Abstract
Microbubbles (MBs) combined with focused ultrasound (FUS) has emerged as a promising noninvasive technique to permeabilize the blood–brain barrier (BBB) for drug delivery into the brain. However, the safety and biological consequences of BBB opening (BBBO) remain incompletely understood. This study aims to investigate the effects of two parameters mediating BBBO: microbubble volume dose (MVD) and mechanical index (MI). High-resolution MRI-guided FUS was employed in mouse brains to assess BBBO by manipulating these two parameters. Afterward, the sterile inflammatory response (SIR) was studied 6 h post-FUS treatment. Results demonstrated that both MVD and MI significantly influenced the extent of BBBO, with higher MVD and MI leading to increased permeability. Moreover, RNA sequencing revealed upregulation of major inflammatory pathways and immune cell infiltration after BBBO, indicating the presence and extent of SIR. Gene set enrichment analysis identified 12 gene sets associated with inflammatory responses that were significantly upregulated at higher MVD or MI. A therapeutic window was established between therapeutically relevant BBBO and the onset of SIR, providing operating regimes to avoid damage from stimulation of the NFκB pathway via TNFɑ signaling to apoptosis. These results contribute to the optimization and standardization of BBB opening parameters for safe and effective drug delivery to the brain and further elucidate the underlying molecular mechanisms driving sterile inflammation.
Keywords: Focused ultrasound, Blood–brain barrier opening, Microbubbles, Sterile inflammatory response, Magnetic resonance imaging
Subject terms: Immunology, Medical research, Neurology, Oncology, Engineering
Introduction
Since the discovery of noninvasive BBBO through MB and FUS, several studies have investigated drug delivery into the brain1,2. This has been reflected in the rising number of clinical trials registered each year3. With the growing number of pre-clinical work utilizing MB + FUS, most attention has been devoted to optimizing BBBO parameters on the ultrasound (US) side, while conventional echocardiography contrast agent MBs with polydisperse size distributions have been employed at or near their clinical dose. As the field moves to more sophisticated MB formulations with longer circulation persistence and monodisperse size distributions, there is a need to elucidate the effects of MB size and number on BBBO. Additionally, the full extent of the sterile inflammatory response (SIR) from BBBO requires characterization, in terms of both ultrasound and MB doses. The safety of BBBO needs to be considered to minimize health concerns and mitigate harmful secondary effects4.
The BBB is crucial in maintaining homeostasis and is the first line of defense against pathogens and noxious insults5. To avoid the passage of undesirable molecules, the BBB is made up of a basal layer of endothelial cells, which selectively excludes hydrophilic molecules larger than 400 Daltons6,7. Tight junction proteins such as occludins, claudins and junctional adhesion molecules exist on endothelial cell membranes and form complexes to fasten together neighboring cells8,9. Additionally, pericytes and astrocytic end feet processes envelop the vasculature, regulating vascular constriction and ensuring proper maintenance of the barrier10–12. Collectively, these constituents form a structure referred to as the neurovascular unit.
Microbubbles are ultrasound responsive colloidal particles that have a gas core encapsulated in a shell (e.g., polymer, phospholipid, protein)13–16. These 1–10 µm diameter spheres experience an isotropic but dynamic pressure field within the ultrasound focus, resulting in volumetric oscillations15. In therapeutic applications, particularly in the brain, ultrasound parameters operate within lower frequencies (F ~ 1 MHz for mice and ~ 0.2–0.5 MHz for humans) to ensure transcranial propagation. Both probability of cavitation and the extent of BBBO are correlated to the mechanical index. Mechanical index can be varied by holding the frequency constant and adjusting the peak negative (MI = )17. With increasing MI, the MB acoustic behavior progresses from mild harmonic oscillations to violent inertial implosions18–20. In the context of BBBO, harmonic MB oscillations were found to pry apart tight junction proteins, creating transient pores in the brain endothelium and allowing blood-borne molecules to extravasate21–24.
As the MB is the acoustic actuator that captures the acoustic energy and uses it to produce localized mechanical work on the endothelium, it is also an important parameter that must considered. Of particular interest is the size and concentration, which can be quantified as the injected microbubble volume dose (MVD, µL/kg)25,26. When matching MVD, MBs of different sizes were found to produce similar pharmacokinetic profiles27,28, acoustic response as measured by passive cavitation detection29, and extent of BBBO25. Thus, MVD and MI serve as relevant MB and ultrasound dosing parameters, respectively, forming two axes to identify a window of safety and efficacy.
Despite technical characterization over the years, one of the most understudied areas is related to the biological consequences of BBBO. On a larger scale, it has been observed that temporarily permeabilizing the BBB with inertial cavitation can induce microhemorrhages, transient edema and even cell death. Upon closer examination, transcriptomic analyses of the parenchymal microenvironment following MB + FUS BBBO have revealed upregulation of inflammatory pathways, mostly notably the NFκB pathway30,31. While occurring in the absence of an active infection, this event has been labeled as the sterile inflammatory response (SIR) and is initiated when damage-associated molecular patterns are released from injured cells32–34. These include ATP, uric acid, DNA and HMBG proteins, which bind to pattern recognition receptors provoking the immune system35,36. Subsequently, proinflammatory cytokines such as TNFɑ and IL2 are released from inflammasomes and stimulate the activation of innate immunity37. Several studies have implicated the hallmark activation of the NFκB pathway in the persistence of SIR; however, the direct mechanism of activation and associated pathways remain unclear38–42. Moreover, microglia, the primary immune cells of the brain, migrate to the source of inflammation and release cytokines, signaling the recruitment of peripheral immune cells and other cell types in the area30,43. Peripheral immune cells such as CD68+ macrophages circulate in the meninges via chemotaxis and migrate to the damaged area to investigate and resolve the inflammatory response30. Despite BBB closure within a 24-h window, peripheral immune cells have been shown to continue extravasating past the BBB44,45.
Here, we report on a study to determine the extent of BBBO and SIR using a combination of MI (0.2–0.6 ) and MVD (0.1–40 µL/kg). By using MRI-guided FUS, we sonicated the right mouse striatum and collected brain samples for bulk RNA sequencing 6 h post-sonication. We defined significant BBBO as a 15% increase in signal intensity over a 1 mm3 volume in contrast enhanced T1-weighted MRI (CE-T1w MRI). The normalized enrichment score (NES) indicated how statistically significant mRNA for a given pathway was increased (upregulation) or decreased (downregulation) following MB + FUS treatment. We propose a definition of SIR according to three classes defined by NES analysis, which describe the statistical significance of a pathway (using an a priori set of genes from published bioinformatics databases like Gene Ontology, KEGG and MSigDB) to the observed phenomenon or phenotype through overrepresentation of specific gene expression (NES > 1.65) (Class I: TNFɑ signaling via NFκB gene set; Class II: both the TNFɑ signaling via NFκB and Inflammatory response gene sets; Class III: TNFɑ signaling via NFκB, Inflammatory response, and a damage-associated marker, apoptosis). Using these criteria, we developed a therapeutic window of ultrasound MI and microbubble MVD between the onset of BBBO and the onset of SIR, for each of the three classes. These windows will help to determine safe and efficacious MB + FUS parameters for BBBO in various applications.
Materials and methods
Animals
All experiments involving animals were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Colorado in CD-1 IGS mice (strain code: 022), and all methods are reported in accordance with ARRIVE guidelines (https://arriveguidelines.org). All mice used were female 9–11-week-old and purchased from Charles River Laboratory.
Microbubble preparation
Lipid-coated microbubbles containing a perfluorobutane (PFB) gas core were synthesized via sonication, as described previously by Feshitan et al.46. A monodispersed population of MBs (3 ± 0.5 µm) was isolated by differential centrifugation. The isolation process, including centrifugation speeds, can be found in Supplemental Fig. S9. Microbubble concentration and number- and volume-weighted size distributions were measured with a Multisizer 3 (Beckman Coulter). Microbubble concentration (, MBs/µL) versus microbubble volume (, µL/MB) was plotted, and the gas volume fraction () was estimated as follows:
where is the index of the sizing bin, 300 bins ranging from 0.7 to 18 µm in diameter. Three independent MB preparations were measured 2 h before FUS treatment to confirm size distributions and concentration. MB stability 1 h after dilution to relevant injection concentration was also measured (Supplemental Fig. S10).
Magnetic resonance imaging
A Bruker BioSpec 9.4/20 Tesla MR Scanner (Bruker, Billerica, MA) with a mouse head RF phase-array coil was used. Mice were placed into an custom designed MRI bed that contained a stereotaxic ear bar. Each MRI session consisted of a 3D-localizer, and T1w MSME (Multi-Spin Multi-Echo) images were acquired in the axial plane (Table S3) 12 min after intravenous injection of 0.4 mmol/kg gadobenate dimeglumine (MultiHance, Bracco, Princeton, NJ). All mice underwent high-resolution 3D T2-turboRARE (Rapid Acquisition with Relaxation Enhancement) scans for anatomical imaging (Table S3). Mice remained stereo-tactically placed on an MRI bed and transferred to the FUS system for treatment. All image acquisition was performed using Bruker ParaVision NEO360 v.3.3 software.
MR image analysis
T1w MRI data sets quantified the extent of BBBO using FIJI (Maryland, USA). All axial slices were analyzed by defining the contralateral hemisphere and determining the mean and standard deviation of voxel intensities. The treated hemisphere was then defined, and all voxels were found above two standard deviations of the contralateral side. The area was determined and multiplied by slice thickness (0.7 mm) to find BBBO volume. The contrast enhancement (CE) was determined by the average intensity within BBBO volume and divided by the average intensity of the control region. Using control images (mice without BBBO), CE variation on the right side was found to be 0 ± 6% (Supplemental Fig. S1), therefore significant opening was defined as a greater than 15% CE.
