Abstract
Murine hepatitis virus (MHV) gene 1, the 22-kb polymerase (pol) gene, is first translated into a polyprotein and subsequently processed into multiple proteins by viral autoproteases. Genetic complementation analyses suggest that the majority of the gene 1 products are required for viral RNA synthesis. However, there is no physical evidence supporting the association of any of these products with viral RNA synthesis. We have now performed immunofluorescent-staining studies with four polyclonal antisera to localize various MHV-A59 gene 1 products in virus-infected cells. Immunoprecipitation experiments showed that these antisera detected proteins representing the two papain-like proteases and the 3C-like protease encoded by open reading frame (ORF) 1a, the putative polymerase (p100) and a p35 encoded by ORF 1b, and their precursors. De novo-synthesized viral RNA was labeled with bromouridine triphosphate in lysolecithin-permeabilized MHV-infected cells. Confocal microscopy revealed that all of the viral proteins detected by these antisera colocalized with newly synthesized viral RNA in the cytoplasm, particularly in the perinuclear region of infected cells. Several cysteine and serine protease inhibitors, i.e., E64d, leupeptin, and zinc chloride, inhibited viral RNA synthesis without affecting the localization of viral proteins, suggesting that the processing of the MHV gene 1 polyprotein is tightly associated with viral RNA synthesis. Dual labeling with antibodies specific for cytoplasmic membrane structures showed that MHV gene 1 products and RNA colocalized with the Golgi apparatus in HeLa cells. However, in murine 17CL-1 cells, the viral proteins and viral RNA did not colocalize with the Golgi apparatus but, instead, partially colocalized with the endoplasmic reticulum. Our results provide clear physical evidence that several MHV gene 1 products, including the proteases and the polymerase, are associated with the viral RNA replication-transcription machinery, which may localize to different membrane structures in different cell lines.
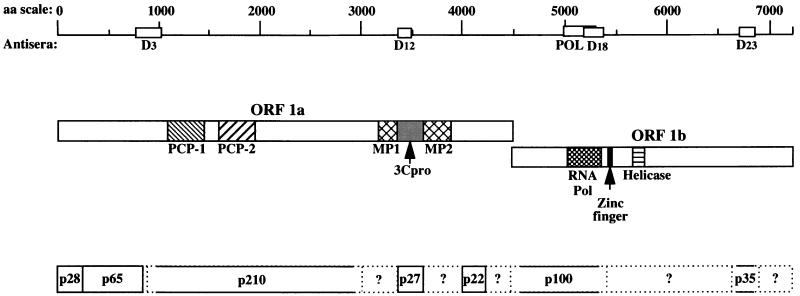
Murine hepatitis virus (MHV), a murine coronavirus, undergoes RNA synthesis by a complex mechanism (27). MHV has a positive-sense single-stranded RNA genome that is 31 kb long and consists of seven or eight genes. The 22-kb gene 1 (also known as the pol gene) is presumed to encode the viral RNA-dependent RNA polymerase, which has been shown by genetic analysis to be responsible for viral RNA synthesis. The pol gene has two open reading frames (ORFs), 1a and 1b, which overlap by 76 nucleotides (29). ORF 1b is in the −1 reading frame with respect to the upstream ORF 1a and is translated following ribosomal frameshifting in the overlap region. The pol gene is first translated into a 750-kDa polyprotein (10, 29), which is subsequently processed by autoproteases to generate a number of proteins containing various functional domains (Fig. 1) (2, 6, 12–15, 20, 33, 34, 45, 48). ORF 1a includes three putative protease motifs, two of which are papain-like cysteine protease motifs (PCP-1 and PCP-2) and one of which is a poliovirus 3C-like protease motif (3Cpro) (22, 29). The functional domains that are predicted to be associated with RNA synthesis have been shown by computer-based motif analyses to be encoded by ORF 1b (8, 21, 22, 24). These include domains for an RNA-dependent RNA polymerase, a nucleoside triphosphate-binding/helicase domain, and a zinc finger nucleic acid-binding domain (29). Genetic complementation analyses suggest that the entire ORF 1a and 1b can be divided into at least five complementation groups, all of which are required for viral RNA synthesis (19, 43).
FIG. 1.
Domains and processed products of the MHV gene 1 polyprotein. Predicted MHV functional domains indicated here were identified by amino acid sequence homology to known viral functional domains (22, 29). Abbreviations: PCP, papain-like cysteine protease; 3Cpro, poliovirus 3C-like protease; MP, membrane-associated protein; HEL, helicase; POL, polymerase. Regions used to generate fusion proteins and ultimately rabbit polyclonal antisera are indicated under the amino acid scale. The processing of ORF 1a products depicted here is based on previous findings (2, 6, 12, 13, 20, 33, 34, 45, 48) and results shown in Fig. 5 and is slightly different from that reported by Denison et al. (15). The dashed lines represent products of uncertain nature.
The processing pathway of MHV gene 1 polyprotein has not yet been fully characterized. A current working model for the predicted cleavage events is presented in Fig. 1. It is clear that the translation of gene 1 results in the synthesis of a polyprotein that is rapidly cleaved by PCP-1 to produce the amino-terminal product, p28 (2, 12, 13, 51). PCP-1 recognizes a conserved cleavage site (RG/V) present in both strains of MHV, MHV-JHM and MHV-A59 (18, 25). PCP-1 also recognizes a downstream cleavage site (A/G) to further process the polyprotein and release a 65-kDa product (p65) adjacent to p28 (6, 17). In vitro processing experiments indicate that PCP-1 may function both in cis and in trans to process the gene 1 polyprotein (7). Most of the remaining processing of the gene 1 polyprotein is carried out by 3Cpro. The mature form of 3Cpro, p27, can be released by cis or trans processing of conserved Q/S cleavage sites (32, 34). In addition, a 22-kDa product downstream of p27 is generated from the polyprotein by the proteolytic activity of 3Cpro (33). A number of other putative 3Cpro cleavage sites have been predicted in ORF 1a and 1b (33). The processing events occurring between p65 and p27 of MHV ORF 1a are controversial. In a study of MHV-A59-infected cells, Denison et al. proposed that a 290-kDa protein, which extends to the p27 cleavage site, is generated and then processed into p240 and p50 (15) and that the cleavage site of p50 is in the PCP-1 domain. In contrast, Schiller et al., in a study of the MHV-JHM strain, identified a 150-kDa protein, which is a precursor of p27 and is expected to include both membrane-associated protein (MP-1 and MP-2) domains (45). Furthermore, a 72-kDa precursor to p65 has also been identified for MHV-JHM (20, 45). The processing of ORF 1b products of MHV-A59 has been studied with in vitro-translated products of viral genomic RNA (14). Five products, p90, p74, p53, p44, and p32, were detected, but the existence and nature of these proteins in virus-infected cells have not been confirmed.
Cysteine and serine protease inhibitors have been used to study the processing of gene 1 polyprotein both in vitro and in vivo. E64d, leupeptin, and zinc chloride have been shown to inhibit viral protein processing (13, 15, 16, 26, 32). E64d is an irreversible cysteine protease inhibitor, which blocks the carboxy-terminal cleavage of p65 by PCP-1 in vitro (26) and, at a concentration of 400 μg/ml, eliminates the proteolytic activity of 3Cpro in infected cells (32). Leupeptin, a reversible inhibitor of trypsin-like serine proteases as well as some cysteine proteases, inhibits the cleavage of p28 by PCP-1 in vitro at 1 to 2 mM (13). However, the cleavage of p28 in vivo is not affected by the presence of up to 2 mM leupeptin (15). At 4 mM, leupeptin only partially inhibits the 3Cpro activity in vivo (32). Zinc chloride partially inhibits the cleavage of p28 by PCP-1 (15) but does not affect the proteolytic activity of 3Cpro in virus-infected cells at 0.1 mM (32). In addition, E64d (26) and leupeptin (16) inhibit MHV RNA replication, at concentrations that inhibit viral protein processing.
Coronavirus RNA and polymerase are present in the membrane fractions of virus-infected cells (9, 47). Immunofluorescence analysis with antisera against some of the MHV ORF 1a products has localized these proteins to the Golgi apparatus in hamster cells transfected with the MHV receptor (3). In contrast, the viral replication complexes of several other RNA viruses, including equine arteritis virus (38, 51, 52), brome mosaic virus (40), and tobacco etch virus (44), are associated with the endoplasmic reticulum (ER) to different extents. Replication complexes of rubella viruses have been identified as virus-modified lysosomes (35). Studies of poliovirus replication have also demonstrated that specific vesicular structures are induced from several intracellular membrane compartments for virus replication during poliovirus infection (4, 46).
