Abstract
The activity of antibodies against filoviruses is poorly understood but has important consequences for vaccine design and passive prophylaxis. To investigate this activity, a panel of recombinant human monoclonal antibodies to Ebola virus antigens was isolated from phage display libraries constructed from RNA from donors who recovered from infection in the 1995 Ebola virus outbreak in Kikwit, Democratic Republic of Congo. Antibodies reactive with nucleoprotein (NP), envelope glycoprotein (GP), and secreted envelope glycoprotein (sGP) were characterized by immunofluorescence and radioimmunoprecipitation assays. Four antibodies reacting strongly with sGP and weakly with GP and two antibodies reacting with NP were not neutralizing. An antibody specific for GP neutralized Ebola virus to 50% at 0.4 μg/ml as the recombinant Fab fragment and to 50% at 0.3 μg/ml (90% at 2.6 μg/ml) as the corresponding whole immunoglobulin G1 molecule. The studies indicate that neutralizing antibodies are produced in infection by Ebola virus although probably at a relatively low frequency. The neutralizing antibody may be useful in vaccine design and as a prophylactic agent against Ebola virus infection.
The filoviruses Ebola virus and Marburg virus (22) cause severe hemorrhagic fever and high mortality in humans. In fatal infections, the host dies with a high viremia within a few days of the onset of symptoms and there is little evidence of any immune response. There are no vaccines or effective treatments for filovirus infection.
We are interested in determining the activity of antibodies (Abs) against filoviruses so that this might be exploited in vaccine design and possibly in prophylactic or therapeutic agents. When immune system-based countermeasures to filoviruses are considered, survivors of infection might provide important information. About 10 to 20% of those infected with Ebola Zaire virus in the 1976 and 1995 outbreaks survived, but it is not clear to what degree the immune system was involved in their recovery (7, 8, 21). Generally, neutralizing Abs are almost certainly not important because they appear late in the convalescent period and then at very low to insignificant titers (22). Cell-mediated immunity may be crucial, but this is unproven. Transfusion of convalescent-phase whole blood to infected patients in the 1995 Ebola virus outbreak in Kikwit, Democratic Republic of Congo, was anecdotally described as conferring increased survival on treated patients, but other explanations for the survival of these patients have been proposed (18a, 20, 24). There are no reports of immunity to Ebola virus infection after a primary infection.
Rodent models of filovirus infection have been developed and used particularly to investigate immune system correlates of protection. Passive transfer of neutralizing Abs protects guinea pigs from Ebola virus and Marburg virus infection (11, 12). Vaccination with recombinant vaccinia virus expressing Ebola virus glycoprotein (GP) confers partial protection in guinea pigs that is not observed with constructs expressing other Ebola virus proteins (10). These studies imply an important role for antibody in protection against filovirus challenge. Other studies suggest that cell-mediated immunity is important. DNA vaccination with constructs expressing either GP or nucleocapsid protein (NP) protects mice from lethal challenge with Ebola virus (27). Protection of guinea pigs by DNA vaccination was correlated with antibody and cell mediated responses to GP (32).
The extent to which the rodent models are representative of human filovirus infection is not known. Considerable viral adaptation may be involved in the model. For instance, Ebola virus must undergo eightfold serial passage through mice to produce a virus lethal for these animals (4). It is therefore important to carry out studies in nonhuman primates. One detailed study has been carried out to evaluate the efficacy of passively administered antibody in protection against Ebola virus in macaques (13). The Ab used was an immunoglobulin G (IgG) preparation from a horse that had been hyperimmunized with Ebola virus (15, 16) and had a high neutralizing-Ab titer as assessed in a plaque reduction assay. The antibody did delay the onset of clinical symptoms and viremia, but 11 of 12 infected monkeys eventually died. As noted by the authors of that study, the polyclonal equine IgG has a number of limitations, suggesting that it may be valuable to investigate the protective and therapeutic benefit of human monoclonal IgGs. The limitations include the inherently rather low specific activity achievable by passive administration of a polyclonal Ab compared to a monoclonal Ab and the unfavorable pharmacokinetics and diminished effector function activity of an equine IgG in macaques. Human IgGs are very similar to macaques IgGs and are expected to show good pharmacokinetics and effector function activity in the macaques (3).
However, although the use of potent neutralizing human Abs to filoviruses could potentially answer a number of questions, it is not clear that such Abs are produced in natural infection as opposed to the hyperimmunization method used to generate equine IgG as described above. Neutralizing-Ab titers in serum of patients recovering from Ebola virus infection are typically low. These could reflect low concentrations of potent neutralizing Abs in serum or higher concentrations of weakly neutralizing Abs. The latter are unlikely to be effective against the virus, given the results of the studies with macaques. On the other hand, potent neutralizing Abs would signal potential approaches for vaccine development and might prove useful in prophylactic or therapeutic reagents.
To investigate the Abs produced in Ebola virus infection in humans, we have constructed Ab phage display libraries from donors who recovered from infection in the 1995 Ebola Zaire virus outbreak in Kikwit, Democratic Republic of Congo. Specific Abs have been affinity selected from these libraries on Ebola virus antigens including whole inactivated virions. One anti-GP Fab has been engineered to a whole IgG1 molecule and shown to potently neutralize Ebola virus in vitro. A preliminary account of the feasibility of isolating specific human Abs from Ebola virus infection-convalescent donors appeared previously (18).