MRI-guided FUS treatment
The experimental setup is shown in Fig. 1D. A single element, geometrically focused transducer (frequency: 1.515 MHz, diameter: 30 mm) was driven by the RK-50 system (FUS Instruments, Toronto, Canada). A single element, geometrically focused transducer (frequency: 0.7575 MHz, diameter: 10 mm) coaxially inside the driving transducer was used for passive cavitation detection. Using the T2-weighted MR image (coronal), the center of the striatum was targeted (Fig. 1D). Ultrasound gel (Aquasonic gel, Clinton Township, MI) was placed on the mouse. An acoustically transparent tank filled with degassed water was placed on top of the gel Fig. 1D. Microbubbles (0.1–40 μL/kg; 0.1 mL) and 0.1 mL MultiHance was injected intravenously through a retroorbital injection via 26 Ga needle. Afterward (within 10 s) FUS was applied. FUS parameters were as follows: 10 ms PL, 1 Hz PRF, 300 s treatment time, and a PNP of 0.308, 0.615, or 0.923 MPa (0.246, 0.492, or 0.738 MPa in situ). Voltage data from the PCD was collected during the entire FUS treatment and analyzed as previously described29. The remaining PCD analysis was done using MATLAB (Massachusetts, USA) including the calculations of harmonic and broadband cavitation doses. Mice were sent back to MRI to complete post-FUS T1-weighted imaging. Groups were divided into n = 3 for all 12 dose levels (3 MIs, 4 MVDs).
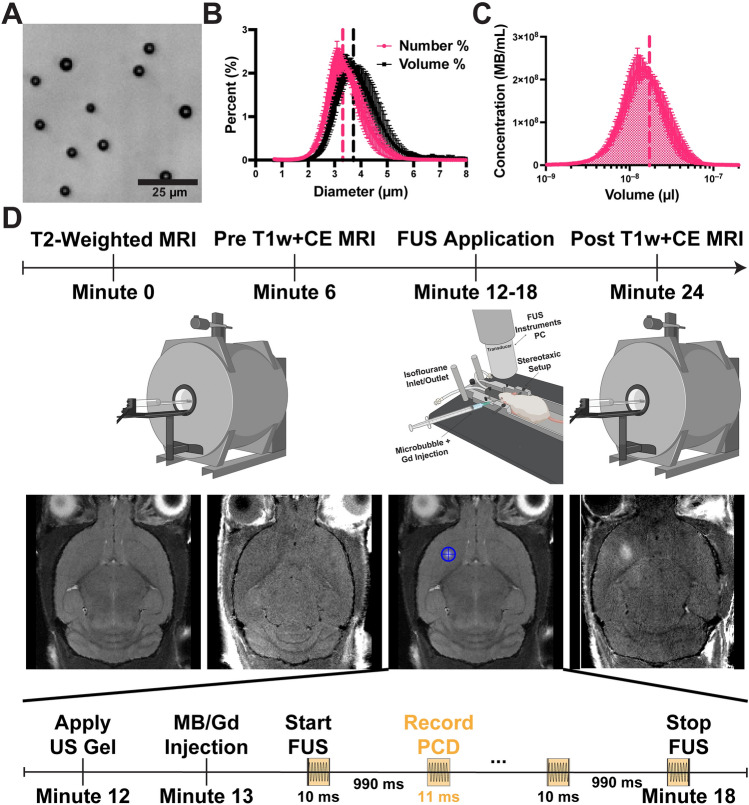
Figure 1.
Microbubble Characterization and Treatment Timeline. (A) Brightfield image of isolated microbubbles (3 µm) in diameter. (B) Number- and volume-weighted size distributions. Vertical dotted lines represent mean values. (C) Microbubble concentration plotted against volume at a basis concentration of 1010 MBs/mL. The shaded region under the curve represents the gas volume fraction. (D) Illustration of the MB + FUS treatment timeline. Initially, mice were imaged using T2w and CE-T1w MRI. Subsequently, they were moved to the RK-50 system to receive MB + FUS treatment. A more detailed timeline is provided at the bottom of panel D. After treatment, mice were imaged again with CE-T1w MRI. Data are presented as mean ± standard deviation (n = 5).
RNA extraction and bulk RNA sequencing
At 6 h post FUS treatment mice were sacrificed via perfusion with 60 mL of ice-cold PBS. Post dissection, the treated site was removed and snap-frozen using liquid nitrogen. Brain samples were weighed and then immediately placed into a cell lysing buffer (Qiagen, Hilden, Germany), and homogenized for 30 s. RNA was isolated and purified using the RNAeasy Kit (74,004, Qiagen). Quality control and library preparation were performed through the Anschutz Genomics Core for sequencing. Poly A selected total RNA paired-end sequencing was conducted at 40 million paired reads (80 million total reads) on a NovaSEQ 6000 sequencer.
Bulk RNA sequencing analysis
FASTQ files were obtained from Anschutz Genomics Core after sequencing. RNA analysis was performed using Pluto (https://pluto.bio). Principal component analysis (PCA)47 was calculated by applying the prcomp() R function to counts per million (CPM)-normalized values for all 40,773 targets in samples from all groups. The data was shifted to be zero-centered. The data was scaled to have unit variance before PCA was computed. Differential expression analysis was performed comparing the groups to the isoflurane-only control group unless otherwise noted in the figure caption. Genes were filtered to include only genes with at least 3 reads counted in at least 20% of samples in any group. Differential expression analysis was then performed with the DESeq2 R package47, which tests for differential expression based on a model using the negative binomial distribution. Log2 fold change was calculated for the comparison of the experiment to the control group. Thus, genes with a positive log2 fold change value had increased expression in treatment samples. Genes with a negative log2 fold change value had increased expression in control samples. Gene set enrichment analysis (GSEA) was done using the fgsea R package and the fgseaMultilevel() function48. The log2 fold change from the experiment versus control differential expression comparison was used to rank genes. Hallmark gene set collection from the Molecular Signatures Database (MSigDB)49,50 was curated using the msigdbr R package.
Statistical analysis
All data collected are presented as mean ± SD. No preprocessing was done to the data except for voltage data collected from the PCD. PCD data were preprocessed as described in Martinez et al.29. All statistical analysis was completed in Prism 9 (GraphPad, California, USA). Star representations of p-values are indicated in captions and less than 0.05 was indicative of statistical significance. An unpaired Student’s t-test and ANOVA/multiple comparisons were used to compare two groups and larger comparisons, respectively. The false discovery rate (FDR) method was applied for multiple testing corrections51. An adjusted p-value of 0.05 was used as the threshold for statistical significance.
Results
Microbubbles have a narrow size distribution
Microbubbles were isolated to a uniform size of 3-µm (± 0.5) diameter, as seen under brightfield microscopy (Fig. 1A). The size distribution of the MBs is monodisperse, with narrow peaks observed in both the number- and volume-weighted distributions (Fig. 1B). The mean diameters for the number- and volume-weighted distributions were 3.3 µm and 3.7 µm, respectively. The 10th and 90th percentiles in diameter were determined to be 2.53 µm and 4.19 µm respectively (Table S1). To ensure consistent MVD injection, each MB batch was analyzed. Figure 1C illustrates the relationship between MB concentration and volume, which was integrated to calculate the gas volume fraction (ϕMB). For a basis concentration of 1010 MBs/mL, the mean ϕMB was determined to be 17 µL/mL (Table S1). MVD is calculated by multiplying ϕMB by the fluid volume dose (mL/kg) injected intravenously into the subject.
Blood–brain barrier opening is dependent on both MVD and MI
The CE-T1wMRI revealed an intact BBB before the FUS procedure, as seen of minimal enhancement after MultiHance injection. Quantification of BBBO was performed on post-FUS CE-T1w MRI, based on the changes in T1 signal intensities in T1w versus CE-T1w MRI as illustrated in Fig. 2A. The CE difference between the contralateral hemispheres before FUS treatment was determined to be 0.05 ± 6% (mean ± standard deviation) (Supplemental Fig. S1). T2w MRI confirmed normal brain morphology.
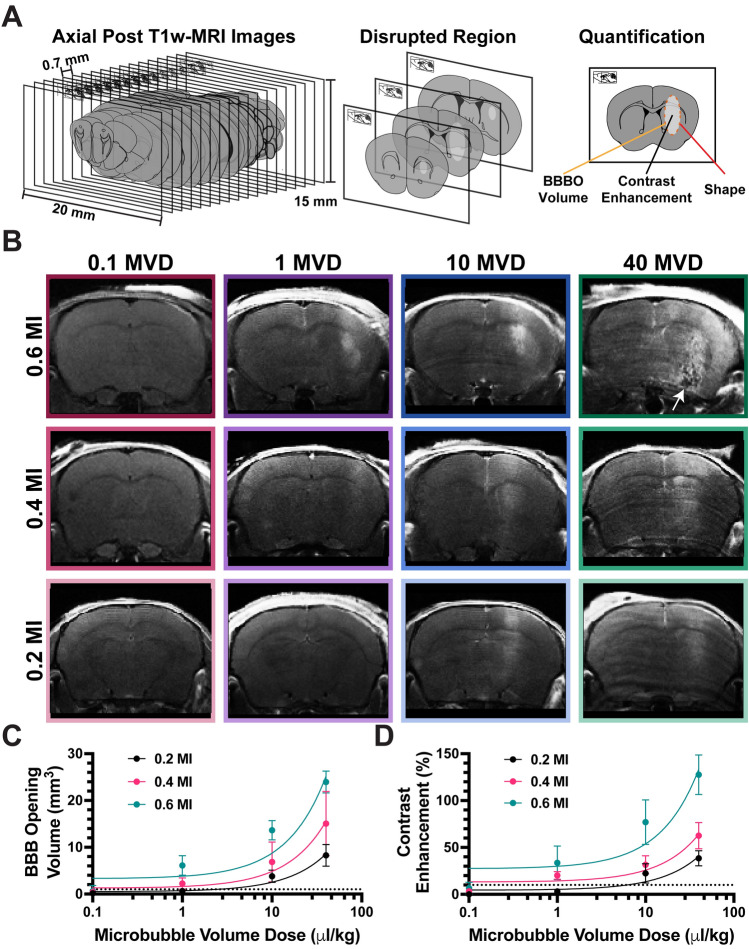
Figure 2.