Despite the genetic evidence implicating most of the MHV gene 1 products in viral RNA synthesis, so far there is no biochemical evidence in support of their roles, partly because of the lack of efficient in vitro RNA replication systems. In this study, we used another approach by performing immunofluorescent-staining studies and confocal microscopy to examine whether MHV ORF 1a and 1b products and viral RNA coincided in MHV-infected cells. Polyclonal antisera against two regions of 1a protein and two regions of 1b protein (Table 1 and Fig. 1) (45), including the polymerase domain, were used to localize the gene 1 products representing several complementation groups. Bromouridine triphosphate (BrUTP) introduced by cell permeabilization in the presence of actinomycin D and subsequent detection with an antibody against BrUTP-labeled RNA was used to identify de novo-synthesized MHV RNA. In both murine (17CL-1) and human (HeLa expressing the MHV receptor, HeLa-MHVR) cells infected with MHV, we found a colocalization of MHV gene 1 products with de novo-synthesized viral RNA, suggesting that these viral proteins are associated with the viral RNA replication-transcription machinery. We also showed that inhibitors of MHV proteases effectively blocked viral RNA synthesis without altering the localization of viral proteins, suggesting that the processing of gene 1 polyprotein is tightly associated with MHV RNA synthesis. In addition, we observed a colocalization of MHV RNA and proteins with different intracellular membrane structures in different cell lines.
TABLE 1.
Antisera generated against MHV-JHM gene 1 proteins
| Antiserum | Amino acid positionsa | Products detected by immunoprecipitation |
|---|---|---|
| Anti-D3b | 784–1037 | p65, p72, p210, p250c |
| Anti-D12b | 3340–3470 | p27, p150c |
| Anti-D18b | 5192–5379 | p100, >p300 |
| Anti-POLd | 4998–5300 | 100-kDa proteine |
| Anti-D23b | 6679–6821 | p35, >p300 |
MATERIALS AND METHODS
Cells and virus.
Murine 17CL-1 fibroblasts (49) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 10% tryptone phosphate broth. HeLa-MHVR cells (39), which are HeLa cells permanently transfected with the MHV receptor, were kindly provided by T. Gallagher, Loyola University, Chicago, Ill. They were grown in DMEM supplemented with 10% FBS and selected in gpt+select media (10% FBS, 2.5 μg of mycophenolic acid per ml, 250 μg of xanthine per ml, 20 μg of hypoxanthine per ml, and 100 μg of G418 per ml in DMEM) at every fifth passage. Cells were plated in eight-well chamber slides 1 day before infection and then infected with MHV strain A59 (41) at a multiplicity of infection of 1 PFU. Viruses were removed after 1 h, and the cells were maintained in their respective growth media containing 1% FBS.
Antisera.
Polyclonal rabbit antisera raised against the D3, D12, D18, D23, and putative POL regions of MHV-JHM gene 1 polyprotein were used in immunofluorescent-staining and immunoprecipitation experiments (Table 1 and Fig. 1). These antisera recognize gene 1 products from cells infected with either MHV-JHM or the closely related strain, MHV-A59 (5). The D3 region (amino acids [aa] 784 to 1037) extends from the C-terminal portion of p65 to the N-terminal portion of p210 (45). Immunoprecipitation experiments have shown that MHV-JHM gene 1a products, p65 and p210, and their respective intermediates, p72 and p250, were all precipitated by the D3 antiserum (45). The antiserum against the D12 region (aa 3340 to 3470) reacts with p27 and its precursor p150 (45). The D18 antiserum was raised against a glutathione S-transferase (GST) fusion protein containing aa 5192 to 5379, which includes the C-terminal portion of the putative RNA polymerase. The D23 antiserum was raised against a GST fusion protein containing a region (aa 6679 to 6821) of unknown function in ORF 1b. The POL antiserum was raised against the putative polymerase domain (aa 4998 to 5300) fused to maltose-binding protein and shown to immunoprecipitate the in vitro-translated polymerase of approximately 100 kDa (37a). The peptide sequences used to generate the D18 and POL antisera overlap by 109 aa. The D18 antiserum efficiently immunoprecipitated p100 from MHV-infected cells but generated a weak signal in immunofluorescent staining (data not shown). In contrast, the POL antiserum generates a strong signal in immunofluorescent staining but does not efficiently immunoprecipitate p100 from MHV-infected cells (data not shown). Therefore, the POL antiserum was used for immunofluorescent staining and the D18 antiserum was used for immunoprecipitation assays. A monoclonal anti-bromodeoxyuridine (BrdU) antibody, which cross-reacts with bromouridine (BrU), was purchased from Boehringer Mannheim Biochemicals (Indianapolis, Ind.). A monoclonal antibody (StressGen, Victoria, Canada) and a polyclonal antibody (Affinity Bioreagents, Golden, Colo.) against a protein, GRP78, resident in the ER were used to stain the ER membrane (37). Rhodamine-conjugated wheat germ agglutinin purchased from Vector Laboratories (Burlingame, Calif.) was used to localize the Golgi apparatus (50). A monoclonal antibody against lysosome-associated membrane protein 1 (LAMP-1), obtained from The Developmental Studies Hybridoma Bank (Iowa City, Iowa), was used to localize lysosomes (11).
Protein synthesis and protease inhibitors.
The protein synthesis inhibitor cycloheximide was purchased from Sigma (St. Louis, Mo.). Three cysteine protease inhibitors, E64d, leupeptin, and zinc chloride, were purchased from Matreya (Pleasant Gap, Pa.), Boehringer Mannheim, and Sigma, respectively. They were added to MHV-infected cells 3 or 1.5 h before permeabilization or fixation of the cells as indicated below.
Cell permeabilization and labeling of de novo-synthesized viral RNA.
Cell permeabilization with lysolecithin and detection of viral RNA synthesis were performed by the method described by Leibowitz and DeVries (30) with some modifications. Unless indicated otherwise, MHV-A59 or mock-infected 17CL-1 or HeLa-MHVR cells were permeabilized at 8.5 or 7 h postinfection, respectively. The cells were washed twice with serum-free medium and treated with 100 μg of lysolecithin per ml for 90 s on ice. The permeabilized cells were then incubated for 10 to 40 min in a transcription buffer containing 0.5 mM BrUTP to label newly synthesized viral RNA. The standard BrUTP labeling time used was 40 min unless noted otherwise. To inhibit cellular RNA synthesis, the cells were treated with 5 μg of actinomycin D per ml 1 h before permeabilization. Experiments were terminated by fixation of the cells.
Immunofluorescent staining.
Cells were washed in phosphate-buffered saline (PBS) and fixed in 4% formaldehyde for 20 min at room temperature and permeabilized in either acetone for 5 min at −20°C or 0.1% Triton X-100 for 30 min at room temperature. To inhibit nonspecific antibody binding, the cells were incubated in 5% bovine serum albumin for 20 min. Primary antibodies were diluted in 5% bovine serum albumin and incubated with cells for 1 h at room temperature. After three washes in PBS, the cells were incubated with fluorochrome-conjugated secondary antibodies for 1 h at room temperature. Fluorescein isothiocyanate (FITC)- or tetramethylrhodamine-5-isothiocyanate (TRITC)-conjugated goat anti-rabbit immunoglobulin G antibodies were obtained from Pierce (Rockford, Ill.). FITC- or TRITC-conjugated goat anti-mouse immunoglobulin G antibodies were purchased from American Qualex (La Mirada, Calif.). The cells were then washed three times in PBS and mounted in Vectashield (Vector Laboratories).
Labeling and immunoprecipitation of viral proteins.
