Abstract
Colorectal cancer (CRC) is a prevalent and aggressive malignancy characterized by a complex tumor microenvironment (TME). Given the variations in the level of adipocyte infiltration in TME, the prognosis may differ among CRC patients. Thus, there is an urgent need to establish a reliable method for identifying adipocyte subtypes in CRC in order to elucidate the impact of adipocyte infiltration on CRC treatment and prognosis. Herein, 144 adipocyte-infiltration-related genes (AIRGs) were identified as predictive markers for the immune-associated features and prognosis of CRC patients. Based on the 144 genes, the unsupervised clustering algorithm identified two distinct clusters of CRC patients with variations in molecular and signaling pathways, clinicopathological characteristics and responses to CRC chemotherapy and immunotherapy. Furthermore, an AIRG prognostic signature was constructed and validated in independent datasets. Overall, this study developed a prognostic signature based on AIRGs in CRC, which may contribute to the development of personalized treatment strategies and enhance prognostic prediction for CRC patients.
Keywords: Colorectal cancer, Adipocyte infiltration, Unsupervised clustering, Immune-associated features, Prognosis
1. Introduction
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related deaths globally [1]. Despite advancements in its diagnosis and treatment, it remains a significant clinical challenge owing to its heterogeneity and variable treatment responses. Currently, the management of CRC involves a combination of surgical resection, chemotherapy, targeted therapy, and immunotherapy, depending on the stage and molecular characteristics of the tumor [2]. The TNM staging system, which considers tumor size, lymph node involvement, and metastasis, provides valuable prognostic information and guides treatment decisions [3]. Additionally, traditional biomarkers in the TME, such as BRAF-V600E, KRAS, and DNA mismatch repair gene, are utilized for monitoring disease progression or response to therapy [4]. Indeed, classical assessment strategies (TNM staging system or conventional biomarkers) play a crucial role in the clinical setting [5]. Nevertheless, their limitations, such as incomplete assessment of TME heterogeneity, interobserver variability, and limited predictive value, cannot be overlooked [6,7]. Hence, subtyping CRC based on clinical characteristics and TME holds significant implications.
Ascribed to the heterogeneity of the TME, CRC development and progression can be affected by surrounding cells and considerably vary. Adipocytes, with the discrepant infiltration in the TME, are implicated in numerous aspects of cancer biology, including cytokine production, promotion of angiogenesis, modulation of immune response, facilitation of tumor progression, and conferring chemotherapy resistance [[8], [9], [10]]. The critical role of adipocyte infiltration in the TME has been validated in breast cancer [11], epithelial ovarian cancer [12], and esophageal cancer [13]. Nonetheless, a comprehensive understanding of their effects in CRC and their specific contributions to disease heterogeneity is still lacking. Therefore, identifying distinct adipocyte subtypes in CRC could offer new insights into potential mechanisms driving CRC progression, as well as developing treatment strategies and improving prognostic prediction.
In this study, two distinct clusters of CRC patients were identified based on 144 screened adipocyte-infiltration-related genes (AIRGs). Notable, significant differences were noted in molecular signaling pathways, immune-associated features, clinicopathological characteristics, and the response to chemotherapy and immunotherapy between the two distinct clusters. Furthermore, a risk model was constructed using clinicopathological features and AIRG signature, demonstrating high prognostic accuracy for CRC. Identification of AIRG subtypes may garner attention to the role of adipocyte infiltration in CRC and provide a potential strategy for guiding chemotherapy or immunotherapy and predicting outcomes in CRC patients.
2. Results
2.1. Adipocyte infiltration was identified as a risk factor related to CRC prognosis
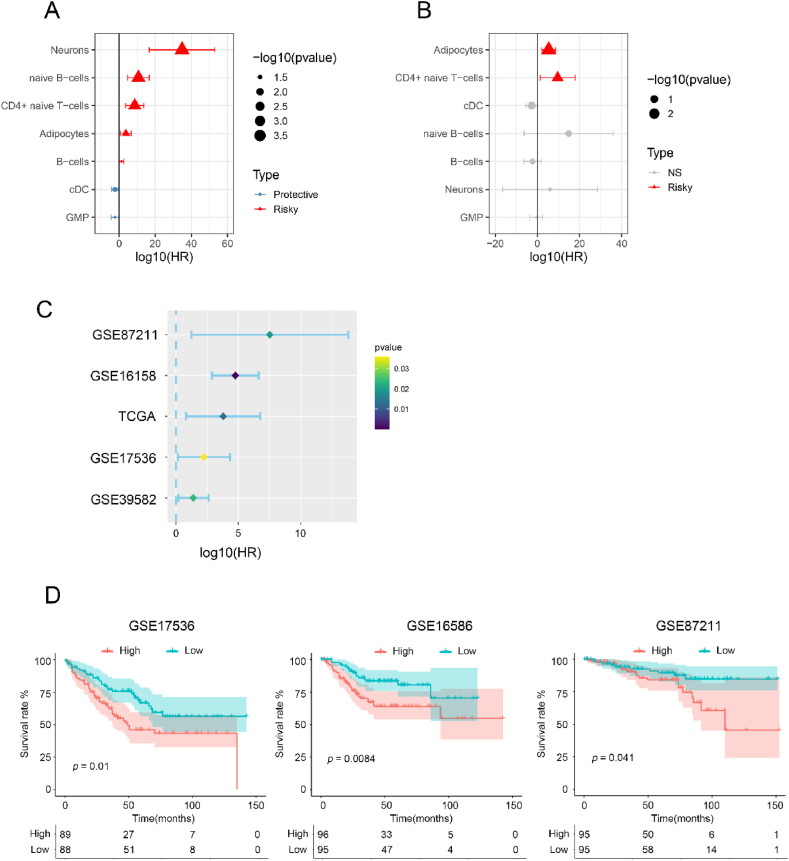
Considering the relevance of different cell type infiltrations in the prognosis of CRC patients, x-cell analysis was employed to assess the degree of immune infiltration in 430 patients with colorectal cancer (CRC) from the TCGA database. Univariate survival analysis identified seven cell types that significantly influenced prognosis (Fig. 1A). Afterward, multivariate analysis revealed that the infiltration of adipocytes and CD4+ cells was negatively correlated with prognosis (Fig. 1B). Adipocytes, the most significantly prominent, were identified in different databases (Fig. 1C–Tables S1–4). In addition, adipocytes infiltration was associated with survival outcomes, as demonstrated by Kaplan-Meier analysis in three independent datasets: GSE17536, GSE16586, and GSE87211 (Fig. 1D). The statistical significance of these results indicates that patients with high adipocyte infiltration have significantly worse survival compared to those with low adipocyte infiltration. Specifically, in GSE17536, the survival difference was highly significant (p = 0.01), reinforcing the negative impact of adipocyte infiltration on survival. Similarly, in GSE16586 and GSE87211, the p-values (0.0084 and 0.041, respectively) underscore the consistent detrimental effect across different cohorts. Taken together, these findings suggest a potential role of adipocyte infiltration in the prognosis of colorectal cancer, highlighting the importance of considering adipocyte levels in patient survival predictions.
Fig. 1.
