Abstract
Typical of many viral fusion proteins, the sequence of the Newcastle disease virus (NDV) fusion protein has several heptad repeat regions. One, HR1, is located just carboxyl terminal to the fusion peptide, while the other, HR2, is located adjacent to the transmembrane domain. The structure and function of a synthetic peptide with a sequence from the region of the NDV HR1 region (amino acids 150 to 173) were characterized. The peptide inhibited fusion with a half-maximal concentration of approximately 2 μM; however, inhibition was observed only if the peptide was added prior to protease activation of the fusion protein. This inhibition was virus specific since the peptide had minimal effect on fusion directed by the Sendai virus glycoproteins. To explore the mechanism of action, the potential HR1 peptide interaction with a previously characterized fusion inhibitory peptide with a sequence from the HR2 domain (J. K. Young, R. P. Hicks, G. E. Wright, and T. G. Morrison, Virology 238:291–304, 1997) was characterized. The results demonstrated an interaction between the two peptides both functionally and directly. First, while the individual peptides each inhibit fusion, equimolar mixtures of the two peptides had minimal effect on fusion, suggesting that the two peptides form a complex preventing their interaction with a target protein. Second, an HR2 peptide covalently linked with biotin was found to bind specifically to HR1 peptide in a Western blot. The structure of the HR1 peptide was analyzed by nuclear magnetic resonance spectroscopy and found to be an α helix.
Infection by paramyxoviruses such as Newcastle disease virus (NDV) is initiated by attachment of virions to cell surfaces followed by fusion of the viral membrane to cell plasma membranes. These steps are directed by the viral glycoproteins, the hemagglutinin-neuraminidase (HN) protein and the fusion (F) protein (reviewed in reference 28). The HN protein is an attachment protein which binds to sialic acid-containing cell receptors. Subsequent fusion is directed by the F protein, although in most systems, the HN protein also plays a necessary but poorly understood role (27).
The F glycoprotein is synthesized as a precursor (F0), which is activated upon proteolytic cleavage to produce disulfide-linked F1 and F2 (28). Several domains important for fusion activity have been identified in the F1 polypeptide. One domain, the fusion peptide, is located at the amino terminus and is thought to insert into the target membrane to initiate membrane fusion (reviewed in reference 24). The F protein also contains two heptad repeat (HR) domains, HR1 and HR2 (5, 9). HR1 is located just carboxyl terminal to the fusion peptide, and HR2, which has a leucine zipper motif, is located adjacent to the transmembrane domain. Mutational analyses of both HR domains have shown that they are important to the fusion activity of the protein (4, 38, 41).
Synthetic peptides with sequences from the HR2 domains of several paramyxoviruses inhibit membrane fusion in a virus-specific manner (29, 37, 48, 52, 55). These studies were stimulated by findings, initially reported by Wild et al., that peptides with sequences from either of two HR regions of the human immunodeficiency virus (HIV) gp41 protein would inhibit HIV fusion (45–47). Furthermore, it has been shown that a mix of peptides with sequences from both gp41 HR regions or protein fragments containing both regions interact to form a six-stranded helical bundle (10, 43, 44). In addition, structural analysis of a soluble form of the simian immunodeficiency virus gp41 has shown the presence of this six-stranded structure in the intact ectodomain of the protein (7). On the basis of these results, it has been proposed that the fusion-active form of the HIV gp41 is folded such that the two HR domains interact and that this interaction may be involved in the close approach of the attack and target membranes (10, 17, 31, 36, 43, 44). It has been suggested that peptides with sequences from either region may inhibit HIV fusion by blocking this interaction (17, 31, 36).
We have previously shown that a synthetic peptide with a sequence from the HR2 region of the NDV F protein will inhibit NDV fusion (55). We were therefore interested in determining if a peptide with a sequence from the HR1 domain of the NDV F protein could inhibit fusion and if such a peptide could interact in a specific manner with the HR2 sequences. We report here that a peptide with a sequence from the HR1 region of the NDV F protein will inhibit fusion if added to cells prior to the cleavage of the F protein. Furthermore, we present two lines of evidence that the HR1 peptide can interact with a peptide with a sequence from the HR2 region of the F protein. Using very different approaches, Ghosh et al. (19) and Joshi et al. (26) suggested an interaction between the HR1 and HR2 regions of Sendai virus and simian virus 5 (SV5), respectively. We also report here the structure of the HR1 peptide determined by nuclear magnetic resonance (NMR) spectroscopy.
MATERIALS AND METHODS
Peptides.
Peptide NDVFhr24, which corresponds to a 24-amino-acid segment (amino acids 150 to 173) of HR1 of the NDV F protein (see Fig. 1B), was obtained from Tufts University School of Medicine Peptide Core Facility, Boston, Mass. The peptide was purified by the Tufts facility to 98% by high-pressure liquid chromatography. This peptide was used for both biological and NMR studies without further purification.
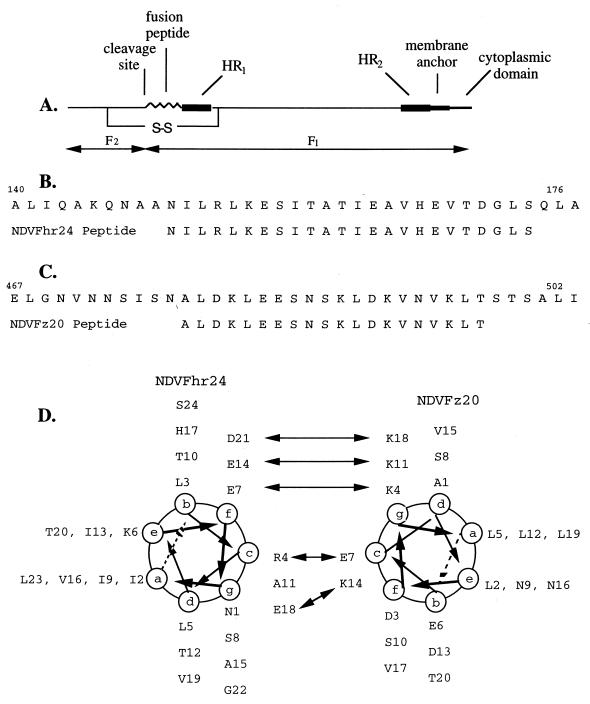
FIG. 1.
Sequence of peptides from the HR1 and HR2 regions of the NDV F protein. (A) Diagram of the fusion protein of NDV, with the approximate locations in the linear sequence of the HR1 and HR2 domains with respect to the fusion peptide sequence, the membrane anchor, and the cytoplasmic domain, as well as the cleavage site which generates F1 and F2 polypeptides. (B) Amino acid sequence from the HR1 region from amino acids 140 to 176 as well as the sequence of the NDVFhr24 (HR1) peptide. (C) Sequences of the NDV F HR2 region from amino acids 467 to 502 and the NDVFz20 (HR2) peptide. (D) Helical structure of NDVFz20 determined previously (55), presented alongside a computer-predicted structure of NDVFhr24. Potential interactions of the charged surfaces of the two helices are shown by arrows.
Peptide NDVFz20, which corresponds to a 20-amino-acid segment (amino acids 478 to 497) of HR2 of the NDV F protein (Fig. 1C), was also obtained from the Tufts University Peptide Core Facility and has been described previously (55).
Peptide NDVFz20-biotin, also obtained from the Tufts University Core Facility, has the sequence of NDVFz20 with the addition of a biotin residue followed by three glycine residues at the amino terminus.
Cells, vectors, and F mutant genes.
Cos-7 cells, obtained from the American Type Culture Collection, were maintained in Dulbecco’s modified Eagle’s medium supplemented with nonessential amino acids, vitamins, penicillin-streptomycin, and 10% fetal calf serum. NDV HN and F genes (derived from strain AV) were expressed in Cos-7 cells by using pSVL (Pharmacia) as previously described (40). Viral genes were inserted into SacI- and XbaI-cut plasmid DNA. Two mutants with mutations of the F gene with altered cleavage sites, mutants F115G and F117L, have been described previously (30, 35).
Sendai virus HN and F genes were the generous gifts of D. Nayak. The genes were inserted into pSVL.
Trypsin digestion of cell surface F0.
Cells were washed twice in OptiMem (BRL/Gibco), incubated at room temperature for 10 min in OptiMem containing 5 or 10 μg of acetylated trypsin per ml, washed in OptiMem containing 20 μg of soybean trypsin inhibitor per ml, washed twice in OptiMem, and then incubated in supplemented Dulbecco’s modified Eagle’s medium.