HeLa-MHVR cells were infected with MHV-A59 at a multiplicity of infection of 1.5 PFU. After 1 h, cells were treated with 2 μg of actinomycin D per ml for the duration of the experiment. At 2.5 h postinfection, the medium was replaced with DMEM lacking methionine and the incubation was continued for 30 min. Proteins were metabolically radiolabeled with [35S]methionine (200 μCi/ml; ICN, Costa Mesa, Calif.) for 2 h followed by a 30-min chase period in complete medium. Cell lysates were prepared at 5.5 h postinfection and subjected to immunoprecipitation with anti-D3, -D12, -D18, and -D23 as described previously (45). The immunoprecipitates were run on a sodium dodecyl sulfate–5 to 10% polyacrylamide gradient gel and detected by autoradiography. E64d (400 μg/ml) was added to the cells at 2 h postinfection and maintained in the medium until cell lysates were prepared at 5.5 h postinfection.
Confocal microscopy.
Confocal microscopy was performed on a Zeiss-210 laser-scanning confocal microscope equipped with an argon laser and a HeNe laser and appropriate filters. The thickness of each digital section obtained by the microscope was 0.3 μm. Each experiment was performed at least three times. For each experiment, approximately 500 cells were examined to ensure that the results were representative. Only cells that were not in syncytia and did not show any visible cytopathic effects were selected for colocalization analysis. Two or more individual cell images were recorded for each experiment. Image analysis was performed with the standard system-operating software provided with the microscope. Fluorescent images were superimposed digitally to allow fine comparison. Colocalization of green (FITC) and red (TRITC) signals in a single pixel produces yellow, while separated signals remain green or red.
RESULTS
Reconstitution of MHV RNA synthesis in permeabilized cells.
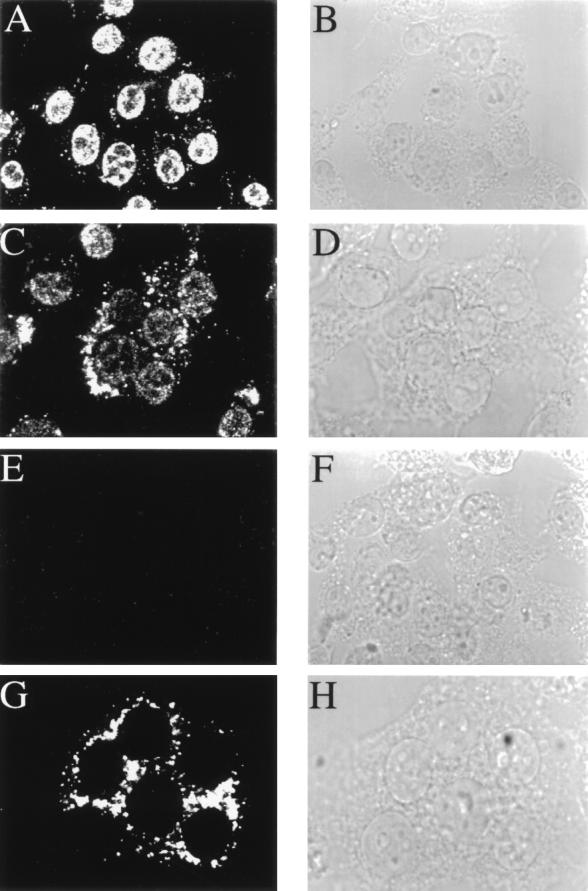
To label newly synthesized viral RNA in virus-infected cells, we reconstituted an in vitro system for MHV RNA synthesis in lysolecithin-permeabilized cells based on a modification of the protocol established by Leibowitz and DeVries (30). Instead of using radiolabeled UTP, viral RNA was labeled by BrUTP incorporation and brominated nucleotides were detected with a monoclonal antibody. In mock-infected 17CL-1 cells, intense nuclear staining was observed, which represents cellular RNA transcription, but the cytoplasmic staining was minimal (Fig. 2A). In MHV-infected 17CL-1 cells, some of which form syncytia, there was a significant increase in cytoplasmic staining, possibly representing viral RNA synthesis; correspondingly, there was a significant decrease in nuclear staining in the majority of BrUTP-labeled cells (Fig. 2C). There appeared to be an inverse relationship between RNA labeling indices in the cytoplasm and the nucleus of infected cells (data not shown), suggesting that host RNA transcription may be inhibited by viral infection, resulting in predominantly viral RNA synthesis. Alternatively, viral RNA synthesis may have depleted BrUTP so that the labeling of cellular RNA synthesis was reduced. Similar experiments were performed on another cell line, DBT; however, this cell line was too fragile to withstand the permeabilization procedure.
FIG. 2.
Detection of de novo-synthesized viral RNA by BrUTP labeling. MHV-A59-infected 17CL-1 cells were permeabilized with lysolecithin, treated with BrUTP at 8.5 h postinfection, and fixed 10 min later. The cells were stained with a monoclonal antibody against BrdU. Representative images of BrUTP labeling are shown for mock-infected (A and E) or MHV-infected (C and G) cells in the presence (E and G) or absence (A and C) of 5 μg of actinomycin D per ml. Actinomycin D was added 1 h before BrUTP labeling. B, D, F, and H are the phase-contrast images of A, C, E, and G, respectively.
To further distinguish the cellular transcription from virus-specific RNA synthesis, 17CL-1 cells were treated with actinomycin D, which blocks host DNA-dependent RNA transcription without affecting MHV-directed, RNA-dependent RNA synthesis. As expected, when cells were preincubated with 5 μg of actinomycin D per ml for 1 h prior to permeabilization and BrUTP labeling, no signal was detected in mock-infected cells (Fig. 2E). In contrast, strong cytoplasmic staining, but no nuclear staining, was observed in MHV-infected cells (Fig. 2G). Thus, the cytoplasmic staining observed in MHV-infected cells in the presence or absence of actinomycin D represents virus-specific RNA. Since BrUTP labeling was carried out for only 15 min, the majority of the labeling observed most probably represents newly synthesized viral RNA, which is closely associated with the viral replication and transcription machinery. Longer labeling times (up to 40 min) did not alter the RNA distribution pattern (data not shown).
We have also performed BrUTP labeling of MHV RNA by introducing BrUTP into cells through N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (DOTAP)-mediated transfection. This procedure is less invasive to cells, and we observed results similar to those obtained by the permeabilization procedure (data not shown). However, only about 5% of the cells could be labeled by DOTAP transfection, in contrast to 60 to 90% for the permeabilization procedure, which allowed for semiquantitative analysis. Thus, the lysolecithin permeabilization procedure was used throughout this study.
Colocalization of MHV gene 1 products with de novo-synthesized viral RNA.