Assessment of BBBO using Gadolinium contrast enhancement. (A) Cartoon depicting post-CE-T1w MRI image analysis. (B) Representative images of post-FUS MRI for each MI/MVD dose, captured at the peak BBBO area. (C) Quantification of BBBO volume (n = 3). Linear regression was performed for each mechanical index, resulting in R squared values of 0.86, 0.71, and 0.91 for 0.2, 0.4, and 0.6 MI, respectively. The X-axis is presented on a logarithmic scale. (D) Quantification of BBBO CE (n = 3). Linear regression was conducted for each mechanical index, resulting in R squared values of 0.79, 0.82, and 0.87 for 0.2, 0.4, and 0.6 MI, respectively. The X-axis is presented on a logarithmic scale. Data are presented as mean ± standard deviation.
Figure 2B shows representative images of BBBO for all twelve doses (MI/MVD). Within these experimental parameters, a clear onset of BBBO occurs at MVDs greater than 1 µL/kg at 0.2 MI, and MVDs greater than 0.1 µL/kg at both 0.4 and 0.6 MI. Irregular morphology was observed at the highest dose, as indicated by loss of brain structure in hypointense areas in T1w MRI (white arrow in Fig. 2B). As the MI or MVD was increased, both the BBBO volume and amplitude increased, as seen by increased relative signal intensity (Fig. 2C,D). Linear trends between brain CE in BBBO (volume and intensity) and the MI/MVD were observed, yielding significant (p < 0.05) trends for all MIs (Fig. 2C,D). Transposed plots comparing volume and CE to MI are shown in Supplemental Fig. S2A,B. Multiple regression analysis (MI + MVD) between BBBO CE resulted in R2 = 0.86, although R2 increased to 0.90 when analyzed as MI*MVD. This was similar for BBBO volume, with R2 moving from 0.89 (MI + MVD) to 0.94 (MI*MVD). Using FIJI, a round and circle score was given to each shape of BBBO to determine any lobe effects at high ultrasound pressures, and no significant differences were found between any dose pair (Supplemental Fig. S3). Based on the significance testing of pre-FUS controls (0.05 ± 6%, Supplemental Fig. S1), significant BBBO after FUS was defined as a 15% CE over a 1 mm3 volume. These intensity and volume thresholds are represented as dotted lines in Fig. 2C,D.
In addition to RNA sequencing, brains were stained fluorescently to visualize immune cell infiltration at the site of MB + FUS treatment. Five markers for immune cells were utilized, including GFAP, Iba1, CD68, CD4 and CD8 staining, astrocytes, microglia, peripheral macrophages, helper T-cells and cytotoxic T-cells (Supplemental Table S2). Representative IHC images were taken of the major effector protein, NFκB, to detect qualitatively the presence of inflammatory hotspots in the region of sonication on the post-FUS T1 images (Supplemental Fig. S4). Additionally, representative IHC images of CD4 (activated immune cells) showed increased signal with increasing MI. Meanwhile, Luxol fast blue staining for myelin (Supplemental Fig. S5) and hematoxylin and eosin staining to observe any changes to gross morphology (Supplemental Fig. S4) for each MI showed no differences between the groups.
Passive cavitation detection demonstrates microbubble activity varies with both MI and MVD
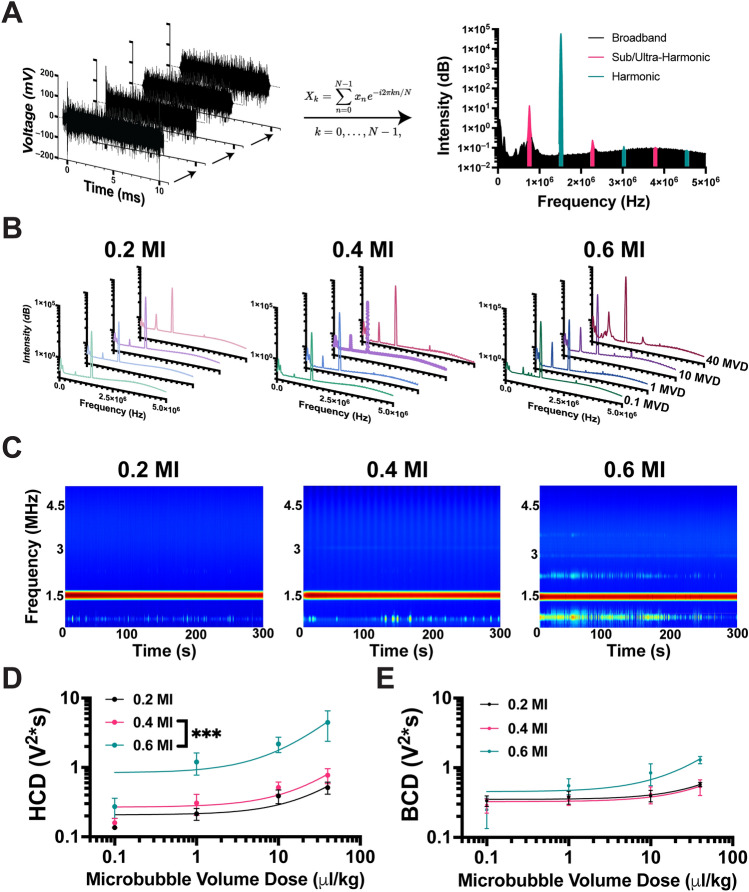
During each sonication, PCD recordings assessed microbubble acoustic activity during treatment. Voltage data obtained from PCD recordings were preprocessed and converted to the frequency domain (Fig. 3A). The frequency content analysis revealed an increase in subharmonic and ultra-harmonic content with higher MVD and MI (Fig. 3B). The presence of significant broadband content was observed only when the MVD exceeded 1 µL/kg at 0.6 MI (Fig. 3B). The frequency content throughout the treatments displayed a slight increase after injection, followed by a decline as MBs were cleared from circulation. This phenomenon was particularly evident at the highest MVD of 40 µL/kg (Fig. 3C). The average harmonic cavitation dose (HCD) was calculated for all doses, and a significant linear trend was observed at all three MIs (p < 0.05, Fig. 3D). Furthermore, there were no significant differences in broadband cavitation doses (BCD) between 0.2 and 0.4 MI at any MVD. However, statistically significant differences were observed at 0.6 MI with MVD higher than 1 µL/kg (Fig. 3D). Transposed plots comparing HCD/BCD to the MI can be found in Supplemental Fig. S6A,B. These values for no FUS and no MB controls are shown in Supplemental Fig. S7. Additionally, Fourier fast transform (FFT) plots of the PCD analysis have been added to Supplemental Fig. S11 to show the effects of retaining versus removing the fundamental frequency on the cavitation dose.
Figure 3.
Assessment of acoustic response using PCD. (A) Illustration of PCD data analysis. Voltage versus time data was cropped to remove the pre-FUS signal, followed by Tukey windowing and high-frequency filtering beyond PCD sensitivity (left). The resulting signal was then converted to the frequency domain using FFT, and the area under the curve (AUC) was quantified for respective regions to determine harmonic cavitation dose (HCD) and broadband cavitation dose (BCD) (right). (B) Average FFT for each FUS pulse during a five-minute treatment at each respective MI/MVD dose. (C) Representative spectrograms for 40 MVD doses are shown throughout the sonication time (300 s). Quantification of harmonic cavitation dose (D) and broadband cavitation dose (E) with respect to MVD and MI (n = 3). Linear regression was performed for each MI, resulting in R squared values for harmonic cavitation dose was 0.71, 0.74, and 0.71 for 0.2, 0.4, and 0.6 MI, respectively. The R-squared values for broadband cavitation dose were 0.73, 0.52, and 0.75 for 0.2, 0.4, and 0.6 MI, respectively. Data are presented as mean ± standard deviation.