Identifying high infiltration of adipocytes associated with worse survival in CRC patients. (A) Univariate and (B) Multivariate survival analysis survival analysis of seven prognosis-related cell types in TCGA cohorts. (C) Identification the impact of adipocytes on the prognosis of CRC in the GSE87211, GSE16158, TCGA, GSE17536 and GSE39582 databases. (D) Survival correlation analysis for adipocytes in GSE17536, GSE16586 and GSE87211 databases.
2.2. Identification of AIRGs
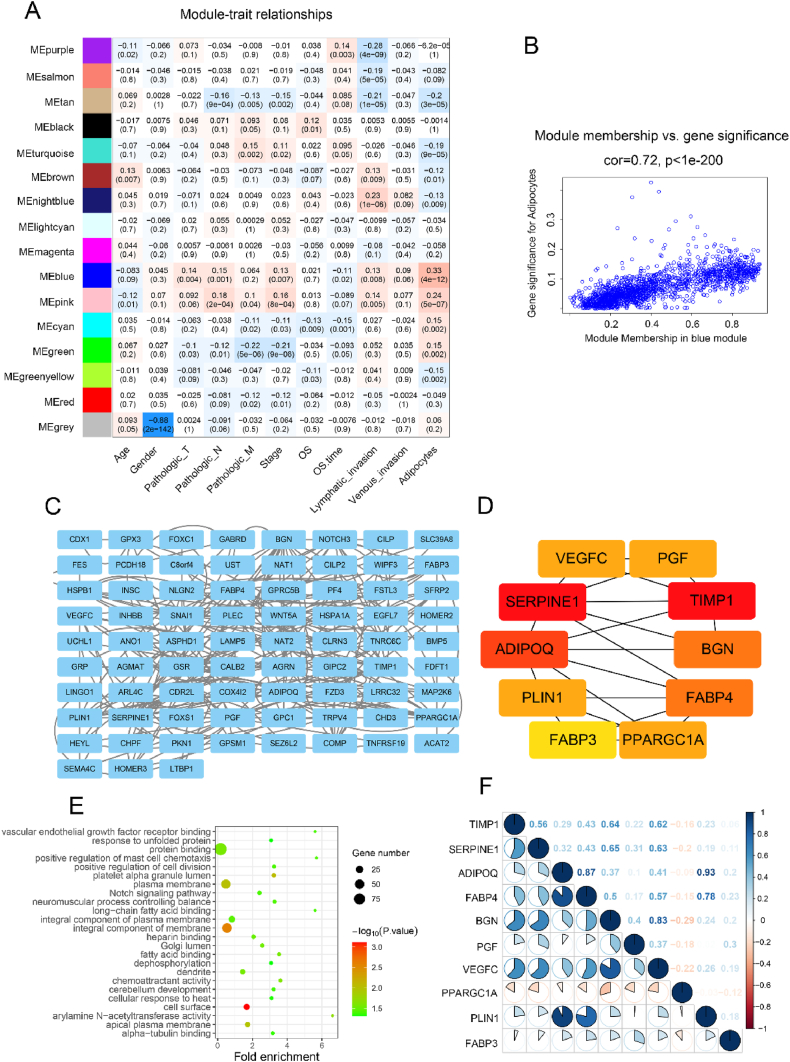
To identify genes associated with adipocyte infiltration, Weighted Gene Co-expression Network Analysis (WGCNA) was performed to analyze the co-expression patterns between invasion and whole-transcriptome profiling data (Fig. S1A). β = 8 was set as the soft threshold (Fig. S1B), and 16 modules were identified (Fig. 2A–S1C). Our analysis revealed significant differences in gene expression levels within the MEblue dataset (Fig. 2B). Thus, the MEblue module highly correlated with adipocytes was chosen for the ensuing analyses (Fig. 2B). To further investigate the biological significance of these 1582 genes in the MEblue module, a Protein-Protein Interaction (PPI) network was constructed using Cytoscape (Fig. 2C). Based on these findings, 10 adipocyte-associated markers were identified as core genes with high betweenness centrality (Fig. 2D), and their correlations were quantified (Fig. 2F). KEGG pathway analysis of genes in the network indicated that they were enriched in tumor invasion (Fig. 2E).
Fig. 2.
Identification of genes associated with adipocyte infiltration. (A) Heatmap of the correlation between the modules and cancer hallmarks. (B) Correlation between MEblue module and adipocytes infiltration. (C) Protein-Protein Interaction network analysis of 144 adipocyte infiltration genes according to MEblue module. (D) Protein-Protein Interaction network of core proteins from (C). (E) The top of 24 KEGG pathways enriched for the 10 core genes. (F) Correlation analysis between 10 core genes.
2.3. AIRGs-based subtypes and clinicopathological analysis
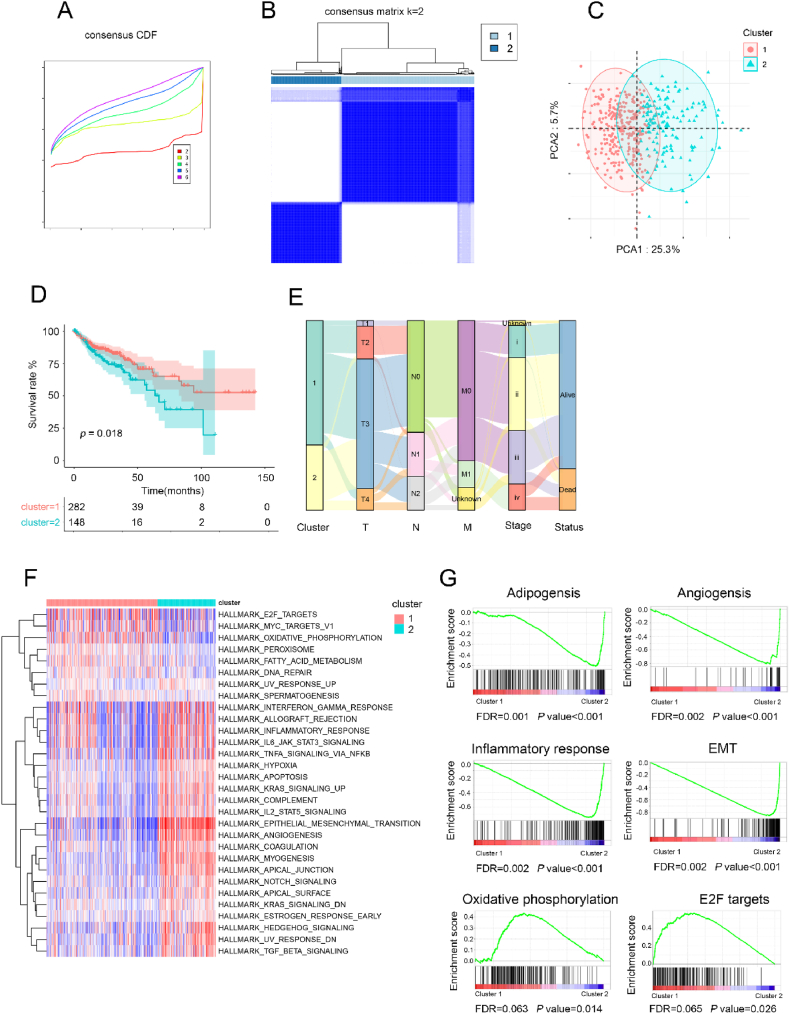
To identify the subtypes of AIRGs in CRC, the Cumulative Distribution Function (CDF) and consensus matrix heatmap suggested that two subgroups might be optimal for CRC patients (Fig. 3A and B). This result was consistent with the finding of principal component analysis (PCA) (Fig. 3C). Furthermore, significant statistical differences were observed in the T stage, N stage, M stage, clinical stage, and survival outcome between the two clusters of CRC patients, demonstrating the rationality of classification for CRC patients based on AIRGs (Fig. 3D and E). Specifically, survival analysis exposed that patients in Cluster 1 had a better prognosis compared to those in Cluster 2. Comparable results were obtained using an independent dataset (Supplementary Materials, Figs. S2A–e). Next, Gene Set Variation Analysis (GSVA) was conducted on the gene sets of Cluster 1 and Cluster 2 to uncover the underlying reasons for the differences between the two subtypes (Fig. 3F). According to the results of Gene Set Enrichment Analysis (GSEA), oxidative phosphorylation and E2F targets were enriched in Cluster 1. On the other hand, adipogenesis, angiogenesis, inflammatory response, and EMT pathways were predominantly enriched in Cluster 2. These results collectively implied that the two categorized AIRG patterns could discriminate the prognosis of CRC patients, with significant differences in biological characteristics.