Transfections.
Transfections with dioleoyl-l-α phosphatidylethanolamine (DOPE) were done essentially as described previously (8, 30). Briefly, Cos-7 cells were plated at 2 × 105 per 35-mm plate and transfected 18 h later (the monolayers were 50 to 80% confluent). For each 35-mm plate, a mixture of 1.0 μg of HN DNA and 1.0 μg of F DNA in 0.1 ml of OptiMem and 10 μl of DOPE in 0.2 ml of OptiMem was incubated at room temperature for 45 min, diluted with 0.7 ml of OptiMem, and added to a plate previously washed with OptiMem. The cells were incubated with the DOPE-DNA mixture for 5 to 7 h, and then 2 ml of Cos-7 cell medium was added.
Fusion assays.
At 48 h posttransfection, the nuclei in 20 to 40 fusion areas were counted to determine the average size at each time point as previously described (30, 40). Values obtained after transfection of the vector alone were subtracted.
Peptide blots.
Peptides were spotted onto Immobilon-P (Millipore Corp.) membranes prewetted in phosphate-buffered saline (PBS). The membranes were air dried for 2 h. After brief washes in methanol and then PBS, the membranes were incubated for 2 h at room temperature or overnight at 4°C in PBS containing 0.5% Tween 20 and 10% nonfat dried milk. The membranes were washed in PBS-Tween 20 and incubated for 3 h at room temperature with NDVFz20-biotin diluted in PBS-Tween 20 and 0.5% nonfat milk. The membranes were washed and then incubated for 2 h at room temperature with neutravidin coupled to horseradish peroxidase (HRP) (Pierce) diluted in PBS-Tween 20 and 0.5% nonfat milk. The membranes were washed extensively, and bound neutravidin was detected with the ECL Western blotting detection reagent system (Amersham).
NMR sample preparation and experiments.
NMR samples were prepared by dissolving 2 mg of the synthetic peptide in 600 μl of 90% 1H2O–10% 2H2O buffered to a pH of 4.0 with 50 mM sodium acetate and 500 mM sodium dodecyl sulfate (SDS) to a final concentration of 1.15 mM (90% purity correction). Sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) was used as an internal chemical shift reference.
All NMR experiments were conducted at Cornell University (laboratory of Linda K. Nicholson, Section of Biochemistry, Molecular and Cell Biology) on a Varian INOVA 600-MHz NMR spectrometer with the use of the 1H channel of a triple-resonance probe (1H, 13C, and 15N). Spectra were processed and interpreted at Colgate University with NMRpipe on a Silicon Graphics Extreme Indigo2 workstation.
Six phase-sensitive watergate-TOCSY (16, 25) experiments were performed every 5°C over a temperature range of 20 to 45°C by using a spin-lock mixing pulse of 80 ms and an MLEV-17 mixing sequence with a 2.5-ms trim pulse at the beginning and end of the MLEV sequence (3). The spectral width in both domains was set to 8,000 Hz. At the beginning of each experiment, 32 dummy scans were collected to allow the system to reach thermal equilibrium. A total of 2K (K = 1,024 points) time-domain datum points for 512 t1 values of 32 scans each were acquired and then zero filled to 4K by 4K, followed by processing with a 90°-shifted sine function in both dimensions. After investigation of each TOCSY (total-correlation spectroscopy) spectrum, it was determined that the experiment performed at 25°C provided the best amide peak-to-peak resolution.
The phase-sensitive watergate-NOESY (6) experiment was performed with a mixing pulse of 200 ms at 25°C. The spectral width in both domains was set to 8,000 Hz. At the beginning of each experiment, 32 dummy scans were collected to allow the system to reach thermal equilibrium. A total of 2K time-domain datum points for 1,024 t1 values of 32 scans each were acquired and then zero filled to 4K by 4K, followed by processing with a 90°-shifted sine function in both dimensions.
Molecular modeling.
The molecular-modeling (simulated-annealing) calculations conducted during this investigation were performed with BioSym software (InsightII, NMRchitect, and Discover) from Molecular Simulations Inc. on a Silicon Graphics Extreme Indigo2 at Colgate University. Simulated-annealing calculations involved a search of all conformational space to find the global minimum-energy conformation of the molecule (13, 53, 54). Structures were generated for NDVFhr24 with the NMR-derived interproton distance restraints and hydrogen bond data and using simulated-annealing calculations involving 16 separate phases. These calculations began with an arbitrary or linear extended conformation to prevent any initial bias in the starting structure toward a particular secondary-structure feature. Phase 1 involved randomization of all atomic coordinates by 10 Å followed by 100 iterations of steepest minimization, using a quadratic potential and very low force fields for each term of the pseudo-energy function. Phase 2 involved additional minimization with 1,500 iterations of conjugate minimization. During these minimization steps (phases 1 and 2, or the preparation stage), the covalent force fields were reduced to 0.02% of their full value and experimental (interproton distances and hydrogen bonds), nonbond, and chiral force fields were reduced to 0.1% of their full value. Phases 3 through 5 involved simulated annealing with restrained dynamics for 30 ps with scaling of the force fields. Molecular dynamics (phase 3, or folding stage) was applied by using weak force fields to allow the potential energy of the system to equal the kinetic energy at 1,000 K. The low values of the force fields allow atoms and bonds to pass through each other (13). In this folding stage, the experimental force fields were scaled up to their full value, covalent and chiral force fields were scaled up to 15% of their full value, and nonbond force fields were scaled up to 0.1% of their full value. Next, the regulation stage (phase 4) scaled up the covalent force fields to their full value and the nonbond force fields to 15% of their full value, so that the coordinates of all the atoms were more tightly held. Phase 5 restrained the molecule significantly by scaling the chiral force fields to their full value, the nonbond force fields to 25% of their full value, and the experimental force fields to twice their full value. Phases 6 through 10 involved cooling of the molecule from 1,000 to 300 K over 10 ps (2 ps for each phase) with the nonbond force fields scaled to their full value. The next two phases (11 and 12) involved 100 iterations of steepest minimization and 1,500 iterations of conjugate minimization while scaling the nonbond force fields to their full value. Phases 13 and 14 involved 100 iterations of steepest minimization and 1,500 iterations of conjugate minimization while the experimental force fields were scaled back to their full value. The final phases (phases 14 and 15) involved 100 iterations of steepest minimization and 1,500 iterations of conjugate minimization with a Lennard-Jones potential (55).
The final structures were analyzed by the superimposition of the backbone atoms to determine if the structures converged to a single family of conformations or if they defined multiple families of conformations. The dihedral angles were then averaged over the number of structures in a family of conformations to determine which, if any, secondary-structure features were defined by the experimental data (13).
RESULTS
Sequence of HR1 synthetic peptide.
The amino-terminal end of paramyxovirus F1 proteins contains two domains, the fusion peptide and an adjacent HR1 region (diagrammed in Fig. 1A) (9). Figure 1B shows the sequence of the NDV HR1 region, from amino acids 140 to 176. Given the idea that the HR1 and HR2 regions of the F protein may interact, we compared the structure of an HR2 peptide (NDVFz20 [Fig. 1C] aligned with the sequence of the entire HR2 region [Fig. 1C]), which had been previously determined by NMR spectroscopy (55), with a computer-generated model of the HR1 region and noticed the possibility of a interaction between the charged surfaces of the two helices, diagrammed in Fig. 1D, with potential interactions indicated by arrows. Furthermore, the results of a mutational analysis of the HR1 region of the intact NDV F protein (40a, 41) showed that the sequence from amino acids 140 to 175 is important in fusion. Based on these considerations, a sequence between amino acids 150 and 173 was chosen for a peptide which might function as an inhibitor of fusion (Fig. 1B, NDVFhr24).
Peptide inhibition of fusion.