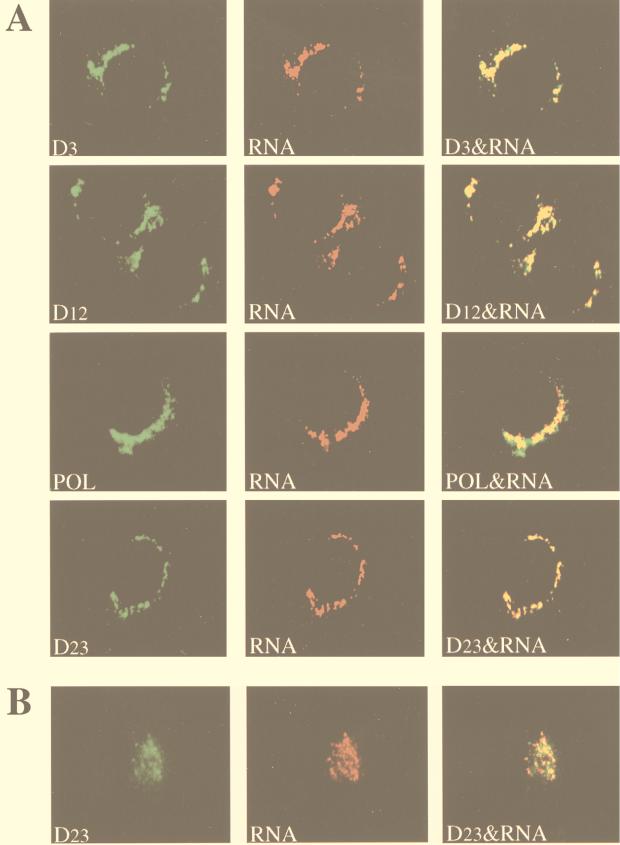
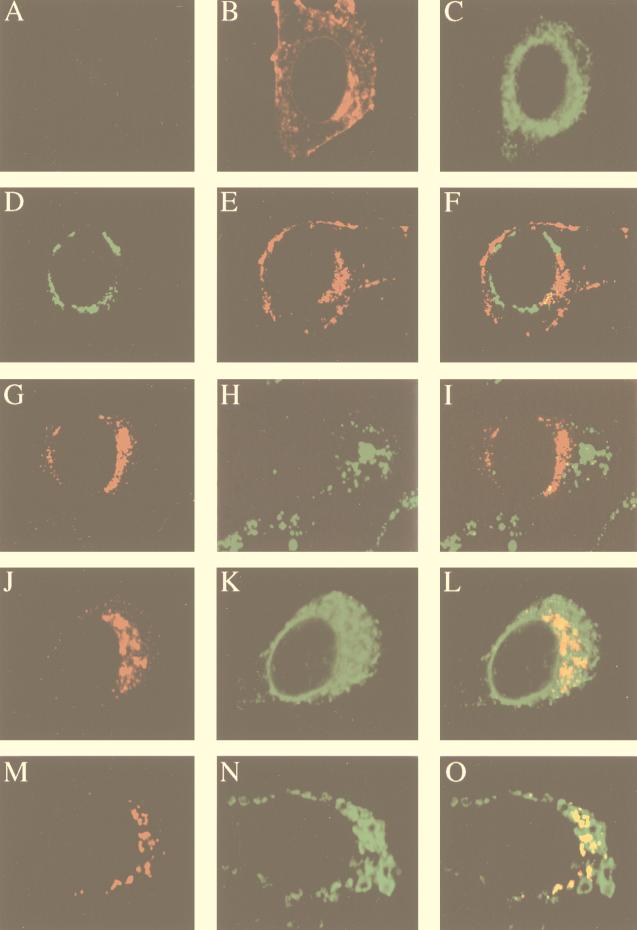
Immunofluorescence analysis with rabbit polyclonal antisera against the D3, D12, POL, and D23 regions of MHV gene 1 polyprotein revealed strong cytoplasmic staining in MHV-infected cells but not in mock-infected cells. Since the virus infection was not synchronized, cells at different stages of infection could be seen at 8.5 h (for 17CL-1) or 7 h (for HeLa-MHVR) postinfection. Fewer than half of the infected cells formed syncytia, and the rest were individual cells without any visible sign of cytopathic effects as a result of MHV infection. As illustrated by the representative images in 17CL-1 cells (Fig. 3A), the staining for all of these viral proteins appeared in discrete spots or patches in the cytoplasm, predominantly in the perinuclear region, of individual virus-infected cells. Cells in syncytia, however, showed more diffuse staining of these proteins in the cytoplasm (data not shown). In all the experiments reported below, only the individual cells not in syncytia were examined. It should be noted that the staining patterns of MHV gene 1 products were similar between the lysolecithin-permeabilized and untreated cells (data not shown), indicating that the lysolecithin treatment did not disrupt the normal cell morphology. Furthermore, the immunofluorescent-staining patterns observed with antisera against D3, D12, POL, and D23 were very similar, suggesting that all of these proteins localized to the perinuclear region, although we could not determine whether they colocalized with each other. None of these antibodies stained uninfected cells (data not shown; see Fig. 7A and 8A).
FIG. 3.
Colocalization of MHV gene 1 products with de novo-synthesized viral RNA. MHV-A59-infected cells were permeabilized and labeled with BrUTP at 8.5 h (17CL-1 cells) or 7 h (HeLa-MHVR cells) postinfection. Actinomycin D (5 μg/ml) was added 1 h before BrUTP labeling. The cells were then double stained with rabbit polyclonal antisera against D3, D12, POL, or D23 regions of gene 1 polyprotein and the mouse monoclonal anti-BrdU antibody (RNA). (A) 17CL-1 cells; (B) HeLa-MHVR cells.
FIG. 7.
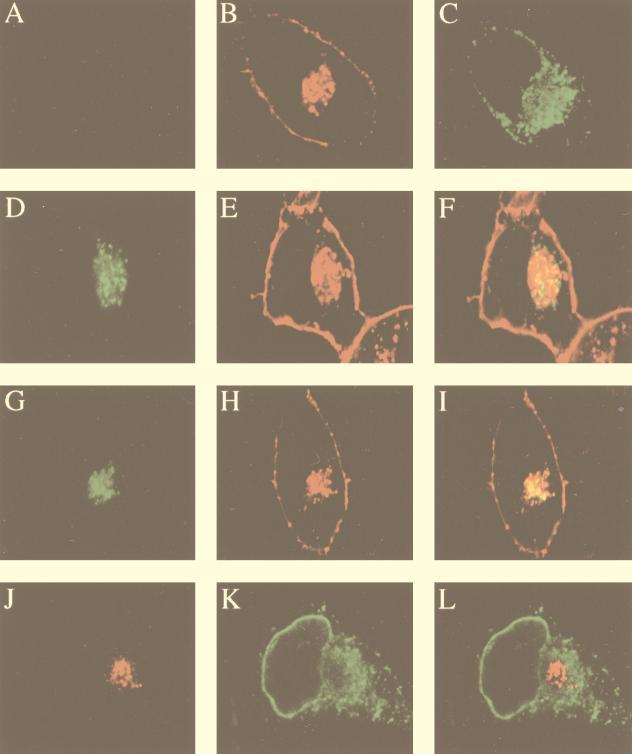
Colocalization of MHV RNA and gene 1 products with the Golgi apparatus in HeLa-MHVR cells. (A) Mock-infected cells were stained with the D3 antiserum; (B and C) mock-infected cells were double stained with WGA (Golgi) (B) and the GRP78 antibody (ER) (C); (D to F) MHV-A59-infected cells were permeabilized and labeled with BrUTP, followed by double staining with the BrdU antibody (RNA) (D) and WGA (E); (G to I) virus-infected cells were double stained with the D3 antiserum (G) and WGA (H); (J to L) virus-infected cells were double stained with antibodies against D3 (J) and GRP78 (ER) (K). Panels F, I, and L are the superimposed images of D and E, G and H, and J and K, respectively.
FIG. 8.
Partial colocalization of MHV gene 1 products with the ER in 17CL-1 cells. (A) Mock-infected cells were stained with the D3 antiserum; (B and C) mock-infected cells were double stained with WGA (Golgi) (B) and the GRP78 antibody (ER) (C); (D to F) MHV-A59-infected cells were double stained with the D3 antiserum (D) and WGA (Golgi) (E); (G to I) virus-infected cells were double stained with antibodies against D3 (G) and LAMP-1 (lysosome) (H); (J to L) virus-infected cells were double stained with antibodies against D3 (J) and GRP78 (ER) (K); (M to O) MHV-A59-infected cells were permeabilized and labeled with BrUTP, followed by double staining with antibodies against BrdU (RNA) (M) and GRP78 (ER) (N). Panels F, I, L, and O are the superimposed images of D and E, G and H, J and K, and M and N, respectively.
We next examined the accumulation sites of these viral proteins with respect to the sites of viral RNA synthesis. Newly synthesized viral RNA was labeled with BrUTP in the presence of actinomycin D and analyzed by dual-labeling immunofluorescence with antibodies against viral proteins and brominated RNA. Representative results of colocalization in 17CL-1 cells are shown in Fig. 3A. Digital superimposition of fluorescent images revealed a significant level of colocalization of MHV gene 1 products and BrUTP-labeled viral RNA, suggesting that all of these proteins are associated with the viral RNA replication-transcription machinery. Since both the processed and precursor proteins were recognized by the antisera, it is conceivable that both protein forms colocalized with viral RNA. The colocalization of MHV gene 1 polyprotein products and viral RNA was also observed in MHV-infected HeLa-MHVR cells. Double staining of one of the proteins (D23) and viral RNA is shown in Fig. 3B. It is interesting that the distribution patterns of both gene 1 products and de novo-synthesized RNA appeared to be different between 17CL-1 and HeLa-MHVR cells, suggesting that they may be associated with different structures in these two cell types (see below).
Time course of viral RNA labeling and protein staining.
