Abstract
Hepatitis A virus (HAV) encodes a single polyprotein which is posttranslationally processed into the functional structural and nonstructural proteins. Only one protease, viral protease 3C, has been implicated in the nine protein scissions. Processing of the capsid protein precursor region generates a unique intermediate, PX (VP1-2A), which accumulates in infected cells and is assumed to serve as precursor to VP1 found in virions, although the details of this reaction have not been determined. Coexpression in transfected cells of a variety of P1 precursor proteins with viral protease 3C demonstrated efficient production of PX, as well as VP0 and VP3; however, no mature VP1 protein was detected. To identify the C-terminal amino acid residue of HAV VP1, we performed peptide sequence analysis by protease-catalyzed [18O]H2O incorporation followed by liquid chromatography ion-trap microspray tandem mass spectrometry of HAV VP1 isolated from purified virions. Two different cell culture-adapted isolates of HAV, strains HM175pE and HM175p35, were used for these analyses. VP1 preparations from both virus isolates contained heterogeneous C termini. The predominant C-terminal amino acid in both virus preparations was VP1-Ser274, which is located N terminal to a methionine residue in VP1-2A. In addition, the analysis of HM175pE recovered smaller amounts of amino acids VP1-Glu273 and VP1-Thr272. In the case of HM175p35, which contains valine at amino acid position VP1-273, VP1-Thr272 was found in addition to VP1-Ser274. The data suggest that HAV 3C is not the protease responsible for generation of the VP1 C terminus. We propose the involvement of host cell protease(s) in the production of HAV VP1.
Hepatitis A virus (HAV) is the sole member of the Hepatovirus genus within the family Picornaviridae. The capsid of HAV encloses a single-stranded RNA genome of about 7.5 kb which is translated into a single polyprotein. The virion proteins VP1 to VP4 and the nonstructural proteins are generated from the polyprotein by a cascade of proteolytic cleavages. Although the overall strategy of virus replication and most of the viral proteins are conserved among all of the picornaviruses, including HAV, the details of polyprotein cleavage differ considerably among the various picornavirus genera. For example, in enteroviruses and rhinoviruses, the primary cleavage that releases the structural protein precursor is mediated by 2A protease at the VP1/2A junction (40, 44). Cardiovirus polyproteins undergo an intramolecular autocatalytic reaction caused by strained peptide bonds in the sequences surrounding the scissile 2A/2B bond, followed by 3C-mediated cleavage between VP1 and 2A (28). A similar reaction occurs in aphthoviruses, whose 2A coding region consists of only a 16-amino-acid sequence homologous to the C terminus of the cardiovirus 2A gene (35). In HAV, viral protease 3C (3Cpro) has been reported to be responsible for all protein scissions (37, 38); the 2A protein has neither inherent nor catalytic proteolytic function (19, 22). The primary cleavage to release the capsid protein from the HAV polyprotein is mediated by 3Cpro between 2A and 2B at amino acids Gln836 and Ala837 (25). 3Cpro is also responsible for the cleavage within the structural proteins between VP3/VP2 and VP2/VP1 (24). This processing of the capsid protein precursor region of HAV generates a unique 38- to 40-kDa intermediate, PX (VP1-2A) (1), which is thought to subsequently yield VP1. Although evidence has been presented that 3Cpro could catalyze scission of a peptide containing VP1-2A sequences (37), the details of VP1 formation from its presumed precursor have not been elucidated, and the terminal amino acids of the resulting VP1 and 2A products have not been identified biochemically. A cleavage site at amino acid residue VP1-300 was initially proposed by sequence alignment analysis (8), which proved to be incorrect (14). Based on the assumption that 3C is the protease responsible for the cleavage within the VP1-2A sequence, several potential 3C cleavage sites were proposed. Modeling of the VP1 protein structure, sequence alignment with other picornaviral capsid proteins, and/or comparative mobilities of recombinant proteins of known sequence favored VP1 amino acid residues Glu273/Ser274 as the likely 3C cleavage site (14, 31). However, no explanation has been offered for the multiple anti-VP1 immunoreactive proteins with molecular masses (29 to 36 kDa) intermediate between those of VP1-2A and VP1 that are observed in HAV-infected cells (1, 7, 45). Some differences in the patterns and/or relative amounts of these products have been detected, presumably depending on the HAV strain and the host cell (14). Additional doubts were raised for HAV strains that contain a Glu-to-Val substitution at the favored VP1/2A cleavage site, resulting in a very unlikely Val/Ser substrate for HAV 3Cpro (21, 30).
We have attempted to determine the C-terminal sequence of HAV VP1 by using peptide sequence analysis by protease-catalyzed [18O]H2O incorporation followed by liquid chromatography ion-trap microspray tandem mass spectrometry (LC-MS/MS) of purified HAV VP1. The technique involves the incorporation of 18O into all α-carboxy groups liberated during the enzyme-catalyzed partial hydrolysis of the protein, followed by MS to identify the C-terminal peptide as the only peptide that did not incorporate any 18O (33, 34, 36, 41, 46). The application of this LC-MS/MS approach has led to the successful identification and sequence analysis of the C-terminal peptides from HAV VP1. Interestingly, our analyses indicate the presence of a mixture of VP1 molecules in the mature hepatitis A virions, containing consecutive amino acid residues from VP1-274 to VP1-272 as C termini.
MATERIALS AND METHODS
HAV cDNA and cloning.
HAV sequences corresponding to various lengths of the coding region were introduced into the plasmid pTM1 (27) to allow transcription control from the T7 promoter and translation under control of the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES). pTM1 contains the EMCV 5′ noncoding region (5′NCR) sequences starting from nucleotide (nt) 260 immediately downstream of the poly(C) tract up to the translation initiation codon.
Constructs encoding various segments of the HAV genome are depicted in Fig. 1. Construct pEHP1-2993 contains HAV cDNA from nt 740 to nt 2993 isolated from pHAV/7 (9) by digestion with AflIII and SacI. This fragment was inserted between the NcoI and SacI sites of the plasmid pTM1. Plasmid pEHP1-2380 was constructed by deletion of the BamHI fragment of pEHP1-2993 from nt 2380 (HAV) to the polylinker BamHI site. The cDNA clone pEHP1-3107 contains the complete HAV P1 region originally predicted by Cohen et al. (8) up to nt 3107, and clone pEHP1-3216 extends to nt 3216 in the HAV coding region. In both of these clones, PCR techniques were used to introduce a stop codon after nt 3107 or 3216, respectively. The generated fragments were inserted between the BamHI site at nt 2380 and the XhoI site in the polylinker of plasmid pEHP1-2993. Construct pEHP1P2 contains almost the complete P1 and P2 regions of HAV, except for the six C-terminal amino acids of P2. These sequences were deleted during cloning of the HAV fragment from nt 740 (AflIII) to nt 4982 (EcoRI) derived from pHAV/7 into the EcoRI-NcoI vector fragment of pTM1.
