Abstract
Following receptor binding and internalization, intracellular trafficking of adenovirus (Ad) among subgroups B and C is different, with significant amounts of Ad serotype 7 (Ad7) (subgroup B) virions found in cytoplasm during the initial hours of infection while Ad5 (subgroup C) virions rapidly translocate to the nucleus. To evaluate the role of the fiber in these differences, we examined intracellular trafficking of Ad5, Ad7, and Ad5f7 (a chimeric vector composed of the Ad5 capsid with the fiber replaced by the Ad7 fiber) by conjugating Ad capsids directly with Cy3 fluorescent dye, permitting the trafficking of the capsids to be examined by fluorescence microscopy. The human lung carcinoma cell line A549 was infected with Cy3-conjugated viruses for 10 min followed by a 1-h incubation. Ad5 virions rapidly translocated to the nucleus (within 1 h of infection), while Ad7 virions were widely distributed in the cytoplasm at the same time point. Interestingly, chimeric Ad5f7 virions behaved similarly to Ad7 but not Ad5. In this regard, the percentages of nuclear localization of Ad5, Ad7, and Ad5f7 at 1 h following infection were 72% ± 4%, 32% ± 6%, and 38% ± 2%, respectively. Consistent with these observations, fluorescence in situ hybridization demonstrated that most of the Ad5 DNA was detected at the nucleus after 1 h, but at the same time point, DNA of Ad7 and Ad5f7 was distributed in both the nucleus and cytoplasm. Quantification of the kinetics of Ad genomic DNA delivery to the nucleus using a fluorogenic probe-based PCR assay (TaqMan PCR) demonstrated that the percentages of nuclear association of Ad5 DNA and Ad5f7 DNA at 1 h postinfection were 80% ± 13% and 43% ± 1%, respectively. Although it has been generally accepted that Ad fiber protein mediates attachment of virions to cells and that fibers dissociate during endocytic uptake, these data suggest that in addition to mediating binding to the cell surface, fiber likely modulates intracellular trafficking as well.
Adenovirus (Ad) has been widely used in gene transfer applications, in part because of its ability to achieve highly efficient targeting of its genome to the nucleus (7, 16, 40, 52). Six subgroups comprising 49 human Ad serotypes have been identified on the basis of their resistance to neutralization and ability to agglutinate red blood cells (40). Ad serotype 5 (Ad5) and Ad2 (subgroup C) have been studied most extensively and have been employed most widely as recombinant gene transfer vectors (7, 52). Although Ad subgroup C vectors demonstrate very efficient gene transfer, their tropism is limited in vitro and in vivo by receptor expression on target cells (2, 3, 15, 22–24, 35, 45, 47–54). One approach to circumvent limited gene transfer resulting from a deficiency of the appropriate Ad receptor on the target cells is to introduce a novel tropism to the capsid. Strategies to accomplish this have included engineering peptide sequences into the fiber protein, using monospecific and bispecific antibodies, and swapping part or all of the fiber protein from one serotype to another (10, 11, 13, 14, 26, 27, 37, 42, 48–51). The “fiber swap” strategy is based on the knowledge that different Ad subgroups have different tropisms, as evidenced by their various pathologies of infection (21). This concept is borne out by the observation that Ad fiber proteins from different Ad subgroups mediate high-affinity attachment to different cell surface receptors on target cells (3, 9, 13, 36, 41, 45).
Efficient gene delivery is achieved in part by binding to the target cell and by efficient intracytoplasmic translocation of Ad virions to the nucleus. Intracellular trafficking of Ad in the target cell is a stepwise process. Ad attaches with high affinity to cell surface receptors via the fiber protein, internalizes by endocytosis into an endocytic vesicle, escapes from the endosome into the cytoplasm, and then translocates to the nucleus of the target cell (16–19, 28, 40). Viruses from different subgroups are known to have different characteristics of intracytoplasmic trafficking in target cells. For example, Ad5 (subgroup C) fibers bind to different cell surface receptors than Ad7 (subgroup B) fibers (13). Further, while Ad5 virions rapidly escape to the cytosol and translocate to the nucleus within 1 h, Ad7 virions are often found clustered in cytoplasm in membranous organelles at the same time point (4, 5, 8). Although it has been generally accepted that the role of the fiber in Ad trafficking is limited to modulating binding to the high-affinity cell surface receptor (17, 43), preliminary studies comparing fluorophore-labeled Ad5 and Ad7 virions suggested that the fiber could play a postinternalization role in delivery of the Ad genome to the nucleus. These preliminary studies have led us to hypothesize that the Ad fiber may, in part, influence the entire process of intracellular trafficking of Ad and that the swapping of the Ad fiber may change the characteristics of Ad translocation beyond the binding process per se.
To evaluate this concept, we compared the intracellular trafficking of Ad5 (subgroup C) and Ad7 (subgroup B) with that of Ad5f7, a chimeric virus composed of the Ad5 capsid with the fiber replaced by the Ad7 fiber (13). To examine the intracellular trafficking of these Ad, viral capsid protein progression to the nucleus was observed by direct conjugation with fluorophores and fluorescence microscopy, and the viral genome was monitored by fluorescence in situ hybridization (FISH) and by subcellular fractionation combined with quantitative real-time PCR. The data show that Ad fiber influences the postinternalization intracellular trafficking of Ad, i.e., that swapping the Ad fiber may change the characteristics of Ad translocation beyond the binding process per se.
MATERIALS AND METHODS
Cells.
The A549 human lung epithelial cell line (CCL-185; American Type Culture Collection [ATCC], Manassas, Va.) was cultured in F-12K medium supplemented with 10% fetal bovine serum, 50 U of penicillin/ml, and 50 μg of streptomycin (GIBCO/BRL Life Technologies, Inc., Gaithersburg, Md.)/ml. Next, 106 A549 cells were seeded in 35-mm-diameter culture plates that were modified by punching a 7-mm-diameter hole in the bottom and resealing it with a cover glass to create a well in which cells could be plated directly on a cover glass for subsequent microscopic observation (28).
Viruses.
Wild-type Ad5 (Ad5wt) and Ad7 (Ad7wt) were obtained from the ATCC (VR-5 and VR-7, respectively). Ad5 subgroup C recombinant Ad vectors, Ad5Null, Ad5GFP, and Ad5CAT, are E1− E3− replication-deficient vectors containing a cytomegalovirus early-intermediate promoter-enhancer and either no expression cassette (null), the green fluorescent protein gene (GFP), or the chloramphenicol acetyltransferase gene (CAT) in the E1 position (12, 20, 25). An Ad7 subgroup B recombinant vector, Ad7CAT, is similar to the Ad5CAT vector but is based on the Ad7 genome (1). Ad5f7 is similar to Ad5CAT but is a chimeric vector in which the Ad5 fiber gene has been replaced with the Ad7 fiber gene (13). The wild-type Ad and the Ad vectors were all propagated in the 293 human embryonic kidney cell line, purified, and stored at −70°C as previously described (38, 39). Stock preparations of Ad were evaluated for particle concentration by recording absorbance at 260 nm and calculating the particle number based on a published extinction coefficient (30).
Characterization of viral stocks included PCR to confirm the identity of hexon and fiber from different serotypes. The hexon similarity between Ad subgroups ranges from 65 to 80% (6). Areas with low homology were selected as PCR probe templates. Based on Ad5 and Ad7 hexon sequences (GenBank accession no. J01966 and Z48571, respectively), the Ad5 hexon primer pair (sense primer, GAACCTCAAATAGGAGAATCT, position 431 of the Ad5 hexon gene; antisense primer, GCAAAATTATTTCCAACTCTT, position 1180 of the Ad5 hexon gene) and Ad7 hexon primer pair (sense primer, CTCAAGTTGGAGAAGAATCAT, position 487 of the Ad7 hexon gene; antisense primer, ATGTTTTCGCTGGTCCTATGC, position 1141 of the Ad7 hexon gene) were synthesized by using a model 392 DNA/RNA synthesizer (Applied Biosystems, Foster City, Calif.). Serotypes of Ad fiber were also confirmed by PCR. Based on Ad5 and Ad7 fiber sequences (GenBank accession no. M18369 and M23696, respectively), the Ad5 fiber primer pairs included sense primer TTAATGCAGGAGATGGGCTTGAAT (position 1488 of the Ad5 fiber gene) and antisense primer ATAAGATGAGCACTTTGAACTGTT (position 1825 of the Ad5 fiber gene). The Ad7 fiber primer pair included sense primer GGGGGTCTTACAATAGATGACACC (position 2376 of the Ad7 fiber gene) and antisense primer TTAAGTGGAGTTTTTAGGGATGAA (position 2804 of the Ad7 fiber gene).
