Abstract
Ferroptosis is a form of iron-related oxidative cell death governed by an integrated redox system, encompassing pro-oxidative proteins and antioxidative proteins. These proteins undergo precise control through diverse post-translational modifications, including ubiquitination, phosphorylation, acetylation, O-GlcNAcylation, SUMOylation, methylation, N-myristoylation, palmitoylation, and oxidative modification. These modifications play pivotal roles in regulating protein stability, activity, localization, and interactions, ultimately influencing both the buildup of iron and lipid peroxidation. In mammalian cells, regulators of ferroptosis typically undergo degradation via two principal pathways: the ubiquitin-proteasome system, which handles the majority of protein degradation, and autophagy, primarily targeting long-lived or aggregated proteins. This comprehensive review aims to summarize recent advances in the post-translational modification and degradation of proteins linked to ferroptosis. It also discusses strategies for modulating ferroptosis through protein modification and degradation systems, providing new insights into potential therapeutic applications for both cancer and non-neoplastic diseases.
Keywords: Ferroptosis, Modification, Degradation, Autophagy, Proteasome
1. Introduction
Cell death is a fundamental process in life, triggered by physiological or pathological stressors. It can generally be categorized into two classes: accidental cell death (ACD) and regulated cell death (RCD) [1]. ACD occurs as an uncontrolled response to severe and unforeseen cellular damage that exceeds the cell's regulatory mechanisms. In contrast, RCD is intricately regulated by intracellular molecules. Apoptosis, primarily mediated by caspases, has been extensively studied as one of the most enduring forms of RCD, contributing to embryonic development, immune system maturation, and tumorigenesis. Recent research has uncovered diverse non-apoptotic forms of RCD characterized by unique morphology, biochemistry, and genetic alterations [2,3]. These non-apoptotic types can be modulated by experimental small molecules or clinical drugs.
Ferroptosis was specified as an iron-dependent, non-apoptotic form of RCD that can be triggered by a few classes of small molecules (e.g., erastin and RSL3) and inhibited by lipophilic antioxidants (e.g., ferrostatin-1) [4]. The process of ferroptosis involves iron-dependent lipid peroxidation, leading to oxidative damage to plasma membrane and the extracellular emission of damage-associated molecular patterns (DAMPs) [5,6]. These DAMPs are pivotal in activating the immune response, serving as signals that notify the immune system of the presence of damaged or stressed cells. Both excessive and deficient ferroptosis are implicated in various diseases and pathological conditions, including cancer, tissue ischemia-reperfusion, inflammatory diseases, neurodegeneration, age-related diseases, and tissue damage (e.g., kidney injury) [[7], [8], [9]]. This discovery has sparked a substantial increase in research on ferroptosis, yielding fresh insights into its molecular and cellular mechanisms, along with its potential applications in disease treatment [[10], [11], [12]].
The regulation of ferroptosis occurs at the transcription, translation, and post-translational levels [13]. Mounting evidence substantiates the importance of post-translational modifications (PTMs) in governing ferroptosis and influencing susceptibility in diverse pathophysiological processes. Eukaryotic cells employ the protein degradation system, primarily the ubiquitin-proteasome system (UPS) and autophagy, to maintain appropriate protein levels. The interplay of PTMs and protein degradation mechanisms offers multiple avenues for intervention in the context of ferroptosis.
This review outlines the fundamental molecular mechanisms of ferroptosis, with a particular focus on the pivotal role of oxidative and antioxidant systems in modulating ferroptosis via PTMs and protein degradation. Finally, we explore the therapeutic implications of targeting PTMs and degradation in diseases related to ferroptosis.
2. The process of ferroptosis
Ferroptosis is characterized by oxidative cell death resulting from the unrestricted peroxidation of polyunsaturated fatty acids (PUFAs) within phospholipids, a critical component of cell membranes [14]. In the following discussion, our focus will be on elucidating the central mechanisms responsible for the formation and mitigation of lipid peroxidation, as they are pivotal in shaping a cell's vulnerability to ferroptosis (Fig. 1).
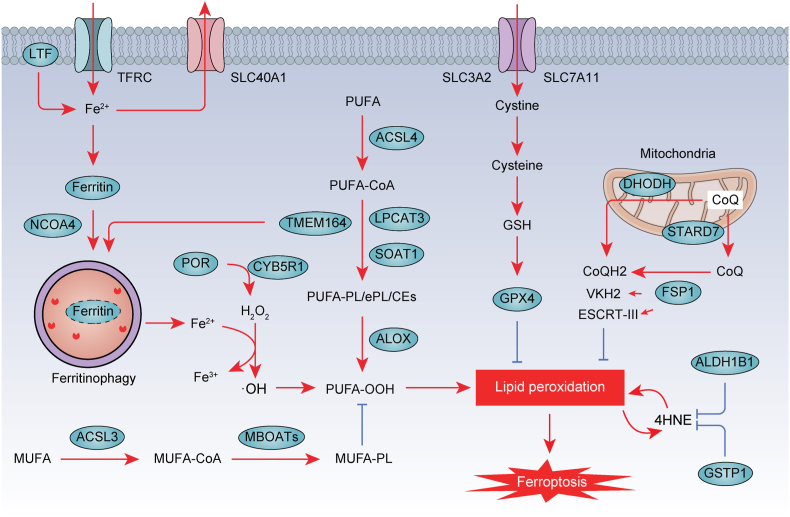
Fig. 1.
The process of ferroptosis. Ferroptosis is a form of nonapoptotic cell death characterized by overwhelming membrane lipid peroxidation and iron accumulation. Ferroptosis involves an imbalance in the oxidation and antioxidant systems. TFRC and LTF mainly promote ferroptosis by transferring Fe2+ into cells, while ferritin and SLC40A1 by storing iron and transporting iron out of cells, respectively. NCOA4-mediated ferritinophagy may play a role in regulating cellular iron levels by targeting ferritin for degradation. Subsequently, Fe2+ generates ROS through Fenton reaction. In particular, ACSL4, LPCAT3, SOAT1, TMEM164, ALOX, and POR pathways mediate the peroxidation of polyunsaturated fatty acids (PUFA), which is necessary for iron-mediated oxidative damage in ferroptosis. To counteract this oxidative stress, antioxidant defense systems have been identified, including SLC7A11-GSH-GPX4 and CoQ. Moreover, phospholipid modifying enzymes ACSL3 and MBOATs inhibit ferroptosis activity by promoting MUFA-PL synthesis. In the intricate network regulating lipid metabolism, ALDH1B1 and GSTP1 emerge as key players in ferroptosis. Their actions restrict the generation of 4-hydroxynonenal (4HNE), effectively acting as suppressors of ferroptosis.
2.1. Iron accumulation
As an abundant essential trace element, iron is crucial for the induction of ferroptosis. Two mechanisms may explain how excess iron contributes to ferroptosis. First, iron can trigger the generation of reactive oxygen species (ROS) via the Fenton reaction. Second, iron also acts as a cofactor for lipid peroxidase enzymes (e.g., lipoxygenase [ALOX]), contributing to oxidative damage in ferroptosis. Mitochondria, a critical metabolic hub, may influence susceptibility to ferroptosis by regulating the balance between mitochondrial iron and lipid peroxidation. Given that iron can accumulate within different subcellular organelles, further investigation is warranted to compare their respective roles in ferroptosis.
Nonetheless, targeting different steps of iron metabolism, including iron uptake (e.g., transferrin receptor [TFRC] and lactotransferrin [LTF]), storage (e.g., ferritin), and efflux (e.g., SLC40A1/ferroportin), can modulate cellular or tissue susceptibility to ferroptosis [15]. Therefore, gaining insight into the threshold and molecular network associated with iron-mediated cell proliferation and cell death (e.g., ferroptosis) is crucial for disease treatment.
2.2. Lipid peroxidation
While the ultimate execution steps of ferroptosis remain poorly understood, lipid peroxidation emerges as a pivotal factor, either directly initiating membrane damage or propelling the activities of downstream membrane-damaging proteins. Lipid peroxidation primarily targets phospholipids containing PUFAs, although lipid sources from organelles (e.g., peroxisomes) and lipophagy (a form of selective autophagy that degrades lipid droplets) also contribute to ferroptosis in a context-dependent manner [[16], [17], [18]]. To identify diverse lipid sources and their associated metabolic pathways in different disease conditions, lipid species profiling through lipidomics is essential [19,20].
As lipid-metabolizing enzymes, acyl-CoA synthetase long-chain family member 4 (ACSL4), lysophosphatidylcholine acyltransferase 3 (LPCAT3), and ALOXs mediate the oxidation of esterified PUFA (e.g., arachidonic acid)-phospholipids during ferroptosis [[21], [22], [23], [24]]. Alongside the ACSL4-LPCAT3 pathway, ACSL4 also activates sterol O-acyltransferase 1 (SOAT1) in pancreas cancer cells, leading to the production of PUFA-cholesteryl esters (CEs) instead of PUFA-PEs, thereby promoting ferroptosis [25]. Transmembrane protein 164 (TMEM164) functions as a pro-ferroptotic enzyme, either playing a direct role in the enrichment of PUFA-ether phospholipids or forming the lipid foundation for the initiation of phagophore formation in the context of autophagy [26,27]. In contrast, acyl-CoA synthetase long-chain family member 3 (ACSL3)-mediated synthesis of monounsaturated fatty acids (MUFA) mitigate the accumulation of lipid ROS and reduce the levels of oxidized PUFA-PEs [28]. Phospholipid-modifying enzymes membrane bound O-acyltransferase domain containing proteins (e.g., MBOAT1 and MBOAT2) act as a ferroptosis suppressor through transferring MUFA into lyso-phosphatidylethanolamine [29]. In addition, the coupling of POR with cytochrome B5 reductase 1 (CYB5R1) facilitates the generation of ROS, which drives lipid peroxidation and subsequent ferroptosis [30,31]. These findings further suggest that various lipid species originating from different synthesis pathways (e.g., de novo lipogenesis) may competitively or synergistically impact the overall extent of lipid peroxidation.
Certain products resulting from lipid peroxidation, notably 4-hydroxynonenal (4HNE) at elevated concentrations, can directly promote ferroptosis through positive feedback mechanisms that enhance lipid peroxidation [[32], [33], [34]]. While these discoveries enhance our understanding of the pathways that initiate lipid peroxidation in ferroptosis, further elucidation of context-dependent signal pathways and the transition from physiological to pathological lipid peroxidation is vital for the development of precise antitumor strategies.
2.3. Antioxidant defense
Major ferroptosis antioxidant systems include GPX4–glutathione (GSH) system axis and ubiquinone (CoQ) system. Classic ferroptosis activators, such as erastin and RSL3, serves as inhibitors of the GPX4–GSH antioxidant system, which plays an essential role in protecting against ferroptosis induced by excessive oxidative stress.
2.3.1. System Xc−-GSH-GPX4 system
GPX4 belongs to the GPX family of proteins and primarily converts phospholipid hydroperoxides to phosphatidyl alcohols [35]. Both cytoplasmic and mitochondrial-localized GPX4 have been shown to confer resistance to ferroptosis [36,37]. Inhibition of GPX4 through genetic or pharmacological means can lead to the buildup of lipid peroxides, consequently triggering ferroptosis [38,39]. GSH, a key cofactor of GPX4, is an antioxidant tripeptide synthesized from three amino acids, glutamate, cysteine, and glycine. Cystine, transported intracellularly via system Xc−, undergoes NADPH-dependent reduction to yield cysteine, a rate-limiting precursor for GSH synthesis. Solute carrier family 7 member 11 (SLC7A11) is a major functional subunit of system Xc−, while solute carrier family 3 member 2 (SLC3A2) anchors SLC7A11 to the plasma membrane. Genetic or pharmacological inhibition of SLC7A11 depletes intracellular GSH, thereby promoting ferroptosis [4].