Fig. 3.
Subtypes, clinicopathological and biological analysis based AIRGs. (A) The CDF curves according to different k values. (B) Consensus matrix heatmap of two clusters (k = 2) and corresponding area. (C) Visualization of consensus Cluster analysis results using PCA. (D) Kaplan - Meier curves for OS of TCGA cohort based adipocyte infiltration genes subtypes. (E) Sankey diagram showing the distribution of clinicopathologic characteristics in two clusters. (F) GSVA analysis of hallmark and KEGG pathway gene sets in cluster 1 and cluster 2 from TCGA datasets. (G) GSEA analysis of adipogenesis, angiogenesis, inflammatory response, EMT, oxidative phosphorylation, and E2F targets in two clusters.
2.4. Differences in immune-associated features between AIRGs-based subtypes
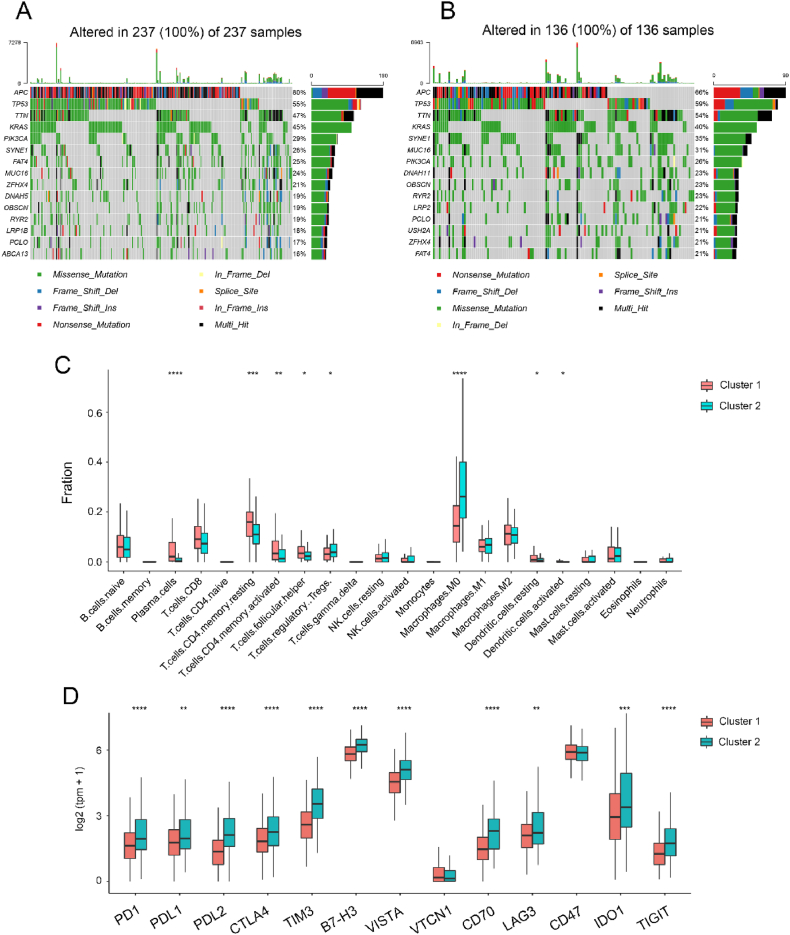
Given the critical role of genomic alterations in CRC progression, differences in gene mutations were examined between the two subtypes. The mutation landscape revealed that the top three mutated genes in both subtypes were APC, TP53, and TTN. Of note, the frequency of TP53 and TTN mutations was higher in Cluster 2, whereas that of APC was higher in Cluster 1 (Fig. 4A and B). Additionally, a comprehensive analysis of the immune infiltration patterns in Clusters 1 and 2 was conducted. The analysis determined a significantly higher infiltration of regulatory T cells (Tregs) and M0 macrophages in Cluster 2 compared to Cluster 1 (Fig. 4C). Moreover, further investigation of immune checkpoints revealed a substantial upregulation in the expression of PD1, PDL1, and CD70 in Cluster 2, indicating the presence of an immunosuppressive microenvironment (Fig. 4D). Taken together, these findings suggest a potential role of immune evasion mechanisms in the progression of colorectal cancer in Cluster 2.
Fig. 4.
Immune-associated features of different adipocyte subgroups. (A) Landscape of genomic aberrations of cluster 1. (B) Landscape of genomic aberrations of cluster 2. (C) Relative proportions of 22 immune cells in different adipocyte subgroups. (D) Gene expression of immune checkpoints between two distinct clusters.
