Abstract
Toxicity and immunity associated with adenovirus backbone gene expression is an important hurdle to overcome for successful gene therapy. Recent efforts to improve adenovirus vectors for in vivo use have focused on the sequential deletion of essential early genes. Adenovirus vectors have been constructed with the E1 gene deleted and with this deletion in combination with an E2a, E2b, or E4 deletion. We report here a novel vector (Av4orf3nBg) lacking E1, E2a, and all of E4 except open reading frame 3 (ORF3) and expressing a β-galactosidase reporter gene. This vector was generated by transfection of a plasmid carrying the full-length vector sequence into A30.S8 cells that express E1 and E2a but not E4. Production was subsequently performed in an E1-, E2a-, and E4-complementing cell line. We demonstrated with C57BL/6 mice that the Av4orf3nBg vector effected gene transfer with an efficiency comparable to that of the Av3nBg (wild-type E4) vector but that the former exhibited a higher level of β-galactosidase expression. This observation suggests that E4 ORF3 alone is able to enhance RNA levels from the β-galactosidase gene when the Rous sarcoma virus promoter is used to drive transgene expression in the mouse liver. In addition, we observed less liver toxicity in mice injected with the Av4orf3nBg vector than those injected with the Av3nBg vector at a comparable DNA copy number per cell. This study suggests that the additional deletion of E4 in an E1 and E2a deletion background may be beneficial in decreasing immunogenicity and improving safety and toxicity profiles, as well as increasing transgene capacity and expression for liver-directed gene therapy.
Adenovirus vectors are currently used for gene transfer in basic research and gene therapy protocols due to their high titers, ability to target a wide range of both dividing and nondividing cells, and mediation of high-level foreign protein expression without replication or integration of the viral genome (4, 18, 36, 37). In spite of this, several factors significantly limit the utility of currently used adenovirus vectors. Data obtained from murine and other animal models have shown that host immune responses to viral and transgene protein products are responsible for eliminating transduced cells and preventing readministration (9, 20–22, 41–44). Progress toward the improvement of these vectors involves strategies to reduce immunogenicity and toxicity associated with viral backbone gene expression.
Two approaches have been taken to disable adenoviral vectors. One is based on the concept that removal of all coding sequences for viral proteins from the vector backbone renders the vector less immunogenic and restricts the immune response to the injected viral capsid proteins. This gutted adenovector (7, 15, 24, 27, 30, 31, 34) is propagated in the presence of helper adenovirus. A second alternative in decreasing adenovirus vector immunogenicity and toxicity is based on a gene attenuation strategy wherein the sequential removal of key early genes is anticipated to reduce expression from other essential genes. This type of vector grows in packaging cell lines that complement the deleted viral genes. Adenovirus vectors have been constructed with the E1 gene deleted (28, 36, 37, 45) and with E1 plus E2a, E2b, or E4 deleted (1, 2, 6, 10, 13, 14, 16, 17, 28, 40). Studies carried out in vitro to evaluate adenovirus vectors with double deletions consistently feature an absence of detectable replication and late gene expression (1, 17). Recently, we extended those studies by semiquantifying the levels of early and late transcripts in an adenovector with a double deletion of E1 and E2a in vitro and in vivo. An analysis of RNA-specific PCR-amplified fragments showed that expression of adenoviral major late gene products was minimal. In contrast, early promoters such as E4, pIX, and E2b were found to actively express high levels of gene transcripts (25).
This molecular evaluation suggests that further elimination of early genes is required to attenuate viral gene expression. Recently, it has been demonstrated that additional deletion of the E4 region in an E1 deletion background has a major influence on the stability of the adenovirus vector genome in addition to prolonging transgene expression (2, 10, 16, 40). The E4 region, which contains seven open reading frames (ORFs), is essential for virus growth, DNA replication, and particle assembly (reviewed in reference 26). In particular, the E4 region encodes E4 ORF3 and E4 ORF6, which share functions required for mRNA splicing and accumulation (5).
In this article we describe the construction of a novel vector (Av4orf3nBg) lacking E1, E2a, and all of E4 except ORF3 and expressing a β-galactosidase reporter gene. This vector was generated by transfection of a plasmid bearing the entire modified vector genome into an A549-based cell line that complements E1 and E2a (17). Production was subsequently performed in an E1-, E2a-, and E4-complementing cell line. To determine the effect of E4 viral gene deletion on vector transduction and liver toxicity, similar doses of either Av4orf3nBg or Av3nBg (with E1 and E2a deleted) (17) were administered to C57BL/6 mice. We demonstrate that the Av4orf3nBg vector exhibits gene transfer with an efficiency comparable to that of the Av3nBg vector but that the former exhibits a higher level of β-galactosidase expression. In addition, we observed less liver toxicity in mice injected with Av4orf3nBg than in mice injected with Av3nBg at comparable DNA copy numbers per cell. This study suggests that the additional deletion of E4 may be useful in limiting cytopathic effects, increasing transgene capacity, and increasing expression over levels expressed by double-deletion vectors in the context of liver-directed gene therapy.
MATERIALS AND METHODS
Plasmid construction.