RNA sequencing indicates differential gene expression varies with MI and MVD
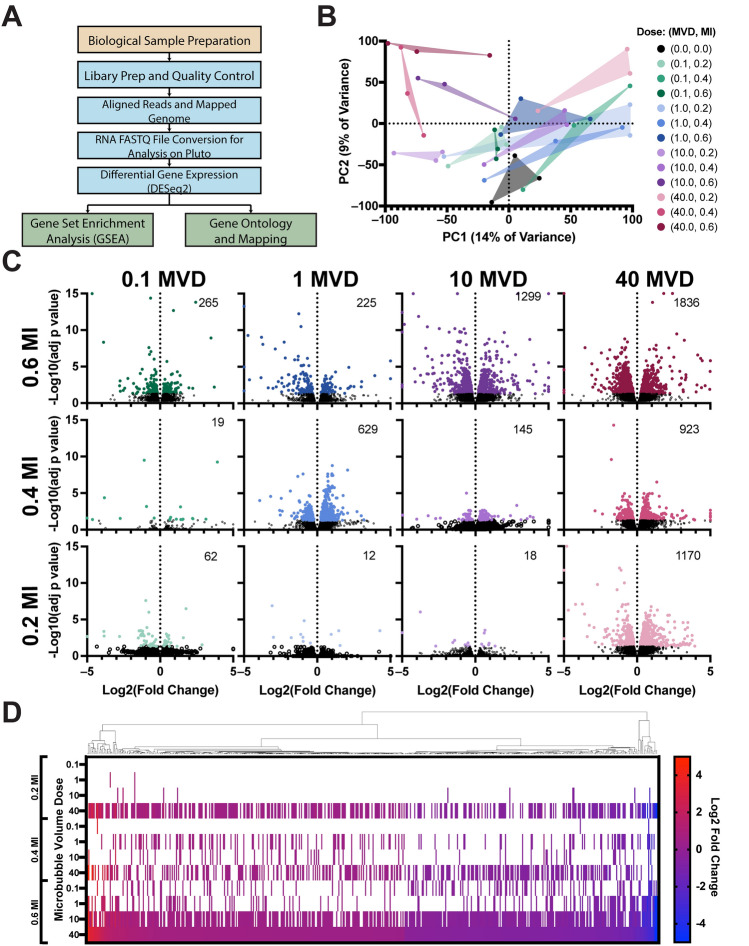
Six hours after MRI-guided FUS treatment, RNA was extracted from the treated brain region. Figure 4A illustrates our RNA sequencing pipeline, highlighting steps from extraction to analysis. To confirm variance between groups, all samples were initially plotted on a principal component analysis (PCA) graph. Figure 4B displays the primary two components with the highest variability, accounting for 14% and 9% of the total variance. Other variability testing was conducted on samples including UMAP and t-SNE plots found in Supplemental Fig. S8A,B.
Figure 4.
Bulk RNA sequencing of treated brain region. (A) Sequential RNA processing steps, from sample preparation to gene set enrichment analysis (GSEA) and mapping. (B) Principal component analysis (PCA) plot displaying all samples in the dataset, including different MI/MVD combinations and isoflurane control samples. (C) Differential gene expression analysis for each MI/MVD dose. The number displayed on the top right of each plot indicates the count of significantly differentially expressed genes. All doses were compared to isoflurane control samples (n = 3). Among the 1,836 differentially expressed genes (DEGs) identified in the highest MI/MVD dose, 536 genes were also found in at least two other doses. (D) Heatmap illustrating the expression of the 536 genes, ranked by log2 fold change in the highest MI/MVD dose. White color represents insignificant differential expression. The Euclidean distance map is shown on top of the heatmap.
Each triplicate sample was then analyzed for differentially expressed genes (DEGs) against the no-FUS control (+Isoflurane) (Fig. 4C). Overall, the DEGs increased with a higher MI or MVD. The highest number of DEGs was observed in the highest MVD and MI dose (40 MVD + 0.6 MI), totaling 1836 genes; the lowest observed number was 12 at the lowest MI and MVD. To understand their significance, the 1836 DEGs from the highest dose were analyzed for similarities with other doses. Among these genes, 536 were also found in at least two other doses, indicating a specific effect of MB + FUS treatment (Fig. 4D). The 536 genes were organized based on their fold change at the highest dose. As the dose was reduced in either MVD or MI, fewer genes in the set showed significant differential expression, as indicated by the white bars. Notably, the lowest dose (0.1 MVD + 0.2 MI) did not exhibit significant differential expression among the 536 genes. Furthermore, the expression levels of these 536 genes remained consistent across all doses. The genes showing higher expression levels in the highest dose (40 MVD + 0.6 MI) also demonstrated higher expression levels in all other doses. Similarly, the less expressed genes maintained their lower expression levels consistently across all doses, following a similar gradient.
Hallmark gene set enrichment analysis reveals inflammatory response after BBBO
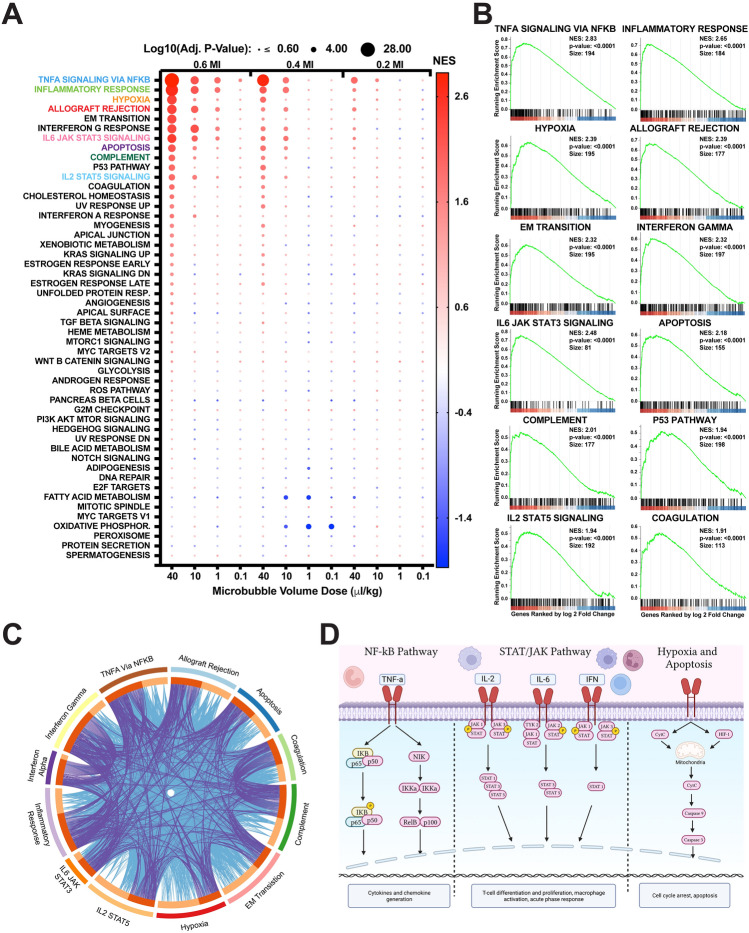
The next step in RNA analysis was to perform GSEA to provide a biological context for the DEGs. To cover a wide range of biological processes, we utilized the 50 hallmark gene sets from the Broad Institute (Cambridge, Massachusetts). Figure 5A illustrates the top gene sets identified across all 12 doses. Notably, there are 12 distinct gene sets that show upregulation at the highest doses and decline as the MI or MVD decreases. These twelve gene sets include, beginning with the most significant: TNFɑ Signaling via NFκB, Inflammatory Response, Hypoxia, Allograft Rejection, Epithelial-Mesenchymal Transition, Interferon Gamma Response, IL6 Jak Stat3 Signaling, Apoptosis, Complement, P53 Pathway, IL2 Stat5 Signaling, and Coagulation. Importantly, all these gene sets are closely associated with inflammatory responses.
Figure 5.
Hallmark gene set enrichment analysis. (A) Dot plot illustrating the 12 MI/MVD doses based on size (adjusted p-value) and color (normalized enrichment score). Gene sets are arranged by adjusted p-value for the highest MI/MVD dose. (B) Enrichment plot presenting the top 12 gene sets identified in (A). The x-axis displays the genes ranked by log2 fold change, with vertical ticks indicating the gene positions within the gene set. The heatmap represents gene expression, with red indicating higher expression in the first group (40 MVD + 0.6 MI), and blue representing higher expression in the isoflurane control group. The green line represents the running enrichment score. (C) Overlapped Circos plot demonstrating relationships between the top 12 enriched gene sets. Purple lines indicate the presence of the same gene in different gene sets, while blue connections represent genes found in similar Gene Ontology (GO) pathways across different gene sets. (D) Schematic representation of the top enriched gene set pathways.
Enrichment plots for all gene sets identified in the highest dose (40 MVD + 0.6 MI) are presented in Fig. 5B. The high peak on the left side of the plots indicates the strong enrichment of these gene sets. Gene set relationships were illustrated via a Circos plot in Fig. 5C, where direct gene connections (represented by purple lines) and connections via Gene Ontology (GO) biological processes (light blue lines) are established between each gene set. Purple lines indicate the presence of the same gene in different gene sets, while blue connections represent genes found in similar Gene Ontology (GO) pathways across different gene sets.
Figure 5D provides another perspective by visualizing the major mechanisms between these gene sets through signaling molecules. Three main groups emerge from the analysis of these gene sets: the initial inflammatory response (TNFɑ signaling via NFκB), the major inflammatory response (which involves more chemokine and cytokine signaling), and damage-associated gene sets (related to apoptosis). Overall, the results of the GSEA highlight the prominent role of inflammatory responses and related pathways in the transcriptional changes observed owing to MB + FUS dose escalation.
