Summary
Connective tissue (CT), which includes tendon and muscle CT, plays critical roles in development, in particular as positional cue provider. Nonetheless, our understanding of fibroblast developmental programs is hampered because fibroblasts are highly heterogeneous and poorly characterized. Combining single-cell RNA-sequencing-based strategies including trajectory inference and in situ hybridization analyses, we address the diversity of fibroblasts and their developmental trajectories during chicken limb fetal development. We show that fibroblasts switch from a positional information to a lineage diversification program at the fetal period onset. Muscle CT and tendon are composed of several fibroblast populations that emerge asynchronously. Once the final muscle pattern is set, transcriptionally close populations are found in neighboring locations in limbs, prefiguring the adult fibroblast layers. We propose that the limb CT is organized in a continuum of promiscuous fibroblast identities, allowing for the robust and efficient connection of muscle to bone and skin.
Subject areas: Biological sciences, Molecular biology, Cell biology, Developmental biology
Graphical abstract

Highlights
-
•
Dissection of connective tissue heterogeneity by scRNA-seq and in situ RNA staining
-
•
In silico fibroblast populations map to distinct fibroblast layers in limbs
-
•
Molecular profiling of fibroblast populations prefiguring adult connective tissue
-
•
Developing fibroblasts switch from providing positional cues to differentiation
Biological sciences; Molecular biology; Cell biology; Developmental biology
Introduction
Although connective tissue (CT) plays critical roles in limb development, our understanding of CT fibroblast differentiation lags behind that of other components of the musculoskeletal system mainly because CT fibroblast populations are highly heterogeneous and poorly characterized.
In Vertebrate limbs, CT corresponds to an interconnected three-dimensional network of fibroblasts and extracellular matrix (ECM) that support the function of the musculo-skeletal system. Muscle attachments include muscle CT (MCT) and tendon.1,2,3 The whole musculoskeletal system is attached to the skin via the hypodermis that is not considered to be part of the dermis per se.4 MCT includes the successive fibroblast layers, epimysium, perimysium, and endomysium that surround individual muscles, muscle fiber bundles, and muscle fibers, respectively.5 Tendon is described as a continuum of MCT linking muscle to bone.5 Tendon is surrounded by a thin fibroblast layer, the epitenon, then by a loose fibroblast sheath, the peritenon. Both are morphologically distinct from the tendon proper. Paratenon includes peritenon and epitenon layers in adult tissues.6,7 MCT and tendon fibroblasts differ by their matrix composition and spatial organization of collagen fibers. Tendons contain collagen fibers displaying a regular organization parallel to the tendon axis, while MCT displays collagen fibers with less specific organization.3 The different matrix compositions of each limb CT prefigure different functions and mechanical properties.
In contrast to the muscle lineage, there is no identified master gene driving the differentiation programs toward dermis, MCT, and tendon, but recognized markers have been identified for each CT type, providing us with robust molecular tools to follow each limb CT type. The bHLH transcription factor TWIST2 (DERMO-1) is expressed in developing dermis8 and has been shown to be sufficient to launch the developmental program leading to skin appendage formation in chicken embryos.9 The bHLH transcription factor scleraxis (SCX) is a recognized marker of developing tendons, and Scx mutant mice show tendon defects.10 The zinc finger transcription factor OSR1 (odd-skipped related-1) identifies a population of developmental fibro-adipogenic progenitors (FAPs) regulating myogenesis11 and controlling the pro-regenerative response of FAPs during muscle regeneration.12,13
Limb CT fibroblasts originate from the lateral plate mesoderm, while myogenic cells are mainly derived from somitic mesoderm.14,15,16 CT is recognized as an important source of extrinsic cues that regulate skeletal muscle differentiation, growth, and patterning.1,2,3,16,17 Osr1 loss-of-function leads to reduced myogenic cell proliferation and survival resulting in limb muscle patterning defects.11 The ablation of tendon cells leads to muscle patterning defect in chicken limbs18,19 and genetic ablation of Scx+ tendon cells to alteration of muscle shapes and attachment sites in mouse limbs.19 Moreover, Osr1 gain- and loss-of-function experiments in chicken mesenchymal limb cells showed that OSR1 favors the MCT differentiation program at the expense of the tendon differentiation program.20 In addition to OSR1, other transcription factors produced by CT fibroblasts have been shown to regulate limb muscle patterning during development, such as Tcf4 (now Tcf7l2), belonging to the TCF/LEF family,21 the T-box genes Tbx3 and Tbx5,22,23 and the homeobox Hoxa11.24 However, Tcf4 and Hoxa11 are also expressed in muscle cells25,26 or in muscle lineage,27 where they could directly act on muscle patterning. It has been recently shown that a subpopulation of CT fibroblasts integrates into muscle fibers at the muscle/tendon interface during development and postnatal stages.27,28,29,30 This unexpected fibroblastic origin of myonuclei at muscle tips close to tendon provides us with a cellular mechanism mediating fibroblast-driven muscle patterning.
Transcriptomic strategies at the level of the cell/nucleus have been used to highlight fibroblast heterogeneity of muscle attachments, providing us with a plethora of CT fibroblast markers in adult tissues. Adult MCT fibroblasts, known as FAPs, are recognized to be highly heterogeneous. Since the first cartography at the cell level of muscle resident cells,31 numerous FAP populations have been identified but with different markers depending on the studies, making it difficult to draw a global picture of the identity and functions of FAP populations.32,33 However, one common feature across these transcriptomic studies is the identification of FAP populations based on low and high levels of molecular markers, such as SCA1 and PDGFRalpha.32,34 Although the hierarchical fibroblast layers surrounding muscle are well identified, the FAP populations described in silico were not assigned to any of these layers in situ.
Similarly to MCT fibroblasts, two to ten tendon fibroblast populations have been identified in adult tendons using transcriptomic analysis of mouse, human, or rat tendons,35,36,37,38 including tendon fibroblasts with progenitor properties.39,40 No universal molecular signature for these in silico fibroblast populations emerged from these studies. However, cells of paratenon and tendon proper show distinct molecular signatures and cellular properties.41,42,43
The studies previously addressed CT biology in the adult. During development, a transcriptomic atlas has been established for digit skeletal elements.44 To date, there is no data concerning fibroblast populations of muscle attachments during development. In this study, we identified CT fibroblast populations, their molecular signatures and developmental trajectories during chicken limb development and growth using single-cell RNA-sequencing and inferred trajectory analyses. With extensive in situ hybridization analysis, we localized these in silico fibroblast populations in fetal limbs and found that they prefigure the mature layers of CT fibroblasts of the musculo-skeletal system.
Results
The dermis, tendon, and MCT branches emerge successively
To exhaustively address fibroblast diversity, we performed single-cell RNA-sequencing (scRNA-seq) analysis on whole chicken forelimbs at successive developmental stages, ranging from E4, the progenitor stage, to E10, the final fibro-muscular pattern. We took advantage of the previously published E4, E6, and E10 datasets28 and generated the E7 and E9 datasets following the same protocol. Clustering analysis on E4, E6, E7, E9, and E10 datasets show that CT fibroblasts segregate from muscle, vessels, blood cells, ectoderm, and neural crest cells at all stages (Figures S1A and S1B for E7 and E9; see Esteves de Lima et al.28 for E4, E6, and E10). The expression of the known CT markers (TWIST2, OSR1, and SCX) confirmed the identity of CT clusters in E7 and E9 whole limb cells (Figures S1C and S1D). To identify fibroblast populations within the limb CT, we bio-informatically extracted CT clusters from the whole limb datasets at each developmental stage and performed unsupervised sub-clustering of these CT datasets (Figure S1E). Plotting the distribution of the cell cycle phases across each CT dataset showed that the proportion of proliferating cells (G2M + S phases) drops from 61% at E4 to 42% at E6 and then stabilizes around 35% up to E10 (Figures S1F and S1G). The five CT datasets were then combined to generate an integrated 5CT dataset containing 24570 cells (Figures 1A and S1H).
Figure 1.
The dermis, tendon, and MCT branches emerge successively
(A) UMAP plot of the integrated 5CT dataset (24,570 cells in total), showing the distribution of the CT datasets of origin.
(B) Dendrogram of the STREAM-derived inferred trajectory for the 5CT dataset, showing the distribution of fibroblasts according to their CT datasets of origin.
(C) Colorimetric in situ hybridization to adjacent and transverse limb sections of E10 chicken embryos with TWIST2, OSR1, and SCX probes (blue staining).
(D) Dendrogram plotting gene expression levels for TWIST2, OSR1, and SCX on the inferred trajectory of the 5CT dataset.
(E) Dendrogram as in (B) showing the association of color-coded DAVID categories with the origin (S0-S1), dermis (S1-S2), tendon (S3-S4), and MCT (S5-S7 and S5-S6) branches. The GO term-associated DAVID categories are: DNA binding/homeobox/transcription in pink; secreted/glycoproteins/extracellular matrix (ECM) in light green; collagens/extracellular matrix (ECM) organization in green; cell adhesion/focal adhesion in brown. For the S3-S4 (tendon) and S5-S6 (MCT) branches, the top 3 associated DAVID categories are ordered by decreasing enrichment scores with the highest score to the left. The other branches are associated with a single DAVID category. See also Figure S1.
In order to visualize the emergence of major CT types during development, trajectory inference using STREAM45 was performed on the integrated 5CT dataset. Three major branches arise successively (Figure 1B). To identify each branch, we plotted the expression of markers for the main CT types, i.e., TWIST2 for dermis,8 OSR1 for MCT,11 and SCX for tendon10 (Figures 1C and 1D). Dermis was the first CT type to emerge at E7, followed by tendon and MCT around E8/E9 (Figures 1B–1D). MCT subsequently split into two branches at E10. We conclude that the dermal, tendon, and MCT types emerge successively during limb development.
CT fibroblasts switch from a positional information to a lineage diversification program at the onset of the fetal period
To identify the main biological activities associated with each branch, we performed a functional enrichment analysis on genes identified by STREAM (Table S1) as having branch-specific expression, using DAVID46,47 (Figure 1E). Categories associated with GO terms such as “DNA binding”, “homeobox” and “transcription regulation” were over-represented in the S0-S1 origin branch. Except for the dermis branch (S1-S2) that was associated with mitosis-related categories, the tendon (S3-S4) and MCT (S5-S7, S5-S6) branches were associated with ECM-related categories. The tendon branch and to a lesser extent the S5-S6 MCT branch exhibited more mature categories of ECM organization and modification as well as cell adhesion compared to the S5-S7 MCT branch associated with ECM production, suggesting that tendon differentiation was more advanced than that of MCT. A switch in biological activities was thus observed at the root of the lineage diversification, indicating that fibroblasts switched from a common homeobox-based program to distinct ECM production programs.