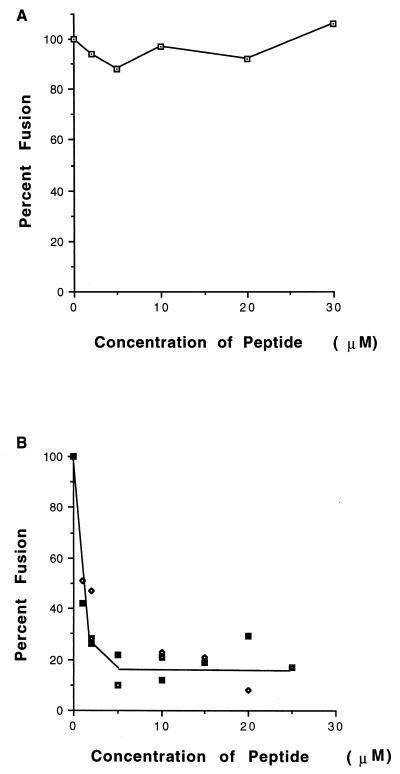
To determine if the NDVFhr24 peptide had fusion-inhibitory activity, the effect of increasing concentrations of peptide on the formation of syncytia was determined. This peptide was not readily soluble in water or in buffers at pH 7. However, it was readily soluble at pH 9. After titration to pH 7, a concentrated stock (2 mg/ml) of peptide remained in solution for several hours. Thus, to measure the inhibitory activity of the peptide, solutions of concentrated peptide at pH 9 were diluted into media in which Cos-7 cells were growing. Syncytium formation directed by expression of the wild-type F protein gene derived from NDV (strain AV) in the presence of the NDV HN protein was not inhibited even at very high concentrations of peptide (Fig. 2A).
FIG. 2.
Inhibition of fusion by the HR1 peptide, NDVFhr24. (A) Cells cotransfected with HN and wild-type F cDNAs as described in Materials and Methods were incubated with 0 to 30 μM NDVFhr24 peptide added with complete medium after transfection. Medium with peptide was replaced every 8 to 12 h. The fusion was measured 48 h after transfection. Values obtained are shown as a percentage of those obtained without peptide and are the average of four experiments. (B) Cells transfected with HN and F115G cDNAs were incubated with 0 to 25 μM NDVFhr24 (HR1) peptide beginning 16 h posttransfection. Medium with peptide was replaced 32 and 45 h posttransfection. At 48 h posttransfection, the cells were incubated with trypsin (5 μg/ml) as described in Materials and Methods. Incubation was continued for 6 h in the presence of peptide and soybean trypsin inhibitor (20 μg/ml), and fusion was determined as described in Materials and Methods. Data points are from three separate experiments.
However, the peptide does inhibit fusion if added before cleavage of the F protein (Fig. 2B). The wild-type F protein has, at the cleavage site, a sequence recognized by furin enzymes in the Golgi membranes and therefore is cleaved intracellularly and inserted at the cell surface in a cleaved form (reviewed in reference 28). To determine if the HR1 peptide, NDVFhr24, could inhibit fusion if added before F-protein cleavage, use was made of a mutant F-protein gene which has an alteration in the cleavage site, F115G. This mutant can be activated by the addition of exogenous trypsin, and syncytium formation rapidly proceeds as previously described (30). If NDVFhr24 peptide was added prior to incubation with trypsin, there was an efficient inhibition of fusion, with 50% inhibition at approximately 2 μM peptide. Identical results were obtained with F117L, another cleavage mutant of the NDV (AV) F-protein gene (not shown) (35). Thus, the HR1 peptide is an inhibitor of fusion.
Inhibition of fusion was not due to an inhibition of cleavage by trypsin in the presence of the HR1 peptide. The extent of cleavage of cell surface F0 by trypsin digestion of cells expressing F117L protein in the presence or absence of peptide was quantitated (Table 1), and no difference in the amount of F1 was detected in the presence or absence of HR1 peptide. In addition, coexpression of HN and F117L did not change the results.
TABLE 1.
Effect of NDVFhr24 on cleavage of F protein
| Peptide | % of F-protein cleavage in cells expressing:
|
||
|---|---|---|---|
| F117L with trypsin | F117L + HN with trypsin | F117L with no trypsin | |
| None | 73 | 70 | 11 |
| NDVFhr24 (30 μM) | 72 | 70 | 10 |
Peptide-mediated inhibition of fusion is virus specific.
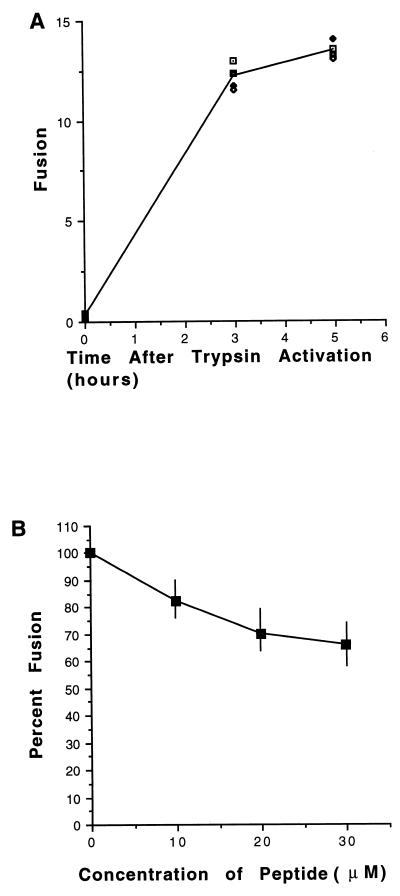
To determine if the HR1 peptide will inhibit fusion directed by other paramyxovirus glycoproteins, we determined the activity of NDVFhr24 on fusion directed by the Sendai virus HN and F proteins. The Sendai virus F protein expressed in tissue culture cells is not cleaved by furin but must be cleaved by the addition of exogenous trypsin (reviewed in reference 28). Cos-7 cells coexpressing the Sendai virus HN and F proteins will not fuse. However, after trypsin-mediated activation, fusion rapidly proceeds, as illustrated in Fig. 3A. To determine the effect of the HR1 peptide on this fusion, Cos-7 cells cotransfected with pSVL-Sendai virus F and pSVL-Sendai virus HN were incubated in the presence of peptide. Fusion was activated by the addition of exogenous trypsin, and the size of syncytia was measured at 3 and 5 h after protease activation. Cells expressing Sendai virus glycoproteins and incubated in the presence of HR1 peptide formed syncytia (Fig. 3B), although the sizes of the syncytia were reduced slightly at very high concentrations of peptide. Thus, the HR1 peptide with the NDV sequence has only a slight effect on Sendai virus fusion.
FIG. 3.
Virus specificity of inhibition. (A) Cells transfected with Sendai virus HN and F cDNAs for 48 h were incubated with trypsin (10 μg/ml) for 10 min followed by soybean trypsin inhibitor (20 μg/ml) and complete medium. Fusion was measured at 0, 3, and 5 h after trypsin activation. Data points are from four separate experiments. (B) Peptide NDVFhr24 was added to cells transfected with Sendai virus HN and F cDNAs at 36 and 45 h posttransfection. At 48 h posttransfection, the cells were incubated with trypsin for 10 min and then further incubated for 3 or 5 h in the presence of soybean trypsin inhibitor and NDVFhr24 peptide. Fusion was measured 0, 3, and 5 h after trypsin activation. The data are the average of three experiments. Actual variation in data is shown by vertical bars.
Fusion in the presence of both HR1 and HR2 peptides.
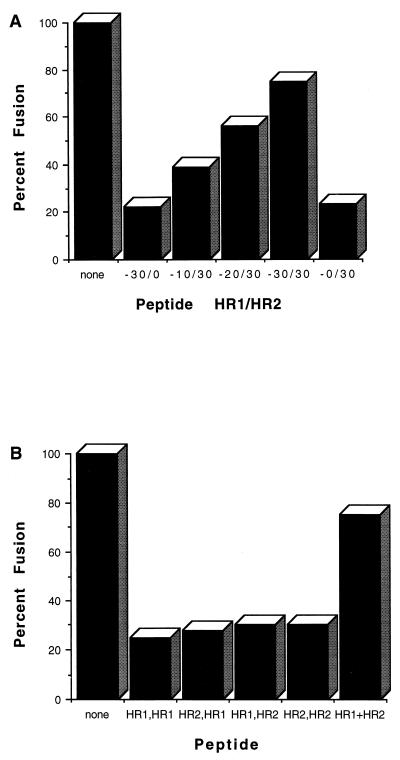
If there is an interaction between the HR1 and HR2 regions of the NDV F protein, peptides from these two regions may also interact, mimicking the structure in the intact protein. If such an interaction occurs, the peptide complex may be unable to interact with domains on the intact protein to inhibit fusion. To test this idea, mixtures of the HR1 and HR2 peptides (NDVFhr24 and NDVFz20) were added to cells expressing F115G or F117L and HN prior to trypsin-mediated activation. As shown in Fig. 4A, the mixtures of peptides had much less inhibitory activity than did of the individual peptides added separately, and an equimolar mix of HR1 and HR2 peptides had the least inhibitory activity. Importantly, mixing of the two peptides did not result in aggregation and precipitation of the peptides (data not shown).