2.5. The correlation between AIRGs subtypes and clinical response of chemotherapy or immunotherapy
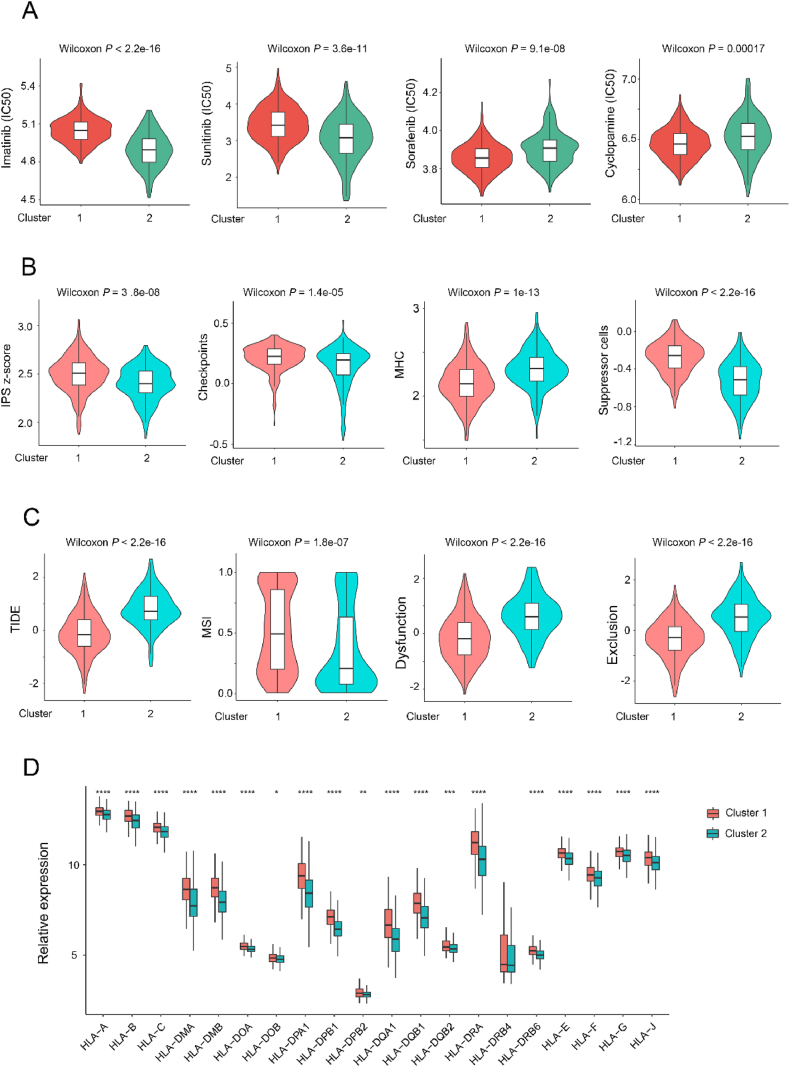
The potential mechanism behind the significant difference in prognosis between the two subtypes was further analyzed. Imatinib and sunitinib had a lower half-maximal inhibitory concentration (IC50) in Cluster 2, whereas sorafenib and cyclopamine had a lower IC50 in Cluster 1, suggesting a potential enhanced treatment response in the corresponding subgroup (Fig. 5A). Interestingly, the majority of conventional chemotherapy drugs exhibited similar efficacy between the two subtypes (Fig. S3A). The IPS score, checkpoint expression, MHC expression, suppression cell levels, TIDE score, MSI status, immune dysfunction, and immune exclusion patterns between two adipocyte subtypes were evaluated to elucidate their response to immune-based therapies (Fig. 5B, C, and S3B). The results uncovered that Cluster 2 was in a more immunosuppressive microenvironment. Following this, MHC-I molecules between the two subtypes were comprehensively analyzed (Fig. 5D). These findings consistently highlight the potential of immunotherapeutic strategies for Cluster 2 colorectal cancer patients.
Fig. 5.
The correlation between AIRGs subtypes and clinical treatment response. (A) Four common therapeutic drugs with differential IC50 between two adipocyte subtypes. (B, C) Violin plot of the IPS score, checkpoints, MHC, suppression cells, TIDE, MSI, dysfunction and exclusion between two adipocyte subtypes. (D) Gene expression of MHC-I molecules between two distinct clusters.
2.6. AIRGs signature for the prediction of CRC prognosis
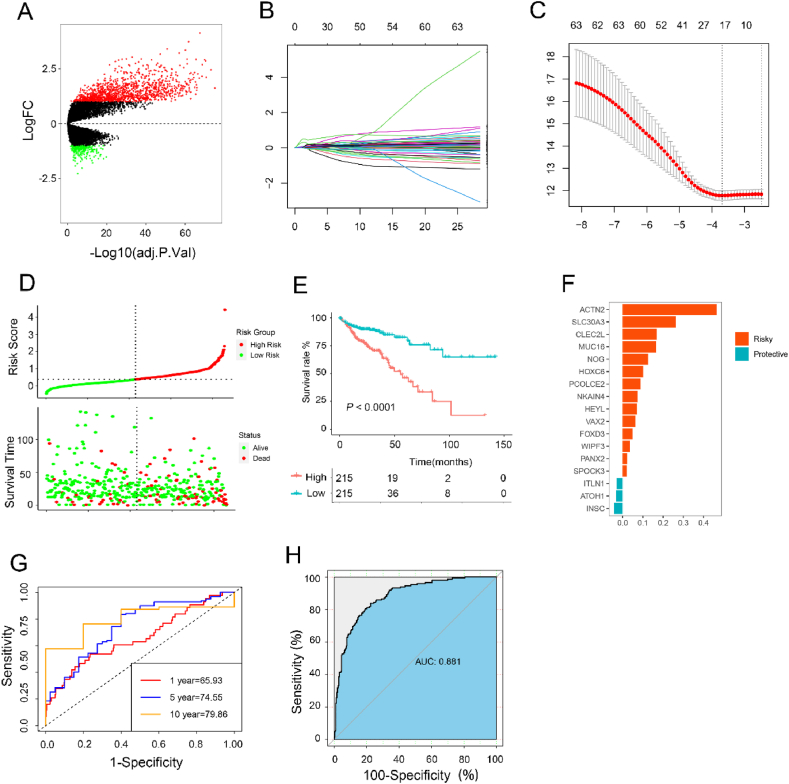
The volcano plot revealed significant differences in gene expression levels between the two subtypes (Fig. 6A). Using these differentially expressed genes, LASSO penalized Cox regression analysis was applied to construct an adipocyte risk signature for CRC patients (Fig. 6B and C). A total of 17 genes (ACTIN2, SLC30A3, CLEC2L, MUC16, NOG, HOXC6, PCOLCE, NKAIN4, HEYL, VAX2, FOXD3, WIPF3, PANX2, SPOCK3, ITLN1, ATOH1, and INSC) was selected to design the novel risk score (Fig. 6F). Afterward, the adipocyte risk signature score was computed using the expression levels of the genes in colorectal cancer (CRC) patients, who were then categorized into a high-risk group or a low-risk group (Fig. 6D). Kaplan-Meier analysis demonstrated that patients with higher risk scores exhibited a poor prognosis (Fig. 6E). At the same time, receiver operating characteristic (ROC) analysis displayed that the area under the curve was 0.881, insinuating that our risk signature could accurately distinguish between the two subtypes (Fig. 6H). Furthermore, our adipocyte infiltration gene signature demonstrated outstanding accuracy in time-dependent ROC analysis, inferring that our signature holds considerable potential as a reliable prognostic tool for assessing long-term survival and disease progression in CRC patients (Fig. 6G). In addition to TCGA, another three independent cohorts (TCGA, GSE38832 and GSE161158) were used for the prognostic value of AIRGs signature (Figure S4, S5 and S6). Real-time PCR was then used to validate the expression of genes related to energy metabolism and immunosuppression in human normal colon epithelial cells (NCM460) and four human colon cancer cell lines (HCT116, LoVo, SW48, DLD1) (Fig. S7).
Fig. 6.
Construction of AIRGs subtypes-based risk score to predict CRC prognosis in TCGA. (A) The volcano plot of differentially expressed genes (DEGs) between the two subtypes (logFC >2 and adjusted p-value <0.05). (B) LASSO coefficient profiles of adipocyte-related prognostic differential expressed genes. (C) 10-fold cross-validation for penalty parameter λ selection in LASSO mode. (D) The distribution of risk score, patients' status, and survival time. (E) Kaplan–Meier survival curves for patients in high- and low-risk groups. (F) LASSO coefficients of 17 adipocyte-related genes. (G) ROC curves for predicting 1-, 5-, and 10-year overall survival by risk score. (H) ROC curves for predicting adipocyte subtypes by risk score.