However, GPX4 inhibitors do not uniformly affect all cells; certain tumor cells display resilience to ferroptosis even when GPX4 is suppressed [40], suggesting the existence of GPX4-independent mechanisms of tumor resistance to ferroptosis. Furthermore, it should also be noted that in some cases, the knockout of GPX4 is unable to induce ferroptosis. For instance, the knockout of GPX4 in red blood cells or myeloid cells can lead to necroptosis and pyroptosis, respectively [41,42]. Although the mechanisms remain unclear, one hypothesis is that GPX4 deficiency may lead to distinct forms of lipid peroxidation, affecting membrane structure and function in a cell-type-dependent manner.
2.3.2. CoQ system
FSP1/AIFM2 and dihydroorotate dehydrogenase (DHODH) can resist ferroptosis through CoQ system independently of GPX4 [43,44]. Apart from its role in mitochondrial electron transfer, CoQ also acts as a scavenger of lipid peroxyl radicals. FSP1 employs an integrated mechanism to inhibit ferroptosis. One facet of this mechanism involves FSP1 functioning as an NADPH-dependent oxidoreductase, reducing CoQ to ubiquinol (CoQH2), thus blocking lipid peroxidation and ferroptosis. Interestingly, FSP1 may produce a cytoplasmic rather than mitochondrial CoQH2 pool. In contrast, CoQ is primarily synthesized in mitochondria, where DHODH serves as another reductase that facilitates the conversion of CoQ to CoQH2 [37]. When GPX4 is inactive, DHODH scavenges mitochondrial lipid peroxidation in a CoQH2-dependent manner, thereby inhibiting ferroptosis [37]. This is in line with the protective role of StAR related lipid transfer domain containing 7 (STARD7)-mediated CoQ transport to the plasma membrane against ferroptosis [45]. Consequently, cytoplasmic FSP1 and mitochondrial DHODH can complement each other, jointly inhibiting lipid peroxidation and exerting a synergistic effect. It's worth noting that recent concerns have been raised regarding the off-target effects of DHODH inhibitors on FSP1 in cancer cells [46]. Despite that, it's important to acknowledge that many compounds exhibit various activities at high concentrations. Furthermore, FSP1 also influences the redox states of vitamin K and the recruitment of membrane repair machinery [47,48]. The expression of FSP1 is positively regulated by transcription factor NFE2L2/NRF2 and m6A methyltransferase METTL3, therefore promoting ferroptosis resistance in cancer cells [49,50]. However, there is currently a lack of systematic research to determine whether these various functions occur sequentially or simultaneously.
3. Protein modification in ferroptosis
PTM refers to chemical modifications that take place on proteins after they have been synthesized or translated from messenger RNA [51]. These modifications can significantly impact the structure, activity, localization, and function of proteins, playing pivotal roles in both cell survival and cell death processes. In this section, we will explore the current understanding of how PTMs function in monitoring ferroptosis through various proteins (Table 1).
Table 1.
Main types of PTM in ferroptosis.
| PTM category | Substrates | Key regulators | Effects of PTM in ferroptosis | Ref |
|---|---|---|---|---|
| Ubiquitination |
FSP1 |
TRIM21, TRIM69, TRIM56 |
K63-linked ubiquitination enhances plasma membrane localization of FSP1 and inhibits ferroptosis; K48-linked ubiquitination enhances degradation of FSP1 and promotes ferroptosis |
[56,143,145] |
| Phosphorylation | DHODH | LOXL3 | Enhances degradation of DHODH and promotes ferroptosis | [66] |
| TFRC | HUWE1, BTRC |
Enhances degradation of TFRC and inhibits ferroptosis | [108,109] | |
| LTF | NEDD4L | Enhances degradation of LTF and promotes ferroptosis | [110] | |
| SLC40A1 | RNF217, USP35 |
Enhances degradation of SLC40A1 and promotes ferroptosis | [111,112] | |
| IRP2 | FBXL5 | Enhances degradation of IRP2 and inhibits ferroptosis | [114] | |
| SLC7A11 | BTRC, HECTD3, NEDD4L, SOCS2, SYVN1, TRIM26, ZRANB1, OTUB1, USP20 |
Enhances degradation of SLC7A11 and promotes ferroptosis | [6,[115], [116], [117], [118], [119], [120], [121], [122]] | |
| GPX4 | FBXO31, NEDD4, NEDD4L, STUB1, TRIM21, TRIM25, TRIM26, TRIM59, LUBAC, OTUB1, OTUD5, USP31 |
Enhances degradation of GPX4 and promotes ferroptosis | [[126], [127], [128], [129], [130], [131], [132], [133], [134]] | |
| ACSL4 | RNF146, NEDD4L, MARCHF6 |
Enhances degradation of ACSL4 and inhibits ferroptosis | [[146], [147], [148]] | |
| GSTP1 | SMURF2 | Enhances degradation of GSTP1 and promotes ferroptosis | [34] | |
| VDAC2/3 | NEDD4, FBXW7 |
Enhances degradation of VDAC2/3 and inhibits ferroptosis | [150,151] | |
| NFE2L2 | KEAP1, MIB1, USP11 |
Enhances degradation of NFE2L2 and promotes ferroptosis | [87,154,155] [156] | |
| HELLS | DCAF8, USP5, USP11 |
Enhances degradation of HELLS and promotes ferroptosis | [[158], [159], [160]] | |
| NCOA4 | TRIM7, FBXW7 |
Enhances degradation of NCOA4 and inhibits ferroptosis | [161,162] | |
| BECN1 | USP11 | Enhances degradation of BECN1 and inhibits ferroptosis | [163] | |
| GPX4 | CKB | Decreases degradation of GPX4 and inhibits ferroptosis | [58] | |
| Acetylation | ACSL4 | PKCBII | Enhances dimerization of ACSL4 and promotes ferroptosis | [63] |
| HSPB1 | PKC | Enhances function of HSPB1 and inhibits ferroptosis | [64] | |
| EIF4E | NA | Enhances formation of EIF4E-ALDH1B1 complex and promotes ferroptosis | [33] | |
| UMPS | AMPK | Enhances function of DHODH and inhibits ferroptosis | [65] | |
| HPCAL1 | PRKCQ | Enhances function of HPCAL1 and promotes ferroptosis | [70] | |
| BECN1 | AMPK | Enhances function of BECN1 and promotes ferroptosis | [71] | |
| TP53 | CREBBP, SIRT1 |
Enhances function of TP53 and promotes ferroptosis | [76,77] | |
| O-GlcNAcylation | ALOX12 | CTH | Enhances function of ALOX12 and promotes ferroptosis | [78] |
| HSPA5 | EP300 | Enhances function of HSPA5 and inhibits ferroptosis | [79] | |
| Histone | KAT5, EHMT2, SIRT1 |
Context-specific effects | [81,83] | |
| SLC7A11 | OGT | Enhances function of SLC7A11 and inhibits ferroptosis | [84] | |
| SUMOylation | YAP1 | OGT | Enhances function of YAP1 and promotes ferroptosis | [85,86] |
| ZEB1 | OGT | Enhances function of ZEB1 and promotes ferroptosis | [85,86] | |
| JUN | OGT | Enhances function of JUN and promotes ferroptosis | [89] | |
| Ferritin | OGT | Enhances binding of ferritin and NCOA4 and promotes ferroptosis | [90] | |
| ACSL4 | SENP1 | Decreases degradation of ACSL4 and promotes ferroptosis | [92] | |
| Methylation | HIF1A | SENP1 | Enhances function of HIF1A and promotes ferroptosis | [93] |
| ACSL4 | CARM1 | Enhances degradation of ACSL4 and inhibits ferroptosis | [97] | |
| N-myristoylation | NFE2L2 | CARM1 | Decreases function of NEF2L2 and promotes ferroptosis | [98] |
| OTUB1 | SETD7 | Decrease binding of OTUB1 and UBE2N, and promotes ferroptosis | [99] | |
| FSP1 | ACSL1 | Enhances plasma membrane localization of FSP1 and inhibits ferroptosis | [43] | |
| Palmitoylation | SLC7A11 | ZDHHC8 | Decreases degradation of SLC7A11 and inhibits ferroptosis | [102] |
| Oxidative modification | PRDX3 | NA | Enhances function of PRDX3 and promotes ferroptosis | [104] |
3.1. Ubiquitination
Ubiquitin, a small 76–amino acid protein, binds to its target substrate, forming a polyubiquitin chain in a process known as ubiquitination. This procedure is catalyzed by a cascade involving E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, and E3 ubiquitin ligase. Ubiquitination occurs predominantly at specific lysine (K) residues, including K6, K11, K27, K29, K33, K48, K63, and methionine-1 (M1). Ubiquitination is a tightly regulated PTM, that can be reversed by deubiquitinating enzymes, which remove ubiquitin molecules from proteins [52]. This PTM has emerged as a critical regulatory mechanism in ferroptosis. The processes of ubiquitination and deubiquitination often determine protein degradation and stability, offering a potential means to modulate the levels of proteins involved in ferroptosis. We will delve into this aspect in the subsequent section on ‘UPS-mediated protein degradation'.
In addition to its traditional role in protein degradation, ubiquitination is linked to various non-degradative functions. Recent research highlights that K63-linked ubiquitination can either promote or hinder the translocation of target proteins to the plasma membrane [53,54]. The function of FSP1 in inhibiting ferroptosis is intricately tied to its membrane localization [43,44,55]. FSP1 undergoes K63-linked ubiquitination and relocates to the plasma membrane to limit ferroptosis [56]. Furthermore, the E3 ubiquitin ligase TRIM21 facilitates K63-linked ubiquitination of FSP1 at Lys 322 and Lys 366. Mutations at these residues disrupt plasma membrane localization and the anti-ferroptosis activity of FSP1 [56].
3.1.1. Phosphorylation
Phosphorylation is a widespread covalent modification in PTM. Protein kinases catalyze the transfer of phosphate groups from ATP or GTP to amino acid residues (e.g., serine, threonine, and tyrosine) on substrate proteins [57]. Protein phosphatases, on the other hand, remove phosphate groups. This dynamic interplay between kinases and phosphatases makes phosphorylation the primary regulatory mechanism in ferroptosis (Fig. 2).