MATERIALS AND METHODS
Sample collection and RNA preparation.
Bone marrow was obtained from two donors (designated K and L) who recovered from infection with Ebola virus during the 1995 outbreak in Kikwit, Democratic Republic of Congo. Donor K became ill on 4 May 1995 and was hospitalized on 7 May. Donor L became ill on 14 April 1995 and was hospitalized on 19 April. Bone marrow from both donors was drawn on 22 August. Serum samples from each donor were drawn concomitantly. Peripheral blood from 10 donors including donors K and L was drawn as well; these samples were drawn between 5 and 8 July 1995.
Bone marrow of donor K was lysed by vigorous mixing with denaturant solution (4.2 M guanidine hydrothiocyanate [Fluka Biochemika, Buchs, Switzerland], 17 mM sodium N-lauroylsarcosine [Sigma, St. Louis, Mo.], 25 mM trisodium citrate [Sigma], 50 mM 2-mercaptoethanol [Sigma]), which had been sent to Zaire in 10-ml aliquots in 50-ml centrifuge tubes (Corning, Corning, N.Y.). Because of difficulties of isolating RNA on-site, we requested that the bone marrow be diluted at least fourfold in the denaturant solution immediately after bone marrow puncture. The samples were held at 4°C and shipped to the United States. Peripheral blood mononuclear cells (PBMC) were isolated from 50 ml of blood and lysed by vigorous mixing with denaturant solution as described for the bone marrow above. Bone marrow from donor L was immediately frozen in dry ice after aspiration and then shipped to the United States on dry ice. This bone marrow was thawed directly in denaturant solution in a laminar-flow biosafety cabinet with the usual precautions.
RNA was prepared by adding 1 ml of 2 M sodium acetate (pH 4.0) to each 10-ml portion of lysate. The samples were extracted with 10 ml of acidic phenol (saturated with 0.1 M citrate buffer [pH 4.3] [Sigma]) and 2 ml of a chloroform-isoamyl alcohol mixture (24:1). After being incubated on ice for 15 min, the samples were centrifuged at 10,000 × g for 20 min at 4°C. RNA was precipitated from the supernatant by the addition of 40 μg of glycogen (Boehringer Mannheim, Indianapolis, Ind.) and 15 ml of 2-propanol (Sigma), overnight incubation at −20°C, and centrifugation at 10,000 × g for 20 min at 4°C. The RNA pellet was redissolved in 3 ml of denaturant solution and reprecipitated for 3 h at −20°C after the addition of an equal volume of 2-propanol. RNA was pelleted in a microcentrifuge, washed twice with 70% ethanol, and resuspended in diethylpyrocarbonate-treated water.
Library construction.
First-strand cDNA was prepared by priming with oligo d(T) with a cDNA kit (Boehringer Mannheim) as recommended by the manufacturer. The IgG1 Fd region and whole κ and λ light chains were then amplified by PCR as described previously (19). From the PBMC RNA, IgG1 κ and λ libraries were prepared after mixing equal amounts of RNA from each of the 10 donors. Heavy-chain Fd and light-chain PCR products were gel purified, electroeluted, and reamplified with extension primers with a 5′ poly(GA) tail to increase restriction enzyme digestion and subsequent cloning efficiency (31). Phage display libraries were constructed in the phage display vector pComb3H as described previously (1, 5). Briefly, the light-chain and heavy-chain PCR fragments were cloned into the SacI-XbaI and XhoI-SpeI restriction sites of the phagemid, respectively. Ligation products were ethanol precipitated and electroporated into Escherichia coli XL1-Blue cells (Stratagene, La Jolla, Calif.). The transformed E. coli cultures were grown in SOC medium and then in SB medium containing 10 μg of tetracycline per ml and 20 μg of carbenicillin per ml, each for 1 h at 37°C. The carbenicillin concentration was increased to 50 μg/ml, and after the cells had grown for 1 h, phage particle assembly was initiated by the addition of VCS-M13 helper phage (5 × 1011 PFU). After an additional 2 h of culture, kanamycin was added to a concentration of 50 μg/ml and the culture was grown overnight at 30°C. Phage was recovered from the cultures by removing bacteria by centrifugation at 4,000 × g and precipitating phage from the supernatant by addition of 4% polyethylene glycol and 0.5 M NaCl and incubation of the mixture on ice for 30 min. After centrifugation, phage pellets were resuspended in 500 μl of phosphate-buffered saline (PBS–4% nonfat dry milk (Bio-Rad, Hercules, Calif.) and centrifuged for 5 min in a microcentrifuge to pellet bacterial debris.
Affinity selection of Ab libraries on Ebola antigens.