2.7. AIRGs-based nomogram improved survival prediction for CRC
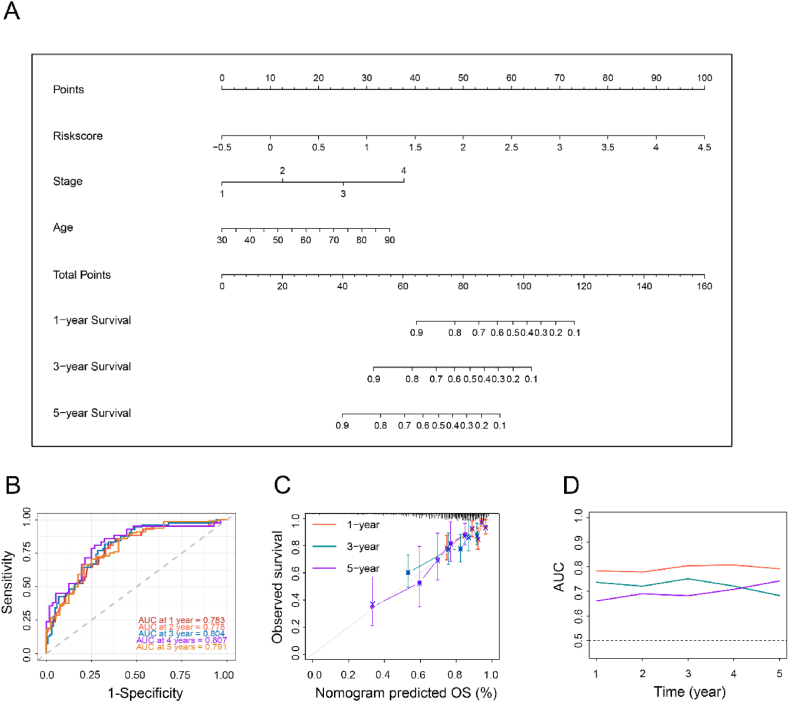
Finally, a nomogram was generated by incorporating four independent prognostic factors to provide a quantitative tool for predicting OS in CRC patients. The nomogram enabled the calculation of an individual's total score based on their specific characteristics, allowing for a personalized assessment of their prognosis (Fig. 7A). Additionally, the calibration plot demonstrated a high degree of concordance between the observed survival probabilities and the predicted survival probabilities derived from the nomogram (Fig. 7C). The time-dependent area under the curve values for predicting 1–5 year survival using the nomogram were 0.783, 0.778, 0.804, 0.807, and 0.791, respectively (Fig. 7B). It is worthwhile emphasizing that the predictive performance of our nomogram outperformed that of the stage and risk scores, demonstrating superior prognostic accuracy (Fig. 7D).
Fig. 7.
Constructing an AIRGs subtypes-based prognostic prediction model. (A) Nomogram developed based on AIRGs subtypes and clinicopathological characteristics. (B) Time-dependent ROC curves at 1, 2, 3, 4 and 5 years of the nomogram. (C) Calibration curves. (D) AUC plotted for different durations of OS for nomogram-based signature, tumor stage, and adipocyte subtypes in TCGA datasets.
3. Discussion
In vertebrates, the infiltration of adipocytes is a prevalent phenotype associated with the pathogenesis of various diseases, including cancer. Adipocyte infiltration is characterized by the accumulation of adipose tissue and has been established to promote tumor growth and progression by affecting energy metabolism, immunosuppressive, and pro-inflammatory effects [[14], [15], [16]].In our study, a series of genes associated with adipocyte infiltration were identified through Weighted Gene Co-expression Network Analysis, and three genes related to energy metabolism and immunosuppression were validated using PCR in both colon cancer cell lines and human colon cancer samples. Notably, significant differences in the expression of these adipocyte-related genes were observed across different samples. Furthermore, our correlation analysis revealed a strong association between high levels of adipocyte infiltration and poor prognosis. Therefore, the degree of adipocyte infiltration can be used to evaluate the prognosis of colon cancer. However, its classification into distinct subtypes and the understanding of their unique characteristics for improved clinical management in colorectal cancer (CRC) patients remains underexplored [17].
Herein, we aimed to identify and characterize distinct adipocyte infiltration-related gene subtypes in CRC patients. These subtypes were termed the low adipocyte infiltration subtype (Cluster 1) and the high adipocyte infiltration subtype (Cluster 2) based on the expression profiles of genes associated with adipocyte infiltration. In the Protein-Protein Interaction (PPI) network, ten core genes (VEGFC, PGF, TIMP1, BGN, FABP4, PPARGC1A, FABP3, PLIN1, ADIPOQ, SERPINE1) were identified that play significant roles in tumor invasion or metastasis. VEGFC (vascular endothelial growth factor C) is crucial for angiogenesis and lymphangiogenesis, promoting tumor metastasis [18]. PGF (placental growth factor) also contributes to angiogenesis and has been linked to poor prognosis in cancer patients [19]. TIMP1 (tissue inhibitor of metalloproteinases 1) inhibits matrix metalloproteinases (MMPs) and regulates extracellular matrix remodeling, influencing tumor invasion and metastasis [20]. BGN (biglycan) is involved in extracellular matrix organization and has been implicated in cancer progression and metastasis [21]. FABP4 (fatty acid-binding protein 4) and FABP3 (fatty acid-binding protein 3) are associated with lipid metabolism and energy homeostasis, and their overexpression is linked to cancer cell proliferation and invasion [22]. PPARGC1A (peroxisome proliferator-activated receptor gamma coactivator 1-alpha) regulates mitochondrial biogenesis and oxidative metabolism, playing a role in cancer cell energy metabolism [23]. PLIN1 (perilipin 1) is involved in lipid droplet formation and adipocyte differentiation, and its dysregulation can affect tumor cell metabolism [24]. ADIPOQ (adiponectin) is an adipokine with anti-inflammatory and insulin-sensitizing properties, and its reduced expression is associated with cancer progression [25]. SERPINE1 (plasminogen activator inhibitor-1) regulates fibrinolysis and extracellular matrix degradation, promoting tumor invasion and metastasis [26]. These core genes collectively contribute to the tumor microenvironment, influencing processes such as angiogenesis, extracellular matrix remodeling, lipid metabolism, and immune response, thereby playing a critical role in tumor invasion and progression.
Our findings revealed significant differences in clinical outcomes, molecular characteristics, tumor microenvironment, and drug sensitivity between these two subtypes. Specifically, patients in Cluster 2 exhibited a poorer prognosis, immunosuppressive features, and decreased responsiveness to immunotherapy. Thereafter, an AIRG-related risk signature and an AIRG-based nomogram were developed for prognostic prediction in CRC patients. Noteworthily, the performance of this nomogram outperformed traditional predictive tools in predicting the prognosis of CRC patients.
The differential gene expression patterns and enriched pathways observed between the subtypes offer valuable insights into the underlying biological processes driving tumor heterogeneity. Cluster 2, characterized by high infiltration of adipocytes, was enriched in pathways associated with adipogenesis, angiogenesis, inflammatory response, and epithelial-mesenchymal transition (EMT) [[27], [28], [29], [30]]. On the other hand, Cluster 1, with low adipocyte infiltration, was enriched in oxidative phosphorylation and E2F target pathways, potentially reflecting a distinct metabolic and proliferative phenotype [[31], [32], [33]]. These findings conjointly signal that adipocyte-rich microenvironments may create a pro-tumorigenic milieu, promoting tumor angiogenesis, inflammation, and invasive properties.