Blood–brain barrier opening by CE-T1w MRI is the best indicator of SIR
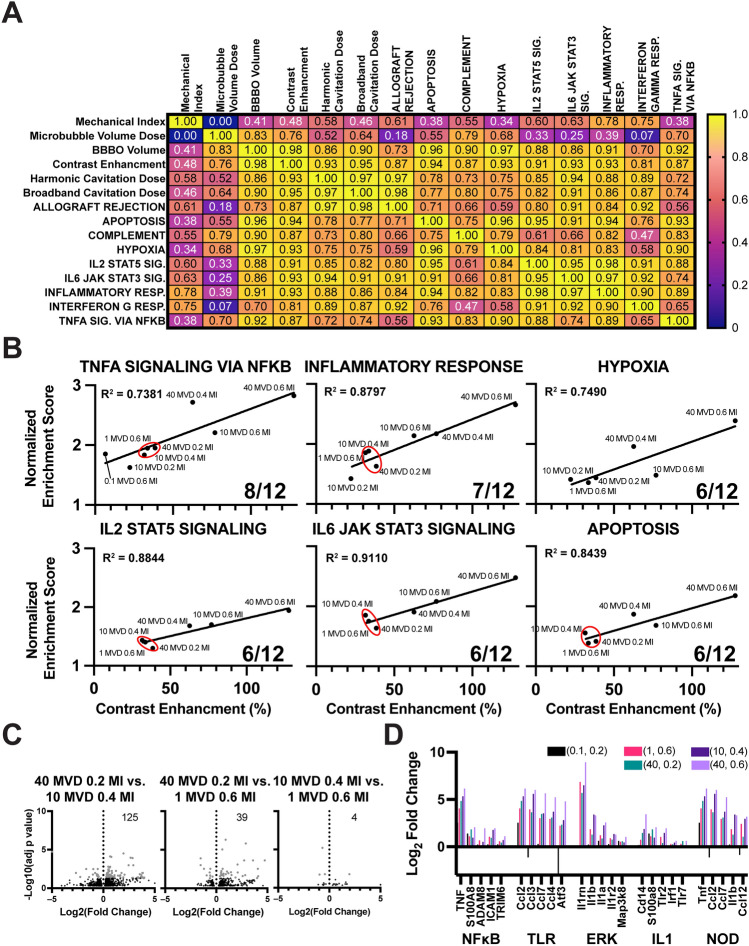
To better understand the relationship between our parameters and BBBO or SIR, we conducted a correlation matrix analysis of all major variables identified. Figure 6A presents this matrix, highlighting both strong and weak correlations. Our analysis revealed a stronger correlation between hyperintense volume and CE in BBBO areas with MVD, as compared to the MI (0.83 vs. 0.76 and 0.41 vs. 0.48, respectively). PCD parameters exhibited consistent relationships (0.46–0.64) with both MVD and MI. In terms of RNA expression, indicated by normalized enrichment scores (NES), correlations varied with MVD or MI. MVD showed a stronger correlation with complement, hypoxia, and TNFɑ signaling via NFκB (R2 = 0.79, 0.68, and 0.70, respectively). On the other hand, MI displayed a stronger correlation with IL2/stat5 signaling, IL6/jak/stat3 signaling, interferon-gamma response, allograft rejection, and inflammatory response (R2 = 0.60, 0.63, 0.75, 0.61, and 0.78, respectively). Our main finding indicated that BBBO volume and BBBO CE were the effects most strongly associated with RNA expression, with all correlations having R2 values greater than 0.86 (excluding allograft rejection and interferon-gamma response). We then plotted our top-represented gene sets against BBBO volume (Fig. 6B). At least half of all doses exhibited significant NES in each gene set, and each relationship demonstrated strong linear correlations.
Figure 6.
Correlation of RNA expression and BBB opening. (A) Correlation heatmap showing the relationship between MI and MVD doses and resulting BBBO volume, contrast enhancement (CE), cavitation doses, and RNA expression. Pearson correlation coefficient (r) values are displayed in each box. The color scale represents the magnitude of the Pearson correlation coefficient. (B) Correlation between normalized enrichment score (NES) scores and BBBO volume. Only pathways from gene sets with more than six doses exhibiting significant NES scores are represented. Linear regression analysis was performed for each plot, and the corresponding R-squared value is shown in the top left corner. The plots indicate the fraction of significant doses in the bottom right. A red circle denotes a small cluster of three doses with similar BBBO volumes. (C) Volcano plot displaying the differentially expressed genes (DEGs) between doses within the small cluster (40 MVD + 0.2 MI, 10 MVD + 0.4 MI, and 1 MVD + 0.6 MI). The number of DEGs is indicated in the top right corner. (D) Bar graph representing the log2 fold change of the lowest dose (0.1 MVD + 0.2 MI), the cluster doses, and the highest dose (40 MVD + 0.6 MI). The x-axis displays five of the most influential genes within five inflammatory signaling families.
A noteworthy observation was the identification of a small cluster of three MVD/MI doses: 1 MVD + 0.6 MI, 10 MVD + 0.4 MI, and 40 MVD + 0.2 MI (red circles). These doses exhibited similar BBBO CE (between 31.6 and 38.5%) and NES in five of six gene sets. To gain further insights, we performed DEG analysis between each dose (Fig. 6C). The volcano plots indicated minimal differences in gene expression, with a limited number of DEGs (4–125 genes). To corroborate these findings, we examined the fold change in highly utilized genes (leading edge genes) within five inflammatory families (Fig. 6D). We observed similar expression levels among the cluster doses, while their expression differed significantly from the lowest and highest doses. Overall, our results suggest that the extent of BBBO plays a pivotal role in the SIR, surpassing the influence of other parameters in isolation.
Therapeutic windows can be defined between BBBO and SIR
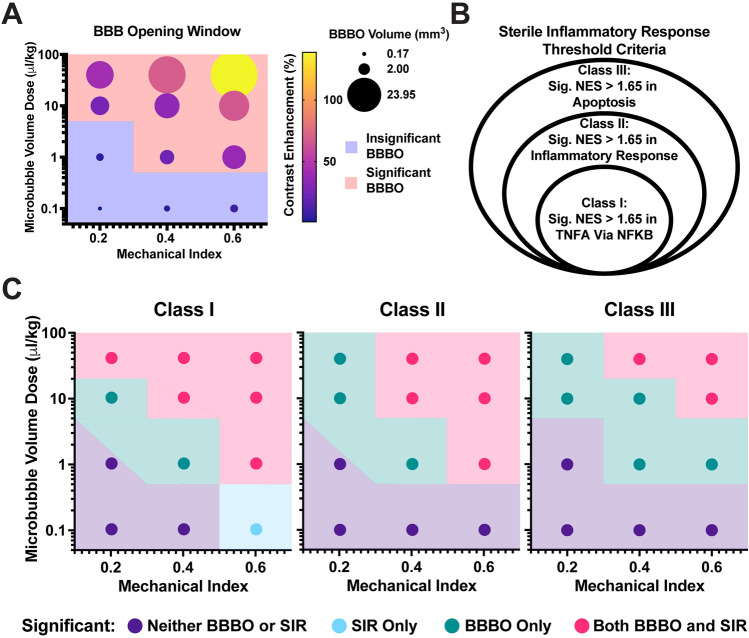
We defined significant BBBO as a minimum of 15% CE in a volume greater than 1 mm3. Figure 7A presents a dot plot depicting all doses and their corresponding BBBO volume (size) and CE (color). With defined thresholds, we observed a region representing insignificant BBBO (blue) and significant BBBO (red).
Figure 7.
Therapeutic window between BBBO and onset of sterile inflammatory response (SIR). (A) Dot plot illustrating the mean blood–brain barrier opening (BBBO) in terms of volume (represented by the size of the dot) and contrast enhancement (indicated by the color of the dot). The grid is plotted based on microbubble volume dose (MVD) and mechanical index (MI). Significant BBBO is defined as 15% contrast enhancement in a 1 mm3 volume. The red region represents significant BBBO, while the blue region represents insignificant BBBO. (B) Classes defined for the onset of the sterile inflammatory response. Each circle represents the criteria required to meet a specific class level. (C) Dot plot displaying the regions of neither significant BBBO nor SIR (purple), BBBO only (green), SIR only (light blue), or significant BBBO and SIR (pink). Each plot represents a different class level for the onset of SIR as defined in (B).
We recognized the gradient of responses and developed different classes of SIR (Fig. 7B). Each class encompassed previous classifications, creating a hierarchical framework. Figure 7C illustrates the therapeutic window between significant BBBO and SIR. In Class I, the least strict classification, significant NES (> 1.65) in the TNFɑ signaling via NFκB gene set serves as the defining criterion. Within this class, a small window of significant BBBO is observed before the onset of SIR (10 MVD + 0.2 MI and 1 MVD + 0.4 MI). The only instance where SIR occurs without significant BBBO is 0.1 MVD + 0.6 MI.
Class II is defined by the onset of significant NES (> 1.65) for both the TNFɑ signaling via NFκB and Inflammatory response; the therapeutic window expands. Here, three doses exhibit significant BBBO without the onset of SIR (10–40 MVD + 0.2 MI and 1 MVD + 0.4 MI).
Finally, Class III is characterized by significant NES (> 1.65) for TNFɑ signaling via NFκB, Inflammatory response, and a damage-associated marker, apoptosis. We observe five doses with significant BBBO but no onset of acute SIR (10–40 MVD + 0.2 MI, 1–10 MVD + 0.4 MI, and 1 MVD + 0.6 MI). These findings delineate the therapeutic window between significant BBBO and the onset of acute SIR, providing valuable insights into the relationship between these two critical factors.
Discussion
FUS with MBs has gained significant attention as a promising approach for non-invasive delivery to the brain52–56. This technique offers modularity with a diverse set of US and MB parameters that can be used, enabling BBBO manipulation and subsequent inflammatory responses. However, defining appropriate thresholds can be challenging. We focused on two key parameters: mechanical index (MI, ) to predict in situ pressure at the focus and microbubble volume dose (MVD, µL/kg) to quantify total injected gas volume. These parameters have demonstrated the crucial roles of US and MBs in determining BBBO17,57. Consistent with previous studies investigating the relationship between BBBO and MI56–60, we observed a similar linear relationship between MI and cavitation activity with the extent of BBBO, as measured by MRI CE. Similarly, as MVD increased, a linear effect on BBBO was observed. Our multiple regression analyses concluded that the strongest relationship occurs when combining the influence of both MI and MVD as the product of these two doses. This synergy, represented through the term, MI*MVD, exhibited a high coefficient of determination (R2 = 0.9), indicating that the combined effect of MI and MVD has a greater impact on BBBO than either parameter alone.
PCD provided valuable insights into MB behavior in real-time during the treatments. The analysis of the voltage data29 determined the HCD/BCD, which showed linear trends with MI and MVD. Notably, the HCD exhibited a powerful relationship with both BBBO volume and CE, confirming its potential as a predictive parameter for assessing BBBO20,42,56,61.
Determining the threshold for significant BBBO is crucial for safe and effective treatments. While various studies have investigated this threshold, the wide array of parameters and metrics utilized makes direct comparisons between experiments challenging. Nonetheless, many studies have reported a threshold in the range of 0.3 to 0.5 MI at different MVDs. We found the threshold for significant BBBO (> 15% CE in a volume of 1 mm3) to be greater than 1 MVD + 0.2 MI and greater than 0.1 MVD + 0.4/ 0.6 MI. Our estimates for a 15% increase in targeted signal intensity are dependent on our specific MRI protocol. It is important to note that our highest dose (40 MVD + 0.6 MI) resulted in irregular tissue morphology illustrated by hyperintense and hypointense regions on T1w + CE MRI. This observation suggests a potential maximum safety threshold at 40 MVD + 0.6 MI at our tested ultrasound parameters (1 MHz, 10 ms PL, 10% D.C).
Bulk RNA sequencing enables us to understand the global transcriptomic activity in cells or tissues. To limit noise from areas outside of our focal region, we performed an MRI-guided dissection of the sonicated region for RNA isolation. Analysis of sequencing data revealed that at higher MB+FUS doses (e.g., 40 MVD + 0.6 MI), more DEGs were present indicating heightened transcriptomic activity. This was not the case when compared to combinations of lower MB+FUS doses (0.1 MVD + 0.2 MI). In Fig. 5D, we compared all DEGs in the highest dose (40 MVD + 0.6 MI) against DEGs in the remaining conditions (11/12). However, when compared against the most common DEGs, no genes were significantly up or downregulated, indicating that the 62 DEGs are not related to inflammation or damage but rather other tangential biological processes.
GSEA was performed on all 12 combinations using the 50-hallmark gene set analysis, which compared gene expression against the 50 most fundamental biological pathways and functions in the body. Out of the 50 pathways analyzed, our DEG expression profile matched similar expression patterns and genes associated with inflammation and damage, with the highest ranked pathway as TNFɑ signaling through NFκB. This finding corroborated results from other studies that identified activation of the NFκB pathway as a primary driving force behind SIR after MB + FUS BBBO30,31,62. Outside of broad inflammatory responses, other pathways that are upregulated are the JAK/STAT and Interferon family. These pathways have been associated with immune recruitment, specifically involving T-cell activation and macrophage chemotaxis towards inflammatory hotspots63,64. This supports the evidence of peripheral immune cell extravasation after BBBO-induced SIR and implicates specific immune cell populations that are responsible for mediating host-specific responses. A third major event observed in the RNA sequencing data was the enrichment of genes in pathways related to hypoxia and apoptosis, indicative of cellular damage. Increased MI is known to inflict cellular damage, as greater mechanical forces exerted on the vessel endothelium have been associated with hemorrhaging65,66. This has similarly been observed in multiple studies56,57, where increased peak negative pressures for BBBO produced cerebral microhemorrhages and damage consistent with traumatic brain injury67. Another study showed transiently altered blood flow and hypoxia were observed with BBBO, and capillary vessel restriction was implicated as the source68. This study along with our observations adds a dimension to the SIR, which considers cellular damage in the potentiation of SIR.
After running the 50-hallmark analysis, we conducted a correlation analysis comparing the most enriched pathways identified by GSEA against tested US and MB parameters. Interestingly, our findings revealed distinct pathways that exhibited a stronger correlation with MVD, such as the activation of the complement system, hypoxia, and NFκB pathway via TNFɑ signaling. Unexpectedly, activation of the NFκB pathway exhibited a more pronounced correlation with MVD, while MI had a stronger relationship with pathways such as allograft rejection, IL2/STAT3, IL6/JAK/STAT3, IFNγ , and inflammatory response. Despite obvious differences observed between MI and MVD, all pathways correlated most highly to BBBO volume (R2 > 0.70) and CE (R2 > 0.81). A study performed by Mathew et al. in 2021 and McMahon et al. in 2017 similarly concluded that contrast enhancement from a gadolinium based contrast agent in CE-T1w MRI is a good predictor of gene expression associated with SIR following MB+FUS treatment31,59.
When NES for the most significantly upregulated pathways were plotted against BBBO CE, a “cluster” of MVD and MI combinations was apparent on the trendlines. This phenomenon was most notably observed with 1.0 MVD + 0.6 MI, 10 MVD + 0.4 MI and 40 MVD + 0.2 MI, which clustered together due to their similar intensities of BBBO and normalized gene enrichment scores. This is seen in 5 of 6 graphs in Fig. 7B, except hypoxia where 10 MVD + 0.4 MI did not have a significant NES, further supporting the notion that the intensity of BBBO is the greatest indicator of bioeffects. Moreover, exchanging higher MI dose with lower MVD, or vice versa, can yield comparable BBBO and SIR.
As defined previously, BBBO was considered significant when 15% CE was achieved in a volume greater than 1 mm3. Applying these criteria in Fig. 7A, regions of significant BBBO and insignificant BBBO were delineated, serving as the basis for developing our therapeutic window. By comparing the RNA sequencing data from the 12 tested parameter combinations, differences in gene expression amongst parameters were analyzed and distilled into three generic categories based on similarities in gene expression and pathway upregulation (Fig. 7B). NFκB signaling has been defined in many studies as a hallmark of the SIR3,30,31,62. Kovacs et al. first defined this as the primary mechanism driving the SIR, especially after MB + FUS BBBO, launching subsequent studies aiming to further investigate the bioeffects30,31. Due to this, we broadly defined “mild” SIR as the activation of the NFκB pathway via TNFɑ signaling (NES > 1.65). Since the SIR acts in concert with adjacent immune processes, we considered a second definition of “moderate” SIR that involved the upregulation of a broad inflammatory response following activation of NFκB (NES > 1.65). Recently, several groups have worked to operate alongside the activation of SIR after MB + FUS69–71 without triggering damage pathways (apoptosis). Likewise, our Class III (“acute”) therapeutic window is defined as the onset of inflammatory responses and activation of NFκB, which avoids activation of apoptosis through cellular damage. This class facilitates a controlled level of inflammation that may be beneficial in neurogenesis, amyloid-beta clearance72, and microglial reprogramming55.
Our categories of SIR above are not standard definitions of SIR but are based on our interpretation of observed patterns in RNA sequencing results. Specific gene sets that were noticed to be enriched alongside the activation of the NFκB pathway via TNFɑ allowed us to formulate distinct definitions of SIR. It is important to acknowledge that while we linked our classification of SIR with specific pathways, such as upregulation of an inflammatory response and apoptosis, these classes may not encompass the full mechanism of the SIR. This limitation arises from the GSEA 50 hallmark pathway, which sorted inflammation observed after SIR into basic categories and indicates the existence of prominent associations within pathways (e.g., inflammatory response and apoptosis) included in our classifications.
Moving forward, further investigations are warranted to address limitations in our study. Firstly, exploring other US parameters such as frequency, pulse length, duty cycle and sonication time, but also MB parameters such as MVD, size and formulation. Secondly, as SIR was only evaluated at one time point (6 h); future studies should explore sampling at different time points after BBBO to assess the temporal extent of SIR. An additional limitation to this study is derived from the nature of bulk RNA sequencing, which cannot differentiate expression profiles from distinct cell populations. Instead, it only evaluates the broad transcriptome input for every cell. Future efforts will provide more tunable therapeutic windows that allow more safe and effective treatments.
In conclusion, our study highlights the substantial impact of both MVD and MI on MB+FUS BBBO and SIR. The combination of MI and MVD exhibited a more pronounced effect on BBBO compared to individual parameters. RNA analysis revealed unique DEGs associated with inflammatory responses and immune recruitment. Our study defines three therapeutic windows bridging together significant BBBO with the onset of SIR in three different levels of severity. This guidance is invaluable for studies aiming to deliver therapeutics without inducing inflammation, thereby ensuring the safe and effective delivery of drugs into the brain.
Supplementary Information
Author contributions
Conceptualization: P.J.M., K.H.S., M.A.B. Methodology: P.J.M., J.J.S., K.H.S., J.D. Investigation: P.J.M., J.J.S., F.G.G., T.M., K.H.S. Visualization: P.J.M., J.J.S., J.S. Funding acquisition: K.H.S., M.A.B. Project administration: A.L.G., M.A.B. Supervision: A.L.G., M.A.B. Writing—original draft: P.J.M., J.J.S. Writing—review and editing: N.E., N.J.S., A.L.G., M.A.B.
Data availability
Sequence data that support the findings of this study have been deposited in the NIH BioProject Archive with the primary accession code PRJNA1100543. Other data are available upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Payton J. Martinez and Jane J. Song.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-67916-8.