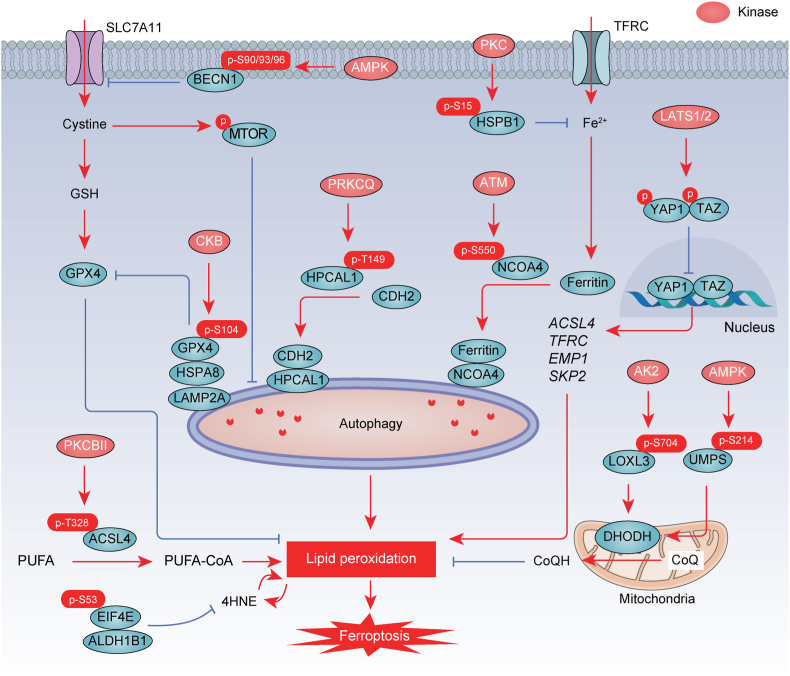
Fig. 2.
Phosphorylation in ferroptosis. Direct phosphorylation of GPX4, ACSL4, BECN1, HPCAL1, PKM2, NCOA4, HSPB1, LOXL3, EIF4E, and UMPS by various kinases is involved in the regulation of ferroptosis. P, phosphorylation.
3.1.2. Phosphorylation of GPX4
Metabolic enzymes can serve as protein kinases, exerting significant control over essential cellular processes in tumor cells. One such enzyme, creatine kinase B (CKB), plays a role in energy metabolism and serves as a downstream of AKT kinase [58]. Phosphorylation of creatine kinase B (CKB) at Thr133 by AKT reduces its affinity for creatine, resulting in decreased metabolic activity [58]. CKB also phosphorylates GPX4 at Ser104, a modification that stabilizes GPX4 by preventing its interaction with the chaperone protein HSPA8/HSC70 and subsequent autophagy-mediated degradation in hepatocellular carcinoma cells [58]. These findings suggest that oncogenic signaling endows CKB with protein kinase activity, which in turn reduces autophagy-dependent ferroptosis [58]. It remains interesting to identify distinct phosphorylation sites that enhance GPX4 functions rather than inhibiting them.
3.1.3. Phosphorylation of hippo pathway effectors
The Hippo pathway is essential for controlling organ size and maintaining tissue homeostasis, and its dysregulation has been implicated in various cancers [59]. In the Hippo pathway, YAP1 and TAZ are mainly phosphorylated by the kinases LATS1/2, which are downstream effectors of MST1/2 [59]. When the Hippo pathway is activated, the transcriptional co-activators YAP1 and TAZ are phosphorylated, leading to their retention in the cytoplasm and subsequent degradation [59]. Cell density determines ferroptosis susceptibility through regulation of the Hippo-YAP1/TAZ pathway [60]. Specifically, high cell density promotes the phosphorylation of YAP1 and TAZ, which blocks their interact with transcription factors to promote the expression of ferroptosis-related genes, including ACSL4, TFRC, EMP1, and SKP2 [[60], [61], [62]]. Therefore, targeting the phosphorylation of Hippo pathway effectors may provide a novel approach for ferroptosis-mediated anti-tumor therapy.
3.1.4. Phosphorylation of metabolic pathways
Protein kinase C beta II (PKCBII) is a key player in lipid signaling at the plasma membrane and appears to function as a sensor of lipid peroxidation [63]. PKCBII-mediated phosphorylation of ACSL4 at Thr328 enhances ACSL4 dimerization and activity, creating a positive feedback loop that promotes ferroptotic cell death [63]. In contrast, PKC-mediated phosphorylation of HSPB1 at Ser 15 restricts cytoskeleton-mediated iron uptake, consequently mitigating ferroptotic cell death in HeLa, U2OS, and LNCaP cancer cells [64]. Thus, understanding the diverse substrates of PKC kinase is crucial for identifying new targets in the context of ferroptosis. Furthermore, phosphorylation of EIF4E at Ser53 promotes ferroptosis through the formation of the aldehyde dehydrogenase 1 family member B1 (ALDH1B1)-EIF4E complex, which hinders ALDH1B1's metabolic function in preventing the production of lipid peroxidation by-product 4HNE in cancer cells [33]. However, the identity of the key upstream kinase responsible for phosphorylating EIF4E at Ser53 remains elusive.
DHODH is involved in pyrimidine metabolism through facilitating the oxidation of dihydroorotate (DHO) to orotate while reducing CoQ. To regulate this metabolic process, activated AMP-activated protein kinase (AMPK) phosphorylates uridine monophosphate synthetase (UMPS) at Ser 214 and enhances pyrimidinosome assembly, which promotes DHODH-mediated anti-ferroptosis response in HeLa cells [65]. Lysyl oxidase like 3 (LOXL3) can translocate into the mitochondria, where it undergoes phosphorylation by adenylate kinase 2 (AK2) kinase at Ser 704 [66]. This phosphorylation event prevents the ubiquitination of DHODH, enhancing the stability of DHODH to resist ferroptosis in liver cancer [66]. Future research may delve into the broader context of pyrimidine metabolism, including its interactions with other metabolic pathways, regulatory mechanisms, and the impact of pyrimidine metabolism on cellular functions and health.
3.1.5. Phosphorylation of autophagy pathways
Autophagy is an intracellular process that performs a crucial role in recycling cellular components, maintaining cellular homeostasis, and promoting cell survival under various stress conditions [67]. Nevertheless, excessive autophagy can result in cellular dysfunction, ultimately leading to cell death. Recent studies suggest that ferroptosis represents a distinctive form of autophagy-dependent cell death in specific contexts [68]. Phosphorylation regulates autophagy-dependent ferroptosis by influencing autophagy receptors (e.g., NCOA4 and hippocalcin like 1 [HPCAL1]), the core autophagy regulator beclin 1 (BECN1, also known as Atg 6 in yeast), and the key upstream kinase, mechanistic target of rapamycin kinase (MTOR). The ATM serine/threonine kinase directly phosphorylates NCOA4 at Ser 550, promoting ferritinophagy and regulating intracellular bioactive iron levels [69]. HPCAL1, another ferroptosis-related autophagy receptor, is activated by protein kinase C theta (PRKCQ) kinase, which phosphorylates HPCAL1 at Thr149. This phosphorylation enhances HPCAL1's ability to mediate cadherin 2 (CDH2) degradation, reducing membrane tension and increasing lipid peroxidation in cancer cells [70]. BECN1, a critical autophagic molecule that forms a complex with PIK3C3/VPS34, undergoes AMPK-mediated phosphorylation at S90/93/96 [71]. This phosphorylation is necessary for the interaction between BECN1 and SLC7A11, promoting ferroptosis in colon adenocarcinoma cells [71]. MTOR, a serine/threonine protein kinase, is inhibited under nutrient-deprived conditions, but activated by cystine, leading to the suppression of autophagy-dependent ferroptosis in pancreatic cancer cells [72]. Although these findings provide important information on the phosphorylation targets of autophagy machinery, a key unanswered question is how to distinguish classical autophagy induced by starvation from non-classical autophagy involved in ferroptosis based on phosphorylation events.
3.2. Acetylation
Emerging evidence suggests that acetylation of various proteins, including tumor protein p53 (TP53), ALOX12, HMGB1, HSPA5, and histones, plays a role in the regulation of ferroptosis (Fig. 3).
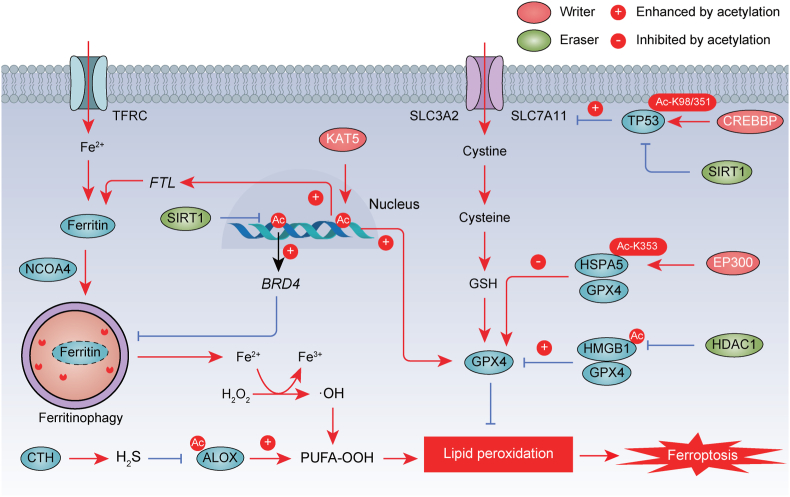
Fig. 3.
Acetylation in ferroptosis. The acetylation of histones, ALOX, TP53, HSPA5, and HMGB1, orchestrated by acetyltransferases and deacetylases, plays a pivotal role in the regulation of ferroptosis. The promotion or inhibition of protein function by acetylation is indicated by a positive (+) or negative (−) sign, respectively. Ac, acetylation.
TP53, known for its role as a transcription factor and a potent tumor suppressor mainly through apoptosis induction, exhibits a distinctive role in ferroptosis regulation when acetylation is altered. The acetylation-deficient mutant, TP533KR (with mutations at K117R, K161R, and K162R), triggers ferroptosis by downregulating SLC7A11, without affecting apoptosis in H1299 lung cancer cells [73]. Interestingly, the loss of acetylation of TP53 at Lys 98 and the 3 KR mutant abolish its ability to downregulate SLC7A11 and induce ferroptosis in H1299 cells [74]. Additionally, TP53 acetylation at Lys 351 is crucial for GINS complex subunit 4 (GINS4)-mediated ferroptosis suppression in lung adenocarcinoma [75]. CREB-binding protein (CREBBP, also known as CBP) acts as a key acetyltransferase for TP53 acetylation, inhibiting SLC7A11 expression at the transcriptional level in the model of acute lung injury [76]. In contrast, sirtuin 1 (SIRT1)-mediated TP53 deacetylation inhibits heat stress-induced ferroptosis in lung epithelial cells [77]. Acetylation finely tunes the activity of ALOX12, thereby enhancing its crucial role in regulating ferroptosis [78]. In contrast, hydrogen sulfide (H2S) synthesized by cystathionine gamma-lyase (CTH, also known as CSE) inhibits acetylation of ALOX12 and protects myoblasts from ferroptosis [78]. Acetyltransferases such as E1A binding protein p300 (EP300) can acetylate HSPA5 at Lys 353, leading to the inhibition of ferroptosis by decreasing GPX4 stability in pancreatic cancer cells [79]. Furthermore, acetylated HMGB1 also interacts with GPX4 and inhibits GPX4's anti-ferroptotic activity in colon cancer cells [80]. Therefore, conducting a comprehensive investigation into the acetylation mechanism of ferroptosis pathway could advance our understanding of cancer and inflammatory-related diseases.
Histone acetylation plays a fundamental role in regulating chromatin structure and gene expression during ferroptosis. The lysine acetyltransferase 5 (KAT5) may promote histone H3 Lys 27 acetylation (H3K27ac) in the GPX4 promoter region, enhancing GPX4 expression [81]. Targeted inhibition of the KAT5-GPX4 axis induces ferroptosis in breast cancer cells [81]. Similarly, KAT5-dependent acetylation occurs in the promoter region of FTL, which mediates iron storage in cells and limits ferroptosis in lung adenocarcinoma [82]. Epigenetic regulation of bromodomain containing 4 (BRD4) expression is mediated by the euchromatic histone lysine methyltransferase 2 (EHMT2, also known as G9a) or the histone deacetylase SIRT1 [83]. Suppression of BRD4 activity or expression can activate ferritinophagy by increasing ATG5 and lysosomal associated membrane protein 1 (LAMP1) expression in lung cancer cells both in vitro and in vivo [83].