The Ebola antigens used for selection (panning) were (i) a γ-irradiated crude supernatant fraction of Ebola Zaire virus 1995-infected Vero E6 cells and (ii) a γ-irradiated Ebola Zaire virus 1976 whole-virion preparation. In both cases, 2 × 106 rads of γ radiation was applied to frozen samples to inactivate the virus. A microtiter plate (Costar, Cambridge, Mass.) was coated overnight at 4°C with antigens, and the subsequent panning was performed as previously described (5, 18). Briefly, the plates were washed and blocked with 4% nonfat dry milk (Bio-Rad) for 1 h at 37°C. The milk solution was shaken out, phage solution was added to each well, and the mixture was incubated for 2 h at 37°C on a rocker platform. The phage solution was removed, and the wells were washed. Bound phage was eluted with glycine buffer (pH 2.2) and neutralized with 2 M Tris base. Eluted phage was reamplified for the next round of panning as previously described (5). The libraries were panned for four or five consecutive rounds with increasing washing stringency (2, 5, and 10 wash steps thereafter, each consisting of a 5-min incubation and vigorous pipetting). Phagemid DNA, isolated after the last round of panning, was digested with NheI and SpeI restriction endonucleases and religated to excise the cpIII gene and obtain plasmids producing soluble Fabs.
Screening of soluble Fab fragments.
Microtiter wells were coated overnight at 4°C with the two Ebola antigens used for panning and a control antigen, ovalbumin (4 μg/ml) (Pierce, Rockford, Ill.). Soluble Fabs were tested by an enzyme-linked immunosorbent assay (ELISA) as described previously (18).
DNA sequencing.
Fabs were analyzed for their DNA sequence with a 373A or 377A automated DNA sequencer (ABI, Foster City, Calif.), using a Taq fluorescent dideoxy terminator cycle-sequencing kit (ABI), as described previously (2).
Immunofluorescence.
To observe the binding of Fabs to live cells infected with Ebola virus, Vero E6 cells infected with Ebola Zaire 1995 were grown under Biosafety Level 4 conditions on 16-well chamber slides for 3 to 4 days. Each well was incubated with dilutions of each Fab (0.5 to 5 μg/ml) in 1% bovine serum albumin–0.05% NaN3–PBS. To avoid nonspecific Ab uptake by the cells, the wells were incubated on ice for 30 min. The wells were then washed with PBS and air dried, and the cells were γ-irradiated and fixed in acetone for 5 min. The cells were then incubated for 1 h at 37°C with a 1:200 dilution of fluorescein isothiocyanate-coupled goat anti-human IgG F(ab′)2 (Jackson) in PBS–1% normal goat serum. After three washes with PBS, the cells were examined by immunofluorescence. Immunofluorescence with fixed cells was performed in a similar manner except that cells were fixed and permeabilized for 5 min in acetone before being incubated with dilutions of Fab (18).
RIPA.
For radioimmunoprecipitation assays (RIPA), [35S]Cys-[35S]Met or [3H]glucosamine was added to Vero E6 cells infected with Ebola Zaire virus or to mock-infected cells at 4 days postinoculation and incubated overnight. Two types of antigens were used: clarified cell lysate (containing soluble proteins) and supernatant (containing soluble extracellular antigens and virions). The cells were lysed in lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% Nonidet P-40 [NP-40], 0.5% deoxycholate, [DOC], 1 mM EDTA, protease inhibitors [Boehringer-Mannheim]) and irradiated frozen for 2 h (with 2 × 106 rads) to inactivate infectious virus. Lysate was cleared by centrifugation at 14,000 × g for 10 min at 4°C. Cell supernatants were cleared by centrifugation at 14,000 × g for 10 min at 4°C to remove debris and diluted 1:2 with 2× lysis buffer. All antigens were treated with 100 μl of protein G-agarose (Boehringer-Mannheim) for 3 h as specified by the manufacturer. Serum (5 to 10 μl) or 1 to 5 μg of human Fabs and monoclonal rabbit and mouse Abs were mixed with 100 μl of precleared antigens. Goat anti-human IgG F(ab′)2 (1:50 dilution) and 50 μl of protein G-agarose were added to the mixture and incubated overnight at 4°C. After being washed twice with 1 ml of RIPA wash buffer 1 (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% DOC, 1 mM EDTA), twice with wash buffer 2 (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, 0.1% NP-40, 0.05% DOC), and once with wash buffer 3 (50 mM Tris-HCl [pH 7.5], 0.1% NP-40, 0.05% DOC), the precipitate was boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer containing β-mercaptoethanol and loaded onto a 10% polyacrylamide gel (Bio-Rad).
Neutralization assay.
All dilutions were made in Eagle’s minimal essential medium supplemented with 5% heat-inactivated fetal bovine serum. The challenge virus was Ebola Zaire virus 1995 in Vero E6 cell culture passage 2, diluted to contain 100 PFU per 0.1 ml. Fab and IgG1 KZ52 were serially diluted (twofold) at 0.5 ml/tube. Virus and antibody were incubated at 37°C for 1 h. Following the incubation, they were placed on ice. A control Ab (IgG1 b12 [anti-human immunodeficiency virus gp120]) (6) was tested at 5 μg/ml, a concentration at which no inhibition in the number of plaques was observed.
Infectious virus remaining in the virus-Ab mixture was quantitated by counting PFU on Vero E6 cell monolayers. A 0.2-ml volume of each mixture was adsorbed to cells grown in 10-cm2 wells of plastic plates (37°C for 1 h). Each mixture was assayed in two wells. Following adsorption, the cells were overlaid with 2 ml of Eagle’s minimal essential medium containing 5% fetal bovine serum, 25 mM HEPES buffer, 50 μg of gentamicin per ml, and 1% agarose. The cells were incubated at 37°C in a humidified CO2 incubator until plaques were visible under an inverted phase microscope (for neutralization tests, this took 10 to 12 days). After incubation, 2 ml of neutral red (1:6,000 final concentration) was added to each well, and the plaques were counted after an additional 24-h incubation (14).