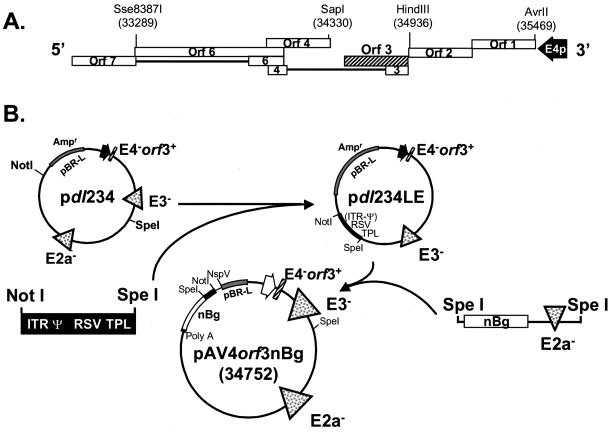
The recombinant adenovirus vector with E1, E2a, and all of E4 except ORF3 deleted, Av4orf3nBg, expressing a nucleus-targeted β-galactosidase reporter was generated with a single plasmid, pAv4orf3nBg, containing the entire adenoviral genome sequence within a pBR-L backbone (Fig. 1). pBR-L is identical to pBR322, except that it is missing all nucleotide sequences between NdeI (position 2297) and EcoRI (position 4286). A multiple-cloning site was created between NdeI and EcoRI by insertion of a DNA linker. This multiple-cloning site contains sites for EcoRI, NspV, ClaI, BamHI, SalI, BclI, PacI, and NdeI. Previous cloning had generated a plasmid in pBR-L that contained the 3′ end of the adenovirus type 5 genome (bp 21562 to 35936) lacking the E2a and E3 regions (17). This plasmid was extended further to contain an additional segment of the adenoviral genome by incorporating sequences from the BamHI site (bp 21562) back to the SalI site (bp 16746), resulting in a plasmid, pdl23. The modification to the E4 region was created by deleting the AvrII (bp 35469)-Sse8387 (bp 33289) fragment (removing all of the E4 coding region except for the E4 promoter) from a plasmid designated pREpac (Fig. 1A). This plasmid contains the right end of adenovirus type 5 dl327 (including the E3 XbaI deletion) from the SnaBI site (bp 25171) to the end of the right inverted terminal repeat (ITR), which was ligated between the SmaI and HindIII sites of pBluescript SK(+) (Stratagene). A PacI site (TTAATTAA) was engineered 152 bp internal to the end of the right ITR by insertion of a 4-bp sequence, TTAA, adjacent to the TTAA sequence already present at this location. The HindIII (bp 34936)-SapI (bp 34330) fragment containing the E4 ORF3 was then cloned into the T4 DNA polymerase blunt-ended AvrII-Sse8387 site to create pREpac+orf3. The wild-type E4 region was removed from pdl23 and replaced by an XbaI-PacI fragment from pREpac+orf3, resulting in plasmid pdl234 (Fig. 1B). An intermediate cloning vector, pdl234LE, was created by ligating the NotI-SpeI fragment from pdl234 (retaining the pBR-L backbone and the E4-modified region) to the left end of the shuttle plasmid pAvS6a (35), to yield a hybrid containing the left ITR, packaging signal (Ψ), Rous sarcoma virus (RSV) promoter, tripartite leader (TPL) nucleotide sequences, regions lacking E3 and E4, and right ITR (Fig. 1B). The resulting SpeI site was then used to insert the 25,633-bp SpeI fragment, derived from the Av3nBg vector, containing the β-galactosidase gene and the region lacking E2a (17). The final construct containing the entire Av4orf3nBg genome in a pBR-L backbone is called pAv4orf3nBg (Fig. 1B).
FIG. 1.
Schematic representation of the adenovirus type 5 E4 region and construction of plasmids for generating the Av4orf3nBg vector. (A) ORFs of the E4 region are indicated by open boxes. ORF3 is represented by a hatched bar. (B) Construction of pAv4orf3nBg. pdl23 is a plasmid that contains the 3′ end of the adenovirus type 5 genome (bp 16746 to 35936) with deletions in the E2a and E3 regions (17). A PacI-XbaI fragment from pdl23 was replaced with a PacI-XbaI fragment containing the E4 promoter and the ORF3 gene (bp 34936 to 34330), generating pdl234. The NotI-SpeI fragment from pdl234 was replaced with a NotI-SpeI fragment derived from pAvS6a (35) to generate pdl234LE, which contains the left-end ITR, Ψ, RSV promoter, TPL, regions with E3 and E4 deleted, and right ITR. The single infectious plasmid, pAv4orf3nBg, was generated by inserting an SpeI fragment of 25,633 bp flanking the nucleus-localizing β-galactosidase reporter transgene (nBg) gene and the region with E2a deleted. E4p, E4 promoter; RSV, heterologous RSV promoter.
Generation and propagation of the Av4orf3nBg recombinant adenovirus.
The Av4orf3nBg vector was generated by cotransfecting 1 μg of NspV-linearized pAv4orf3nBg and 1 μg of pREpac plasmids with Lipofectamine (GIBCO-BRL, Gaithersburg, Md.) into 0.3 μM dexamethasone-induced AE1-2a cells. AE1-2a is a stable cell line, derived from A549 cells, which contains the adenovirus type 5 E1 and E2a genes inducibly expressed from separate glucocorticoid-responsive promoters (17). Cell cultures were maintained in Richter’s CM medium (BioWhittaker) supplemented with 5% fetal bovine serum. Transfected cells were lysed after 7 days by five cycles of freezing and thawing, and the lysate was used to infect a fresh, dexamethasone-induced monolayer of A70.S54 cells (first amplification), which became available after the initial transfection. A70.S54 is a stable cell line derived from AE1-2a cells which additionally expresses adenovirus type 5 E4 genes (this cell line will be described elsewhere). After 7 days, the infected cells were lysed and the crude viral lysate (CVL) was used to isolate individual plaques for analysis.
Virus production and purification.
CVL from infected A70.S54 cells was used to infect a fresh, induced monolayer of A70.S54 cells. After 14 days, individual plaques representing Av4orf3nBg were recovered and virus was purified. This plaque-purified material was subsequently used to prepare large-scale stocks of virus in A70.S54 cells. The vector Av3nBg (lacking E1 and E2a) was propagated and purified as previously described (17). Both vectors used in this study lacked E3, a region not required for viral replication, and contained the same β-galactosidase expression cassette.
Structural characterization of the Av4orf3nBg vector.
Adenovector DNA, obtained from CVLs of purified and amplified plaques, was analyzed by HindIII restriction enzyme digestion. The resulting fragments were fractionated by agarose gel electrophoresis and visualized with ethidium bromide. Additionally, DNAs obtained from CVL of A70.S54 cells infected with Av3nBg or Av4orf3nBg and DNA from plasmid pAv4orf3nBg were digested with the HindIII restriction enzyme, fractionated on a 1% agarose gel, transferred to a Zeta-Probe GT nylon membrane, and hybridized with 32P-labeled DNA probes containing sequences specific for E4 ORF3, E4 ORF6, or the entire viral genome (HindIII-digested pAv4orf3nBg DNA probe).
In vitro gene transfer efficiencies.
The Av4orf3nBg vector was compared to the Av3nBg vector for its ability to transfer and express a β-galactosidase-encoding transgene in A549 cells in vitro. To ensure that equal amounts (equivalent to 10 particles per cell) of each vector were used for infection, the two adenovirus vectors were quantified by determining optical density at 260 nm (29). Cells were recovered 48 h after gene transfer by scraping them into a lysis buffer, and β-galactosidase activity was determined by the Tropix GalactoLight chemiluminescent-reporter assay.
Animal studies.
Five- to six-week-old female C57BL/6 mice purchased from Harlan-Sprague-Dawley were used. Mice (between four and six per group) were injected via the tail vein with 200 μl of a low or high dose of the Av3nBg or Av4orf3nBg vector (6 × 1010 or 3 × 1011 particles per mouse, respectively). Blood was obtained at the time points indicated in Fig. 8 for determination of levels of alanine aminotransferase (ALT) in sera. Animals were sacrificed at 7 days, and the livers obtained were utilized for hematoxylin-eosin, β-galactosidase, Southern blot, and Northern blot analyses.