While most studies have primarily examined acetylation alterations in individual proteins, the comprehensive regulatory role of acetylation in diverse proteins during the biological process of ferroptosis remains unclear. Advances in mass spectrometry have enabled precise identification of acetylation modification sites. It will be crucial to elucidate the enzyme-substrate relationships and differentiate between enzymatic and non-enzymatic acetylation mechanisms to enhance our understanding of acetylation's function and mechanism in ferroptosis. However, the development of specific therapeutic strategies to modulate acetylation, rather than other PTMs, presents a challenge in precision tumor therapy.
3.3. O-GlcNAcylation
O-GlcNAcylation involves the enzymatic processes of adding and removing O-GlcNAc, mainly mediated by two types of enzymes, namely O-linked N-acetylglucosamine (GlcNAc) transferase (OGT) and O-GlcNAcase (OGA), respectively. O-GlcNAcylation is an emerging and promising mechanism for regulating ferroptosis, including its modulation of system Xc−, transcription factors, and autophagy pathways (Fig. 4). OGT O-GlcNAcylates SLC7A11 at Ser 26, which acts as a critical modification for SLC7A11's activity in cystine transport [84]. O-GlcNAcylation of YAP1 at Thr241 and ZEB1 at Ser 555 enhance their transcriptional activity [85,86]. Consequently, high glucose levels have the potential to increase susceptibility to ferroptosis by facilitating O-GlcNAcylation of YAP1 and ZEB1 in hepatocellular carcinoma and pancreatic cancer cells, respectively [85,86]. Furthermore, O‐GlcNAcylation of KEAP1 at Ser104 promotes ubiquitination-mediated degradation of NFE2L2, an anti-ferroptotic transcription factor [87], in response to glucose changes [88]. However, O-GlcNAcylated JUN acts as an impediment to ferroptosis by promoting GSH synthesis in hepatocellular carcinoma [89]. Additionally, the de-O-GlcNAcylation of ferritin at Ser 179 enhances its interaction with the ferritinophagy receptor NCOA4 [90]. Specifically, pharmacological or genetic inhibition of O-GlcNAcylation promotes ferritinophagy and mitophagy, leading to the induction of ferroptosis through increasing the accumulation of labile iron in U2OS or HT29 cancer cells [90]. These findings reveal a novel connection between dynamic O-GlcNAcylation and ferroptosis, although the precise crosstalk mechanism between ferritinophagy and mitophagy requires further elucidation.
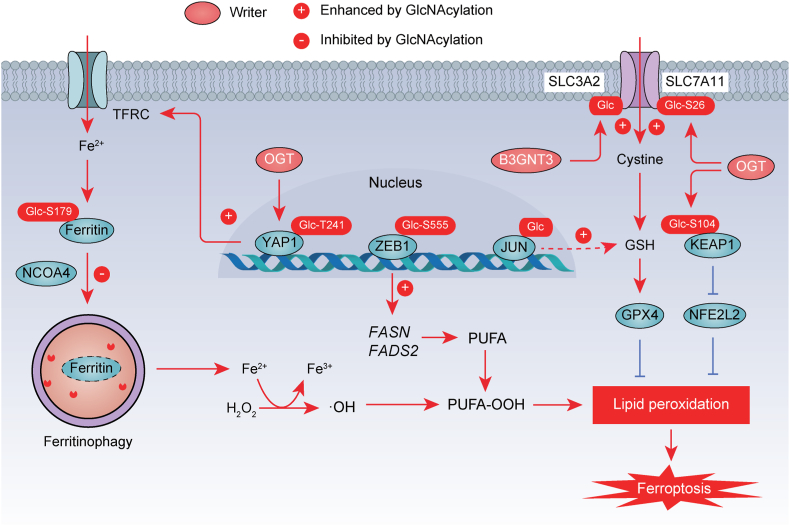
Fig. 4.
O-GlcNAcylation in ferroptosis. O-GlcNAcylation mediated by O-linked N-acetylglucosamine transferase (OGT) plays a crucial role in regulating ferroptosis through modifying ferritin, YAP1, ZEB1, JUN, SLC3A2, and SLC7A11. The promotion or inhibition of protein function by acetylation is indicated by a positive (+) or negative (−) sign, respectively. The promotion or inhibition of protein function by O-GlcNAcylation is indicated by a positive (+) or negative (−) sign, respectively. Ac, acetylation.
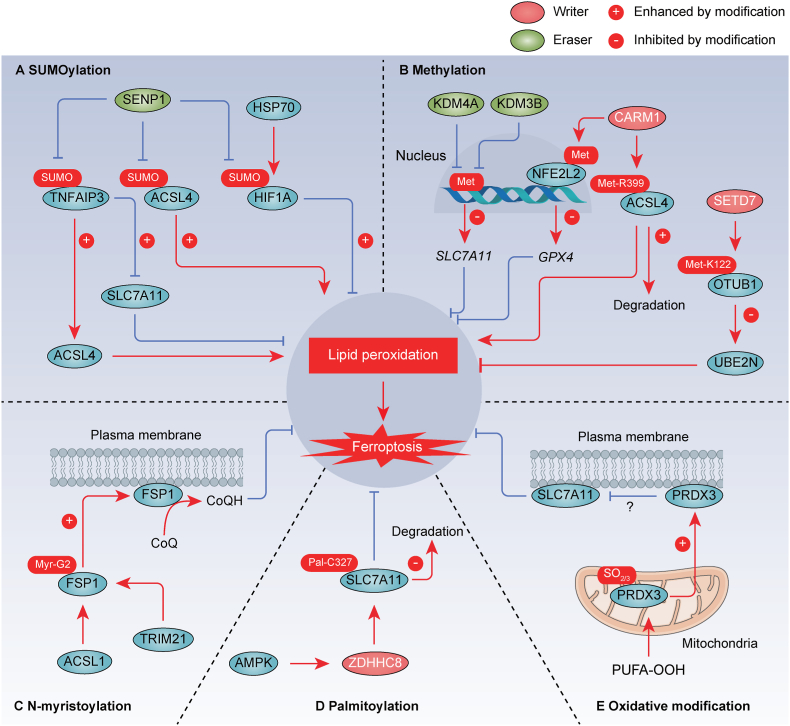
3.4. SUMOylation
PTM mediated by small ubiquitin-like modifier (SUMO) is recognized as a regulatory mechanism for dynamically modulating ferroptosis (Fig. 5A). The SUMO specific peptidase 1 (SENP1) regulates deubiquitinating enzyme TNFAIP3 (also known as A20) through deSUMOylation, thereby inhibiting ferroptosis by influencing the interaction of TNFAIP3 with ACSL4 and SLC7A11 in lung cancer cells [91]. Similarly, SENP1 reduces the SUMOylation of ACSL4, resulting in ACSL4 degradation and ferroptosis inhibition in head and neck squamous cell carcinoma [92]. Furthermore, hypoxia-induced upregulation of SENP1 expression mediates deSUMOylation of hypoxia inducible factor 1 subunit alpha (HIF1A) and ACSL4 in H9c2 embryonic rat cardiomyocytes, leading to the inhibition of ferroptosis [93]. HSP70 inhibits ferroptosis by increasing HIF1A SUMOylation, which contributes to lung cancer recurrence following radiofrequency ablation [94]. Conversely, genetic inhibition of HSP70 can reduce HIF1A SUMOylation, inhibiting lung cancer cell proliferation and migration [94]. The intricate interplay between SUMOylation and ubiquitination in the context of ferroptosis, where SUMOylation can either enhance or impede ubiquitination, remains to be fully elucidated. Additionally, gaining structural insights into SUMOylation enzymes and substrates is crucial for an enhanced understanding of the molecular mechanisms underlying ferroptosis.
Fig. 5.
SUMOylation, methylation, N-myristoylation, palmitoylation, and oxidative modification in ferroptosis. A. SUMOylation modifies various regulatory factors (e.g., TNFAIP3/A20, ACSL4, and HIF1A) of ferroptosis. B. Methylation serves as a modifying mechanism for a range of proteins, including histone, NFE2L2/NRF2, ACSL4, and OTUB1, influencing the processes of ferroptosis. C. N-myristoylation of FSP1 at Gly2 recruits it to the plasma membrane, where it functions as an oxidoreductase to generate CoQH2 parallel to GSH system. D. ZDHHC8 catalyzes the S-palmitoylation of SLC7A11 specifically at C327 and diminishes the ubiquitination levels to stabilize SLC7A11. E. PRDX3 yields cysteine oxidation to sulfinic acid (-SO2) and sulfonic acid (-SO3), which induces its translocation from mitochondria to plasma membrane, where it inhibits cystine uptake. The promotion or inhibition of protein function by the modification is indicated by a positive (+) or negative (−) sign, respectively. SUMO, SUMOylation. Met, methylation. Myr, N-myristoylation. Pal, palmitoylation.
3.5. Methylation
Methylation is a modification that affects both histone and non-histone proteins. Recent studies have unveiled a potential connection between ferroptosis and protein methylation (Fig. 5B). Lysine demethylase 4A (KDM4A) inhibits ferroptosis by demethylating H3K9me3 in the promoter region of SLC7A11 in osteosarcoma [95]. Additionally, lysine demethylase 3B (KDM3B) upregulates SLC7A11 expression by binding to the activating transcription factor 4 (ATF4), thereby preventing erastin-induced ferroptosis in HT-1080 cancer cells [96]. Coactivator associated arginine methyltransferase 1 (CARM1, also known as PRMT4) directly interacts with ACSL4 and inhibits ferroptosis by increasing ACSL4 methylation levels at Arg 339 in colorectal cancer cells [97]. Consequently, methylated ACSL4 can undergo ubiquitination and degradation induced by the E3 ubiquitin ligase ring finger protein 25 (RNF25) [97]. However, CARM1 enhances sensitivity to ferroptosis in cardiomyocytes by interacting with NFE2L2 (best known as NRF2) and promoting its methylation, which limits NFE2L2's transcriptional activity on GPX4 [98]. Moreover, SETD7, a lysine monomethylase, directly interacts with OTUB1 to catalyze the methylation of OTUB1 at Lys 122, disrupting the interaction between OTUB1 and the anti-ferroptosis protein ubiquitin conjugating enzyme E2 N (UBE2N/UBC13) in H1299 lung cancer cells [99]. Of note, ensuring that methylation-targeting ferroptotic therapies selectively affect cancer cells without harming healthy cells is a significant challenge. Methylation processes are vital for normal cellular functions, so specificity is crucial to minimize side effects.
3.6. N-myristoylation
Lipidation is a covalent modification of proteins by lipid molecules, regulating various aspects of physiology, including membrane transport, protein secretion, autophagy, and apoptosis [100]. N-myristoylation is an evolutionarily conserved lipidation process that plays a critical role in the localization of proteins, their stability, and signal transduction. Emerging evidence indicates that N-myristoylation plays a role in regulating ferroptosis (Fig. 5C). N-myristoylation of FSP1 at Gly2 directs it to the plasma membrane, where it functions as an oxidoreductase, contributing to CoQH2 production alongside the GSH system [43]. Furthermore, acyl-CoA synthetase long-chain family member 1 (ACSL1) enhances ferroptosis resistance in ovarian cancer by increasing the N-myristoylation of FSP1 and facilitating its translocation to the cell membrane [101]. A potent inhibitor targeting myristoylated FSP1 prevents its localization to the plasma membrane, resulting in ferroptosis in cancer cells [55]. While the protective role of N-myristoylation of FSP1 against ferroptosis is evident, the specific cellular enzyme responsible for this modification remains to be identified.