Preparation of IgG1 KZ52.
To convert Fab KZ52 to a whole IgG molecule, the heavy-chain variable gene fragment and the light-chain gene of KZ52 were cloned into a eukaryotic expression vector containing the human IgG1 constant-region gene and the protein was expressed in CHO cells as described previously (6). IgG1 KZ52 was purified by protein A column chromatography (Pharmacia).
RESULTS
Ab library characterization.
Two IgG1 κ libraries were constructed from bone marrow of convalescent donors (K and L) and contained a diversity of 6 × 106 and 2.2 × 106 clones, respectively. IgG1 κ and IgG1 λ libraries (designated E10κ and E10λ, respectively) constructed from pooled RNA of peripheral-blood lymphocytes from 10 convalescent donors including donors K and L both contained a diversity of 5 × 106 clones.
Isolation of specific Fabs from the libraries by affinity selection against Ebola antigens.
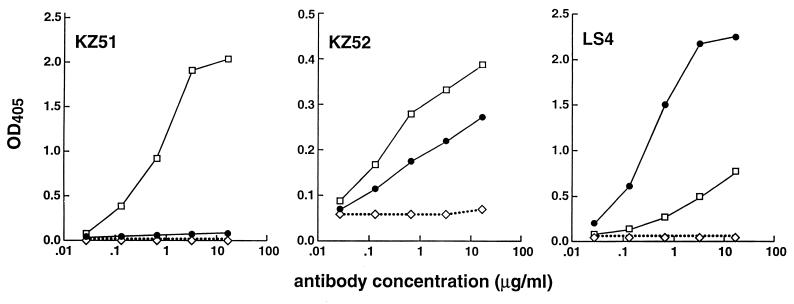
The libraries were panned against γ-irradiated preparations of whole virions (Ebola Zaire virus) and crude supernatants from cultures of infected cells. The former contained all the viral structural proteins, and the latter was greatly enriched for secreted GP (sGP) (see Fig. 3). Specific Fabs were identified by a strong ELISA reactivity with the selecting antigen and a low reactivity with a control antigen (ovalbumin) (Fig. 1).
FIG. 3.
RIPA to show the specificities of anti-Ebola Zaire virus recombinant Fabs. (A) Immunoprecipitations are shown for Fab KZ51 (lanes 1 to 3), Fab ELZ510 (lanes 4 to 6), Fab KS14 (lanes 7 to 9), and convalescent-phase human serum (lanes 10 and 11). For reference, a [35S]Cys-[35S]Met-labeled virion preparation is shown in lane 12. The first lane shown for each Fab is the immunoprecipitate from [35S]Cys-[35S]Met-labeled Ebola virus-infected cell lysate, the second lane is from 35S-labeled crude Ebola virus supernatant, and the third lane is from 35S-labeled lysates from uninfected cells. KZ51 and ELZ510 precipitated NP (97 kDa) from 35S-labeled Ebola virus-infected cell lysate (lanes 1 and 4, respectively) and 35S-labeled crude Ebola virus supernatant (lanes 2 and 5, respectively). (B) Immunoprecipitations are shown for Fab KZ52 (lanes 1 to 3), Fab KS518 (lanes 4 to 6), and Fab LZ51 (lanes 7 to 9). For each Fab, immunoprecipitations are shown from [3H]glucosamine-labeled Ebola virus-infected cell lysate, 3H-labeled Ebola virus supernatants, and 3H-labeled uninfected cell lysates, respectively. KZ52, KS518, and LZ51 immunoprecipitated a band of 120 kDa corresponding to GP1 from [3H]glucosamine-labeled Ebola virus-infected cell lysate (lanes 1, 4, and 7, respectively). KZ52 also immunoprecipitated a band of 120 kDa from [3H]glucosamine-labeled Ebola virus supernatant (lane 2). The other two Fabs predominantly immunoprecipitated a band of 50 kDa, corresponding to sGP from this supernatant (lanes 5 and 8), with only a faint band at 120 kDa. (C) Longer-exposure autoradiograms obtained to reveal the immunoprecipitation of GP2. Exposure of autoradiograms for 2.5 months (compared to 1 week for panel B) reveals immunoprecipitation of a band corresponding to GP2 by Fabs KZ52, KS518, and LZ51 from Ebola virus-infected cell supernatants (lanes 1, 4, and 7, respectively) and from crude infected-cell supernatants (lanes 2, 5, and 8, respectively). Lanes 3, 6, and 9 are uninfected-cell supernatant controls.
FIG. 1.
Reactivity of selected human anti-Ebola virus Fabs with a γ-irradiated whole-virion preparation (open squares) and with irradiated crude infected-cell supernatants (solid circles) determined by ELISA. Ovalbumin (4 μg/ml) is included as a control antigen (open diamonds). OD405, optical density at 405 nm.
