Abstract
Deep stromal invasion is an important pathological factor associated with the treatments and prognosis of cervical cancer patients. Accurate determination of deep stromal invasion before radical hysterectomy (RH) is of great value for early clinical treatment decision-making and improving the prognosis of these patients. Machine learning is gradually applied in the construction of clinical models to improve the accuracy of clinical diagnosis or prediction, but whether machine learning can improve the preoperative diagnosis accuracy of deep stromal invasion in patients with cervical cancer was still unclear. This cross-sectional study was to construct three preoperative diagnostic models for deep stromal invasion in patients with early cervical cancer based on clinical, radiomics, and clinical combined radiomics data using the machine learning method. We enrolled 229 patients with early cervical cancer receiving RH combined with pelvic lymph node dissection (PLND). The least absolute shrinkage and selection operator (LASSO) and the fivefold cross-validation were applied to screen out radiomics features. Univariate and multivariate logistic regression analyses were applied to identify clinical predictors. All subjects were divided into the training set (n = 160) and testing set (n = 69) at a ratio of 7:3. Three light gradient boosting machine (LightGBM) models were constructed in the training set and verified in the testing set. The radiomics features were statistically different between deep stromal invasion < 1/3 group and deep stromal invasion ≥ 1/3 group. In the training set, the area under the curve (AUC) of the prediction model based on radiomics features was 0.951 (95% confidence interval (CI) 0.922–0.980), the AUC of the prediction model based on clinical predictors was 0.769 (95% CI 0.703–0.835), and the AUC of the prediction model based on radiomics features and clinical predictors was 0.969 (95% CI 0.947–0.990). The AUC of the prediction model based on radiomics features and clinical predictors was 0.914 (95% CI 0.848–0.980) in the testing set. The prediction model for deep stromal invasion in patients with early cervical cancer based on clinical and radiomics data exhibited good predictive performance with an AUC of 0.969, which might help the clinicians early identify patients with high risk of deep stromal invasion and provide timely interventions.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10278-023-00906-w.
Keywords: Cervical cancer, Deep stromal invasion, Radiomics, MRI
Introduction
Cervical cancer is the fourth most common malignancy and the fourth leading cause of cancer-associated death in females [1]. Previous evidence has reported that there will be 604,000 new cases of cervical cancer and 342,000 deaths in 2020, posing a serious threat to global women’s health all over the world [2]. Over the past few decades, increasing number of cervical cancer patients were detected at an early stage due to the spread of cervical cancer screening [3]. Deep stromal invasion is an important pathological factor associated with the treatments and prognosis of cervical cancer patients [4, 5]. Patients with moderate or 1/3 deep stromal invasion were recommended to receive adjuvant radiotherapy after radical hysterectomy (RH), especially for cervical cancer patients with vascular infiltration and other risk factors [5, 6]. At present, the diagnosis of deep stromal invasion is mainly confirmed by postoperative pathology data [7]. Accurate determination of deep stromal invasion before RH is of great value for early clinical treatment decision-making and improving the prognosis of these patients.
Magnetic resonance imaging (MRI) is a routine imaging examination method for diagnosis, staging, and monitoring of cervical cancer [8]. Currently, studies based on MRI features or quantitative imaging parameters were extracted by naked eyes, which can observe limited visual image gray scale, and some microscopic imaging features related to clinical results may be lost, hampering the accurate representation of tumor heterogeneity [9, 10]. The visual assessment of MRI features by trained radiologists is prone to interobserver variability and lacks generalizability across different institutions [11]. Radiomics is an emerging technology with quantitative features extracted from radiographic medical images by data-characterization algorithms, which is designed to develop prognostic prediction tools and treatment decision support tools in cancers [12]. The predictive value of radiomics using MRI data for preoperative lymph node metastasis, vascular invasion, and parastatal invasion of early cervical cancer has been confirmed previously [10, 13, 14]. Recently, Ren et al. constructed a MRI-based radiomics model to predict the preoperative deep stromal invasion, and the AUC of the model based on radiomics features constructed by logistics regression was 0.879, and combined with clinical features, the AUC was 0.886 [15]. Nonetheless, the predictive values of prediction models for preoperative deep stromal invasion in patients with early cervical cancer still need improving.
The conventional logistic regression model can only explore the linear associations, and nonlinear associations cannot be solved; the accuracy of the prediction models was not always good [16]. Lack of high-quality dataset algorithm training and development and proper validation using more updated methods might be major drawbacks in current clinical practices to predict preoperative deep stromal invasion in patients with early cervical cancer. In order to improve the accuracy of clinical diagnosis or prediction, machine learning is gradually applied in the construction of clinical models, which showed better effects than traditional models such as logistic regression [17, 18]. Machine learning involves the utilization of computer algorithms to derive predictive models from data, and these algorithms ascertain mathematical functions that elucidate the relationships between features within a given dataset [19]. Lately, increasing studies revealed that the integration of radiomics and machine learning enabled the development of classification models for targeted diagnosis of various diseases [19, 20]. However, there was no study combining radiomics and machine learning methods to construct prediction models for preoperative diagnosis of deep stromal invasion in patients with early cervical cancer. Light gradient boosting machine (GBM) is one of the machine learning methods that can reduce calculation time and allow missing values for prediction, which is more advantageous than the conventional logistic regression model [21]. Compared to deep learning and other traditional machine learning algorithms, LightGBM showed better generalization ability [22]. Whether LightGBM can improve the preoperative diagnosis accuracy of deep stromal invasion in patients with cervical cancer based on radiomics data was still unclear.
In the present study, the machine learning method was used to construct three preoperative diagnostic models for deep stromal invasion in patients with early cervical cancer based on clinical, radiomics, and clinical combined radiomics data, respectively. The predictive efficacy of different models was compared. The findings might help identify a novel tool for risk stratification of deep stromal invasion in patients with early cervical cancer in a quicker and more accurate manner. This might help guide the clinicians to make proper treatment adjustments for these patients and improve their prognosis.
Methods
Study Design and Population
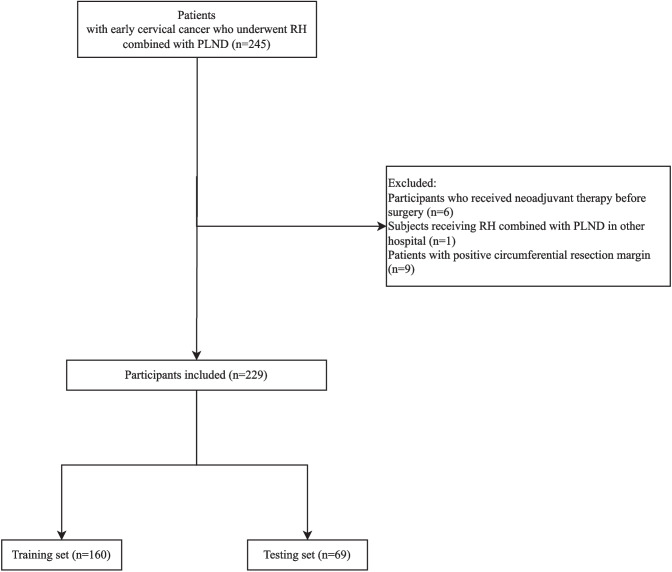
This cross-sectional study enrolled 245 patients with early cervical cancer receiving RH combined with pelvic lymph node dissection (PLND) in the local hospital. The inclusion criteria were as follows: (1) patients’ age ≥ 18 years old, (2) patients with primary cervical cancer confirmed by pathology, (3) patients receiving RH combined with PLND, (4) patients who underwent MRI examination within 2 weeks before surgery, (5) patients with complete clinical data. The exclusion criteria were (1) patients with other malignant tumors, (2) patients undergoing palliative tumor resection, (3) pregnant or lactating women, (4) patients who received neoadjuvant therapy before surgery, and (5) MRI data does not meet the requirements of post-processing. After excluding participants who received neoadjuvant therapy before surgery, subjects receiving RH combined with PLND in other hospital, and patients with positive circumferential resection margin, 229 patients were included. This study was approved by the Ethics Committee of the local hospital. Informed consent was obtained from all individual participants included in the study.