3.7. Palmitoylation
Palmitoylation, a reversible form of lipidation, involves the enzymatic attachment of a 16-carbon fatty acid to a specific protein. In glioblastoma, the SLC7A11 protein undergoes S-palmitoylation, a critical modification essential for its stability [102]. The zinc finger DHHC-type palmitoyltransferase 8 (ZDHHC8) catalyzes the S-palmitoylation of SLC7A11, specifically at Cys 327, reducing ubiquitination levels and thereby stabilizing SLC7A11 [102]. This enzymatic process is facilitated by AMPK, which directly phosphorylates ZDHHC8 at Ser 299 [102]. Consequently, ZDHHC8 knockdown disrupts glioblastoma growth by promoting ferroptosis [102]. These findings underscore the pivotal role of S-palmitoylation in the regulation of ferroptosis during tumorigenesis (Fig. 5D). Overall, palmitoylation serves as a mechanism for precisely modulating the behavior and function of cellular proteins, especially those linked to cellular membranes or engaged in membrane-related processes, such as the regulation of SLC7A11-mediated cysteine uptake at the cell membrane.
3.8. Oxidative modification
ROS (e.g., H2O2) has the capacity to directly oxidize protein thiols, resulting in the formation of sulfinic acid (RSO2H) and sulfonic acid (RSO3H) oxidation states [103]. This PTM can lead to alterations in protein function. A recent study has identified hyperoxidized peroxiredoxin 3 (PRDX3) as a potential marker for ferroptosis in chronic liver diseases [104]. PRDX3 undergoes specific hyperoxidation during ferroptosis, distinguishing it from apoptosis, necroptosis, and cuproptosis [104]. Furthermore, hyperoxidized PRDX3 induces ferroptosis by translocating from the mitochondria to the plasma membrane, where it inhibits cystine uptake (Fig. 5E) [104]. Additional research is required to ascertain whether hyperoxidized PRDX3 inhibits the function of system Xc−. Although the precise reasons for the selective induction of PRDX3 oxidation over other PRDX family members remain unclear, high-throughput proteomics approaches may provide potential avenues for the discovery of newly oxidized protein targets.
4. Protein degradation in ferroptosis
4.1. UPS
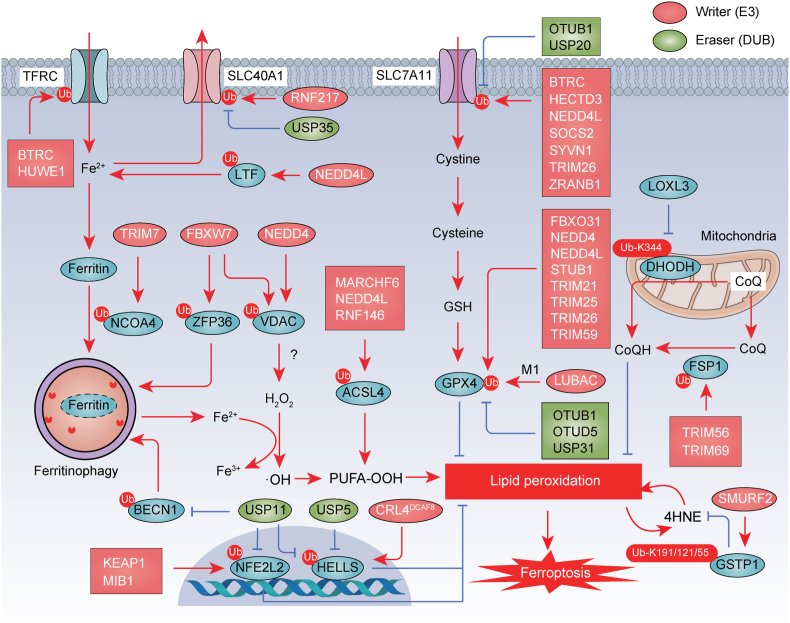
Protein degradation mediated by the UPS plays a multifaceted role in regulating ferroptosis. The proteasome, a crucial protein-degrading machinery, is essential for maintaining proper proteostasis and mitigating cellular stress. Interestingly, ferroptosis inducer RSL3 inhibits proteasome activity, resulting in the accumulation of ubiquitination [105]. However, NFE2L1 (also known as NRF1) and PARK7 (also known as DJ-1) exert protective roles against ferroptosis by preserving proteasomal activity [105,106]. Moreover, the UPS takes center stage in the orchestration of the ferroptotic process by governing the degradation of key signaling molecules and modulators involved in ferroptosis, as discussed in detail below (Fig. 6) [107].
Fig. 6.
UPS-mediated protein degradation in ferroptosis. The ubiquitination of proteins is important for the initiation of ferroptosis, which is regulated by various E3 ubiquitin ligases and deubiquitinating enzymes (DUBs). The ubiquitin-proteasome system (UPS) is involved in the degradation and stability of key regulators governing iron metabolism (e.g., TFRC, LTF, and SLC40A1), autophagy (e.g., NCOA4, and ZFP36, and BECN1), antioxidant system (e.g., SLC7A11, GPX4, FSP1, DHODH, and GSTP1), and transcription (e.g., NFE2L2/NRF2 and HELLS/LSH), as well as other proteins, including ACLS4 and VDAC. Ub, ubiquitination.
4.1.1. UPS-mediated regulation of iron metabolism
Some of the key proteins involved in regulating iron metabolism during ferroptosis, including TFRC, LTF, SLC40A1, and iron-regulatory proteins (IRPs), are subject to regulation by the UPS. In the context of ferroptosis, the E3 ubiquitin ligase HUWE1 specifically targets TFRC for ubiquitination and subsequent proteasomal degradation, thereby reducing iron accumulation and suppressing ferroptosis in models of acute liver injury [108]. Additionally, tribbles pseudokinase 2 (TRIB2), a member of the tribbles family, facilitates the ubiquitination of TFRC via the E3 ubiquitin ligase BTRC (also known as βTrCP), resulting in decreased labile iron levels in hepatocellular carcinoma cells [109]. The E3 ubiquitin ligase NEDD4L, belonging to the HECT family, targets LTF for degradation, leading to increased intracellular iron levels, which, in turn, promote the accumulation of ROS and downstream ferroptosis in pancreatic cancer cells [110]. Furthermore, the E3 ubiquitin ligase RNF217 mediates the ubiquitination and subsequent degradation of SLC40A1, thereby enhancing tissue injury responses to iron overload in the liver [111]. Conversely, the deubiquitinating enzyme USP35 interacts with SLC40A1 to maintain the protein stability of iron transporters, preventing the onset of ferroptosis in lung cancer [112].
The iron response element binding proteins IRP1 and IRP2 play master roles in post-transcriptionally regulating iron homeostasis. IRPs promote the translation of TFRC while inhibiting the synthesis of SLC40A1 and ferritin genes FTH1/FTL. IRP2, but not IRP1, is subject to regulation at the protein level through UPS-mediated degradation [113]. Mechanistically, the E3 ubiquitin ligase FBXL5 mediates the ubiquitination and subsequent proteasomal degradation of IRP2, thereby mitigating ferroptosis-associated acute kidney injury [114]. Further examination is required to determine whether targeting IRP2 degradation is a viable approach in cancer cells.
4.1.2. UPS-mediated regulation of system Xc−-GSH-GPX4 pathway
The maintenance of SLC7A11 protein stability is tightly regulated by the UPS. Several E3 ubiquitin ligases, including BTRC [115], HECTD3 [116], NEDD4L [117], SOCS2 [118], SYVN1 (also known as HRD1) [119], TRIM26 [120], and ZRANB1 [121], have been identified as positive regulators of ferroptosis by promoting ubiquitin proteasomal degradation of SLC7A11. These complex regulatory mechanisms have a context-dependent impact on the progression of ferroptosis-mediated conditions, such as hepatic fibrosis, colorectal cancer, breast cancer, and hepatocellular carcinoma. Conversely, the deubiquitinating enzymes OTUB1 and ubiquitin specific peptidase 20 (USP20) mediate the deubiquitination of SLC7A11, thereby stabilizing SLC7A11 levels in cancer cells [6,122]. Additionally, hepatocellular carcinoma ferroptosis associative lncRNA (HEPFAL) accelerates ferroptosis in hepatocellular carcinoma cells, whereas lncRNA LINC00578 inhibits ferroptosis by modulating the ubiquitination of SLC7A11 in pancreatic cancer cells [123,124]. In contrast, fascin actin-bundling protein 1 (FSCN1) directly interacts with SLC7A11, enhancing its degradation by UPS in breast cancer cells [125]. However, it is noteworthy that drugs specifically designed to induce the degradation of SLC7A11 have not yet been reported.
The degradation of GPX4 is subject to stringent regulation by ubiquitination, a process mediated by multiple E3 ubiquitin ligases, including FBXO31 [126], NEDD4 [127], NEDD4L [128,129], STUB1 [130], TRIM21 [131], TRIM25 [132], TRIM26 [133], and TRIM59 [134]. Among them, TRIM25 selectively degrades GPX4 in pancreatic cancer cells, as opposed to immune cells, attributed to its specific and elevated expression in cancer cells [132]. Additionally, the linear ubiquitin chain assembly complex (LUBAC) serves as a pivotal positive regulator of GPX4 stability through linear ubiquitination in MEFs, playing a protective role in ferroptosis [135]. Furthermore, p21, a regulator of cell cycle progression at G1, has been reported to enhance M1-linked ubiquitination of GPX4, induced by LUBAC, leading to the stability of GPX4 in osteoarthritic chondrocytes [136]. Broad-spectrum inhibition of deubiquitinating enzymes results in the ubiquitination and degradation of GPX4, culminating in ferroptotic cell death in tumor cells [137]. Specifically, the deubiquitinating enzymes OTUB1 [138], OTU deubiquitinase 5 (OTUD5) [32,139], and ubiquitin specific peptidase 31 (USP31) [140] remove ubiquitination, inducing subsequent proteasomal degradation of GPX4, thus inhibiting ferroptosis-mediated tissue injury or tumor progression. Additional studies have demonstrated that SMG9 nonsense mediated mRNA decay factor and trypsin, a proteolytic enzyme, promote ferroptosis by directly interacting with GPX4 and facilitating its degradation in cancer cells [141,142]. Nevertheless, to date, there is no available evidence elucidating the UPS-mediated degradation mechanism of GPX4 within different organelles. Understanding the direct or indirect degradation pathways of GPX4 is crucial for the development of selective ferroptotic activators. Since multiple E3 ligases contribute to GPX4 degradation, the identification of upstream signaling pathways may be key to understanding this mechanism's complexity.