Fluorophore-conjugated Ad.
Wild-type Ad and Ad vectors were conjugated with Cy3 fluorescent dye (Amersham Life Science, Arlington Heights, Ill.) or 5 (and 6)-carboxyfluorescein, succinimidyl ester (Molecular Probes, Eugene, Oreg.), to allow assessment by fluorescence microscopy. To accomplish this, Ad (1012 particles/ml) were reacted with Cy3 dye in 0.1 M sodium carbonate buffer (pH 9.3) (at 20% of the concentration recommended by the manufacturer, as previously described [28]) or 40 μg of carboxyfluorescein/ml in 0.1 M sodium bicarbonate buffer (pH 8.3). After incubation (23°C for 30 min), the conjugation reaction was stopped by adding 0.2% glycine. Labeled virus was separated from the excess, unconjugated dye by dialysis against a mixture of 10% glycerol, 50 mM Tris-HCl (pH 7.8), 150 mM NaCl2, and 10 mM MgCl2 at 4°C overnight. The amount of dye incorporated into Cy3-Ad and carboxyfluorescein-Ad was measured by recording absorbance at 552 and 495 nm, respectively, using a spectrophotometer (model DU640; Beckman, Fullerton, Calif.). Glycerol was added to fluorophore-conjugated virus to a final concentration of 30%, and the preparations were stored at −20°C.
Fluorescence microscopy.
Samples were observed by using a Nikon Microphot SA microscope equipped with ×60 and ×100 PlanApo objective lenses and a cooled CCD camera (Princeton Instruments, Trenton, N.J.) as previously described (28). Image manipulation and digital image analysis were performed with Metamorph imaging software (Universal Imaging, West Chester, Pa.). For quantitative studies, three to six fields per condition were selected randomly, acquired by the ×60 objective lens, and used for digital image analysis. Each field contained between 6 and 11 cells. Fluorescence intensities over 3% of dynamic range of the camera following background subtraction were collected and digitally analyzed.
Binding specificity.
To evaluate the specificity of binding of the Ad to A549 cells, the cells were rinsed three times with binding buffer (modified Eagle’s medium supplemented with 1% bovine serum albumin–10 mM HEPES [pH 7.3]) and infected with a 100-fold excess of competitor virus (unlabeled Ad5 or Ad7; 1012 particles/ml) for 30 min at 37°C. After preincubation, a mixture of Cy3-Ad (1010 particles/ml) and unlabeled virus (1012 particles/ml) was applied to the cells and incubated for 30 min at 37°C. At the end of the incubation, cells were washed three times with phosphate-buffered saline (PBS) (pH 7.4) and fixed with 4% paraformaldehyde (23°C for 15 min). Then, cells were treated with 1 μg of the DNA dye 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes)/ml in PBS with 0.1% Triton X-100 for 5 min at 23°C. The cover glass was detached from culture plate and mounted on a glass slide with antifade solution (Slowfade; Molecular Probes).
Internalization rate.
To assess the internalization rate of the virus preparations, an experiment was designed based on the principle that fluorescence associated with surface-bound Ad is quenched by horseradish peroxidase–3,3′-diaminobenzidine (HRP-DAB) precipitation in the presence of H2O2 (29). A549 cells were rinsed three times with binding buffer and then infected with carboxyfluorescein-Ad (1011 particles/ml) at 4°C for 1 h or at 37°C for 10 min. After a 10-min infection at 37°C, the cells were washed three times with binding buffer to wash out unbound virus and incubated with binding buffer for 0 to 40 min at 37°C. After infection at 4°C or infection with incubation at 37°C as indicated, the cells were washed three times with ice-cold PBS and incubated with HRP (10 μg/ml; Pierce, Rockford, Ill.) and metal-enhanced DAB (Pierce) in PBS or in stable peroxide buffer (H2O2; Pierce) for 1 h at 4°C. At the end of the incubation, the cells were washed five times with ice-cold PBS and fixed with ice-cold 4% paraformaldehyde for 30 min. The samples were evaluated by fluorescence microscopy. Virus number per cell in single optical sections was estimated by the ratio of unquenched (internalized) Ad to total Ad (including internalized and surface-bound Ad) by using the DAPI nuclear stain to determine the number of cells per field. Background-subtracted digital images (see above) were quantified by using an object-counting algorithm in the MetaMorph image analysis software.
Intracellular trafficking of the Ad capsid.
To follow intracellular trafficking of the Ad capsid, A549 cells were rinsed three times with binding buffer and infected with Cy3-Ad (1011 particles/ml) at 37°C for 10 min. After infection, the cells were washed three times with PBS and fixed with 4% paraformaldehyde or washed three times with binding buffer and incubated for 1 h at 37°C prior to fixation. Cells were then stained with DAPI and mounted on glass slides as described above. The percentage of nuclear localization was estimated in single optical sections following background subtraction by calculating the ratio of nuclear Cy3 fluorescence identified by overlap with nuclear DAPI staining and total cell-associated fluorescence intensity. Cy3 intensity was measured by using a gray scale integration algorithm in the image analysis software.
Colocalization of carboxyfluorescein-Ad and Cy3-α2M.
To demonstrate the intracellular location of Ad prior to nuclear localization, we used the serum protease inhibitor α2-macroglobulin (α2M) as a marker of endocytic compartments. A549 cells were rinsed three times with binding buffer and infected with carboxyfluorescein-Ad (1011 particles/ml) at 37°C for 10 min. After infection, the cells were washed three times with binding buffer and incubated with 10 μg of Cy3-conjugated α2M/ml (kindly provided by F. R. Maxfield, Cornell University) at 37°C. After a 45-min incubation, the cells were washed with binding buffer three times and incubated without Cy3-α2M for 15 min. Cells were examined with a Zeiss LSM510 confocal laser scanning microscope with LSM510 software.
Localization of Ad genome.
To monitor the localization of the Ad genome, FISH was used. Ad5 and Ad7 DNA probes were synthesized, incorporating digoxigenin (DIG)-coupled dUTP by nick translation. DNA purified from Ad5Null or Ad7wt was used as the template; agarose gel assessment of the average probe size was 400 bp. A549 cells were cultured on Silane-prep slides (Sigma, St. Louis, Mo.) at 37°C overnight. After the slides were rinsed three times with binding buffer, Cy3-Ad (1011 particles/ml) was applied to the cells, which were then incubated at 37°C for 10 min. After incubation, the slides were washed three times with PBS and immediately fixed or washed three times with binding buffer and incubated at 37°C for 1 to 8 h. The slides were then washed three times with PBS, fixed with Histochoice (Amresco, Solon, Ohio) for 30 min at 23°C, and dehydrated in a graded alcohol series (50, 70, 95, and 100%). Prehybridization solution (70% formamide–2× SSC [300 mM NaCl–30 mM sodium citrate; pH 7.0]) was applied to the sample on the slide glass, which was overlaid with a plastic coverslip, denatured at 70°C for 12 min, and then immediately dehydrated in a graded alcohol series at 4°C. The DIG-labeled Ad DNA probes were denatured with hybridization solution (a mixture of 50% formamide, 2% Denhardt’s solution, 10% dextran sulfate, and 1× SSC) at 70°C for 30 min, applied to the sample, overlaid with a plastic coverslip, and hybridized at 37°C overnight. After hybridization, the sample slides were washed three times for 5 min in 50% formamide–2× SSC at 43°C and subsequently washed three times for 5 min in 0.1× SSC at 60°C. In situ DIG-labeled Ad DNA was detected by immunofluorescence staining. The samples were incubated with fluorescein isothiocyanate (FITC)–sheep anti-DIG Fab fragment (1 μg/ml; Boehringer Ingelheim, Mannheim, Germany) at 37°C for 30 min. DAPI staining to detect the positions of nuclei was performed.