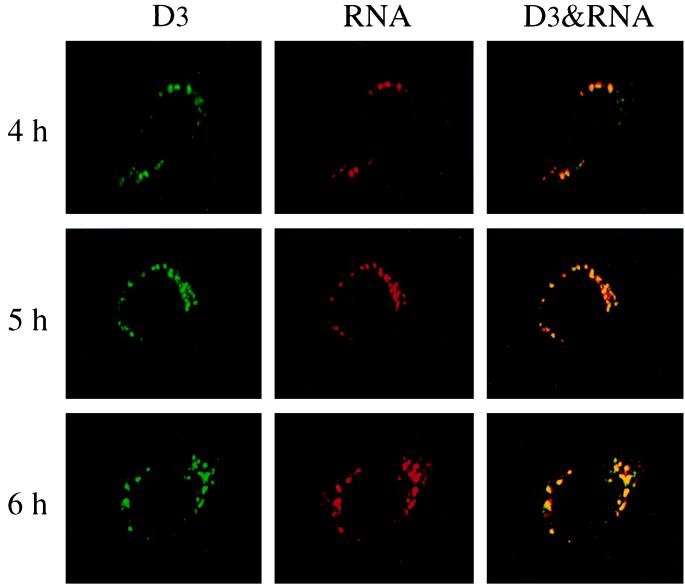
At 8.5 and 7 h postinfection for 17CL-1 and HeLa-MHVR cells, respectively, syncytium formation could be observed. To rule out the possibility that the patterns of viral RNA and gene 1 proteins observed at this time point were influenced by the possible cytopathic effects induced by MHV infection, we performed a kinetic study of BrUTP labeling and immunofluorescent staining of viral proteins at different time points postinfection. RNA labeling was performed at hourly intervals from 2 to 7 h postinfection in the presence of actinomycin D. Clear-cut BrUTP labeling of viral RNA was not detectable until 4 h postinfection for 17CL-1 (Fig. 4) and 3 h postinfection for HeLa-MHVR cells (data not shown). Both the labeling intensity and the number of cells labeled increased with the length of infection. The accumulation of viral proteins was also detected at the same time points as BrUTP labeling in the same cells. Furthermore, confocal microscopy revealed significant colocalization of MHV gene 1 products with newly synthesized viral RNA at any time points examined (Fig. 4). There was no significant difference in the patterns of RNA and protein localization between the early and late time points of infection.
FIG. 4.
Kinetic study of MHV RNA synthesis. MHV-A59-infected 17CL-1 cells were permeabilized and subjected to BrUTP labeling at 4, 5, and 6 h postinfection in the presence of 5 μg of actinomycin D per ml. Actinomycin D was added 1 h before BrUTP labeling. The cells were then double stained with the D3 antiserum and mouse monoclonal BrdU antibody (RNA) as described in the Materials and Methods.
Inhibition of MHV RNA synthesis by protease inhibitors.
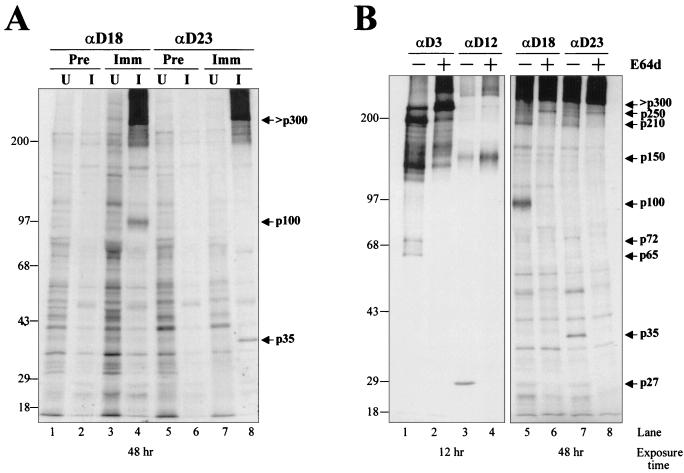
Several cysteine protease inhibitors, i.e., E64d, leupeptin and zinc chloride, have been shown to inhibit MHV PCP-1 and 3Cpro activities, which are responsible for the processing of gene 1 polyprotein (13, 15, 16, 26, 32). We wanted to determine the effects of these inhibitors on viral RNA synthesis and gene 1 protein distribution. We first confirmed the inhibition of MHV gene 1 polyprotein processing by E64d by immunoprecipitation with antisera against D3, D12, D18, and D23 in HeLa-MHVR cells infected with MHV-A59. The D18 antiserum was used in lieu of the POL antiserum in this study because the POL antiserum did not work well in immunoprecipitation and because the D18 and POL proteins overlap by 109 amino acids. As shown in Fig. 5A, anti-D18 and anti-D23, but not the rabbit preimmune serum, immunoprecipitated ORF 1b proteins p100 and p35, respectively, as well as a precursor predicted to be larger than 300 kDa in virus-infected cells. In the absence of E64d, the D3 antiserum immunoprecipitated p65, p72, p210, and p250 whereas the D12 antiserum precipitated p27 and its precursor, p150 (Fig. 5B). These processed products were of the same sizes as those of MHV-JHM, as previously described (45). In the presence of 400 μg of E64d per ml, none of the processed products, including p65, p72, p27, p100, and p35, were detected by their corresponding antisera, indicating that E64d almost completely inhibited the processing of MHV gene 1 polyprotein (Fig. 5B).
FIG. 5.
(A) Immunoprecipitation of MHV ORF 1b proteins by anti-D18 and anti-D23. HeLa-MHVR cells were infected with MHV-A59 and treated with 2 μg of actinomycin D per ml at 1 h postinfection for the duration of the experiment. At 2.5 h postinfection, the medium was replaced for 30 min with DMEM lacking methionine. Proteins were metabolically radiolabeled with [35S]methionine for 2 h, followed by a 30-min chase period in complete medium. Cell lysates were prepared at 5.5 h postinfection and subjected to immunoprecipitation as described previously (45). (B) Inhibition of the processing of MHV gene 1 polyprotein by E64d. E64d (400 μg/ml) was added to the cells at 2 h postinfection and maintained in the medium until the cell lysates were prepared at 5.5 h postinfection. The immunoprecipitated viral proteins are indicated by arrows.
E64d (26) and leupeptin (16) inhibit MHV RNA synthesis at concentrations that inhibit protein processing. We further examined the effects of these protease inhibitors on protein localization. When added to 17CL-1 cells at 3 h (5.5 h postinfection) or 1.5 h (7 h postinfection) before permeabilization and BrUTP labeling (8.5 h postinfection), all three protease inhibitors completely inhibited MHV RNA synthesis at concentrations of 400 μg/ml for E64d (Fig. 6F), 10 mM for leupeptin, and 0.1 mM for zinc chloride (data not shown). These results indicate that continuous protein processing is necessary for viral RNA synthesis, suggesting that the processed products may be unstable. Interestingly, the staining patterns of the gene 1 products examined were not significantly altered by the treatment with these protease inhibitors (compare Fig. 6A and C). These results suggest that the processed protein products were not translocated elsewhere after cleavage. Since these viral proteins were shown to colocalize with viral RNA (Fig. 3), it is likely that the processing of MHV gene 1 polyprotein occurs at or near the site of viral RNA synthesis.
FIG. 6.
Inhibition of MHV RNA synthesis by protein synthesis and protease inhibitors. MHV-A59-infected cells were untreated (A and D) or treated with 10 μg of cycloheximide per ml (B and E) or 400 μg of E64d per ml (C and F) 1.5 h before permeabilization and BrUTP labeling. The cells were labeled with BrUTP for 40 min and double stained with the D3 (A to C) and BrdU (RNA) (D to F) antibodies.
To confirm that the processed gene 1 products were unstable, we treated cells with cycloheximide, which inhibits MHV RNA synthesis when added at any time after virus infection (42). When 17CL-1 cells were treated with 10 μg of cycloheximide per ml for 1.5 h before permeabilization and BrUTP labeling, MHV RNA synthesis was completely inhibited (Fig. 6E). Significantly, gene 1 products were also completely undetectable (Fig. 6B), indicating that both the precursor polyprotein and the processed proteins were degraded within 1.5 h. This result indicates that in the presence of protease inhibitors, the antisera against gene 1 proteins must be detecting the newly synthesized precursor protein but not the processed products. These results, when combined, indicate that both continuous protein synthesis and processing of MHV gene 1 polyprotein are required for viral RNA synthesis. Furthermore, the precursor and processed proteins are probably localized in the same region. Similar results were obtained in HeLa-MHVR cells when exposed to the inhibitors of protein synthesis and proteases (Fig. 6). Although only the staining with the D3 antiserum is presented in Fig. 6, antisera against D12, POL, and D23 produced very similar results (data not shown).
