have other effects. Our study also showed that the U83 viral chemokine induced the chemotactic activity of the THP-1 cell line. However, further studies are needed to identify its functional receptor and delineate its in vivo function. The biological roles of the viral chemokine in herpesviruses are not yet known. Mononuclear cells are infected with HHV-6 in vivo, resulting in acute, chronic, and latent infections (39). It is possible that such a virally encoded chemokine acts as chemokine receptor agonist and recruits particular uninfected leukocytes for further viral infection and/or for transmission of the virus or that it stimulates proliferation of these uninfected target cells, hence priming them for productive viral infection. However, it was predicted that the U83 protein of HHV-6A might not be secreted. In this case, it is unlikely to exert U83 chemokine functions. A better understanding of the role of the HHV-6B viral chemokine U83 in viral pathogenesis may allow us to develop better antiviral strategies to reduce the burden of herpesvirus-associated diseases.
ACKNOWLEDGMENTS
We acknowledge S. Nagata, Osaka University, for his gift of pEF-Fc and pEF-Fas plasmids as well as helpful discussions. We also thank T. Imai and O. Yoshie for helpful discussions.
REFERENCES
- 1.Ablashi D V, Balachandran N, Josephs S F, Hung C L, Krueger G R, Kramarsky B, Salahuddin S Z, Gallo R C. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology. 1991;184:545–552. doi: 10.1016/0042-6822(91)90424-a. [DOI] [PubMed] [Google Scholar]
- 2.Aubin J T, Collandre H, Candotti D, Ingrand D, Rouzioux C, Burgard M, Richard S, Huraux J M, Agut H. Several groups among human herpesvirus 6 strains can be distinguished by Southern blotting and polymerase chain reaction. J Clin Microbiol. 1991;29:367–372. doi: 10.1128/jcm.29.2.367-372.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubin J T, Agut H, Collandre H, Yamanishi K, Chandran B, Montagnier L, Huraux J M. Antigenic and genetic differentiation of the two putative types of human herpes virus 6. J Virol Methods. 1993;41:223–234. doi: 10.1016/0166-0934(93)90129-f. [DOI] [PubMed] [Google Scholar]
- 4.Baggiolini M, Dewald B, Moser B. Human chemokines: an update. Annu Rev Immunol. 1997;15:675–705. doi: 10.1146/annurev.immunol.15.1.675. [DOI] [PubMed] [Google Scholar]
- 5.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff C, Endo Y, Collins P D, Takeuchi Y, Reeves J D, Schweickart V L, Siani M A, Sasaki T, Williams T J, Gray P W, Moore P S, Chang Y, Weiss R A. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science. 1997;278:290–294. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 7.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrigan D R, Drobyski W R, Russler S K, Tapper M A, Knox K K, Ash R C. Interstitial pneumonitis associated with human herpesvirus-6 infection after marrow transplantation. Lancet. 1991;338:147–149. doi: 10.1016/0140-6736(91)90137-e. [DOI] [PubMed] [Google Scholar]
- 9.Chandran B, Tirawatnapong S, Pfeiffer B, Ablashi D V. Antigenic relationships among human herpesvirus-6 isolates. J Med Virol. 1992;37:247–254. doi: 10.1002/jmv.1890370403. [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Bacon K B, Li L, Garcia G E, Xia Y, Lo D, Thompson D A, Siani M A, Yamamoto T, Harrison J K, Feng L. In vivo inhibition of CC and CX3C chemokine-induced leukocyte infiltration and attenuation of glomerulonephritis in Wistar-Kyoto (WKY) rats by vMIP-II. J Exp Med. 1998;188:193–198. doi: 10.1084/jem.188.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 12.Clark-Lewis I, Kim K S, Rajarathnam K, Gong J H, Dewald B, Moser B, Baggiolini M, Sykes B D. Structure-activity relationships of chemokines. J Leukoc Biol. 1995;57:703–711. doi: 10.1002/jlb.57.5.703. [DOI] [PubMed] [Google Scholar]