FIG. 1.
Schematic representation of subgenomic HAV constructs used to examine the capsid protein cleavage mediated by HAV 3C. The constructs encode various lengths of HAV protein sequences up to the nucleotide position indicated on the right of each HAV diagram, downstream of the EMCV IRES. All plasmids contain a T7 RNA polymerase promoter upstream of the EMCV sequence. The predicted molecular mass (MW) of each uncleaved protein is indicated to the right. A diagram of the HAV polyprotein is shown on top for comparison.
For expression of the HAV 3C protease, the plasmid pTM-3C was generated by PCR to engineer HAV 3C nucleotide sequences surrounded by an AUG codon and a stop codon. This fragment was cloned directly into the NcoI and XhoI sites of pTM1. Plasmids pE5H-P3 and pE5H-P3μ3C have been described previously (23). pE5H-P3 contains the entire P3 coding region of HAV, including sequences corresponding to the eight C-terminal amino acid residues of 2C; pE5H-P3μ3C contains a HindIII linker insertion at nt 5864 in the 3C coding region that inactivates the 3C protease activity (22).
At least six independent plasmid isolates of each construct were confirmed by analysis of restriction enzyme digests, protein expression, and immunoblotting.
Transient expression assay.
Simultaneous infection with recombinant vaccinia virus vTF7-3 (16) and transfection with plasmid DNA were described previously (43). Briefly, six-well plates with BS-C-1 (African green monkey kidney) cells were washed with Dulbecco’s modified Eagle’s medium (DMEM) and 4 μg of plasmid DNA, 8 μl of Lipofectin reagent (Gibco BRL), and approximately 10 PFU of vTF7-3 per cell was added in 500 μl of DMEM to each well. Incubation took place for 3 h at 37°C, after which 2 ml of DMEM supplemented with 10% fetal bovine serum (FBS) was added, and incubation continued at 37°C for 16 h. An aliquot of one-fourth of the transfected cell material was analyzed by Western blot for HAV-specific protein expression.
Western blot analysis.
HAV-specific proteins were identified by Western immunoblotting with rabbit antiserum raised to HAV VP1 (18) and HAV VP4 (42) as primary antibodies and alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G-Fc (Promega) as a secondary antibody as previously described (42).
Cells and virus.
BS-C-1 cells and cells from subclone 11-1 of the FRhK-4 cell line (17) were grown in DMEM supplemented with 10% FBS at 37°C. HAV HM175pE, a cell culture-adapted variant derived from an early passage of HM175 (kindly provided by L. Binn, Walter Reed Army Institute of Research, Washington, D.C.), which was subsequently propagated in our laboratory, was used to infect BS-C-1 cells. HM175p35 (8) was used to infect 11-1 cells. Eighty percent confluent cell monolayers were infected by replacement of the cell culture medium with unpurified, frozen-thawed cells that had been infected for 10 days with the particular HM175 strain. The cells were incubated for 2 h at 34.5°C, after which DMEM supplemented with 10% FBS was added to each flask and incubation continued at 34.5°C. The cells were split 1:3 7 days postinfection. An aliquot was used for immunofluorescence microscopy to determine the percentage infected (26). Infection of 100% of the cells was usually reached 10 to 14 days postinfection.
Purification of virus and viral proteins.
BS-C-1 or 11-1 cells infected with HAV and grown in 36 T150 flasks were harvested 10 to 14 days postinfection. The cells were trypsinized and then washed once with DMEM supplemented with 10% FBS to inhibit the trypsin and twice with DMEM without supplements. The cell pellets were combined and resuspended in 5 volumes of RSB (10 mM Tris-HCl [pH 7.5], 10 mM NaCl and 1.5 mM MgCl2). Cells were lysed by addition of 10% Nonidet P-40 to a final concentration of 1% and three cycles of freezing and thawing. The suspension was adjusted to 1% sodium dodecyl sulfate (SDS) and treated with 200 U of DNase I (Boehringer Mannheim) in 5 mM MgCl2 for 30 min at room temperature. Cell debris was removed by centrifugation at 16,000 × g for 10 min at 20°C. The virus was concentrated by centrifugation at 150,000 × g for 3 h at 20°C, resuspended in NET buffer (100 mM NaCl, 20 mM Tris-HCl [pH 7.5], 1 mM EDTA) containing 1% Brij 58, and again clarified by centrifugation (16,000 × g, 10 min, 4°C). Three milliliters of clarified supernatant was used to dissolve 2.25 g of CsCl. The final volume was adjusted to 5 ml with NET buffer containing 1% Brij 58. The material was subjected to two rounds of CsCl isopycnic ultracentrifugation at 190,000 × g for 22 h at 4°C. Fractions of 190 μl were collected from the top of the gradient and stored at 4°C. Five microliters of each fraction was analyzed on an SDS–12% polyacrylamide gel at an acrylamide/bisacrylamide ratio of 29:1 and analyzed by silver staining with purified poliovirus as a marker. HAV-positive fractions were pooled, diluted 1:10 in phosphate-buffered saline (PBS), and concentrated by centrifugation at 150,000 × g for 3 h at 4°C. The virus pellet was resuspended in 40 μl of PBS and stored at −70°C.
The HAV capsid proteins were separated in an SDS–12% polyacrylamide gel, stained with Coomassie brilliant blue R250, and washed extensively with water and the HAV VP1 band was excised from the gel and used for amino acid sequencing.
Enzymatic cleavage of proteins in the presence of [18O]H2O.
Proteins were proteolytically cleaved in the presence of 50% [18O]H2O (Isotec, Marrisburg, Ohio) prior to analysis by LC-MS/MS based on the procedure first described by Rose et al. (33) and later modified by others (34, 36, 46). In the work presented here, the method was further modified: following SDS-polyacrylamide gel electrophoresis (PAGE), protein bands were washed by a method based on that of Hellman et al. (20). The last washing step was performed with 50 mM ammonium acetate (pH 6.5), enriched in 50% [18O]H2O, and 50% acetonitrile. The gel slice was then completely dried. One tube of trypsin (Promega, Madison, Wis.) was reconstituted with 50 μl of 50 mM acetic acid to give a concentration of 17.4 pmol/μl. An aliquot of 5 μl was further diluted with an additional 15 μl of 50 mM ammonium acetate (pH 6.5) prepared with 50% [18O]H2O, resulting in a final trypsin concentration of 4.35 pmol/μl. The gel was rehydrated with 5 μl of this trypsin solution and 5-μl aliquots of 50 mM ammonium acetate (pH 6.5), enriched in 50% [18O]H2O, and the buffer was continuously added until the gel was fully reswollen. After incubation at 37°C overnight, the reaction was stopped by addition of 1/10 volume of 10% trifluoroacetic acid (TFA). The peptides were extracted according to the protocol of Hellman et al. (20). No 18O-enriched buffer was utilized for the extraction of the peptides.
MS.