Radiomic Features Extraction
T2-weighted images and contrast-enhanced T1-weighted imaging were exported from the workstation of image storage and transmission system in Digital Imaging and Communications in Medicine format. A semi-automatic threshold classification method was used to select region of interest (ROI) of MRI using the 3D region growing GrowCut algorithm from the medical image analysis and visualization Slicer platform (3D-Slicer; version 4.3.1). Given a set of initial label points, the 3D-Slicer algorithm can automatically segment the remaining images through cellular automation, which achieves reliable and reasonably fast segmentation of moderately difficult objects in 2D and 3D using an iterative labeling procedure resembling competitive region growing [23]. Since the MRI were collected from different devices, the images were normalized before extraction, and all images were unified into a resolution of 1 × 1 mm by means of interpolation. ROI covered the entire tumor region. For each patient, a total of 2632 features (T2-weighted images + T1-weighted imaging) were extracted using the “PyRadiomics” package implemented in Python 3.11.1 (Supplementary Table 1). The features included first-order features (n = 18), texture features derived from texture matrices including grey-level co-occurrence matrix (n = 24), grey-level run length matrix (n = 16), grey-level size zone matrix (n = 16), grey-level dependence matrix (n = 14), neighboring gray tone difference matrix (n = 5) and shape-based (n = 14), wavelet transform features including first-order features (n = 144), grey-level co-occurrence matrix (n = 192), grey-level dependence matrix (n = 112), grey-level run length matrix (n = 128), grey-level size zone matrix (n = 128) and neighboring gray tone difference matrix (n = 40), and local binary pattern including first-order features (n = 90), grey-level co-occurrence matrix (n = 120), grey-level dependence matrix (n = 70), grey-level run length matrix (n = 80), and grey-level size zone matrix (n = 80) and neighboring gray tone difference matrix (n = 25).
Clinical Variables
Age (years), body mass index (BMI, kg/m2), menopausal status (premenopausal, perimenopause or postmenopausal), the International Federation of Gynecology and Obstetrics (FIGO) staging (IA, IIA, IB, or IIB), marital status (married or unmarried), preterm birth history (yes or no), reproductive history (primipara or meningopara), history of abortion (yes or no), histological subtype (adenocarcinoma, squamous cell carcinoma or other), complicated with other diseases (yes or no), red blood cell (RBC), white blood cell (WBC), platelet (PLT), neutrophil percentage (NEU; %), lymphocyte percentage (LYM; %), monocyte percentage (MONO; %), eosinophil percentage (EOS; %), basophil percentage (BASO; %), NEU (109/L), LYM (109/L), MONO (109/L), EOS (109/L), BASO (109/L), tumor size, carcinoembryonic antigen (CEA; normal or abnormal; ng/mL), squamous cell carcinoma antigen (SCC-Ag; normal or abnormal; ng/mL), carbohydrate antigen-125 (CA125; normal or abnormal; ng/mL), and carbohydrate antigen-199 (CA199; normal or abnormal; U/mL) were analyzed.
Building Prediction Classifiers
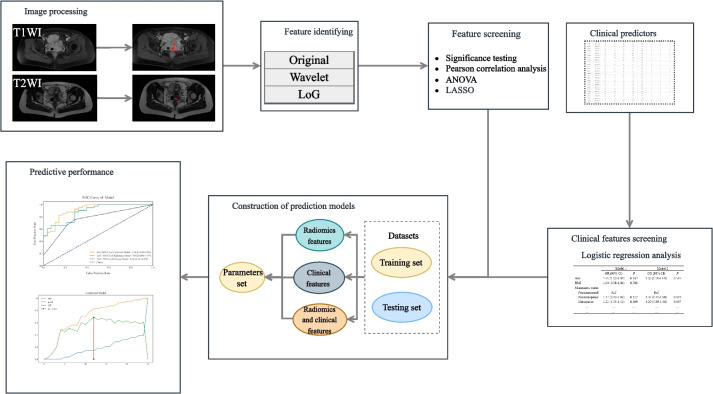
The radiomics features were extracted after image segmentation on the original MRI image to delineate the ROI, and features with statistical significance (P < 0.05) were included (SciPy tool in Python version 1.10.0). Then Pearson’s correlation coefficient was applied; when the Pearson correlation coefficient between the two features > 0.85, the features with higher P-value were excluded (Pandas tool in Python version 1.5.3). Further, the analysis of variance (ANOVA) was applied to select the top 15 radiomics features with high variance (scikit-learn tool in Python version 1.2.1). Next, the least absolute shrinkage and selection operator (LASSO) and the fivefold cross-validation were applied to further screen out features (coefficent ≠ 0). Univariable and multivariate logistic regression analyses were applied to identify clinical predictors associated with the deep stromal invasion in patients with early cervical cancer, and variables with statistical association with deep stromal invasion in patients with early cervical cancer were included as clinical predictors (P < 0.05). All subjects were randomly divided into the training set (n = 160) and testing set (n = 69) at a ratio of 7:3. Three LightGBM models were constructed in the training set: a radiomics model constructed with radiomics features alone (model 1), a clinical model constructed with clinic features alone (model 2), and a combined model constructed with the combination of radiomics features and clinical predictor (model 3). The parameters set for training each model are shown in Table 1. During the training of each model, optuna ultra parameter optimization tool was adopted to optimize the parameters, the optimized model was used to verify in the training set, and the corresponding evaluation indexes were calculated. The predictive performances of the models were verified in the testing set. The proposed model’s whole architecture is exhibited in Fig. 1. The pseudocode for the proposed work was shown as follows:


Table 1.
The parameters used for training each prediction model
| Parameters | Radiomics features | Clinical features | Radiomics and clinical features |
|---|---|---|---|
| boosting_type | gbdt | gbdt | gbdt |
| objective | binary | binary | binary |
| metric | auc | auc | auc |
| max_depth | 6 | 8 | 5 |
| num_leaves | 12 | 14 | 13 |
| min_data_in_leaf | 15 | 7 | 7 |
| max_bin | 127 | 31 | 31 |
| feature_fraction | 0.9 | 0.7 | 0.7 |
| bagging_fraction | 0.6 | 0.9 | 0.8 |
| lambda_l1 | 0.006587301 | 0.057721404 | 0.002437294 |
| lambda_l2 | 0.029924765 | 0.000530493 | 0.000600306 |
| learning_rate | 0.01 | 0.01 | 0.01 |
| random_state | 3000 | 3000 | 3000 |
Fig. 1.
The proposed model’s whole architecture
Measurement of the Performance of the Prediction Model
The proposed model more accurately predicted the deep stromal invasion in patients with early cervical cancer. The robustness of the model was assessed in the training set and the testing set. F1 score, accuracy, sensitivity, specificity, negative predictive value (NPV), positive predictive value (PPV), and area under the curve (AUC) were employed to evaluate the predictive values of the models. The receiver operator characteristic (ROC) curves and Kolmogorov–Smirnov (KS) curves were plotted.