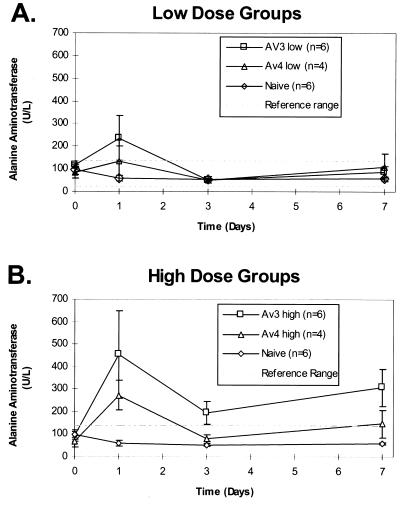
FIG. 8.
Comparison of levels of liver injury between mice infected with the Av3nBg vector and those infected with the Av3orf3nBg vector. Serum samples from mice infused with a low dose (A) or a high dose (B) of the Av3nBg or Av4orf3nBg vector were harvested at the indicated time points and analyzed for ALT concentrations. Numbers in parentheses are the numbers of animals per point. For reference, the average normal levels of ALT are shown.
Histological analysis.
Liver tissue sections were fixed in 10% neutral buffered Formalin for 24 h, processed by routine methods on a Sakura VIP tissue processor, and embedded in paraffin, followed by hematoxylin and eosin staining.
In vivo gene transfer efficiencies.
Immunohistochemical staining of β-galactosidase was performed by the avidin-biotin immunoperoxidase method. Briefly, sections were deparaffinized, rehydrated, and pretreated with 0.3% hydrogen peroxide solution for 30 min at room temperature (RT) to quench endogenous peroxidase activity. The pretreated sections were then incubated with 2% normal goat serum for 30 min at RT. After being blocked, the slides were incubated with a polyclonal antibody to β-galactosidase (Cortex Biochem, Inc.) diluted 1:2,000 in phosphate-buffered saline (PBS) for 1 h at RT, rinsed in PBS, and then incubated with a biotinylated goat anti-rabbit antibody for 30 min at RT. Sections were then washed in PBS and incubated with avidin DH-biotinylated horseradish peroxidase H (Vectastain Elite ABC kit) for 30 min at RT. After a PBS wash, the reaction was visualized with diaminobenzidine tetrahydrochloride as the peroxidase substrate. Sections were counterstained with hematoxylin and coverslipped. In addition, liver tissues were processed for quantification of β-galactosidase activity by following the protocol used for in vitro samples.
Southern blotting.
Total genomic DNA was isolated from mouse liver with a QIAmp tissue kit (Qiagen) and further incubated with RNase. DNA concentrations were determined spectrophotometrically. A total of 10 μg of each DNA sample was digested with BamHI and subjected to electrophoresis on a 0.8% agarose gel, stained with ethidium bromide, and transferred to a Zeta-Probe GT membrane (Bio-Rad). The copy number control standards were prepared by adding 600 and 60 pg of pAv4orf3nBg plasmid DNA, equivalent to 10 and 1 vector copies per cell, respectively, to 10 μg of BamHI-digested genomic DNA from an uninfected animal. The 32P-labeled probe, prepared by random oligonucleotide priming, contained β-galactosidase cDNA sequences. The relative amount of DNA was determined by PhosphorImager analysis as the ratio between sample and control signals.
Northern blot analysis.
Total RNA was extracted from liver tissue by the RNAzol B method (Teltest, Inc.). RNA concentration was quantified by determining the A260. Total RNA (15 μg) was electrophoresed through a 0.8% agarose–7% formaldehyde gel, transferred to nylon membrane, and bound to the filter by UV cross-linking. A DNA probe was radiolabeled by random oligonucleotide priming of a lacZ cDNA template. The band intensities were quantified with a Molecular Dynamics PhosphorImager SF.
RESULTS
Construction of Av4orf3nBg plasmid.
Av4orf3nBg, a new-generation vector lacking E1, E2a, and all of E4 except ORF3 and expressing a nucleus-targeted β-galactosidase reporter, was constructed in several steps. E4 ORF3 was maintained in the vector backbone for two reasons. First, we had a cell line that complemented only E1 and E2a functions (17) and transient-transfection experiments with 293 cells demonstrated that expression of E4 ORF3 alone could support growth of a virus with E4 deleted, dl1011 (5), although not as well as a virus with the entire E4. Second, previous studies suggested that E4 gene products helped maintain vector transgene expression (2, 6, 10).
In developing the next phase of construction, we took advantage of an observation made by other investigators, namely, that an entire adenovirus genome could be propagated as a bacterial plasmid. After release from the plasmid by restriction digestion and transfection into cells, the cloned viral genome could produce infectious virus (19, 39). Our strategy is outlined in Fig. 1. The NotI-SpeI fragment (1,020 bp) from plasmid pAvS6a containing the 5′ terminus, ITR, packaging signal, RSV promoter, and TPL was subcloned into pdl234 to produce pdl234LE, which contained the E3 and E4 deletions, as well as the right ITR. To generate the final plasmid, pAv4orf3nBg, pdl234LE was cut with SpeI and ligated with a SpeI fragment of 25,633 bp prepared from the Av3nBg adenoviral vector (17) that carried the β-galactosidase reporter gene, major late promoter/L4 elements, and the E2a deletion. High yields of unrearranged pAv4orf3nBg were obtained with Escherichia coli Stbl2 (GIBCO-BRL).
Generation of the Av4orf3nBg vector.
pAv4orf3nBg linearized with NspV was cotransfected with pREpac (E4+) into dexamethasone-induced AE1-2a cells (17). Plasmid pREpac was used to provide E4 proteins during virus replication in the E1- and E2a-complementing cell line. The cells were incubated for 14 days, and the resulting CVL was used to infect A70.S54 cells (which complement E1, E2A, and E4). When all the cells demonstrated cytopathic effects, virus was harvested and a stock was grown by reinfecting fresh A70.S54 cells. The vector growth was efficient, resulting in approximately 700 viral particles/cell with purified vector stocks in the range of 2 × 1011 to 4 × 1011 particles/ml as quantified by determining the optical density at 260 nm.