Quantitative analysis of Ad DNA delivery.
To quantify delivery of the Ad genomic DNA to the nucleus over time, Ad DNA and β-actin DNA of A549 cells were quantified by real-time PCR (TaqMan PCR detection; Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). A549 cells (2 × 106) cultured in a 10-cm-diameter culture plate were infected with Ad (2 × 109 particles/ml, 103 particles/cell) at 37°C for 10 min, washed three times with binding buffer, and incubated for 0 to 8 h at 37°C. At each time point, the cells were collected by the addition of trypsin (0.5 mg/ml for 5 min at 37°C) (GIBCO) followed by washing with PBS. Half of the cells were used for the extraction of total cellular DNA. The remaining cells were used for nuclear isolation as described by Muggeridge and Fraser (31). Briefly, cells were suspended with reticulocyte standard buffer solution (10 mM Tris-HCl [pH 7.5]–10 mM NaCl–3 mM MgCl2), broken in a Dounce homogenizer with a loose pestle, incubated on ice for 10 min, and brought to 1.5% (vol/vol) with respect to the nonionic detergent IGEPAL CA-630 (Sigma). Samples were overlaid onto a 2× sample volume of 0.33 M sucrose in 10 mM Tris-HCl [pH 8.0]–5 mM MgCl2, and the homogenate was centrifuged at 500 × g for 10 min. The pellet was collected as the nuclear fraction. The DNA of Ad and A549 cells was purified from total cell lysate and from the nuclear fraction by using the QIAamp blood kit (QIAGEN Inc., Santa Clara, Calif.). The Ad E2 region and β-actin gene of A549 cell were amplified and quantified by TaqMan PCR. A fluorogenic probe (CCGCCTCGGCTTGTCACATTTTTC) was designed to anneal to the target between the sense primer (AATAAACAAGTTCCCGGATCGAT) and the antisense primer (GCACATAGGAGAGATGAGCTTCC) in the E2 region of the Ad genome. The fluorogenic probe contains both a reporter dye (6-carboxyfluorescein; 5′ end) and a quencher dye (6-carboxy-N,N,N′,N′-tetramethylrhodamine; 3′ end). During the extension phase of the PCR cycle, the 5′-3′ exonuclease activity of Taq polymerase cleaves the hybridized fluorogenic probe, resulting in an increase in fluorescence emission from the reporter dye that quantitatively reports the amount of PCR product. Samples were amplified for 40 cycles in a Perkin-Elmer model 7700 sequence detection system with continuous monitoring of the fluorescence. Data were processed by the SDS 1.6 software package (Perkin-Elmer). For human β-actin detection, β-actin probes (Perkin-Elmer) were used. To control for differences in DNA recovery from samples, the data are presented as ratios of Ad genome DNA to β-actin DNA.
Statistical analysis.
All data are presented as means ± standard errors of the means. Statistical evaluations were carried out by using the two-tailed Student’s t test.
RESULTS
Characterization of fluorophore-conjugated Ad.
For all Ad used in this study, the dye/capsomere ratio of Cy3- or carboxyfluorescein-conjugated Ad was 0.4 to 1.5. In this range, the fluorophore did not affect Ad infectivity as assessed by plaque assay. The molecular identity of viral stocks was further verified by PCR assessment with primers specific for Ad5 or Ad7 hexon. The product length of amplified nucleotide with Ad5 and Ad5f7 was 770 bp, corresponding to the predicted Ad5 hexon template, while the product length with Ad7 was 675 bp, corresponding to the predicted Ad7 hexon template (not shown). The product lengths of the amplified Ad fiber genes were, as expected, 452 bp for Ad5 and 361 bp for Ad7 (not shown). There was no contamination of the chimeric virus by parental stocks.
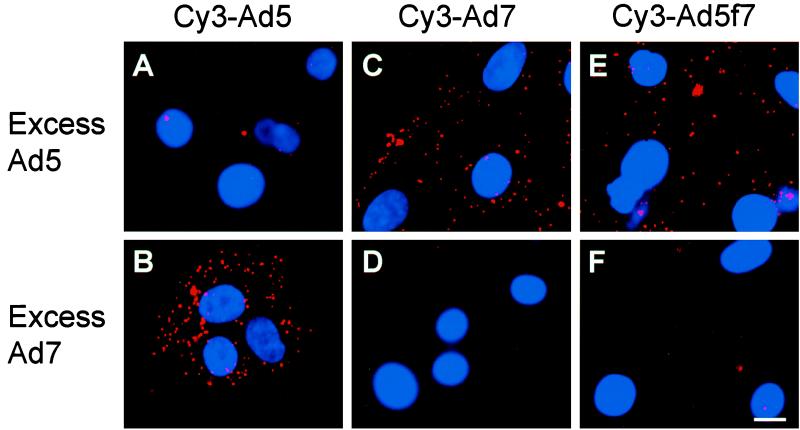
To confirm the binding specificity of Cy3-Ad5, Cy3-Ad7, and Cy3-Ad5f7, each was incubated with a 100-fold excess of unlabeled Ad5 or Ad7. Cy3-Ad5 binding to the target cells was blocked by excess unlabeled Ad5 (Fig. 1A) but not by excess Ad7 (Fig. 1B). In parallel studies, the binding of Ad7 was blocked by excess unlabeled Ad7 (Fig. 1D) but not by Ad5 (Fig. 1C). These data are consistent with previous reports that subgroup B and C Ad utilize different cell surface receptors (9, 13, 36, 41). The Ad5f7 chimera vector, identical to Ad5 but with an Ad7 fiber, was blocked by excess unlabeled Ad7 (Fig. 1F) but not by Ad5 (Fig. 1E). These observations confirmed that Ad5f7 interacts with target cells via the engineered chimeric Ad7 fiber as previously noted by using radioactively labeled Ad5f7 (13).
FIG. 1.
Competition of unlabeled Ad5 or Ad7 with Cy3-Ad5, Cy3-Ad7, or Cy3-Ad5f7 for entry and translocation in A549 cells. A549 cells were incubated (30 min at 37°C) with unlabeled competitor virus (Ad5 or Ad7, each at 1012 particles/ml). Cy3-Ad (1010 particles/ml) mixed with unlabeled Ad (1012 particles/ml) was then added, and the incubation continued for 30 min at 37°C. Shown are Cy3-Ad (red) and DAPI-stained nuclei (blue) evaluated by fluorescence microscopy. (A) Cy3-Ad5 in the presence of excess unlabeled Ad5. (B) Cy3-Ad5 in the presence of excess Ad7. (C) Cy3-Ad7 in the presence of Ad5. (D) Cy3-Ad7 in the presence of Ad7. (E) Cy3-Ad5f7 in the presence of Ad5. (F) Cy3-Ad5f7 in the presence of Ad7. Bar = 10 μm.
Internalization rates for fluorophore-conjugated Ad serotypes.