Colocalization of MHV RNA and gene 1 products with intracellular membrane structures.
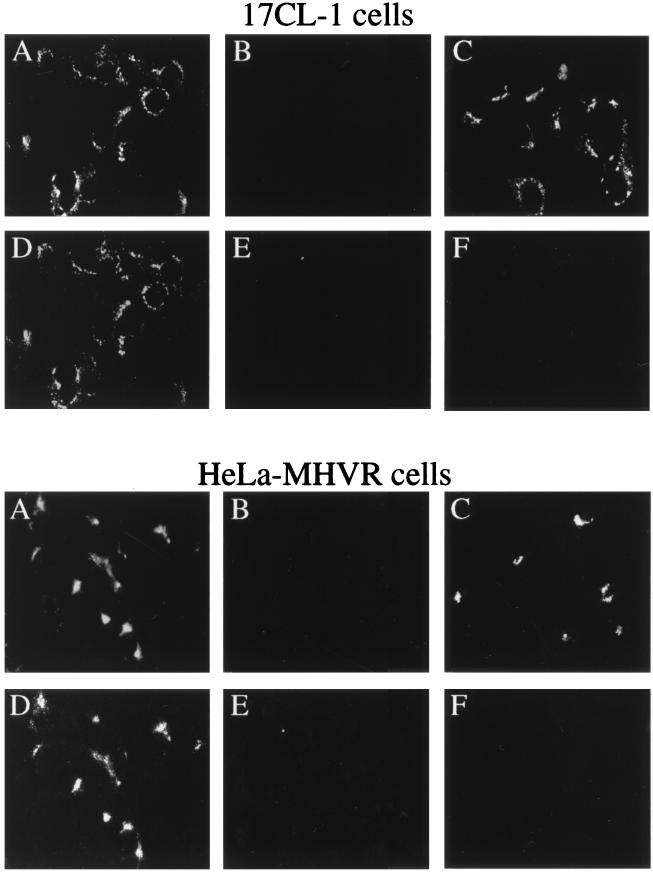
Coronavirus replication or transcription complexes are present in membrane fractions of virus-infected cells (9, 47). The perinuclear cytoplasmic distribution of both BrUTP labeling and staining of gene 1 products shown above is suggestive of their possible association with the intracellular membrane compartments. To test this possibility, antibodies against an ER-resident protein (GRP78) (37), a lysosomal membrane protein (LAMP-1) (11), and wheat germ agglutinin (WGA), which stains the distal face of the Golgi stack as well as the cell surface (50), were used to localize the respective membrane structures. Dual-labeling immunofluorescence analysis was carried out with these markers together with antibodies against BrUTP or MHV gene 1 products. In mock-infected cells, the Golgi and ER staining (Fig. 7A to C and 8A to C) was similar to that in virus-infected cells (Fig. 7D to L and 8D to O), suggesting that there were no gross changes of the cellular membrane structures in infected cells at the time points when this study was performed. In HeLa-MHVR cells, both viral RNA and MHV gene 1 products were localized to the Golgi apparatus (Fig. 7D to I). In contrast, the viral RNA and protein staining and the ER staining were almost mutually exclusive (Fig. 7J to L). These results suggest that the Golgi apparatus or a Golgi-derived membrane structure is probably the site of the viral RNA replication-transcription machinery. There was no apparent difference in WGA staining between MHV-infected and mock-infected HeLa-MHVR cells (Fig. 7A to C) up to at least 7 h postinfection. Surprisingly, when similar studies were performed on the murine 17CL-1 cells, a different pattern emerged: neither MHV RNA nor viral gene 1 products colocalized with the Golgi apparatus (Fig. 8D to F). It is notable that the Golgi structure in 17CL-1 cells appears to be significantly different from that in HeLa-MHVR cells. The viral RNA and proteins also did not colocalize with lysosomes labeled by the LAMP-1 antibody (Fig. 8G to I). However, immunofluorescent staining of the viral proteins (Fig. 8J to L) and BrUTP labeling of the RNA (Fig. 8M to O) in 17CL-1 cells showed partial colocalization of viral proteins and RNA with the ER. These findings suggest that MHV RNA synthesis in 17CL-1 cells may occur on a structure that consists, in part, of the ER membrane. A similar membrane localization pattern of MHV gene 1 products, detected by antisera against D3, D12, POL, and D23, was seen in DBT cells (data not shown). These results, when combined, suggest that MHV RNA synthesis occurs on a membrane structure in virus-infected cells; however, the origin of this membrane structure may be different in different cell lines.
DISCUSSION
The present study used immunofluorescent staining and confocal microscopy to show that several MHV-A59 gene 1a and 1b products, including the proteases and the putative polymerase, colocalized with de novo-synthesized viral RNA, suggesting that these gene 1 products are involved in viral RNA synthesis. This result is consistent with the previous findings by genetic analyses that both 1a and 1b proteins are required for MHV RNA synthesis (19, 43). Furthermore, our results confirmed and extended the finding that MHV RNA synthesis is associated with intracellular membranes in the cytoplasm of virus-infected cells and that the origin of the membrane structure involved in MHV RNA synthesis may be different in different cell lines.
In this study, newly synthesized MHV RNA was labeled with BrUTP in lysolecithin-permeabilized cells. Viral RNA synthesis could be detected as early as 10 min after the addition of BrUTP, and there was no difference in the staining pattern of viral RNA between labeling for 10 min and labeling for 40 min. Furthermore, the putative viral polymerase gene product (p100) and several other gene 1 products were localized to the same perinuclear region as viral RNA in infected cells. Therefore, it is likely that the BrUTP labeling observed in this study represents the site of MHV RNA synthesis.
Our immunoprecipitation study presented the first description of ORF 1b products detected from MHV-infected cells. Using anti-D18, we identified a 100-kDa protein that is proposed to contain the MHV polymerase motif (29). The MHV p100 product is consistent with that reported for the polymerase domain of infectious bronchitis virus (p100) (31) and human coronavirus 229E (p105) (23). Using anti-D23, which was generated against a region near the C-terminal end of ORF 1b, we identified a processed product of 35 kDa. It is currently unclear whether p35 is related to the p32 or p44 products previously identified from in vitro-translated products of MHV-A59 genomic RNA (14). In addition to the processed products, we found that both anti-D18 and anti-D23 immunoprecipitated a large viral protein (>300 kDa), which probably represents the unprocessed ORF 1b precursor. Anti-D3 immunoprecipitated p65, p210, p250, and, to our surprise, p72, from MHV-A59-infected HeLa-MHVR cells (Fig. 5B, lane 1). Although we consistently detected these products from MHV-JHM-infected DBT and 17CL-1 cells, we could not detect p72 from MHV-A59-infected DBT cells (45). As noted previously, we found that only about 10 to 20% of DBT cells are infectable with MHV at a given time point (45) whereas 100% of HeLa-MHVR cells are infectable (39). This high level of simultaneous infection in HeLa-MHVR cells permits a substantial increase in the amount of polymerase polyprotein products present in cell lysates. We believe that increased concentration of MHV polymerase protein in HeLa-MHVR cells permits the detection of p72 from MHV-A59-infected cells in the present study.
The present study demonstrates that cysteine and serine protease inhibitors, i.e., E64d, leupeptin, and zinc chloride, are effective inhibitors of MHV RNA synthesis when added to cells 1.5 h before BrUTP labeling, even though the viral gene 1 products were still detected by immunofluorescent staining and radiolabeling. This result suggests that the uncleaved precursor protein of MHV polymerase cannot support viral RNA synthesis and that the proteolytic activity is required to generate an active polymerase. Both E64d and leupeptin inhibit the proteolytic activity of 3Cpro (26, 32), which is believed to be responsible for the cleavage of many of the ORF 1a and 1b proteins. Zinc chloride, however, does not appear to inhibit 3Cpro (32) but, rather, blocks the cleavage of p28 by PCP-1 (15). The inhibition of MHV RNA synthesis by zinc chloride suggests that a processed p28 is required for viral RNA synthesis. However, no temperature-sensitive mutations have been mapped to this region; thus, its functional significance has not been confirmed by complementation analysis. Alternatively, the processing of p28 may be necessary for the subsequent processing of other MHV gene 1 products. Based on the kinetics of protein processing, the MHV gene 1 polyprotein is probably processed sequentially (15). Since p28 is the first protein to be cleaved from the polyprotein, it is possible that the retention of p28 at the N terminus of the polyprotein prevents cleavage of the rest of the polyprotein, thus affecting the viral RNA synthesis indirectly.