The accuracy assessment parameter is calculated:
SN, sensitivity; TP, true positive; TN, true negative; FP, false positive; FN, false negative; PRE, TP/(TP + FP).
Statistical Analysis
The measurement data of normal distribution were expressed as mean and standard deviation (Mean (SD)), and t test was used to compare the differences between the two groups. Median and quartiles were used to describe the distribution of non-normally-distributed measurement data, and Wilcoxon rank sum test was used to compare the difference between the two groups. The enumeration data were displayed using the number of cases and percentages, and the chi-square test was used to compare differences between groups. The radiomics features were extracted, and features were selected via Pearson’s correlation coefficient, ANOVA, LASSO regression analysis, and the fivefold cross-validation. Univariable and multivariate logistic regression analyses were applied to identify clinical predictors associated with the deep stromal invasion in patients with early cervical cancer. All subjects were randomly split into the training set (n = 160) and testing set (n = 69) at a ratio of 7:3. Three LightGBM models were constructed in the training set: model 1 included radiomics features, model 2 included clinical predictors, and model 3 included radiomics features and clinical predictors. The models were verified in the testing set. The ROC and KS curves were plotted. The confidence level was alpha = 0.05. R (Institute for Statistics and Mathematics, Vienna, Austria) was used for data analysis. Python 3.11.1 was used for radiomics features extraction and model construction.
Results
Identification of Predictors in the Models for Deep Stromal Invasion in Patients with Early Cervical Cancer
In total, 245 patients with early cervical cancer who underwent RH combined with PLND in the local hospital were enrolled. Among them, participants who received neoadjuvant therapy before surgery (n = 6), subjects receiving RH combined with PLND in other hospital (n = 1), and patients with positive circumferential resection margin (n = 9) were excluded. Finally, 229 patients were included. The screen process of participants is shown in Fig. 2.
Fig. 2.
The screen process of the participants
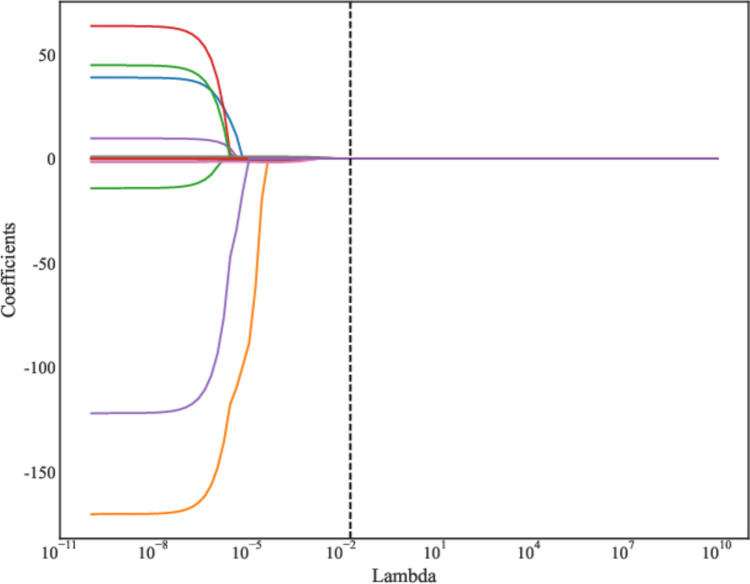
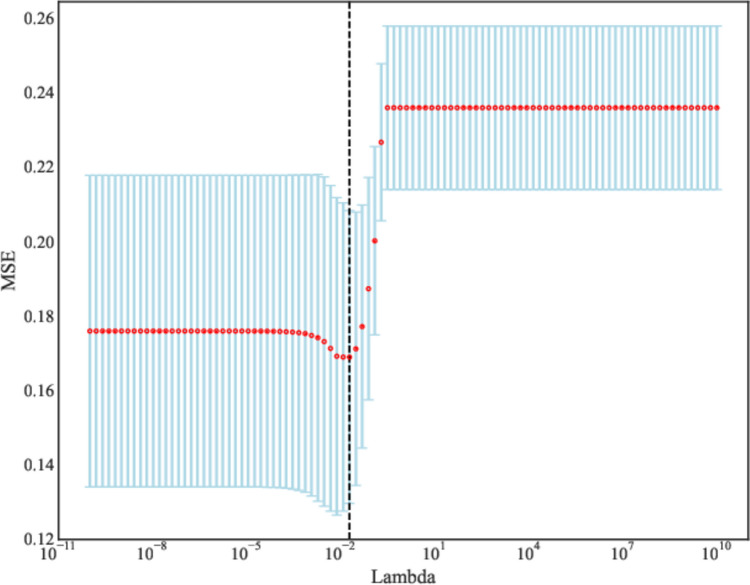
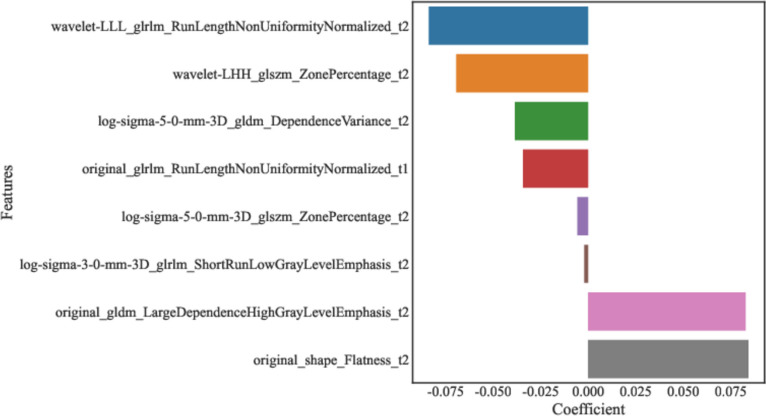
A total of 2632 features were extracted from MRI, and those with statistical significance (P < 0.05) were kept. When the Pearson correlation coefficient between the two features were > 0.85, the features with higher P-value were excluded. Further, the ANOVA was applied to select the top 15 radiomics features with high variance. Finally, LASSO regression analysis was applied to screen out the features (Fig. 3, Table 2). We used fivefold cross-validation to find the optimal value of regularization parameter lambda with mean square error, and MSE was changed with lambda. The optimal lambda value was used for variable selection and was 0.019179102616724848 (Fig. 4). The coefficients of features finally included are exhibited in Table 2 and Fig. 5.
Fig. 3.
The results of LASSO regression analysis for radiomics features
Table 2.
The radiomics features associated with deep stromal invasion in patients with early cervical cancer screened by LASSO
| Features | Coefficient |
|---|---|
| wavelet-LLL_glrlm_RunLengthNonUniformityNormalized_t2 | −0.08381 |
| wavelet-LHH_glszm_ZonePercentage_t2 | −0.06938 |
| log-sigma-5–0-mm-3D_gldm_DependenceVariance_t2 | −0.03851 |
| original_glrlm_RunLengthNonUniformityNormalized_t1 | −0.03422 |
| log-sigma-5–0-mm-3D_glszm_ZonePercentage_t2 | −0.00558 |
| log-sigma-3–0-mm-3D_glrlm_ShortRunLowGrayLevelEmphasis_t2 | −0.00195 |
| original_gldm_LargeDependenceHighGrayLevelEmphasis_t2 | 0.082895 |
| original_shape_Flatness_t2 | 0.084346 |
LASSO least absolute shrinkage and selection operator
Fig. 4.
The optimal Lambda value of LASSO regression analysis
Fig. 5.