- 13.Cone R W, Hackman R C, Huang M L, Bowden R A, Meyers J D, Metcalf M, Zeh J, Ashley R, Corey L. Human herpesvirus 6 in lung tissue from patients with pneumonitis after bone marrow transplantation. N Engl J Med. 1993;329:156–161. doi: 10.1056/NEJM199307153290302. [DOI] [PubMed] [Google Scholar]
- 14.Dairaghi D J, Greaves D R, Schall T J. Abduction of chemokine elements by herpesviruses. Semin Virol. 1998;8:377–376. [Google Scholar]
- 15.Damon I, Murphy P M, Moss B. Broad spectrum chemokine antagonistic activity of a human poxvirus chemokine homolog. Proc Natl Acad Sci USA. 1998;95:6403–6407. doi: 10.1073/pnas.95.11.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drobyski W R, Dunne W M, Burd E M, Knox K K, Ash R C, Horowitz M M, Flomenberg N, Carrigan D R. Human herpesvirus-6 (HHV-6) infection in allogeneic bone marrow transplant recipients: evidence of a marrow-suppressive role for HHV-6 in vivo. J Infect Dis. 1993;167:735–739. doi: 10.1093/infdis/167.3.735. [DOI] [PubMed] [Google Scholar]
- 17.Drobyski W R, Knox K K, Majewski D, Carrigan D R. Brief report: fatal encephalitis due to variant B human herpesvirus-6 infection in a bone marrow-transplant recipient. N Engl J Med. 1994;330:1356–1360. doi: 10.1056/NEJM199405123301905. [DOI] [PubMed] [Google Scholar]
- 18.Gompels U A, Carrigan D R, Carss A L, Arno J. Two groups of human herpesvirus 6 identified by sequence analyses of laboratory strains and variants from Hodgkin’s lymphoma and bone marrow transplant patients. J Gen Virol. 1993;74:613–622. doi: 10.1099/0022-1317-74-4-613. [DOI] [PubMed] [Google Scholar]
- 19.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 20.Gong J-H, Clark-Lewis I. Antagonists of monocyte chemoattractant protein-1 identified by modification of functionally critical NH2-terminal residues. J Exp Med. 1995;181:631–640. doi: 10.1084/jem.181.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedrick J A, Zlotnik A. Identification and characterization of a novel beta chemokine containing six conserved cysteines. J Immunol. 1997;159:1589–1593. [PubMed] [Google Scholar]
- 22.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isegawa Y, Ping Z, Nakano K, Sugimoto N, Yamanishi K. Human herpesvirus 6 open reading frame U12 encodes a functional beta-chemokine receptor. J Virol. 1998;72:6104–6112. doi: 10.1128/jvi.72.7.6104-6112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Josephs S F, Salahuddin S Z, Ablashi D V, Schachter F, Wong-Staal F, Gallo R C. Genomic analysis of the human B-lymphotropic virus (HBLV) Science. 1986;234:601–603. doi: 10.1126/science.3020691. [DOI] [PubMed] [Google Scholar]
- 25.Kledal T N, Rosenkilde M M, Coulin F, Simmons G, Johnsen A H, Alouani S, Power C A, Luttichau H R, Gerstoft J, Clapham P R, Clark-Lewis I, Wells T N C, Schwartz T W. A broad-spectrum chemokine antagonist encoded by Kaposi’s sarcoma-associated herpesvirus. Science. 1997;277:1656–1659. doi: 10.1126/science.277.5332.1656. [DOI] [PubMed] [Google Scholar]
- 26.Knox K K, Carrigan D R. Active HHV-6 infection in the lymph nodes of HIV-infected patients: in vitro evidence that HHV-6 can break HIV latency. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;11:370–378. doi: 10.1097/00042560-199604010-00007. [DOI] [PubMed] [Google Scholar]