Positive clones were sequenced to reveal relatedness. Table 1 shows that from library K two distinct Fabs (indicated by the prefix KZ) were isolated by selection against the virion preparation and four were isolated by selection against the infected-cell supernatant preparation (prefix KS). However, the clone KZ52 selected by panning against the virion preparation was identical in sequence to the clone KS56 isolated by panning against the supernatant preparation. Two distinct Fabs, with identical heavy-chain but different light-chain sequences, were isolated from library L by panning against the supernatant preparation (LS4) and the virion preparation (LZ51). A single Fab was selected from the pooled PBMC libraries (E10κ and E10λ) by panning against the virion preparation.
TABLE 1.
CDR3 sequences of Fabs to Ebola Zaire virus
| Fab | HCDR3 sequencea | LCDR3 sequencea |
|---|---|---|
| Library K; IgG1 κ | ||
| KZ51 | EVVVVPTAPTHNFYYYMDV | AHRGGWPLS |
| KS56, KZ52 | EGPRATGYSMADVFDI | QQYYSAPLT |
| KS14 | NEMSYDILTGPGDYYLDS | EESYSAVFT |
| KS5 | KYYSRLDV | QQYFATVWT |
| KS518 | RGSITLHREGNWFDP | QQRGNWPPIT |
| Library L; IgG1 κ | ||
| LS4 | TLSFAEVLYMDVFDI | MPGTHWPPT |
| LZ51 | TLSFAEVLYMDVFDI | QQANTFPFT |
| Library E10; IgG1 λ | ||
| ELZ510 | GYCSSTSCPPLFDS | ATWADSLSGVV |
HCDR3 and LCDR3 indicate heavy-chain and light-chain complementarity-determining region, respectively.
The reactivity of specific Fabs against viral preparations other than the selecting antigen was explored. Three distinct reactivity profiles were apparent as exemplified by the Fabs in Fig. 1. Fabs KZ51 and ELZ510, obtained by panning against the virion preparation, showed no reactivity with the supernatant preparation. Fab KZ52, also obtained by panning against the virion preparation, had a unique reactivity pattern. In addition to virion binding, it showed significant cross-reactivity with the supernatant preparation. Fabs LS4, KS14, KS518, and LZ51, obtained by panning against the supernatant preparation, showed some weak cross-reactivity with the virion preparation. These profiles can be readily interpreted in terms of the antibody specificities determined below and summarized in Table 2.
TABLE 2.
Immunofluorescence and immunoprecipitation of Ebola virus-specific Fabs
| Fab | Immunofluorescence
|
Specificity deduced from immunoprecipitation with:
|
Primary antigen specificity | ||
|---|---|---|---|---|---|
| Live | Fixed | Viral lysates | Crude viral supernatants | ||
| KZ51 | ± | ++ | NP | NP | NP |
| ELZ510 | ± | ++ | NP | NP | NP |
| KZ52 | +++ | ++ | GP | GP | GP |
| KS14 | − | ++ | GP | sGP >> GP | sGP |
| KS5 | + | + | NDa | ||
| KS518 | − | − | GP | sGP >> GP | sGP |
| LS4 | ++ | ++ | GP | sGP >> GP | sGP |
| LZ51 | ++ | ++ | GP | sGP >> GP | sGP |
ND, not determined.
Binding of Fabs to live and fixed Ebola virus-infected cells as confirmed by immunofluorescence.
The specificity of the selected Fabs for Ebola antigens was confirmed by immunofluorescence to detect Fab binding to live and fixed Ebola virus-infected cells. All Fabs reacted with fixed infected cells but not with uninfected control cells. Four reactivity patterns with live Ebola virus-infected cells were observed among the Fabs tested. Fab KZ52 reacted strongly with live infected cells (Fig. 2A), giving a staining pattern that was indistinguishable from that of human convalescent-phase serum. Fabs LZ51, LS4, KS518, and KS5 showed a weak “intercellular” staining pattern of live infected cells (Fig. 2B), and LZ51 showed a spotty cytoplasmic pattern on fixed infected cells, suggestive of Golgi-like staining (Fig. 2E). Fabs ELZ510 and KZ51 reacted only with a few live infected cells (less than 1 per field of 100 [Fig. 2C]), which may represent disrupted cells. Consistent with this, these Fabs did react well with fixed cells and showed a distinctive cytoplasmic staining pattern (Fig. 2F). Fab KS14 did not stain live infected cells but did stain fixed infected cells (data not shown).
FIG. 2.
Reactivity of selected anti-Ebola virus Fabs with live and fixed Ebola virus-infected Vero E6 cells shown by immunofluorescence. Staining is shown for Fab KZ52 on live (A) and fixed (D) Ebola virus-infected cells, for Fab LZ51 on live (B) and fixed (E) Ebola virus-infected cells, and for ELZ510 on live (C) and fixed (F) Ebola virus-infected cells. No binding to uninfected Vero E6 cells was observed (data not shown).
RIPA of Fab reactivity.
Two types of antigens were used in RIPA: cell lysates which contained all the structural viral proteins (Fig. 3A, lane 10) and a crude supernatant antigen rich in sGP but also containing virions (26). Fabs of three broad specificities were identified (summarized in Table 2): those that reacted with NP, those that reacted primarily with sGP, and one that reacted with GP. The RIPA reactivity of Fabs KZ51 and ELZ510 suggested that they were specific to NP in that they specifically precipitated a band of 97 kDa both from Ebola virus-infected cell lysates and from crude supernatants labeled with [35S]Cys-[35S]Met but not from mock-infected cells (Fig. 3A, lanes 1 to 6). This 97-kDa protein was not observed when [3H]glucosamine-labeled antigens were used (results not shown). KZ51 immunoprecipitated a 97-kDa band from infected-cell lysates of three other Ebola virus subtypes (Reston, Sudan, and Ivory Coast) in addition to the Zaire subtype. ELZ510 showed reactivity only with the Ivory Coast subtype in addition to the Zaire virus (data not shown).