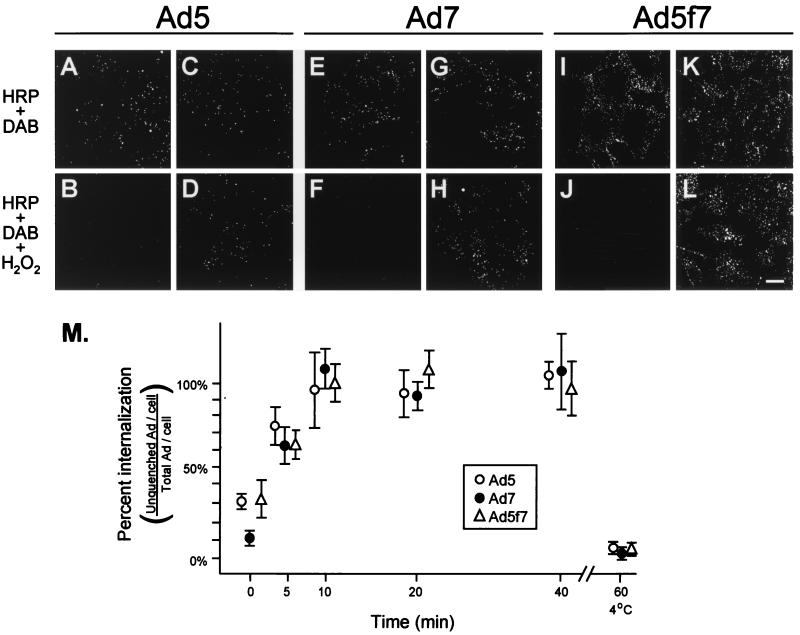
One potential difference in viral trafficking following a change in tropism may be the rate of viral internalization following binding. Using a novel fluorescence-based internalization assay, the internalization rates of Ad5, Ad7, and Ad5f7 were compared. The assay takes advantage of the ability of precipitated DAB to quench fluorophore (29). In this experimental setting, DAB was precipitated by the addition of HRP and H2O2 to culture medium, resulting in selective quenching of surface-bound fluorophore, i.e., permitting assessment of the proportion of fluorophore-conjugated Ad that is on the cell surface compared to the proportion that is internalized. To test the efficiency of the assay, carboxyfluorescein-conjugated Ad were bound to the surface of A549 cells for 1 h at 4°C, conditions that do not permit viral internalization. Dishes were treated in parallel with either HRP and DAB or HRP, DAB, and H2O2, a key substrate in HRP-conjugated precipitation of DAB. Cultures treated with H2O2 exhibited a nearly undetectable carboxyfluorescein-Ad pattern, while cultures treated with HRP and DAB but not H2O2 had clear carboxyfluorescein staining patterns (Fig. 2). The ratio of internalized fluorophore to total fluorophore was quantified by using digital image analysis to assess the number of fluorescein puncta in the presence or absence of H2O2. At 4°C, carboxyfluorescein associated with cell-surface-bound Ad was quenched completely by HRP-DAB precipitation in the presence of H2O2 (Fig. 2A, B, E, F, I, and J). In contrast, almost no quenching was observed when A549 cells were infected for 10 min at 37°C, washed, and incubated an additional 10 min at 37°C (Fig. 2C, D, G, H, K, and L). By calculating the ratio of internalized Ad to total Ad at various incubation times at 37°C following a 10-min infection, curves reflecting the internalization rates of Ad5, Ad7, and Ad5f7 were generated (Fig. 2M). Together, these data demonstrate that there is no difference in the kinetics of internalization among Ad5, Ad7, and Ad5f7.
FIG. 2.
Internalization rates of carboxyfluorescein-Ad5, -Ad7, and -Ad5f7. Carboxyfluorescein-Ad (1011 particles/ml) was incubated with A549 cells at 4°C to evaluate only surface binding or at 37°C to allow binding and internalization of Ad. At each time point, cells were washed, fixed, and treated with HRP and DAB, with or without H2O2. Extracellular fluorophore was quenched by precipitation of DAB by HRP in the presence of H2O2. (A) A549 cells incubated with carboxyfluorescein-Ad5 (4°C for 1 h) and treated with HRP-DAB without H2O2. (B) Same as panel A, but treated with HRP-DAB in the presence of H2O2. (C) A549 cells infected with carboxyfluorescein-Ad5 at 37°C for 10 min, washed, and incubated for an additional 10 min at 37°C, then treated with HRP-DAB without H2O2. (D) Same as panel C, but treated with HRP-DAB in the presence of H2O2. (E) A549 cells incubated with carboxyfluorescein-Ad7 (4°C for 1 h) and treated with HRP-DAB without H2O2. (F) Same as panel E, but treated with HRP-DAB in the presence of H2O2. (G) A549 cells infected with carboxyfluorescein-Ad7 at 37°C for 10 min, washed, and incubated for an additional 10 min at 37°C, then treated with HRP-DAB without H2O2. (H) Same as panel G, but treated with HRP-DAB in the presence of H2O2. (I) A549 cells incubated with carboxyfluorescein-Ad5f7 (4°C for 1 h) and treated with HRP-DAB without H2O2. (J) Same as panel I, but treated with HRP-DAB in the presence of H2O2. (K) A549 cells infected with carboxyfluorescein-Ad5f7 at 37°C for 10 min, washed, and incubated for an additional 10 min at 37°C, then treated with HRP-DAB without H2O2. (L) Same as panel K, but treated with HRP-DAB in the presence of H2O2. Bar = 10 μm. (M) Percentage of internalized Ad per cell relative to the total amount of Ad per cell. Levels of internalized Ad5 (●), Ad7 (○), and Ad5f7 (▵) were determined by digital image analysis (see Materials and Methods). Data for zero internalization time correspond to surface-labeled cells (4°C binding for 1 h).
Intracellular trafficking of Cy3-Ad.
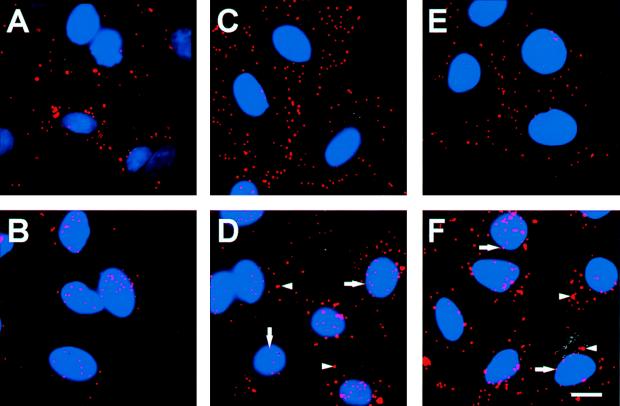
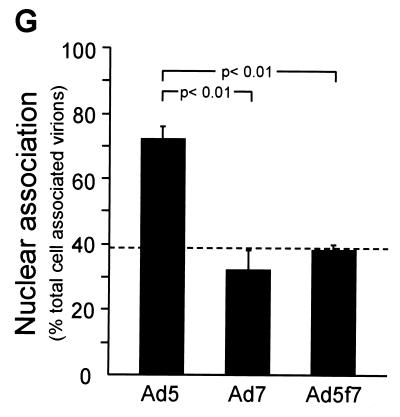
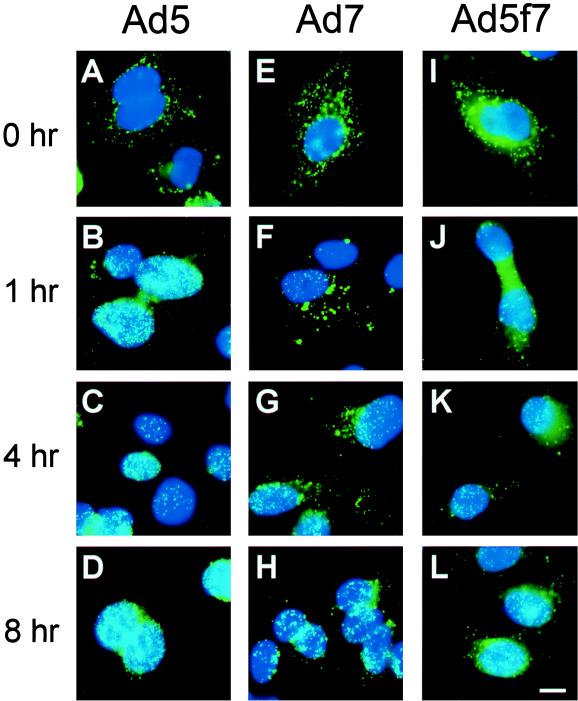
The efficiency with which fluorophore-conjugated Ad targeted the nucleus following internalization was evaluated by examining the intracellular distribution of fluorophore-Ad 1 h after infection. As noted above, Cy3-Ad5 bound avidly to cells following a 10-min infection period at 37°C (Fig. 3A). Most Cy3-Ad5 translocated to the nucleus within 1 h of infection (Fig. 3B). In contrast, despite good binding to target cells (Fig. 3C), large amounts of Cy3-Ad7 were found in the cytosol 1 h after infection (Fig. 3D). Strikingly, the same observation was made with Cy3-Ad5f7 (Fig. 3E and F); i.e., with the only difference being an Ad7 fiber, the Ad5f7 virus behaved as an Ad7 virus, not as an Ad5 virus. The percentages of nuclear localization of Ad5, Ad7, and Ad5f7 at 1 h following infection were 72% ± 4%, 32% ± 6%, and 38% ± 2%, respectively (Fig. 3G) (P < 0.01, Ad5 versus Ad7; P < 0.01, Ad5 versus Ad5f7; P > 0.1, Ad7 versus Ad5f7). The values for Ad7 and Ad5f7 were comparable to the overlap of randomly distributed Cy3-Ad immediately following infection with the nuclear stain and thus represent the background values for this time point. Immediately following infection, all three fluorophore-conjugated viruses displayed a uniform punctate appearance at the cell surface (Fig. 3A, C, and E). While Cy3-Ad5 retained the uniform punctate appearance throughout translocation to the nucleus (Fig. 3B), the cytoplasmic stain for both Cy3-Ad7 and Cy3-Ad5f7 included small puncta similar to those immediately following infection as well as larger, brighter puncta that may have reflected the accumulation of vectors in intracellular organelles (Fig. 3D and F). Cy3-Ad7 and Cy3-Ad5f7 were observed at the nuclear perimeter most often as small puncta.