MS was performed with a Finnigan MAT LCQ ion trap mass spectrometer. The instrument was equipped with a Finnigan MAT electrospray ion source modified for microelectrospray as previously described (10, 12, 13). The electrospray source was coupled to a gradient, capillary high-performance liquid chromatography system as described below. The LCQ ion trap mass spectrometer was operated under manual control in the Tune Plus view with the automatic gain control (AGC) active unless stated otherwise. The AGC targets were full MS − 1e+008, MSn − 2e+007, and zoom MS − 5e+006. The default maximal injection time of 100 ms was increased as necessary over the ions of interest to achieve the AGC targets. The numbers of “micro scans” collected were 3, 3, and 5 for full MS, MSn, and zoom MS, respectively. The relative collision energy for collision-induced dissociation (CID) was optimized for each ion in real time following a mass-to-charge (m/z) and charge-dependent relationship as a guide. In a few cases, manual control of the ion injection time was performed to improve the quality of weaker CID product ion spectra.
LC-MS.
In the initial MS analysis, tryptic peptide ions were fractionated by m/z ratio. LC-MS analyses with the LCQ ion trap mass spectrometer were performed with an integrated online microspray interface (13) in which the column is contained within the microspray needle (41). The 150-μm-inner-diameter by 350-μm-outer diameter online microspray needles were pulled by using a model P-2000 laser-based micropipette puller (Sutter Instrument Co., Novato, Calif.) to a terminal inner diameter of approximately 5 μm. The programmed values for heat, filament, velocity, delay, and pull strength were 400, 0, 30, 125, and 0, respectively. The program cycled twice before separation occurred. The final assembly and packing of the integrated microspray needle with Vydac C18 reverse-phase support was performed as described previously (13). LC-MS analyses were performed with the Apple Macintosh controlled gradient loop microcapillary HPLC system developed by Davis et al. (11) and previously described (12). Samples were analyzed with a linear gradient from 2% to 92% buffer B (A, 0.1% TFA in water; B, 90% acetonitrile, 0.07% TFA in water [vol/vol]). The delivery of the gradient was monitored by precolumn UV detection at 200 nm with an ABI 759A UV/VIS spectrophotometer (Applied Biosystems, Inc., Foster City, Calif.).
MS/MS data interpretation.
MS/MS spectra are obtained after CID to induce one dissociation event per peptide ion. Thus, a daughter ion spectrum is obtained. Since dissociation of a peptide can be induced at any peptide bond, from either the NH2 or COOH termini, the spectrum can be analyzed to determine the peptide sequence for either terminus, thus identifying the peptide. MS/MS spectra derived from putative C-terminal peptides were evaluated and interpreted manually. MS/MS spectra generated from internal peptides were interpreted by correlation with the OWL nonredundant composite protein sequence database version 26.0 by using the SEQUEST database searching program (15). Following the automatic data interpretation, the MS/MS spectra were evaluated by hand to confirm the peptide sequence assignment.
RESULTS
Transcleavage of HAV protein precursor.
To examine the products of cleavage by HAV 3Cpro on HAV capsid protein substrates, we performed transient transfection of BS-C-1 cells with plasmid DNAs expressing a series of partial HAV polyprotein substrates in the presence or absence of a second plasmid expressing HAV 3C or an inactive mutated form of 3C. HAV 3C proteins were expressed from pE5H-P3 or pE5H-P3μ3C, which produced all P3 region proteins, including 3C and 3CD, or from pTM-3C, which produced only 3C proteins. Simultaneous infection with recombinant vaccinia virus vTF7-3 provided T7 RNA polymerase to transcribe the different HAV RNAs. Viral capsid proteins were detected by Western immunoblot analysis with antiserum against HAV VP1 or HAV VP4. All HAV constructs used in this assay were derived from pHAV/7 (9) and therefore contain Val at position VP1-273.
Expression of proteins from the different HAV constructs, depicted in Fig. 1, in the presence or absence of active HAV 3C derived from pE5H-P3, was examined by immunoblotting with antiserum against VP1, as shown in Fig. 2A and B. For comparison, HAV proteins derived from HAV-infected BS-C-1 cells were analyzed on the same gels (both panels, lane 2). All constructs produced uncleaved proteins of the expected size in the absence of HAV 3C: P1-2380, ∼60 kDa; P1-2993, ∼82 kDa; P1P2, ∼155 kDa (Fig. 2A, lanes 3, 5, and 7); and P1-3107, ∼87 kDa and P1-3216, ∼90 kDa (Fig. 2B, lanes 3 and 5). Cotransfection with pE5H-P3 (3C) generated cleavage products detectable with antiserum against HAV VP1 in all protein substrates except for P1-2380. The expected VP1-specific cleavage product derived from P1-2380 would be 6 kDa, which was not likely retained on the SDS–10% polyacrylamide gel. Expression of all proteins was decreased when the HAV VP1-containing precursor constructs were cotransfected with pE5H-P3, possibly due to less efficient transfection when two plasmid DNAs were used at the same time. An immunoreactive protein of about 120 kDa was detected in all samples cotransfected with pE5H-P3, which is most likely due to an unspecific reaction of the antibody.
FIG. 2.
Immunoblot analysis of HAV polyproteins expressed from cDNA in the presence of HAV 3C in BS-C-1 cells. Transient transfection was performed in the presence of recombinant vaccinia virus vTF7-3, which carries the gene for T7 RNA polymerase. Proteins P1-2380, P1-2993, and P1P2 were expressed from plasmids pEHP1-2380, pEHP1-2993, and pEHP1P2, respectively, in the absence (A and C, lanes 3, 5, and 7) or presence (A and C, lanes 4, 6, and 8) of HAV 3C expressed from pE5H-P3. VP1-specific products were detected with HAV VP1 antiserum (A), and VP4-specific products were detected with HAV VP4 antiserum (C). (B) VP1-immunoreactive proteins P1-3107 and P1-3216 expressed from plasmids pEHP1-3107 and pEHP1-3216, respectively, in the absence (lanes 3 and 5) or presence (lanes 4 and 6) of HAV 3C expressed from pE5H-P3. Extract from mock-transfected BS-C-1 cells as a negative control (NC) and extract from HAV-infected cells as a positive control (HAV) were analyzed on the same gel (all panels). Immunoreactive proteins VP1 (A and B), PX (A), and VP0VP3 and VP0 (C) are indicated to the right; molecular masses of marker proteins are indicated in kilodaltons to the left (all panels).
In a parallel set of experiments, 3C protein sequences were provided by cotransfection with pTM-3C, which contains only the 3C coding sequence rather than the P3 precursor. The cleavage products obtained from all capsid precursor proteins were similar to those induced by pE5H-P3 (data not shown), confirming that 3CD or other P3 precursors manifested no additional or unique substrate recognition or proteolytic activities, different from 3C (data not shown). This conclusion was drawn previously by other investigators (32).