Fabs KZ52, KS518, and LZ51 immunoprecipitated a 120-kDa band corresponding to GP1 from [3H]glucosamine-labeled Ebola virus-infected cell lysates but not from mock-infected controls (Fig. 3B). Similarly, Fab KS14 immunoprecipitated a 120-kDa band corresponding to GP1 from [35S]Cys-[35S]Met-labeled Ebola virus cell lysate. However, whereas KZ52 precipitated a strong band of 120 kDa from infected-cell supernatants (rich in soluble GP and virions), the other three Fabs immunoprecipitated a strong band in the 50-kDa region corresponding to sGP (Fig. 3B; Fig. 3A, lane 8). A longer exposure of the autoradiograms indicated a band at about 24 kDa, corresponding to GP2, in infected-cell lysates and supernatants (Fig. 3C).
Fab LS4, which has an identical heavy chain to LZ51, showed a similar immunoprecipitation profile to LZ51 (data not shown). Fab KS5 did not immunoprecipitate any clearly identifiable bands (data not shown).
The reactivity of Fab KZ51 and Fab KZ52 to NP and GP, respectively, was further confirmed by comparing their ELISA reactivity with the γ-irradiated virion preparation directly coated on wells or captured on the lectin wheat germ agglutinin (WGA). Direct coating immobilizes all proteins in the preparation, whereas by indirect coating via the lectin, only glycoproteins are immobilized and contaminating free NP in the purified virion preparation is removed. Thus, Fab KZ51 binding to the directly coated wells was much greater than Fab KZ52 binding but fell to background levels for the WGA-captured preparation. In contrast, KZ52 binding to the WGA-captured virion preparation was greater than to the directly coated virion preparation (data not shown), consistent with the reactivity of KZ52 with GP.
Neutralizing activity of Ebola virus-specific Fabs.
Fabs were tested for their ability to neutralize the virus in a plaque reduction assay with Ebola Zaire virus. KZ52 showed 50% neutralization at 0.4 μg/ml (8 nM) (Fig. 4). None of the other Fabs isolated showed any neutralizing ability. Interestingly, KZ52 was the Fab shown above by immunofluorescence to react most effectively with live virus-infected cells. The two Fabs, LS4 and LZ51, showing lower but still relatively strong reactivity with live infected cells, were nonneutralizing at 1 μg/ml.
FIG. 4.
Neutralization of Ebola virus by Fab KZ52 (◊) and IgG1 KZ52 (○). Neutralization of Ebola Zaire 1995 virus was measured in a plaque reduction assay as described in the text.
Fab KZ52 was then engineered to a whole human IgG1 molecule and expressed in CHO cells, and the neutralization assay was repeated. As shown in Fig. 4, whole IgG1 KZ52 neutralized about fourfold more effectively than Fab KZ52: 50% at 0.3 μg/ml (2 nM) and 90% at 2.6 μg/ml (17 nM). A control antibody, IgG1 b12 (anti-HIV gp120), showed no inhibition in the number of plaques (data not shown).
DISCUSSION
A panel of human monoclonal Abs to Ebola Zaire virus has been generated and characterized in this study, and these Abs reveal aspects of the humoral response to the virus. Two Abs (KZ51 and ELZ510) that immunoprecipitate NP from viral preparations were isolated. These Abs react with fixed but not live infected cells. One of the Abs cross-reacts with NP from four different Ebola virus subtypes, and its binding to NP was inhibited by 10 of 10 Ebola Zaire virus-positive sera tested (data not shown). This suggests that the Ab recognizes a conserved immunodominant epitope on NP and that it may be useful in a competition format in determining seropositivity. For example, the host species of Ebola virus is not known, and a major problem is the availability of detection reagents for a diverse species set. An assay in which test sera compete with an Ab to a conserved immunodominant viral epitope could circumvent this problem.
The remaining Abs were reactive with GP and/or sGP. These two proteins result from unconventional features of GP gene organization and transcription. The primary gene product is sGP, which is encoded in a single reading frame (0 frame). GP is encoded in two reading frames (0 and −1 frames), and expression of GP occurs only when the two frames are connected through a transcriptional editing event (25, 28). Recent studies (26, 29, 30) have revealed that GP and sGP are structurally distinct. Maturation of GP involves cleavage by the enzyme furin into two glycoproteins (GP1 and GP2) which are linked by disulfide bonding. Mature GP is composed of trimers of GP1-GP2 heterodimers. On the other hand, sGP is secreted from infected cells almost exclusively in the form of a homodimer linked by a disulfide bond. Most of the Abs generated here showed a strong reactivity with sGP and a weak reactivity with GP. All of these Abs reacted with fixed infected cells but to various degrees with live infected cells. It seems likely that these Abs were elicited by sGP, which, because of its abundance, could be expected to be a major immunogen during natural infection. The cross-reactivity with GP, albeit relatively weak, suggests that there are related structural elements between the two proteins. This may be detrimental to the development of an optimal antibody response to mature GP, since abundant B cells expressing Ig receptor for sGP could compete for binding to mature GP. Activation of these B cells would produce an Ab of only moderate affinity for mature GP and therefore probably with only weak binding to virions.