FIG. 3.
Intracellular distribution of Cy3-Ad5, Cy3-Ad7, and Cy3-Ad5f7 following binding and internalization. A549 cells were infected with Cy3-Ad (1011 particles/ml, 10 min at 37°C), washed, and either immediately fixed or incubated for 1 h at 37°C prior to fixation. Cy3-Ad (red) and DAPI-stained nuclei (blue) were evaluated. (A) Cy3-Ad5, 10-min infection. (B) Cy3-Ad5, 10-min infection plus a 1-h incubation. (C) Cy3-Ad7, 10-min infection. (D) Cy3-Ad7, 10-min infection plus a 1-h incubation. (E) Cy3-Ad5f7, 10-min infection. (F) Cy3-Ad5f7, 10-min infection plus a 1-h incubation. Bar = 10 μm. Arrows indicate small puncta. Arrowheads indicate larger aggregates of vector. (G) Percentage of nuclear localization relative to total cell-associated virions at 1 h following infection. The dashed line represents the background value (see Results).
Intracellular location of Ad7 and Ad5f7 prior to nuclear localization.
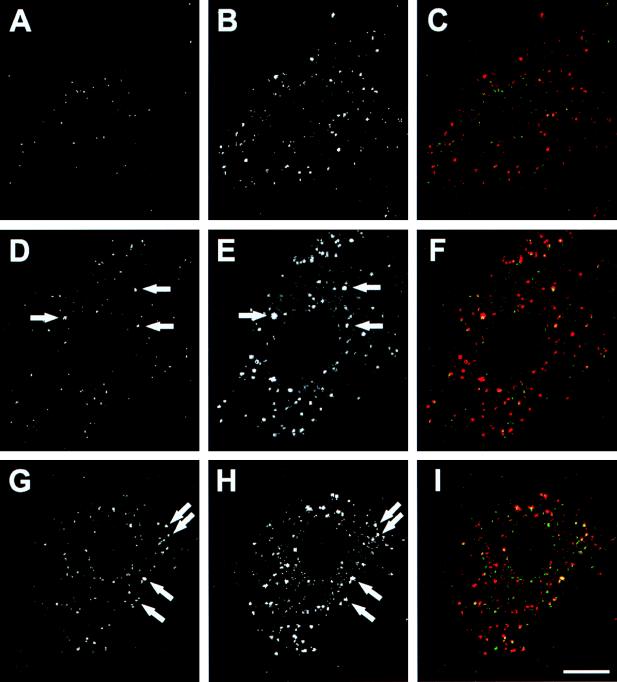
The localization of Ad in cytoplasm was evaluated by comparing Ad localization with the location of α2M, an endocytic marker that occupies sorting endosomes, late endosomes, and lysosomes (53). After a 1-h incubation, most of Ad5 was around the nuclei without colocalization with α2M (Fig. 4A to C). In contrast, a significant proportion of Ad7 and Ad5f7 was colocalized with α2M in the cytoplasm (Fig. 4D to I). These results suggested that Ad7 and Ad5f7 remained in endocytic compartments as late as 1 h after infection.
FIG. 4.
Colocalization of carboxyfluorescein-Ad with Cy3-α2M. A549 cells were infected with carboxyfluorescein-Ad (1011 particles/ml, 10 min at 37°C), washed, and incubated with Cy3-conjugated α2M (10 μg/ml, 45 min at 37°C), then washed and incubated for 15 min at 37°C. Carboxyfluorescein-Ad (green) and Cy3-α2M (red) were examined by confocal laser scanning microscopy. (A) Ad5. (B) α2M, same field as shown in panel A. (C) Combined image of panels A and B. (D) Ad7. (E) α2M, same field as shown in panel D. (F) Combined image of panels D and E. (G) Ad5f7. (H) α2M, same field as shown in panel G. (I) Combined image of panels G and H. Arrows highlight examples of colocalization. Bar = 10 μm.
Localization of the Ad genome.
Trafficking of Cy3-Ad provided information on the intracellular fate of the fluorophore covalently conjugated capsid proteins. The fate of the Ad genome was independently assessed by FISH. Consistent with Cy3-Ad capsid observations, FISH analysis using a DIG-labeled Ad DNA probe demonstrated that most of the Ad5 DNA was in the cytoplasm immediately after infection (Fig. 5A) and at the nucleus after 1 h (Fig. 5B to D). In contrast, DNA of Ad7 and Ad5f7 was detected both in the nucleus and cytoplasm as long as 4 h following infection (Fig. 5E to G and I to K). By 8 h, however, the majority of both Ad7 and Ad5f7 genomes became localized to the nucleus (Fig. 5H and L). Thus, in regard to postinternalization intracellular trafficking, both fluorophore labeling of capsid and detection of the Ad genome by FISH demonstrated that the Ad5f7 chimera behaves as Ad7, not Ad5.
FIG. 5.
FISH evaluation of the Ad genome following binding and internalization of Ad5, Ad7, and Ad5f7. The various Ad (1011 particles/ml) were incubated with A549 cells (10 min at 37°C) and washed, and the incubation was continued at 37°C for 0, 1, 4, or 8 h. The cells were then hybridized with a DIG-labeled Ad DNA probe. After hybridization, DIG-Ad DNA was stained with FITC–anti-DIG antibody. The Ad DNA (green) and DAPI-stained nuclei (blue) are shown. (A) Ad5, 10-min infection. (B) Ad5, 10-min infection plus a 1-h incubation. (C) Ad5, 10-min infection plus a 4-h incubation. (D) Ad5, 10-min infection plus an 8-h incubation. (E) Ad7, 10-min infection. (F) Ad7, 10-min infection plus a 1-h incubation. (G) Ad7, 10-min infection plus a 4-h incubation. (H) Ad7, 10-min infection plus an 8-h incubation. (I) Ad5f7, 10-min infection. (J) Ad5f7, 10-min infection plus a 1-h incubation. (K) Ad5f7, 10-min infection plus a 4-h incubation. (L) Ad5f7, 10-min infection plus an 8-h incubation. Bar = 10 μm.
Kinetics of Ad DNA delivery to the nucleus.
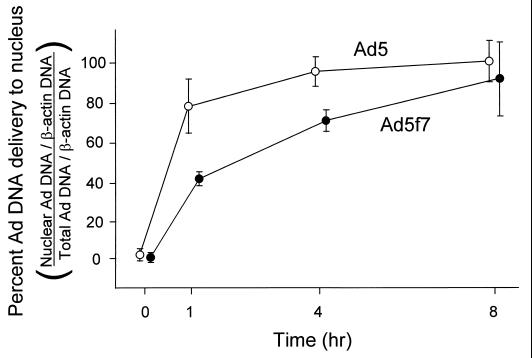
To examine further the rate of DNA transfer to the nucleus, real-time quantitative PCR (TaqMan PCR) using primers specific for the Ad genome E2 region was performed. Ad DNA was quantified at various time points from both total cell lysate and isolated nuclei, and Ad genome delivery to nuclei was calculated as the ratio of Ad DNA in a nuclear subcellular fraction to Ad DNA in total cell lysate. To control for variation in the recovery of DNA from cell lysate and nuclei, data were expressed as the ratios of Ad DNA to cellular DNA as measured by using probes specific for the β-actin gene. Most of the Ad5 DNA was delivered to the nucleus within 1 h (Fig. 6). In contrast, the rate of Ad5f7 genome accumulation in the nucleus was significantly lower (Fig. 6) (Ad5 versus Ad5f7; 1 h, P < 0.01; 4 h, P < 0.01), but eventually the same degree of nuclear localization (8 h, P > 0.5). The half time for accumulation of Ad5 DNA in the nucleus was 40 min, while the Ad5f7 genome exhibited a half time of 110 min.