FIG. 4.
Inhibition of fusion by mixtures of HR1 and HR2 peptides. (A) Cells transfected with HN and F115G cDNAs were incubated with either NDVFhr24 (HR1), NDVFz20 (HR2), or different mixtures (amount of HR1/amount of HR2) of the two peptides as described in the legend to Fig. 2B. Amounts of each peptide are given in micromoles. At 48 h posttransfection, the cells were incubated with trypsin. Incubation was continued for 6 h, and fusion was determined as described in Materials and Methods. Values obtained are shown as a percentage of those obtained without peptide. The results of one experiment are shown. Similar results were obtained in two other experiments with the F117L cleavage mutant. (B) Monolayers of transfected cells (F115G and HN DNAs) were incubated with equimolar amounts of both peptides (30 μM) but added sequentially (the order is indicated by peptide added first, peptide added second). The first peptide was added 28 and 36 h posttransfection. The second peptide was added 45 h posttransfection. In the last column (HR1+HR2), the peptides were added together. Trypsin activation was accomplished as described in the legend to Fig. 2 at 48 h posttransfection.
If the loss of inhibitory activity in mixtures of the two peptides was due to the formation of a complex between the two peptides, sequential addition of the peptides to the cells should still result in significant inhibition of fusion, since the first peptide added would interact with the intact protein and be unavailable to interact with the second peptide when added. Indeed, addition of HR1 peptide followed by the addition of HR2 peptide resulted in fusion inhibition, as did the sequential addition of HR2 peptide and then HR1 peptide (Fig. 4B).
Binding of HR2 peptide to HR1 peptide.
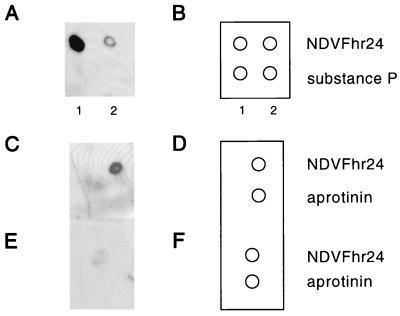
The finding that mixtures of HR1 and HR2 peptides showed less inhibition of fusion than did either peptide alone suggested that the two peptides formed a complex which blocks the interaction of each of the peptides to its target. To determine directly if the two peptides interact, a modification of a Western blotting protocol was used. The HR1 peptide, NDVFhr24, was bound to an Immobilon-P membrane. In addition, two nonspecific peptides, substance P and aprotinin, were bound (diagrammed in Fig. 5B and D). The membrane was incubated with HR2 peptide that was covalently linked at its amino terminus to biotin (NDVFz20-biotin). Any NDVFz20-biotin peptide binding to the membrane was detected by using the biotin binding molecule neutravidin, which was coupled to HRP. As shown in Fig. 5A and C, the HR2 peptide, NDVFz20-biotin, bound to the HR1 peptide, NDVFhr24, but did not bind to the nonspecific peptides, substance P and aprotinin. That the binding of the HR2 peptide to the HR1 peptide was specific is also indicated by results shown in Fig. 5C to F. The binding of NDVFz20-biotin was competed by excess NDVFz20 peptide untagged with biotin.
FIG. 5.
Interaction of HR2 peptide (NDVFz20-biotin) with HR1 peptide (NDVFhr24). (A, C, and E) Densitometer scan of chemiluminescent detection of the binding of NDVFz20-biotin to peptide bound to Immobilon-P membranes. (B, D, and F) Diagrams of the peptides bound to the membrane. (A and B) NDVFhr24 (HR1), 10 μg (lane 1) and 1 μg (lane 2) and substance P were bound to a membrane. The membrane was incubated in a solution of NDVFz20-biotin (0.1 μg/ml), washed, and incubated in a solution of neutravidin-HRP (0.1 μg/ml) as described in Materials and Methods. (C and D) NDVFhr24 (1 μg) and 1 μg of aprotinin were bound to membranes. The membranes were then incubated with 1 μg of NDVFz20-biotin per ml. (E and F) NDVFhr24 (1 μg) and 1 μg of aprotinin were bound to membranes. The membranes were then incubated with 1 μg of NDVFz20-biotin per ml and 10 μg of NDVFz20 per ml as a competitor.
NMR assignments.
To determine if the NDVFhr24 was indeed an α helix as predicted by computer analysis of the primary sequence, the structure of the peptide was determined by NMR spectroscopy. NMR analysis of small peptides is usually carried out in solutions of pH 6 or less. As noted above, the peptide was soluble in aqueous solutions only at pH 9. This pH causes a significant loss of structural information due to the increased 1H-2H exchange rate of the amide protons. This problem can be overcome by studying the peptide in a hydrophobic environment such as micelles. Ideal hydrophobic or membrane model systems are derived from phospholipid vesicles. However, the long correlation times associated with these systems limit their application to high-resolution NMR experiments. SDS micelles offer reasonable line widths and have been used extensively as a simple hydrophobic environment for the investigation of polypeptides (1, 12, 32, 33, 53). The solubility properties associated with this peptide suggest that this region of the protein either may be buried in the interior of the protein or membranes or may be interacting with another domain in the protein. Therefore, the structure determined in the presence of micelles becomes relevant since it mimics the hydrophobic environment found in a protein interior as well as in membranes.
Assignments of the 1H spectra of the NDVFhr24 peptide (Fig. 1C) in SDS solution were made by using the technique of sequence-specific resonance assignments developed by Wüthrich (50). The assignments were made by the interactive interpretation of the two-dimensional TOCSY (16, 25) and NOESY (42) spectra at 25°C. From the TOCSY spectrum, a set of resonances can be assigned to a particular spin system (amino acid). Cross-peaks in the TOCSY spectrum arise as a result of through-bond interactions (15). For example, an amide 1H magnetization will travel through the bond to the other 1H atoms in its own spin system until interrupted by a unprotonated position (16). However, it was not possible to make specific assignments of like (I2, I9, I13) or similar (L,I,V and D,N) amino acids by using the TOCSY spectrum because of the common occurrence of these residues in this peptide. Therefore, the NOESY (nuclear Overhauser enhancement spectroscopy) experiment was used to make the sequence-specific assignments shown in Table 2.
TABLE 2.
1H chemical shift assignments and ACSTD of 1.15 mM NDVFhr24 at pH 4.0 in 750 mM SDS as obtained from the TOCSY and NOESY spectra at 25°C
| Chemical shift (ppm) of:
|
ACSTD (ppb/°C) | ||||
|---|---|---|---|---|---|
| HN | α | β | Others | ||
| Asn1 | 4.43 | 3.11, 3.28 | NH2, 6.88, 7.73 | ||
| Ile2 | 8.55 | 4.15 | 2.01 | γCH2, 1.33, 1.58 | −6.8 |
| γCH3, 0.96 | |||||
| δCH3, 0.96 | |||||
| Leu3 | 7.85 | 4.09 | 1.75 | γCH, 1.61 | −5.8 |
| δCH3, 0.87, 0.95 | |||||
| Arg4 | 7.90 | 4.19 | 1.91 | γCH2, 1.67 | −4.0 |
| δCH2, 3.20 | |||||
| NH, 7.07 | |||||
| Leu5 | 7.76 | 4.18 | 1.82 | γCH, 1.72 | −4.0 |
| dCH3, 0.95 | |||||
| Lys6 | 8.29 | 3.87 | 1.88, 2.01 | γCH2, 1.39, 1.50 | −8.8 |
| δCH2, 1.70 | |||||
| ɛCH2, 2.94 | |||||
| NH3, 7.49 | |||||
| Glu7 | 8.26 | 4.17 | 2.17, 2.21 | γCH2, 2.55, 2.60 | −7.2 |
| Ser8 | 8.24 | 4.40 | 3.89, 4.05 | −6.4 | |
| Ile9 | 8.47 | 3.77 | 2.01 | γCH2, 1.14, 1.76 | −6.0 |
| γCH3, 0.92 | |||||
| δCH3, 0.83 | |||||
| Thr10 | 8.16 | 3.83 | 4.24 | γCH3, 1.24 | −5.6 |
| Ala11 | 8.03 | 4.13 | 1.52 | −5.6 | |
| Thr12 | 7.91 | 3.89 | 4.32 | γCH3, 1.15 | −6.4 |
| Ile13 | 8.31 | 3.52 | 1.95 | γCH2, 1.06 | −6.4 |
| γCH3, 0.84 | |||||
| δCH3, 0.84 | |||||
| Glu14 | 8.09 | 4.08 | 2.11, 2.21 | γCH2, 2.54, 2.66 | −4.0 |
| Ala15 | 7.94 | 4.20 | 1.58 | −4.0 | |
| Val16 | 8.32 | 3.65 | 2.19 | γCH3, 0.91, 1.06 | −6.0 |
| His17 | 8.39 | 4.54 | 3.30, 3.35 | 2H, 8.63 | −6.4 |
| 4H, 7.33 | |||||
| Glu18 | 8.08 | 4.10 | 2.21, 2.29 | γCH2, 2.53, 2.65 | −2.0 |
| Val19 | 7.87 | 3.99 | 2.30 | γCH3, 0.98, 1.10 | −1.0 |
| Thr20 | 8.20 | 4.15 | 4.34 | γCH3, 1.24 | −9.8 |
| Asp21 | 8.10 | 4.64 | 2.90 | −6.8 | |
| Gly22 | 8.00 | 3.94, 4.01 | −8.4 | ||
| Leu23 | 7.74 | 4.39 | 1.72 | γCH, 1.60 | −1.4 |
| δCH3, 0.88 | |||||
| Ser24 | 8.06 | 4.47 | 3.87, 3.94 | −8.0 | |
ACSTD.