4.1.3. UPS-mediated regulation of CoQ pathway
E3 ubiquitin ligases play a significant role in regulating the degradation of FSP1 and DHODH involved in ferroptosis. The E3 ubiquitin ligase TRIM69 facilitates the proteasomal degradation of FSP1 through K48-linked ubiquitination, but this process is counteracted by high density lipoprotein binding protein (HDLBP), which stabilizes the long noncoding RNA plexin B2 (PLXNB2, also known as lncFAL) [143]. CD36, a scavenger receptor, positively regulates the ubiquitination of FSP1 at Lys 16 and Lys 24, leading to FSP1 degradation and the promotion of ferroptosis in acute kidney injury [144]. Tripartite motif containing 56 (TRIM56) mediates the ubiquitination and degradation of FSP1, contributing to the ferroptosis-inducing activity of sorafenib in hepatocellular carcinoma cells [145]. In contrast, ACSL1 inhibits the proteasomal degradation of FSP1 by promoting its N-myristylation in ovarian cancer cells [101]. Additionally, DHODH is subject to ubiquitination at Lys 344, which promotes its proteasome-mediated degradation in liver cancer cells [66]. LOXL3 suppresses ferroptosis to enhance liver cancer chemoresistance by reducing DHODH ubiquitination [66]. Overall, the activation of various UPS pathways can regulate CoQ levels by degrading FSP1 or DHODH. However, the cell type-dependent UPS pathway responsible for their degradation remains unknown.
4.1.4. UPS-mediated degradation of ACSL4
The E3 ubiquitin ligase RNF146 interacts with ACSL4, and facilitates the ubiquitination and degradation of ACSL4 [146]. Accordingly, the overexpression of RNF146 prevents oxygen-glucose deprivation/reperfusion-induced ferroptosis [146]. Consistently, the activating transcription factor 3 (ATF3) activates the transcription of RNF146, leading to the inhibition of neuron ferroptosis in ischemic stroke model [146]. Similarly, E3 ubiquitin ligases NEDD4L and MARCHF6 orchestrate the ubiquitination and degradation of ACSL4, mitigating ferroptotic cell death in myocardial ischemia-reperfusion injury and tumor models, respectively [147,148]. Cytochrome P450 family 1 subfamily B member 1 (CYP1B1) suppresses ferroptosis and enhances resistance to anti-PD-1 therapy by degrading ACSL4 in colorectal cancer [149]. Thus, exploring ACSL4 ubiquitination provides a promising foundation for developing therapeutic strategies in the context of ferroptosis.
4.1.5. UPS-mediated degradation of GSTP1
Glutathione S-transferases (GSTs), a group of detoxification enzymes, play a crucial role in counteracting hydrophobic electrophilic molecules. Glutathione S-transferase pi 1 (GSTP1) has been identified as a defense enzyme against ferroptosis by catalyzing the conjugation of GSH with 4HNE to detoxify it and reduce lipid hydroperoxides [34]. The E3 ubiquitin ligase SMURF2 is responsible for ubiquitination of GSTP1 at Lys 191/121/55 and degradation of GSTP1 in the process of ferroptosis [34]. Pharmacological or genetic inhibition of GSTP1 sensitized cancer cell responses to ferroptosis inducers both in vitro and in vivo [34]. In contrast, overexpression of GSTP1 confers resistance to immune checkpoint blockade [34]. These findings, along with prior research [32,33], collectively highlight the role of the 4HNE detoxification systems as a cellular defense mechanism against ferroptosis.
4.1.6. UPS-mediated degradation of VDAC
The voltage-dependent anion channels (VDAC) are multifunctional channel proteins situated on the outer mitochondrial membrane and consist of three isoforms: VDAC1, VDAC2, and VDAC3. These isoforms facilitate the exchange of metabolites and ions between mitochondria and the cytoplasm in eukaryotic cells. VDACs have emerged as potential critical targets of erastin, and the knockdown of VDAC2 or VDAC3 effectively inhibits erastin-induced ferroptosis [150]. The E3 ubiquitin ligase NEDD4 regulates VDAC2/3 through ubiquitination, leading to their degradation and reduced melanoma cell sensitivity to erastin-induced ferroptosis [150]. Additionally, the E3 ubiquitin ligase FBXW7 modulates the protein levels of VDAC3 via ubiquitination in gastric cancer cells [151]. Nevertheless, the precise mechanism by which VDACs govern mitochondrial dysfunction and various cell death (including ferroptosis) remains to be fully elucidated.
4.1.7. UPS-mediated regulation of KEAP1-NFE2L2 pathway
The transcription factor NFE2L2 plays a central role in the regulation of ferroptosis primarily by expressing genes involved in the antioxidant system. NFE2L2 activation leads to the upregulation of ferroptosis-resistant genes like SLC7A11, enhancing cellular resistance to ferroptosis [152]. Simultaneously, NFE2L2 regulates genes associated with iron metabolism, influencing intracellular ferric ion levels [153]. The E3 ubiquitin ligase KEAP1 targets NFE2L2 for proteasomal degradation [87,154]. Sequestosome 1 (SQSTM1, also known as p62), an autophagy receptor, binds to KEAP1, displacing NFE2L2 and preventing its proteasomal degradation, thus protecting hepatocellular carcinoma against ferroptosis [87]. Furthermore, the MIB E3 ubiquitin protein ligase 1 (MIB1) promotes NFE2L2 degradation independently of Notch signaling, increasing lung cancer cell susceptibility to ferroptosis inducers [155]. In contrast, the deubiquitinating enzyme ubiquitin specific peptidase 11 (USP11) reduces cell sensitivity to ferroptosis by deubiquitinating NFE2L2 and stabilizing its expression in lung cancer cells [156]. Additionally, KEAP1 degradation is regulated by the UPS, affecting the ferroptosis process. The E3 ubiquitin ligase TRIM25 promotes KEAP1 ubiquitination, inducing temozolomide resistance in glioma cells by inhibiting ferroptotic cell death [157]. These mechanisms collectively oversee and adapt the antioxidant system to modulate ferroptosis.
4.1.8. UPS-mediated degradation of HELLS
Lymphoid-specific helicase (HELLS, also known as LSH) plays a crucial role in various aspects of epigenetic regulation, including histone modifications. HELLS inhibits ferroptosis by sequestering unstable iron and limiting the production of lipid ROS [158]. However, the ferroptosis inducer erastin destabilizes HELLS [158]. This destabilization is facilitated by the cullin-RING E3 ubiquitin ligase complex Cullin-RING ligase 4 (CRL4)-DDB1 and CUL4 associated factor 8 (DCAF8), which mediates polyubiquitination and subsequent proteasomal degradation of HELLS in cancer cells [158]. Furthermore, the deubiquitinating enzymes ubiquitin specific peptidase 5 (USP5) and USP11 inhibit ferroptosis by mediating the deubiquitination and stabilization of HELLS in hepatocellular carcinoma and colorectal cancer, respectively [159,160].
4.1.9. UPS-mediated degradation of autophagy regulator
The ubiquitination of autophagy regulatory components is essential for both the positive and negative regulation of autophagy. The direct involvement of UPS-mediated degradation of autophagy regulators in ferroptosis has been reported in several studies. For instance, the E3 ubiquitin ligase TRIM7 interacts directly with NCOA4 and ubiquitinates it via K48-linked chains, thus inhibiting ferritinophagy-mediated ferroptosis in glioblastoma cells [161]. Similarly, the E3 ubiquitin ligase FBXW7 reduces the levels of ZFP36 ring finger protein, which affects the stability of autophagy related 16 like 1 (ATG16L1) and regulates ferroptosis in hepatocellular carcinoma [162]. Furthermore, the deubiquitinating enzyme USP11 promotes autophagy-dependent ferroptosis by stabilizing BECN1, leading to ferroptosis-mediated spinal cord ischemia-reperfusion injury [163]. These findings underscore the role of ubiquitination in fine-tuning autophagy-dependent ferroptosis.
4.2. Autophagy
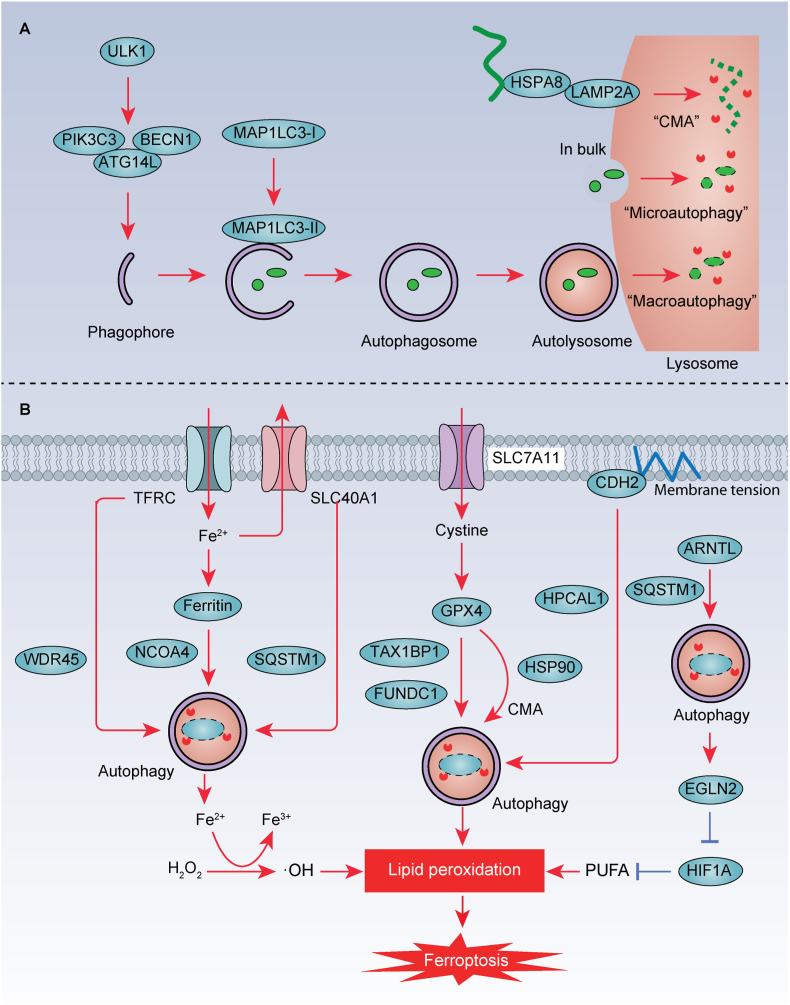
Autophagy is a system for lysosome-dependent degradation that involves the packaging of autophagic cargo within double-membrane vacuoles called 'autophagosomes' (Fig. 7A). These autophagosomes eventually fuse with lysosomes, leading to cargo digestion. In addition to macroautophagy, there are two other types of autophagy processes: microautophagy and chaperone-mediated autophagy (CMA). Microautophagy and CMA differ from macroautophagy as they involve the direct or indirect delivery of cargo to lysosomes without the use of vesicles. CMA is highly selective and targets specific proteins for degradation with the assistance of chaperone proteins HSPA8 and HSP90. The lysosome membrane receptor protein LAMP2A recognizes the KFERQ motif exposed by binding proteins and guides the target proteins into lysosomes for degradation.
Fig. 7.
Autophagy-mediated degradation in ferroptosis. A. The process of autophagy, including macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA). B. Selective autophagy is involved in regulating important players in iron metabolism (e.g., TFRC, ferritin, and SLC40A1), antioxidant system (e.g., GPX4), membrane tension (e.g., CDH2), and lipid metabolism (e.g., ARNTL) in the process of ferroptotic cell death. Furthermore, CTSB and cysteine released by lysosome can promote and inhibit ferroptosis, respectively.
In the following sections, we will provide concrete examples of how the degradation of autophagic substrate proteins regulates ferroptotic cell death (Fig. 7B) [164].