Interestingly, although MHV RNA synthesis was completely inhibited by protease inhibitors (Fig. 6F), the localization of MHV gene 1 products was not altered, as indicated by their colocalization with the same membrane structure in the presence and absence of protease inhibitors (compare Fig. 6A and C). Therefore, the subcellular localization of the MHV replicase complex is independent of complete proteolytic processing. However, ongoing proteolytic processing is clearly required for viral RNA synthesis, since the addition of protease inhibitors rapidly reduced viral RNA synthesis to an undetectable level within 1.5 h (Fig. 6F). In addition, continued protein synthesis is required for viral RNA synthesis (Fig. 6E). Indeed, all of the viral gene 1 products were completely undetectable at 1.5 h after cycloheximide treatment, indicating that both the precursor and processed gene 1 products were rapidly degraded (Fig. 6B). This is consistent with the previous findings in cells treated with cycloheximide (42), anisomycin (36), or E64d (26). Interestingly, the protease inhibitor E64d blocks processing to the mature form of 3Cpro (p27); however, a 150-kDa precursor is detected. This indicates that the proteolytic processing event responsible for cleavage of p150 is not sensitive to E64d. Furthermore, p250 and a high-molecular-mass protein (>300 kDa) are also detected in the presence of the drug. Although these polyprotein intermediates are present in MHV-infected, E64d-treated cells, new RNA synthesis is inhibited. These results demonstrate that intermediates in proteolytic processing are not sufficient for mediating detectable levels of MHV RNA synthesis. They also suggest that the processing of MHV gene 1 polyprotein is likely to be tightly associated with viral RNA synthesis. We cannot, however, rule out the possibility that the inhibitors of protein synthesis and protease also inhibit the synthesis or processing of cellular proteins that are involved in the regulation of MHV RNA synthesis.
The finding that MHV autoproteases, the papain-like proteases (detected by anti-D3) and 3Cpro (detected by anti-D12), colocalized with newly synthesized viral RNA also supports this notion. These proteases are associated with viral RNA synthesis most probably because they process the MHV polyprotein to generate active polymerases. They may also directly participate in viral RNA synthesis in a role similar to that of the poliovirus 3C protease, whose precursor protein (3CD) binds the 5′ end of the viral RNA genome to form an initiation complex for RNA synthesis (1).
The coronavirus replication-transcription complexes are closely associated with the membrane fractions of virus-infected cells (9, 47), but the exact origin of the membrane structure has not been determined. In the present study, MHV RNA and gene 1 products were found to colocalize with the Golgi apparatus in HeLa-MHVR cells, which is consistent with the previous study of several MHV gene 1a products in hamster cells expressing the MHV receptor (3). However, our study also showed that viral RNA and proteins were associated with a different perinuclear membrane compartment in 17CL-1 cells (Fig. 8) and DBT cells (data not shown), although the nature of this membrane is still not clear. One possible candidate is the ER, which was shown to partially colocalize with MHV RNA and gene 1 products (Fig. 8). Studies of other RNA viruses, including equine arteritis virus (38, 51, 52), brome mosaic virus (40), and tobacco etch virus (44), have identified the ER as the site of viral RNA synthesis. More remarkably, the poliovirus replication complex is associated with a unique membrane compartment that is derived from membranes of the ER, Golgi, and other cellular membranes (4, 46). Thus, it is possible that MHV also assembles a replication complex on virus-induced vesicles originating from several cellular membranes such as the ER. This possibility is consistent with the finding that MHV RNA and gene 1 proteins colocalized with only a very small fraction of the ER in 17CL-1 cells (Fig. 8), suggesting that they are in a subcompartment of the ER. Since there are only two regions on the MHV gene 1 polyprotein that are predicted to contain membrane-spanning domains (Fig. 1) (29), most of the processed proteins may not attach to the membrane directly but may form a replication complex with either viral (i.e., the membrane-associated protein domains encoded by ORF 1a) or cellular membrane proteins. Indeed, it has previously been shown that in mild detergent (0.1% Nonidet P-40), the MHV gene 1 products can be immunoprecipitated as a complex by the antisera against D3 and D12 used in this study (45), suggesting that the processed viral proteins are still associated with each other.
Fragmentation of the Golgi apparatus has been observed in MHV-A59-infected L2 murine fibroblasts and 17CL-1 cells, but only at a much later time point, 24 h postinfection (28). In fact, the same study has also indicated that an intact Golgi apparatus for a period of 4 to 16 h postinfection is required for MHV replication (28). Our results revealed no apparent change of the Golgi apparatus in virus-infected HeLa-MHVR and 17CL-1 cells at 7 and 8.5 h postinfection, respectively, compared to mock-infected cells (Fig. 7 and 8). Therefore, the Golgi apparatus we observed at or before 8.5 h postinfection was not fragmented or rearranged by virus infection, although it is possible that minor changes within the Golgi apparatus were not detected by our immunostaining method. There is an apparent difference between the WGA-labeled Golgi apparatus in HeLa-MHVR and 17CL-1 cells (Fig. 7 and 8). The Golgi staining appeared as a compact triangular structure in the juxtanuclear region of HeLa-MHVR cells and as a less well defined structure in 17CL-1 cells. The observed difference may be attributed to the nature of these two cell lines. HeLa cells are human cervical carcinoma cells of epithelial origin and have a well-defined Golgi apparatus for their secretory purposes. In contrast, 17CL-1 is a nonsecretory murine embryonic fibroblast cell line. This difference may partially account for the finding that MHV gene 1 products are associated with the Golgi apparatus in HeLa-MHVR cells but not in 17CL-1 cells.
Taken together, the results of present study demonstrated that MHV gene 1a and 1b products are associated with viral RNA replication-transcription machinery. One of the important functions of these products is proteolytic cleavage, which may occur at the site of viral RNA synthesis. This study further demonstrates the membrane association of viral RNA synthesis; however, the origin of membrane structure involved may be different in different cell lines. Further investigation of the formation of RNA replication and transcription complexes should be helpful in defining the mechanism of MHV RNA synthesis.
ACKNOWLEDGMENTS
We thank Ernesto Barron for his assistance in confocal microscopy. The LAMP-1 monoclonal antibody developed by Thomas August was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by Department of Biological Sciences, University of Iowa, Iowa City, Iowa.