Amide 1H atoms are directly affected by changes in temperature which cause a change in the chemical shift of the amide and can be related to intramolecular hydrogen bonding (39). An amide 1H that is involved in an intramolecular hydrogen bond is shielded from the solvent and therefore shows a low temperature dependence (small change in chemical shift), while solvent-accessible amide 1H atoms show a high temperature dependence (large change in chemical shift). At higher temperatures, some of the amide protons of NDVFhr24 overlapped, making the temperature dependence hard to determine from the one-dimensional spectra. Therefore, six TOCSY spectra (obtained every 5°C) were acquired over a temperature range of 20 to 45°C to unambiguously assign the amide chemical shifts (55). A plot of the chemical shift against temperature resulted in a linear dependence for each amide proton (21). The slope of this linear dependence is the ACSTD (amide chemical shift temperature dependence), and the values are shown in Table 2 for NDVFhr24 (HR1). The low temperature dependence (less than −5 ppb/°C) for the amide 1H atoms of R4, L5, E14, A15, E18, V19, and L23 suggests the presence of seven intramolecular hydrogen bonds.
NOE connectivity and αH chemical shift index.
Wüthrich et al. have reported that the observation of a grouping of specific medium-range NOEs can be used to determine the existence of secondary structural features such as an α-helix or a β-turn (51). The NOEs, obtained from the NOESY spectrum, that are important to characterize the secondary structure of NDVFhr24 are shown in Fig. 6. These NOEs suggest that the secondary structure of NDVFhr24 may be helical due the occurrence of strong sequential dNN(i, i + 1), medium-range dNN(i, i + 3), dαN(i, i + 3), and dαβ(i, i + 3) and weak dαN(i, i + 4) connectivities. In particular, the 14 medium-range dαβ(i, i + 3) NOEs strongly suggest that this peptide exists in a helical structure, since this NOE is observed only in helices (13, 21, 50). The low occurrence of dαN(i, i + 2) and dNN(i, i + 2) connectivities strongly suggest that NDVFhr24 is α helical.
FIG. 6.
Structural NOEs of NDVFhr24. NOEs obtained from the NOESY spectrum of NDVFhr24 are shown. The bars show the position and strength of each NOE. The pattern suggests a helical structure.
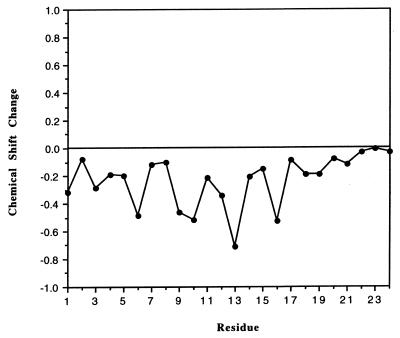
Wishart et al. have reported that a deviation of an αH chemical shift from its random-coil value can provide an insight into the secondary structure (49). A chemical shift that deviates in a positive direction indicates a possible β-strand structure, and a negative deviation indicates a possible helical structure. The deviation of the αH chemical shifts for NDVFhr24 is given in Fig. 7, which shows that the NDVFhr24 peptide gives a negative shift in the αH chemical shift index, suggesting a helical structure.
FIG. 7.
Change in alpha protein chemical shifts from random-coil values for NDVFhr24. All alpha protons shift negative, suggesting a helical structure. See the text for details.
Molecular modeling.
A total of 100 structures were generated for NDVFhr24 by using the NMR-derived distance constraints and intramolecular hydrogen bond data by simulated-annealing calculations involving 16 separate phases, as described in Materials and Methods. A total of 212 NOEs were assigned from the NOESY spectrum collected at a mixing time of 200 ms. Of these NOEs, 81 represented intraresidue interactions, 82 represented i to i + 1 interactions, and 49 represented medium-range (i to i + 2, i to i + 3, and i to i + 4) interactions. These interactions were converted to interproton distances by using the strong, medium, and weak designations of Clore et al. (13). This method involves the visual inspection of the NOESY spectrum and assignment of each cross-peak intensity to an interproton distance of 1.9 to 2.7 Å for strong, 1.9 to 3.3 Å for medium, and 1.9 to 4.0 Å for weak (Fig. 6).
As a first step, 20 structures of NDVFhr24 were generated by using only interproton distance data to determine if a secondary structure existed. It was evident from the resulting structures that a stable helix was present throughout the peptide. The seven intramolecular hydrogen bonds determined from the ACSTD study (Table 2) were assigned based on these structures. Hydrogen bonds within a reverse turn represent an i to i + 3 interaction, while those in a helical structure represent an i to i + 4 interaction (14, 51). Therefore, the six hydrogen bonds were defined from the amide 1H of L5, E14, A15, E18, V19, and L23 to the backbone carbonyl oxygen of N1, T10, A11, E14, A15, and V19 respectively. One i to i + 3 hydrogen bond was defined from the amide 1H of R4 to the carbonyl oxygen of N1, since it is not possible for an i to i + 4 hydrogen bond. All hydrogen bonds were assigned a distance of 2.5 Å. Another 20 structures generated by using interproton distance and hydrogen bond data converged to one family of conformations. Therefore, 100 structures were generated for a final analysis by using the protocols in Materials and Methods. Twenty randomly selected structures of NDVFhr24 are shown in Fig. 8A, with all backbone atoms superimposed, showing convergence to a helical structure. The average energy of the resulting structures was 270 kcal/mol. The root mean squared deviations of the backbone atoms for the 100 structures range from 0.01 to 0.70 Å.
FIG. 8.