The coefficients of features screened out by LASSO regression analysis
As presented in Table 3, age, postmenopausal, FIGO-IIA, LYM, tumor size, SCC-Ag, and CA125 might be associated with deep stromal invasion in patients with early cervical cancer. Multivariate logistical regression analysis revealed that FIGO-IIA (OR = 2.43, 95% CI 1.36–4.37), FIGO-IB (OR = 1.87, 95% CI 1.05–3.33) and FIGO-IIB (OR = 3.42, 95% CI 1.28–9.15), and SCC-Ag (OR = 1.38, 95% CI 1.19–1.59) were correlated with deep stromal invasion in patients with early cervical cancer.
Table 3.
Clinical predictors for deep stromal invasion in patients with early cervical cancer
| Univariate | Multivariable | |||
|---|---|---|---|---|
| Characteristics | Odd ratio | P | Odd ratio | P |
| Age (years) | 1.01 (1.00–1.02) | 0.012 | 1.00 (0.99–1.01) | 0.585 |
| BMI | 1.00 (0.98–1.03) | 0.780 | ||
| Menopausal status | ||||
| Premenopausal | Ref | Ref | ||
| Perimenopause | 1.37 (0.92–2.03) | 0.122 | 1.37 (0.95–1.96) | 0.091 |
| Postmenopausal | 1.22 (1.05–1.42) | 0.009 | 1.20 (0.99–1.46) | 0.067 |
| FIGO staging | ||||
| IA | ||||
| IIA | 2.46 (1.26–4.79) | 0.009 | 2.43 (1.36–4.37) | 0.003 |
| IB | 1.78 (0.93–3.40) | 0.085 | 1.87 (1.05–3.33) | 0.035 |
| IIB | 2.72 (0.89–8.30) | 0.081 | 3.42 (1.28–9.15) | 0.016 |
| Marital status | ||||
| Married | Ref | |||
| Unmarried | 1.45 (0.56–3.76) | 0.446 | ||
| Preterm birth history | ||||
| No | Ref | |||
| Yes | 0.53 (0.21–1.37) | 0.192 | ||
| Reproductive history | ||||
| Primipara | Ref | |||
| Meningopara | 0.89 (0.55–1.43) | 0.621 | ||
| History of abortion | ||||
| No | Ref | |||
| Yes | 0.93 (0.80–1.09) | 0.379 | ||
| Histological subtype | ||||
| Adenocarcinoma | Ref | |||
| Squamous cell carcinoma | 1.10 (0.90–1.34) | 0.356 | ||
| Other | 1.16 (0.82–1.64) | 0.406 | ||
| Complicated with other diseases | ||||
| No | Ref | |||
| Yes | 0.96 (0.81–1.15) | 0.677 | ||
| RBC | 0.91 (0.80–1.04) | 0.165 | ||
| WBC | 1.02 (0.99–1.05) | 0.208 | ||
| PLT | 1.00 (1.00–1.00) | 0.193 | ||
| NEU% | 1.01 (1.00–1.01) | 0.111 | ||
| LYM% | 0.99 (0.98–1.00) | 0.046 | 0.99 (0.99–1.00) | 0.060 |
| MONO% | 0.98 (0.94–1.03) | 0.471 | ||
| EOS% | 1.02 (0.98–1.06) | 0.343 | ||
| BASO% | 1.14 (0.85–1.52) | 0.378 | ||
| NEU | 1.03 (0.99–1.06) | 0.140 | ||
| LYM | 0.97 (0.86–1.09) | 0.586 | ||
| MONO | 0.91 (0.76–1.09) | 0.320 | ||
| EOS | 1.23 (0.76–2.00) | 0.407 | ||
| BASO | 1.54 (0.37–6.43) | 0.556 | ||
| Tumor size | 1.01 (1.01–1.02) | < 0.001 | 1.00 (1.00–1.01) | 0.165 |
| CEA | ||||
| Normal | Ref | |||
| Abnormal | 1.18 (0.93–1.49) | 0.174 | ||
| SCC-Ag | ||||
| Normal | Ref | |||
| Abnormal | 1.52 (1.32–1.75) | < 0.001 | 1.38 (1.19–1.59) | < 0.001 |
| CA125 | ||||
| Normal | Ref | Ref | ||
| Abnormal | 1.31 (1.02–1.68) | 0.033 | 1.21 (0.96–1.53) | 0.105 |
| CA199 | ||||
| Normal | Ref | |||
| Abnormal | 1.15 (0.91–1.44) | 0.242 | ||
BMI body mass index, FIGO International Federation of Gynecology and Obstetrics, RBC red blood cell, WBC white blood cell, PLT platelet, NEU neutrophil, LYM lymphocyte, MONO monocyte, EOS eosinophil, BASO basophil, CEA carcinoembryonic antigen, SCC-Ag squamous cell carcinoma antigen, CA125 carbohydrate antigen-125, CA199 carbohydrate antigen-199
Construction of the Prediction Models for Deep Stromal Invasion in Patients with Early Cervical Cancer
All the samples were randomly divided into the training set (n = 160) and the testing set (n = 69). There was no statistical difference between the data in the training set and testing set according to the results of equilibrium test (all P > 0.05) (Table 4). The numbers of samples with deep stromal invasion < 1/3 and deep stromal invasion ≥ 1/3 in different dataset are presented in Table 5. The percentages of patients with different FIGO staging and abnormal SCC-Ag (11.86% vs 54.46%) were statistically different between deep stromal invasion < 1/3 group and deep stromal invasion ≥ 1/3 group. The radiomics features were also statistically different between deep stromal invasion < 1/3 group and deep stromal invasion ≥ 1/3 group (Table 6).
Table 4.
Comparisons of the variables in the training set and the testing set
| Variables | Training set (n = 160) | Testing set (n = 69) | P |
|---|---|---|---|
| FIGO staging | 0.930 | ||
| IA | 2 (1.25) | 1 (1.45) | |
| IIA | 30 (18.75) | 13 (18.84) | |
| IB | 127 (79.38) | 55 (79.71) | |
| IIB | 1 (0.62) | 0 (0.00) | |
| SCC-Ag (abnormal) | 62 (38.75) | 33 (47.83) | 0.257 |
| wavelet.LHH_glszm_ZonePercentage_t2 (median [IQR]) | 0.00 [0.00, 0.00] | 0.00 [0.00, 0.00] | 0.495 |
| wavelet.LLL_glrlm_RunLengthNonUniformityNormalized_t2 (median [IQR]) | 0.11 [0.08, 0.14] | 0.11 [0.08, 0.15] | 0.905 |
| original_glrlm_RunLengthNonUniformityNormalized_t1 (median [IQR]) | 0.09 [0.07, 0.12] | 0.09 [0.07, 0.12] | 0.739 |
| original_shape_Flatness_t2 (mean (SD)) | 0.56 (0.13) | 0.56 (0.15) | 0.911 |
| original_gldm_LargeDependenceHighGrayLevelEmphasis_t2 (median [IQR]) | 2117.57 [1826.07, 2412.38] | 2186.58 [1891.59, 2399.83] | 0.443 |
| log.sigma.3.0.mm.3D_glrlm_ShortRunLowGrayLevelEmphasis_t2 (median [IQR]) | 0.11 [0.10, 0.13] | 0.11 [0.10, 0.13] | 0.609 |
| log.sigma.5.0.mm.3D_gldm_DependenceVariance_t2 (median [IQR]) | 27.99 [25.43, 30.79] | 28.30 [25.92, 30.64] | 0.808 |
| log.sigma.5.0.mm.3D_glszm_ZonePercentage_t2 (median [IQR]) | 0.00 [0.00, 0.00] | 0.00 [0.00, 0.00] | 0.573 |
| Deep stromal invasion ≥ 1/3 | 101 (63.12) | 41 (59.42) | 0.598 |
SD standard deviation, FIGO International Federation of Gynecology and Obstetrics, SCC-Ag squamous cell carcinoma antigen
Table 5.