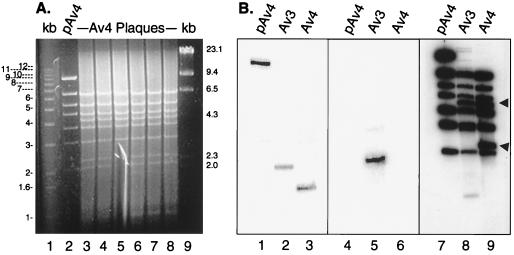
The genome of Av4orf3nBg (32,083 bp) was structurally characterized by restriction enzyme digestion and Southern blot analysis (Fig. 2). HindIII digestion provided the expected fragment pattern for the entire genome in six independent adenovirus vector isolates (Fig. 2A, lanes 3 to 8). The same fragments were visualized by Southern blot analysis (Fig. 2B, lane 9); of these, the 4.2- and 2.3-kb fragments (lane 9, arrows) are characteristic of the adenoviral genome compared to that of the plasmid DNA and represent the ends of the adenovirus vector. In the plasmid DNA these fragments were not observed; instead, an expected plasmid fragment of 9.2 kb was detected (Fig. 2B, lane 7). This restriction pattern difference suggests that after transfection, the plasmid DNA was able to serve as the template and package the viral genome without carryover of extra nucleotides.
FIG. 2.
Structural characterization of the Av4orf3nBg vector. (A) Adenovector DNAs obtained from crude lysates of purified and amplified adenoviral plaques were analyzed by HindIII restriction enzyme digestion. The DNA fragment products were fractionated by agarose gel electrophoresis and visualized with ethidium bromide (lanes 3 to 8). The HindIII-digested plasmid pAv4orf3nBg (pAv4) was included as a control (lane 2). The sizes of marker DNA fragments (1-kb ladder and λ-HindIII; GIBCO-BRL) are indicated (lanes 1 and 9). Av4, cells infected with the Av4orf3nBg vector. (B) Adenovector DNA obtained from crude lysates of cells infected with the Av3nBg (Av3) or Av4orf3nBg (Av4) vector and DNA from plasmid pAv4 were digested with the HindIII restriction enzyme, fractionated on a 1% agarose gel, transferred to a nylon filter, and hybridized with a 32P-labeled ORF3 DNA probe (lanes 1 to 3), a 32P-labeled ORF6 DNA probe (lanes 4 to 6), or a 32P-labeled HindIII-digested pAv4 probe (lanes 7 to 9). Arrows indicate fragments representing the left and right ends of the adenovirus vector.
Evaluation of the E4 region of Av4orf3nBg by PCR (data not shown) and by Southern blotting confirmed the expected deletion. Thus, the HindIII restriction digestion gave the expected 2.3-kb fragment with Av4orf3nBg (Fig. 2B, lane 9), rather than the 2.3- and 1.0-kb fragments corresponding to the wild-type E4 region present in the Av3nBg vector (Fig. 2B, lane 8).
Because the transfection contained both the pAv4orf3nBg and the pREpac plasmid (which contains the entire E4 region) there was the potential of incorporating this entire region in the Av4orf3nBg vector by recombination. Thus, a further Southern blot analysis of the E4 region was carried out after HindIII digestion with probes specific for E4 ORF3 and E4 ORF6 nucleotide sequences. In agreement with the previous results, the E4 ORF3 probe hybridized with the 9.2-kb fragment of pAv4orf3nBg, the 2.9-kb fragment of Av3nBg, and the 2.3-kb fragment of Av4orf3nBg (Fig. 2B, lanes 1, 2, and 3, respectively). As predicted, the E4 ORF6 probe recognized only the 2.9-kb fragment of the Av3nBg vector (Fig. 2B, lane 5). Altogether, these results demonstrated that the E4 region of Av4orf3nBg contained only E4 ORF3 and that the adenovector was free of any wild-type E4 sequences. In addition, HindIII fragments of 6.5 and 3.7 kb were observed in both the Av3nBg (Fig. 2B, lane 8) and Av4orf3nBg (Fig. 2B, lane 9) vectors; these fragments verified the E2a deletion (6.5 kb) and the E3 deletion (3.7 kb). PCR evaluation of the E2a and E3 regions also confirmed the expected deletions (data not shown).
Quantitative analysis of in vitro β-galactosidase expression.
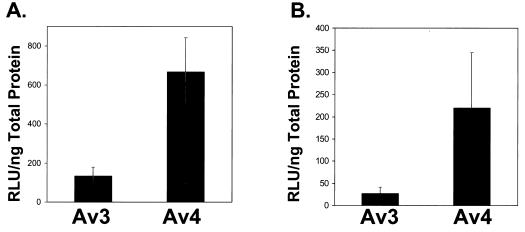
A remarkable feature that emerged from infection with the adenoviral vectors with E1 and E4 deleted was that expression of a reporter gene under the control of the cytomegalovirus (CMV) or RSV promoter was influenced by E4 protein expression (2, 6, 10). Thus, a quantitative β-galactosidase assay was used to evaluate transgene expression in a noncomplementing A549 cell background, by comparing the β-galactosidase expression of the Av3nBg vector (E4+) to that of the Av4orf3nBg vector. Under the conditions used in this experiment (10 particles/cell) there was about fivefold more β-galactosidase detected in cells transduced with Av4orf3nBg than in those transduced with Av3nBg (Fig. 3A).
FIG. 3.
Av4orf3nBg vector gene transfer efficiency in vitro (A) and in vivo (B). (A) Noncomplementing A549 cells were infected with either the Av3nBg (Av3) or the Av4orf3nBg (Av4) vector at equivalent doses of 10 particles/cell based on the optical density measurements of the adenoviral vectors at 260 nm. β-Galactosidase activity in cell lysates was determined 48 h postinfection by the Tropix GalactoLight chemiluminescent-reporter assay. The results are expressed as relative light units (RLU) of β-galactosidase activity per nanogram of total cellular protein. The data are means ± standard deviations of three separate determinations for three different production lots of each vector type. (B) Livers of C57BL/6 mice infected with a low dose of Av3nBg or Av4orf3nBg were evaluated quantitatively for β-galactosidase expression at 7 days postinjection by the same procedure described for panel A. The data are means ± standard deviations of three separate determinations with three different animals.
Recombinant adenovirus-mediated transduction and transgene expression in mouse liver.