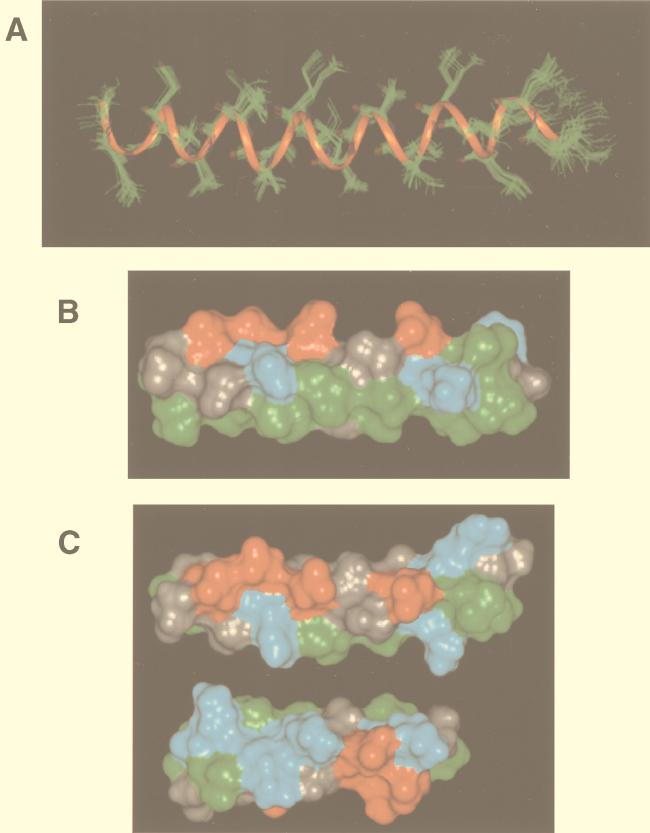
Representations of the structure of NDVFhr24 determined by NMR. (A) Twenty randomly selected structures of NDVFhr24 from simulated-annealing calculations using interproton distance and intramolecular hydrogen bond data as described in Materials and Methods superimposed showing backbone residues only. A ribbon is shown through the backbone atoms to highlight the helical structure. (B) Representative structure of NDVFhr24 showing solid surfaces. The hydrophobic face of the peptide is shown along the bottom of the structure. Green indicates hydrophobic residues, red indicates acidic residues, and blue indicates basic residues. (C) The top structure is a representative structure of NDVFhr24 (HR1) showing solid surfaces. The charged face of the helix is shown. The bottom structure shows the charged face of the NDVFz20 (HR2) peptide. Rotation of both peptides along the long axis and toward the center would result in alignment of the positive charges with the negative charges.
Peptide secondary structure.
To characterize the secondary structural features of the NDVFhr24 peptide, the individual backbone dihedral angles were averaged over the 100 structures obtained from molecular modeling and are given in Table 3. Dihedral angles that vary by ±20° or less are considered sufficient to compare with ideal dihedral angles of known secondary structural features (14). For NDVFhr24, all dihedral angles vary by only ±10.6° or less with the exception of N1 (Ψ = 69.2 ± 130.3) and S24 (φ = −29.2 ± 87.7), consistent with the greater degree of motion expected at each terminus. These dihedral angles correspond to a right-handed α helix (φ = −57, Ψ = −47) (14, 51).
TABLE 3.
Backbone dihedral angles for NDVFhr24 averaged over the 100 structures from molecular modeling
| Residue | Angle | Residue | Angle | |
|---|---|---|---|---|
| N1 | φ = —a | I13 | φ = −60.0 ± 5.5 | |
| Ψ = 69.2 ± 130.3 | Ψ = −30.3 ± 8.0 | |||
| I2 | φ = −60.4 ± 2.7 | E14 | φ = −67.3 ± 6.6 | |
| Ψ = −42.3 ± 4.0 | Ψ = −36.9 ± 5.1 | |||
| L3 | φ = −59.0 ± 2.0 | A15 | φ = −64.1 ± 4.0 | |
| Ψ = −42.7 ± 1.8 | Ψ = −42.4 ± 7.1 | |||
| R4 | φ = −54.5 ± 1.2 | V16 | φ = −61.6 ± 3.3 | |
| Ψ = −46.5 ± 1.8 | Ψ = −40.9 ± 5.4 | |||
| L5 | φ = −59.1 ± 2.0 | H17 | φ = −61.5 ± 5.3 | |
| Ψ = −38.9 ± 2.1 | Ψ = −29.0 ± 6.8 | |||
| K6 | φ = −66.5 ± 1.3 | E18 | φ = −73.9 ± 3.3 | |
| Ψ = −48.5 ± 5.4 | Ψ = −30.8 ± 5.4 | |||
| E7 | φ = −60.9 ± 4.4 | V19 | φ = −71.3 ± 6.0 | |
| Ψ = −29.1 ± 6.8 | Ψ = −33.8 ± 6.9 | |||
| S8 | φ = −69.7 ± 5.4 | T20 | φ = −65.8 ± 6.4 | |
| Ψ = −33.7 ± 6.0 | Ψ = −43.6 ± 5.4 | |||
| I9 | φ = −65.9 ± 4.7 | D21 | φ = −58.3 ± 2.8 | |
| Ψ = −35.9 ± 2.8 | Ψ = −31.7 ± 5.3 | |||
| T10 | φ = −67.1 ± 3.0 | G22 | φ = −58.3 ± 8.1 | |
| Ψ = −34.7 ± 3.8 | Ψ = −45.8 ± 6.3 | |||
| A11 | φ = −62.5 ± 2.4 | L23 | φ = −73.7 ± 5.9 | |
| Ψ = −26.6 ± 7.1 | Ψ = −66.2 ± 10.6 | |||
| T12 | φ = −72.9 ± 4.2 | S24 | φ = −29.2 ± 87.7 | |
| Ψ = −49.9 ± 3.5 | Ψ = — |
—, not possible.
DISCUSSION
Peptide structure.
As noted above, the structure of the NDVFhr24 peptide was determined in SDS micelles since the peptide was not soluble in aqueous solutions compatible with NMR analysis. SDS micelles have been used extensively in the investigation of polypeptide structure (1, 12, 32, 33, 53). The solubility properties associated with this peptide suggest that this sequence within the intact F protein may be buried in the interior of the protein or in membranes or, alternatively, may be interacting with another domain in the protein. Therefore, the structure determined in the presence of micelles becomes relevant, since micelles mimic the hydrophobic environment found in a protein interior as well as in membranes.
NMR analysis of the structure of this peptide yielded results consistent with a typical α helix. A solid surface of a representative calculated structure is given in Fig. 8B, showing the hydrophobic face of the peptide. The opposite or charged face of the peptide is shown in Fig. 8C (top structure). Also shown in Fig. 8C (bottom structure) is a solid-surface representation of the structure of the HR2 peptide, NDVFz20, previously reported (55). Comparisons of the two structures show that, indeed, there is the potential for an interaction between the charged surfaces of the HR1 and HR2 peptides, as suggested by the computer modeling described above. Interestingly, a similar comparison of computer-derived models of HR1 and HR2 domains of several other paramyxovirus fusion proteins also shows such a potential for interactions of charged surfaces.
Peptide inhibition of fusion.
Membrane fusion is thought to occur in a series of steps including attachment of the attack membrane to the target membrane, activation of a fusion protein, close approach of the attack and target membranes, hemifusion, pore formation, and pore expansion (24, 34). In most paramyxovirus systems, the coexpression of the HN and F proteins is both necessary and sufficient for these steps (reviewed in 24). NDV attachment is minimally mediated by the HN protein (28). Paramyxovirus fusion protein activation requires the cleavage of the F0 protein to form a disulfide-linked F1-F2 complex (28). In addition, other changes in the F protein, probably mediated in some way by the HN protein, are required since a cleaved F protein alone will not direct fusion in most systems (2). Close approach requires that the attack and target membranes be pulled together. How this step occurs is not clear; however, it has been proposed that the HIV env protein accomplishes this step by the interaction of two distant domains in the gp41 protein, an interaction thought to be initiated by the attachment of the env to its receptor (11, 31).
Candidate domains for such an interaction in the paramyxovirus fusion protein are two heptad repeat regions (HR domains) initially recognized by Chambers et al. (9). HR1 is located just carboxyl terminal to the fusion peptide and may therefore be initially located near the target membrane. HR2 is adjacent to the transmembrane domain and is therefore located near the attack membrane. Thus, an interaction of these two domains during the fusion process could cause the close approach of the two membranes. Mutational analyses of both regions of the several F proteins including the NDV F protein have shown that alterations in the sequences of both these regions inhibit fusion, suggesting that they play a central role in the fusion process (4, 38, 41).
We and others have previously shown that a peptide with a sequence from the F-protein HR2 region can inhibit fusion mediated by the coexpression of the HN and F proteins (4, 29, 37, 48, 52, 55). Here, we have shown that a peptide with a sequence from the NDV HR1 domain, from amino acids 150 to 174, will also inhibit fusion directed by the NDV F and HN proteins. The inhibitory activity of the peptide is comparable to that of the peptide derived from the HR2 region. Both peptides show 50% inhibition at approximately 2 μM. Previously, Rapaport et al. (37) showed that a peptide with a sequence from the Sendai virus F HR1 region did not inhibit fusion. However, this peptide was derived from a sequence slightly more carboxyl terminal than the one characterized here. Lambert et al. reported that peptides from HR1 sequences in the respiratory syncytial virus, human parainfluenza virus 3, and measles virus HR1 regions inhibit fusion, but the inhibition was not further characterized (29). Joshi et al. have recently reported that a much larger peptide, with a sequence from amino acids 129 to 184, inhibits SV5 fusion, although not as efficiently as a peptide from the HR2 region of the SV5 F protein (26).