One Ab, KZ52, showed strong reactivity with GP and no reactivity with sGP. This Ab stained live infected cells particularly strongly and neutralized the virus effectively at nanomolar concentrations. It may have been elicited by virion-bound GP or alternately by secreted or shed GP1, as has been recently described in experiments performed under tissue culture conditions (30). The activity of KZ52 establishes the principle that Abs elicited in natural infection can neutralize a filovirus. The poor neutralization of Ebola virus by convalescent-phase sera (20), however, would indicate that such Abs are probably produced at relatively low frequency. By comparison with other viruses, the potency of neutralization of KZ52 is within the range that may lead to protection in passive-immunization studies. A dose of 10 mg/kg would produce a concentration of Ab in serum 40-fold higher than the 90% in vitro neutralization titer; alternately, a 1:40 dilution of serum should produce 90% neutralization. This is the type of efficacy that has been effective for other viruses (9, 17, 23). Passive-immunization studies in rodents and macaques will reveal whether Ebola virus is typical in this regard.
ACKNOWLEDGMENTS
We are grateful to the inhabitants of Kikwit, Democratic Republic of Congo, for their cooperation.
This work is a contribution from The Scripps Research Institute Emerging Diseases Research Center and is supported by a grant from NIH (AI39808). T.M. acknowledges financial support by the Department of Academic Affairs of the Scripps Clinic and Research Foundation.
REFERENCES
- 1.Barbas C F, III, Wagner J. Synthetic human antibodies selecting and evolving functional protein. Methods Companion Methods Enzymol. 1995;8:94–103. [Google Scholar]
- 2.Binley J M, Ditzel H J, Barbas C F, Sullivan N, Sodroski J, Parren P W H I, Burton D R. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res Hum Retroviruses. 1996;12:911–924. doi: 10.1089/aid.1996.12.911. [DOI] [PubMed] [Google Scholar]
- 3.Blancher A, Roubinet F, Blanchard N, Byrne P, Broly H, Ducos J, Socha W W, Ruffie J. Survival of human monoclonal anti-Rho (D) antibodies in the rhesus monkey. J Med Primatol. 1992;21:328–331. [PubMed] [Google Scholar]
- 4.Bray M, Davis K, Geisbert T, Schmaljohn C, Huggins J. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J Infect Dis. 1999;179:S248–S258. doi: 10.1086/514292. [DOI] [PubMed] [Google Scholar]
- 5.Burton D R, Barbas C F, Persson M A A, Koenig S, Chanock R M, Lerner R A. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton D R, Pyati J, Koduri R, Sharp S J, Thornton G B, Parren P W H I, Sawyer L S W, Hendry R M, Dunlop N, Nara P L, Lamacchia M, Garratty E, Stiehm E R, Bryson Y J, Cao Y, Moore J P, Ho D D, Barbas C F. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Outbreak of Ebola viral hemorrhagic fever—Zaire. Morbid Mortal Weekly Rep. 1995;44:381–382. [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. Update: outbreak of Ebola viral hemorrhagic fever—Zaire. Morbid Mortal Weekly Rep. 1995;44:399. [PubMed] [Google Scholar]
- 9.Crowe J E, Jr, Murphy B R, Chanock R M, Williamson R A, Barbas C F, Burton D R. Recombinant human RSV monoclonal antibody Fab is effective therapeutically when introduced directly into the lungs of respiratory synctial virus-infected mice. Proc Natl Acad Sci USA. 1994;91:1386–1390. doi: 10.1073/pnas.91.4.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilligan K J, Geisbert J B, Jahrling P B, Anderson K. Assessment of protective immunity conferred by recombinant vaccinia viruses to guinea pigs challenged with Ebola virus. Vaccines. 1997;97:87–92. [Google Scholar]
- 11.Hevey M, Negley D, Geisbert J, Jahrling P B, Schmaljohn A. Antigenicity and vaccine potential of Marburg virus glycoprotein expressed by baculovirus recombinants. Virology. 1997;239:206–216. doi: 10.1006/viro.1997.8883. [DOI] [PubMed] [Google Scholar]
- 12.Jahrling P B, Geisbert J, Swearengen J R, Jaax G P, Lewis T, Huggins J W, Schmidt J J, LeDue J W, Peters C J. Passive immunization of Ebola virus-infected cynomologus monkeys with immunoglobulin from hyperimmune horses. Arch Virol. 1996;11(Suppl.):135–140. doi: 10.1007/978-3-7091-7482-1_12. [DOI] [PubMed] [Google Scholar]
- 13.Jahrling P B, Geisbert T W, Geisbert J B, Swearengen J R, Bray M, Jaax N K, Huggins J W, LeDuc J W, Peters C J. Evaluation of immune globulin and recombinant interferon α-2b for treatment of experimental Ebola virus infections. J Infect Dis. 1999;179:S224–S234. doi: 10.1086/514310. [DOI] [PubMed] [Google Scholar]