This work was supported by National Institutes of Health research grant AI19244 (to Michael M. C. Lai) and AI32065 (to Susan C. Baker). M.M.C.L. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker S C, Shieh C K, Soe L H, Chang M F, Vannier D M, Lai M M. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J Virol. 1989;63:3693–3699. doi: 10.1128/jvi.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi W, Pinon J D, Hughes S, Bonilla P J, Holmes K V, Weiss S R, Leibowitz J L. Localization of mouse hepatitis virus open reading frame 1A derived proteins. J Neurovirol. 1998;4:594–605. doi: 10.3109/13550289809114226. [DOI] [PubMed] [Google Scholar]
- 4.Bienz K, Egger D, Pfister T, Troxler M. Structural and functional characterization of the poliovirus replication complex. J Virol. 1992;66:2740–2747. doi: 10.1128/jvi.66.5.2740-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonilla P J, Gorbalenya A E, Weiss S R. Mouse hepatitis virus strain A59 RNA polymerase gene ORF 1a: heterogeneity among MHV strains. Virology. 1994;198:736–740. doi: 10.1006/viro.1994.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonilla P J, Hughes S A, Pinnon J D, Weiss S R. Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site. Virology. 1995;209:489–497. doi: 10.1006/viro.1995.1281. [DOI] [PubMed] [Google Scholar]
- 7.Bonilla P J, Hughes S A, Weiss S R. Characterization of a second cleavage site and demonstration of activity in trans by the papain-like proteinase of the murine coronavirus mouse hepatitis virus strain A59. J Virol. 1997;71:900–909. doi: 10.1128/jvi.71.2.900-909.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boursnell M E G, Brown T D K, Foulds I J, Green P F, Tomley F M, Binns M M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- 9.Brayton P R, Lai M M C, Patton C D, Stohlman S A. Characterization of two RNA polymerase activities induced by mouse hepatitis virus. J Virol. 1982;42:847–853. doi: 10.1128/jvi.42.3.847-853.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brierley I, Boursnell M E G, Binns M M, Bilimoria B, Blok V C, Brown T D K, Inglis S C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J W, Murphy T L, Willingham M C, Pastan I, August J T. Identification of two lysosomal membrane glycoproteins. J Cell Biol. 1985;101:85–95. doi: 10.1083/jcb.101.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denison M, Perlman S. Identification of putative polymerase gene product in cells infected with murine coronavirus A59. Virology. 1987;157:565–568. doi: 10.1016/0042-6822(87)90303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denison M R, Perlman S. Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. J Virol. 1986;60:12–18. doi: 10.1128/jvi.60.1.12-18.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denison M R, Zoltick P W, Leibowitz J L, Pachuk C J, Weiss S R. Identification of polypeptides encoded in open reading frame 1b of the putative polymerase gene of the murine coronavirus mouse hepatitis virus A59. J Virol. 1991;65:3076–3082. doi: 10.1128/jvi.65.6.3076-3082.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denison M R, Zoltick P W, Hughes S A, Giangreco B, Olson A L, Perlman S, Leibowitz J L, Weiss S R. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology. 1992;189:274–284. doi: 10.1016/0042-6822(92)90703-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denison M R, Ross T, Gombold J. Inhibition of mouse hepatitis virus A59 replication by the protease inhibitor, leupeptin. Ithaca, N.Y: American Society of Virology, Cornell University; 1992. [Google Scholar]
- 17.Denison M R, Hughes S A, Weiss S R. Identification and characterization of a 65-kDa protein processed from the gene 1 polyprotein of the murine coronavirus MHV-A59. Virology. 1995;207:316–320. doi: 10.1006/viro.1995.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong S, Baker S C. Determinants of the p28 cleavage site recognized by the first papain-like cysteine proteinase of murine coronavirus. Virology. 1994;204:541–549. doi: 10.1006/viro.1994.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu K, Baric R S. Map locations of mouse hepatitis virus temperature-sensitive mutants: confirmation of variable rates of recombination. J Virol. 1994;68:7458–7466. doi: 10.1128/jvi.68.11.7458-7466.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao H Q, Schiller J J, Baker S C. Identification of the polymerase polyprotein products p72 and p65 of the murine coronavirus MHV-JHM. Virus Res. 1996;47:101–109. doi: 10.1016/S0168-1702(96)01368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbalenya A E, Blinov V M, Donchenko A P, Koonin E V. An NTP-binding motif is the most conserved sequence in a highly diverged monophyletic group of proteins involved in positive strand RNA viral replication. J Mol Evol. 1989;28:256–258. doi: 10.1007/BF02102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Coronavirus genome: prediction of putative functional domains in the nonstructural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grotzinger C, Heusipp G, Ziebuhr J, Harms U, Suss J, Siddell S G. Characterization of a 105-kDa polypeptide encoded in gene 1 of the human coronavirus HCV 229E. Virology. 1996;222:227–235. doi: 10.1006/viro.1996.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodgman T C. A new superfamily of replicative proteins. Nature. 1988;333:22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- 25.Hughes S A, Bonilla P J, Weiss S R. Identification of the murine coronavirus p28 cleavage site. J Virol. 1995;69:809–813. doi: 10.1128/jvi.69.2.809-813.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J C, Spence R A, Currier P F, Lu X, Denison M R. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology. 1995;208:1–8. doi: 10.1006/viro.1995.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavi E, Wang Q, Weiss S R, Gonatas N K. Syncytia formation induced by coronavirus infection is associated with fragmentation and rearrangement of the Golgi apparatus. Virology. 1996;221:325–334. doi: 10.1006/viro.1996.0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee H J, Shieh C K, Gorbalenya A E, Koonin E V, La Monica N, Tuler J, Bagdzhadzhyan A, Lai M M C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leibowitz J, DeVries J R. Synthesis of virus-specific RNA in permeabilized murine coronavirus-infected cells. Virology. 1988;166:66–75. doi: 10.1016/0042-6822(88)90147-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu D X, Brierley I, Tibbles K W, Brown T D K. A 100-kilodalton polypeptide encoded by open reading frame (ORF) 1b of the coronavirus infectious bronchitis virus is processed by ORF 1a products. J Virol. 1994;68:5772–5780. doi: 10.1128/jvi.68.9.5772-5780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu X, Lu Y, Denison M R. Intracellular and in vitro-translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV-A59. Virology. 1996;222:375–382. doi: 10.1006/viro.1996.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu X T, Sims A C, Denison M R. Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro. J Virol. 1998;72:2265–2271. doi: 10.1128/jvi.72.3.2265-2271.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Y, Lu X, Denison M R. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J Virol. 1995;69:3554–3559. doi: 10.1128/jvi.69.6.3554-3559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magliano D, Marshall J A, Bowden D S, Vardaxis N, Meanger J, Lee J Y. Rubella virus replication complexes are virus-modified lysosomes. Virology. 1998;240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- 36.Mahy B W J, Siddell S, Wege H, ter Meulen V. RNA-dependent RNA polymerase activity in murine coronavirus-infected cells. J Gen Virol. 1983;64:103–111. doi: 10.1099/0022-1317-64-1-103. [DOI] [PubMed] [Google Scholar]
- 37.Munro S, Pelham H R B. An hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- 37a.Oh, J.-W., and M. M. C. Lai. Unpublished data.
- 38.Pedersen K W, van der Meer Y, Roos N, Snijder E J. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao P V, Gallagher T M. Intracellular complexes of viral spike and cellular receptor accumulate during cytopathic murine coronavirus infections. J Virol. 1998;72:3278–3288. doi: 10.1128/jvi.72.4.3278-3288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Restrepo-Hartwig M A, Ahlquist P. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robb J A, Bond C W. Pathogenic murine coronaviruses. I. Characterization of biological behavior in vitro and virus-specific intracellular RNA of strongly neurotropic JHMV and weakly neurotropic A59V viruses. Virology. 1979;94:352–370. doi: 10.1016/0042-6822(79)90467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawicki S G, Sawicki D L. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J Virol. 1986;57:328–334. doi: 10.1128/jvi.57.1.328-334.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaad M C, Stohlman S A, Egbert J, Lum K, Fu K, Wei T, Baric R S. Genetics of mouse hepatitis virus transcription: identification of cistrons which may function in positive and negative strand RNA synthesis. Virology. 1990;177:634–645. doi: 10.1016/0042-6822(90)90529-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaad M C, Jensen P E, Carrington J C. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 1997;16:4049–4059. doi: 10.1093/emboj/16.13.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schiller J J, Kanjanahaluethai A, Baker S C. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology. 1998;242:288–302. doi: 10.1006/viro.1997.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlegel A, Giddings T H, Ladinsky M S, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sethna P B, Brian D A. Coronavirus genomic and subgenomic minus-strand RNAs copartition in membrane-protected replication complexes. J Virol. 1997;71:7744–7749. doi: 10.1128/jvi.71.10.7744-7749.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soe L H, Shieh C K, Baker S C, Chang M F, Lai M M C. Sequence and translation of the murine coronavirus 5′-end genomic RNA reveals the N-terminal structure of the putative RNA polymerase. J Virol. 1987;61:3968–3976. doi: 10.1128/jvi.61.12.3968-3976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sturman L S, Takemoto K K. Enhanced growth of a murine coronavirus in transformed mouse cells. Infect Immun. 1972;6:501–507. doi: 10.1128/iai.6.4.501-507.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tartakoff A M, Vassalli P. Lectin-binding sites as markers of Golgi subcompartments: proximal-to-distal maturation of oligosaccharides. J Cell Biol. 1983;97:1243–1248. doi: 10.1083/jcb.97.4.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Meer Y, van Tol H, Locker J K, Snijder E J. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J Virol. 1998;72:6689–6698. doi: 10.1128/jvi.72.8.6689-6698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Dinten L C, Wassenaar A L, Gorbalenya A E, Spaan W J M, Snijder E J. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J Virol. 1996;70:6625–6633. doi: 10.1128/jvi.70.10.6625-6633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]