In contrast to results described for other paramyxovirus systems (26, 29), we have found that the NDV HR1 peptide will not inhibit fusion unless the peptide is added before the cleavage of surface-expressed F protein. Cleavage of our wild-type F protein (derived from virulent NDV) is mediated by the host cell protease furin, located in the trans-Golgi membranes (22, 23, 28). Thus, the protein expressed at the surface is cleaved. Fusion of cells expressing this F protein is not inhibited by the HR1 peptide. However, alteration of the F-protein furin recognition site by mutagenesis results in the expression of F0 at the cell surface, and the mutant F0 can be readily cleaved upon the addition of exogenous trypsin, resulting in synchronized fusion (30, 35). Addition of the HR1 peptide prior to trypsin results in inhibition of fusion. The HR1 peptide does not, however, block the cleavage of the F protein. This result suggests that the target of the HR1 peptide is accessible only prior to cleavage or, alternatively, that binding of the HR1 prior to F protein cleavage may inhibit a conformational change in a protein that occurs upon cleavage.
We have also found that the inhibition mediated by the HR1 peptide is virus specific. We were able to explore this question by using Sendai virus glycoproteins, since the Sendai virus F protein must also be cleaved by the addition of exogenous trypsin (28). The peptide only minimally affects fusion directed by the Sendai virus HN and F proteins. Thus, it is likely that the HR1 peptide binds to a target protein via interactions with sequence motifs unique to a specific protein.
The target of the HR1 peptide is potentially the lipid bilayer, host cell proteins, HN protein, or F protein. Indeed, Ghosh et al. have reported that peptides from the HR1 and HR2 regions of the Sendai virus F protein will bind to lipid bilayers (18, 19). However, such an interaction is not likely to be directly responsible for fusion inhibition, since the inhibition is virus specific. Similar arguments can be made in ruling out a host cell protein as primary target. While our data cannot exclude the HN protein as a target for the peptide, we explored potential interactions of the HR1 peptide with the HR2 peptide because of results with gp41-derived peptides (31, 45–47). Our results clearly show that the HR1 peptide can interact specifically with the HR2 sequence. This result was obtained in two different assays. One directly measured the binding of the HR2 peptide tagged with biotin to the HR1 peptide. This interaction was specific for the HR1 peptide and was competed by the addition of untagged HR2 peptide. The second method was a functional assay of the interaction of the two peptides. Importantly, our results show that equimolar mixture of the two peptides minimally inhibited fusion. Furthermore, relief of inhibition was maximal when the peptides were present in equimolar amounts, suggesting a 1:1 complex. These results are consistent with the notion that the two peptides form a complex which inhibits the interaction of either peptide with the intact protein and therefore relieves the inhibition observed with each peptide when added alone.
Using very different methods, Ghosh et al. (19) have also reported data suggesting that peptides from the HR1 and HR2 regions of Sendai virus interact. Joshi et al. (26) have also reported that peptides from HR1 and HR2 regions of the SV5 F protein coassemble. However, their complex still inhibits fusion, in contrast to the results reported here for the NDV system. The reasons for this different result are unclear, although the difference may indicate that the SV5 complex may be somewhat different than the NDV complex described here. Furthermore, the peptides used by Joshi et al. were much larger than those described here, and the two peptides were of different lengths, in contrast to those described here, which differed in length by only 4 amino acids.
Our results do not address the precise form of the complex between the two peptides. By direct analogy to the HIV env structure, Joshi et al. (26) proposed that the HR1 and HR2 peptides form a six-stranded complex, with the HR1 forming a trimer and three HR2 peptides lying on the outside. Indeed, because of the hydrophobic face of the HR1 peptide, it is likely to form an oligomeric structure in aqueous solution, and, by gel filtration, we have obtained results consistent with the proposal that HR1 forms an oligomer (unpublished data). However, the NDV HR2 peptide may also form an oligomer in an aqueous environment. The results of NMR analyses suggest an ordered helical structure with a hydrophobic face which should promote oligomer formation. Indeed, gel filtration analysis has yielded results consistent with a trimer (unpublished data). Furthermore, Ghosh et al. have reported that an HR2 peptide with sequences from the Sendai virus HR2 region self-assembles in aqueous solution (19). Thus the HR1-HR2 complex described here may result in an association of an oligomer of HR1 and an oligomer of HR2. Indeed, our choice of a sequence for the HR1 peptide was governed by a potential interaction which could occur between the charged surfaces of the two peptides (Fig. 8C), surfaces that would be exposed if the peptides formed oligomeric structures with their hydrophobic faces in the interior.
Predictions of the structures of the inhibitory forms of the peptides and their potential interactions in the context of the cell membrane are considerably complicated by the report that peptides with sequences from both the HR1 and HR2 regions of the Sendai virus F protein can associate with lipid (18, 19). The results presented in this paper are consistent with the idea that the peptides bind, as monomers, parallel to the bilayer, with the hydrophobic face of the peptides buried in the membrane. Such an interaction would promote the dissociation of any oligomers in the presence of the cell membrane and allow for alternative associations of the peptides with each other as well as with the intact protein. Indeed, our results reported here show that the HR1 peptide retains an ordered α-helical structure in a hydrophobic environment. Furthermore, any conclusions about structures in the fusion-active form of the intact F protein as well as any prefusion conformation should also be tempered by the presence of other possible HR domains in the F protein that may be involved in fusion. For example, Ghosh et al. (18, 20) reported that peptides from other HR domains within the Sendai virus F protein sequence had inhibitory activity and potential to interact with peptides from the HR1 and HR2 domains (18–20).
ACKNOWLEDGMENTS
This publication was made possible by grant AI30572 from the National Institutes of Health.
We thank Debi Nayak for the Sendai virus cDNA clones and Linda Nicholson for the use of the NMR spectrometer.