The numbers of samples with deep stromal invasion < 1/3 and deep stromal invasion ≥ 1/3 in different dataset
| Datasets | Sample size | Deep stromal invasion < 1/3 | Deep stromal invasion ≥ 1/3 |
|---|---|---|---|
| Total | 229 | 87 | 142 |
| Training set | 160 | 59 | 101 |
| Testing set | 69 | 28 | 41 |
Table 6.
Comparisons of variables of patients between deep stromal invasion < 1/3 group and deep stromal invasion ≥ 1/3 group
| Variables | Deep stromal invasion < 1/3 (n = 59) | Deep stromal invasion ≥ 1/3 (n = 101) | P |
|---|---|---|---|
| FIGO staging | 0.002 | ||
| IA | 2 (3.39) | 0 (0.00) | |
| IIA | 3 (5.08) | 27 (26.73) | |
| IB | 54 (91.53) | 73 (72.28) | |
| IIB | 0 (0.00) | 1 (0.99) | |
| SCC-Ag (abnormal) (%) | 7 (11.86) | 55 (54.46) | < 0.001 |
| wavelet.LHH_glszm_ZonePercentage_t2 (median [IQR]) | 0.00 [0.00, 0.00] | 0.00 [0.00, 0.00] | < 0.001 |
| wavelet.LLL_glrlm_RunLengthNonUniformityNormalized_t2 (median [IQR]) | 0.14 [0.13, 0.17] | 0.10 [0.07, 0.12] | < 0.001 |
| original_glrlm_RunLengthNonUniformityNormalized_t1 (median [IQR]) | 0.11 [0.10, 0.14] | 0.08 [0.07, 0.10] | < 0.001 |
| original_shape_Flatness_t2 (mean (SD)) | 0.49 (0.14) | 0.60 (0.11) | < 0.001 |
| original_gldm_LargeDependenceHighGrayLevelEmphasis_t2 (median [IQR]) | 1827.87 [1358.60, 2036.52] | 2300.27 [2067.12, 2464.69] | < 0.001 |
| log.sigma.3.0.mm.3D_glrlm_ShortRunLowGrayLevelEmphasis_t2 (median [IQR]) | 0.13 [0.11, 0.15] | 0.11 [0.09, 0.13] | < 0.001 |
| log.sigma.5.0.mm.3D_gldm_DependenceVariance_t2 (median [IQR]) | 30.71 [28.31, 34.02] | 26.73 [24.52, 29.08] | < 0.001 |
| log.sigma.5.0.mm.3D_glszm_ZonePercentage_t2 (median [IQR]) | 0.00 [0.00, 0.00] | 0.00 [0.00, 0.00] | < 0.001 |
FIGO International Federation of Gynecology and Obstetrics, SCC-Ag squamous cell carcinoma antigen
Evaluation of the Predictive Performance of the Prediction Models for Deep Stromal Invasion in Patients with Early Cervical Cancer
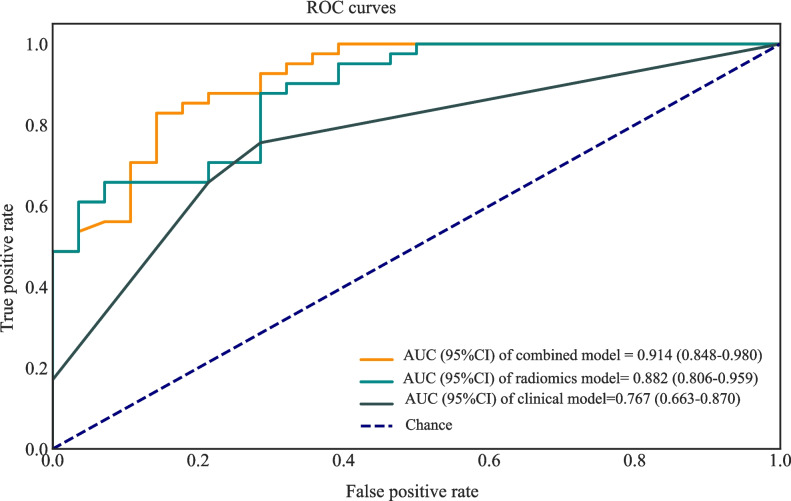
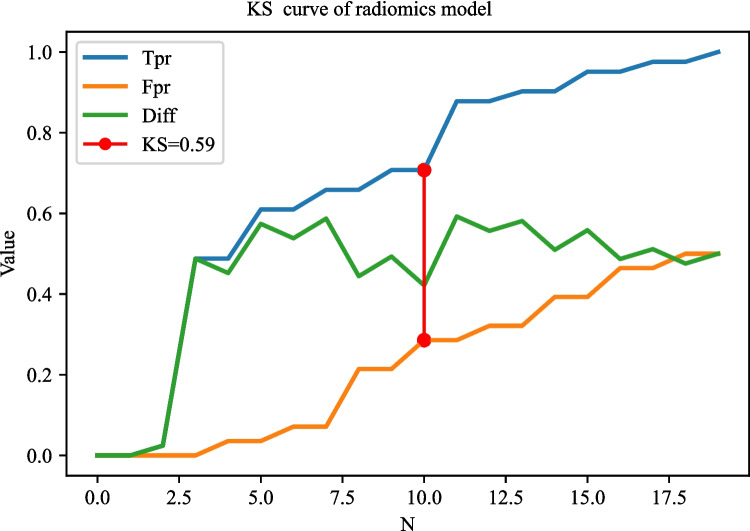
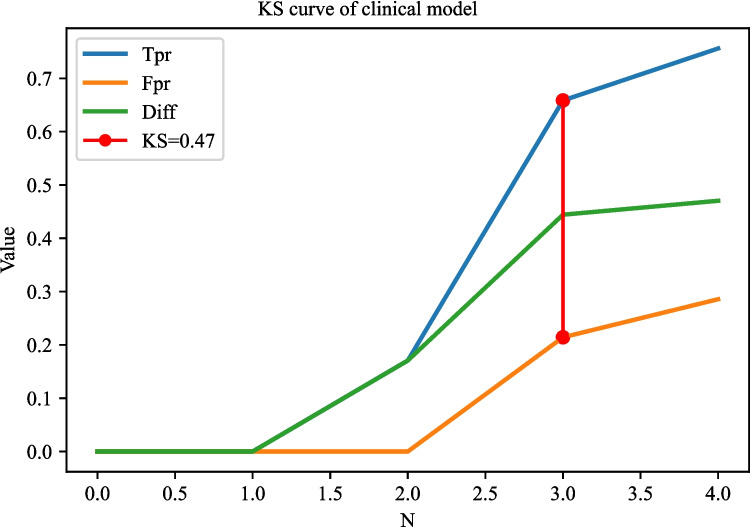
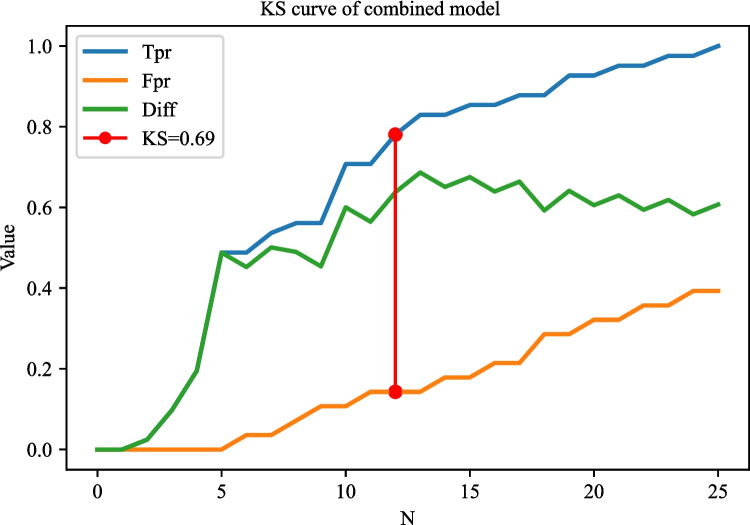
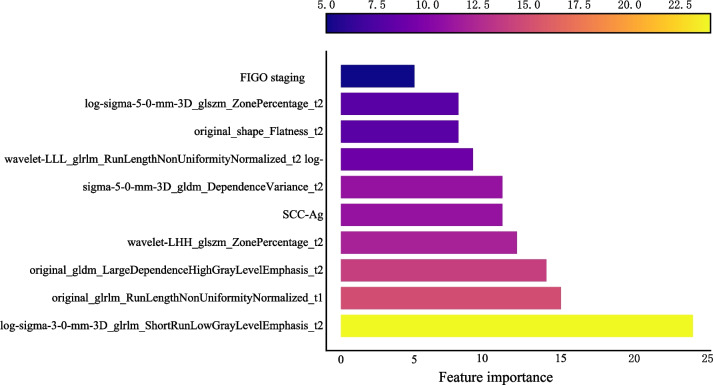
The AUC of the prediction model based on radiomics features was 0.951 (95% CI 0.922–0.980) in the training set. The AUC of the prediction model based on clinical predictors was 0.769 (95% CI 0.703–0.835) in the training set. The AUC of the prediction model based on radiomics features and clinical predictors was 0.969 (95% CI 0.947–0.990) in the training set (Table 7). The AUC of the prediction model based on radiomics features and clinical predictors was 0.914 (95% CI 0.848–0.980) in the testing set (Table 7, Fig. 6). The KS curves of the prediction models based on radiomics features, clinical predictors, and radiomics features combined with clinical predictors were plotted. The KS test was used to assess the agreement between the predicted and actual probabilities of deep stromal invasion and higher KS values indicating greater ability of the model to discriminate the samples. Generally, KS > 0.2 denotes a strong risk differentiation ability of the model developed. The KS values of the prediction models based on radiomics features, clinical predictors, and radiomics features combined with clinical predictors were 0.59 (Fig. 7), 0.47 (Fig. 8), and 0.69 (Fig. 9), respectively. The variable importance of all the predictors in the prediction model based on radiomics features combined with clinical predictors is presented in Fig. 10.
Table 7.
The predictive values of the models
| Models | Cut-off | Sensitivity | Specificity | PPV | NPV | F1 score | Accuracy | AUC (95% CI) |
|---|---|---|---|---|---|---|---|---|
| Training set | ||||||||
| Radiomics features | 0.546 | 0.871 | 0.881 | 0.926 | 0.800 | 0.892 | 0.875 | 0.951 (0.922–0.980) |
| Clinical predictors | 0.630 | 0.683 | 0.847 | 0.885 | 0.610 | 0.771 | 0.744 | 0.769 (0.703–0.835) |
| Radiomics and clinical predictors | 0.623 | 0.901 | 0.932 | 0.958 | 0.846 | 0.929 | 0.912 | 0.969 (0.947–0.990) |
| Testing set | ||||||||
| Radiomics features | 0.571 | 0.878 | 0.714 | 0.818 | 0.800 | 0.833 | 0.812 | 0.882 (0.806–0.959) |
| Clinical predictors | 0.630 | 0.756 | 0.714 | 0.795 | 0.667 | 0.775 | 0.739 | 0.767 (0.663–0.870) |
| Radiomics and clinical predictors | 0.633 | 0.829 | 0.857 | 0.895 | 0.774 | 0.861 | 0.841 | 0.914 (0.848–0.980) |
NPV negative predictive value, PPV positive predictive value, AUC area under the curve
Fig. 6.
The ROC curves showing the AUCs of different models in the testing set
Fig. 7.
The KS curves of the prediction model based on radiomics features
Fig. 8.
The KS curves of the prediction model based on clinical predictors
Fig. 9.
The KS curves of the prediction model based on radiomics features combined with clinical predictors
Fig. 10.
The variable importance of all the predictors in the prediction model based on radiomics features combined with clinical predictors
Discussion
The present study constructed three preoperative diagnostic models for deep stromal invasion in patients with early cervical cancer based on clinical, radiomics, and clinical combined radiomics data based on machine learning method. The model combined with radiomics features and clinical predictors showed better predictive performance than the prediction models based on radiomics features or clinical predictors. The findings might provide an effective tool to help clinicians early identify patients with the deep stromal invasion and guide the treatments accordingly.
Previously, there were several prediction models based on MRI data for deep stromal invasion in patients with cervical cancer. Song et al. constructed a prediction model based on amide proton transfer weighted imaging combined with dynamic contrast-enhanced MRI and found that Ktrans + SCC-Ag had the AUC of 0.819 for predicting deep stromal invasion in patients with IB1-IIA1 cervical cancer [24]. Another prospective multicenter study constructed a preoperative prediction model for deep stromal invasion in women with invasive cervical cancer using 2D and 3D ultrasound and showed an AUC of 0.93 [25]. These models mostly constructed based on the conventional logistic regression model, which can only explore the linear associations, and the predictive ability still needs improvement [16]. To use the machine learning algorithm to train and validate the prediction model might help improve the predictive accuracy of deep stromal invasion in patients with early cervical cancer. In our study, the model based on radiomics features had an AUC of 0.951, and the AUC of the model based on radiomics features and clinical predictors was 0.969. The models presented better predictive performance for deep stromal invasion in patients with early cervical cancer than previous models. The detailed information on database, computational complexity, and reliability of our model and previous prediction models are exhibited in Table 8. MRI had the advantages of relatively low cost, high spatial resolution and contrast of pelvic tissues and organs, and no radiation [26, 27]. MRI was highly individual specific and non-invasive, which has been applied to clinical decision support for the improvement of the screening accuracy, diagnosis, and prognosis prediction [28]. The prediction model in our study was constructed using LightGBM, which used histogram-based segmentation algorithm instead of presort traversal algorithm to reduce the number of features by gradient-based one-side sampling (GOSS) and exclusive feature bundling (EFB) [29]. LightGBM had higher efficiency and accuracy [30] and better generalization ability [22]. The model combining LightGBM methods and MRI in the current study might provide a convenient and easy tool for early identification of those at a high risk of deep stromal invasion in patients with early cervical cancer. The accuracy for predicting deep stromal invasion in patients with early cervical cancer was improved compared to previous models, which might help guide the treatments options of these patients with high risk of deep stromal invasion, and early interventions might improve their prognosis.
Table 8.
Comparisons of our prediction model and previous prediction models for deep stromal invasion in patients with early cervical cancer
| Models | Samples (n) | Database | Space complexity | AUC | Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| Our model (LightGBM) | 229 | Guangzhou Panyu Central Hospital | Memory cost O (nfeature × ndata) | 0.969 | 0.901 | 0.932 |
| Calculation of split gain O (nbin × nfeature) | ||||||
| Exclusive feature bundling O (ndata × nfeature) → O (ndata × nbundle) | ||||||
| Pálsdóttir et al. (logistic regression) | 104 | Lund University Hospital and Karolinska University Hospital | O (nfeature) | 0.930 | 0.905 | 0.972 |
| Ren et al. (logistic regression) | 234 | Peking Union Medical College Hospital | O (nfeature) | 0.886 | 0.879 | 0.846 |
AUC area under the curve, LightGBM light gradient boosting machine
MRI is a vital exam for the initial assessment of loco-regional involvement of cervical cancer. In previous studies, multiple studies found that MRI was applied to evaluate the early response to radiochemotherapy before image-guided brachytherapy in patients with locally advanced cervical cancer [31]. Multiparametric MRI–derived radiomics was also applied for the prediction of disease-free survival in early-stage squamous cervical cancer [32]. Multimodal MRI was reported to have good diagnostic value for the discrimination of metastatic and non-metastatic pelvic lymph nodes in cervical cancer [33]. Another prospective preliminary study applied the synthetic MRI to evaluate the prognostic factors in cervical cancer [34]. These studies gave support to the results of this study, which elucidated that MRI-derived radiomics features were important predictors for deep stromal invasion in patients with early cervical cancer. Cancer staging is an essential index for the diagnosis, prognosis, and treatment of cervical cancer [35]. The FIGO staging system was widely applied in cervical cancer [36], which was reported to be associated with the treatment outcomes in early-stage cervical cancer patients [37]. Herein, the FIGO staging system was also found to be an important predictor for deep stromal invasion in patients with early cervical cancer. Another predictor for deep stromal invasion in patients with early cervical cancer in this study was SCC-Ag. This was allied by previous evidence. SCC-Ag was used in outcome prediction after concurrent chemo-radiotherapy and treatment decisions for patients with cervical cancer [38]. SCC-Ag changes in patients with locally advanced cervical cancer were one of the parameters of prognostic evaluation [39].
The current study compared the predictive abilities of three preoperative diagnostic models using the machine learning method for preoperative non-invasive diagnosis of deep stromal invasion in patients with early cervical cancer based on clinical, radiomics, and clinical combined radiomics data, respectively. The predicting performance of the model for deep stromal invasion in patients with early cervical cancer based on clinical combined radiomics data was good. The findings might provide a tool to help clinicians identify deep stromal invasion in patients with early cervical cancer and formulate treatment strategies accordingly. There were several limitations in this study. Firstly, the participants were from a single center, and there might be selection bias. Secondly, the MRI images were collected from different devices, which might have a potential impact on the stability of radiomics features. Therefore, the images were normalized before feature extraction, and all images were unified to a resolution of 1 × 1 mm. The standardization process was considered a useful way to promote good feature robustness in cervical cancer. In recent years, more and more deep learning methods such as automated in-depth feature learning algorithm [40] and a deep convolutional neural network-based approach [41] were widely applied for disease prediction and prognosis evaluation. These methods are unsupervised active learning, which increase efficiency and accuracy of diseases and prognosis prediction including cancers [42]. The future of applied deep learning in cervical cancer might help integrate medical images and clinical data to construct more reliable prediction models. In the future, more well-designed studies using deep learning methods were needed to verify the results in this study.
Conclusions
The AUC values of the prediction model for deep stromal invasion in patients with early cervical cancer based on clinical and radiomics data were 0.969 in the training set and 0.914 in the testing set, which exhibited good predictive performance than previous prediction models. The prediction model might help the clinicians early and accurately identify patients with high risk of deep stromal invasion and provide timely interventions.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
None.
Author Contribution
HY and ZX designed the study. HY wrote the manuscript. GH, ZY, and YC collected, analyzed, and interpreted the data. ZX critically reviewed, edited, and approved the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by National Natural Science Foundation of China [No. 82171931], the Science and Technology Program of Guangzhou [Nos. 201903010032 and 202102080572], and the Panyu Science and Technology Program of Guangzhou [Nos. 2019-Z04-01, 2019-Z04-23, and 2022-Z04-013].
Data Availability
Data will be made available on request.
Declarations
Ethics Approval
This study was approved by the Ethics Committee of The First Affiliated Hospital of Jinan University and Guangzhou Panyu Central Hospital (ZN2022-5).
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.D'Oria O, Corrado G, Laganà AS, Chiantera V, Vizza E, Giannini A: New Advances in Cervical Cancer: From Bench to Bedside. International journal of environmental research and public health 19, 2022 [DOI] [PMC free article] [PubMed]
- 2.Sung H, et al.: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians, 2021 [DOI] [PubMed]
- 3.Gavinski K, DiNardo D: Cervical Cancer Screening. The Medical clinics of North America 107:259-269, 2023 10.1016/j.mcna.2022.10.006 [DOI] [PubMed] [Google Scholar]
- 4.Biewenga P, et al.: Prognostic model for survival in patients with early stage cervical cancer. Cancer 117:768-776, 2011 10.1002/cncr.25658 [DOI] [PubMed] [Google Scholar]
- 5.Cao L, Wen H, Feng Z, Han X, Zhu J, Wu X: Role of adjuvant therapy after radical hysterectomy in intermediate-risk, early-stage cervical cancer. International journal of gynecological cancer : official journal of the International Gynecological Cancer Society 31:52-58, 2021 10.1136/ijgc-2020-001974 [DOI] [PubMed] [Google Scholar]
- 6.Cibula D, et al.: The European Society of Gynaecological Oncology/European Society for Radiotherapy and Oncology/European Society of Pathology guidelines for the management of patients with cervical cancer. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology 127:404-416, 2018 10.1016/j.radonc.2018.03.003 [DOI] [PubMed] [Google Scholar]
- 7.Zhu J, Cao L, Wen H, Bi R, Wu X, Ke G: The clinical and prognostic implication of deep stromal invasion in cervical cancer patients undergoing radical hysterectomy. Journal of Cancer 11:7368-7377, 2020 10.7150/jca.50752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matani H, Patel AK, Horne ZD, Beriwal S: Utilization of functional MRI in the diagnosis and management of cervical cancer. Front Oncol 12:1030967, 2022 10.3389/fonc.2022.1030967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Q, Zheng D, Shi L, Liu M, Wang M, Shi D: Differentiating metastatic from nonmetastatic lymph nodes in cervical cancer patients using monoexponential, biexponential, and stretched exponential diffusion-weighted MR imaging. Eur Radiol 27:5272-5279, 2017 10.1007/s00330-017-4873-1 [DOI] [PubMed] [Google Scholar]
- 10.Wu Q, et al.: Radiomics analysis of magnetic resonance imaging improves diagnostic performance of lymph node metastasis in patients with cervical cancer. Radiotherapy and Oncology 138:141-148, 2019 10.1016/j.radonc.2019.04.035 [DOI] [PubMed] [Google Scholar]
- 11.Lefebvre TL, et al: Development and Validation of Multiparametric MRI-based Radiomics Models for Preoperative Risk Stratification of Endometrial Cancer. Radiology 305(2):375-386, 2022 10.1148/radiol.212873 [DOI] [PubMed] [Google Scholar]
- 12.Gillies RJ, Kinahan PE, Hricak H: Radiomics: Images Are More than Pictures, They Are Data. Radiology 278:563-577, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, et al.: MR-Based Radiomics Nomogram of Cervical Cancer in Prediction of the Lymph-Vascular Space Invasion preoperatively. Journal of magnetic resonance imaging : JMRI 49:1420-1426, 2019 10.1002/jmri.26531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T, et al.: Preoperative prediction of parametrial invasion in early-stage cervical cancer with MRI-based radiomics nomogram. Eur Radiol 30:3585-3593, 2020 10.1007/s00330-019-06655-1 [DOI] [PubMed] [Google Scholar]
- 15.Ren J, et al.: MRI-based radiomics analysis improves preoperative diagnostic performance for the depth of stromal invasion in patients with early stage cervical cancer. Insights into imaging 13:17, 2022 10.1186/s13244-022-01156-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoltzfus JC: Logistic regression: a brief primer. Academic emergency medicine : official journal of the Society for Academic Emergency Medicine 18(10):1099-1104, 2011 10.1111/j.1553-2712.2011.01185.x [DOI] [PubMed] [Google Scholar]
- 17.Lam LHT, Chu NT, Tran TO, Do DT, Le NQK: A Radiomics-Based Machine Learning Model for Prediction of Tumor Mutational Burden in Lower-Grade Gliomas. Cancers 14, 2022 [DOI] [PMC free article] [PubMed]
- 18.Yang R, Xiong X, Wang H, Li W: Explainable Machine Learning Model to Prediction EGFR Mutation in Lung Cancer. Frontiers in oncology 12:924144, 2022 10.3389/fonc.2022.924144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gitto S, et al.: MRI radiomics-based machine learning classification of atypical cartilaginous tumour and grade II chondrosarcoma of long bones. EBioMedicine 75:103757, 2022 10.1016/j.ebiom.2021.103757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu L, et al.: Machine learning-based radiomics analysis of preoperative functional liver reserve with MRI and CT image. BMC medical imaging 23(1):94, 2023 10.1186/s12880-023-01050-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuno T, Sahashi Y, Kawahito S, Takahashi M, Iwagami M, Egorova NN: Prediction of in-hospital mortality with machine learning for COVID-19 patients treated with steroid and remdesivir. Journal of medical virology 94(3):958-964, 2022 10.1002/jmv.27393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye Z, Ouyang D: Prediction of small-molecule compound solubility in organic solvents by machine learning algorithms. Journal of cheminformatics 13(1):98, 2021 10.1186/s13321-021-00575-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egger J, et al.: GBM volumetry using the 3D Slicer medical image computing platform. Scientific reports 3:1364, 2013 10.1038/srep01364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song Q, et al.: Amide proton transfer weighted imaging combined with dynamic contrast-enhanced MRI in predicting lymphovascular space invasion and deep stromal invasion of IB1-IIA1 cervical cancer. Front Oncol 12:916846, 2022 10.3389/fonc.2022.916846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pálsdóttir K, Fischerova D, Franchi D, Testa A, Di Legge A, Epstein E: Preoperative prediction of lymph node metastasis and deep stromal invasion in women with invasive cervical cancer: prospective multicenter study using 2D and 3D ultrasound. Ultrasound in obstetrics & gynecology : the official journal of the International Society of Ultrasound in Obstetrics and Gynecology 45:470-475, 2015 10.1002/uog.14643 [DOI] [PubMed] [Google Scholar]
- 26.Rauch GM, et al.: Optimization of MR imaging for pretreatment evaluation of patients with endometrial and cervical cancer. Radiographics : a review publication of the Radiological Society of North America, Inc 34:1082–1098, 2014 [DOI] [PMC free article] [PubMed]
- 27.Park JJ, Kim CK, Park SY, Park BK: Parametrial invasion in cervical cancer: fused T2-weighted imaging and high-b-value diffusion-weighted imaging with background body signal suppression at 3 T. Radiology 274:734-741, 2015 10.1148/radiol.14140920 [DOI] [PubMed] [Google Scholar]
- 28.Chen Q, et al.: Radiomics in precision medicine for gastric cancer: opportunities and challenges. European radiology 32:5852-5868, 2022 10.1007/s00330-022-08704-8 [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Li Y: LightGBM Indoor Positioning Method Based on Merged Wi-Fi and Image Fingerprints. Sensors (Basel, Switzerland) 21, 2021 [DOI] [PMC free article] [PubMed]
- 30.Wang W: Random Forest and LightGBM-Based Human Health Check for Medical Device Fault Detection. Journal of healthcare engineering 2022:2847112, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Cordoba A, et al.: Prognostic impact of tumor size reduction assessed by magnetic resonance imaging after radiochemotherapy in patients with locally advanced cervical cancer. Front Oncol 12:1046087, 2022 10.3389/fonc.2022.1046087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, Gu HL, Zhang XL, Tian ZF, Xu XQ, Tang WW: Multiparametric magnetic resonance imaging-derived radiomics for the prediction of disease-free survival in early-stage squamous cervical cancer. European radiology 32:2540-2551, 2022 10.1007/s00330-021-08326-6 [DOI] [PubMed] [Google Scholar]
- 33.Xu J, Ma Y, Mei H, Wang Q: Diagnostic Value of Multimodal Magnetic Resonance Imaging in Discriminating Between Metastatic and Non-Metastatic Pelvic Lymph Nodes in Cervical Cancer. International journal of general medicine 15:6279-6288, 2022 10.2147/IJGM.S372154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, et al.: Application of synthetic magnetic resonance imaging and DWI for evaluation of prognostic factors in cervical carcinoma: a prospective preliminary study. The British journal of radiology 96:20220596, 2023 10.1259/bjr.20220596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soares LC, Junior JCD, de Souza RJ, de Oliveira MAP: Critical analysis of the FIGO 2018 cervical cancer staging. Journal of the Turkish German Gynecological Association 23:325-326, 2022 10.4274/jtgga.galenos.2022.2022-1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng Q, Wang W, Liu X, Wang D, Zhang F: Nomograms Predicting Survival of Cervical Cancer Patients Treated With Concurrent Chemoradiotherapy Based on the 2018 FIGO Staging System. Front Oncol 12:870670, 2022 10.3389/fonc.2022.870670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaur S, Garg H, Nandwani M, Kalita M, Bansal S, Singh R: Influence of New FIGO 2018 Staging on Treatment Outcomes in Early-Stage Cervical Cancer: A Single-Center Study. South Asian journal of cancer 11:213-217, 2022 10.1055/s-0042-1743422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu J, Wang W, Wang Y, Liu C, Wang P: The role of squamous cell carcinoma antigen (SCC Ag) in outcome prediction after concurrent chemoradiotherapy and treatment decisions for patients with cervical cancer. Radiation oncology (London, England) 14:146, 2019 [DOI] [PMC free article] [PubMed]
- 39.Chen W, et al.: Prognostic value of tumor measurement parameters and SCC-Ag changes in patients with locally-advanced cervical cancer. Radiation oncology (London, England) 17:6, 2022 [DOI] [PMC free article] [PubMed]
- 40.Mahmood T, Li J, Pei Y, Akhtar F: An Automated In-Depth Feature Learning Algorithm for Breast Abnormality Prognosis and Robust Characterization from Mammography Images Using Deep Transfer Learning. Biology 10(9), 2021 [DOI] [PMC free article] [PubMed]
- 41.Mahmood T, Li J, Pei Y, Akhtar F, Rehman MU, Wasti SH: Breast lesions classifications of mammographic images using a deep convolutional neural network-based approach. PloS one 17(1):e0263126, 2022 10.1371/journal.pone.0263126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tran KA, Kondrashova O, Bradley A, Williams ED, Pearson JV, Waddell N: Deep learning in cancer diagnosis, prognosis and treatment selection. Genome medicine 13(1):152, 2021 10.1186/s13073-021-00968-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.