4.2.1. Autophagy-mediated regulation of iron metabolism
Iron metabolism regulators, such as ferritin, TFRC, and SLC40A1 (also known as ferroportin-1), are targeted for degradation through autophagy to modulate ferroptosis. NCOA4 serves as a selective autophagy receptor for the degradation of ferritin, a process known as ferritinophagy. Inhibition of classical autophagy-related genes (e.g., ATG3, ATG5, and ATG7) or the use of classical autophagy inhibitors (e.g., chloroquine, bafilomycin A1, and 3-methyladenine) prevents lipid peroxidation and ferroptosis in mouse embryonic fibroblasts (MEFs) and cancer cells [165,166]. Activation of NCOA4 promotes ferritin degradation through autophagy, leading to increased intracellular Fe2+ levels and susceptibility to ferroptosis [165,166]. Aldehyde dehydrogenase 1 family member A3 (ALDH1A3) interacts with microtubule-associated protein 1 light chain 3 beta (MAP1LC3B) and facilitates ferritinophagy in glioblastoma cells [167]. Additionally, the suppression of the deubiquitinating enzyme USP8 enhances ferritinophagy by increasing SQSTM1 ubiquitination, while NFE2L2 deletion hinders ferritinophagy [153,168]. TFRC is also targeted by autophagy, and genetic or pharmacological autophagy inhibition leads to TFRC accumulation [169]. Mutation of WD repeat domain 45 (WDR45), a key protein involved in autophagosome formation, reduces autophagic degradation of TFRC, promoting ferroptosis by increasing iron accumulation [169]. WDR45 is associated with β-propeller protein-associated neurodegeneration. SLC40A1 has emerged as a novel autophagy substrate that contributes to ferroptosis tolerance [68]. Erastin-induced reductions in SLC40A1 protein expression in various cancer cells can be reversed through genetic or pharmacological autophagy inhibition [68]. Mechanistically, autophagy receptor SQSTM1 mediates SLC40A1 degradation during ferroptosis in cancer cells [68]. These findings shed light on the interplay between autophagy and iron homeostasis [68,170].
4.2.2. Autophagic degradation of GPX4
In addition to UPS, GPX4 can also undergo degradation through CMA or autophagy. GPX4 contains putative KEFRQ-like motifs that are recognized by CMA. In this context, HSPA8 interacts with GPX4 and LAMP2A, facilitating CMA-mediated GPX4 degradation [171]. Antimony, a metallic element, promotes the formation of complexes involving HSPA8, HSP90, LAMP2A, and GPX4, thereby stimulating ferroptosis, mainly through GPX4 degradation mediated by CMA [172]. Copper, another metal, contributes to ferroptosis by activating autophagy-mediated GPX4 degradation [173,174]. Copper directly interacts with GPX4, leading to the formation of GPX4 aggregates that are subsequently cleared through autophagy, mediated by the autophagic receptor Tax 1 binding protein 1 (TAX1BP1) [173]. Consequently, copper chelators reduce ferroptosis-associated acute pancreatitis, while copper ions increase ferroptosis-mediated tumor inhibition in pancreatic cancer [173]. The mitochondrial outer-membrane protein FUNDC1 facilitates the mitochondrial translocation of GPX4 and its subsequent autophagic degradation, ultimately leading to hepatic ferroptosis and fibrotic injury [175]. Additionally, acid sphingomyelinase (ASM), a key enzyme in sphingomyelin metabolism, promotes autophagic GPX4 degradation and subsequent ferroptosis. However, the specific autophagic receptor responsible for this process remains unknown [176].
4.2.3. Autophagic degradation of ARNTL by clockophagy
The circadian rhythm is an endogenous system that oscillates with a period to control various cellular processes, including cell death. The transcription factor ARNTL (also known as BMAL1), a crucial component of the mammalian circadian clock, regulates the expression of a set of circadian genes, including PER and CRY [177]. SQSTM1, acting as an autophagy receptor, mediates ARNTL degradation, a process known as clockophagy [178]. Specifically, ferroptosis activators (e.g., RSL3 and FIN56) reduce the protein stability of ARNTL by activating SQSTM1-mediated clockophagy [178]. Furthermore, the degradation of ARNTL facilitates ferroptosis by enhancing the expression of egl-9 family hypoxia inducible factor 2 (EGLN2) and destabilizing the pro-survival factor HIF1A in cancer cells [178]. Consequently, ARNTL protects against experimental acute pancreatitis by blocking ferroptosis [96]. Further validation is required to determine whether clockophagy plays a role in circadian rhythms-related disorders.
4.2.4. Autophagic degradation of CDH2
Cadherins, including CDH1, CDH2, and CDH5, are a class of transmembrane glycoproteins that rely on Ca2+ to mediate intercellular adhesion and play crucial roles in normal tissue development and various diseases. It has been reported that cell-cell contacts mediated by CDH2 promote membrane tension, inhibiting ferroptosis [70]. Membrane tension is the result of various mechanical and biochemical processes that can impact the integrity, stability, and functionality of the cellular membrane. Mechanistically, HPCAL1 acts as an autophagy receptor, selectively inducing autophagic degradation of CDH2 in cancer cells through the interaction of the LC3-interacting region (LIR) motif of HPCAL1 with MAP1LC3B [70]. This process requires Thr149 phosphorylation of HPCAL1 induced by PRKCQ [70]. Further research is required to ascertain whether inducing autophagy-dependent ferroptosis could be a strategy to inhibit cancer with high CDH2 expression.
5. Strategic targeting for protein modification and degradation in ferroptosis
The operational mechanisms of ferroptosis have been elucidated in various biological and pathological scenarios, encompassing its involvement in suppressing tumors and causing damage to organs [7]. Accumulating investigations propose that pharmacological inhibition of ferroptosis could offer advantages for specific pathological conditions, including ischemia/reperfusion injury, neurodegeneration, and inflammation damage. Furthermore, induction of ferroptosis is known to suppress tumor growth and increase the immunogenicity of dying cancer cells. Protein modification and degradation pathways present promising targets for the development of drugs capable of intervening in ferroptosis. Next, we will focus on discussing the ferroptosis inhibitors or inducers that targeting the UPS and autophagy, as well as their therapeutic effects in diseases.
5.1. Ferroptosis inducers
The emerging anti-tumor strategy revolves around inducing ferroptotic cell death in cancer cells. This is achieved by promoting the degradation of antioxidant proteins, especially GPX4, through two key cellular processes: the UPS and autophagy.
Certain small-molecule compounds play a crucial role in this strategy. They can either facilitate the connection between E3 ubiquitin ligases and their substrates or inhibit specific deubiquitinating enzymes, which ultimately leads to increased ubiquitination of the target protein, followed by its proteasomal degradation. An example of such a compound is N6F11, a small molecule known for selectively triggering ferroptosis in cancer cells while sparing immune cells. Mechanistically, N6F11 promotes K48-linked ubiquitination of GPX4 at Lys 48 and its subsequent degradation by activating TRIM25 [132,179].
Another innovative approach involves the use of Proteolysis Targeting Chimeras (PROTAC), which are heterobifunctional molecules designed to bind to both the target protein and the E3 ubiquitin ligase. This interaction triggers the degradation of the target protein by the proteasome machinery. Researchers have successfully employed PROTAC technology to design and synthesize a series of GPX4 degraders, enabling precise degradation of GPX4 to facilitate ferroptosis [[180], [181], [182]]. Additionally, compounds like DMOCPTL, bufotalin, and timosaponin AIII effectively induce ferroptosis in cancer cells by promoting UPS-mediated degradation of GPX4 [[183], [184], [185]].
Furthermore, inhibitors of deubiquitinating enzymes, such as PR‐619, PdPT, and NCI677397, have been shown to increase the ubiquitination of GPX4, leading to its destabilization in various types of cancer cells [137,186,187]. Despite these advancements, it's important to note that no drugs utilizing these strategies have progressed to clinical trials yet.
An alternative approach for inducing ferroptosis in tumor cells is by triggering autophagy to eliminate specific anti-ferroptosis proteins or by inducing selective autophagy. FIN56, for instance, is a specific ferroptosis inducer that works by inducing autophagic degradation of GPX4 [188,189]. Moreover, FIN56 inhibits CoQ production in the mevalonate pathway, enhancing cellular sensitivity to ferroptosis induction [189]. Classical ferroptosis inducers like erastin (an SLC7A11 inhibitor) and RSL3 (a GPX4 inhibitor) also induce autophagic degradation of anti-ferroptosis proteins, and this effect can be further enhanced by copper ions [178,190,191]. Multiple pathways, such as AMPK, MTOR, BECN1, and ROS, are involved in the regulation of autophagy. Certain agents like amentoflavone, dihydroartemisinin, curcumin, and chrysin can activate or disrupt these pathways, thereby inducing autophagy-dependent ferroptosis in tumor cells [[192], [193], [194], [195]]. These findings lay a promising foundation for future clinical development in this field.
Additionally, another potential avenue for regulating ferroptotic cell death involves targeting non-ubiquitination modifications of proteins. A notable example is icFSP1, which binds to the myristoylated form of FSP1, promoting phase condensation [55]. As a result, icFSP1-induced subcellular relocalization of FSP1 promotes ferroptosis in cancer cells and hinder tumor growth in vivo [55].
5.2. Ferroptosis inhibitors
While less frequently discussed, ferroptosis inhibitors associated with protein modification and degradation hold substantial untapped potential. For instance, the natural product salvianolic acid B has been found to impede the proteasomal degradation of GPX4, effectively inhibiting ferroptosis. This action has shown promise in ameliorating myocardial ischemia-reperfusion injury in vivo [196]. Similarly, Ginkgolide B has demonstrated its potential in improving diabetic nephropathy by protecting the kidneys from ferroptosis-induced damage. It achieves this by reducing the ubiquitination of GPX4 in a mouse model [197].
In the context of ferroptosis inhibition, classical autophagy inhibitors like chloroquine, bafilomycin A1, and 3-methyladenine have exhibited a protective role. They play a crucial part in scenarios such as alcohol-induced ferroptosis in liver epithelial cells or the 6-hydroxydopamine model of Parkinson's disease [198,199]. Furthermore, the HSP90 inhibitor 2-amino-5-chloro-N,3-Dimethylbenzamide (CDDO) primarily attenuates ferroptosis through blocking HSP90-mediated CMA and the autophagic degradation of GPX4 in mice hippocampal neurons (HT-22) cells [171]. Another small compound, iHPCAL1, directly interacts with HPCAL1, an autophagy receptor. It inhibits the function of HPCAL1 in mediating the autophagic degradation of CDH2, thereby offering protection against ferroptosis-induced acute pancreatitis [70].
However, it's important to note that the comparative benefits of these drugs in comparison to traditional antioxidants are yet to be fully assessed.
6. Conclusions and perspectives
Ferroptosis, a complex cellular process, involves a myriad of proteins and diverse protein modifications. A deep comprehension of the landscape of protein modification and degradation in ferroptosis offers important insights into the underlying molecular mechanisms.
In the pursuit of unraveling these complexities, ongoing research focused on identifying novel therapeutic targets related to protein modification or degradation holds immense promise. This research could potentially pave the way for the development of targeted drugs or interventions tailored to combat ferroptosis-associated diseases. It is noteworthy that specific protein modification or degradation profiles, such as hyperoxidized PRDX3 or downregulated GPX4, may emerge as biomarkers for diagnosis and treatment efficacy of ferroptosis-related diseases.
Nevertheless, navigating the intricacies of protein modification and degradation in the context of ferroptosis poses several formidable challenges.