Moreover, the mutation landscape analysis exposed subtype-specific gene mutations. In other words, TP53 mutations were more frequently observed in Cluster 2, whereas APC mutations were more prevalent in Cluster 1. As a key transcription factor, TP53 is involved in cell cycle arrest and tumor suppression [34]. Thus, TP53 mutation in Cluster 2 may contribute to the loss of the tumor-suppressive function and influence CRC progression. In Cluster 1, the frequency of APC mutations was higher, and patients had a more favorable prognosis. These findings showcase the potential contribution of different mutational processes and driver genes in shaping the characteristics of adipocyte-related subtypes. Consequently, identifying these subtype-specific mutations not only enhances our understanding of CRC biology but also raises the possibility of subtype-specific targeted therapies.
Immune infiltration analysis provided insights into the immune landscape of the adipocyte-related subtypes. Compelling evidence suggests that M0 macrophages in tumors are associated with a worse prognosis [35,36]. Cluster 2 exhibited higher infiltration of immunosuppressive Tregs and M0 macrophages, along with upregulated expression of immune checkpoint markers such as PD1, PDL1, and CD70. Excessive accumulation of Tregs may limit the efficacy of immunotherapy, which is frequently associated with a poor prognosis [37]. The differential response to conventional chemotherapy drugs and the enhanced sensitivity to imatinib observed in Cluster 2 further highlights the potential of personalized treatment approaches based on subtype-specific characteristics. These findings suggest an immunosuppressive microenvironment in patients in Cluster 2, which may facilitate immune evasion and reduced response to immunotherapy/chemotherapy.
In summary, adipocyte infiltration in colorectal cancer (CRC) has significant clinical implications for prognosis and response to treatment. High levels of adipocyte infiltration are associated with poor prognosis, as demonstrated by worse survival outcomes in patients with high adipocyte infiltration. The immunosuppressive and pro-inflammatory effects of adipocyte infiltration can contribute to an unfavorable tumor microenvironment, promoting cancer progression and resistance to therapy. Our study highlights that patients with high adipocyte infiltration (Cluster 2) exhibit decreased responsiveness to immunotherapy and a distinct drug sensitivity profile. These patients showed a differential response to conventional chemotherapy drugs and enhanced sensitivity to imatinib, suggesting the need for personalized treatment approaches. Understanding the clinical relevance of adipocyte infiltration can aid in the development of more effective therapeutic strategies and improve prognostic assessments for CRC patients. Future studies should aim to validate these findings in independent cohorts and explore the functional roles of specific adipocyte-related genes and pathways in CRC progression and therapeutic response.
4. Materials and methods
4.1. Data preprocessing
Five accessible CRC gene expression datasets (GSE87211, GSE16158, GSE17536, GSE39582, and GSE17536) along with pertinent clinical information were retrievede from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The RNA-sequencing and somatic mutation data of TCGA-COAD were obtained from the UCSC public database (https://xenabrowser.net/). Patients with incomplete survival data were excluded from this study. The FPKM values of TCGA-COAD were converted to transcripts per kilobase million (TPM) for further analysis. The somatic mutation data of TCGA-COAD patients were examined using the R package "maftools". All expression profiles were processed using the normalization and log2 transformation methods described in a previous study [38].
4.2. Unsupervised clustering analysis of AIRGs
The most recent compilation of 1582 adipocyte-related genes was retrieved from the MEblue module. Then, 144 adipocyte-related genes were selected through univariate Cox analysis. Based on the expression levels of these 144 genes, Nonnegative Matrix Factorization (NMF) clustering analysis was applied to categorize 430 CRC samples from the TCGA dataset. In order to validate the clustering accuracy, a consensus clustering algorithm was applied for unsupervised clustering analysis. Ultimately, this unsupervised clustering approach yielded two distinct subtypes.
4.3. Clinicopathological features between the AIRGs-related subtypes
To evaluate the clinical relevance of the two subtypes identified by unsupervised clustering, the chi-square test was employed to investigate the relationship between molecular subtypes and clinicopathological characteristics. The clinicopathological features considered in this analysis included age, gender, survival status, venous invasion, lymphatic invasion, and tumor stage.
4.4. Immune infiltration analysis
In order to uncover differences in TME characteristics between the AIRGs-related subtypes, two algorithms, namely CIBERSORT [39] and MCP-counter [40], were employed to quantify the relative or absolute abundance of immune cell populations in CRC patients. Additionally, the estimate package was utilized to quantify mesenchymal cells and immune cells within malignant tumor tissues. This scoring system, which relies on a single-sample gene set enrichment analysis, generates three scores: stromal score, immune score, and estimate score [41]. The "estimate" R package was employed to calculate the immune score and stromal score for each patient.
4.5. Differentially expressed genes (DEGs) identification
The differentially expressed genes (DEGs) between the high adipocyte and low adipocyte subtypes were determined utilizing the "limma" R package. Differentially expressed genes with an adjusted p-value <0.05 and a fold change of 1 were considered statistically significant. To gain insights into the functional implications of DEGs, functional enrichment analysis was conducted using the David database (https://david.ncifcrf.gov/).
4.6. Construction of an AIRGs signature
In order to evaluate the adipocyte modification patterns for each CRC patient, the adipocyte score was calculated. Univariate Cox regression analysis was conducted on the differentially expressed genes (DEGs) to identify genes significantly associated with overall survival (OS). Risk scores were calculated using a 10-fold cross-validated LASSO regression, based on the adipocyte-related prognostic genes. An adipocyte gene signature, referred to as the adipocyte score, was generated using the 16 genes and their correlation coefficients. Subsequently, CRC patients were categorized into two subtypes based on their median adipocyte score. Survival analysis was conducted using the Kaplan-Meier method to compare outcomes between the two subtypes. The predictive ability of survival was evaluated using the receiver operating characteristic (ROC) analysis.
4.7. Cell culture and quantitative real-time polymerase chain reaction (RT-qPCR)
Normal colonic epithelial cells (NCM460), and human colorectal cancer cells (HCT116, LoVo, SW480, and DLD1) were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco's Modified Eagle's Medium (DMEM) containing 10 % fetal bovine serum, 100 unit/mL penicillin and 100 μg/mL streptomycin in a cell incubator at 37 °C and 5 % CO2. Total RNA was extracted from cells or tissues using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA).Total RNA was reverse transcribed to cDNA using a RT reagent kit (Vazyme Biotech, Nanjing, China). The RT-qPCR was performed using a SYBR-Green assays (Vazyme Biotech, Nanjing, China) on a StepOnePlusTM real-time PCR instrument (Thermo Fisher Scientific, Inc., USA) with 3 replicates. The mRNA expression level of ITLN1, ATOH1 and INSC was normalized with β-Actin and the data were calculated through the 2-ΔΔCt method. The primer sequences from 5′ to 3′ of 4 genes are listed below. ITLN1: TAACACTGAGCACCACTGCAT (forward), GCTGCTGCTGTAACCAACAT (reverse). ATOH1: AACAGCAAAACTTCGCCTCG (forward), ACTTGCCTCATCCGAGTCAC (reverse). INSC: CGCATCATAGCCAAGGTGGA (forward), GAAGCTACTGAGGTGCTGGG (reverse). β-Actin: ACAGAGCCTCGCCTTTGCC (forward), GATATCATCATCCATGGTGAGCTGG (reverse).
4.8. Additional bioinformatic and statistical analysis
Principal component analysis (PCA) was employed to visualize differences among the different groups. Pathway enrichment analysis between the two subtypes was conducted using the "GSVA" and "GSEA" packages. The "pRophetic" package was utilized to predict the half-inhibitory concentration (IC50) values of CRC therapeutics. To assess the potential response to immune checkpoint blockade treatment, the Tumor Immune Dysfunction and Exclusion (TIDE) [42] and Immunophenoscore (IPS) [43] algorithms were employed. All statistical analyses were performed using R version 4.0.3. The Wilcoxon test was used to compare specific variables (such as risk score, adipocyte-related genes, and adipocyte-related gene clusters) between the two groups. The chi-square test was employed to compare categorical variables. A p-value less than 0.05 was considered statistically significant.
5. Conclusion
In short, a novel gene signature was constructed based on AIRGs in CRC patients, who were classified into two subtypes. Moreover, significant differences were noted between the two subtypes, with variations in clinical outcomes, molecular and signaling pathway characteristics, immune microenvironment status, and chemotherapy responses. Briefly, the findings derived from the AIRG signature lay a theoretical basis for elucidating the function of adipocyte infiltration in TME.
Ethics approval and consent to participate
This study was conducted according to the Helsinki human subject doctrine and was approved by the Union Hospital, Tongji Medical College, Huazhong University of Science and Technology review board and Ethics Committee (IEC Approval Letter No.2021-0159), written consents to participate was acquired from all the patients.
Data availability statement
Data associated with our study had not been deposited in publicly available repositories. Data will be made available on request.
CRediT authorship contribution statement
Daan Fu: Writing – original draft, Visualization, Validation, Software, Methodology, Funding acquisition, Formal analysis, Data curation. Tianhao Zhang: Writing – original draft, Software, Methodology, Investigation. Jia Liu: Writing – review & editing, Validation. Bingcheng Chang: Writing – review & editing, Validation. Qingqing Zhang: Validation, Writing – review & editing. Yuyan Tan: Supervision, Resources. Xiangdong Chen: Supervision, Resources. Lulu Tan: Writing – review & editing, Supervision, Conceptualization.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (82303765), Open Foundation of Hubei Key Laboratory of Regenerative Medicine and Multi-disciplinary Translational Research (Grant No. 2022zsyx008).
Footnotes
Supplementary data to this article can be found online at.https://doi.org/10.1016/j.heliyon.2024.e33616.
Contributor Information
Yuyan Tan, Email: tyytyz@sina.com.
Xiangdong Chen, Email: xdchen@hust.edu.cn.
Lulu Tan, Email: tanll94@163.com.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Dekker E., Tanis P.J., Vleugels J.L.A., Kasi P.M., Wallace M.B. Colorectal cancer. Lancet. 2019;394:1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 3.Weiser M.R. AJCC 8th edition: colorectal cancer. Ann. Surg Oncol. 2018;25:1454–1455. doi: 10.1245/s10434-018-6462-1. [DOI] [PubMed] [Google Scholar]
- 4.Carethers J.M., Jung B.H. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology. 2015;149:1177–1190 e3. doi: 10.1053/j.gastro.2015.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruni D., Angell H.K., Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer. 2020;20:662–680. doi: 10.1038/s41568-020-0285-7. [DOI] [PubMed] [Google Scholar]
- 6.Sadanandam A., Lyssiotis C.A., Homicsko K., Collisson E.A., Gibb W.J., Wullschleger S., Ostos L.C., Lannon W.A., Grotzinger C., Del Rio M., Lhermitte B., Olshen A.B., Wiedenmann B., Cantley L.C., Gray J.W., Hanahan D. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med. 2013;19:619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bramsen J.B., Rasmussen M.H., Ongen H., Mattesen T.B., Orntoft M.W., Arnadottir S.S., Sandoval J., Laguna T., Vang S., Oster B., Lamy P., Madsen M.R., Laurberg S., Esteller M., Dermitzakis E.T., Orntoft T.F., Andersen C.L. Molecular-subtype-specific biomarkers improve prediction of prognosis in colorectal cancer. Cell Rep. 2017;19:1268–1280. doi: 10.1016/j.celrep.2017.04.045. [DOI] [PubMed] [Google Scholar]
- 8.Saha A., Kolonin M.G., DiGiovanni J. Obesity and prostate cancer - microenvironmental roles of adipose tissue. Nat. Rev. Urol. 2023;20:579–596. doi: 10.1038/s41585-023-00764-9. [DOI] [PubMed] [Google Scholar]
- 9.Silva A., Faria G., Araújo A., Monteiro M.P. Impact of adiposity on staging and prognosis of colorectal cancer. Crit. Rev. Oncol. Hematol. 2020;145 doi: 10.1016/j.critrevonc.2019.102857. [DOI] [PubMed] [Google Scholar]
- 10.Lin A., Qi C., Li M., Guan R., Imyanitov E.N., Mitiushkina N.V., Cheng Q., Liu Z., Wang X., Lyu Q., Zhang J., Luo P. Deep learning analysis of the adipose tissue and the prediction of prognosis in colorectal cancer. Front. Nutr. 2022 May 11;9 doi: 10.3389/fnut.2022.869263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Q., Li B., Li Z., Li J., Sun S., Sun S. Cancer-associated adipocytes: key players in breast cancer progression. J. Hematol. Oncol. 2019;12:95. doi: 10.1186/s13045-019-0778-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morigny P., Boucher J., Arner P., Langin D. Lipid and glucose metabolism in white adipocytes: pathways, dysfunction and therapeutics. Nat. Rev. Endocrinol. 2021;17(5):276–295. doi: 10.1038/s41574-021-00471-8. [DOI] [PubMed] [Google Scholar]
- 13.Bhat A.A., Nisar S., Maacha S., Carneiro-Lobo T.C., Akhtar S., Siveen K.S., Wani N.A., Rizwan A., Bagga P., Singh M., Reddy R., Uddin S., Grivel J.C., Chand G., Frenneaux M.P., Siddiqi M.A., Bedognetti D., El-Rifai W., Macha M.A., Haris M. Cytokine-chemokine network driven metastasis in esophageal cancer; promising avenue for targeted therapy. Mol. Cancer. 2021;20:2. doi: 10.1186/s12943-020-01294-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zoico E., Rizzatti V., Darra E., Budui S.L., Franceschetti G., Vinante F., Pedrazzani C., Guglielmi A., De Manzoni G., Mazzali G., Rossi A.P., Fantin F., Zamboni M. Morphological and functional changes in the peritumoral adipose tissue of colorectal cancer patients. Obesity. 2017;25(Suppl 2):S87–S94. doi: 10.1002/oby.22008. [DOI] [PubMed] [Google Scholar]
- 15.Del Corno M., Conti L., Gessani S. Innate lymphocytes in adipose tissue homeostasis and their alterations in obesity and colorectal cancer. Front. Immunol. 2018;9:2556. doi: 10.3389/fimmu.2018.02556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Corno M., Vari R., Scazzocchio B., Varano B., Masella R., Conti L. Dietary fatty acids at the crossroad between obesity and colorectal cancer: fine regulators of adipose tissue homeostasis and immune response. Cells. 2021;10 doi: 10.3390/cells10071738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rask-Andersen M., Ivansson E., Hoglund J., Ek W.E., Karlsson T., Johansson A. Adiposity and sex-specific cancer risk. Cancer Cell. 2023;41:1186–1197 e4. doi: 10.1016/j.ccell.2023.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y., Xue R., Li Y., Ran W., Chen Y., Luo Z., Zhang K., Zhang R., Wang J., Fang M., Chen C., Lou M. Impaired meningeal lymphatics and glymphatic pathway in patients with white matter hyperintensity. Adv. Sci. 2024 doi: 10.1002/advs.202402059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izadpanah A., Daneshimehr F., Willingham K., Barabadi Z., Braun S.E., Dumont A., Mostany R., Chandrasekar B., Alt E.U., Izadpanah R. Targeting TRAF3IP2 inhibits angiogenesis in glioblastoma. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.893820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu S.Y., Wang Y.C., Zuchini R., Lan K.Y., Liu H.S., Lan S.H. Secretory autophagy-promoted cargo exocytosis requires active RAB37. Autophagy. 2024;20:933–934. doi: 10.1080/15548627.2023.2210446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Appunni S., Rubens M., Ramamoorthy V., Anand V., Khandelwal M., Biglycan A. Sharma. An emerging small leucine-rich proteoglycan (SLRP) marker and its clinicopathological significance. Mol. Cell. Biochem. 2021;476:3935–3950. doi: 10.1007/s11010-021-04216-z. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z., Gao Z., Li B., Li J., Ou Y., Yu X., Zhang Z., Liu S., Fu X., Jin H., Wu J., Sun S., Sun S., Wu Q. Lipid-associated macrophages in the tumor-adipose microenvironment facilitate breast cancer progression. OncoImmunology. 2022;11 doi: 10.1080/2162402X.2022.2085432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian L., Zhu Y., Deng C., Liang Z., Chen J., Chen Y., Wang X., Liu Y., Tian Y., Yang Y. Peroxisome proliferator-activated receptor gamma coactivator-1 (PGC-1) family in physiological and pathophysiological process and diseases. Signal Transduct. Targeted Ther. 2024;9:50. doi: 10.1038/s41392-024-01756-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandel-Brehm C., Vazquez S.E., Liverman C., Cheng M., Quandt Z., Kung A.F., Parent A.F., Miao A., Disse E., Cugnet-Anceau C., Dalle S., Orlova E., Frolova E., Alba D., Michels A., Oftedal B.E., Lionakis M.S., Husebye E.S., Agarwal A.K., Li X., Zhu C., Li Q., Oral E., Brown R., Anderson M.S., Garg A., DeRisi J.L. Autoantibodies to perilipin-1 define a subset of acquired generalized lipodystrophy. Diabetes. 2023;72:59–70. doi: 10.2337/db21-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C., Zhang J., Dionigi G., Liang N., Guan H., Sun H. Adiponectin inhibits the progression of obesity-associated papillary thyroid carcinoma through autophagy. Endocrinology. 2024;165 doi: 10.1210/endocr/bqae030. [DOI] [PubMed] [Google Scholar]
- 26.Czekay R.P., Higgins C.E., Aydin H.B., Samarakoon R., Subasi N.B., Higgins S.P., Lee H., Higgins P.J. SERPINE1: role in cholangiocarcinoma progression and a therapeutic target in the desmoplastic microenvironment. Cells. 2024;13:796. doi: 10.3390/cells13100796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M., Wu B., Li L., Lv C., Tian Y. Reprogramming of cancer-associated fibroblasts combined with immune checkpoint inhibitors: a potential therapeutic strategy for cancers. Biochim. Biophys. Acta Rev. Canc. 2023 doi: 10.1016/j.bbcan.2023.188945. [DOI] [PubMed] [Google Scholar]
- 28.Zeng Q., Mousa M., Nadukkandy A.S., Franssens L., Alnaqbi H., Alshamsi F.Y., Safar H.A., Carmeliet P. Understanding tumour endothelial cell heterogeneity and function from single-cell omics. Nat. Rev. Cancer. 2023;23:544–564. doi: 10.1038/s41568-023-00591-5. [DOI] [PubMed] [Google Scholar]
- 29.Morrison H.A., Trusiano B., Rowe A.J., Allen I.C. Negative regulatory NLRs mitigate inflammation via NF-kappaB pathway signaling in inflammatory bowel disease. Biomed. J. 2023 doi: 10.1016/j.bj.2023.100616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y., Zhang Z. OVOL2: an epithelial lineage determiner with emerging roles in energy homeostasis. Trends Cell Biol. 2023;33:824–833. doi: 10.1016/j.tcb.2023.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Oliveira M.P., Liesa M. The role of mitochondrial fat oxidation in cancer cell proliferation and survival. Cells. 2020;9 doi: 10.3390/cells9122600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wai Hon K., Zainal Abidin S.A., Othman I., Naidu R. Insights into the role of microRNAs in colorectal cancer (CRC) metabolism. Cancers. 2020;12 doi: 10.3390/cancers12092462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kassab A., Gupta I., Moustafa A.A. Role of E2F transcription factor in oral cancer: recent insight and advancements. Semin. Cancer Biol. 2023;92:28–41. doi: 10.1016/j.semcancer.2023.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Mijit M., Caracciolo V., Melillo A., Amicarelli F., Giordano A. Role of p53 in the regulation of cellular senescence. Biomolecules. 2020;10 doi: 10.3390/biom10030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X., Wu S., Yang Y., Zhao M., Zhu G., Hou Z. The prognostic landscape of tumor-infiltrating immune cell and immunomodulators in lung cancer. Biomed. Pharmacother. 2017;95:55–61. doi: 10.1016/j.biopha.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Zou J., Chen R. An M0 macrophage-related prognostic model for hepatocellular carcinoma. BMC Cancer. 2022;22:791. doi: 10.1186/s12885-022-09872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka A., Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017;27:109–118. doi: 10.1038/cr.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan Y., Tan L., Wang L., Zou D., Liu J., Lu X., Fu D., Wang G., Wang L., Wang Z. The expression pattern of hypoxia-related genes predicts the prognosis and mediates drug resistance in colorectal cancer. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.814621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen B., Khodadoust M.S., Liu C.L., Newman A.M., Alizadeh A.A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becht E., Giraldo N.A., Lacroix L., Buttard B., Elarouci N., Petitprez F., Selves J., Laurent-Puig P., Sautes-Fridman C., Fridman W.H., de Reynies A. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshihara K., Shahmoradgoli M., Martinez E., Vegesna R., Kim H., Torres-Garcia W., Trevino V., Shen H., Laird P.W., Levine D.A., Carter S.L., Getz G., Stemke-Hale K., Mills G.B., Verhaak R.G. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., Li Z., Traugh N., Bu X., Li B., Liu J., Freeman G.J., Brown M.A., Wucherpfennig K.W., Liu X.S. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., Hackl H., Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with our study had not been deposited in publicly available repositories. Data will be made available on request.