The 3C-mediated cleavage of protein P1-2993 generated a protein which migrated at a rate similar to but slightly faster than that of VP1 (Fig. 2A, compare lane 6 to lane 2). Proteins from the construct containing the complete P1P2 region of HAV showed no apparent cleavage in the absence of HAV 3C (Fig. 2A, lane 7); as expected from previous reports that HAV 2A, present in the P1P2 construct, has no proteolytic activity (23, 38). When expressed in the presence of HAV 3C, a VP1-immunospecific cleavage product of about 38 kDa was produced from P1P2 (Fig. 2A, lane 8). This protein comigrates with the PX protein in HAV-infected cells (data not shown). The proteins analyzed from cells productively infected with HAV showed almost complete processing of PX to VP1 in this experiment (lane 2). Very little or no protein representing the size of VP1 was produced from P1P2 (Fig. 2A, lane 8). Transcleavage of the protein P1-3107 by HAV 3C generated a 3C-mediated VP1-immunospecific cleavage product larger than HAV VP1 and smaller than HAV PX (about 33 kDa) (Fig. 2B, lane 4). The P1-3107 protein contains the originally predicted VP1 sequence (8), which was subsequently shown to be larger than VP1 (14). No further cleavage of this protein was induced by 3C. A VP1-immunospecific protein of approximately 37 kDa was derived from P1-3216 in the presence of HAV 3C (Fig. 2B, lane 6) which migrated slightly faster than HAV PX (data not shown). Again, HAV-VP1 was not detected. Thus, none of the P1 substrates that extended into the P2 region were cleaved by 3C sequences at the VP1/2A junction.
To verify that the 3C protease expressed from pE5H-P3 exhibited the correct functional activity on the capsid precursor proteins used in this study, an immunoblot analysis with antiserum against HAV VP4 was also performed with P1-2380, P1-2993, and P1P2 to visualize other products of the cleavage reactions. The expected cleavage product VP0 and the intermediate VP0VP3 were produced from all precursor capsid proteins (Fig. 2C, lanes 4, 6, and 8). The mutated HAV 3C derived from pE5H-P3μ3C failed to cleave any HAV capsid precursors (data not shown).
The above transcleavage assays show that HAV 3C did not cleave a variety of HAV capsid precursor proteins of different lengths specifically between VP1 and 2A, even while catalyzing efficient and accurate proteolysis between VP0/VP3 and VP3/VP1. These data support the conclusions drawn from in vitro translation studies performed in our laboratory previously (23), but contrast with conclusions drawn from other studies (31, 32, 38).
Preparation of HAV VP1 for C-terminal sequence analysis.
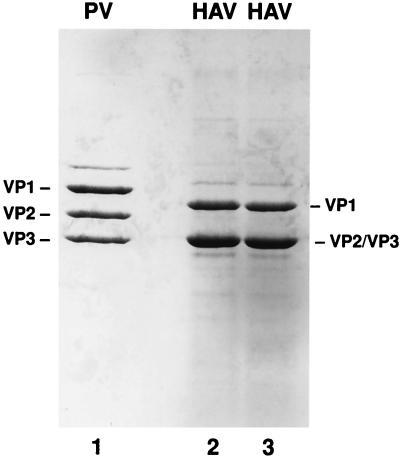
HAV HM175pE grown in BS-C-1 cells and HM175p35 grown in 11-1 cells were harvested 10 to 14 days postinfection and subjected to CsCl isopycnic gradient centrifugation in order to obtain purified virions for analysis of the C-terminal amino acids of HAV VP1. Gradient fractions were analyzed by SDS-PAGE followed by silver staining to identify HAV-containing fractions. The HAV-positive fractions were combined, and virus was collected by sedimentation. Virions were dissociated in SDS, and proteins were separated on an SDS–12% polyacrylamide gel followed by Coomassie blue staining for visualization and excision of HAV VP1. Figure 3, lanes 2 and 3, shows the capsid proteins of purified HAV virions. Purified poliovirus (PV) was separated on the same gel (lane 1), and its VP1 was also excised from the gel as a protein control with a known C terminus for the sequence analysis.
FIG. 3.
Polyacrylamide-SDS gel analysis of HAV capsid proteins. HM175p35 was propagated in 11-1 cells and purified by CsCl isopycnic gradient centrifugation. Capsid proteins were separated by SDS-PAGE (lanes 2 and 3) in comparison with PV (lane 1). The gel was stained with Coomassie blue for excision of capsid protein VP1. PV capsid proteins are indicated to the left, and HAV capsid proteins are indicated to the right.
Determination of C-terminal peptides by incorporation of [18O]H2O upon proteolytic cleavage.
The approach taken for the identification of the C-terminal peptide of HAV VP1 is based on the incorporation of 18O into all internal α-carboxy groups liberated during the trypsin-catalyzed partial hydrolysis of the protein, excluding 18O incorporation into the C-terminal peptide. Figure 4 illustrates the reaction. The mass of all internal peptides will be shifted by 2 atomic mass units (amu) by the 18O incorporation. The experiment can be carried out by performing two parallel digests of the proteins, one of which contains buffer prepared with regular water, the other containing buffer prepared with [18O]H2O. Comparative mass spectral analysis of the resulting peptides would identify the C-terminal peptide as the only one whose mass was not shifted in the digest performed with [18O]H2O. In order to simplify the experiment and to conserve sample material, only one digestion was carried out in the presence of 50% 18O-enriched buffer. In this case, the mass spectral analysis was expected to reveal a characteristic isotope pattern indicating the partial incorporation of 18O for all peptides other than the C-terminal peptide.
FIG. 4.
Principle of 18O incorporation into the internal α-carboxyl group upon proteolytic cleavage of the polypeptide chain. In the presence of 50% 18O-enriched buffer, 18O incorporation is excluded from the C-terminal peptide.
Analysis of PV VP1 and identification of its C-terminal peptide.
To verify the feasibility of this approach, a control experiment was performed with the poliovirus capsid protein VP1, since the complete sequence and the C-terminal amino acid residues are known. Following separation of the capsid proteins by SDS-PAGE, the excised gel band containing VP1 was subjected to in-gel trypsin digestion in the presence of 50% 18O-enriched buffer as described in Materials and Methods. Figure 5A shows the reverse-phase chromatogram of the resulting peptides. Although 50% of all internal peptides are expected to contain 18O at their COOH terminus and 50% contain 16O at their COOH terminus, the retention times of both species of the same peptide remain the same during reverse-phase chromatography.
FIG. 5.
LC-MS/MS analysis of capsid protein VP1. A reverse-phase chromatogram of the resulting peptides derived from the tryptic digests of PV VP1 (A) and HAV VP1 (B) is shown. The asterisks and bar indicate the regions which identified the C-terminal peptides in further analyses.