- 27.Koch A E, Polverini P J, Kunkel S L, Harlow L A, DiPietro L A, Elner V M, Elner S G, Strieter R M. Interleukin-8 as a macrophage-derived mediator of angiogenesis. Science. 1992;258:1798–1801. doi: 10.1126/science.1281554. [DOI] [PubMed] [Google Scholar]
- 28.Kondo K, Kondo T, Okuno T, Takahashi M, Yamanishi K. Latent human herpesvirus 6 infection of human monocytes/macrophages. J Gen Virol. 1991;72:1401–1408. doi: 10.1099/0022-1317-72-6-1401. [DOI] [PubMed] [Google Scholar]
- 29.Kosuge H, Isegawa Y, Yamanishi K. Nucleotide sequence analysis of a 30-kilobase-pair region of human herpesvirus-6B (HHV-6B) genome and strain-specific variations in major immediate-early genes. Virus Res. 1997;52:1–14. doi: 10.1016/s0168-1702(97)00099-3. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence G L, Chee M, Craxton M A, Gompels U A, Honess R W, Barrell B G. Human herpesvirus 6 is closely related to human cytomegalovirus. J Virol. 1990;64:287–299. doi: 10.1128/jvi.64.1.287-299.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lusso P, Markham P D, Tschachler E, di Marzo Veronese F, Salahuddin S Z, Ablashi D V, Pahwa S, Krohn K, Gallo R C. In vitro cellular tropism of human B-lymphotropic virus (human herpesvirus-6) J Exp Med. 1988;167:1659–1670. doi: 10.1084/jem.167.5.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacDonald M R, Li X, Virgin H W. Late expression of a β-chemokine homolog by murine cytomegalovirus. J Virol. 1997;71:1671–1678. doi: 10.1128/jvi.71.2.1671-1678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyoshi I, Taguchi H, Kubonishi I, Yoshimoto S, Ohtsuki Y, Shiraishi Y, Akage T. Type C virus-producing cell lines derived from adult T-cell leukemia. GANN Monogr Can Res. 1982;28:219–228. [Google Scholar]
- 34.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322–5323. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 36.Mukai T, Isegawa Y, Yamanishi K. Identification of the major capsid protein gene of human herpesvirus 7. Virus Res. 1995;37:55–62. doi: 10.1016/0168-1702(95)00022-i. [DOI] [PubMed] [Google Scholar]
- 37.Murphy P M. Molecular piracy of chemokine receptors by herpesviruses. Infect Agents Dis. 1994;3:137–154. [PubMed] [Google Scholar]
- 38.Nagira M, Imai T, Hieshima K, Kusuda J, Ridanpaa M, Takagi S, Nishimura M, Kakizaki M, Nomiyama H, Yoshie O. Molecular cloning of a novel human CC chemokine secondary lymphoid-tissue chemokine that is a potent chemoattractant for lymphocytes and mapped to chromosome 9p13. J Biol Chem. 1997;272:19518–19524. doi: 10.1074/jbc.272.31.19518. [DOI] [PubMed] [Google Scholar]
- 39.Pellett P E, Black J B. Human herpesvirus 6. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2587–2608. [Google Scholar]
- 40.Raport C J, Schweickart V L, Eddy J R, Shows T B, Gray P W. The orphan G-protein-coupled receptor-encoding gene V28 is closely related to genes for chemokine receptors and is expressed in lymphoid and neural tissues. Gene. 1995;163:295–299. doi: 10.1016/0378-1119(95)00336-5. [DOI] [PubMed] [Google Scholar]
- 41.Salahuddin S Z, Ablashi D V, Markham P D, Josephs S F, Sturzenegger S, Kaplan M, Halligan G, Biberfeld P, Wong-Staal F, Kramarsky B, Gallo R C. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science. 1986;234:596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- 42.Schagger H, Von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 43.Schall T J, Bacon K, Toy K J, Goeddel D V. Selective attraction of monocytes and T lymphocytes of the memory phenotype by cytokine RANTES. Nature. 1990;347:669–671. doi: 10.1038/347669a0. [DOI] [PubMed] [Google Scholar]
- 44.Schirmer E C, Wyatt L S, Yamanishi K, Rodriguez W J, Frenkel N. Differentiation between two distinct classes of viruses now classified as human herpesvirus 6. Proc Natl Acad Sci USA. 1991;88:5922–5926. doi: 10.1073/pnas.88.13.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 46.Simmons G, Clapham P R, Picard L, Offord R E, Rosenkilde M M, Schwartz T W, Buser R, Wells T N C, Proudfoot A E. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 47.Singh N, Carrigan D R. Human herpesvirus-6 in transplantation: an emerging pathogen. Ann Intern Med. 1996;124:1065–1071. doi: 10.7326/0003-4819-124-12-199606150-00007. [DOI] [PubMed] [Google Scholar]
- 48.Steeper T A, Horwitz C A, Ablashi D V, Salahuddin S Z, Saxinger C, Saltzman R, Schwartz B. The spectrum of clinical and laboratory findings resulting from human herpesvirus-6 (HHV-6) in patients with mononucleosis-like illnesses not resulting from Epstein-Barr virus or cytomegalovirus. Am J Clin Pathol. 1990;93:776–783. doi: 10.1093/ajcp/93.6.776. [DOI] [PubMed] [Google Scholar]
- 49.Suda T, Nagata S. Purification and characterization of the Fas-ligand that induces apoptosis. J Exp Med. 1994;179:873–879. doi: 10.1084/jem.179.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi K, Sonoda S, Higashi K, Kondo T, Takahashi H, Takahashi M, Yamanishi K. Predominant CD4 T-lymphocyte tropism of human herpesvirus 6-related virus. J Virol. 1989;63:3161–3163. doi: 10.1128/jvi.63.7.3161-3163.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeda K, Nakagawa N, Yamamoto T, Inagi R, Kawanishi K, Isegawa Y, Yamanishi K. Prokayotic expression of an immediate-early gene of human herpesvirus 6 and analysis of its viral antigen expression on human cells. Virus Res. 1996;42:193–200. doi: 10.1016/0168-1702(96)01287-7. [DOI] [PubMed] [Google Scholar]
- 52.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1) Int J Cancer. 1980;26:171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 54.von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 55.Wang J M, McVicar D W, Oppenheim J J, Kelvin D J. Identification of RANTES receptors on human monocytic cells: competition for binding and desensitization by homologous chemotactic cytokines. J Exp Med. 1993;177:699–705. doi: 10.1084/jem.177.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wells T N, Schwartz T W. Plagiarism of the host immune system: lessons about chemokine immunology from viruses. Curr Opin Biotechnol. 1997;8:741–748. doi: 10.1016/s0958-1669(97)80129-2. [DOI] [PubMed] [Google Scholar]
- 57.Wyatt L S, Balachandran N, Frenkel N. Variations in the replication and antigenic properties of human herpesvirus 6 strains. J Infect Dis. 1990;162:852–857. doi: 10.1093/infdis/162.4.852. [DOI] [PubMed] [Google Scholar]
- 58.Xu L, Rahimpour R, Ran L, Kong C, Biraggn A, Andrews J, Derries M, Wang J-M, Kelvin D J. Regulation of CCR2 chemokine receptor mRNA stability. J Leukoc Biol. 1997;62:653–660. doi: 10.1002/jlb.62.5.653. [DOI] [PubMed] [Google Scholar]
- 59.Yamamoto T, Mukai T, Kondo K, Yamanishi K. Variation of DNA sequence in immediate-early gene of human herpesvirus 6 and variant identification by PCR. J Clin Microbiol. 1994;32:473–476. doi: 10.1128/jcm.32.2.473-476.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;i:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]