Homeobox genes are associated with positional information during the establishment of the three limb axes,48,49 while extracellular matrix production is associated with CT fibroblast differentiation.3 To explore further this switch in biological activities, we analyzed the expression patterns of positional genes and lineage genes in silico and in situ (Figure 2). At E4, the positional genes (LMX1B for dorsal, MSX2 for anterior, HOXA11 and HOXA13 for distal, HOXD11 for posterior, and MEIS2 for proximal) showed regionalized and complementary expression patterns on the E4 UMAP (Figures 2A–2E), indicative of a clustering of E4 limb cells according to positional cues. At the same stage, SCX and OSR1 expression pattern did not show obvious regionalization in the E4 UMAP (Figure 2B), while these lineage genes were expressed in dorsal and ventral limb regions, as previously shown (Figures 2C and 2D;20,50). SCX- and OSR1-expressing cells therefore did not segregate according to their lineage cues but rather to limb positional information cues. After E4, when the dorsoventral and antero-posterior axes are established, the proximodistal axis is still under construction as limbs still grow along this axis. MEIS2 and HOXA13, as proxies of proximal and distal cues, respectively,51,52 showed regionalized and complementary expression patterns up to E7 (Figure 2E). Due to the emergence of regions devoid of MEIS2 and HOXA13 expression, MEIS2 and HOXA13 complementary expression pattern was lost after E7, although still being mutually exclusive at E9 and E10. At E6, we observed the emergence of fibroblast populations with no proximodistal cue (Figure 2E, see ellipses). These regions correspond to overlapping expression of OSR1, SCX, and TWIST2 on E6/E7 UMAPs (Figures 2F and 2G). In E6 limbs, at the onset of spatial organization of the fibro-muscular system, SCX and OSR1 expression was partially overlapping in and around muscle masses (Figures 2H–2J). After E7, SCX, OSR1, and TWIST2 showed regionalized expression patterns on the E9 and E10 UMAPs (Figures 2F and 2G). The perfect complementary expression of SCX and OSR1 on the E10 UMAP and in limbs53 is consistent with the transcriptional repression of SCX expression by OSR1 in limb cell cultures.20 We conclude that from E7, i.e., the onset of fetal period, fibroblasts cluster according to their lineage identity and no longer to their positional information.
Figure 2.
CT fibroblasts switch from a positional information program to a lineage diversification program around E7
(A) Feature plots showing the distribution of cells positive for the positional markers LMX1B, MSX2, HOXA11, and HOXD11 in the E4 CT dataset.
(B) Feature plots showing the distribution of cells positive for the lineage markers SCX and OSR1 in the E4 CT dataset.
(C and D) Colorimetric in situ hybridization with SCX (C) or OSR1 (D) probes to longitudinal limb sections of E4 embryos.
(E) Feature plots showing the distribution of MEIS2+ cells (proximal marker) and HOXA13+ cells (distal marker) in E4, E6, E7, E9, and E10 CT datasets.
(F) Feature plots showing the distribution of SCX+ cells (green dots), OSR1+ cells (red dots), and SCX+/OSR1+ cells (yellow dots) in E4, E6, E7, E9, and E10 CT datasets.
(G) Feature plots showing the distribution of TWIST2+ cells in E4, E6, E7, E9, and E10 CT datasets. Ellipses in (E–G) delineate UMAP regions with no positional information but emerging lineage information.
(H–J) Colorimetric in situ hybridization with SCX (H and I) or OSR1 (J) probes (dark blue) to transverse limb sections of E6 chicken embryos. (H) Myosin immunodetection (green) and DAPI staining (light blue) were overlaid over SCX detection by in situ hybridization (dark blue). (I and J) Ventral muscle masses are delineated with white dashed lines. u, ulna, r radius.
Dermis, MCT, and tendon are composed of molecularly distinct populations at E10
To identify potential fibroblast populations within the limb CT types, we focused on the E10 stage since the final fibro-muscular pattern is set at this stage. Unsupervised clustering analysis of the E10 CT dataset revealed nine distinct clusters (Figure 3A) and provided us with marker lists for each cluster, i.e., lists of differentially expressed genes in one given cluster compared to the remaining clusters (Table S2). The analysis of TWIST2, OSR1, and SCX expression on the E10 UMAP, combined with the localization of the nine E10 clusters on the 5CT trajectory, allowed us to attribute each cluster to a dermis, MCT, or tendon identity (Figures 3A–3C). We identified two dermal clusters (clusters 2 and 3), four MCT clusters (clusters 7, 1, 0, and 8), and three tendon clusters (clusters 4, 6, and 5). In this dataset, dermis, MCT, and tendon fibroblasts corresponded to roughly 1/4, 1/2, and 1/4 of all limb CT fibroblasts, respectively (Figure S1E). The heatmap of the top 10 markers of each cluster, ranked by fold-change and ordered by CT type, showed that the clusters of dermis and MCT types exhibited specific molecular signatures, while the signatures of tendon clusters could be established but were more promiscuous (Figure 3D). For each cluster, module scores were calculated as the corrected average expression levels of specific cluster markers (in pink, Table S2) and plotted for each cluster. In order to enrich for markers with low to no expression outside the cluster of interest, specific markers were selected on the basis of their association with a unique cluster (in great majority) combined to a low pct.2 value (fraction of marker-expressing cells in clusters other than the cluster of interest <0.25), except for cluster 7 (pct.2 < 0.30) due its short marker list. For each cluster (Figures 4D, 4E, 5A–5D, 6A, 6G, and 6K), the highest levels of the score overlapped with the cluster of interest (see Figure 3A for comparison), showing that the combined expression of the selected markers is sufficient to define the cluster and therefore constitute an in silico molecular signature. We conclude that dermis, MCT, and tendon types are subdivided in fibroblast populations with distinct but overlapping molecular signatures.
Figure 3.
Dermis, MCT, and tendon are composed of molecularly distinct populations at E10
(A) UMAP plot showing the distribution of the CT fibroblast clusters at E10.
(B) Feature plots showing the distribution of TWIST2+, OSR1+, and SCX+ fibroblasts across CT clusters at E10.
(C) Dendrogram of the 5CT inferred trajectory, highlighting the distribution of E10 clusters. Clusters from the other stages are grouped in light blue. (A) and (B) combined allow to position the dermis, MCT, and tendon types on (C).
(D) Heatmap showing the relative expression of the top 10 markers across all cells for each of the nine CT clusters, ordered by CT types at E10. Upregulated genes in yellow, downregulated genes in purple.
(E) Dotplot showing the average expression levels and the fraction of expressing cells for a selection of genes in each of the 9 CT clusters, ordered by CT types. See also Figure S2.
Figure 4.
Dermal fibroblasts are divided in two molecularly distinct populations with no obvious spatial regionalization
(A and B) TWIST2 expression on violin plot showing log-normalized expression levels across clusters (A) and in chicken limbs with in situ hybridization to transverse limb sections of E10 chicken embryos (B).
(C) Cell numbers and associated percentage of dermal markers in clusters 2 and 3.
(D and E) Feature plots showing the distribution of the module score for cluster 2 (D) and cluster 3 (E) in the E10 CT dataset.
(F) Violin plots showing the log-normalized expression levels of HTRA1, BCL11B, and SST genes across clusters.
(G–L) Colorimetric in situ hybridization to adjacent limb transverse sections of E10 chicken embryos with HTRA1 (G and J), BCL11B (H and K), and SST (I and L) probes (blue). (J–L) are high magnifications of dorso-posterior dermis regions from sections shown in (G–I). Limb sections are oriented dorsal to the top and posterior to the left. u, ulna; r, radius.
Figure 5.
MCT clusters map to concentric fibroblast layers
(A–D) Feature plots showing the distribution of the module score for cluster 7 (A), cluster 1 (B), cluster 0 (C), and cluster 8 (D) in the E10 CT dataset.
(E–H) Colorimetric in situ hybridization to transverse limb sections at the level of the middle of the forearm of E10 chicken embryos with CHODL (E), ANXA2 (F), ALDH1A2 (G), and EPHA3 (H) probes (blue).
(I–L) Fluorescent in situ hybridization to transverse limb sections hybridized with CHODL (I), ANXA2 (J), ALDH1A2 (K), and EPHA3 (L) probes (red) combined with myosin immunostaining (green) and DAPI staining (blue). Limb muscles shown in (I–L) are labeled with black asterisks in limb sections of (E–H), respectively. (I) Arrows point to CHODL gene expression in the hypodermis.
(M) Double fluorescent in situ hybridization to transverse limb sections focused on the FCU muscle (ventro-posterior muscle) with CHODL (cluster 7, green) and ANXA2 (cluster 1, red) probes, combined with myosin immunolabeling (gray). The white arrow on high magnification points to the hypodermis (green CHODL labeling), while the arrowhead points to epimysium (red ANXA2 labeling).
(N) Percentage of CHODL+, ANXA2+, and CHODL+/ANXA2+ fibroblasts among the CHODL-ANXA2 population.
(O) Double fluorescent in situ hybridization to transverse limb sections focused to the FCU muscle with ANXA2 (cluster 1, green) and ALDH1A2 (cluster 0, red) probes, combined with myosin immunolabeling (gray).
(P) Percentage of ANXA2+, ALDH1A2+, and ALDH1A2+/ANXA2+ fibroblasts among the ANXA2-ALDH1A2 population. Limb sections are oriented dorsal to the top and posterior to the left. t, tendon, u, ulna; r, radius. See also Figures S3–S6.
Figure 6.
Fetal tendons are divided in three fibroblast populations: peritenon, enthesis/perichondrium, and tendon proper
(A) Feature plot showing the distribution of the module score for clusters 4 in the E10 CT dataset.
(B) Violin plot showing the log-normalized expression levels of ADGRG2 gene across clusters.
(C,C′) Double in situ hybridization with ADGRG2 probe (red) and CHODL probe (to label tendons, in green) to transverse limb sections at distal level (C) and longitudinal limb sections (C′) of E10 chicken embryos, combined with myosin immunolabeling (gray) and DAPI staining (blue). (C,C′) Arrows point to ADGRG2 expression (red) surrounding tendons (green).
(D and E) Double fluorescent in situ hybridization to adjacent transverse limb sections at E10 with ADGRG2 (red)/CHODL (green) (D) and ANXA2 (red)/CHODL (green) probes (E) combined with myosin immunolabeling (gray) and DAPI staining (blue).
(F,F′) Double fluorescent in situ hybridization to adjacent transverse limb sections at E10 with ADGRG2 (red)/CHODL (green) (F) and ANXA2 (red)/CHODL (green) probes (F′) combined with myosin immunolabeling (gray) and DAPI staining (blue). (D-F′) White arrows point to ADGRG2 (D and F) and ANXA2 (E,F′) expression surrounding tendons and muscles.
(G) Feature plot showing the distribution of the module score for cluster 6 in the E10 CT dataset.
(H) Colorimetric in situ hybridization to transverse limb sections at E10 with MYLK probe (blue), focused on tendon attachment to cartilage element.
(I and J) Fluorescent in situ hybridization to longitudinal tendon section with NET1 (a cluster 6 marker) probe (red) (I) and to transverse limb section with COL12A1 probe (red) (J) combined with myosin immunolabeling (green) and DAPI staining (blue). (I and J) Arrows point to tendon attachments to cartilage elements labeled with NET1 (I) and COL12A1 (J).
(K) Feature plot showing the distribution of the module score for cluster 5 in the E10 CT dataset.
(L and M) Double fluorescent in situ hybridization to transverse limb sections at E10 (L) and focused on FCU muscle (M) with SCX (red) and TNMD (green) probes, combined with myosin immunolabeling (gray).
(N) Percentage of SCX+, TNMD+, and SCX+/TNMD+ fibroblasts among the SCX-TNMD population.
(O and P) Fluorescent in situ hybridization with the NET1 probe (red) combined with myosin immunolabeling (green) to transverse limb sections (O) and to longitudinal muscle sections (P, DAPI staining in blue) of E10 chicken embryos. Arrows point to NET1 expression in tendons (O) and to tendon attachment close to muscle (P).
(Q) Cell numbers and associated percentage of tendon markers in cluster 5.
(R and S) Double fluorescent in situ hybridization to transverse limb sections at E10, focused on FCU muscle (R) and FCU tendon (S) with NET1 (green) and TNMD (red) probes, combined with myosin imunolabeling (gray).
(T) Percentage of NET1+, TNMD+, and NET1+/TNMD+ fibroblasts among the NET1-TNMD population. See also Figure S7.
In order to map the in silico clusters in fetal limbs, we selected up to four specific markers per cluster from the score-selected marker list (Table S2) and characterized their expression patterns by in situ hybridization to E10 limb sections. On top of association with a single cluster and low pct.2 value (<0.25), specific markers were chosen for their high pct.1 value (fraction of marker-expressing cells in the cluster of interest, variable value), in order to select for specific and robust markers. The dotplot shows a mix of top 5 markers, of single-cluster markers and of markers selected for ISH using the aforementioned criteria, ranked by fold-change and ordered by CT type, to illustrate the following general features (Figure 3E). Specific markers were expressed in at most 30%–55% of the cells in the cluster of interest. Markers expressed in 90%–100% of the cells for a given cluster, such as collagens, were non-specific markers, i.e., they were expressed in a large fraction of cells in multiple clusters. For example, COL1A1 and COL3A1 were listed as markers for clusters 5-6 and 0-1-5, respectively (Table S2) and were also found in high proportions in other clusters (Figure S2A), consistent with collagen widespread expression in limbs.54,55 In the following sections, we will describe each cluster, grouped by CT type, following the neighborhood order of the E10 UMAP and starting from the dermis clusters.
Dermal fibroblasts are divided in two molecularly distinct populations with no obvious spatial regionalization
The dermis type was divided into two transcriptionally distinct populations (clusters 2 and 3), which was reminiscent of the two dermis layers, the upper papillary and lower reticular dermis.4 TWIST2, a general marker for Dermis (clusters 2 and 3), was expressed in dermis of feather buds and inter buds, while being excluded from the ectoderm (Figures 4A–4C). The HTRA1 (HtrA serine peptidase 1) gene, a specific marker of cluster 2 (expressed in 52% of cluster 2 fibroblasts) showed a higher expression in inter-bud dermis than in feather bud dermis (Figures 4C–4F, 4G, and 4J). The BCL11B gene, a specific marker of cluster 3 (38% of cluster 3 fibroblasts), displayed enriched expression in dorsal dermis (Figures 4C–4F, 4H, and 4K). The SST (somatostatin) gene, another specific marker of cluster 3 expressed in 42% of cluster 3 fibroblasts, was expressed in sub-regions of the feather bud dermis (Figures 4C–4F, 4I, and 4L). Analysis of additional cluster 2 markers, TNMD and POSTN (Figures 6L and S6E), did not show any regionalization. Although the expression patterns of certain genes were highly regionalized, they did not allow us to assign clusters to specific regions such as the papillary and reticular dermis layers or the inter bud and feather bud dermis or the dorsal and ventral dermis. This is consistent with other scRNA-seq studies that did not identify obvious molecular signatures for dermis regions.4,56
MCT clusters map to concentric fibroblast layers
The MCT type was divided into four fibroblast populations, clusters 7, 1, 0, and 8 (Figure 3A). The four MCT clusters corresponded to nearly half of limb CT fibroblasts and included the two largest fibroblast clusters (clusters 0 and 1) (Figure S1E). PDGFRA, PRRX1, and TCFL7L1 are routinely used to generate mouse Cre lines to specifically study MCT.2,29 However, we observed that PDGFRA, PRRX1, and TCFL7L1 displayed a widespread expression both in silico and in situ with a noticeable PRRX1 expression in tendon in silico populations and in limb tendons (Figures S2B–S2J). We therefore show that those classical markers are unsuitable not only to discriminate MCT fibroblast populations but also to specifically study MCT in chicken fetal limbs.
In situ hybridization for selected markers allowed us to assign the four MCT clusters to distinct locations in limbs (Table S2; Figure 5). CHODL (chondrolectin), ZFHX4 (zinc finger homeobox 4), and CCN3 (cellular communication network 3) are specific markers of cluster 7, although being expressed also in other clusters (Table S2; Figures S3A, S3E, and S3I). Their expression overlapped in a fibroblast layer underneath the dermis and surrounding the entire musculo-skeletal system (Figures 5E–5I and S3), in a region that anatomically corresponds to the hypodermis. We conclude that cluster 7 fibroblasts correspond to the hypodermis.
ANXA2 (the calcium regulated phospholipid-binding protein annexin A2) was the main specific marker for cluster 1 (Table S2; Figure S4A). ANXA2, along with the additional specific markers, AHNAK2, KLF2, and CD34, was expressed in a fibroblast layer delineating individual muscles and their associated tendons (Figures 5F–5J and S4). Double ANXA2/AHNAK2 fluorescent in situ hybridization confirmed that they were expressed within the same peripheral layer (Figures S4R and S4S). Cluster 1 fibroblasts delineate each individual limb muscle and thus prefigure the future epimysium.
The enzyme necessary for retinoic acid (RA) synthesis, ALDH1A2, was the main specific marker for cluster 0 (Table S2; Figure S5A). ALDH1A2 transcripts labeled interstitial fibroblasts within limb muscles and were excluded from myosin+ cells (Figures 5G–5K and S5). Three other specific markers for cluster 0 were expressed in the same location: HIC1 (hyper methylated in cancer1), a marker of quiescent fibroblasts in skeletal muscle,57 and the inhibitors of the FGF and Wnt signaling pathways, SPRY2 and FRZB (Figure S5). We conclude that cluster 0 interstitial fibroblasts correspond to the future endomysium.
Cluster 8 was the smallest fibroblast population and in close proximity to cluster 0 on the E10 UMAP (Figure 3A). Consistently, as for cluster 0, specific markers for cluster 8 were expressed in interstitial fibroblasts but enriched in a subset of limb muscles. The main specific marker EPHA3, along with the additional POSTN and CXCL12 markers, were enriched in interstitial fibroblasts of the ANC muscle and of the posterior part of the FCU muscle (Table S2; Figures 5H, 5L, and S6). The ventral part of the EMR muscle shows enriched POSTN and CXCL12, but not EPHA4, expression (Figures S6D–S6F). We could not correlate the cluster 8 molecular signature to slow and fast muscle types.58
To analyze the interface between MCT fibroblast populations, we performed double fluorescence in situ hybridization for CHODL (cluster 7) and ANXA2 (cluster 1) and for ANXA2 (cluster 1) and ALDH1A2 (cluster 0) and quantified the double-positive cells in silico. With both approaches, we find very little overlap between markers of distinct MCT clusters (Figures 5M–5P), confirming that these clusters correspond to distinct fibroblast populations that are spatially segregated.
We conclude that the future hypodermis (cluster 7), epimysium (cluster 1), and endomysium (cluster 0) are molecularly distinct in E10 chicken limbs. The MCT populations display a concentric and hierarchical organization in fetal limbs that prefigures the mature fibroblast layers in adult skeletal muscle.
Fetal tendons are divided in three fibroblast populations: peritenon, enthesis/perichondrium, and tendon proper
Three clusters (clusters 4, 5, and 6) were assigned to tendon identity (Figure 3A). Consistent with the promiscuity of their molecular signatures (Figure 3D), we identified markers common to the three tendon clusters in the marker lists, exampled with COL12A1 (coding for the alpha1 chain of the FACIT type XII collagen), PTN (coding for the secreted molecule pleiotrophin), and TGFBI (coding for the secreted molecule, Transforming growth factor β induced) (Table S2; Figures S7A–S7C). COL12A1, PTN, and TGFBI genes displayed similar expression domains associated with tendons as compared to SCX expression in limbs (Figures S7A–S7H). This is consistent with collagen XII role in tendon structure and function59 and PTN expression in developing limb tendons.60 Because the addition of all COL12A1, PTN, and TGFBI gene combinations corresponded to 93% of cluster 4/5/6 fibroblasts (Figure S7H), we reasoned that their combined expression domains include the regions of the three tendon populations.
As for the MCT cluster 7, cluster 4 displayed very few specific markers (Table S2). ADGRG2 (adhesion G protein coupled receptor G2), a specific marker of cluster 4 (Figure 6B), was located in regions surrounding tendons (Figure 6C and 6C′). ADGRG2 was also observed delineating muscles in a similar manner to cluster 1 (epimysium) markers, exampled here with ANXA2 (Figures 6D–6F and 6F′). Double fluorescent in situ hybridization against ADGRG2 (cluster 4) and ANXA2 (cluster 1) confirmed that both genes overlap in the fibroblast layer delineating muscle and associated tendons (Figures S7J and S7K). Double fluorescent in situ hybridization against ADGRG2 (cluster 4) and CHODL (cluster 7, also expressed in tendons, Figure 5E) showed that ADGRG2 was expressed underneath the cluster 7 hypodermis layer (Figure 6D). These results combined with the recent identification of ADGRG as a marker of horse peritenon43 led us to conclude that cluster 4 fibroblasts correspond to the peritenon surrounding tendons and that peritenon and epimysium overlap when tendon interfaces with muscle.
Expression of specific markers of cluster 6 fibroblasts converged on perichondrium and associated tendon attachments. MYLK (myosin light-chain kinase), COL8A1, and PRRX1 known to be involved in mouse perichondrium development,61 were strongly expressed in the perichondrium (Table S2; Figures 6H, S2D, S2I, and S7L–S7Q). NET1 (coding for neuroepithelial cell-transforming gene 1 protein) showed an expression in tendons close to the perichondrium similarly to COL12A1, a marker common to all tendon clusters (Figures 6I, 6J, S7A, and S7D). We conclude that cluster 6 fibroblasts correspond to perichondrium and tendon attachments to perichondrium, namely the enthesis.
Specific markers for cluster 5 pointed to recognized tendon genes involved in tendon development and homeostasis (Table S2). The top three markers of cluster 5 were known tendon-associated collagens, COL1A1, COL12A1, and COL14A1,54,62,63 albeit being listed as markers for other tendon clusters (Table S2). Other markers of cluster 5 known to be involved in tendon function included POSTN (coding for the secreted extracellular matrix protein, periostin)64 and SPARC (coding for the secreted protein acidic and cysteine rich).45 One striking observation was that the main recognized tendon markers, SCX and TNMD, were not expressed in the same tendon region; TNMD transcripts appeared to be enriched in the tendon core, while SCX transcripts were more homogenously distributed within tendons (Figures 6L–6N). In addition to being a specific marker of enthesis and perichondrium (cluster 6) (Table S2), NET1 was expressed in cluster 5 tendon fibroblasts and in limb tendons (Figures 6O–6Q). NET1 expression was enriched in tendon fibroblasts in contact with or very close to muscle cells (Figure 6P). Double fluorescent in situ hybridization confirmed that NET1+ cells were enriched in apposition to muscle cells and did not co-localize with TNMD+ cells that were more centrally located (Figures 6R and 6S), while NET1+ cells overlapped with SCX+ cells in this area (Figures S7R and S7S). In silico quantification of double-positive cells further confirmed the little overlap between NET1+ and TNMD+ cells (7% of NET1-TNMD population) (Figure 6T). We conclude that cluster 5 contains the fibroblasts of the tendon proper. Tendon proper fibroblasts are heterogeneous with fibroblasts located in the tendon core and others at the tendon periphery.
We conclude that the fetal tendon fibroblasts are divided into 3 populations, peritenon (cluster 4), enthesis/perichondrium (cluster 6), and tendon proper (cluster 5), which prefigure the mature tendon organization.
CT fibroblast clusters are identified by a mosaic of partially overlapping gene expression patterns
Each fibroblast cluster is associated with a list of specific markers. However, as stated previously, a specific marker for a given cluster is expressed in at most 30%–55% of fibroblasts (Table S2; Figures 4C, 6N, 6Q, S3M, S3N, S4Q, S4R, S5Q, S5R, S6M, S6N, S7H, S7I, S7P, and S7Q). In addition, only 30% (on average) of the cluster fibroblasts are double-positive for any given couple of specific markers. However, the addition of all the possible specific marker combinations accounts for 90% (on average) of the cluster cells. Accordingly, the combined expression of specific markers recapitulates the corresponding cluster (Figures 4D, 4E, 5A–5D, 6A, 6G, and 6K). This shows that a fibroblast cluster corresponds to a combination of genes with partially overlapping expression patterns. Fibroblast clusters are thus inherently heterogeneous in terms of gene expression. In conclusion, these fibroblast populations cannot be defined by the expression of one key gene but by the combined expression of multiple genes.
MCT and tendon fibroblast populations successively emerge from a common population at the onset of the fetal period in silico
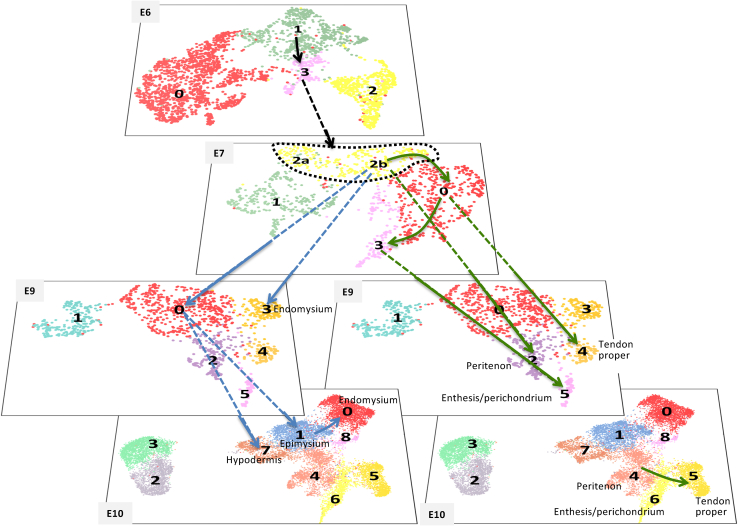
In order to identify the developmental origin of individual E10 CT clusters and their time of emergence, we turned to inferred analysis with STREAM.65 The 5CT inferred trajectory was not exploitable as it was not resolutive enough, and it included the E4 stage that did not contain lineage-relevant information. We thus performed trajectory inference analysis with STREAM65 on datasets resulting from the integration or the merge of two consecutive CT datasets between E6 and E10 (Figures 7 and S8). The analysis of the inferred trajectory for each combined dataset enabled us to establish lineage links between individual clusters of successive stages (Figures S8A–S8C), which when compiled, leads to an in silico lineage tree of individual clusters across time (Figure S8D). A lineage link between clusters x and y corresponds to a transition on the trajectory where cluster x gives way to/is relayed by cluster y along the trajectory (Figure S8, named arrows). To better visualize the lineage tree of MCT and tendon clusters, we duplicated the lineage tree for the E9 and E10 stages and highlighted the MCT- and tendon-related trajectories (Figure 7).
Figure 7.
MCT and tendon fibroblast populations successively emerge from a common population at the onset of the fetal period
Representation of the CT cluster lineage tree derived from the analysis of STREAM trajectories performed on the E6-E7, E7-E9, and E9-E10 combined CT datasets (see Figure S8). UMAP plots showing the distribution of CT clusters for the E6, E7, E9, and E10 CT datasets. This schematic only includes the MCT- and tendon-related lineage links. To illustrate that tendon and MCT lineages segregate from E7, the cluster lineage tree was separated in two branches and the E9 and E10 UMAP plots were duplicated. Black arrows refer to lineage links before MCT-tendon lineage segregation. Blue arrows refer to MCT-related lineage links. Green arrows refer to tendon-related lineage links. The cluster identity is indicated as early as it can be identified. See also Figures S8 and S9.
At E7, the E7-cluster 2b was identified as the common origin for the MCT and tendon types (Figures 7 and S8, see arrows m and l for MCT and k and g for tendon). Consistently, E7 was the stage where the proportion of OSR1+/SCX+ double-positive cells was the highest (Figures S9A and S9B) and the highest proportion of those OSR1+/SCX+ double-positive cells was found in E7-cluster 2b (Figures S9C and S9D). Notably, apart from the high values found only in single-expressing cells, the expression level distributions for SCX and OSR1 are similar in single-versus double-expressing cells (Figure S9B). These results support the existence of a common progenitor for both CT types in limbs, which is consistent with Scx- or Osr1-derived fibroblasts contributing to MCT or tendon, respectively.28 At E9, the endomysium (E10-cluster 0) was the first MCT cluster to emerge from E7-cluster 2b (Figures 7 and S8, arrow m). At E10, the last MCT clusters to emerge were the epimysium (E10-cluster 1) and hypodermis (E10-cluster 7), both deriving from the splitting of E9-cluster 0 (Figures 7 and S8, arrows n and o). These lineage links are consistent with the emergence of the MCT branch at E9 and the splitting of the MCT branch into two sub-branches at E10 identified on the 5CT trajectory (Figure 3C). Concomitantly to MCT cluster emergence, the enthesis/perichondrium (E10-cluster 6) was the first tendon cluster to emerge between E7 and E9 as E7-cluster 3 deriving from E7-cluster 0 (Figure S8, arrows i and j). By E9, the other two tendon clusters, peritenon (E10-cluster 4) and tendon proper (E10-cluster 5), have emerged from the E7-cluster 2b (arrow k) and the E7-cluster 0 (arrow h), respectively (Figures 7 and S8 arrows k and h). At E10, our trajectory inference analysis suggests lineage relationships between already-segregated lineages. Epimysium (cluster E10-1) contributes to endomysium (cluster E10-0), while peritenon (cluster E10-4) contributes to enthesis/perichondrium (cluster E10-6), itself contributing to tendon proper (cluster E10-5) (Figures 7 and S8 arrows p and q). These lineage links could correspond to lineage progression or lineage switching, which is difficult to decipher given the promiscuity of the population signatures.
We conclude that, in silico, the MCT and tendon fibroblast populations successively emerge from a common population at the onset of the fetal period.
Discussion
In this work, we showed that limb fibroblasts successively differentiate into CT populations after losing their positional information at the onset of fetal period. When the final fibro-muscular pattern is set, transcriptionally distinct but overlapping fibroblast populations are regionalized in limbs, prefiguring the mature fibroblast layers associated with adult skeletal muscle.
Limb fibroblasts provide positional cues and differentiate in a sequential manner
Fibroblasts provide the positional information cues to the limb cells during the establishment of the three limb axes.48,49 This is clear from our gene profiling analysis as fibroblasts cluster according to positional cues up to E7. At E7, fibroblasts switch to a differentiation program leading to CT types, indicating that they fulfill their two roles (provider of positional cues and differentiation) in a sequential manner. MCT fibroblasts are recognized to have a role in muscle bundle pre-patterning until E7.18 After this switch, they seem to lose their positional cues. At least, this positional program is no longer dominant in their transcriptional identity. However, we do observe residual HOX expression at E10 with HOXA13 expression in cluster 4/peritenon (Figure 2D). Peritenon fibroblasts are known to display stemness features as compared to tendon proper fibroblasts and are required for tendon repair after injury.66,67 HOXA13 expression in peritenon fibroblasts could be associated with residual positional information maintained in immature fibroblasts only48 or with the differentiation of the peritenon lineage; these possibilities are not mutually exclusive.
CT fibroblasts differentiate without continuous recruitment of progenitors
As stated previously, the second phase of limb CT development corresponds to differentiation and lineage diversification. A striking observation was that at each stage, CT clusters correspond to different populations. At each developmental stage, we did not identify clusters corresponding to successive differentiation steps of a given population. This is different from the muscle differentiation process, where at each developmental stage, the different steps of myogenesis from progenitors to differentiated cells are observed as clusters in scRNA-seq analysis of limb muscle cells.68 For CT, one has to combine successive developmental stages to visualize the progression of each CT fibroblast population along its differentiation path. CT fibroblast clustering therefore highlights lineage diversification rather than fibrogenesis progression. This shows that muscle and CT form and grow in two different ways. Muscle grows by persistent recruitment of progenitors into the myogenic program, while CT progresses from one differentiation step to the next in cohorts, exhausting populations from the previous differentiation steps. CT growth may occur with cell proliferation for each fibroblast population at different differentiation steps. Indeed, the distribution of cell cycle phases shows a persistent mix of cell cycle phases up to E10. Notably, CT fibroblasts start to differentiate and diversify at E7, the transition stage from embryonic to fetal development when muscle contraction starts in chicken embryos.
Delineating and interstitial fibroblasts display distinct transcriptional identities in line with their functions
At fetal stages, chicken limb MCT contains two main fibroblast populations, delineating and interstitial fibroblasts that prefigure the future epimysium and endomysium of adult skeletal muscle (Figure 8). Furthermore, inferred trajectories show that these two MCT populations derive from a common origin. We further show in chicken limbs that interstitial fibroblasts (endomysium) emerge before delineating fibroblasts (epimysium). Nonetheless, once established, the peripheral layer (epimysium) contributes to the more central layer (endomysium) (Figures 7 and S8). We believe that the perimysium (surrounding muscle bundles) is not present at fetal stages, since muscle bundles are not formed yet. Whether perimysium originates from epimysium or endomysium is not known.
Figure 8.
Spatial arrangement of fibroblast populations prefigures the mature fibroblast layers associated with adult skeletal muscle
(A) UMAP plot showing the distribution of CT clusters at E10. Specific markers for which in situ hybridization was performed are assigned to clusters.
(B) Schematic summarizing the location of fibroblast populations on limb transverse section at E10. The same color code was used for the in silico clusters and the fibroblast populations in limbs, except for cluster 8, color-coded as cluster 0, for the sake of clarity. Dermal fibroblast populations (clusters 2 and 3) were located at the limb periphery underneath the ectoderm. Underneath the dermis, the hypodermis (cluster 7) is the fibroblast layer that surrounds the whole musculoskeletal system. The fibroblast layers delineating tendon and muscles are the peritenon (Cluster 4) and epimysium (cluster 1) in continuity of each other. In addition to the peritenon fibroblasts, tendon fibroblasts were divided into two transcriptionally distinct clusters that correspond to enthesis /perichondrium (cluster 6) and tendon proper (cluster 5). The interstitial fibroblasts (clusters 0 and 8) in between muscle fibers (in white) prefigure the future endomysium.
The function of the outermost CT sheath of muscle is to constrain muscle in size and shape and to prevent friction between muscles during contraction.5 One main marker of these fetal delineating/epimysial fibroblasts is ANXA2. ANXA2 has been shown to be associated with membrane repair, cell-to-cell contact, membrane ruffling and cytoskeletal scaffolding,69,70 consistent with a function in maintaining/constraining muscle size. Moreover, ANXA2 partner proteins, such as the scaffold protein AHNAK271 and the Ca2+-binding protein S100A6,72 are also markers of delineating fibroblasts (Table S2). ANXA2 promotes FAP differentiation toward adipogenesis in the context of limb girdle dystrophy.73 CD34, a marker of delineating fibroblasts, is also a marker of adipose stem cells.74 Altogether, this is consistent with the epimysium as being a reservoir of adipocyte progenitors.34 Interestingly, we found that delineating fibroblast population (cluster 1) has a common origin with hypodermis fibroblast population (cluster 7) that is also recognized to contain white adipose tissue progenitors.4
The interstitial/endomysial fibroblasts (cluster 0) found in between muscle fibers have a retinoic acid (RA) signature, with ALDH1A2 as the main marker of these fibroblasts. This enzyme is required for RA function, suggestive of RA activity in fetal interstitial fibroblasts. RA has been associated with FAP differentiation toward fibroblasts (and not adipocytes), while loss of RA signaling drives FAPs toward adipogenesis.75 RA regulates matrix production and is seen as a therapeutic option to fight against fibrosis in skin disorders and to promote organ repair.76 HIC1, another marker of interstitial fibroblasts, is a downstream target of RA77 and has been shown to maintain muscle-associated fibroblasts in quiescence in adult normal and regenerative muscles.57 Inhibitors of FGF (Sprouty2) and Wnt (FRZB) signaling are also markers of interstitial fibroblasts. Interestingly, as for RA, Wnt inhibitors are also considered as a therapeutic option against fibrosis.78 We therefore see the hypodermis and epimysium external layers as a source of adipogenic progenitors, while the endomysium contains fibrogenic progenitors.
Fetal tendon fibroblasts at different maturation states are spatially segregated
Fetal tendon contains three main fibroblast populations, the peritenon (cluster 4), enthesis/perichondrium (cluster 6), and tendon proper (cluster 5) that prefigure adult tendon organization (Figure 8). Enthesis/perichondrium (cluster 4) is the first population to emerge at E7, consistent with enthesis function that is recognized to drive tendon position along cartilage elements.79,80 Enthesis/perichondrium fibroblast population is associated with adhesion molecules, exemplified by the adhesion molecule NET1. NET1 participates in cardiac fibrosis by promoting collagen synthesis in fibroblasts via the activation of Wnt/β-catenin and TGF/Smads signaling pathways.81 Within the tendon proper (cluster 5), we identified a regionalization of tendon fibroblasts with the classical tendon markers SCX and TNMD. Because TNMD is associated with differentiated tendon fibroblasts while SCX expression is associated with immature tendon fibroblasts,82,83 we assume that tendon fibroblasts located in the tendon core are more advanced in differentiation than those in the tendon periphery. Peritenon (cluster 4), the outermost tendon layer, is seen as a type of synovial sheath allowing tendon gliding and is recognized to concentrate blood vessels and innervation necessary for tendon function,5 consistent with the specific marker GHGB (chromogranin B), a secreted peptide of the neuroendocrine system.84 Tendon progenitors recruited for tendon repair are concentrated in the peritenon versus tendon proper,66,67 suggestive of an immature state of this fibroblast layer. Consistently, at E10, peritenon (cluster 4) provides cells to the other two tendon clusters 6 and 5 (Figure S8D).
A continuum in fibroblast identities to bridge tissues of highly different nature
Strikingly, each in silico fibroblast cluster maps to a specific in situ location in limbs, which supports the biological relevance of this scRNA-seq approach (Figure 8). As stated in the results, a fibroblast cluster corresponds to a mosaic of partially overlapping gene expression patterns. In addition, a gene identified as a marker for a given cluster by the differential gene expression analysis often shows weaker expression, outside of this cluster, both in silico and in situ. Along the same line, except for the dermis clusters that appear more distant, CT clusters are grouped together, highlighting their transcriptional proximity. Finally, the neighborhood relationships are conserved between the in silico and in situ analyses. Clusters that appear as neighbors on the E10 UMAP correspond to populations that are neighbors in limbs. This means that fibroblast populations that are close in transcriptional identity are also close in space. This shows that at the limb level, there is a continuum of promiscuous fibroblast identities reflecting the interconnected 3D network of fibroblasts associated with muscles. Consistently, we identified a molecular continuity between the outermost layers of tendon (peritenon) and MCT (epimysium). We propose that this continuum in identities underlies a continuum in mechanical properties that is instrumental to CT function. CT bridges muscle to bone or muscle to skin that are mechanically different. Our hypothesis is that it would do so by opposing CT layers with mechanical properties that would be close to that of muscle and progressively evolve toward that of bone, for example. The characterization of mechanical properties of fibroblast populations awaits further studies.
Limitations of the study
The trajectory inference analysis was performed on datasets spaced by one or two days of development. Given the known developmental pace of chicken limbs and the difference in trajectory continuity observed between datasets spaced by one or two days, this study would have benefited from additional time points (E5 and E8, in particular) to reach a better resolution in terms of event dating and lineage tree.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal IgG2b anti-MyHC (MF20) | DSHB | Cat. # MF 20 |
| Biological samples | ||
| E4, E6, E7, E9, E10 chicken forelimbs | This paper | N/A |
| Critical commercial assays | ||
| Single Cell 3′ Reagent Kit v3 | 10X Genomics | PN-1000121 |
| Riboprobe kit | Promega | P1460 |
| Kit GoTaq green master mix | Promega | M7122 |
| Kit TSA+ Cyanin 3 or 5 system | AKOYA biosciences | NEL744001KT ; NEL745001KT |
| Deposited data | ||
| E4, E6 and E10 scRNAseq datasets | Esteves de Lima et al.,28 | GEO: GSE166981 |
| E7 and E9 scRNAseq datasets | This paper | GEO : GSE261503 |
| Experimental models: Organisms/strains | ||
| Gallus Gallus | EARL Les Bruyères, Dangers, France | JA 57 strain |
| Oligonucleotides | ||
| Primers for PCR Probes, see Table S4 | This paper | N/A |
| Software and algorithms | ||
| Seurat package (v3.2.3) | Stuart et al., 201985 | https://satijalab.org/seurat/ |
| STREAM pipeline (v1.0) | Chen et al., 201965 | https://github.com/pinellolab/STREAM |
| DAVID version 202144,46 | Sherman et al. 202246, Huang da et al. 200947 | https://david.ncifcrf.gov/ |
| Other | ||
| 10X Chromium Chip | 10X Genomics | 1000073 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Delphine Duprez (delphine.duprez@sorbonne-universite.fr).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Chicken embryos
Fertilized chicken (Gallus gallus, RRID:NCBITaxon_9031) eggs from commercial sources (JA 57 strain: Morizeau, Dangers, France) were stored at 14°C upon arrival and then incubated at 38.5°C in a humidified incubator until appropriate stages. Embryos were staged according to the number of days in ovo (E). Limb buds were dissected at E4, E6, E7, E9 and E10 stages. All experiments on chicken embryos were performed before E14 and consequently are not submitted to a licensing committee, in accordance with European guidelines and regulations. Sex determination is not classically performed for chicken embryos.
Method details
Seurat clustering analysis
Sample preparation
The sample collection and scRNAseq protocol from E7 and E9 limb cells were performed as described for E4, E6 and E10 by Esteves de Lima et al.28
Both whole forelimbs from one E7 embryo and one E9 embryo were dissected in PBS. Ectoderm was removed by trypsine treatment (Sigma T4674) at room temperature for 2 min (E7) and 3 min (E9). Cell dissociation was achieved by collagenase treatment (12,5 μl of collagenase (Sigma C0130) in 1 ml of PBS/10% SVF/2mM EDTA/1% PS buffer) at 37°C for 7 min (E7) and 10 min (E9). Reaction was stopped by adding 9 mL of PBS/10% SVF/2mM EDTA/1% PS. To retrieve dissociated cells, the reaction solution was centrifuged for 5 min at 1200 rpm, the supernatant filtered on Sysmex 04-004-2326 filters and centrifuged again for 5 min at 1200 rpm. The pelleted cells were then resuspended in 1 mL of PBS/10% SVF/2mM EDTA/1% PS buffer. Cartilage tissues were poorly dissociated and therefore excluded from the samples.
Cell concentration was adjusted to 5000 cells/μl in the buffer. 5000 cells per conditions were loaded into the 10x Chromium Chip with the Single Cell 3′ Reagent Kit v3 according to the manufacturer’s protocol. Libraries were then sequenced by pair with a HighOutput flowcel using an Illumina Nextseq 500 with the following mode (150 HO): 28 base-pairs (bp) (Read1), 125 bp (Read 2) and 8 bp (i7 Index). A minimum of 50 000 reads per cell were sequenced and analysed with Cell Ranger Single Cell Software Suite 3.0.2 by 10x Genomics. Raw base call files from the Nextseq 500 were demultiplexed with the cellranger mkfastq pipeline into library-specific FASTQ files. The FASTQ files for each library were then processed independently with the cellranger count pipeline. This pipeline used STAR21 to align cDNA reads to the Gallus gallus genome (Sequence: GRCg6a, Gallus gallus reference).
Seurat clustering of whole limb datasets
The Seurat package (v3.2.3)85 under R (v4.3.1)86 was used to perform downstream clustering analysis on scRNAseq data87 (see Table S3 for parameter and cut-off values). Cells went through a classical Quality Control using the number of detected genes per cell (nFeatures), the number of mRNA molecules per cell (nCounts) and the percentage of expression of mitochondrial genes (pMito) as cut-offs. Outliers on a nFeature vs nCount plot were manually identified and removed from the dataset. Potential doublets were identified by running the Scrublet algorithm88 and then removed from the dataset. Gene counts for cells that passed the above selections were normalized to the total expression and Log-transformed with the NormalizeData function of Seurat using the nCount median as scale factor. Highly variable genes were detected with the FindVariableFeatures function (default parameters). Using highly variable genes as input, principal component analysis was performed on the scaled data in order to reduce dimensionality. Cell cycle effect was regressed out using the ScaleData function. Statistically significant principal components were determined by using the JackStrawPlot and the ElbowPlot functions. Cell clusters were generated with the FindNeighbors/FindClusters functions (default parameters except for the number of selected PCs). Different clustering results were generated at different resolutions and for different sets of PCs. Non-linear dimensional reduction (UMAP) and clustering trees using Clustree89 were used to visualize clustering results and select for the most robust and relevant result. Markers for each cluster were found using the FindAllMarkers function of Seurat (using highly variable genes as an input, default parameters otherwise) that ran Wilcoxon rank sum tests (p-val adjusted < 0.05). The clusters identified as CT clusters by the differential expression of classical CT markers (SCX, OSR1, TWIST2) represented the majority of limb cells, in addition to other clusters encompassing the expected cell populations present in developing limb tissues such as muscle, vessels/blood, neural crest cells and ectoderm (Figures S1A and S1B;28).
Seurat clustering of CT subsets
For all datasets, we selected the CT cluster cells and extracted them from the post-Quality Control object to generate a CT subset of the whole limb dataset. The sizes of CT datasets were 4654 cells (E4), 2869 cells (E6), 1229 cells (E7), 1328 cells (E9) and 14490 cells (E10). CT datasets were then processed again through the pipeline described above, from the Normalization to clustering and marker identification steps. The CT datasets were analyzed using classical Seurat tools such as Feature plots, Violin plots, Dimplots, Heatmaps and Dotplots. Scores of selected markers for each cluster were calculated using the AddModuleScore function. Gene expression was defined by ‘gene log-normalized count>0’. The R package ggplot2 v3.4.490 was used to generate custom feature plots highlighting gene co-expression. Population intersection plots were generated with the R package UpSetR v1.4.0.91
Merge and RPCA integration of CT subsets
In order to conduct trajectory inference analyses, individual post-clustering CT datasets were combined into single datasets either using the Seurat Merge tool (E7-E9 merge) or the RPCA integration strategy after merging (5CT, E6-E7, E9-10 integrations) to deal with the batch effect observed between the E4/E6/E10 and the E7/E9 batches. Before RPCA integration, merged CT datasets were individually normalized, highly variable genes identified and used to select integration features with the SelectIntegrationFeatures function (default parameters). Scaling followed by PCA with cell cycle gene regression were performed on the integration features. Anchor cells were identified with the FindIntegrationAnchors function, using the integration features and the RPCA reduction (default parameters). CT datasets were integrated with the IntegrateData function, using the integration features and anchor cells (default parameters). The merged or RPCA-integrated objects were then processed through the pipeline described above, from the Normalization (merge) or the Scaling (RPCA) steps to the non-linear dimension reduction (UMAP) step before being imported into the STREAM environment. For the 5CT dataset, we believe that the little overlap observed between some of the samples on the UMAP (Figure S1H) are due to biological differences and not to residual batch effect, because the extent of the overlap correlates with the time interval between consecutive stages rather than with a transition between different experimental batches.
STREAM trajectory inference
The STREAM pipeline (v1.0)65 under Python (v3.7.9) was used to perform trajectory inference analysis on integrated/merged CT datasets (see Table S3 for parameter values). The merged E7-E9 and the integrated E6-E7 and E9-E10 datasets were imported into the STREAM environment under the h5ad format to build the trajectory on the Seurat UMAP dimension reduction. To perform differential gene expression analysis on top of trajectory inference, the integrated 5CT dataset was imported under the 10X format and we generated an anndata object combining the unmodified expression matrix (RNA slot) of the 5CT Seurat object with a dimension reduction matrix derived from the integrated expression matrix of the 5CT Seurat object. The dimension reduction matrix was calculated on the integration features with the dimension_reduction function (method = 'mlle', n_neighbor = 250). Trajectories were inferred with the elastic_principal_graph function, using the mlle reduction for 5CT and the umap reduction for the other datasets. Trajectories were visualized as a dendrogram, color-coded with the Seurat cluster labels or gene expression levels, with the plot_stream function. Variable genes and then branch-associated marker ( z-score > 1.0; p value < 0.01) were identified from the unmodified expression matrix by the select_variable_genes and detect_leaf_markers functions (default parameters) for the 5CT.
DAVID analysis of the branch-associated markers
GO analysis of the marker lists associated with the Dermis, MCT, SCX and origin branches was performed using the Functional Annotation Clustering tool from DAVID (Database for Annotation, Visualization and Integrated Discovery, version 2021, DAVID Knowledgebase v2023q4).46,47 DAVID clusters GO terms into categories. The selected categories (enrichment score > 2.0) were associated with the following GO terms (p value < 0.05): Mitosis; DNA binding / Homeobox / Transcription; Secreted / Glycoproteins / Extra Cellular Matrix; Collagens / Extra Cellular Matrix organization; Cell adhesion/ Focal adhesion.
In situ hybridization
Forelimbs of E6 or E10 chicken embryos were fixed in 4% paraformaldehyde (Sigma- Aldrich) overnight at 4°C, then processed in 7.5%/15% gelatin/sucrose (Sigma-Aldrich) for cryostat sectioning (14 μm thick sections, CM3050S Leica cryostat), as previously described.28,68,92 Transverse or longitudinal limb sections were performed. Alternating serial sections were hybridized with probe 1, probe 2, probe 3 and probe 4 to allow comparison of expression domains on adjacent sections of the same limb.
Digoxigenin-labeled or Fluorescein-labeled RNA probes were prepared. The SCX, OSR1, KLF2 and SPRY2 probes were synthetized using the Riboprobe kit (Promega P1460) and the linearization/polymerization enzymes EcoR1/T3, NotI/T3, EcoR1/T3 and EcoR1/T7, respectively, from published plasmids.53,93 The other probes were prepared as PCR probes with the primers designed with DNA Star listed in Table S4 and using the Kit GoTaq green master mix (Promega M7122) (PCR program: TM = 55°C, 40 cycles).
Single colorimetric or fluorescent in situ hybridization were performed as described.28,68 The slides were incubated with hybridization buffer (50% formamide, 10X Salt 1X, Dextran Sulfate 10%, Yeast RNA 1 mg/mL, 50X Denhardt 1X, Tween 0,2%) with DIG-labeled RNA probes overnight at 65°C in a wet chamber. The slides were then washed twice with 0.2× SSC for 30 min at 65°C. After 5 min in MABT buffer (MAB 1X and 0.1% Tween-20), they were then blocked for 1 h in buffer containing 1× MABT, 10% blocking reagent (Roche) and 20% goat serum. For colorimetric in situ hybridization, slides were incubated in the same buffer with anti-DIG-AP antibodies (1:2000, Roche) overnight at 4°C. Labelling was revealed with NBT-BCIP solution. For fluorescent in situ hybridization, slides were incubated in the same buffer with anti-DIG-POD antibodies (1:500, Roche) overnight at 4°C. Labelling was revealed using the kit TSA-Plus Cyanin-3 (AKOYA).
Double fluorescent in situ hybridization were performed as described.94 The slides were incubated with hybridization buffer (50% formamide, 10X Salt 1X, Dextran Sulfate 10%, Yeast RNA 1 mg/mL, 50X Denhardt 1X, Tween 0,2%) with DIG-labeled RNA probe1 and Fluorescein-labeled RNA probe2 overnight at 65°C in a wet chamber. The slides were then washed twice with 0.2× SSC for 30 min at 65°C. After 5 min in MABT buffer [MAB 1X and 0.1% Tween-20], they were then blocked for 1 h in buffer containing 1× MABT, 10% blocking reagent (Roche) and 20% goat serum, then incubated in the same buffer with anti-DIG-POD antibodies (1:500, Roche) for 3 h minimum and revealed using the kit TSA-Plus Cyanin-3 (AKOYA). The first reaction was stopped with 2 % H2O2 in 1X PBS for 30 min. Slides were re-incubated in blocking buffer (1× MABT, 10% blocking reagent (Roche) and 20% goat serum) and then incubated in the same buffer with anti-Fluorescein-POD (1:1000, Roche) for 3 h minimum and revealed using the kit TSA-Plus Cyanin-5 (AKOYA).
Immunohistochemistry
In situ hybridization was followed with an immunohistochemistry with the MF20 monoclonal antibody to label sarcomeric myosin heavy chains. Undiluted MF20 antibody (DSHB cat. # MF20, undiluted supernatant) was incubated overnight at 4°C and revealed by an IgG2b Goat-anti-mouse secondary antibody conjugated to Alexa 488, Alexa 555 or Alexa 647 (Life Cat. # A21141, Cat.#A21147, Cat. # A-21242) in PBS for 1h at room temperature. Nuclei were visualized with DAPI (Sigma-Aldrich D9542) staining. Slides were mounted in Immuno-Mount (Thermo Fisher ref 99990402). The monoclonal antibody MF20, developed by A. Kawakami, was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biology Iowa City, IA 52242, USA.
Image capture
After immunohistochemistry or in situ hybridization experiments, images were obtained using a Zeiss apotome epifluorescence microscope or a Nikon eclipse E800 microscope, with the possibility to combine colorimetric and fluorescence labeling, or a leica DMR microscope for colorimetric images only.
Quantification and statistical analysis
The statistical analysis for Figure S9B was performed using the function stat_compare_means (method = wilcox.test) from the R package ggpubr v0.6.0.95 The details are described in the Figure S9B legend. scRNA-seq was performed in monoplicate.
No statistical method was used to predetermine sample size. Unless stated in figure legend, in situ hybridization and immunohistochemistry experiments have been performed on at least 3 limbs of different embryos.
No data was excluded in this study.
Randomization and blinding are not relevant in this study since all embryos are wild-type.
Acknowledgments
We thank Marie-Claire Delfini for critical reading the manuscript. We are grateful to Lale Alpar for artwork. Sequencing and genome mapping of the scRNA-seq datasets were performed by the GENOM’IC platform at the Cochin Institute (Paris). This work was supported by the CNRS, Inserm, SU, AFM_2019_MyoFibro (n° 22234), ANR_AAPG_2020_LimbCT, ANR_AAPG_2022_MyoDom, AFM_2022_MyoReg (n° 24414). ARTbio was supported by the CNRS, SU, the Institut Français de Bioinformatique and by a grant from the SIRIC CURAMUS.
Author contributions
Conceptualization: E.H. an D.D.; methodology: E.H., C.B., M.-A.B., L.B., and D.D.; software: E.H., L.B., and T.G.; validation: E.H., C.B., M.-A.B., and D.D.; formal analysis: E.H. and D.D.; investigation: E.H., C.B., and D.D.; resources: E.H., C.B., M.-A.B., and D.D.; data curation: E. H. and D.D.; writing – original draft: E.H. and D.D.; writing – review and editing: E.H. and D.D.; visualization: E.H., C.B., M.-A.B., and D.D.; supervision: E.H. and D.D.; project administration: E.H. and D.D.; funding acquisition: D.D.
Declaration of interests
The authors declare no competing interests.
Published: June 19, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110305.
Contributor Information
Estelle Hirsinger, Email: estelle.hirsinger@sorbonne-universite.fr.
Delphine Duprez, Email: delphine.duprez@sorbonne-universite.fr.
Supplemental information
Excel file of the differentially expressed genes in each trajectory branch, ordered by decreasing zscore. The ultimate_gene_names have been identified by combined data-mining of the Ensembl and NCBI Entrez databases.
Excel file of the differentially expressed genes in each cluster, ordered by decreasing fold-change. The ultimate_gene_names have been identified by combined data-mining of the Ensembl and NCBI Entrez databases. Pct.1 is the fraction of marker-expressing cells in the cluster of interest. Pct.2 is the fraction of marker-expressing cells in the other clusters. In the column “cluster” is listed the cluster the marker is associated with for a given row. In the column “cluster list” are listed all the clusters this marker is associated with.
References
- 1.Helmbacher F., Stricker S. Tissue cross talks governing limb muscle development and regeneration. Semin. Cell Dev. Biol. 2020;104:14–30. doi: 10.1016/j.semcdb.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Sefton E.M., Kardon G. Connecting muscle development, birth defects, and evolution: An essential role for muscle connective tissue. Curr. Top. Dev. Biol. 2019;132:137–176. doi: 10.1016/bs.ctdb.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nassari S., Duprez D., Fournier-Thibault C. Non-myogenic Contribution to Muscle Development and Homeostasis: The Role of Connective Tissues. Front. Cell Dev. Biol. 2017;5:22. doi: 10.3389/fcell.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch M.D., Watt F.M. Fibroblast heterogeneity: implications for human disease. J. Clin. Invest. 2018;128:26–35. doi: 10.1172/JCI93555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purslow P.P. The Structure and Role of Intramuscular Connective Tissue in Muscle Function. Front. Physiol. 2020;11:495. doi: 10.3389/fphys.2020.00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjamin M., Kaiser E., Milz S. Structure-function relationships in tendons: a review. J. Anat. 2008;212:211–228. doi: 10.1111/j.1469-7580.2008.00864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nourissat G., Berenbaum F., Duprez D. Tendon injury: from biology to tendon repair. Nat. Rev. Rheumatol. 2015;11:223–233. doi: 10.1038/nrrheum.2015.26. [DOI] [PubMed] [Google Scholar]
- 8.Li L., Cserjesi P., Olson E.N. Dermo-1: a novel twist-related bHLH protein expressed in the developing dermis. Dev. Biol. 1995;172:280–292. doi: 10.1006/dbio.1995.0023. [DOI] [PubMed] [Google Scholar]
- 9.Hornik C., Krishan K., Yusuf F., Scaal M., Brand-Saberi B. cDermo-1 misexpression induces dense dermis, feathers, and scales. Dev. Biol. 2005;277:42–50. doi: 10.1016/j.ydbio.2004.08.050. [DOI] [PubMed] [Google Scholar]
- 10.Murchison N.D., Price B.A., Conner D.A., Keene D.R., Olson E.N., Tabin C.J., Schweitzer R. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development. 2007;134:2697–2708. doi: 10.1242/dev.001933. [DOI] [PubMed] [Google Scholar]
- 11.Vallecillo-García P., Orgeur M., Vom Hofe-Schneider S., Stumm J., Kappert V., Ibrahim D.M., Borno S.T., Hayashi S., Relaix F., Hildebrandt K., et al. Odd skipped-related 1 identifies a population of embryonic fibro-adipogenic progenitors regulating myogenesis during limb development. Nat. Commun. 2017;8:1218. doi: 10.1038/s41467-017-01120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotsaris G., Qazi T.H., Bucher C.H., Zahid H., Pöhle-Kronawitter S., Ugorets V., Jarassier W., Börno S., Timmermann B., Giesecke-Thiel C., et al. Odd skipped-related 1 controls the pro-regenerative response of fibro-adipogenic progenitors. NPJ Regen. Med. 2023;8:19. doi: 10.1038/s41536-023-00291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stumm J., Vallecillo-García P., Vom Hofe-Schneider S., Ollitrault D., Schrewe H., Economides A.N., Marazzi G., Sassoon D.A., Stricker S. Odd skipped-related 1 (Osr1) identifies muscle-interstitial fibro-adipogenic progenitors (FAPs) activated by acute injury. Stem Cell Res. 2018;32:8–16. doi: 10.1016/j.scr.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Chevallier A., Kieny M., Mauger A. Limb-somite relationship: origin of the limb musculature. J. Embryol. Exp. Morphol. 1977;41:245–258. [PubMed] [Google Scholar]
- 15.Christ B., Jacob H.J., Jacob M. Experimental analysis of the origin of the wing musculature in avian embryos. Anat. Embryol. 1977;150:171–186. doi: 10.1007/BF00316649. [DOI] [PubMed] [Google Scholar]
- 16.Duprez D. Signals regulating muscle formation in the limb during embryonic development. Int. J. Dev. Biol. 2002;46:915–925. [PubMed] [Google Scholar]
- 17.Kardon G. Development of the musculoskeletal system: meeting the neighbors. Development. 2011;138:2855–2859. doi: 10.1242/dev.067181. [DOI] [PubMed] [Google Scholar]
- 18.Kardon G. Muscle and tendon morphogenesis in the avian hind limb. Development. 1998;125:4019–4032. doi: 10.1242/dev.125.20.4019. [DOI] [PubMed] [Google Scholar]
- 19.Ono Y., Schlesinger S., Fukunaga K., Yambe S., Sato T., Sasaki T., Shukunami C., Asahara H., Inui M. Scleraxis-lineage cells are required for correct muscle patterning. Development. 2023;150 doi: 10.1242/dev.201101. [DOI] [PubMed] [Google Scholar]
- 20.Stricker S., Mathia S., Haupt J., Seemann P., Meier J., Mundlos S. Odd-skipped related genes regulate differentiation of embryonic limb mesenchyme and bone marrow mesenchymal stromal cells. Stem Cells Dev. 2012;21:623–633. doi: 10.1089/scd.2011.0154. [DOI] [PubMed] [Google Scholar]
- 21.Kardon G., Harfe B.D., Tabin C.J. A Tcf4-positive mesodermal population provides a prepattern for vertebrate limb muscle patterning. Dev. Cell. 2003;5:937–944. doi: 10.1016/s1534-5807(03)00360-5. [DOI] [PubMed] [Google Scholar]
- 22.Hasson P., DeLaurier A., Bennett M., Grigorieva E., Naiche L.A., Papaioannou V.E., Mohun T.J., Logan M.P.O. Tbx4 and tbx5 acting in connective tissue are required for limb muscle and tendon patterning. Dev. Cell. 2010;18:148–156. doi: 10.1016/j.devcel.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colasanto M.P., Eyal S., Mohassel P., Bamshad M., Bonnemann C.G., Zelzer E., Moon A.M., Kardon G. Development of a subset of forelimb muscles and their attachment sites requires the ulnar-mammary syndrome gene Tbx3. Dis. Model. Mech. 2016;9:1257–1269. doi: 10.1242/dmm.025874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swinehart I.T., Schlientz A.J., Quintanilla C.A., Mortlock D.P., Wellik D.M. Hox11 genes are required for regional patterning and integration of muscle, tendon and bone. Development. 2013;140:4574–4582. doi: 10.1242/dev.096693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy M.M., Lawson J.A., Mathew S.J., Hutcheson D.A., Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asfour H., Hirsinger E., Rouco R., Zarrouki F., Hayashi S., Swist S., Braun T., Patel K., Relaix F., Andrey G., et al. Inhibitory SMAD6 interferes with BMP-dependent generation of muscle progenitor cells and perturbs proximodistal pattern of murine limb muscles. Development. 2023;150 doi: 10.1242/dev.201504. [DOI] [PubMed] [Google Scholar]
- 27.Flynn C.G.K., Ginkel P.R.V., Hubert K.A., Guo Q., Hrycaj S.M., McDermott A.E., Madruga A., Miller A.P., Wellik D.M. Hox11-expressing interstitial cells contribute to adult skeletal muscle at homeostasis. Development. 2023;150 doi: 10.1242/dev.201026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esteves de Lima J., Blavet C., Bonnin M.A., Hirsinger E., Comai G., Yvernogeau L., Delfini M.C., Bellenger L., Mella S., Nassari S., et al. Unexpected contribution of fibroblasts to muscle lineage as a mechanism for limb muscle patterning. Nat. Commun. 2021;12:3851. doi: 10.1038/s41467-021-24157-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yaseen W., Kraft-Sheleg O., Zaffryar-Eilot S., Melamed S., Sun C., Millay D.P., Hasson P. Fibroblast fusion to the muscle fiber regulates myotendinous junction formation. Nat. Commun. 2021;12:3852. doi: 10.1038/s41467-021-24159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan R., Zhang H., Ma Y., Lin R., Zhou B., Zhang T., Fan C., Zhang Y., Wang Z., Fang T., et al. Discovery of Muscle-Tendon Progenitor Subpopulation in Human Myotendinous Junction at Single-Cell Resolution. Research (Wash D C) 2022;2022 doi: 10.34133/2022/9760390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giordani L., He G.J., Negroni E., Sakai H., Law J.Y.C., Siu M.M., Wan R., Corneau A., Tajbakhsh S., Cheung T.H., Le Grand F. High-Dimensional Single-Cell Cartography Reveals Novel Skeletal Muscle-Resident Cell Populations. Mol. Cell. 2019;74:609–621.e6. doi: 10.1016/j.molcel.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 32.Giuliani G., Rosina M., Reggio A. Signaling pathways regulating the fate of fibro/adipogenic progenitors (FAPs) in skeletal muscle regeneration and disease. FEBS J. 2022;289:6484–6517. doi: 10.1111/febs.16080. [DOI] [PubMed] [Google Scholar]
- 33.Fukada S.I., Uezumi A. Roles and Heterogeneity of Mesenchymal Progenitors in Muscle Homeostasis, Hypertrophy, and Disease. Stem Cell. 2023;41:552–559. doi: 10.1093/stmcls/sxad023. [DOI] [PubMed] [Google Scholar]
- 34.Giuliani G., Vumbaca S., Fuoco C., Gargioli C., Giorda E., Massacci G., Palma A., Reggio A., Riccio F., Rosina M., et al. SCA-1 micro-heterogeneity in the fate decision of dystrophic fibro/adipogenic progenitors. Cell Death Dis. 2021;12:122. doi: 10.1038/s41419-021-03408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Micheli A.J., Swanson J.B., Disser N.P., Martinez L.M., Walker N.R., Oliver D.J., Cosgrove B.D., Mendias C.L. Single-cell transcriptomic analysis identifies extensive heterogeneity in the cellular composition of mouse Achilles tendons. Am. J. Physiol. Cell Physiol. 2020;319:C885–C894. doi: 10.1152/ajpcell.00372.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kendal A.R., Layton T., Al-Mossawi H., Appleton L., Dakin S., Brown R., Loizou C., Rogers M., Sharp R., Carr A. Multi-omic single cell analysis resolves novel stromal cell populations in healthy and diseased human tendon. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-70786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu W., Yang R., Li J. Single-cell and spatial transcriptomics reveal changes in cell heterogeneity during progression of human tendinopathy. BMC Biol. 2023;21:132. doi: 10.1186/s12915-023-01613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steffen D., Mienaltowski M., Baar K. Spatial gene expression in the adult rat patellar tendon. Matrix Biol. 2023;19–20 doi: 10.1016/j.mbplus.2023.100138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin Z., Hu J.J., Yang L., Zheng Z.F., An C.R., Wu B.B., Zhang C., Shen W.L., Liu H.H., Chen J.L., et al. Single-cell analysis reveals a nestin(+) tendon stem/progenitor cell population with strong tenogenic potentiality. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1600874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harvey T., Flamenco S., Fan C.M. A Tppp3(+)Pdgfra(+) tendon stem cell population contributes to regeneration and reveals a shared role for PDGF signalling in regeneration and fibrosis. Nat. Cell Biol. 2019;21:1490–1503. doi: 10.1038/s41556-019-0417-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mienaltowski M.J., Adams S.M., Birk D.E. Tendon proper- and peritenon-derived progenitor cells have unique tenogenic properties. Stem Cell Res. Ther. 2014;5:86. doi: 10.1186/scrt475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mienaltowski M.J., Cánovas A., Fates V.A., Hampton A.R., Pechanec M.Y., Islas-Trejo A., Medrano J.F. Transcriptome profiles of isolated murine Achilles tendon proper- and peritenon-derived progenitor cells. J. Orthop. Res. 2019;37:1409–1418. doi: 10.1002/jor.24076. [DOI] [PubMed] [Google Scholar]
- 43.Pechanec M.Y., Mienaltowski M.J. Decoding the transcriptomic expression and genomic methylation patterns in the tendon proper and its peritenon region in the aging horse. BMC Res. Notes. 2023;16:267. doi: 10.1186/s13104-023-06562-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feregrino C., Sacher F., Parnas O., Tschopp P. A single-cell transcriptomic atlas of the developing chicken limb. BMC Genom. 2019;20:401. doi: 10.1186/s12864-019-5802-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang T., Wagner A., Gehwolf R., Yan W., Passini F.S., Thien C., Weissenbacher N., Lin Z., Lehner C., Teng H., et al. Load-induced regulation of tendon homeostasis by SPARC, a genetic predisposition factor for tendon and ligament injuries. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abe5738. [DOI] [PubMed] [Google Scholar]
- 46.Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., Imamichi T., Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 48.Pineault K.M., Wellik D.M. Hox genes and limb musculoskeletal development. Curr. Osteoporos. Rep. 2014;12:420–427. doi: 10.1007/s11914-014-0241-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McQueen C., Towers M. Establishing the pattern of the vertebrate limb. Development. 2020;147 doi: 10.1242/dev.177956. [DOI] [PubMed] [Google Scholar]
- 50.Havis E., Bonnin M.A., Esteves de Lima J., Charvet B., Milet C., Duprez D. TGFβ and FGF promote tendon progenitor fate and act downstream of muscle contraction to regulate tendon differentiation during chick limb development. Development. 2016;143:3839–3851. doi: 10.1242/dev.136242. [DOI] [PubMed] [Google Scholar]
- 51.Nelson C.E., Morgan B.A., Burke A.C., Laufer E., DiMambro E., Murtaugh L.C., Gonzales E., Tessarollo L., Parada L.F., Tabin C. Analysis of Hox gene expression in the chick limb bud. Development. 1996;122:1449–1466. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
- 52.Mercader N., Leonardo E., Piedra M.E., Martínez-A C., Ros M.A., Torres M. Opposing RA and FGF signals control proximodistal vertebrate limb development through regulation of Meis genes. Development. 2000;127:3961–3970. doi: 10.1242/dev.127.18.3961. [DOI] [PubMed] [Google Scholar]
- 53.Orgeur M., Martens M., Leonte G., Nassari S., Bonnin M.A., Börno S.T., Timmermann B., Hecht J., Duprez D., Stricker S. Genome-wide strategies identify downstream target genes of chick connective tissue-associated transcription factors. Development. 2018;145 doi: 10.1242/dev.161208. [DOI] [PubMed] [Google Scholar]
- 54.Lejard V., Blais F., Guerquin M.J., Bonnet A., Bonnin M.A., Havis E., Malbouyres M., Bidaud C.B., Maro G., Gilardi-Hebenstreit P., et al. EGR1 and EGR2 involvement in vertebrate tendon differentiation. J. Biol. Chem. 2011;286:5855–5867. doi: 10.1074/jbc.M110.153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gaut L., Duprez D. Tendon development and diseases. Wiley Interdiscip. Rev. Dev. Biol. 2016;5:5–23. doi: 10.1002/wdev.201. [DOI] [PubMed] [Google Scholar]
- 56.Vorstandlechner V., Laggner M., Kalinina P., Haslik W., Radtke C., Shaw L., Lichtenberger B.M., Tschachler E., Ankersmit H.J., Mildner M. Deciphering the functional heterogeneity of skin fibroblasts using single-cell RNA sequencing. Faseb. J. 2020;34:3677–3692. doi: 10.1096/fj.201902001RR. [DOI] [PubMed] [Google Scholar]
- 57.Scott R.W., Arostegui M., Schweitzer R., Rossi F.M.V., Underhill T.M. Hic1 Defines Quiescent Mesenchymal Progenitor Subpopulations with Distinct Functions and Fates in Skeletal Muscle Regeneration. Cell Stem Cell. 2019;25:797–813.e9. doi: 10.1016/j.stem.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duprez D., Lapointe F., Edom-Vovard F., Kostakopoulou K., Robson L. Sonic hedgehog (SHH) specifies muscle pattern at tissue and cellular chick level, in the chick limb bud. Mech. Dev. 1999;82:151–163. doi: 10.1016/s0925-4773(99)00040-4. [DOI] [PubMed] [Google Scholar]
- 59.Izu Y., Adams S.M., Connizzo B.K., Beason D.P., Soslowsky L.J., Koch M., Birk D.E. Collagen XII mediated cellular and extracellular mechanisms regulate establishment of tendon structure and function. Matrix Biol. 2021;95:52–67. doi: 10.1016/j.matbio.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mittapalli V.R., Christ B., Pröls F., Scaal M. Pleiotrophin is expressed in avian somites and tendon anlagen. Histochem. Cell Biol. 2009;132:413–422. doi: 10.1007/s00418-009-0612-2. [DOI] [PubMed] [Google Scholar]
- 61.ten Berge D., Brouwer A., Korving J., Martin J.F., Meijlink F. Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development. 1998;125:3831–3842. doi: 10.1242/dev.125.19.3831. [DOI] [PubMed] [Google Scholar]
- 62.Guerquin M.J., Charvet B., Nourissat G., Havis E., Ronsin O., Bonnin M.A., Ruggiu M., Olivera-Martinez I., Robert N., Lu Y., et al. Transcription factor EGR1 directs tendon differentiation and promotes tendon repair. J. Clin. Invest. 2013;123:3564–3576. doi: 10.1172/JCI67521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Havis E., Bonnin M.A., Olivera-Martinez I., Nazaret N., Ruggiu M., Weibel J., Durand C., Guerquin M.J., Bonod-Bidaud C., Ruggiero F., et al. Transcriptomic analysis of mouse limb tendon cells during development. Development. 2014;141:3683–3696. doi: 10.1242/dev.108654. [DOI] [PubMed] [Google Scholar]
- 64.Rolnick K.I., Choe J.A., Leiferman E.M., Kondratko-Mittnacht J., Clements A.E.B., Baer G.S., Jiang P., Vanderby R., Chamberlain C.S. Periostin modulates extracellular matrix behavior in tendons. Matrix Biol. 2022;16 doi: 10.1016/j.mbplus.2022.100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen H., Albergante L., Hsu J.Y., Lareau C.A., Lo Bosco G., Guan J., Zhou S., Gorban A.N., Bauer D.E., Aryee M.J., et al. Single-cell trajectories reconstruction, exploration and mapping of omics data with STREAM. Nat. Commun. 2019;10:1903. doi: 10.1038/s41467-019-09670-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dyment N.A., Liu C.F., Kazemi N., Aschbacher-Smith L.E., Kenter K., Breidenbach A.P., Shearn J.T., Wylie C., Rowe D.W., Butler D.L. The paratenon contributes to scleraxis-expressing cells during patellar tendon healing. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mienaltowski M.J., Adams S.M., Birk D.E. Regional differences in stem cell/progenitor cell populations from the mouse achilles tendon. Tissue Eng. Part A. 2013;19:199–210. doi: 10.1089/ten.TEA.2012.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Esteves de Lima J., Blavet C., Bonnin M.A., Hirsinger E., Havis E., Relaix F., Duprez D. TMEM8C-mediated fusion is regionalized and regulated by NOTCH signalling during foetal myogenesis. Development. 2022;149 doi: 10.1242/dev.199928. [DOI] [PubMed] [Google Scholar]
- 69.Lim H.I., Hajjar K.A. Annexin A2 in Fibrinolysis, Inflammation and Fibrosis. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22136836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Méndez-Barbero N., San Sebastian-Jaraba I., Blazquez-Serra R., Martin-Ventura J.L., Blanco-Colio L.M. Annexins and cardiovascular diseases: Beyond membrane trafficking and repair. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pascal A., Gallaud E., Giet R., Benaud C. Annexin A2 and Ahnak control cortical NuMA-dynein localization and mitotic spindle orientation. J. Cell Sci. 2022;135 doi: 10.1242/jcs.259344. [DOI] [PubMed] [Google Scholar]
- 72.Cheng C.W., Rifai A., Ka S.M., Shui H.A., Lin Y.F., Lee W.H., Chen A. Calcium-binding proteins annexin A2 and S100A6 are sensors of tubular injury and recovery in acute renal failure. Kidney Int. 2005;68:2694–2703. doi: 10.1111/j.1523-1755.2005.00740.x. [DOI] [PubMed] [Google Scholar]
- 73.Hogarth M.W., Defour A., Lazarski C., Gallardo E., Diaz Manera J., Partridge T.A., Nagaraju K., Jaiswal J.K. Fibroadipogenic progenitors are responsible for muscle loss in limb girdle muscular dystrophy 2B. Nat. Commun. 2019;10:2430. doi: 10.1038/s41467-019-10438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raajendiran A., Ooi G., Bayliss J., O'Brien P.E., Schittenhelm R.B., Clark A.K., Taylor R.A., Rodeheffer M.S., Burton P.R., Watt M.J. Identification of Metabolically Distinct Adipocyte Progenitor Cells in Human Adipose Tissues. Cell Rep. 2019;27:1528–1540.e7. doi: 10.1016/j.celrep.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 75.Zhao L., Son J.S., Wang B., Tian Q., Chen Y., Liu X., de Avila J.M., Zhu M.J., Du M. Retinoic acid signalling in fibro/adipogenic progenitors robustly enhances muscle regeneration. EBioMedicine. 2020;60 doi: 10.1016/j.ebiom.2020.103020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang S., Yu J., Kane M.A., Moise A.R. Modulation of retinoid signaling: therapeutic opportunities in organ fibrosis and repair. Pharmacol. Ther. 2020;205 doi: 10.1016/j.pharmthera.2019.107415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kristensen L.S., Raynor M.P., Candiloro I., Dobrovic A. Methylation profiling of normal individuals reveals mosaic promoter methylation of cancer-associated genes. Oncotarget. 2012;3:450–461. doi: 10.18632/oncotarget.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burgy O., Königshoff M. The WNT signaling pathways in wound healing and fibrosis. Matrix Biol. 2018;68–69:67–80. doi: 10.1016/j.matbio.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 79.Blitz E., Viukov S., Sharir A., Shwartz Y., Galloway J.L., Pryce B.A., Johnson R.L., Tabin C.J., Schweitzer R., Zelzer E. Bone ridge patterning during musculoskeletal assembly is mediated through SCX regulation of Bmp4 at the tendon-skeleton junction. Dev. Cell. 2009;17:861–873. doi: 10.1016/j.devcel.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blitz E., Sharir A., Akiyama H., Zelzer E. Tendon-bone attachment unit is formed modularly by a distinct pool of Scx- and Sox9-positive progenitors. Development. 2013;140:2680–2690. doi: 10.1242/dev.093906. [DOI] [PubMed] [Google Scholar]
- 81.Li T., Xiong X., Wang Y., Li Y., Liu Y., Zhang M., Li C., Yu T., Cao W., Chen S., et al. Neuroepithelial cell-transforming 1 promotes cardiac fibrosis via the Wnt/β-catenin signaling pathway. iScience. 2023;26 doi: 10.1016/j.isci.2023.107888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shukunami C., Takimoto A., Oro M., Hiraki Y. Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev. Biol. 2006;298:234–247. doi: 10.1016/j.ydbio.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 83.Shukunami C., Takimoto A., Nishizaki Y., Yoshimoto Y., Tanaka S., Miura S., Watanabe H., Sakuma T., Yamamoto T., Kondoh G., Hiraki Y. Scleraxis is a transcriptional activator that regulates the expression of Tenomodulin, a marker of mature tenocytes and ligamentocytes. Sci. Rep. 2018;8:3155. doi: 10.1038/s41598-018-21194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Helle K.B. Chromogranins A and B and secretogranin II as prohormones for regulatory peptides from the diffuse neuroendocrine system. Results Probl. Cell Differ. 2010;50:21–44. doi: 10.1007/400_2009_26. [DOI] [PubMed] [Google Scholar]
- 85.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.R Core Team . R Foundation for Statistical Computing; 2018. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 87.Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wolock S.L., Lopez R., Klein A.M. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst. 2019;8:281–291.e9. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zappia L., Oshlack A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. GigaScience. 2018;7 doi: 10.1093/gigascience/giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wickham H. Springer; 2016. ggplot2: Elegant Graphics for Data Analysis; pp. 1–182. [DOI] [Google Scholar]
- 91.Conway J.R., Lex A., Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Esteves de Lima J., Bonnin M.A., Birchmeier C., Duprez D. Muscle contraction is required to maintain the pool of muscle progenitors via YAP and NOTCH during fetal myogenesis. Elife. 2016;5 doi: 10.7554/eLife.15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eloy-Trinquet S., Wang H., Edom-Vovard F., Duprez D. Fgf signaling components are associated with muscles and tendons during limb development. Dev. Dyn. 2009;238:1195–1206. doi: 10.1002/dvdy.21946. [DOI] [PubMed] [Google Scholar]
- 94.Wilmerding A., Rinaldi L., Caruso N., Lo Re L., Bonzom E., Saurin A.J., Graba Y., Delfini M.C. HoxB genes regulate neuronal delamination in the trunk neural tube by controlling the expression of Lzts1. Development. 2021;148 doi: 10.1242/dev.195404. [DOI] [PubMed] [Google Scholar]
- 95.Kassambara A. ggpubr: 'ggplot2' Based Publication Ready Plots. 2023. https://rpkgs.datanovia.com/ggpubr/ R package version 0.6.0.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Excel file of the differentially expressed genes in each trajectory branch, ordered by decreasing zscore. The ultimate_gene_names have been identified by combined data-mining of the Ensembl and NCBI Entrez databases.
Excel file of the differentially expressed genes in each cluster, ordered by decreasing fold-change. The ultimate_gene_names have been identified by combined data-mining of the Ensembl and NCBI Entrez databases. Pct.1 is the fraction of marker-expressing cells in the cluster of interest. Pct.2 is the fraction of marker-expressing cells in the other clusters. In the column “cluster” is listed the cluster the marker is associated with for a given row. In the column “cluster list” are listed all the clusters this marker is associated with.
Data Availability Statement
-
•
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.