References
- 1.Wu, S.-K., Tsai, C.-L., Huang, Y. & Hynynen, K. Focused ultrasound and microbubbles-mediated drug delivery to brain tumor. Pharmaceutics13(1), 15. 10.3390/pharmaceutics13010015 (2020). 10.3390/pharmaceutics13010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitusova, K. et al. Overcoming the blood–brain barrier for the therapy of malignant brain tumor: current status and prospects of drug delivery approaches. J. Nanobiotechnol.20(1), 412. 10.1186/s12951-022-01610-7 (2022). 10.1186/s12951-022-01610-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung, O. et al. Neuroinflammation associated with ultrasound-mediated permeabilization of the blood–brain barrier. Trends Neurosci.45(6), 459–470. 10.1016/j.tins.2022.03.003 (2022). 10.1016/j.tins.2022.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Todd, N. et al. Secondary effects on brain physiology caused by focused ultrasound-mediated disruption of the blood–brain barrier. J. Controll. Releas.324, 450–459. 10.1016/j.jconrel.2020.05.040 (2020). 10.1016/j.jconrel.2020.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menaceur, C., Gosselet, F., Fenart, L. & Saint-Pol, J. The blood–brain barrier, an evolving concept based on technological advances and cell–cell communications. Cells11(1), 133. 10.3390/cells11010133 (2021). 10.3390/cells11010133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow, B. W. & Gu, C. The Molecular constituents of the blood–brain barrier. Trends Neurosci.38(10), 598–608. 10.1016/j.tins.2015.08.003 (2015). 10.1016/j.tins.2015.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardridge, W. M. The blood–brain barrier: Bottleneck in brain drug development. Neurotherapeutics2(1), 3–14. 10.1602/neurorx.2.1.3 (2005). 10.1602/neurorx.2.1.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luissint, A.-C., Artus, C., Glacial, F., Ganeshamoorthy, K. & Couraud, P.-O. Tight junctions at the blood brain barrier: Physiological architecture and disease-associated dysregulation. Fluids Barriers CNS9(1), 23. 10.1186/2045-8118-9-23 (2012). 10.1186/2045-8118-9-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siegenthaler, J. A., Sohet, F. & Daneman, R. ‘Sealing off the CNS’: Cellular and molecular regulation of blood–brain barriergenesis. Curr. Opin. Neurobiol.23(6), 1057–1064. 10.1016/j.conb.2013.06.006 (2013). 10.1016/j.conb.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mills, W. A. et al. Astrocyte plasticity in mice ensures continued endfoot coverage of cerebral blood vessels following injury and declines with age. Nat. Commun.13(1), 1794. 10.1038/s41467-022-29475-2 (2022). 10.1038/s41467-022-29475-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, L. S. et al. Pericytes and Neurovascular function in the healthy and diseased brain. Front. Cell. Neurosci.13, 282. 10.3389/fncel.2019.00282 (2019). 10.3389/fncel.2019.00282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubotera, H. et al. Astrocytic endfeet re-cover blood vessels after removal by laser ablation. Sci. Rep.9(1), 1263. 10.1038/s41598-018-37419-4 (2019). 10.1038/s41598-018-37419-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klibanov, A. L. Targeted delivery of gas-filled microspheres, contrast agents for ultrasound imaging. Adv. Drug Deliv. Rev.37(1–3), 139–157. 10.1016/s0169-409x(98)00104-5 (1999). 10.1016/s0169-409x(98)00104-5 [DOI] [PubMed] [Google Scholar]
- 14.Unger, E. C., Hersh, E., Vannan, M. & McCreery, T. Gene delivery using ultrasound contrast agents. Echocardiography18(4), 355–361. 10.1046/j.1540-8175.2001.00355.x (2001). 10.1046/j.1540-8175.2001.00355.x [DOI] [PubMed] [Google Scholar]
- 15.Ferrara, K., Pollard, R. & Borden, M. Ultrasound microbubble contrast agents: Fundamentals and application to gene and drug delivery. Annu. Rev. Biomed. Eng.9(1), 415–447. 10.1146/annurev.bioeng.8.061505.095852 (2007). 10.1146/annurev.bioeng.8.061505.095852 [DOI] [PubMed] [Google Scholar]
- 16.Borden, M. A. & Song, K.-H. Reverse engineering the ultrasound contrast agent. Adv. Colloid Interface Sci.262, 39–49. 10.1016/j.cis.2018.10.004 (2018). 10.1016/j.cis.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apfel, R. E. & Holland, C. K. Gauging the likelihood of cavitation from short-pulse, low-duty cycle diagnostic ultrasound. Ultrasound Med. Biol.17(2), 179–185. 10.1016/0301-5629(91)90125-G (1991). 10.1016/0301-5629(91)90125-G [DOI] [PubMed] [Google Scholar]
- 18.Miller, D. L. Overview of experimental studies of biological effects of medical ultrasound caused by gas body activation and inertial cavitation. Prog. Biophys. Mol. Biol.93(1–3), 314–330. 10.1016/j.pbiomolbio.2006.07.027 (2007). 10.1016/j.pbiomolbio.2006.07.027 [DOI] [PubMed] [Google Scholar]
- 19.Choi, J. J., Pernot, M., Small, S. A. & Konofagou, E. E. Noninvasive, transcranial and localized opening of the blood-brain barrier using focused ultrasound in mice. Ultrasound Med. Biol.33(1), 95–104. 10.1016/j.ultrasmedbio.2006.07.018 (2007). 10.1016/j.ultrasmedbio.2006.07.018 [DOI] [PubMed] [Google Scholar]
- 20.O’Reilly, M. A. & Hynynen, K. Blood–brain barrier: Real-time feedback-controlled focused ultrasound disruption by using an acoustic emissions-based controller. Radiology263(1), 96–106. 10.1148/radiol.11111417 (2012). 10.1148/radiol.11111417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tung, Y.-S., Vlachos, F., Feshitan, J. A., Borden, M. A. & Konofagou, E. E. The mechanism of interaction between focused ultrasound and microbubbles in blood–brain barrier opening in mice. J Acoust. Soc. Am.130(5), 3059–3067. 10.1121/1.3646905 (2011). 10.1121/1.3646905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMahon, D., O’Reilly, M. A. & Hynynen, K. Therapeutic agent delivery across the blood–brain barrier using focused ultrasound. Annu. Rev. Biomed. Eng.23(1), 89–113. 10.1146/annurev-bioeng-062117-121238 (2021). 10.1146/annurev-bioeng-062117-121238 [DOI] [PubMed] [Google Scholar]
- 23.Poon, C., McMahon, D. & Hynynen, K. Noninvasive and targeted delivery of therapeutics to the brain using focused ultrasound. Neuropharmacology120, 20–37. 10.1016/j.neuropharm.2016.02.014 (2017). 10.1016/j.neuropharm.2016.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konofagou, E. E. Optimization of the ultrasound-induced blood–brain barrier opening. Theranostics2(12), 1223–1237. 10.7150/thno.5576 (2012). 10.7150/thno.5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song, K.-H. et al. Microbubble gas volume: A unifying dose parameter in blood–brain barrier opening by focused ultrasound. Theranostics7(1), 144–152. 10.7150/thno.15987 (2017). 10.7150/thno.15987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song, K.-H., Harvey, B. K. & Borden, M. A. State-of-the-art of microbubble-assisted blood-brain barrier disruption. Theranostics8(16), 4393–4408. 10.7150/thno.26869 (2018). 10.7150/thno.26869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sirsi, S., Feshitan, J., Kwan, J., Homma, S. & Borden, M. Effect of microbubble size on fundamental mode high frequency ultrasound imaging in mice. Ultrasound Med. Biol.36(6), 935–948. 10.1016/j.ultrasmedbio.2010.03.015 (2010). 10.1016/j.ultrasmedbio.2010.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro-Becerra, J. A., Song, K.-H., Martinez, P. & Borden, M. A. Microbubble size and dose effects on pharmacokinetics. ACS Biomater. Sci. Eng.8(4), 1686–1695. 10.1021/acsbiomaterials.2c00043 (2022). 10.1021/acsbiomaterials.2c00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez, P., Bottenus, N. & Borden, M. Cavitation characterization of size-isolated microbubbles in a vessel phantom using focused ultrasound. Pharmaceutics14(9), 1925. 10.3390/pharmaceutics14091925 (2022). 10.3390/pharmaceutics14091925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovacs, Z. I. et al. Disrupting the blood–brain barrier by focused ultrasound induces sterile inflammation. Proc. Natl. Acad. Sci. U.S.A.10.1073/pnas.1614777114 (2017). 10.1073/pnas.1614777114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMahon, D. & Hynynen, K. Acute inflammatory response following increased blood–brain barrier permeability induced by focused ultrasound is dependent on microbubble dose. Theranostics7(16), 3989–4000. 10.7150/thno.21630 (2017). 10.7150/thno.21630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong, T., Liu, L., Jiang, W. & Zhou, R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol.20(2), 95–112. 10.1038/s41577-019-0215-7 (2020). 10.1038/s41577-019-0215-7 [DOI] [PubMed] [Google Scholar]
- 33.Rock, K. L., Latz, E., Ontiveros, F. & Kono, H. The sterile inflammatory response. Annu. Rev. Immunol.28(1), 321–342. 10.1146/annurev-immunol-030409-101311 (2010). 10.1146/annurev-immunol-030409-101311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen, C.-J. et al. Identification of a key pathway required for the sterile inflammatory response triggered by dying cells. Nat. Med.13(7), 851–856. 10.1038/nm1603 (2007). 10.1038/nm1603 [DOI] [PubMed] [Google Scholar]
- 35.Chen, G. Y. & Nuñez, G. Sterile inflammation: Sensing and reacting to damage. Nat. Rev. Immunol.10(12), 826–837. 10.1038/nri2873 (2010). 10.1038/nri2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feldman, N., Rotter-Maskowitz, A. & Okun, E. DAMPs as mediators of sterile inflammation in aging-related pathologies. Ageing Res. Rev.24, 29–39. 10.1016/j.arr.2015.01.003 (2015). 10.1016/j.arr.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 37.Keyel, P. A. How is inflammation initiated? Individual influences of IL-1, IL-18 and HMGB1. Cytokine69(1), 136–145. 10.1016/j.cyto.2014.03.007 (2014). 10.1016/j.cyto.2014.03.007 [DOI] [PubMed] [Google Scholar]