First, the intricate interplay between different protein modifications adds complexity, making it challenging to establish a comprehensive understanding of the mechanisms governing ferroptosis. As noted above, both K63-linked ubiquitination and N-myristoylation can drive the plasma membrane translocation process of FSP1. Given the pivotal role of N-myristoylation in membrane anchoring, further investigations are necessary to delve into the potential influence of ubiquitination on FSP1 N-myristoylation.
Second, a single protein (e.g., ACSL4) may undergo various modifications, including ubiquitination, phosphorylation, methylation, and SUMOylation. The regulatory mechanisms of different modifications on ACLS4 in various diseases remain mysterious. Furthermore, another protein, like GPX4, may experience degradation via both autophagy and the UPS. The balancing mechanism of these two degradation systems within cells is still worth exploring. One potential pathway is that, as mentioned above, UPS can degrade autophagy regulators, thereby fine-tuning the process of autophagy. Achieving precise control of specific protein modifications and degradation is challenging, necessitating the development of highly targeted intervention strategies.
Third, the roles of certain novel modifications (e.g., lactylation, succinylation, crotonylation, and malonylation) in diseases are currently under careful investigation. Exploring the connection between these modifications and ferroptosis is one of the future research directions. However, current analytical technologies may not be exhaustive enough to fully characterize all facets of protein modification, especially at the in vivo level. Some modifications may prove challenging to detect or quantify accurately using existing methods, which could limit the discovery of novel biomarkers and therapeutic targets.
To address these challenges effectively, innovative approaches and advancements in experimental techniques are imperative. Collaborative endeavors across disciplines, involving researchers in biochemistry, molecular biology, and bioinformatics, will be essential to tackle the intricacies of ferroptosis comprehensively. Additionally, improved technological advancements (e.g., mass spectrometry techniques and high-throughput screening methods) have the potential to alleviate the current limitations in characterizing protein modifications.
Funding
This work was supported by the National Natural Science Foundation of China (82272660), the Plan on Enhancing Scientific Research in GMU (02-410-2302289XM), and the Basic and Applied Basic Research Project of the Guangzhou Basic Research Program (202201011411).
CRediT authorship contribution statement
Yuan Wang: Writing – original draft. Ding Yan: Writing – original draft. Jinbao Liu: Writing – review & editing. Daolin Tang: Writing – review & editing. Xin Chen: Writing – review & editing, Conceptualization.
Declaration of competing interest
The authors declare no competing interests.
Contributor Information
Daolin Tang, Email: daolin.tang@utsouthwestern.edu.
Xin Chen, Email: chenxin@gzhmu.edu.cn.
Data availability
No data was used for the research described in the article.
References
- 1.Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P., Alnemri E.S., Altucci L., Amelio I., Andrews D.W., Annicchiarico-Petruzzelli M., Antonov A.V., Arama E., Baehrecke E.H., Barlev N.A., Bazan N.G., Bernassola F., Bertrand M.J.M., Bianchi K., Blagosklonny M.V., Blomgren K., Borner C., Boya P., Brenner C., Campanella M., Candi E., Carmona-Gutierrez D., Cecconi F., Chan F.K., Chandel N.S., Cheng E.H., Chipuk J.E., Cidlowski J.A., Ciechanover A., Cohen G.M., Conrad M., Cubillos-Ruiz J.R., Czabotar P.E., D'Angiolella V., Dawson T.M., Dawson V.L., De Laurenzi V., De Maria R., Debatin K.M., DeBerardinis R.J., Deshmukh M., Di Daniele N., Di Virgilio F., Dixit V.M., Dixon S.J., Duckett C.S., Dynlacht B.D., El-Deiry W.S., Elrod J.W., Fimia G.M., Fulda S., Garcia-Saez A.J., Garg A.D., Garrido C., Gavathiotis E., Golstein P., Gottlieb E., Green D.R., Greene L.A., Gronemeyer H., Gross A., Hajnoczky G., Hardwick J.M., Harris I.S., Hengartner M.O., Hetz C., Ichijo H., Jaattela M., Joseph B., Jost P.J., Juin P.P., Kaiser W.J., Karin M., Kaufmann T., Kepp O., Kimchi A., Kitsis R.N., Klionsky D.J., Knight R.A., Kumar S., Lee S.W., Lemasters J.J., Levine B., Linkermann A., Lipton S.A., Lockshin R.A., Lopez-Otin C., Lowe S.W., Luedde T., Lugli E., MacFarlane M., Madeo F., Malewicz M., Malorni W., Manic G., Marine J.C., Martin S.J., Martinou J.C., Medema J.P., Mehlen P., Meier P., Melino S., Miao E.A., Molkentin J.D., Moll U.M., Munoz-Pinedo C., Nagata S., Nunez G., Oberst A., Oren M., Overholtzer M., Pagano M., Panaretakis T., Pasparakis M., Penninger J.M., Pereira D.M., Pervaiz S., Peter M.E., Piacentini M., Pinton P., Prehn J.H.M., Puthalakath H., Rabinovich G.A., Rehm M., Rizzuto R., Rodrigues C.M.P., Rubinsztein D.C., Rudel T., Ryan K.M., Sayan E., Scorrano L., Shao F., Shi Y., Silke J., Simon H.U., Sistigu A., Stockwell B.R., Strasser A., Szabadkai G., Tait S.W.G., Tang D., Tavernarakis N., Thorburn A., Tsujimoto Y., Turk B., Vanden Berghe T., Vandenabeele P., Vander Heiden M.G., Villunger A., Virgin H.W., Vousden K.H., Vucic D., Wagner E.F., Walczak H., Wallach D., Wang Y., Wells J.A., Wood W., Yuan J., Zakeri Z., Zhivotovsky B., Zitvogel L., Melino G., Kroemer G. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang D., Kang R., Berghe T.V., Vandenabeele P., Kroemer G. The molecular machinery of regulated cell death. Cell Res. 2019;29:347–364. doi: 10.1038/s41422-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan J., Ofengeim D. A guide to cell death pathways. Nat. Rev. Mol. Cell Biol. 2023 doi: 10.1038/s41580-023-00689-6. [DOI] [PubMed] [Google Scholar]
- 4.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S., Morrison B., 3rd, Stockwell B.R. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai E., Han L., Liu J., Xie Y., Kroemer G., Klionsky D.J., Zeh H.J., Kang R., Wang J., Tang D. Autophagy-dependent ferroptosis drives tumor-associated macrophage polarization via release and uptake of oncogenic KRAS protein. Autophagy. 2020;16:2069–2083. doi: 10.1080/15548627.2020.1714209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen Q., Liu J., Kang R., Zhou B., Tang D. The release and activity of HMGB1 in ferroptosis. Biochem. Biophys. Res. Commun. 2019;510:278–283. doi: 10.1016/j.bbrc.2019.01.090. [DOI] [PubMed] [Google Scholar]
- 7.Tang D., Chen X., Kang R., Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021;31:107–125. doi: 10.1038/s41422-020-00441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen F., Kang R., Tang D., Liu J. Ferroptosis: principles and significance in health and disease. J. Hematol. Oncol. 2024;17:41. doi: 10.1186/s13045-024-01564-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berndt C., Alborzinia H., Amen V.S., Ayton S., Barayeu U., Bartelt A., Bayir H., Bebber C.M., Birsoy K., Bottcher J.P., Brabletz S., Brabletz T., Brown A.R., Brune B., Bulli G., Bruneau A., Chen Q., DeNicola G.M., Dick T.P., Distefano A., Dixon S.J., Engler J.B., Esser-von Bieren J., Fedorova M., Friedmann Angeli J.P., Friese M.A., Fuhrmann D.C., Garcia-Saez A.J., Garbowicz K., Gotz M., Gu W., Hammerich L., Hassannia B., Jiang X., Jeridi A., Kang Y.P., Kagan V.E., Konrad D.B., Kotschi S., Lei P., Le Tertre M., Lev S., Liang D., Linkermann A., Lohr C., Lorenz S., Luedde T., Methner A., Michalke B., Milton A.V., Min J., Mishima E., Muller S., Motohashi H., Muckenthaler M.U., Murakami S., Olzmann J.A., Pagnussat G., Pan Z., Papagiannakopoulos T., Pedrera Puentes L., Pratt D.A., Proneth B., Ramsauer L., Rodriguez R., Saito Y., Schmidt F., Schmitt C., Schulze A., Schwab A., Schwantes A., Soula M., Spitzlberger B., Stockwell B.R., Thewes L., Thorn-Seshold O., Toyokuni S., Tonnus W., Trumpp A., Vandenabeele P., Vanden Berghe T., Venkataramani V., Vogel F.C.E., von Karstedt S., Wang F., Westermann F., Wientjens C., Wilhelm C., Wolk M., Wu K., Yang X., Yu F., Zou Y., Conrad M. Ferroptosis in health and disease. Redox Biol. 2024;75 doi: 10.1016/j.redox.2024.103211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J., Fulda S., Gascon S., Hatzios S.K., Kagan V.E., Noel K., Jiang X., Linkermann A., Murphy M.E., Overholtzer M., Oyagi A., Pagnussat G.C., Park J., Ran Q., Rosenfeld C.S., Salnikow K., Tang D., Torti F.M., Torti S.V., Toyokuni S., Woerpel K.A., Zhang D.D. Ferroptosis: a regulated cell death nexus linking metabolism. Redox Biology, and Disease, Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X., Kang R., Kroemer G., Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 2021;18:280–296. doi: 10.1038/s41571-020-00462-0. [DOI] [PubMed] [Google Scholar]
- 12.Jiang X., Stockwell B.R., Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021;22:266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stockwell B.R. Ferroptosis turns 10: emerging mechanisms, physiological functions, and therapeutic applications. Cell. 2022;185:2401–2421. doi: 10.1016/j.cell.2022.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai E., Chen X., Linkermann A., Jiang X., Kang R., Kagan V.E., Bayir H., Yang W.S., Garcia-Saez A.J., Ioannou M.S., Janowitz T., Ran Q., Gu W., Gan B., Krysko D.V., Zhu X., Wang J., Krautwald S., Toyokuni S., Xie Y., Greten F.R., Yi Q., Schick J., Liu J., Gabrilovich D.I., Liu J., Zeh H.J., Zhang D.D., Yang M., Iovanna J., Kopf M., Adolph T.E., Chi J.T., Li C., Ichijo H., Karin M., Sankaran V.G., Zou W., Galluzzi L., Bush A.I., Li B., Melino G., Baehrecke E.H., Lotze M.T., Klionsky D.J., Stockwell B.R., Kroemer G., Tang D. A guideline on the molecular ecosystem regulating ferroptosis. Nat. Cell Biol. 2024 doi: 10.1038/s41556-024-01360-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X., Yu C., Kang R., Tang D. Iron metabolism in ferroptosis. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.590226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou Y., Henry W.S., Ricq E.L., Graham E.T., Phadnis V.V., Maretich P., Paradkar S., Boehnke N., Deik A.A., Reinhardt F., Eaton J.K., Ferguson B., Wang W., Fairman J., Keys H.R., Dančík V., Clish C.B., Clemons P.A., Hammond P.T., Boyer L.A., Weinberg R.A., Schreiber S.L. Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature. 2020;585:603–608. doi: 10.1038/s41586-020-2732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui W., Liu D., Gu W., Chu B. Peroxisome-driven ether-linked phospholipids biosynthesis is essential for ferroptosis. Cell Death Differ. 2021;28:2536–2551. doi: 10.1038/s41418-021-00769-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai