ABSTRACT
SARS-CoV-2 variants of concern (VOCs) differentially trigger neutralizing and antibody-dependent cellular cytotoxic (ADCC) antibodies with variable cross-reactivity. Omicron BA.4/5 was approved for inclusion in bivalent vaccination boosters, and therefore the antigenic profile of antibodies elicited by this variant is critical to understand. Here, we investigate the ability of BA.4/5-elicited antibodies following the first documented (primary) infection (n = 13) or breakthrough infection after vaccination (n = 9) to mediate neutralization and FcγRIIIa signaling across multiple SARS-CoV-2 variants including XBB.1.5 and BQ.1. Using a pseudovirus neutralization assay and a FcγRIIIa crosslinking assay to measure ADCC potential, we show that unlike SARS-CoV-2 Omicron BA.1, BA.4/5 infection triggers highly cross-reactive functional antibodies. Cross-reactivity was observed both in the absence of prior vaccination and in breakthrough infections following vaccination. However, BQ.1 and XBB.1.5 neutralization and FcγRIIIa signaling were significantly compromised compared to other VOCs, regardless of prior vaccination status. BA.4/5 triggered FcγRIIIa signaling was significantly more resilient against VOCs (<10-fold decrease in magnitude) compared to neutralization (10- to 100-fold decrease). Overall, this study shows that BA.4/5 triggered antibodies are highly cross-reactive compared to those triggered by other variants. Although this is consistent with enhanced neutralization and FcγRIIIa signaling breadth of BA.4/5 vaccine boosters, the reduced activity against XBB.1.5 supports the need to update vaccines with XBB sublineage immunogens to provide adequate coverage of these highly antibody evasive variants.
IMPORTANCE
The continued evolution of SARS-CoV-2 has resulted in a number of variants of concern. Of these, the Omicron sublineage is the most immune evasive. Within Omicron, the BA.4/5 sublineage drove the fifth wave of infection in South Africa prior to becoming the dominant variant globally. As a result this spike sequence was approved as part of a bivalent vaccine booster, and rolled out worldwide. We aimed to understand the cross-reactivity of neutralizing and Fc mediated cytotoxic functions elicited by BA.4/5 infection following infection or breakthrough infection. We find that, in contrast to BA.1 which triggered fairly strain-specific antibodies, BA.4/5 triggered antibodies that are highly cross-reactive for neutralization and antibody-dependent cellular cytotoxicity potential. Despite this cross-reactivity, these antibodies are compromised against highly resistant variants such as XBB.1.5 and BQ.1. This suggests that next-generation vaccines will require XBB sublineage immunogens in order to protect against these evasive variants.
KEYWORDS: SARS-CoV-2, variant of concern, Omicron BA.4, BA.5, BA.1, XBB sublineage, neutralization, breakthrough infection, antibody-dependent cellular cytotoxicity
INTRODUCTION
SARS-CoV-2 variants of concern (VOCs) which contain mutations within the spike gene have been associated with viral escape from neutralization (1–4), and thus reduced vaccine efficacy (5). Omicron BA.1 emerged in November 2021 and contains >30 mutations in the spike region, resulting in further reduced neutralization titers (6). Omicron has since evolved into several sublineages showing even more extensive immune escape, including BA.2, BA2.12.1, BA.4, and BA.5 (7–9). The BA.4 and BA.5 sublineages, which share the same spike mutations but differ from one another in non-structural proteins as well as the nucleocapsid and matrix genes, drove the fifth wave of infection in South Africa, and were subsequently detected worldwide (10). Within spike, BA.4 and BA.5 are genetically similar to BA.2 but contain the 69–70 deletion, and additional substitutions in the receptor-binding domain, namely L452R, F486V, and the R493 reversion to the ancestral Wuhan sequence. Therefore, compared to BA.1 and BA.2, BA.4 showed increased neutralization resistance to vaccinee and convalescent sera, and monoclonal antibodies (4, 11, 12). Subsequently, two highly immune evasive Omicron subvariants BQ1.1 and XBB.1.5 arose from BA.4/5 and BA.2, respectively, and circulated globally, evading both vaccine and infection elicited humoral immunity (13, 14). More recently the emergence of XBB.1.16, EG.5.1, and the extremely mutated BA.2.86 continue to threaten vaccine efficacy (15).
In contrast to neutralizing antibodies (nAbs), Fc effector function, or the ability of antibodies to recruit cytotoxic functions through their binding to Fc receptor or complement proteins, has typically been preserved against VOCs (16–19). However, we recently found the crosslinking of FcγRIIIa/CD16 and BA.4, the first stage of antibody-dependent cellular cytotoxicity (ADCC), was the lowest in convalescent plasma, suggesting that these functions are vulnerable to certain mutations in the spike (20). Given the association between Fc effector function and vaccine protection in animal models (21–25) as well as reduced COVID-19 mortality and severity (26, 27), Fc-mediated function is an important parameter to consider for future vaccine design.
In addition to conferring variable escape from antibodies, mutations in spike also result in the elicitation of qualitatively different responses after infection by different VOCs. We and others have shown that each SARS-CoV-2 variant triggers different profiles of nAbs and Fc effector functions (2, 3, 16, 17, 20, 28, 29). For example, the Beta variant triggered humoral responses with increased cross-reactivity, and consequently became a focus for the development of second-generation vaccines (16, 28, 30–34). In contrast, Omicron BA.1 triggered more strain-specific nAbs and Fc effector functions with limited cross-reactivity in individuals without prior infection (17). This likely reflects its significant divergence from the wild-type variant and suggests that BA.1 may not be an optimal insert for next-generation vaccines alone (17, 29). In line with this, ancestral Wuhan-Hu-1 and Omicron BA.1 bivalent vaccines have shown superior neutralizing activity only against BA.1 related lineages but are comparable for other variants relative to Wuhan-Hu-1 vaccines alone (35, 36), suggesting that for increased breadth, other variant-based vaccines may be preferable. BA.4/5 vaccine boosters also showed significantly improved neutralization of the BA.4 variant compared to ancestral vaccines but remain compromised by currently circulating variants such as XBB.1.5 (37).
Here, we assessed neutralization and ADCC potential (as measured by FcγRIIIa signaling) triggered by BA.4 infection in previously vaccinated and unvaccinated individuals and compared the degree of cross-reactivity with plasma from previous waves of infection in South Africa, caused by D614G, Beta, Delta, or BA.1. We confirm that unvaccinated individuals with sequence confirmed BA.4/5 infection (or those who were infected when BA.4 was dominant, and presumed infected with BA.4/5), show cross-reactive neutralization and ADCC potential responses, to the same degree as Beta elicited infection. Similarly, following BA.4/5 breakthrough infections (BTIs), responses were cross-reactive although of higher magnitude compared to non-BTIs. In addition, we note the significant immune evasion of new Omicron sublineages such as XBB.1.5 and BQ 1.1 even in the case of BA4/5 BTI. This suggests that vaccine boosters will likely need to be updated with circulating XBB lineages in order to provide neutralization and Fc effector function breadth against these immune evasive variants.
RESULTS
BA.4 triggered neutralizing antibodies are highly cross-reactive
Plasma from individuals infected during the fifth wave of the COVID-19 pandemic in South Africa was used to assess neutralization cross-reactivity. Samples were obtained from 34 hospitalized individuals from the Tshwane District Hospital recruited between 9 and 30 May 2022, at a median of 2 days (range 0–9 days) after a positive PCR test (Table S1). During this period, national sequencing showed that Omicron BA.4/5 was responsible for >90% of infections (10). Of these participants, 29 had matched nasal swabs available. Whole-genome sequencing data with more than 50% genome coverage was generated for 21/29 samples and 62% (13/21) of these could be assigned a clade and lineage. Clade and lineage assignments reflected that 85% (11/13) and 15% (2/13) of these Omicron sequences were BA.4 and BA.5, respectively. Eleven individuals had previously been vaccinated with either one dose of Ad26.CoV2.S (n = 2) or two doses of BNT162b2 (n = 8) at least 2 months (range 56–163 days) prior to infection (Table S1). One individual was only vaccinated with one dose of BNT162b2. Twenty-three individuals were unvaccinated and had no history of previous symptomatic COVID-19 infection either confirmed by interview or with the absence of any record in the national database of previous infection (Table S1).
Plasma from unvaccinated individuals were screened against the sequence matched BA.4/5 pseudovirus using a lentiviral pseudovirus neutralization assay and 43% (10/23) showed no detectable neutralization (Fig. S1). We note that this is a higher proportion of non-responders than we previously observed in the fourth wave of infections caused by BA.1 (20% non-responders) (17), perhaps reflecting milder disease and a consequent lack of humoral response. Indeed, 9/10 unvaccinated non-responders were hospitalized for other reasons and were incidental SARS-CoV-2 detections, with mild or asymptomatic infections. In contrast, a lower fraction of the 12 vaccinated individuals were non-responders (n = 2). This suggests that vaccination is reliably able to induce higher magnitude neutralizing antibodies than infection.
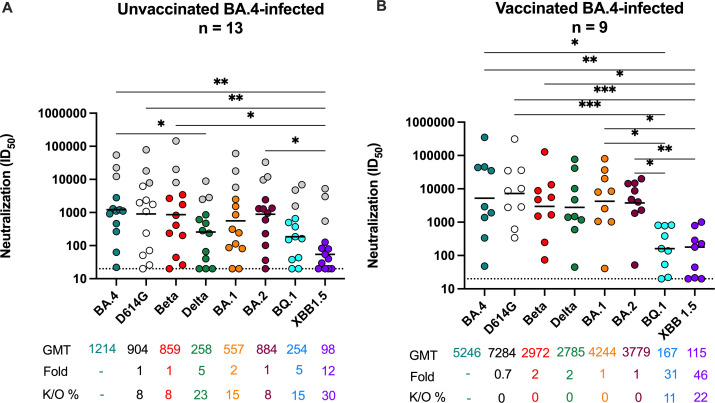
We next assessed cross-neutralization to the ancestral D614G, Beta, Delta, Omicron BA.1, BA.2 and BA.4, BQ.1, and XBB.1.5, excluding non-responders (individuals who failed to mount autologous responses to BA.4/5) (Fig. 1A). In unvaccinated individuals, we observed the highest titers against the matched BA.4/5 spike [geometric mean titer (GMT) of 1,214 in those with detectable neutralizing titers]. These moderate titers are consistent with the mild/asymptomatic infections these individuals experienced (Table S1). Against VOCs, the fold reduction in neutralization titers was variable, with titers against D614G, Beta, and BA.2 not significantly lower than against BA.4/5 (GMT of 904, 859, and 884, respectively). Titers against BA.1 were twofold lower (GMT of 557) and those against Delta were significantly lower, with a GMT of 258. In line with other studies (14, 38), BQ.1 and XBB.1.5 were the most neutralization evasive variants tested, with 5- and 12-fold reductions in titers compared to BA.4/5, respectively. Of the 13, we noted three individuals (highlighted in gray) who despite reporting no previous vaccination or SARS-CoV-2 infection showed high titers across the VOCs, consistent with the cross-reactivity seen following BTI (17, 39). This is likely the consequence of a previous asymptomatic/unknown infection, but we could not confirm this by nucleocapsid antibody testing, as no pre-infection samples had been stored. However, exclusion of these samples did not result in a significant change in the pattern of cross-reactivity.
Fig 1.
Omicron BA.4/5 triggers cross-variant neutralizing antibodies which are broadened by prior vaccination. Neutralization titer (ID50) of Omicron BA.4/5-infected plasma against D614G, Beta, Delta, Omicron BA.1, BA.2, BA.4, BQ.1, and XBB.1.5 pseudoviruses shown for (A) unvaccinated individuals (n = 13) or (B) individuals vaccinated with either one dose of Ad26.CoV.2S or two doses of BNT162b2 (n = 9) before infection. Lines indicate geometric mean titer (GMT), also represented below the plot with fold decrease and knock-out (K/O) of activity for other variants as a percentage relative to Omicron BA.4. Dotted lines indicate the limit of detection of the assay. Statistical significance across variants is shown by Friedman test with Dunns correction. All data are representative of two independent experiments. Gray dots indicate individuals that have consistently high titers across all VOCs, but no evidence of prior infection/vaccination.
We and others have previously shown that BTI following vaccination results in increased neutralization titers (17, 39, 40). Here, in nine previously vaccinated individuals with BTI, we observed significantly boosted titers against BA.4/5 with a GMT of 5,246 (Fig. 1B), fourfold higher than observed in unvaccinated individuals. In contrast to unvaccinated participants where autologous titers were highest, in BTI titers were highest against D614G (GMT 7,284), not BA.4, consistent with prior exposure to the vaccine insert (Wuhan-1). Against Beta, Delta, BA.1, and BA.2, we observed a one- to twofold reduction in titers, none of which were significant, with GMTs of 2,972 for Beta to 4,244 for BA.1 (Fig. 1B). However, against the highly resistant BQ.1 and XBB.1.5 variants, neutralization titers were reduced 31- and 46-fold, respectively, compared to BA.4/5. Overall, in previously vaccinated BTIs, neutralizing responses triggered by BA.4/5 BTI triggered high titers of cross-reactive antibodies that nonetheless show reduced activity against currently circulating VOCs.
BA.4/5-elicited antibodies show cross-reactive FcγRIIIa signaling
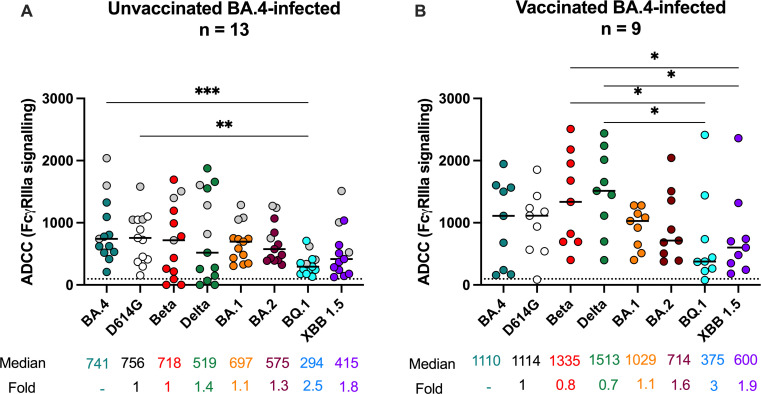
Given the role of ADCC in reduced disease severity and vaccine-elicited protection (25, 27), we investigated the ADCC cross-reactivity of BA.4/5-elicited antibodies as measured by FcγRIIIa signaling, a proxy for lysis. In unvaccinated individuals, FcγRIIIa signaling was similar and highest against BA.4/5 and ancestral D614G (median relative RLU 741 and 756, respectively) (Fig. 2A). For the other VOCs, FcγRIIIa signaling against Beta, Delta, BA.1, BA.2, and XBB.1.5 was lower but not significantly so (1- to 1.8-fold), while against BQ.1 FcγRIIIa signaling was significantly lower (2.5-fold) compared to both BA.4/5 and D614G. In contrast to neutralization where titers were similar between BQ.1 and XBB.1.5, FcγRIIIa signaling levels were lower against BQ.1 than XBB.1.5. In support of this, correlations between neutralization and FcγRIIIa signaling were strong against D614G but weaker for BQ.1 (Fig. S2A). The loss of both FcγRIIIa signaling and neutralization activity against BQ.1 and XBB.1.5 did not however coincide with a detectable difference in IgG binding to these spikes relative to D614G similar to what has been observed for other variants (28). This suggests that the two functions target these VOCs and their defining mutations slightly differently.
Fig 2.
Omicron BA.4/5 triggers cross-variant FcγRIIIa signaling antibodies in unvaccinated and BTI individuals. ADCC potential as measured by FcγRIIIa (CD16) cross-linking of Omicron BA.4/5-infected plasma against D614G, Beta, Delta, Omicron BA.1, BA.2, BA.4, BQ.1, and XBB.1.5 spike shown for (A) unvaccinated individuals (n = 13) or (B) individuals vaccinated with either one dose of Ad26.CoV.2S or two doses of BNT162b2 (n = 9). Lines indicate median relative light units (RLUs) also represented below the plot with fold decrease as a percentage relative to Omicron BA.4. Statistical significance across variants is shown by Friedman test with Dunn’s correction. All data are representative of two independent experiments. The dotted line represents the limit of detection of the assay, calculated on the highest mean of the tested VOCs plus 3 standard deviations of the mean.
In plasma from vaccinated donors, BA.4/5 BTI elicited a similar pattern of FcγRIIIa signaling cross-reactivity compared to unvaccinated individuals (Fig. 2B). However, BTI elicited 1.2- and 2.9-fold higher FcγRIIIa signaling levels across VOCs compared to unvaccinated individuals. This was similar to the ranges seen for BA.1 BTI compared to unvaccinated BA.1-infected individuals (1.1- to 2.6-fold higher), as we reported elsewhere (17). D614G-specific FcγRIIIa signaling levels were not significantly different from BA.4/5 levels (median relative RLUs 1,114 and 1,110, respectively). While FcγRIIIa signaling against Beta and Delta (1,335 and 1,513) was higher than BA.4/5, this was not significant. However, FcγRIIIa signaling was significantly lower against BQ.1 and XBB.1.5, compared to other VOCs. These data show that overall, BA.4/5 infection elicited cross-reactive ADCC potential activity which was broadened by vaccination, and as with neutralization was compromised against BQ.1 and XBB.1.5.
BA.4 elicits antibodies with higher cross-reactive neutralizing and FcγRIIIa signaling responses than previous VOCs
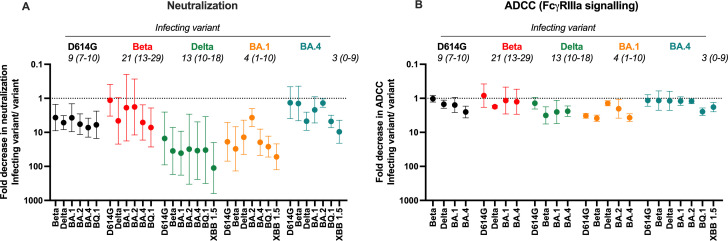
To compare the breadth of antibodies triggered by BA.4/5 infection in unvaccinated individuals, we performed a meta-analysis of previously described responses in infections caused by each of the waves in South Africa (D614G, Beta, Delta, BA.1, and BA.4/5) (16, 17, 29, 41). To assess cross-reactivity, we measured the ratio of the neutralization titer (Fig. 3A) or ADCC potential (Fig. 3B) against the infecting (autologous) virus over that of each VOC, with values <1 representing loss of cross-reactivity against specific variants. We show that while Beta and BA.4/5 triggered neutralizing responses that are highly cross-reactive (close to 1), the Delta and BA.1 variants triggered responses with less cross-reactivity, with 10- to 100-fold reductions in titer against many VOCs (Fig. 3A). Against all waves, BQ.1 and XBB.1.5 show the most substantial fold differences relative to the infecting variant. FcγRIIIa signaling showed a similar pattern to neutralization with Beta and BA.4 showing the most cross-reactivity. However, in line with previous studies, FcγRIIIa signaling showed far smaller fold changes than neutralization (less than 10-fold), an indication that Fc effector function is preserved against VOCs (16, 19). This further reflects the ability of ADCC potential-eliciting antibodies to target more diverse epitopes on the spike as compared to neutralizing antibodies. Overall, BA.4/5 elicits a highly cross-reactive response, not unlike those of Beta elicited antibodies (28).
Fig 3.
Comparison of cross-reactivity of antibodies elicited by infection D614G, Beta, Delta, BA.1, and BA.4 infection in previously unvaccinated individuals. Fold decrease in (A) neutralization and (B) ADCC potential for each VOC represented as a ratio of the titer against the infecting variant (D614G, Beta, Delta, BA.1, and BA.4/5) over that titer against unmatched VOCs. Shown are geometric means of the ratios, with error bars denoting the 95% CI. All data are representative of two independent experiments. Median days post-infection for each group in indicated in italics with ranges indicated in brackets.
DISCUSSION
While several studies have explored the ability of BA.4/5-elicited antibodies to neutralize VOCs (14, 38, 42–44), we compare this to plasma from infections by other VOCs to assess the value of BA.4/5-specific vaccine boosters. We also measured the ADCC potential of BA.4/5-elicited plasma, in order to understand the Fc effector cross-reactivity, given its role in vaccine protection (21–25). Here, we show that humoral responses triggered by BA.4/5 show higher neutralization and ADCC potential cross-reactivity overall, comparable to that induced by Beta. Following BTI infections, responses were of similar cross-reactivity but of higher magnitude compared to non-BTIs. Despite higher titers following BTI, both ADCC potential and neutralization were significantly escaped by immune evasive Omicron sublineages XBB.1.5 and BQ.1.
In this study, we confirm the extreme neutralization resistance of both XBB.1.5 and BQ.1 to antibodies from unvaccinated and vaccinated individuals, as reported elsewhere in the following BA.4/5/5 infection or BTI (14, 38, 43, 44). We, like others, show that XBB.1.5 is more resistant to BA.4/5-elicited antibodies than BQ.1 (38, 43, 44), illustrating the continued emergence of concerning mutations that continue to evade humoral responses. While BA.4/5 triggered FcγRIIIa signaling was extensively escaped by XBB.1.5 and BQ.1, this was particularly true for BQ.1, even following BTI. A similar pattern of evasion was seen in a study that measured ADCC against BQ 1.1 and XBB.1 in individuals who were boosted with a bivalent Wu/BA.4/5 vaccine or broke through the vaccination with an Omicron sublineage variant infection (45). The contrasting pattern between FcγRIIIa signaling and neutralization likely reflects potential differential targeting between the two functions, as we have previously shown (16); however, this requires further epitope mapping.
We and others have shown that BTI significantly increases the cross-reactivity and levels of both neutralizing and ADCC antibodies (17, 20, 40, 46). We show here that this is also true for BA.4/5 BTI responses. However, in contrast to infections by other VOCs, titers against BQ.1 and XBB.1.5 were similar in vaccinated individuals with BA.4/5 BTIs and unvaccinated individuals. This Indicates how significantly these variants compromise neutralization, regardless of hybrid immune status through vaccination prior to infection. While BQ.1 and XBB.1.5 also showed compromised FcγRIIIa signaling, it was elevated in vaccinated BTIs compared with unvaccinated individuals, indicative of the relative preservation of ADCC-mediating antibodies against most SARS-CoV-2 variants (16, 19, 20).
Mutations in the spike imprint different responses following infection, with variants triggering unique antigenic profiles of both neutralizing antibodies and Fc effector functions (2, 3, 16, 17, 20, 28, 29). In our study, we show that BA.4/5-elicited antibodies have the highest neutralization titers against the autologous BA.4/5 variant, similar to what was observed in BA.5 infected hamsters (38). However, in another study, unvaccinated individuals infected with BA.4/5 showed the highest neutralization titers against the ancestral D614G variant (14), likely as a result of unreported previous infections with other variants. As SARS-CoV-2 seropositivity was estimated at 79% in South Africa shortly after the fourth wave driven by BA.1, we cannot rule out previous asymptomatic infection, and it is highly likely that many individuals had prior infection despite reporting no COVID-19 symptoms. We find, similar to others, BA.4/5-elicited plasma shows high neutralization titers against BA.2, from which BA.4/5 only differs by three amino acids (38). In comparison to BA.1 infections, we also found that BA.4/5 resulted in a higher proportion of individuals without detectable neutralization titers. As neutralization titer correlates with disease severity, the lack of neutralizing responses may be as a result of BA.4/5-infected individuals being less likely to develop severe disease than BA.1-infected individuals in South Africa (47).
For ADCC potential, we show that BA.4/5 infection elicits antibodies capable of similar function against the autologous variant, ancestral D614G and Beta. This is in contrast to BA.1 triggered antibodies which showed significantly lower ADCC potential against ancestral D614G and Beta (17). In addition to XBB.1.5 and BQ.1, which are more resistant to BA.4/5-elicited ADCC antibodies, Delta showed reduced sensitivity to FcγRIIIa signaling. This reflects unique antigenic imprinting that also influences Fc effector functions. BA.4/5 triggered neutralization and ADCC potential primarily showed similar antigenic profiles, with Delta showing both compromised neutralization and FcγRIIIa signaling levels. However, this may not be true for all variants, with significantly lower Fc effector function mounted against BA.2 compared to BA.1 by BNT162b2 elicited antibodies, despite similar neutralization profiles (48). Similarly, there are differences between Fc effector functions, with antibody dependent cellular phagocytosis (ADCP) showing a profile distinct from ADCC potential following BA.1 infection (17), indicative of differential targeting of the spike between functions. Additionally, overall we confirm that FcγRIIIa signaling cross-reactivity was preserved against variants of concern (<10-fold difference from the infecting variant) compared to neutralization (10- to 100-fold difference), supporting the significant role in protection that ADCC may have in the absence of neutralization.
We found that BA.4 elicited antibodies are highly cross-reactive in both neutralization and FcγRIIIa signaling compared to antibodies elicited by other VOCs, including ancestral D614G, Beta, Delta, and Omicron BA.1. While it is likely that this increased cross-reactivity compared to BA.1 infection may result from prior undetected SARS-CoV-2 infections, this immune profile is now representative of the South African population, and much of the world (49). Nonetheless, these data suggest that in the context of highly immune evasive variants such as XBB.1.5, immunization with BA.4/5 is unlikely to result in adequate cross-reactivity to prevent infection.
The U.S. FDA and European Medicines Agency authorized the emergency use of the BNT162b2 bivalent BA.4/5-vaccine in September 2022. In line with our study, a booster dose of a bivalent BA.4/5 vaccine resulted in higher neutralization titers for BA.4/5, BQ.1, and XBB compared to the original mRNA vaccines (50–52). However, compared to a BA.1/Wu bivalent vaccine, neutralization escape was similar against Omicron sublineages, although titers were higher against BA.4 in individuals vaccinated with BA.4/5/Wu bivalent vaccine (53). Similarly, in this study, we also show that despite much higher cross-reactivity, BA.4/5-infected individuals, even those with hybrid immunity show significant losses against emergent variants. Indeed, individuals vaccinated with three doses of the ancestral variant vaccine followed by a fourth dose of the BA.4/5 vaccine also show compromised neutralization against BQ.1 and XBB 1.5.1 (54). Early data suggest a small advantage in vaccine effectiveness with BA.4/5 bivalent vaccines over original or BA.1 bivalent vaccine in preventing hospitalization and death (55, 56). However, given the pace at which new variants emerge, new vaccine boosters are currently recommended to target the XBB lineages.
Overall, these data suggest that, in the absence of prior vaccination, unlike BA.1, the BA.4/5 spike elicits antibodies that are highly cross-reactive for VOCs. Serological mapping studies and isolation of monoclonal antibodies from BA.4-infected individuals will provide insights into the targets of these cross-reactive neutralizing and ADCC responses. These data support the use of particular variants to elicit more cross-reactive neutralization and Fc effector function. As such it remains crucial to continue monitoring variants as they emerge in the context of hybrid immunity. However, given the significant decrease noted against currently circulating variants such as XBB.1.5 and BQ.1 even for BA4/5 triggered antibodies, next-generation vaccines will likely require the use of XBB sublineage immunogens.
Limitations of the study
We acknowledge that the numbers of samples in each group are small. Furthermore, although we have extensive clinical follow-up, it is likely that convalescent donors had experienced previous undocumented asymptomatic infection which could alter the breadth of humoral responses. Finally, viral sequences were available only for a subset of samples in each wave, though the samples were collected when each variant dominated infections during that particular wave.
MATERIALS AND METHODS
Human subjects
Plasma samples from the first SARS-CoV-2 wave (D614G-infected, n = 10) were obtained from a previously described cohort across various sites in South Africa prior to September 2020 (3). Second-wave samples (Beta-infected, n = 10) were obtained from a cohort of patients admitted to Groote Schuur Hospital, Cape Town on December 2020 to January 2021 (28). Third-wave samples (Delta-infected, n = 10) were obtained from the Steve Biko Academic Hospital, Tshwane from patients admitted in July 2021 (29). Samples infected in the fourth COVID-19 wave of infection in South Africa (Omicron BA.1-infected, n = 20) were collected from participants enrolled to the Pretoria COVID-19 study cohort following admission to Tshwane District Hospital (Pretoria, South Africa) between 25 November 2021 and 20 December 2021 (17). Participants from the fifth wave (BA.1-infected, n = 34) were also admitted to Tshwane District Hospital (Pretoria, South Africa) in May 2022 (Table S1). For this cohort, only those that showed neutralization titers (responders) were used for formal analysis. In all waves, samples were collected when more than 90% of SARS-CoV-2 cases in South Africa were caused by the respective variants. Sequence confirmation was only available for a subset of samples but all the samples that were sequenced corresponded to the appropriate variant for that wave. All samples were from HIV-uninfected individuals who were above 18 years of age. Ethical clearance was obtained for each cohort from Human Research Ethics Committees from the University of Pretoria (247/2020) and the University of Cape Town (R021/2020). All patients had PCR confirmed SARS-CoV-2 infection before blood collection. Written informed consent was obtained from all participants.
Cell lines
Human embryo kidney HEK293T cells were cultured at 37°C, 5% CO2, in Dulbecco’s modified Eagle’s medium containing 10% heat-inactivated fetal bovine serum (Gibco BRL Life Technologies) and supplemented with 50 µg/mL gentamicin (Sigma). Cells were disrupted at confluence with 0.25% trypsin in 1 mM EDTA (Sigma) every 48–72 h. HEK293T/ACE2.MF cells were maintained in the same way as HEK293T cells but were supplemented with 3 µg/mL puromycin for selection of stably transduced cells. HEK293F suspension cells were cultured in 293 Freestyle media (Gibco BRL Life Technologies) and cultured in a shaking incubator at 37°C, 5% CO2, 70% humidity at 125 rpm maintained between 0.2 and 0.5 million cells/mL. Jurkat-Lucia NFAT-CD16 cells were maintained in IMDM media with 10% heat-inactivated fetal bovine serum (Gibco, Gaithersburg, MD), 1% Penicillin Streptomycin (Gibco, Gaithersburg, MD), and 10 µg/mL of Blasticidin and 100 µg/mL of Zeocin were added to the growth medium every other passage.
Spike plasmid and lentiviral pseudovirus production
The SARS-CoV-2 Wuhan-1 spike, cloned into pCDNA3.1 was mutated using the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies) and NEBuilder HiFi DNA Assembly Master Mix (NEB) to include D614G (original) or lineage defining mutations for Beta (L18F, D80A, D215G, 242–244del, K417N, E484K, N501Y, D614G, and A701V), Delta (T19R, 156–157del, R158G, L452R, T478K, D614G, P681R, and D950N), Omicron BA.1 (A67V, 69–70del, T95I, G142D, 143–145del, 211del, L212I, 214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, and L981F), Omicron BA.2 (T19I, L24S, 25–27del, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, and N969K), Omicron BA.4 (T19I, L24S, 25–27del, 69–70del, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, and N969K), BQ.1 (BA.4 mutations with K444T and N460K) or XBB.1.5 (T19I, L24S, 25–27del, V83A, G142D, 144del, H146Q, Q183E, V213E, G252V, G339H, R346T, L368I, S371F, S373P, S373F, T376A, D405N, R408S, K417N, N440K, V445P, G446S, N460K, S477N, T478K, E484A, F486P, F490S, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, and N969K). Pseudotyped lentiviruses were prepared by co-transfecting HEK293T cell line with the SARS-CoV-2 ancestral variant spike (D614G), Beta, Delta, Omicron BA.1, Omicron BA.2, Omicron BA.4, BQ.1, or XBB.1.5 spike plasmids in conjunction with a firefly luciferase encoding lentivirus backbone (HIV-1 pNL4.luc) plasmid as previously described (20). Culture supernatants were clarified of cells by 0.45 µM filter and stored at −70°C.
Pseudovirus neutralization assay
For the neutralization assay, plasma samples were heat-inactivated and clarified by centrifugation. Heat-inactivated plasma samples from vaccine recipients were incubated with the SARS-CoV-2 pseudotyped virus for 1 h at 37°C, 5% CO2. Subsequently, 1 × 104 HEK293T cells engineered to over-express ACE-2 (293T/ACE2.MF) [kindly provided by M. Farzan (Scripps Research)] were added and incubated at 37°C, 5% CO2 for 72 h upon which the luminescence of the luciferase gene was measured. Titers were calculated as the reciprocal plasma dilution (ID50) causing 50% reduction of relative light units (RLUs). AIRU946-A6 was used as a positive control.
SARS-CoV-2 antigens
For enzyme-linked immunosorbent assay (ELISA), SARS-CoV-2 D614G full spike protein was expressed in Human Embryonic Kidney (HEK) 293F suspension cells by transfecting the cells with the respective expression plasmid. After incubating for 6 days at 37°C, 70% humidity, and 10% CO2, proteins were first purified using a nickel resin, followed by size-exclusion chromatography. Relevant fractions were collected and frozen at −80°C until use. BQ.1 and XBB 1.5 full spikes were purchased from SinoBiological.
SARS-CoV-2 spike enzyme-linked immunosorbent assay
A total of 2 μg/mL of spike protein was used to coat 96-well, high-binding plates and incubated overnight at 4°C. The plates were incubated in a blocking buffer consisting of 5% skimmed milk powder, 0.05% Tween 20, 1× PBS. Plasma samples were diluted to 1:100 starting dilution in a blocking buffer and added to the plates. IgG secondary antibody was diluted to 1:3,000 or 1:1,000, respectively, in blocking buffer and added to the plates followed by TMB substrate (Thermofisher Scientific). Upon stopping the reaction with 1 M H2SO4, absorbance was measured at a 450 nm wavelength. Palivizumab was used as a negative control.
FcγRIIIa (CD16) signaling assay
The ability of plasma antibodies to cross-link and signal through FcγRIIIa (CD16) and spike-expressing cells was measured as a proxy for ADCC. For spike assays, HEK293T cells were transiently transfected with 5 mg of native SARS-CoV-2 spike plasmids using PEI- MAX 40,000 (Polysciences) and incubated for 2 days at 37°C. AIRU946-A6, which binds to different soluble spike variants comparably as determined by ELISA, was used to confirm similar amounts of spike expression on the surface of the cells across variants through the detection by anti-IgG PE staining measured by flow cytometry. Palivizumab against all variants, untransfected cells and transfected cells not incubated with mAb were used as negative controls. Subsequently, spike transfected cells were incubated with heat inactivated plasma (1:100 final dilution) or monoclonal antibodies (final concentration of 100 µg/mL) in RPMI 1640 media supplemented with 10% FBS 1% Pen/Strep (Gibco, Gaithersburg, MD) for 1 h at 37 C. Jurkat-Lucia NFAT-CD16 cells (Invivogen) (2 × 105 cells/well) were added and incubated for 24 h at 37C, 5% CO2. About 20 µL of supernatant was then transferred to a white 96-well plate with 50 µL of reconstituted QUANTI-Luc secreted luciferase and read immediately on a Victor 3 luminometer with 1 s integration time. Normalized RLUs of a no antibody control were subtracted as background. Palivizumab was used as a negative control, CR3022 was used as a positive control, and P2B-2F6 was used to differentiate the Beta from the D614G variant. 084-7D was used as a positive control for Omicron BA.1 and Beta. AIRU946-E4 and AIRU946-A6 were used as additional positive controls showing similar activity across variants. A positive threshold was set using 20 SARS-CoV-2 negative plasma samples from prior to the pandemic as shown in Fig. S3. To induce the transgene 1× cell stimulation cocktail (Thermofisher Scientific, Oslo, Norway) and 2 µg/mL ionomycin in R10 was added as a positive control to confirm sufficient expression of the Fc receptor. RLUs for spikes were normalized to each other and between runs using AIRU946-A6. All samples were run head to head in the same experiment as were all variants tested. Samples were run in duplicate and repeated a minimum of two times.
SARS-CoV-2 whole-genome sequencing
Viral RNA was extracted using the MagNA Pure 96 DNA and Viral Nucleic Acid kit on the MagNA Pure 96 system (Roche Diagnostics) as per the manufacturer’s instructions. Quantitative PCR was performed with the TaqPath COVID-19 CE-IVD RT-PCR Kit (Thermo Fisher Scientific) to obtain mean cycle threshold (Ct) values for the extracts, for SARS-CoV-2 N, S, and ORF1ab genes, according to the manufacturer’s instructions. The Ct values were used to split extracts into batches of n = 8 per batch, with a maximum Ct range of 5 within a batch. Two batches were included per run. Extracts with <1,000 copies were sequenced according to the low viral titer protocol. Batches were sequenced using the Ion AmpliSeq SARS-CoV-2 Insight Research Assay on an Ion Torrent Genexus Integrated Sequencer (Thermo Fisher Scientific). Each run completed within 24 h, and included automated cDNA synthesis, library preparation, templating preparation, and sequencing. The Ion AmpliSeq SARS-CoV-2 Insight Research Assay includes human expression controls for quality control. Final SARS-CoV-2 amplicons displayed lengths ranging from 125 to 275 bp.
SARS-CoV-2 genome assemblies
Fastq files were uploaded to the SARS-CoV-2 RECoVERY pipeline (Reconstruction of Coronavirus Genomes & Rapid Analysis, v3.1) available on Galaxy ARIES (https://aries.iss.it) for genome assembly (24). Final consensus genomes were downloaded and aligned to the reference MN908947 using NextAlign via the NextClade web portal (https://clades.nextstrain.org/, v2.0.0). Genomes were viewed in Nextclade for the presence of unknown frameshifts. Further inspection of alignments using Aliview v1.27 (https://ormbunkar.se/aliview/) revealed that these were due to single nucleotide indels and as such were likely sequencing artifacts; these were corrected to fix the open reading frames. Known frameshifts were not corrected. Nextstrain clade assignments were obtained using NextClade v2.0.0 and pangolin lineage assignments were obtained using the pangolin command line tool (pangolin v4.0.6, using the command pangolin --skip-scorpio <fasta > to avoid overwriting of BA.4 and BA.5 calls, https://github.com/cov-lineages/pangolin/issues/449). All sequences were further manually inspected to confirm lineage assignments. For all sequences that remained unassigned, clade assignments were updated to match.
Statistical analyses
Analyses were performed in Prism (v9; GraphPad Software Inc, San Diego, CA, USA). Nonparametric tests were used for all comparisons. The Mann-Whitney and Wilcoxon tests were used for unmatched and paired samples, respectively. The Friedman test with Dunn’s correction for multiple comparisons was used for matched comparisons across variants. All correlations reported are nonparametric Spearman’s correlations. P values less than 0.05 were considered statistically significant.
ACKNOWLEDGMENTS
The authors thank Z van der Walt, T de Villiers, P Rheeder, A Malan, W van Hougenhouck-Tulleken for clinical support. P.L.M. is supported by the South African Research Chairs Initiative of the Department of Science and Innovation and National Research Foundation of South Africa, the SA Medical Research Council SHIP program, the Centre for the AIDS Program of Research (CAPRISA).
This work was supported, in whole or in part, by the Bill & Melinda Gates Foundation (INV-030570). Under the grant conditions of the Foundation, a Creative Commons Attribution 4.0 Generic License has already been assigned to the Author Accepted Manuscript version that might arise from this submission. The Sequencing activities for NICD (CRDM) are supported by the African Society of Laboratory Medicine (ASLM) and Africa Centers for Disease Control and Prevention through a sub-award from the Bill and Melinda Gates Foundation (grant number INV-018978).
Contributor Information
Penny L. Moore, Email: pennym@nicd.ac.za.
Mark T. Heise, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA
DATA AVAILABILITY
Whole-genome sequencing data of SARS-CoV-2 isolates has been deposited in the Global Initiative on Sharing All Influenza Data (GISAID) database and accession numbers are indicated in Table S1.
All data are available in this manuscript.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.00678-24.
Table S1; Figures S1 to S3.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Cele S, Jackson L, Khoury DS, Khan K, Moyo-Gwete T, Tegally H, San JE, Cromer D, Scheepers C, Amoako DG, et al. 2022. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 602:654–656. doi: 10.1038/s41586-021-04387-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cele S, Gazy I, Jackson L, Hwa S-H, Tegally H, Lustig G, Giandhari J, Pillay S, Wilkinson E, Naidoo Y, Karim F, Ganga Y, Khan K, Bernstein M, Balazs AB, Gosnell BI, Hanekom W, Moosa M-Y, Lessells RJ, de Oliveira T, Sigal A, Network for Genomic Surveillance in South Africa, COMMIT-KZN Team . 2021. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature 593:142–146. doi: 10.1038/s41586-021-03471-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, Lambson BE, de Oliveira T, Vermeulen M, van der Berg K, Rossouw T, Boswell M, Ueckermann V, Meiring S, von Gottberg A, Cohen C, Morris L, Bhiman JN, Moore PL. 2021. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med 27:622–625. doi: 10.1038/s41591-021-01285-x [DOI] [PubMed] [Google Scholar]
- 4. Tuekprakhon A, Nutalai R, Dijokaite-Guraliuc A, Zhou D, Ginn HM, Selvaraj M, Liu C, Mentzer AJ, Supasa P, Duyvesteyn HME, et al. 2022. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 185:2422–2433. doi: 10.1016/j.cell.2022.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collie S, Champion J, Moultrie H, Bekker L-G, Gray G. 2022. Effectiveness of BNT162b2 vaccine against Omicron variant in South Africa. N Engl J Med 386:494–496. doi: 10.1056/NEJMc2119270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Viana R, Moyo S, Amoako DG, Tegally H, Scheepers C, Althaus CL, Anyaneji UJ, Bester PA, Boni MF, Chand M, et al. 2022. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 603:679–686. doi: 10.1038/s41586-022-04411-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evans JP, Zeng C, Qu P, Faraone J, Zheng Y-M, Carlin C, Bednash JS, Zhou T, Lozanski G, Mallampalli R, Saif LJ, Oltz EM, Mohler PJ, Xu K, Gumina RJ, Liu S-L. 2022. Neutralization of SARS-CoV-2 Omicron sub-lineages BA.1, BA.1.1, and BA.2. Cell Host Microbe 30:1093–1102. doi: 10.1016/j.chom.2022.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hachmann NP, Miller J, Collier AY, Ventura JD, Yu J, Rowe M, Bondzie EA, Powers O, Surve N, Hall K, Barouch DH. 2022. Neutralization escape by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, and BA.5. N Engl J Med 387:86–88. doi: 10.1056/NEJMc2206576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Qu P, Faraone J, Evans JP, Zou X, Zheng Y-M, Carlin C, Bednash JS, Lozanski G, Mallampalli RK, Saif LJ, Oltz EM, Mohler PJ, Gumina RJ, Liu S-L. 2022. Neutralization of the SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 subvariants. N Engl J Med 386:2526–2528. doi: 10.1056/NEJMc2206725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tegally H, Moir M, Everatt J, Giovanetti M, Scheepers C, Wilkinson E, Subramoney K, Makatini Z, Moyo S, Amoako DG, et al. 2022. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat Med 28:1785–1790. doi: 10.1038/s41591-022-01911-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Q, Guo Y, Iketani S, Nair MS, Li Z, Mohri H, Wang M, Yu J, Bowen AD, Chang JY, Shah JG, Nguyen N, Chen Z, Meyers K, Yin MT, Sobieszczyk ME, Sheng Z, Huang Y, Liu L, Ho DD. 2022. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 608:603–608. doi: 10.1038/s41586-022-05053-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, et al. 2022. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 608:593–602. doi: 10.1038/s41586-022-04980-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uraki R, Ito M, Furusawa Y, Yamayoshi S, Iwatsuki-Horimoto K, Adachi E, Saito M, Koga M, Tsutsumi T, Yamamoto S, Otani A, Kiso M, Sakai-Tagawa Y, Ueki H, Yotsuyanagi H, Imai M, Kawaoka Y. 2023. Humoral immune evasion of the omicron subvariants BQ.1.1 and XBB. Lancet Infect Dis 23:30–32. doi: 10.1016/S1473-3099(22)00816-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qu P, Faraone JN, Evans JP, Zheng Y-M, Carlin C, Anghelina M, Stevens P, Fernandez S, Jones D, Panchal AR, Saif LJ, Oltz EM, Zhang B, Zhou T, Xu K, Gumina RJ, Liu S-L. 2023. Enhanced evasion of neutralizing antibody response by Omicron XBB.1.5, CH.1.1, and CA.3.1 variants. Cell Rep 42:112443. doi: 10.1016/j.celrep.2023.112443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Q, Guo Y, Zhang RM, Ho J, Mohri H, Valdez R, Manthei DM, Gordon A, Liu L, Ho DD. 2023. Antibody neutralization of emerging SARS-CoV-2: EG.5.1 and XBC.1.6. bioRxiv. doi: 10.1101/2023.08.21.553968 [DOI] [PubMed]
- 16. Richardson SI, Manamela NP, Motsoeneng BM, Kaldine H, Ayres F, Makhado Z, Mennen M, Skelem S, Williams N, Sullivan NJ, Misasi J, Gray GG, Bekker L-G, Ueckermann V, Rossouw TM, Boswell MT, Ntusi NAB, Burgers WA, Moore PL. 2022. SARS-CoV-2 Beta and Delta variants trigger Fc effector function with increased cross-reactivity. Cell Rep Med 3:100510. doi: 10.1016/j.xcrm.2022.100510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Richardson SI, Madzorera VS, Spencer H, Manamela NP, van der Mescht MA, Lambson BE, Oosthuysen B, Ayres F, Makhado Z, Moyo-Gwete T, Mzindle N, Motlou T, Strydom A, Mendes A, Tegally H, de Beer Z, Roma de Villiers T, Bodenstein A, van den Berg G, Venter M, de Oliviera T, Ueckermann V, Rossouw TM, Boswell MT, Moore PL. 2022. SARS-CoV-2 Omicron triggers cross-reactive neutralization and Fc effector functions in previously vaccinated, but not unvaccinated, individuals. Cell Host Microbe 30:880–886. doi: 10.1016/j.chom.2022.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bartsch YC, Tong X, Kang J, Avendaño MJ, Serrano EF, García-Salum T, Pardo-Roa C, Riquelme A, Cai Y, Renzi I, Stewart-Jones G, Chen B, Medina RA, Alter G. 2022. Omicron variant Spike-specific antibody binding and Fc activity are preserved in recipients of mRNA or inactivated COVID-19 vaccines. Sci Transl Med 14:eabn9243. doi: 10.1126/scitranslmed.abn9243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaplonek P, Fischinger S, Cizmeci D, Bartsch YC, Kang J, Burke JS, Shin SA, Dayal D, Martin P, Mann C, Amanat F, Julg B, Nilles EJ, Musk ER, Menon AS, Krammer F, Saphire EO, Alter G, Andrea Carfi . 2022. mRNA-1273 vaccine-induced antibodies maintain Fc effector functions across SARS-CoV-2 variants of concern. Immunity 55:355–365. doi: 10.1016/j.immuni.2022.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richardson SI, Kgagudi P, Manamela NP, Kaldine H, Venter EM, Pillay T, Lambson BE, van der Mescht MA, Hermanus T, Balla SR, de Beer Z, de Villiers TR, Bodenstein A, van den Berg G, du Pisanie M, Burgers WA, Ntusi NAB, Abdullah F, Ueckermann V, Rossouw TM, Boswell MT, Moore PL. 2023. Antibody-dependent cellular cytotoxicity against SARS-CoV-2 Omicron sub-lineages is reduced in convalescent sera regardless of infecting variant. Cell Rep Med 4:100910. doi: 10.1016/j.xcrm.2022.100910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McMahan K, Yu J, Mercado NB, Loos C, Tostanoski LH, Chandrashekar A, Liu J, Peter L, Atyeo C, Zhu A, et al. 2021. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 590:630–634. doi: 10.1038/s41586-020-03041-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, Nkolola JP, Liu J, Li Z, Chandrashekar A, et al. 2020. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369:806–811. doi: 10.1126/science.abc6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gorman MJ, Patel N, Guebre-Xabier M, Zhu AL, Atyeo C, Pullen KM, Loos C, Goez-Gazi Y, Carrion R, Tian J-H, et al. 2021. Fab and Fc contribute to maximal protection against SARS-CoV-2 following NVX-CoV2373 subunit vaccine with Matrix-M vaccination. Cell Rep Med 2:100405. doi: 10.1016/j.xcrm.2021.100405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mercado NB, Zahn R, Wegmann F, Loos C, Chandrashekar A, Yu J, Liu J, Peter L, McMahan K, Tostanoski LH, et al. 2020. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 586:583–588. doi: 10.1038/s41586-020-2607-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mackin SR, Desai P, Whitener BM, Karl CE, Liu M, Baric RS, Edwards DK, Chicz TM, McNamara RP, Alter G, Diamond MS. 2023. Fc-γR-dependent antibody effector functions are required for vaccine-mediated protection against antigen-shifted variants of SARS-CoV-2. Nat Microbiol 8:569–580. doi: 10.1038/s41564-023-01359-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Atyeo C, Fischinger S, Zohar T, Slein MD, Burke J, Loos C, McCulloch DJ, Newman KL, Wolf C, Yu J, Shuey K, Feldman J, Hauser BM, Caradonna T, Schmidt AG, Suscovich TJ, Linde C, Cai Y, Barouch D, Ryan ET, Charles RC, Lauffenburger D, Chu H, Alter G. 2020. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity 53:524–532. doi: 10.1016/j.immuni.2020.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zohar T, Loos C, Fischinger S, Atyeo C, Wang C, Slein MD, Burke J, Yu J, Feldman J, Hauser BM, Caradonna T, Schmidt AG, Cai Y, Streeck H, Ryan ET, Barouch DH, Charles RC, Lauffenburger DA, Alter G. 2020. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell 183:1508–1519. doi: 10.1016/j.cell.2020.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moyo-Gwete T, Madzivhandila M, Makhado Z, Ayres F, Mhlanga D, Oosthuysen B, Lambson BE, Kgagudi P, Tegally H, Iranzadeh A, et al. 2021. Cross-reactive neutralizing antibody responses elicited by SARS-CoV-2 501Y.V2 (B.1.351). N Engl J Med 384:2161–2163. doi: 10.1056/NEJMc2104192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moyo-Gwete T, Madzivhandila M, Mkhize NN, Kgagudi P, Ayres F, Lambson BE, Manamela NP, Richardson SI, Makhado Z, van der Mescht MA, de Beer Z, de Villiers TR, Burgers WA, Ntusi NAB, Rossouw T, Ueckermann V, Boswell MT, Moore PL. 2022. Shared N417-dependent epitope on the SARS-CoV-2 Omicron, Beta, and Delta plus variants. J Virol 96:e00558-23. doi: 10.1128/jvi.00558-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Keeton R, Richardson SI, Moyo-Gwete T, Hermanus T, Tincho MB, Benede N, Manamela NP, Baguma R, Makhado Z, Ngomti A, et al. 2021. Prior infection with SARS-CoV-2 boosts and broadens Ad26.COV2.S immunogenicity in a variant-dependent manner. Cell Host Microbe 29:1611–1619. doi: 10.1016/j.chom.2021.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sheward DJ, Mandolesi M, Urgard E, Kim C, Hanke L, Perez Vidakovics L, Pankow A, Smith NL, Castro Dopico X, McInerney GM, Coquet JM, Karlsson Hedestam GB, Murrell B. 2021. Beta RBD boost broadens antibody-mediated protection against SARS-CoV-2 variants in animal models. Cell Rep Med 2:100450. doi: 10.1016/j.xcrm.2021.100450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanofi . 2022. Sanofi-GSK next-generation COVID-19 booster delivers strong immune response against variants of concern, including Omicron
- 33. CureVac . 2022. CureVac and GSK’s bivalent second-generation mRNA vaccine candidate shown to be highly effective against SARS-CoV-2 variants in preclinical study
- 34. Launay O, Cachanado M, Luong Nguyen LB, Ninove L, Lachâtre M, Ben Ghezala I, Bardou M, Schmidt-Mutter C, Lacombe K, Laine F, et al. 2022. Immunogenicity and safety of beta-adjuvanted recombinant booster vaccine. N Engl J Med 387:374–376. doi: 10.1056/NEJMc2206711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chalkias S, Harper C, Vrbicky K, Walsh SR, Essink B, Brosz A, McGhee N, Tomassini JE, Chen X, Chang Y, Sutherland A, Montefiori DC, Girard B, Edwards DK, Feng J, Zhou H, Baden LR, Miller JM, Das R. 2022. A bivalent Omicron-containing booster vaccine against Covid-19. N Engl J Med 387:1279–1291. doi: 10.1056/NEJMoa2208343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winokur P, Gayed J, Fitz-Patrick D, Thomas SJ, Diya O, Lockhart S, Xu X, Zhang Y, Bangad V, Schwartz HI, et al. 2023. Bivalent Omicron BA.1–adapted BNT162b2 booster in adults older than 55 years. N Engl J Med 388:214–227. doi: 10.1056/NEJMoa2213082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Springer DN, Bauer M, Medits I, Camp JV, Aberle SW, Burtscher C, Höltl E, Weseslindtner L, Stiasny K, Aberle JH. 2023. Bivalent COVID-19 mRNA booster vaccination (BA.1 or BA.4/BA.5) increases neutralization of matched Omicron variants. npj Vaccines 8:110. doi: 10.1038/s41541-023-00708-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mykytyn AZ, Rosu ME, Kok A, Rissmann M, van Amerongen G, Geurtsvankessel C, de Vries RD, Munnink BBO, Smith DJ, Koopmans MPG, Lamers MM, Fouchier RAM, Haagmans BL. 2023. Antigenic mapping of emerging SARS-CoV-2 Omicron variants BM.1.1.1, BQ.1.1, and XBB.1. Lancet Microbe 4:e294–e295. doi: 10.1016/S2666-5247(22)00384-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kitchin D, Richardson SI, Mescht MA, Motlou T, Mzindle N, Moyo-Gwete T, Makhado Z, Ayres F, Manamela NP, Spencer H, Lambson B, Oosthuysen B, Mennen M, Skelem S, Williams N, Ntusi NAB, Burgers WA, Gray GG, Bekker L-G, Boswell MT, Rossouw TM, Ueckermann V, Moore PL. 2021. Ad26.COV2.S breakthrough infections induce high titers of antibodies capable of neutralizing variants of concern. medRxiv. doi: 10.1101/2021.11.08.21266049 [DOI] [PMC free article] [PubMed]
- 40. Walls AC, Sprouse KR, Bowen JE, Joshi A, Franko N, Navarro MJ, Stewart C, Cameroni E, McCallum M, Goecker EA, Degli-Angeli EJ, Logue J, Greninger A, Corti D, Chu HY, Veesler D. 2022. SARS-CoV-2 breakthrough infections elicit potent, broad, and durable neutralizing antibody responses. Cell 185:872–880. doi: 10.1016/j.cell.2022.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scheepers C, Everatt J, Amoako DG, Tegally H, Wibmer CK, Mnguni A, Ismail A, Mahlangu B, Lambson BE, Richardson SI, et al. 2021. Emergence and phenotypic characterization of C.1.2, a globally detected lineage that rapidly accumulated mutations of concern. medRxiv. doi: 10.1101/2021.08.20.21262342 [DOI] [Google Scholar]
- 42. Faraone JN, Qu P, Evans JP, Zheng Y-M, Carlin C, Anghelina M, Stevens P, Fernandez S, Jones D, Lozanski G, Panchal A, Saif LJ, Oltz EM, Gumina RJ, Liu S-L. 2023. Neutralization escape of Omicron XBB, BR.2, and BA.2.3.20 subvariants. Cell Rep Med 4:101049. doi: 10.1016/j.xcrm.2023.101049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen J-J, Li L-B, Peng H-H, Tian S, Ji B, Shi C, Qian C, Jiang W-G, Liu M-C, Li T-T, Shen Y, Fang L-Q, Wang G-L. 2023. Neutralization against XBB.1 and XBB.1.5 after omicron subvariants breakthrough infection or reinfection. Lancet Reg Health West Pac 33:100759. doi: 10.1016/j.lanwpc.2023.100759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Q, Iketani S, Li Z, Liu L, Guo Y, Huang Y, Bowen AD, Liu M, Wang M, Yu J, Valdez R, Lauring AS, Sheng Z, Wang HH, Gordon A, Liu L, Ho DD. 2023. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 186:279–286. doi: 10.1016/j.cell.2022.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Addetia A, Piccoli L, Case JB, Park Y-J, Beltramello M, Guarino B, Dang H, Pinto D, Scheaffer SM, Sprouse K, et al. 2023. Therapeutic and vaccine-induced cross-reactive antibodies with effector function against emerging Omicron variants. Immunology. doi: 10.1101/2023.01.17.523798 [DOI]
- 46. Kitchin D, Richardson SI, van der Mescht MA, Motlou T, Mzindle N, Moyo-Gwete T, Makhado Z, Ayres F, Manamela NP, Spencer H, Lambson B, Oosthuysen B, Kaldine H, du Pisanie M, Mennen M, Skelem S, Williams N, Ntusi NAB, Burgers WA, Gray GG, Bekker L-G, Boswell MT, Rossouw TM, Ueckermann V, Moore PL. 2022. Ad26.COV2.S breakthrough infections induce high titers of neutralizing antibodies against Omicron and other SARS-CoV-2 variants of concern. Cell Rep Med 3:100535. doi: 10.1016/j.xcrm.2022.100535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wolter N, Jassat W, Walaza S, Welch R, Moultrie H, Groome MJ, Amoako DG, Everatt J, Bhiman JN, Scheepers C, et al. 2022. Clinical severity of SARS-CoV-2 Omicron BA.4 and BA.5 lineages compared to BA.1 and Delta in South Africa. Nat Commun 13:5860. doi: 10.1038/s41467-022-33614-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bartsch YC, Cizmeci D, Kang J, Gao H, Shi W, Chandrashekar A, Collier AY, Chen B, Barouch DH, Alter G. 2023. Selective SARS-CoV2 BA.2 escape of antibody FC/FC-receptor interactions. iScience 26:106582. doi: 10.1016/j.isci.2023.106582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Madhi SA, Kwatra G, Myers JE, Jassat W, Dhar N, Mukendi CK, Nana AJ, Blumberg L, Welch R, Ngorima-Mabhena N, Mutevedzi PC. 2022. Population immunity and Covid-19 severity with Omicron variant in South Africa. N Engl J Med 386:1314–1326. doi: 10.1056/NEJMoa2119658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Davis-Gardner ME, Lai L, Wali B, Samaha H, Solis D, Lee M, Porter-Morrison A, Hentenaar IT, Yamamoto F, Godbole S, Liu Y, Douek DC, Lee FE-H, Rouphael N, Moreno A, Pinsky BA, Suthar MS. 2023. Neutralization against BA.2.75.2, BQ.1.1, and XBB from mRNA bivalent booster. N Engl J Med 388:183–185. doi: 10.1056/NEJMc2214293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kurhade C, Zou J, Xia H, Liu M, Chang HC, Ren P, Xie X, Shi P. 2023. Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Nat Med 29:344–347. doi: 10.1038/s41591-022-02162-x [DOI] [PubMed] [Google Scholar]
- 52. Zou J, Kurhade C, Patel S, Kitchin N, Tompkins K, Cutler M, Cooper D, Yang Q, Cai H, Muik A, Zhang Y, Lee D-Y, Şahin U, Anderson AS, Gruber WC, Xie X, Swanson KA, Shi P-Y. 2023. Neutralization of BA.4–BA.5, BA.4.6, BA.2.75.2, BQ.1.1, and XBB.1 with bivalent vaccine. N Engl J Med 388:854–857. doi: 10.1056/NEJMc2214916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Branche AR, Rouphael NG, Losada C, Baden LR, Anderson EJ, Luetkemeyer AF, Diemert DJ, Winokur PL, Presti RM, Kottkamp AC, et al. 2023. Immunogenicity of the BA.1 and BA.4/BA.5 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) bivalent boosts: preliminary results from the COVAIL randomized clinical trial. Clin Infect Dis 77:560–564. doi: 10.1093/cid/ciad209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rössler A, Netzl A, Knabl L, Bante D, Wilks SH, Borena W, von Laer D, Smith DJ, Kimpel J. 2023. Characterizing SARS-CoV-2 neutralization profiles after bivalent boosting using antigenic cartography. Nat Commun 14:5224. doi: 10.1038/s41467-023-41049-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Andersson NW, Thiesson EM, Baum U, Pihlström N, Starrfelt J, Faksová K, Poukka E, Meijerink H, Ljung R, Hviid A. 2023. Comparative effectiveness of the bivalent BA.4-5 and BA.1 mRNA-booster vaccines in the Nordic countries. medRxiv. doi: 10.1101/2023.01.19.23284764 [DOI] [PMC free article] [PubMed]
- 56. Lin D-Y, Xu Y, Gu Y, Zeng D, Wheeler B, Young H, Sunny SK, Moore Z. 2023. Effectiveness of bivalent boosters against severe Omicron infection. N Engl J Med 388:764–766. doi: 10.1056/NEJMc2215471 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1; Figures S1 to S3.
Data Availability Statement
Whole-genome sequencing data of SARS-CoV-2 isolates has been deposited in the Global Initiative on Sharing All Influenza Data (GISAID) database and accession numbers are indicated in Table S1.
All data are available in this manuscript.