- 38.Chen, Y., Yousaf, M. N. & Mehal, W. Z. Role of sterile inflammation in fatty liver diseases. Liver Res.2(1), 21–29. 10.1016/j.livres.2018.02.003 (2018). 10.1016/j.livres.2018.02.003 [DOI] [Google Scholar]
- 39.Ratajczak, M. Z. et al. Sterile inflammation of brain, due to activation of innate immunity, as a culprit in psychiatric disorders. Front. Psychiatry9, 60. 10.3389/fpsyt.2018.00060 (2018). 10.3389/fpsyt.2018.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otani, K. & Shichita, T. Cerebral sterile inflammation in neurodegenerative diseases. Inflamm. Regener.40(1), 28. 10.1186/s41232-020-00137-4 (2020). 10.1186/s41232-020-00137-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serkova, N. J. Nanoparticle-based magnetic resonance imaging on tumor-associated macrophages and inflammation. Front. Immunol.8, 590. 10.3389/fimmu.2017.00590 (2017). 10.3389/fimmu.2017.00590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji, R. et al. Cavitation-modulated inflammatory response following focused ultrasound blood–brain barrier opening. J. Controll. Releas.337, 458–471. 10.1016/j.jconrel.2021.07.042 (2021). 10.1016/j.jconrel.2021.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinharay, S. et al. In vivo imaging of sterile microglial activation in rat brain after disrupting the blood–brain barrier with pulsed focused ultrasound: [18F]DPA-714 PET Study. J. Neuroinflammation16(1), 155. 10.1186/s12974-019-1543-z (2019). 10.1186/s12974-019-1543-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poon, C., Pellow, C. & Hynynen, K. Neutrophil recruitment and leukocyte response following focused ultrasound and microbubble mediated blood–brain barrier treatments. Theranostics11(4), 1655–1671. 10.7150/thno.52710 (2021). 10.7150/thno.52710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchetti, L. & Engelhardt, B. Immune cell trafficking across the blood-brain barrier in the absence and presence of neuroinflammation. Vasc. Biol.2(1), H1–H18. 10.1530/VB-19-0033 (2020). 10.1530/VB-19-0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feshitan, J. A., Chen, C. C., Kwan, J. J. & Borden, M. A. Microbubble Size isolation by differential centrifugation. J. Colloid Interface Sci.329(2), 316–324. 10.1016/j.jcis.2008.09.066 (2009). 10.1016/j.jcis.2008.09.066 [DOI] [PubMed] [Google Scholar]
- 47.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15(12), 550. 10.1186/s13059-014-0550-8 (2014). 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korotkevich, G., Sukhov, V., Budin, N., Shpak, B., Artyomov, M. N. & Sergushichev, A. Fast Gene Set Enrichment Analysis; preprint; Bioinformatics, 2016. 10.1101/060012.
- 49.Liberzon, A. et al. The molecular signatures database hallmark gene set collection. Cell Syst.1(6), 417–425. 10.1016/j.cels.2015.12.004 (2015). 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian, A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A.102(43), 15545–15550. 10.1073/pnas.0506580102 (2005). 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.)57(1), 289–300 (1995). 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 52.Alli, S. et al. Brainstem blood brain barrier disruption using focused ultrasound: A demonstration of feasibility and enhanced doxorubicin delivery. J. Controll. Releas.281, 29–41. 10.1016/j.jconrel.2018.05.005 (2018). 10.1016/j.jconrel.2018.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arvanitis, C. D. et al. Mechanisms of enhanced drug delivery in brain metastases with focused ultrasound-induced blood-tumor barrier disruption. Proc. Natl. Acad. Sci. U.S.A.10.1073/pnas.1807105115 (2018). 10.1073/pnas.1807105115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bing, C. et al. Characterization of different bubble formulations for blood–brain barrier opening using a focused ultrasound system with acoustic feedback control. Sci. Rep.8(1), 7986. 10.1038/s41598-018-26330-7 (2018). 10.1038/s41598-018-26330-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaves, C. et al. Characterization of the blood–brain barrier integrity and the brain transport of SN-38 in an orthotopic xenograft rat model of diffuse intrinsic pontine glioma. Pharmaceutics12(5), 399. 10.3390/pharmaceutics12050399 (2020). 10.3390/pharmaceutics12050399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chu, P.-C. et al. Focused ultrasound-induced blood-brain barrier opening: Association with mechanical index and cavitation index analyzed by dynamic contrast-enhanced magnetic-resonance imaging. Sci. Rep.6(1), 1–13 (2016). 10.1038/srep33264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McDannold, N., Vykhodtseva, N. & Hynynen, K. Blood–brain barrier disruption induced by focused ultrasound and circulating preformed microbubbles appears to be characterized by the mechanical index. Ultrasound Med. Biol.34(5), 834–840. 10.1016/j.ultrasmedbio.2007.10.016 (2008). 10.1016/j.ultrasmedbio.2007.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arsiwala, T. A. et al. Characterization of passive permeability after low intensity focused ultrasound mediated blood–brain barrier disruption in a preclinical model. Fluids Barriers CNS19(1), 72. 10.1186/s12987-022-00369-1 (2022). 10.1186/s12987-022-00369-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mathew, A. S., Gorick, C. M. & Price, R. J. Multiple regression analysis of a comprehensive transcriptomic data assembly elucidates mechanically- and biochemically-driven responses to focused ultrasound blood–brain barrier disruption. Theranostics11(20), 9847–9858. 10.7150/thno.65064 (2021). 10.7150/thno.65064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mathew, A. S. et al. Transcriptomic response of brain tissue to focused ultrasound-mediated blood–brain barrier disruption depends strongly on anesthesia. Bioeng. Transl. Med.6(2), e10198. 10.1002/btm2.10198 (2021). 10.1002/btm2.10198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun, T. et al. Acoustic cavitation-based monitoring of the reversibility and permeability of ultrasound-induced blood–brain barrier opening. Phys. Med. Biol.60(23), 9079–9094. 10.1088/0031-9155/60/23/9079 (2015). 10.1088/0031-9155/60/23/9079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi, H. J. et al. The new insight into the inflammatory response following focused ultrasound-mediated blood–brain barrier disruption. Fluids Barriers CNS19(1), 103. 10.1186/s12987-022-00402-3 (2022). 10.1186/s12987-022-00402-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu, X., Li, J., Fu, M., Zhao, X. & Wang, W. The JAK/STAT Signaling pathway: From bench to clinic. Sig. Transduct. Target Ther.6(1), 402. 10.1038/s41392-021-00791-1 (2021). 10.1038/s41392-021-00791-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jain, M. et al. Role of JAK/STAT in the neuroinflammation and its association with neurological disorders. Ann. Neurosci.28(3–4), 191–200. 10.1177/09727531211070532 (2021). 10.1177/09727531211070532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fan, C.-H. et al. Detection of intracerebral hemorrhage and transient blood-supply shortage in focused-ultrasound-induced blood–brain barrier disruption by ultrasound imaging. Ultrasound Med. Biol.38(8), 1372–1382. 10.1016/j.ultrasmedbio.2012.03.013 (2012). 10.1016/j.ultrasmedbio.2012.03.013 [DOI] [PubMed] [Google Scholar]
- 66.Tsai, H.-C. et al. Safety Evaluation of frequent application of microbubble-enhanced focused ultrasound blood–brain-barrier opening. Sci. Rep.8(1), 17720. 10.1038/s41598-018-35677-w (2018). 10.1038/s41598-018-35677-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kovacs, Z. I. et al. MRI and histological evaluation of pulsed focused ultrasound and microbubbles treatment effects in the brain. Theranostics8(17), 4837–4855. 10.7150/thno.24512 (2018). 10.7150/thno.24512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chu, P.-C. et al. Neuromodulation accompanying focused ultrasound-induced blood–brain barrier opening. Sci. Rep.5(1), 15477. 10.1038/srep15477 (2015). 10.1038/srep15477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu, H.-L. et al. Low-pressure pulsed focused ultrasound with microbubbles promotes an anticancer immunological response. J. Transl. Med.10(1), 221. 10.1186/1479-5876-10-221 (2012). 10.1186/1479-5876-10-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Curley, C. T., Sheybani, N. D., Bullock, T. N. & Price, R. J. Focused ultrasound immunotherapy for central nervous system pathologies: Challenges and opportunities. Theranostics7(15), 3608–3623. 10.7150/thno.21225 (2017). 10.7150/thno.21225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bathini, P. et al. Acute Effects of focused ultrasound-induced blood–brain barrier opening on anti-pyroglu3 abeta antibody delivery and immune responses. Biomolecules12(7), 951. 10.3390/biom12070951 (2022). 10.3390/biom12070951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leinenga, G. & Götz, J. Scanning ultrasound removes amyloid-β and restores memory in an Alzheimer’s disease mouse model. Sci. Transl. Med.7(278), 278ra33. 10.1126/scitranslmed.aaa2512 (2015). 10.1126/scitranslmed.aaa2512 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that support the findings of this study have been deposited in the NIH BioProject Archive with the primary accession code PRJNA1100543. Other data are available upon request.