FIG. 6.
Quantitative evaluation of DNA delivery to the nucleus at various times after a 10-min infection. A549 cells were infected with Ad5 or Ad5f7 (103 particles/cell) at 37°C for 10 min. The cells were washed and then incubated for 0 to 8 h at 37°C. The cells were then harvested, and DNA was extracted from the total cells or isolated nuclei. Ad DNA and A549 DNA were quantified by TaqMan PCR using probes for the Ad E2 region and for the cellular β-actin gene. Data are percentages of Ad DNA delivered to the nucleus, i.e., a ratio of Ad genome copies to β-actin gene copies from the nuclear fraction versus those from the total cell lysate. Shown are DNA analyses of Ad5 (○) and Ad5f7 (●). Data are mean values and standard errors from three experiments for each vector.
Comparison of Ad capsid and Ad DNA localization.
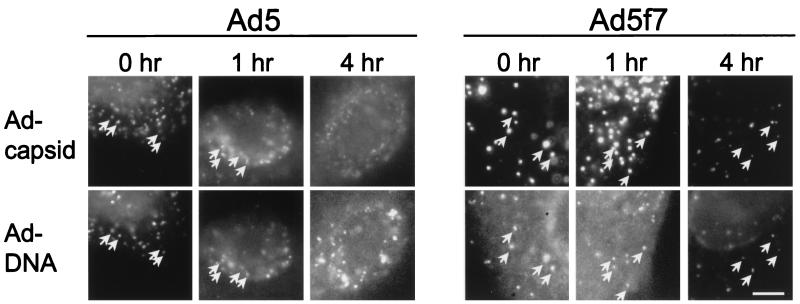
To confirm that fluorophore-conjugated capsid localization and Ad genome localization reflected the same population of vectors, Ad capsid (detected with Cy3 fluorescence) and Ad DNA (detected with FITC–anti-DIG) were simultaneously located in A549 cells at various time points after infection. Immediately following infection, Ad5 and Ad5f7 capsids showed good colocalization with the Ad genome (Fig. 7). After 1 h, Ad5 capsid and its colocalized genome were primarily at the periphery of the nucleus. In contrast, Ad5f7 capsids and their colocalized genome were found both in the cytoplasm and at the nucleus. Ad5 DNA was completely dissociated from its capsid within 4 h following infection, while Ad5f7 DNA retained a significant level of colocalization with its capsid at 4 h.
FIG. 7.
Comparison of localization of the Cy3-Ad capsid and Ad DNA localization (FISH) for Ad5 and Ad5f7 at various times after infection. The Ad capsids (upper panels) were detected by Cy3 fluorescence, and Ad DNA (lower panel) was detected with FITC–anti-DIG after hybridization with DIG-Ad DNA probe. Micrographs show an individual nucleus with adjacent cytoplasm. Arrows indicate colocalization of Ad capsid and DNA. Ad5 DNA was dissociated from its capsid within 4 h following infection, while Ad5f7 DNA was still colocalized with its capsid at 4 h. Bar = 5 μm.
DISCUSSION
Ad5 (subgroup C) has been widely used as a gene therapy vector due to the efficient manner in which it delivers its genome to the nuclei of target cells. However, efficient gene transfer and expression occurs in cells expressing the coxsackievirus-Ad receptor (CAR) (3, 45). The requirement for expression of the Ad5 receptor on the target cell represents a hurdle to genetic modification of cells that lack CAR, leading to the strategy of modifying the tropism of Ad5 vectors (10, 11, 13, 14, 26, 27, 37, 42, 48–51). This strategy presupposes that intracellular trafficking of Ad5 will not be affected by modified tropism. Utilizing an Ad5 vector with modified tropism by virtue of expression of the Ad7 (subgroup B) fiber protein in place of Ad5 fiber (13), the consequences of modified tropism on intracellular trafficking of an Ad5 vector were examined. The two parental Ad exhibited significant differences in intracellular trafficking. Cy3-Ad5 capsids showed efficient targeting of the nucleus within 1 h after infection, with the majority of the Ad5 genome detected in association with the nucleus 1 h after infection by both FISH on fixed cells and TaqMan PCR following subcellular fractionation. In contrast, at the same time point, Cy3-Ad7 capsids displayed a predominantly cytoplasmic distribution, with the Ad7 genome progressively accumulating in the nucleus over an 8-h period. Strikingly, the chimeric Ad5f7 vector exhibited intracellular trafficking characteristics similar to those of Ad7 rather than Ad5, suggesting that substitution of the Ad7 fiber had a significant impact on intracellular trafficking of the Ad5 capsid and delivery of the Ad5 genome to the nucleus.
Intracellular trafficking of Ad5 and Ad7.
The present study showed that intracellular trafficking of Ad5 capsids and that of Ad7 capsids were distinct in several respects. The differences in Ad5 and Ad7 trafficking occurred distal to the internalization step, given the observation that Ad5 and Ad7 internalized into A549 cells at comparable rates. The half time for internalization, approximately 5 min for each vector, was comparable to internalization half times previously reported for subgroup C Ad (17, 28, 44, 46). In contrast, Cy3-Ad5 capsid progressed to the nucleus more rapidly than that of Cy3-Ad7. The distribution of capsid in the cytoplasm also differed, with Ad5 exhibiting a homogeneous punctate distribution throughout the cytoplasm whereas Ad7 initially exhibited a homogeneous punctate distribution that evolved into a pattern including cytoplasmic aggregations of heterogenous size. Importantly, Ad7 at the perimeter of the nucleus often exhibited a uniform punctate appearance similar to the appearance of Ad7 immediately following infection, suggesting that Ad7 capsids were released from aggregates prior to their encounter with the nuclear envelope. These data are consistent with previous observations comparing the intracellular distribution of subgroup B and C Ad following infection. Working with HeLa cells, Dales and coworkers (4, 5, 8) noted that the principal morphological distinction between the two subgroups was that subgroup B Ad accumulated in large, membranous organelles during infection whereas multiple subgroup C virions were rarely found within the same membranous organelle. Similarly, working with A549 cells, Defer et al. (9) observed that subgroup C Ad caused a more complete release of cointernalized particles to the cytoplasm, suggesting escape from an early, common endocytic compartment, whereas subgroup B Ad was less efficient in releasing cointernalized particles, suggesting escape from a later, more specialized endocytic compartment. Furthermore, our data obtained by confocal laser scanning microscopy showed significant colocalization of Ad7 and Ad5f7 with a marker of endocytic compartments, α2M. This fact supported the concept that a significant population of subgroup B Ad and the chimeric vector remained in endocytic compartments 1 h after infection. Taken together, the present report and previous studies with several cell types (4, 5, 8, 9) establish the fact that subgroups B and C follow different intracellular trafficking routes.
The fact that Ad5 and Ad7 capsids showed distinct routes to the nucleus predicted that the Ad genome would also differ in the kinetics of approach to the nucleus. Using FISH analysis, the Ad5 genome’s arrival at the nucleus clearly preceded the arrival of the Ad7 genome, with the majority of the Ad5 genome showing nuclear localization at 1 h while Ad7 required 8 h to achieve the same nuclear localization. Quantitative determination of Ad genome delivery to the nucleus performed using TaqMan PCR showed that, as predicted by the FISH observations, the Ad5 genome encountered the nucleus with more rapid kinetics than Ad7. Ad5 showed a half time of 40 min for nuclear localization, a value comparable to those previously reported for the half time of dissociation of hexon protein from the Ad genome (17). In contrast, the Ad7 genome localized to the nucleus with slower kinetics, exhibiting a half time of approximately 2 h. The progressive accumulation of the Ad7 genome at the nucleus assessed by FISH demonstrated that the Ad7 capsids were eventually effective in accomplishing delivery of DNA to the nucleus, albeit with slower kinetics, despite the failure of the pulse of incoming virus to localize to the nucleus in a coherent manner, as observed with Ad5.