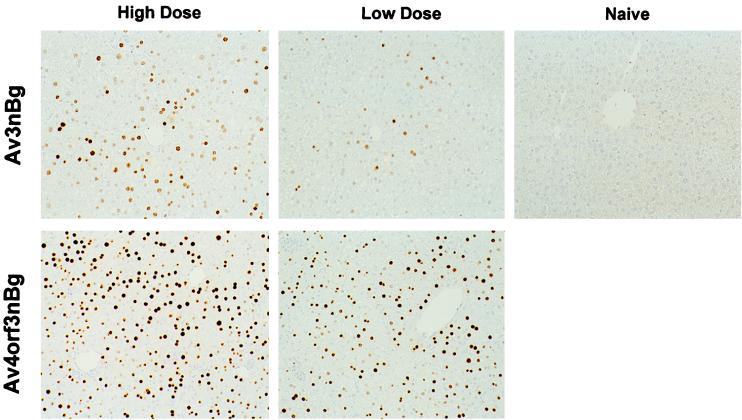
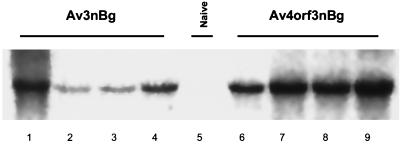
Based on the previous results that E4 ORF3 influences the activity of the RSV promoter in vitro, we wanted to determine if E4 ORF3 would have a similar effect in vivo. To determine the profiles of transgene expression, paired low and high doses of Av4orf3nBg and Av3nBg (6 × 1010 and 3 × 1011 particles, respectively) were administered to C57BL/6 mice by tail vein injection. At 7 days postinjection, animals were sacrificed and liver tissues were analyzed for vector content. We detected, using Southern blot analysis that probed for the β-galactosidase gene, similar DNA copy numbers in animals receiving either the Av3nBg or the Av4orf3nBg vector (Fig. 4). Similarly, the efficiencies of transduction based on β-galactosidase staining of liver sections were not significantly different for any of the adenoviral vectors tested when equivalent numbers of particles were used. Only the intensity of the staining increased in cells transduced with the Av4orf3nBg vector (Fig. 5). In fact, a quantification of RSV-promoted β-galactosidase expression from the Av4orf3nBg vector showed a fivefold increase compared to the expression from cells infected with the Av3nBg vector (Fig. 3B). Using Northern blot analysis, we then examined whether the β-galactosidase expression in vivo was directly correlated to the amount of β-galactosidase mRNA present in liver tissues (Fig. 6). Mice that received the Av4orf3nBg vector showed on average a fivefold increase in transgene transcription accumulation compared with that of mice receiving the Av3nBg vector. Together, these results support the concept that E4 ORF3 modulates expression of the RSV promoter and further suggest that a gene product(s) of E4 other than ORF3 may repress RSV promoter activity in vivo.
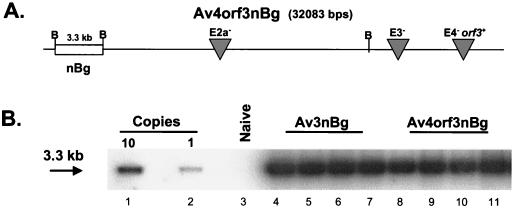
FIG. 4.
In vivo detection of Av3nBg and Av4orf3nBg transgene sequences. (A) Av4orf3nBg DNA is represented schematically. Shown are the locations of the E2a, E3, and E4 deletions and of the heterologous nucleus-localizing β-galactosidase reporter transgene as well as its BamHI digestion pattern. (B) Total liver DNA of C57BL/6 mice infected with a high dose (3 × 1011 viral particles) of the infectious Av3nBg (lanes 4 to 7) or Av4orf3nBg (lanes 8 to 11) vector was isolated on day 7 following gene transfer and digested with BamHI. The Southern blot was probed with a BamHI 32P-labeled fragment encoding the β-galactosidase gene. Lanes 1 and 2 contain pAv4orf3nBg plasmid DNAs equivalent to 10 copies and 1 copy per cell, respectively. The control sample included liver DNA of naive mice (lane 3).
FIG. 5.
Recombinant adenovirus-mediated transgene expression in mouse liver. A low or high dose of the Av4orf3nBg or Av3nBg vector (6 × 1010 or 3 × 1011 viral particles, respectively) were injected into the tail vein of C57BL/6 mice. Seven days postinjection animals were sacrificed and liver tissues were evaluated for β-galactosidase expression by immunohistochemical staining.
FIG. 6.
In vivo detection of Av3nBg and Av4orf3nBg transgene activities. Total liver RNA of C57BL/6 mice infected with a high dose (3 × 1011 viral particles) of the infectious Av3nBg (lanes 1 to 4) or Av4orf3nBg (lanes 6 to 9) vector was isolated on day 7 following gene transfer, transferred to a nylon membrane, and hybridized with a 32P-labeled lacZ probe. The control sample included liver RNA extracted from a naive mouse (lane 5).
Reduced mouse liver toxicity with the Av4orf3nBg vector.
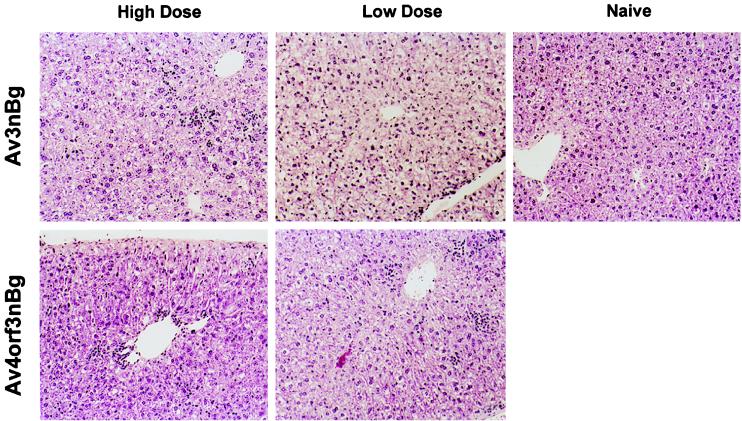
Previous analysis of early gene transcripts in vitro and in vivo indicated that the E4 promoter was functional in an Av3nBg vector lacking E1 and E2a. The hypothesis in the present study was that an additional E4 deletion in the Av3nBg vector would impart lowered expression of early and late viral proteins and further attenuate viral gene expression; the result could be to improve the in vivo utility of this vector by reducing toxicity and the host immune response and increasing the duration of transgene expression. Histochemical analysis of hematoxilin- and eosin-stained liver sections of mice 7 days postinjection of high and low doses of Av3nBg and Av4orf3nBg are shown in Fig. 7. In a dose-dependent manner, Av3nBg caused vacuolization, nuclear pleomorphism, and a loss of tissue architecture. These histopathologic changes were not observed with Av4orf3nBg. However, both vectors produced mild inflammatory changes characterized by mononuclear cell infiltrates.
FIG. 7.
Pathological response in liver following Av3nBg or Av4orf3nBg vector administration. A low or high dose of the Av4orf3nBg or Av3nBg vector (6 × 1010 or 3 × 1011 viral particles, respectively) was injected into the tail veins of mice. Morphological observation of liver sections prepared at 7 days postinjection was carried out after hematoxylin and eosin staining. For consistency, liver samples from the same mice used for the samples in Fig. 4 are shown here.