Mass spectral analysis of individual peptide peaks from Fig. 5A are shown in Fig. 6. All internal peptides generated a characteristic isotope pattern, as illustrated in Fig. 6A and B for two peptide peaks resolved in Fig. 5A. The isotope distribution observed for these peptides indicates the presence of two species. One contains 16O; the other has exchanged one 16O with one 18O, resulting in a net mass increase of two mass units for 50% of the molecules (m/z of 456.2 and 458.2 and m/z of 1,032.5 and 1,034.5, respectively). In contrast, the isotope distribution of the polypeptide at m/z 612.1 reveals no 18O incorporation and is therefore identified as the putative C-terminal peptide of the protein (Fig. 6, compare panel C with panels A and B). All panels also show the presence of peaks with increased masses of 1 mass unit, indicating significant amounts of other enriched stable isotopes (13C, 15N, etc.) in the preparations. These are represented by peaks of 457.2 and 459.3 (Fig. 6A), peaks of 1,033.4 and 1,035.4 (Fig. 6B), and the peak of 613.1 (panel C). Other peaks (m/z 460.3 in panel A and m/z 1,036.4 in panel B) indicate smaller amounts of peptide increased by two mass units.
FIG. 6.
Mass spectral analysis of individual peptide peaks from the reverse-phase chromatogram of PV VP1 (Fig. 5A). The ion signals of two representative peptides for which 50% of the molecules have incorporated 18O are shown ([M−H]+, 456.2 [A] and 1,032.5 [B]). The isotope distribution in panels A and B indicates the mixture of two peptide species, one containing 16O (peaks of 456.2 and 1,032.5); the other species has exchanged one 16O for one 18O resulting in a net mass increase by two mass units for 50% of the molecules (peaks of 458.2 and 1,034.5). In contrast, the isotope distribution of the [M−H]+ ion at m/z 612.1 reveals no 18O incorporation (C) and therefore is the carboxy-terminal peptide.
The putative C-terminal peptide was subjected to MS/MS analysis in which the ion is subjected to CID, inducing one fragmentation per peptide. This analysis identified the sequence as the expected C-terminal peptide sequence as shown in Fig. 7. The sequence AspLeuThrThrTyr (Fig. 7) can be derived from the MS/MS spectrum by subjecting the data to the automatic analysis routine SEQUEST as well as by manual interpretation (for a review, see reference 39). The parent mass of the C-terminal peptide in Fig. 6C is 612.1. During CID of this peptide, parts of a b-ion and y-ion series can be observed. The b-ions are NH2-terminal fragments derived from successive removal of amino acids from the COOH terminus, e.g., b5 is the entire peptide minus water, b4 lacks Tyr, etc. Ions designated y are COOH-terminal fragments derived from successive removal of amino acids from the NH2 terminus, e.g., y3 lacks Asp and Leu, etc. Due to the presence of two Thr residues in the peptide, additional losses of water can be observed for several fragment ions, resulting, for example, in b5 minus water at 576.2 (Fig. 7).
FIG. 7.
MS/MS analysis of the putative C-terminal peptide of PV VP1. The MS/MS spectrum of the [M−H]+ ion at m/z 612.1 was generated during the LC-MS/MS analysis of the PV VP1 tryptic digestion mixture. The fragmentation pattern reveals the C-terminal peptide expected sequence. The b-ions are NH2-terminal fragments derived from successive removal of amino acids from the COOH terminus, and the y-ions are COOH-terminal fragments derived from successive removal of amino acids from the NH2 terminus.
Analysis of HAV VP1 and identification of its C-terminal peptides.
HAV VP1 isolated from HM175pE was analyzed in the same fashion as described above for PV VP1. Following SDS-PAGE of capsid proteins, the VP1 band was excised and subjected to proteolytic digestion in the presence of 50% 18O-enriched buffer as described in Materials and Methods. The digestion mixtures were analyzed by LC-MS/MS. The chromatogram of HAV VP1 tryptic peptides is shown in Fig. 5B. Analysis of the resulting peptides revealed successful 50% incorporation of [18O]H2O in all peptides generated by internal proteolytic cleavage. During LC-MS/MS analysis of this digestion mixture, three doubly charged peptide ions were identified as putative C-terminal fragments in the high-resolution scan. All three ions indicated no incorporation of 18O (insets, Fig. 8), and the masses calculated from the doubly charged ions observed are 1,359.2 (Fig. 8A), 1,272.2 (Fig. 8B), and 1,143.2 (Fig. 8C), respectively. The three doubly charged ions were selected during the LC-MS analysis for MS/MS, and the collected spectra are displayed in Fig. 8. Upon interpretation of the spectra, the sequences were identified as APLNSNAM*LPTES, APLNSNAM*LPTE, and APLNSNAM*LPT (Fig. 8 and 9). The star depicts the oxidation of the methionine residue which was observed in all three peptides. The same three peptides without methionine oxidation eluted with an expected approximately 2-min delay in retention time (41). MS/MS analysis of these peptides revealed the same sequences as those for the peptides containing oxidized methionine (data not shown).
FIG. 8.
MS/MS analysis of the putative C-terminal peptides of HAV VP1 derived from HM175pE. Three ion signals corresponding to C-terminal peptides of HAV VP1 were identified. The isotopic distribution of the doubly charged ions is shown in the insets of panels A, B, and C. The sequences were derived by MS/MS analysis. The mass difference of 87 amu of the y-ion series generated from the peptide in panel A to the y-ion series generated from the peptide in panel B denotes the lack of a serine residue in the peptide (marked in red). The mass difference of 129 amu similarly observed during the CID analysis of the peptides displayed in panels B and C denotes loss of a glutamate residue in panel C (marked in green).
FIG. 9.
C-terminal sequence of capsid protein VP1 determined by trypsin-catalyzed 18O incorporation followed by LC-MS/MS. An asterisk indicates that oxidation of the methionine residue was detected in all HAV VP1 peptides. The predominant C-terminal amino acid (aa) of HAV VP1 detected in both cases was Ser274.
The MS/MS analyses revealed the same NH2-terminal fragments (b-ions) (b11, b9, and b8) in all three peptides, indicating that the N-terminal sequence (APLNSNAMLPT) of the three peptides is identical. Since no b-ion larger than b11 can be observed, no further sequence assignment could be made. The differences in the three peptide sequences can be assigned, however, by closer examination of the COOH-terminal fragment (y-ion) series. By comparison of the MS/MS spectrum in Fig. 8A with the MS/MS spectrum in Fig. 8B, a shift of the y-ion series by 87 amu can be observed. This indicates that the peptide in Fig. 8B is shorter by one serine residue but otherwise identical in sequence to the peptide in panel A. The mass difference of 87 amu observed for the parent masses correlates well with this finding. If Fig. 8B is compared to Fig. 8C, a shift of the y-ion series of an additional 129 amu can be observed, indicating that the last peptide is shorter by one glutamate residue, again consistent with the difference between the parental masses. Since the N-terminal sequences of all three peptides are identical, the sequence differences of the three peptides must be located in the two last C-terminal amino acids. This is confirmed by the fact that the offset of the y-ion series by 87 amu and 129 amu, respectively, can be found for the complete y-ion series. A sequence difference located in the middle of the molecule would be accompanied by identical masses of smaller y-ions.