REFERENCES
- 1.Almeida F C L, Opella S J. fd coat protein structure in membrane environments: structural dynamics of the loop between the hydrophobic transmembrane helix and the amphipathic in-plane helix. J Mol Biol. 1993;270:481–495. doi: 10.1006/jmbi.1997.1114. [DOI] [PubMed] [Google Scholar]
- 2.Bagai S, Lamb R A. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J Virol. 1995;69:6712–6719. doi: 10.1128/jvi.69.11.6712-6719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bax A, Davis D G. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J Magn Reson. 1985;65:355–360. [Google Scholar]
- 4.Buckland R, Malvoisin E, Beauverger P, Wild F. A leucine zipper structure present in the measles virus fusion protein is not required for its tetramerization but is essential for fusion. J Gen Virol. 1992;73:1703–1707. doi: 10.1099/0022-1317-73-7-1703. [DOI] [PubMed] [Google Scholar]
- 5.Buckland R, Wild F. Leucine zipper motif extends. Nature (London) 1989;338:547. doi: 10.1038/338547a0. [DOI] [PubMed] [Google Scholar]
- 6.Bull T E. ROESY relaxation theory. J Magn Reson. 1988;80:470–481. [Google Scholar]
- 7.Caffrey M, Cai M, Kaufman J, Stahl S J, Wingfield P T, Covell D G, Gronenborn A M, Clore G M. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 1998;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell M J. Lipofection reagents prepared by a simple ethanol injection technique. Bio/Technology. 1995;18:1027–1032. [PubMed] [Google Scholar]
- 9.Chambers P, Pringle C R, Easton J J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J Gen Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 10.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 11.Chan D C, Kim P S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 12.Chang D K, Cheng S F, Chien W J. The amino-terminal fusion domain peptide of human immunodeficiency virus type 1 gp41 inserts into the sodium dodecyl sulfate micelle primarily as a helix with a conserved glycine at the micelle-water interface. J Virol. 1997;71:6593–6602. doi: 10.1128/jvi.71.9.6593-6602.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clore G M, Gronenborn A M, Brunger A T, Karplus M. Solution conformation of a heptadecapeptide comprising the DNA binding helix F of the cyclic AMP receptor protein of Escherichia coli. Combined use of 1H nuclear magnetic resonance and restrained molecular dynamics. J Mol Biol. 1985;186:435–455. doi: 10.1016/0022-2836(85)90116-0. [DOI] [PubMed] [Google Scholar]
- 14.Creighton T E. Proteins: structure and molecular properties. W. H. New York, N.Y: Freeman & Co.; 1984. [Google Scholar]
- 15.Davis D G, Bax A. Assignment of complex 1H NMR spectra vis two-dimensional homonuclear Hartmenn-Hahn spectroscopy. J Am Chem Soc. 1985;107:2820–2821. [Google Scholar]
- 16.Eich G, Bodenhausen G, Ernst R R. Exploring nuclear spin systems by relayed magnetization transfer. J Am Chem Soc. 1982;104:3731. [Google Scholar]
- 17.Furuta R A, Wild C T, Weng Y, Weiss C D. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh J K, Ovadia M, Shai Y. A leucine zipper motif in the ectodomain of Sendai virus fusion protein assembles in solution and in membranes and specifically binds biologically-active peptides and the virus. Biochemistry. 1997;36:15451–15462. doi: 10.1021/bi971152i. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh J K, Peisajavich S G, Ovadia M, Shai Y. Structure-function study a a heptad repeat positioned near the transmembrane domain of Sendai virus fusion protein which blocks virus-cell fusion. J Biol Chem. 1998;273:27182–27190. doi: 10.1074/jbc.273.42.27182. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh J K, Shai Y. A peptide derived from a conserved domain of Sendai virus fusion protein inhibits virus-cell fusion. J Biol Chem. 1998;273:7252–7259. doi: 10.1074/jbc.273.13.7252. [DOI] [PubMed] [Google Scholar]
- 21.Gierasch L M, Lacy J E, Thompson K F, Rockwell A L, Watnick P I. Conformations of model peptides in membrane-mimetic environments. Biophys J. 1982;37:275–284. doi: 10.1016/S0006-3495(82)84676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gotoh B, Ogasawara T, Toyoda T, Inocencio N M, Hamaguchi M, Nagai Y. An endoprotease homologous to the blood clotting factor X as a determinant of viral tropism in chick embryo. EMBO J. 1990;9:4189–4195. doi: 10.1002/j.1460-2075.1990.tb07643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gotoh B, Ohnishii Y, Inocencio N M, Esaki E, Nakayama K, Barr P J, Thomas G, Nagai Y. Mammalian subtilisin-related proteinases in cleavage activation of the paramyxovirus fusion glycoprotein: superiority of furin/PACE to PC2 or PC1/PC3. J Virol. 1992;66:6391–6397. doi: 10.1128/jvi.66.11.6391-6397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Virus-cell and cell-cell fusion. Annu Rev Cell Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 25.Hicks R P, Young J K. Magnetization transfer via isotropic mixing: an introduction to the HOHAHA experiment. Concepts Magn Reson. 1994;6:115–130. [Google Scholar]
- 26.Joshi S B, Dutch R E, Lamb R A. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
- 27.Lamb R A. Paramyxovirus fusion: a hypothesis of changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 28.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1177–1206. [Google Scholar]
- 29.Lambert D M, Barney S, Lambert A L, Guthrie K, Medinas R, Davis D E, Bucy T, Erickson J, Merutka G, Petteway S R. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci USA. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Sergel T, Razvi E, Morrison T. Effect of cleavage mutants on syncytium formation directed by the wild-type fusion protein of Newcastle disease virus. J Virol. 1998;72:3789–3795. doi: 10.1128/jvi.72.5.3789-3795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthews T J, Wild C, Chen C-H, Bolognesi D P, Greenberg M L. Structural rearrangements in the transmembrane glycoprotein after receptor binding. Immunol Rev. 1994;140:93–104. doi: 10.1111/j.1600-065x.1994.tb00866.x. [DOI] [PubMed] [Google Scholar]
- 32.McDonnell P A, Opella S J. Effect of detergent concentration on multidimensional solution NMR spectra of membrane proteins in micelles. J Magn Reson Ser B. 1993;102:120–125. [Google Scholar]
- 33.McDonnell P A, Shon K, Kim Y, Opella S J. fd coat protein structure in membrane environments. J Mol Biol. 1993;233:447–463. doi: 10.1006/jmbi.1993.1523. [DOI] [PubMed] [Google Scholar]
- 34.Monck J R, Fernandez J M. The exocytotic fusion pore. J Cell Biol. 1992;119:1395–1404. doi: 10.1083/jcb.119.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrison T, McQuain C, Sergel T, McGinnes L, Reitter J. The role of the amino terminus of F1 of the Newcastle disease virus fusion protein in cleavage and fusion. Virology. 1993;193:997–1000. doi: 10.1006/viro.1993.1214. [DOI] [PubMed] [Google Scholar]
- 36.Munoz-Barroso I, Durell S, Sakaguchi K, Apella E, Blumenthal R. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J Cell Biol. 1998;140:315–323. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rapaport D, Ovadia M, Shai Y. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 1995;14:5524–5531. doi: 10.1002/j.1460-2075.1995.tb00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reitter J, Sergel T, Morrison T. Mutational analysis of the leucine zipper motif in the Newcastle disease virus fusion protein. J Virol. 1995;69:5995–6004. doi: 10.1128/jvi.69.10.5995-6004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rose G D, Gierasch L M, Smith J A. Turns in peptides and proteins. Adv Protein Chem. 1985;37:1–106. doi: 10.1016/s0065-3233(08)60063-7. [DOI] [PubMed] [Google Scholar]
- 40.Sergel T, McGinnes L W, Peeples M E, Morrison T G. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology. 1993;193:717–726. doi: 10.1006/viro.1993.1180. [DOI] [PubMed] [Google Scholar]
- 40a.Sergel, T., et al. Unpublished data.
- 41.Sergel-Germano T, McQuain C, Morrison T. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J Virol. 1994;68:7654–7658. doi: 10.1128/jvi.68.11.7654-7658.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.States D J, Haberhorn R A, Rubin D J. A two dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J Magn Reson. 1982;48:286–292. [Google Scholar]
- 43.Tan K, Liu J-H, Wang J-H, Shen S, Lu M. Atomic structure of a thermostable subdomain of the HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiessenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp-41. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 45.Wild C, Dubay J W, Greenwell T, Baird J T, Oas T G, McDanal C, Hunter E, Mathews T. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc Natl Acad Sci USA. 1994;91:12676–12680. doi: 10.1073/pnas.91.26.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wild C, Oas T, McDanal C, Bolognesi D, Mathews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc Natl Acad Sci USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wild C T, Shugars D C, Greenwell T K, McDanal C B, Mathews T J. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wild T F, Buckland R. Inhibition of measles virus infection and fusion with peptides corresponding to the leucine zipper region of the fusion protein. J Gen Virol. 1997;78:107–111. doi: 10.1099/0022-1317-78-1-107. [DOI] [PubMed] [Google Scholar]
- 49.Wishart D S, Sykes B D, Richards F M. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 50.Wüthrich K. NMR of proteins and nucleic acids. New York, N.Y: John Wiley & Sons, Inc.; 1986. [Google Scholar]
- 51.Wüthrich K, Billeter M, Braun W. Polypeptide secondary structure determined by nuclear magnetic resonance observation of short proton-proton distances. J Mol Biol. 1984;180:715–740. doi: 10.1016/0022-2836(84)90034-2. [DOI] [PubMed] [Google Scholar]
- 52.Yao Q, Compans R W. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology. 1996;223:103–112. doi: 10.1006/viro.1996.0459. [DOI] [PubMed] [Google Scholar]
- 53.Young J K, Anklin C, Hicks R P. NMR and molecular modeling investigations of the neuropeptide substance P in the presence of 15 mM sodium dodecyl sulfate micelles. Biopolymers. 1994;34:1449–1462. doi: 10.1002/bip.360341102. [DOI] [PubMed] [Google Scholar]
- 54.Young J K, Hicks R P. NMR and molecular modeling investigations of the neuropeptide bradykinin in three different solvent systems: DMSO, 9:1 dioxane/water, and in the presence of 7.4 mM lysophosphatidylcholine micelles. Biopolymers. 1994;34:611–623. doi: 10.1002/bip.360340504. [DOI] [PubMed] [Google Scholar]
- 55.Young J K, Hicks R P, Wright G E, Morrison T G. Analysis of a peptide inhibitor of paramyxovirus (NDV) fusion using biological assays, NMR, and molecular modeling. Virology. 1997;238:291–304. doi: 10.1006/viro.1997.8834. [DOI] [PubMed] [Google Scholar]