Intracellular trafficking of the Ad5f7 chimera.
Particle for particle, the chimeric Ad5f7 vector has previously been shown to be comparable to Ad5 and Ad7 vectors with respect to transgene expression as assessed by CAT assay 12 to 72 h after infection (1, 13). The differences in intracellular trafficking of the Ad5 capsid as a result of the presence of the Ad7 fiber protein, therefore, do not affect the ultimate efficacy of the vector over time. However, modifying the tropism does slow the process of genome delivery to the nucleus, a parameter that may be important for some gene transfer applications. Further, given the multiple potential endocytic routes within the cell (32), it is conceivable that introduction of a novel tropism might induce trafficking of modified vectors into inhospitable, degradative environments such as lysosomes and thus modify the presentation of the virus to host defense immune recognition systems in a different fashion. With the establishment of a precedent for modified intracellular trafficking of an Ad vector shown here, investigators of vectors with modified tropism should consider the concept of intracellular trafficking as well as the binding-internalization step.
Role of fiber.
Data in the present study demonstrating the influence of the Ad7 fiber on Ad5 trafficking in the Ad5f7 chimeric vector suggest a new role for the Ad fiber protein during Ad infection. The Ad fiber protein imparts high-affinity binding of the Ad capsid to the plasma membrane of target cells (33, 34). Biochemical studies of Ad2 indicate that the fiber and penton base protein are rapidly released from the body of the capsid following internalization (17, 43). Data presented here suggest that the Ad fiber protein, at least in some Ad serotypes, influences postinternalization Ad trafficking. The observation that the Ad7 fiber modifies the Ad5f7 vector such that it trafficks more like an Ad7 virus suggests that the fiber may play a role in directing the virus into a specific intracellular environment. One possibility is that the accumulation of Ad7 and Ad5f7 is a result of nondissociation of fiber after internalization. If the utilization of a different receptor is instrumental in dictating the resulting trafficking pattern, then future attempts to alter Ad tropism for gene transfer applications may encounter similar intracellular differences in trafficking, some of which may be detrimental (or advantageous) to genome delivery to the nucleus.
ACKNOWLEDGMENTS
We thank Noboru Sato, Chisa Hidaka, and Mannix Quitoriano for helpful discussions; Tarek El-Sawy and Ivan Silva for assistance with the quantitative PCR assays; Stephanie Rempel for technical assistance; and N. Mohamed for help in preparing the manuscript.
These studies were supported in part by the National Institutes of Health, National Heart, Lung and Blood Institute, grants P01 HL51746 and P01 HL59312; the Will Rogers Memorial Fund, Los Angeles, Calif.; the Cystic Fibrosis Foundation, Bethesda, Md.; and GenVec, Inc., Rockville, Md. P.L.L. is supported in part by NIH grant R29 AI-42250.
REFERENCES
- 1.Abrahamsen K, Kong H-L, Mastrangeli A, Brough D, Lizonova A, Crystal R G, Falck-Pedersen E. Construction of an adenovirus type 7a E1a− vector. J Virol. 1997;71:8946–8951. doi: 10.1128/jvi.71.11.8946-8951.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai M, Harfe B, Freimuth P. Mutations that alter an Arg-Gly-Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell-rounding activity and delay virus reproduction in flat cells. J Virol. 1993;67:5198–5205. doi: 10.1128/jvi.67.9.5198-5205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 4.Chardonnet Y, Dales S. Early events in the interaction of adenoviruses with HeLa cells. I. Penetration of type 5 and intracellular release of the DNA genome. Virology. 1970;40:462–477. doi: 10.1016/0042-6822(70)90189-3. [DOI] [PubMed] [Google Scholar]
- 5.Chardonnet Y, Dales S. Early events in the interaction of adenoviruses with HeLa cells. II. Comparative observations on the penetration of types 1, 5, 7, and 12. Virology. 1970;40:478–485. doi: 10.1016/0042-6822(70)90190-x. [DOI] [PubMed] [Google Scholar]
- 6.Crawford-Miksza L, Schnurr D P. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J Virol. 1996;70:1836–1844. doi: 10.1128/jvi.70.3.1836-1844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crystal R G. Transfer of genes to humans: early lessons and obstacles to success. Science. 1995;270:404–410. doi: 10.1126/science.270.5235.404. [DOI] [PubMed] [Google Scholar]
- 8.Dales S. An electron microscope study of the early association between two mammalian viruses and their roles. J Cell Biol. 1962;13:303–322. doi: 10.1083/jcb.13.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Defer C, Belin M T, Caillet-Boudin M L, Boulanger P. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J Virol. 1990;64:3661–3673. doi: 10.1128/jvi.64.8.3661-3673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas J T, Rogers B E, Rosenfeld M E, Michael S I, Feng M, Curiel D T. Targeted gene delivery by tropism-modified adenoviral vectors. Nat Biotechnol. 1996;14:1574–1578. doi: 10.1038/nbt1196-1574. [DOI] [PubMed] [Google Scholar]
- 11.Feng M, Jackson W H, Jr, Goldman C K, Rancourt C, Wang M, Dusing S K, Siegal G, Curiel D T. Stable in vivo gene transduction via a novel adenoviral/retroviral chimeric vector. Nat Biotechnol. 1997;15:866–870. doi: 10.1038/nbt0997-866. [DOI] [PubMed] [Google Scholar]
- 12.Frey B M, Hackett N R, Bergelson J M, Finberg R, Crystal R G, Moore M A S, Rafii S. High-efficiency gene transfer into ex vivo expanded human hematopoietic progenitors and precursor cells by adenovirus vectors. Blood. 1998;91:2781–2792. [PubMed] [Google Scholar]
- 13.Gall J, Kass-Eisler A, Leinwand L, Falck-Pedersen E. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J Virol. 1996;70:2116–2123. doi: 10.1128/jvi.70.4.2116-2123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldman C K, Rogers B E, Douglas J T, Sosnowski B A, Ying W, Siegal G P, Baird A, Campain J A, Curiel D T. Targeted gene delivery to Kaposi’s sarcoma cells via the fibroblast growth factor receptor. Cancer Res. 1997;57:1447–1451. [PubMed] [Google Scholar]
- 15.Goldman M J, Wilson J M. Expression of αvβ5 integrin is necessary for efficient adenovirus-mediated gene transfer in the human airway. J Virol. 1995;69:5951–5958. doi: 10.1128/jvi.69.10.5951-5958.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greber U F, Kasamatsu H. Nuclear targeting of adenovirus and simian virus SV40. Trends Cell Biol. 1996;6:189–195. doi: 10.1016/0962-8924(96)10016-7. [DOI] [PubMed] [Google Scholar]
- 17.Greber U F, Willetts M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–486. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- 18.Greber U F, Webster P, Weber J, Helenius A. The role of the adenovirus protease on virus entry into cells. EMBO J. 1996;15:1766–1777. [PMC free article] [PubMed] [Google Scholar]
- 19.Greber U F, Suomalainen M, Stidwill R P, Boucke K, Ebersold M W, Helenius A. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 1997;16:5998–6007. doi: 10.1093/emboj/16.19.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hersh J, Crystal R G, Bewig B. Modulation of gene expression after replication-deficient, recombinant adenovirus-mediated gene transfer by the product of a second adenovirus vector. Gene Ther. 1995;2:124–131. [PubMed] [Google Scholar]
- 21.Horwitz M S. Adenoviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers, Inc.; 1996. pp. 2149–2171. [Google Scholar]
- 22.Huang S, Endo R I, Nemerow G R. Upregulation of integrins αvβ3 and αvβ5 on human monocytes and T lymphocytes facilitates adenovirus-mediated gene delivery. J Virol. 1995;69:2257–2263. doi: 10.1128/jvi.69.4.2257-2263.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang S, Kamata T, Takada Y, Ruggeri Z M, Nemerow G R. Adenovirus interaction with distinct integrins mediates separate events in cell entry and gene delivery to hematopoietic cells. J Virol. 1996;70:4502–4508. doi: 10.1128/jvi.70.7.4502-4508.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaner R J, Worgall S, Leopold P L, Stolze E, Milano E, Hidaka C, Ramalingam R, Hackett N R, Singh R, Bergelson J M, Finberg R W, Falck-Pedersen E, Crystal R G. Modification of the genetic program of human alveolar macrophages by adenovirus vectors in vitro is feasible but inefficient, limited in part by the low level of expression of the coxsackie/adenovirus receptor. Am J Respir Cell Mol Biol. 1999;20:361–370. doi: 10.1165/ajrcmb.20.3.3398. [DOI] [PubMed] [Google Scholar]
- 25.Kass-Eisler A, Falck-Pedersen E, Alvira M, Rivera J, Buttrick P M, Wittenberg B A, Cipriani L, Leinwand L A. Quantitative determination of adenovirus-mediated gene delivery to rat cardiac myocytes in vitro and in vivo. Proc Natl Acad Sci USA. 1993;90:11498–11502. doi: 10.1073/pnas.90.24.11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krasnykh V, Dmitriev I, Mikheeva G, Miller C R, Belousova N, Curiel D T. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J Virol. 1998;72:1844–1852. doi: 10.1128/jvi.72.3.1844-1852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krasnykh V N, Mikheeva G V, Douglas J T, Curiel D T. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J Virol. 1996;70:6839–6846. doi: 10.1128/jvi.70.10.6839-6846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leopold P L, Ferris B, Grinberg I, Worgall S, Hackett N R, Crystal R G. Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum Gene Ther. 1998;9:367–378. doi: 10.1089/hum.1998.9.3-367. [DOI] [PubMed] [Google Scholar]
- 29.Mayor S, Sabharanjak S, Maxfield F R. Cholesterol-dependent retention of GPI-anchored proteins in endosomes. EMBO J. 1998;17:4626–4638. doi: 10.1093/emboj/17.16.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mittereder N, March K L, Trapnell B C. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70:7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muggeridge M I, Fraser N W. Chromosomal organization of the herpes simplex virus genome during acute infection of the mouse central nervous system. J Virol. 1986;59:764–767. doi: 10.1128/jvi.59.3.764-767.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukherjee S, Ghosh R N, Maxfield F R. Endocytosis. Physiol Rev. 1997;77:759–803. doi: 10.1152/physrev.1997.77.3.759. [DOI] [PubMed] [Google Scholar]
- 33.Persson R, Wohlfart C, Svensson U, Everitt E. Virus-receptor interaction in the adenovirus system: characterization of the positive cooperative binding of virions on HeLa cells. J Virol. 1985;54:92–97. doi: 10.1128/jvi.54.1.92-97.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Philipson L, Lonberg-Holm K, Pettersson U. Virus-receptor interaction in an adenovirus system. J Virol. 1968;2:1064–1075. doi: 10.1128/jvi.2.10.1064-1075.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickles R J, Barker P M, Ye H, Boucher R C. Efficient adenovirus-mediated gene transfer to basal but not columnar cells of cartilaginous airway epithelia. Hum Gene Ther. 1996;7:921–931. doi: 10.1089/hum.1996.7.8-921. [DOI] [PubMed] [Google Scholar]
- 36.Roelvink P W, Kovesdi I, Wickham T J. Comparative analysis of adenovirus fiber-cell interaction: adenovirus type 2 (Ad2) and Ad9 utilize the same cellular fiber receptor but use different binding strategies for attachment. J Virol. 1996;70:7614–7621. doi: 10.1128/jvi.70.11.7614-7621.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers B E, Douglas J T, Ahlem C, Buchsbaum D J, Frincke J, Curiel D T. Use of a novel cross-linking method to modify adenovirus tropism. Gene Ther. 1997;4:1387–1392. doi: 10.1038/sj.gt.3300541. [DOI] [PubMed] [Google Scholar]
- 38.Rosenfeld M A, Siegfried W, Yoshimura K, Yoneyama K, Fukayama M, Stier L E, Paakko P K, Gilardi P, Stratford-Perricaudet L D, Perricaudet M, Jallat S, Pavirani A, Lecocq J-P, Crystal R G. Adenovirus-mediated transfer of a recombinant α1-antitrypsin gene to the lung epithelium in vivo. Science. 1991;252:431–434. doi: 10.1126/science.2017680. [DOI] [PubMed] [Google Scholar]
- 39.Rosenfeld M A, Yoshimura K, Trapnell B C, Yoneyama K, Rosenthal E R, Dalemans W, Fukayama M, Bargon J, Stier L E, Stratford-Perricaudet L, Perricaudet M, Guggino W B, Pavirani A, Lecocq J-P, Crystal R G. In vivo transfer of the human cystic fibrosis transmembrane conductance regulator gene to the airway epithelium. Cell. 1992;68:143–155. doi: 10.1016/0092-8674(92)90213-v. [DOI] [PubMed] [Google Scholar]
- 40.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers, Inc.; 1996. pp. 2111–2148. [Google Scholar]
- 41.Stevenson S C, Rollence M, White B, Weaver L, McClelland A. Human adenovirus serotypes 3 and 5 bind to two different cellular receptors via the fiber head domain. J Virol. 1995;69:2850–2857. doi: 10.1128/jvi.69.5.2850-2857.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson S C, Rollence M, Marshall-Neff J, McClelland A. Selective targeting of human cells by a chimeric adenovirus vector containing a modified fiber protein. J Virol. 1997;71:4782–4790. doi: 10.1128/jvi.71.6.4782-4790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sussenbach J S. Early events in the infection process of adenovirus type 5 in HeLa cells. Virology. 1967;33:567–574. doi: 10.1016/0042-6822(67)90056-6. [DOI] [PubMed] [Google Scholar]
- 44.Svensson U. Role of vesicles during adenovirus 2 internalization into HeLa cells. J Virol. 1985;55:442–449. doi: 10.1128/jvi.55.2.442-449.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomko R P, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang K, Huang S, Kapoor-Munshi A, Nemerow G. Adenovirus internalization and infection require dynamin. J Virol. 1998;72:3455–3458. doi: 10.1128/jvi.72.4.3455-3458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 48.Wickham T J, Roelvink P W, Brough D E, Kovesdi I. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat Biotechnol. 1996;14:1570–1573. doi: 10.1038/nbt1196-1570. [DOI] [PubMed] [Google Scholar]
- 49.Wickham T J, Lee G M, Titus J A, Sconocchia G, Bakacs T, Kovesdi I, Segal D M. Targeted adenovirus-mediated gene delivery to T cells via CD3. J Virol. 1997;71:7663–7669. doi: 10.1128/jvi.71.10.7663-7669.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wickham T J, Segal D M, Roelvink P W, Carrion M E, Lizonova A, Lee G M, Kovesdi I. Targeted adenovirus gene transfer to endothelial and smooth muscle cells by using bispecific antibodies. J Virol. 1996;70:6831–6838. doi: 10.1128/jvi.70.10.6831-6838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wickham T J, Tzeng E, Shears II L L, Roelvink P W, Li Y, Lee G M, Brough D E, Lizonova A, Kovesdi I. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J Virol. 1997;71:8221–8229. doi: 10.1128/jvi.71.11.8221-8229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson J M. Adenoviruses as gene-delivery vehicles. N Engl J Med. 1996;334:1185–1187. doi: 10.1056/NEJM199605023341809. [DOI] [PubMed] [Google Scholar]
- 53.Yamashiro D J, Tycko B, Fluss S R, Maxfield F R. Segregation of transferrin to a mildly acidic (pH 6.5) para-Golgi compartment in the recycling pathway. Cell. 1984;37:789–800. doi: 10.1016/0092-8674(84)90414-8. [DOI] [PubMed] [Google Scholar]
- 54.Zabner J, Zeiher B G, Friedman E, Welsh M J. Adenovirus-mediated gene transfer to ciliated airway epithelia requires prolonged incubation time. J Virol. 1996;70:6994–7003. doi: 10.1128/jvi.70.10.6994-7003.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]