The above C-terminal LC-MS/MS analysis of HAV HM175pE identified three peptides with consecutive amino acids Thr, Glu, and Ser as C-terminal ends of HAV VP1. Alignment with the amino acid sequence of HAV VP1 localized these C-terminal peptides to VP1-Ala262 through VP1-Thr272, VP1-Ala262 through VP1-Glu273, and VP1-Ala262 through VP1-Ser274 (Fig. 9). In contrast, the analysis of PV VP1 showed a single peptide ending in Tyr at the C terminus of VP1 (Fig. 9). The HAV strain HM175pE contains the amino acid Pro at position VP1-271 and Glu at position VP1-273. Some cell culture-adapted HAV strains derived from HM175, however, contain VP1-271Ser and VP1-273Val at these positions. To determine the influence of a different amino acid within the identified C terminus of HAV VP1, we also performed C-terminal sequencing by LC-MS/MS of purified virions of HAV HM175p35, the variant with the Pro271Ser and Glu273Val substitutions. The LC-MS/MS analysis of HM175p35 again identified multiple amino acid residues at the C terminus of HAV VP1 (Fig. 9). In contrast to the C termini revealed from HM175pE, only two peptides with VP1-274Ser and VP1-272Thr, respectively, representing the C terminus of VP1 were identified. The amino acid valine at position VP1-273 in this HAV variant does not seem to present a suitable target for proteolysis or else does not generate a stable product and is rapidly further processed. In both strains of HAV, the predominant species of VP1 molecules present in mature virions contained Ser274 as their C termini.
DISCUSSION
Most of the cleavages that generate the functional HAV proteins from their polyprotein precursors have been identified and are mediated by the viral protease 3C. The primary scission of the HAV polyprotein occurs between 2A and 2B (25), and PX (VP1-2A) accumulates as a major protein intermediate (1). The process by which VP1 is released from PX is still unclear. No autocatalytic activity of 2A has been observed. Demonstration that 3Cpro could catalyze cleavage of a peptide containing VP1-2A sequences in vitro provided support for the general assumption that 3C, the only identified viral protease, mediated the release of VP1 from PX (37, 38). Based on that assumption, a likely cleavage site between residues Glu273 and Ser274 of VP1 was predicted (14, 31), although never confirmed biochemically. The HAV 3C protease shows high specificity for a glutamine residue followed by a small amino acid residue (30). Substitution of Gln by Glu, as was proposed for the VP1/2A junction, is predicted to form a substrate that will be cleaved by HAV 3C, albeit very poorly (3). Indeed, the existence of HAV strains with a Val substitution for Glu273 raised questions about the suitability of Val/Ser as a scissile bond for 3Cpro; subsequent studies showed it to be less efficiently cleaved by 3C in vitro (31). There are no apparent differences in virus replication rates or protein patterns between the different variants. In addition, the observation of a series of proteins intermediate in mobility between PX and VP1 that are present in infected cells remained unexplained by the model that 3C-mediated cleavage of PX generated VP1.
In the present study, coexpression of several HAV capsid precursor proteins with HAV 3C or total P3 proteins failed to effect cleavage at the VP1/2A junction, although production of VP0 and VP3 was readily detected. These results were consistent with previous studies in our laboratory that examined cleavages following translation in vitro of HAV constructs (23), but appeared to contradict results from studies performed by others (31, 38). During virus infection, cleavage of PX appears to occur only quite late in the morphogenesis pathway of virion particles (1, 4, 6, 47); thus, the processing of VP1-2A that occurs in infected cells might require an assembled conformation of substrate, and the model reactions studied in vitro or in transfected cells may not mimic the relevant biological reaction.
The identification of the C-terminal amino acid(s) of proteins is still one of the more challenging tasks in bioanalyses. Chemical C-terminal sequence analysis analogous to the N-terminal sequence analysis of protein (Edman degradation) has been attempted for a number of years (2). However, the approach bears several disadvantages, including the fact that certain amino acids cannot be identified and that large amounts of protein are required. Earlier attempts to sequence the C terminus of HAV VP1 by using the chemical approach failed (unpublished results); in retrospect, difficulties were likely due to the C-terminal heterogeneity and consequent inadequate sample loading. Another strategy to reveal the C-terminal sequence of a polypeptide is the use of carboxy-terminal-specific proteases in combination with mass spectral analysis (29). Although this is a reasonably sensitive approach for the C-terminal sequence analysis of pure peptides or smaller proteins, it is generally unsuitable for larger biomolecules and molecules in mixtures. In addition, certain amino acids are resistant to carboxy-terminal proteolysis catalyzed by carboxypeptidases. We also attempted mass spectral analysis of intact HAV virion VP1; the resolution of the mass analysis was not sufficient to determine single masses for the individual species of the protein. Heterogeneity of the C terminus in addition to the partial oxidation of possibly all methionine residues contributed to the difficulty in determining the mass(es) of the whole protein.
Thus, we settled on a biochemical characterization of VP1 cleavage products for identification of the C terminus of HAV VP1 from purified virions by LC-MS/MS following partial proteolytic hydrolysis in the presence of 18O-enriched water. This approach successfully identified the final carboxy-terminal amino acid residues of VP1 molecules that are present in mature viruses.
The protease-catalyzed [18O]H2O incorporation followed by LC-MS/MS and identification of peptide sequences represents a suitable alternative to the methods mentioned above. Through the application of microspray LC-MS/MS, protein amounts in the femtomole range become amenable to C-terminal analysis. The sensitivity of this approach, which is further demonstrated by the fact that three different C-terminal peptides of VP1 could be identified in the case of HM175pE, is superior. All terminal peptides were found in their nonoxidized and oxidized forms, resulting in the presence of six C-terminal peptides. In addition, the protein sample need not be highly purified, as long as it resolves as a single band on an SDS-polyacrylamide gel. In cases in which the full sequence of the protein of interest is known, the analysis can be carried out with mixtures as well, since only the peptides of known structure will be examined for 18O incorporation. The relevant peptide which shows no 18O incorporation can quickly be selected from the remaining peptides.
The LC-MS/MS analysis identified a mixture of three C-terminal peptides derived from HAV HM175pE, which contained three consecutive amino acids, VP1-Thr272, VP1-Glu273, and VP1-Ser274, as C-terminal residues of HAV VP1. The analysis of HAV HM175p35, which contains Val at position VP1-273, identified two C-terminal peptides with VP1-Thr272 and VP1-Ser274 as C-terminal amino acids. In both cases, the predominant species of VP1 terminated with Ser274, which is one residue downstream of the previously predicted 3C cleavage site. These results make it highly unlikely that 3Cpro was responsible for generating the C terminus of VP1, since Ser274/Met275 does not have properties of a 3C cleavage substrate and the viral protease is not known to produce heterogeneous ends.
We propose that 3Cpro cleaves the HAV polyprotein at the 2A/2B junction to yield P1-2A and, subsequently, at the VP3/VP1 junction to generate VP1-2A, which participates in initial assembly events. During virus particle formation, cellular proteases may trim 2A sequences from the precursor to generate a series of VP1-containing molecules. The precise substrate for trimming is not known. Those that undergo sufficient trimming (to Ser274) appear to accumulate as stable virion particles. Trimming of the C-terminus of VP1 by cellular proteases has been reported previously for mengovirus (5). Following 3Cpro cleavage at a Glu/Ser junction, three amino acid residues are removed during virus assembly, generating Leu274 as the C terminus of the mature mengovirus VP1.
The data reported here on the heterogeneous C-terminal amino acid residues of HAV VP1 molecules in mature virus capsids do not provide direct information about the processing mechanism or the enzyme(s) responsible for generating the C terminus of VP1. The data do, however, rule out the previously predicted Glu273/Ser274 bond as a cleavage junction that generates VP1, since the Ser274 residue is still present in the majority of virion VP1 molecules.
ACKNOWLEDGMENTS
We are grateful to S. U. Emerson and R. H. Purcell, in whose laboratory some of the experimental work was conducted. S. U. Emerson generously provided cells and virus for some of the experiments.
This work was supported by Public Health Service grants AI26350, AI17386, CA33572, and RR06217 from the National Institutes of Health.
REFERENCES
- 1.Anderson D A, Ross B C. Morphogenesis of hepatitis A virus: isolation and characterization of subviral particles. J Virol. 1990;64:5284–5289. doi: 10.1128/jvi.64.11.5284-5289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baily J M, Shenoy N R, Ronk M, Shively J E. Automated carboxy-terminal sequence analysis of peptides. Protein Sci. 1992;1:68–80. doi: 10.1002/pro.5560010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann E M, Mosimann S C, Chernaia M M, Malcolm B A, James M N G. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J Virol. 1997;71:2436–2448. doi: 10.1128/jvi.71.3.2436-2448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop N E, Anderson D A. Hepatitis A virus subviral particles: purification, accumulation, and relative infectivity of virions, provirions and procapsids. Arch Virol. 1997;142:2147–2160. doi: 10.1007/s007050050232. [DOI] [PubMed] [Google Scholar]
- 5.Boege U, Scraba D G. Mengo virus maturation is accompanied by C-terminal modification of capsid protein VP1. Virology. 1989;168:409–412. doi: 10.1016/0042-6822(89)90284-5. [DOI] [PubMed] [Google Scholar]
- 6.Borovec S V, Anderson D A. Synthesis and assembly of hepatitis A virus-specific proteins in BS-C-1 cells. J Virol. 1993;67:3095–3102. doi: 10.1128/jvi.67.6.3095-3102.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho M W, Ehrenfeld E. Rapid completion of the replication cycle of hepatitis A virus subsequent to reversal of guanidine inhibition. Virology. 1991;180:770–780. doi: 10.1016/0042-6822(91)90090-x. [DOI] [PubMed] [Google Scholar]
- 8.Cohen J I, Ticehurst J R, Purcell R H, Buckler-White A, Baroudy B M. Complete nucleotide sequence of wild-type hepatitis A virus: comparison with different strains of hepatitis A virus and other picornaviruses. J Virol. 1987;61:50–59. doi: 10.1128/jvi.61.1.50-59.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen J I, Ticehurst J R, Feinstone S M, Rosenblum B, Purcell R H. Hepatitis A virus cDNA and its RNA transcripts are infectious in cell culture. J Virol. 1987;61:3035–3039. doi: 10.1128/jvi.61.10.3035-3039.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis M T, Stahl D C, Hefta S A, Lee T D. A microscale electrospray interface for on-line, capillary liquid chromatography/tandem mass spectrometry of complex peptide mixtures. Anal Chem. 1995;67:4549–4556. doi: 10.1021/ac00120a019. [DOI] [PubMed] [Google Scholar]
- 11.Davis M T, Stahl D C, Lee T D. Low flow high-performance liquid chromatography delivery system designed for tandem capillary liquid chromatography-mass spectrometry. J Am Soc Mass Spectrom. 1995;6:571–577. doi: 10.1016/1044-0305(95)00192-G. [DOI] [PubMed] [Google Scholar]
- 12.Davis M T, Lee T D. Variable flow liquid chromatography-tandem mass spectrometry and the comprehensive analysis of complex protein digest mixtures. J Am Soc Mass Spectrom. 1997;8:1059–1069. [Google Scholar]
- 13.Davis M T, Lee T D. Rapid protein identification using a microscale electrospray LC/MS system on an ion trap mass spectrometer. J Am Soc Mass Spectrom. 1998;9:194–201. doi: 10.1016/S1044-0305(97)00282-1. [DOI] [PubMed] [Google Scholar]
- 14.Dotzauer A, Vallbracht A, Keil G M. The proposed gene for VP1 of HAV encodes for a larger protein than that observed in HAV-infected cells and virions. Virology. 1995;213:671–675. doi: 10.1006/viro.1995.0040. [DOI] [PubMed] [Google Scholar]
- 15.Eng J K, McCormack A L, Yates J R., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 16.Fuerst T R, Niles E G, Studier F W, Moss B. Eucaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Funkhouser A W, Purcell R H, D’Hondt E, Emerson S U. Attenuated hepatitis A virus: genetic determinants of adaptation to growth in MRC-5 cells. J Virol. 1994;68:148–157. doi: 10.1128/jvi.68.1.148-157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harmon S A, Johnston J M, Ziegelhoffer T, Richards O C, Summers D F, Ehrenfeld E. Expression of hepatitis A virus capsid sequences in insect cells. Virus Res. 1988;10:273–280. doi: 10.1016/0168-1702(88)90022-6. [DOI] [PubMed] [Google Scholar]
- 19.Harmon S A, Updike W, Jia X-Y, Summers D F, Ehrenfeld E. Polyprotein processing in cis and in trans by hepatitis A virus 3C protease cloned and expressed in Escherichia coli. J Virol. 1992;66:5242–5247. doi: 10.1128/jvi.66.9.5242-5247.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hellman U, Wernstedt C, Góñez J, Heldin C-H. Improvement of an “in-gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal Biochem. 1995;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]
- 21.Jewell D A, Swietnicki W, Dunn B M, Malcolm B A. Hepatitis A virus 3C proteinase substrate specificity. Biochemistry. 1992;31:7862–7869. doi: 10.1021/bi00149a017. [DOI] [PubMed] [Google Scholar]
- 22.Jia X-Y, Scheper G, Brown D, Updike W, Harmon S, Richards O, Summers D, Ehrenfeld E. Translation of hepatitis A virus RNA in vitro: aberrant internal initiations influenced by 5′noncoding region. Virology. 1991;182:712–722. doi: 10.1016/0042-6822(91)90612-f. [DOI] [PubMed] [Google Scholar]
- 23.Jia X-Y, Summers D F, Ehrenfeld E. Primary cleavage of the HAV capsid protein precursor in the middle of the proposed 2A coding region. Virology. 1993;193:515–519. doi: 10.1006/viro.1993.1157. [DOI] [PubMed] [Google Scholar]
- 24.Kusov Y Y, Sommergruber W, Schreiber M, Gauss-Müller V. Intermolecular cleavage of hepatitis A virus (HAV) precursor protein P1-P2 by recombinant HAV proteinase 3C. J Virol. 1992;66:6794–6796. doi: 10.1128/jvi.66.11.6794-6796.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin A, Escriou N, Chao S-F, Girard M, Lemon S M, Wychowski C. Identification and site-directed mutagenesis of the primary (2A/2B) cleavage site of the hepatitis A virus polyprotein: functional impact on the infectivity of HAV RNA transcripts. Virology. 1995;213:213–222. doi: 10.1006/viro.1995.1561. [DOI] [PubMed] [Google Scholar]
- 26.Mathiesen L R, Feinstone S M, Purcell R H, Wagner J A. Detection of hepatitis A antigen by immunofluorescence. Infect Immun. 1977;18:524–530. doi: 10.1128/iai.18.2.524-530.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 28.Palmenberg A C. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 29.Patterson D H, Tarr G E, Regnier F E, Martin S A. C-terminal ladder sequencing via matrix assisted laser adsorption mass spectrometry coupled with carboxypeptidase Y time-dependent and concentration-dependent digestions. Anal Chem. 1995;67:3971–3978. doi: 10.1021/ac00117a024. [DOI] [PubMed] [Google Scholar]
- 30.Petithory J R, Masiarz F R, Kirsch J F, Santi D V, Malcolm B A. A rapid method for determination of endoproteinase substrate specificity: specificity of the 3C proteinase from hepatitis A virus. Proc Natl Acad Sci USA. 1991;88:11510–11514. doi: 10.1073/pnas.88.24.11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Probst C, Jecht M, Gauss-Müller V. Proteinase 3C-mediated processing of VP1-2A of two hepatitis A virus strains: in vivo evidence for cleavage at amino acid position 273/274 of VP1. J Virol. 1997;71:3288–3292. doi: 10.1128/jvi.71.4.3288-3292.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Probst C, Jecht M, Gauss-Müller V. Processing of proteinase precursors and their effect on hepatitis A virus particle formation. J Virol. 1998;72:8013–8020. doi: 10.1128/jvi.72.10.8013-8020.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rose K, Simona M G, Offord R E, Prior C P, Otto B, Thatcher D R. A new mass-spectrometric C-terminal sequencing technique finds a similarity between γ-interferon and α2-interferon and identifies a proteolytically clipped γ-interferon that retains full antiviral activity. Biochem J. 1983;215:273–277. doi: 10.1042/bj2150273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose K, Savoy L-A, Simona M G, Offord R E, Wingfield P. C-terminal peptide identification by fast atom bombardment mass spectrometry. Biochem J. 1988;250:253–259. doi: 10.1042/bj2500253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan M D, King A M Q, Thomas G P. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J Gen Virol. 1991;72:2727–2732. doi: 10.1099/0022-1317-72-11-2727. [DOI] [PubMed] [Google Scholar]
- 36.Schnölzer M, Jedrzejewski P, Lehmann W D. Protease-catalyzed incorporation of 18O into peptide fragments and its application for protein sequencing by electrospray and matrix-assisted laser desorption/ionization mass spectrometry. Electrophoresis. 1996;17:945–953. doi: 10.1002/elps.1150170517. [DOI] [PubMed] [Google Scholar]
- 37.Schultheiss T, Kusov Y Y, Gauss-Müller V. Proteinase 3C of hepatitis A virus (HAV) cleaves the HAV polyprotein P2-P3 at all sites including VP1/2A and 2A/2B. Virology. 1994;198:275–281. doi: 10.1006/viro.1994.1030. [DOI] [PubMed] [Google Scholar]
- 38.Schultheiss T, Sommergruber W, Kusov Y, Gauss-Müller V. Cleavage specificity of purified recombinant hepatitis A virus 3C proteinase on natural substrates. J Virol. 1995;69:1727–1733. doi: 10.1128/jvi.69.3.1727-1733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siuzdak G. The emergence of mass spectrometry in biomedical research. Proc Natl Acad Sci USA. 1994;91:11290–11297. doi: 10.1073/pnas.91.24.11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sommergruber W, Zorn M, Blaas D, Fessl F, Volkmann P, Maurer-Fogy I, Pallai P, Merluzzi V, Matteo M, Skern T, Kuechler E. Polypeptide 2A of human rhinovirus type 2: identification as a protease and characterization by mutational analysis. Virology. 1989;169:68–77. doi: 10.1016/0042-6822(89)90042-1. [DOI] [PubMed] [Google Scholar]
- 41.Swiderek K M, Davis M T, Lee T D. The identification of peptide modifications derived from gel-separated proteins using electrospray triple quadrupole and ion trap analyses. Electrophoresis. 1998;19:989–997. doi: 10.1002/elps.1150190614. [DOI] [PubMed] [Google Scholar]
- 42.Tesar M, Harmon S A, Summers D F, Ehrenfeld E. Hepatitis A virus polyprotein synthesis initiates from two alternative AUG codons. Virology. 1992;186:609–618. doi: 10.1016/0042-6822(92)90027-m. [DOI] [PubMed] [Google Scholar]
- 43.Tesar M, Jia X-Y, Summers D F, Ehrenfeld E. Analysis of a potential myristoylation site in hepatitis A virus capsid protein VP4. Virology. 1993;194:616–626. doi: 10.1006/viro.1993.1301. [DOI] [PubMed] [Google Scholar]
- 44.Toyoda H, Nicklin M J H, Murray M G, Anderson C W, Dunn J J, Studier F W, Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986;45:761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- 45.Updike W S, Tesar M, Ehrenfeld E. Detection of hepatitis A virus proteins in infected BS-C-1 cells. Virology. 1991;185:411–418. doi: 10.1016/0042-6822(91)90789-e. [DOI] [PubMed] [Google Scholar]
- 46.Whaley B, Caprioli R M. Identification of nearest-neighbor peptides in protease digests by mass spectrometry for construction of sequence-ordered tryptic maps. Biol Mass Spectrom. 1991;20:210–214. doi: 10.1002/bms.1200200409. [DOI] [PubMed] [Google Scholar]
- 47.Winokur P L, McLinden J H, Stapleton J T. The hepatitis A virus polyprotein expressed by a recombinant vaccinia virus undergoes proteolytic processing and assembly into viruslike particles. J Virol. 1991;65:5029–5036. doi: 10.1128/jvi.65.9.5029-5036.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]