The toxicities of both Av3nBg and Av4orf3nBg were also quantified by determining levels of ALT in serum. Both vectors at low and high doses resulted in elevation of the ALT levels at 24 h, with a subsequent decrease by day 3 (Fig. 8). With the higher vector dose of Av3nBg the ALT levels increased significantly above those of uninjected control animals at day 7 (Fig. 8B). In contrast, ALT levels in Av4orf3nBg-injected animals were not different from those in controls at day 7.
DISCUSSION
Based on previous studies demonstrating that the E4 promoter is still active in an E1 and E2a deletion vector (25), we reasoned that an additional E4 deletion would further attenuate viral gene expression and toxicity compared to levels produced by double-deletion vectors. Thus, six of the seven ORFs were deleted from the E4 region, with the exception of ORF3. This ORF was chosen for its ability to functionally complement a complete E4 region deletion such as that in the dl1011 mutant virus (5). A recombinant adenovirus vector with E1, E2a, and E4 deletions and expressing a nucleus-targeted β-galactosidase reporter gene was generated with a single plasmid system to transfect an A549-based cell line that expresses E1 and E2a genes. Several reports have described the use of a plasmid containing a copy of the adenovirus genome to rescue the virus after transfection (e.g., see references 19 and 39). In these previous cases, plasmids were linearized before transfection to release an adenovirus genome as an intact linear molecule with exposed ITRs. In the present study, we also linearized the plasmid, but the right and left ITRs were flanked by 2.0 and 0.5 kb, respectively, of extra nucleotides. Virus production was subsequently performed in a triple-complementation (E1, E2a, and E4) cell line, in which a viral titer similar to those obtained with Av3-type vectors (17) was achieved. DNA analysis performed on six randomly selected amplified virus plaques and on total crude lysates confirmed that the cloned viral genome was able to generate a stable and full-length genome containing the appropriate deleted regions. The fact that no rearrangements in DNA structure were observed between virus stock and virus plaques suggests that little or no detectable sequence instability is associated with this procedure. This and other reports clearly demonstrate that this approach to generating adenoviral vectors is faster and less tedious than the more traditional methods which rely on homologous recombination in situ. The level and duration of in vivo transgene expression measured from the RSV- and CMV-promoted expression cassette in the adenovirus vectors appears to correlate with the structure of the vector backbone. An interesting observation was made with adenovirus vectors lacking E1 and E4 in which the presence of E4 gene products seemed to regulate the expression from both RSV and CMV promoters (2, 6, 10). Transgene expression was observed only with a vector that had a wild-type E4 region and not with vectors that contained either a complete E4 deletion or a deletion of all of E4 except ORF6 (2). Our results with the Av4orf3nBg vector indicate that ORF3 alone is able to enhance expression of β-galactosidase when the RSV promoter is used to drive transgene expression in the mouse liver. We observed that the level of β-galactosidase expression was higher in animals receiving the Av4orf3nBg than in those receiving the Av3nBg (wild-type E4) vector. These observations suggest that an E4 product(s) competes with ORF3 to affect expression or the accumulation of β-galactosidase in vivo. The molecular basis for ORF3 activation must await further evaluation; however, preliminary analysis of β-galactosidase mRNA accumulation suggests that ORF3 may act either directly or indirectly on the activity of the RSV promoter. Both E1-E2a and E1-E4 double-deletion vectors have lower toxicity profiles than a first-generation vector for liver-directed gene therapy. Studies performed with murine models show reduced liver damage as determined by serum transaminase levels. Based on the results of our experiment, we speculate that the toxicity was biphasic. The first ALT elevation observed for both vectors at day 1 may simply involve the vector entering the liver, and ALT levels would be the same with both vectors, or alternatively, this early hepatocellular toxicity may not be related to capsid proteins but rather to nonspecific immune mechanisms (27). The levels of ALT at day 7, which were lower with Av4orf3nBg, may be related to reduced backbone gene expression. This pattern of ALT elevation correlates well with the reduced cytopathic effect observed in animal livers following vector administration. Tail vein injection of Av3nBg in C57BL/6 mice resulted in liver vacuolization, nuclear pleomorphism, and loss of tissue architecture; none of the mice that received the Av4orf3nBg vector demonstrated these pathological changes. This result was expected by virtue of the known effects of E4 viral proteins on cell toxicity. Among the many activities of E4 proteins are oncogenesis, blocking of p53 function, modulation of cellular transcription factors, induction of the cell cycle, and modification of protein phosphatase 2a activity (11, 23, 32, 33). Removal of these functions from the adenovirus vector backbone will undoubtedly have an impact on cell viability. However, since E4 ORF3 is active in the adenovirus vector backbone, the effect of this protein needs to be evaluated in the context of long-term vector persistence, especially on cell modulation, since ORF3 alone may play a role in activating cellular RSV-like promoters. In addition, recent data suggest that E4 ORF3 is directly associated with the nuclear matrix, affecting the distribution of essential transcription-replication factors in the nucleus (8, 12).
The host immune response following in vivo gene transfer with adenovirus vectors is directed to both viral antigens and encoded transgene products (38). The contribution of a specific viral or transgene antigen to the host immune response is very complex and seems to be species and strain dependent. Virus late gene products are dominant antigens in C57BL/6 mice, whereas the transgene product dominates in the C3H mice (3, 16). In this study both the Av3nBg and the Av4orf3nBg vector behave similarly in C57BL/6 mice by their ability to induce mononuclear cell infiltrates. The interpretation of this result is not immediately evident because β-galactosidase expression is much higher in animals injected with the Av4orf3nBg vector than in animals injected with an E4-containing Av3nBg vector. This observation may imply that transgene-directed cellular immune responses are more predominant in adenovirus vectors whose virus-specific gene expression is significantly reduced than in those whose virus-specific gene expression is stable. Interestingly, C57BL/6 mice administered an Av4nBg vector lacking E1, E2a, and E4 (attenuated by a complete removal of the E4 promoter and expressing a β-galactosidase gene from an RSV promoter) showed a lower level of transgene expression, a lower toxicity, and a smaller amount of cell infiltrates than mice administered the Av4orf3nBg vector (data not shown). The above results exemplify the relationship between E4 viral proteins and promoter regulation but more importantly suggest that attenuation of E4 proteins in the vector backbone may reduce host antiviral responses to the transduced cells. The existence of an interplay between the immune responses to transgene and viral antigens was documented in this model of liver-directed gene transfer (10) and illustrates the impact that a reporter gene may harbor when adenovirus vector backbone modifications are being evaluated in vivo. Thus, a key issue in the further evaluation of a triple-deletion adenovirus vector is to demonstrate persistent transgene expression of a nonimmunogenic transgene in several mouse strains and in large-animal models.
We describe in this study the construction of an adenovirus vector lacking E1, E2a, and all of E4 except ORF3. This adenovirus vector can be propagated to a high titer in a cell line that complements the deleted viral functions, facilitating scale-up and purification steps. The vector has several clear advantages over a double-deletion vector: (i) additional deletion of E4 in a vector with previous E1 and E2a deletions improves the safety and toxicity profile of the vector and should decrease immunogenicity, (ii) rescue of replication-competent adenovirus is unlikely, and (iii) the shortened adenovirus vector backbone allows for larger insertions of foreign DNA sequences. In addition, identification of ORF3 as the only E4 gene product required to increase RSV promoter activity has important implications for the design of other recombinant vectors. The impact of this protein in long-term gene expression should be carefully investigated.
ACKNOWLEDGMENTS
We thank Robert Jambou for critical review of the manuscript. We also thank Russette Lyons and Christoph Wey for assistance with the animal procedures and Christine Mech for preparing liver sections and staining.
REFERENCES
- 1.Amalfitano A, Hauser M A, Hu H, Serra D, Begy C R, Chamberlain J S. Production and characterization of improved adenovirus vectors with the E1, E2b, and E3 genes deleted. J Virol. 1998;72:926–933. doi: 10.1128/jvi.72.2.926-933.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armentano D, Zabner J, Sacks C, Sookdeo C C, Smith M P, St. George J A, Wadsworth S C, Smith A E, Gregory R J. Effect of the E4 region on the persistence of transgene expression from adenovirus vectors. J Virol. 1997;71:2408–2416. doi: 10.1128/jvi.71.3.2408-2416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr D, Tubb J, Fergunson D, Scaria A, Lieber A, Wilson C, Perkin J, Kay M A. Strain related variations in adenovirally mediated transgene expression from mouse hepatocytes in vivo: comparison between immunocompetent and immunodeficient inbred strains. Gene Ther. 1995;2:151–155. [PubMed] [Google Scholar]
- 4.Berkner K L. Development of adenovirus vectors for the expression of heterologous genes. BioTechniques. 1988;6:616–629. [PubMed] [Google Scholar]
- 5.Bridge E, Ketner G. Redundant control of adenovirus late gene expression by early region 4. J Virol. 1989;63:631–638. doi: 10.1128/jvi.63.2.631-638.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brough D E, Hsu C, Kulesa V A, Lee G M, Cantolupo L J, Lizonova A, Kovesdi I. Activation of transgene expression by early region 4 is responsible for a high level of persistent transgene expression from adenovirus vectors in vivo. J Virol. 1997;71:9206–9213. doi: 10.1128/jvi.71.12.9206-9213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burcin M M, Schieder G, Kochaneck S, Tsai S Y, O’Malley B W. Adenovirus-mediated regulable target gene expression in vivo. Proc Natl Acad Sci USA. 1999;96:355–360. doi: 10.1073/pnas.96.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho T, Seeler J-S, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4orf3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai Y, Schwartz E M, Gu D, Zhang W W, Sarvetnick N, Verma I M. Cellular and humoral immune responses to adenoviral vectors containing factor IX gene: tolerization of factor IX and vector antigens allows for long-term expression. Proc Natl Acad Sci USA. 1995;92:1401–1405. doi: 10.1073/pnas.92.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dedieu J F, Vigne E, Torrent C, Julien C, Mahfouz I, Caillaud J M, Aubailly N, Orsini C, Guillaume J M, Opolon P, Delaere P, Perricaudet M, Yeh P. Long-term gene delivery into the livers of immunocompetent mice with E1/E4-defective adenoviruses. J Virol. 1997;71:4626–4637. doi: 10.1128/jvi.71.6.4626-4637.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcription activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 12.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 13.Engelhardt J F, Ye X, Doranz B, Wilson J M. Ablation of E2A in recombinant adenoviruses improves transgene persistence and decreases inflammatory response in mouse liver. Proc Natl Acad Sci USA. 1994;91:6196–6200. doi: 10.1073/pnas.91.13.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelhardt J F, Litzky L, Wilson J M. Prolonged transgene expression in cotton rat lung with recombinant adenoviruses defective in E2a. Hum Gene Ther. 1994;5:1217–1229. doi: 10.1089/hum.1994.5.10-1217. [DOI] [PubMed] [Google Scholar]
- 15.Fisher K J, Choi H, Burda J, Chen S-J, Wilson J M. Recombinant adenovirus deleted of all DNA viral genes for gene therapy of cystic fibrosis. Virology. 1996;217:11–22. doi: 10.1006/viro.1996.0088. [DOI] [PubMed] [Google Scholar]
- 16.Gao G P, Yang Y, Wilson J M. Biology of adenovirus vectors with E1 and E4 deletions for liver-directed gene therapy. J Virol. 1996;70:8934–8943. doi: 10.1128/jvi.70.12.8934-8943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorziglia M I, Kadan M J, Yei S, Lim J, Lee G M, Luthra R, Trapnell B C. Elimination of both E1 and E2a from adenovirus vectors further improves prospects for in vivo human gene therapy. J Virol. 1996;70:4173–4178. doi: 10.1128/jvi.70.6.4173-4178.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham F L, Prevec L. Adenovirus based expression vectors and recombinant vaccines. In: Ellis R W, editor. Vaccines: new approaches to immunological problems. London, United Kingdom: Butterworth-Heinemann; 1992. pp. 363–390. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D, Gluzman Y. Rescue of functional replication origins from embedded configurations in a plasmid carrying the adenovirus genome. Mol Cell Biol. 1984;4:302–309. doi: 10.1128/mcb.4.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joos K, Hildegund C J, Wilson J M. Cytotoxic T-lymphocyte target proteins and their major histocompatibility complex class I restriction in response to adenovirus vectors delivered to mouse liver. J Virol. 1998;72:2945–2954. doi: 10.1128/jvi.72.4.2945-2954.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kajiwara K, Byrnes A P, Charlton H M, Wood M J, Wood K J. Immune responses to adenoviral vectors during gene transfer in the brain. Hum Gene Ther. 1997;8:253–265. doi: 10.1089/hum.1997.8.3-253. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan J M, Armentano D, Sparer T E, Wynn S G, Peterson P A, Wadsworth S C, Couture K K, Pennington S E, St. George J A, Smith L R. Characterization of factors involved in modulating persistence of transgene expression from recombinant adenovirus in the mouse lung. Hum Gene Ther. 1997;8:45–56. doi: 10.1089/hum.1997.8.1-45. [DOI] [PubMed] [Google Scholar]
- 23.Kleinberger T, Shenk T. Adenovirus E4orf4 protein binds to protein phosphatase 2A, and the complex down regulates E1A-enhanced junB transcription. J Virol. 1993;67:7556–7560. doi: 10.1128/jvi.67.12.7556-7560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kochaneck S, Clemens P R, Mitani K, Chen H-H, Chan S, Caskey C T. A new adenoviral vector: replacement of all viral coding sequences with 28 kb of DNA independently expressing both full-length dystrophin and β-galactosidase. Proc Natl Acad Sci USA. 1996;93:5731–5736. doi: 10.1073/pnas.93.12.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lapcevich, C., Q. Kang, and M. Gorziglia. Unpublished results.
- 26.Leppard K N. E4 gene function in adenovirus, adenovirus vector and adeno-associated virus infections. J Gen Virol. 1997;78:2131–2138. doi: 10.1099/0022-1317-78-9-2131. [DOI] [PubMed] [Google Scholar]
- 27.Lieber A, He C-Y, Kirillova I, Kay M A. Recombinant adenoviruses with large deletions generated by Cre-mediated excision exhibit different biological properties compared with first-generation vectors in vitro and in vivo. J Virol. 1996;70:8944–8960. doi: 10.1128/jvi.70.12.8944-8960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lusky M, Christ M, Rittner K, Dieterle A, Dreyer D, Mourot B, Schultz H, Stoeckel F, Pavarani A, Mehtali M. In vitro and in vivo biology of recombinant adenovirus vectors with E1, E1/E2a, or E1/E4 deleted. J Virol. 1998;72:2022–2032. doi: 10.1128/jvi.72.3.2022-2032.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maizel J V, White D O, Scarff M D. The polypeptides of adenovirus. II. Soluble proteins, cores, top components and structure of the virion. Virology. 1968;36:126–136. doi: 10.1016/0042-6822(68)90122-0. [DOI] [PubMed] [Google Scholar]
- 30.Mitani K, Graham F L, Caskey C T, Kochanek S. Rescue, propagation, and partial purification of a helper virus-dependent adenovirus vector. Proc Natl Acad Sci USA. 1995;92:3854–3858. doi: 10.1073/pnas.92.9.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morsy M A, Gu M, Motzel S, Zhao J, Lin J, Su Q, Allens H, Franlin L, Parks R J, Graham F L, Kochaneck S, Bett A J, Caskey C T. An adenoviral vector deleted for all viral coding sequences results in enhanced safety and extended expression of a leptin transgene. Proc Natl Acad Sci USA. 1998;95:7866–7871. doi: 10.1073/pnas.95.14.7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nevels M, Rubenwolf S, Spruss T, Wolf H, Dobner T. The adenovirus E4orf6 protein can promote E1A/E1β-induced focus formation by interfering with p53 tumor suppressor function. Proc Natl Acad Sci USA. 1997;94:1206–1211. doi: 10.1073/pnas.94.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nevins J R. Utilization of cellular transcription factors for adenovirus-induced transcription. Virus Res. 1991;20:1–10. doi: 10.1016/0168-1702(91)90056-2. [DOI] [PubMed] [Google Scholar]
- 34.Schiedner G, Morral N, Parks R J, Wu Y, Koopmans S C, Langston C, Graham F L, Beaudet A L, Kochanek S. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat Genet. 1998;18:180–183. doi: 10.1038/ng0298-180. [DOI] [PubMed] [Google Scholar]
- 35.Smith T A G, Mehaffey M G, Kayda D B, Saunders J M, Yei S, Trapnell B C, McClelland A, Kaleko M. Adenovirus mediated expression of therapeutic plasma levels of human factor IX in mice. Nat Genet. 1993;5:397–402. doi: 10.1038/ng1293-397. [DOI] [PubMed] [Google Scholar]
- 36.Trapnell B C. Adenoviral vectors for gene transfer. Adv Drug Delivery Rev. 1993;12:185–199. [Google Scholar]
- 37.Trapnell B C, Gorziglia M. Gene therapy using adenoviral vectors. Curr Opin Biotechnol. 1994;5:617–625. doi: 10.1016/0958-1669(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 38.Tripathy S K, Black H B, Goldwasser E, Leiden J M. Immune responses to transgene-encoded proteins limit the stability of gene expression after injection of replication-defective adenovirus vectors. Nat Med. 1996;2:545–550. doi: 10.1038/nm0596-545. [DOI] [PubMed] [Google Scholar]
- 39.Vrati S, Macavoy E S, Xu Z Z, Smole C, Boyle D B, Both G W. Construction and transfection of ovine adenovirus genomic clones to rescue modified viruses. Virology. 1996;220:200–203. doi: 10.1006/viro.1996.0300. [DOI] [PubMed] [Google Scholar]
- 40.Wang Q, Greenburg C, Bunch D, Farson D, Finer M H. Persistent transgene expression in mouse liver following in vivo gene transfer with dlE1/dlE4 adenovirus vectors. Gene Ther. 1997;4:393–400. doi: 10.1038/sj.gt.3300404. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y, Ertl H C J, Wilson J M. MHC class I-restricted cytotoxic T lymphocytes to viral antigens destroy hepatocytes in mice infected with E1-deleted recombinant adenoviruses. Immunity. 1994;1:433–442. doi: 10.1016/1074-7613(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 42.Yang Y, Nunes F A, Berencsi K, Furth E E, Gonczol E, Engelhardt J F, Wilson J M. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc Natl Acad Sci USA. 1994;91:4407–4411. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, Nunes F A, Berencsi K, Gonczol E, Engelhardt J F, Wilson J M. Inactivation of E2a in recombinant adenoviruses improves the prospect for gene therapy in cystic fibrosis. Nat Genet. 1994;7:362–369. doi: 10.1038/ng0794-362. [DOI] [PubMed] [Google Scholar]
- 44.Yang Y, Li Q, Ertl H J C, Wilson J M. Cellular and humoral immune responses to viral antigens create barriers to lung-directed gene therapy with recombinant adenovirus. J Virol. 1995;69:2004–2015. doi: 10.1128/jvi.69.4.2004-2015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yei S, Mittereder N, Wert S, Whitsett J A, Wilmott R W, Trapnell B C. In vivo evaluation of the safety of adenovirus-mediated transfer of the human cystic fibrosis transmembrane conductance regulator cDNA to the lung. Hum Gene Ther. 1994;5:731–744. doi: 10.1089/hum.1994.5.6-731. [DOI] [PubMed] [Google Scholar]