- 14.Jahrling P B, Hesse R A, Eddy G A, Johnson K M, Callis R T, Stephen E L. Lassa virus infection of rhesus monkeys: pathogenesis and treatment with ribavirin. J Infect Dis. 1980;141:580–589. doi: 10.1093/infdis/141.5.580. [DOI] [PubMed] [Google Scholar]
- 15.Krasnyanskii V P, Mikhailov V V, Borisevich I V, Gradoboev V N, Evseev A A, Pshenichnov V A. Preparation of hyperimmune horse serum to Ebola virus. Vopr Virusol. 1994;39:91–92. . (In Russian.) [PubMed] [Google Scholar]
- 16.Kudoyarova-Zubavichene N M, Sergeyev N N, Chepurnov A A, Netesov S V. Preparation and use of hyperimmune serum for prophylaxis and therapy of Ebola virus infections. J Infect Dis. 1999;179(Suppl. 1):S218–S223. doi: 10.1086/514294. [DOI] [PubMed] [Google Scholar]
- 17.Lodmell D L, Ray N B, Parnell M J, Ewalt L C, Hanlon C A, Shaddock J H, Sanderlin D S, Rupprecht C E. DNA immunization protects nonhuman primates against rabies virus. Nat Med. 1998;4:949–952. doi: 10.1038/nm0898-949. [DOI] [PubMed] [Google Scholar]
- 18.Maruyama T, Parren P W H I, Sanchez A, Rensink I, Rodriguez L L, Khan A, Peters C J, Burton D R. Recombinant human monoclonal antibodies to Ebola virus. J Infect Dis. 1999;179:S235–S239. doi: 10.1086/514280. [DOI] [PubMed] [Google Scholar]
- 18a.Mupapa, K., M. Massamba, K. Kibadi, K. Kumula, A. Bwaka, M. Kipasa, R. Colebunders, and J. J. Muyembe-Tamfum, on behalf of the International Scientific and Technical Committee. 1999. Treatment of Ebola hemorrhagic fever with blood transfusions from convalescent patients. J. Infect. Dis. 179:(Suppl. 1):S18–S23. [DOI] [PubMed]
- 19.Persson M A, Caothien R H, Burton D R. Generation of diverse high-affinity human monoclonal antibodies by repertoire cloning. Proc Natl Acad Sci USA. 1991;88:2432–2436. doi: 10.1073/pnas.88.6.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters C J, LeDuc J W. An introduction. Ebola: the virus and the disease. J Infect Dis. 1999;179(Suppl. 1):ix–xvi. doi: 10.1086/514322. [DOI] [PubMed] [Google Scholar]
- 21.Peters C J, Sanchez A, Feldmann H, Rollin P E, Nichol S, Ksiazek T G. Filoviruses as emerging pathogens. Semin Virol. 1994;5:147–154. [Google Scholar]
- 22.Peters C J, Sanchez A, Rollin P E, Ksiazek T G, Murphy F A. Filoviridae: Marburg and Ebola viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press, Inc.; 1996. pp. 1161–1176. [Google Scholar]
- 23.Prince G A, Horswood R L, Chanock R M. Quantitative aspects of passive immunity to respiratory syncytial virus infection in infant cotton rats. J Virol. 1985;55:517–520. doi: 10.1128/jvi.55.3.517-520.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadek R F, Khan A S, Stevens G, Peters C J. Ebola hemorrhagic fever, Democratic Republic of Congo, 1995: determinants of survival. J Infect Dis. 1999;179(Suppl. 1):S24–S27. doi: 10.1086/514311. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez A, Trappier S G, Mahy B W J, Peters C J, Nichol S T. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcription editing. Proc Natl Acad Sci USA. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez A, Yang Z-Y, Xu L, Nabel G J, Crews T, Peters C J. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J Virol. 1998;72:6442–6447. doi: 10.1128/jvi.72.8.6442-6447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanderzanden L, Bray M, Fuller D, Roberts T, Custer D, Spik K, Jahrling P B, Huggins J, Schmaljohn A, Schmaljohn C. DNA vaccines expressing either the GP or NP genes of Ebola virus protect mice from lethal challenge. Virology. 1998;246:134–144. doi: 10.1006/viro.1998.9176. [DOI] [PubMed] [Google Scholar]
- 28.Volchkov V E, Becker S, Volchkova V A, Ternovoj V A, Kotov A N, Netesov S V, Klenk H-D. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology. 1995;214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- 29.Volchkov V E, Feldmann H, Volchkova V A, Klenk H-D. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci USA. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volchkov V E, Volchkova V A, Slenczka W, Klenk H-D, Feldmann H. Release of viral glycoproteins during Ebola virus infection. Virology. 1998;245:110–119. doi: 10.1006/viro.1998.9143. [DOI] [PubMed] [Google Scholar]
- 31.Williamson R A, Burioni R, Sanna P P, Partridge L J, Barbas C F, Burton D R. Human monoclonal antibodies against a plethora of viral pathogens from single combinatorial libraries. Proc Natl Acad Sci USA. 1993;90:4141–4145. doi: 10.1073/pnas.90.9.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu L, Sanchez A, Yang Z-Y, Zaki S R, Nabel E G, Nichol S T, Nabel G J. Immunization for Ebola virus infection. Nat Med. 1998;4:37–42. doi: 10.1038/nm0198-037. [DOI] [PubMed] [Google Scholar]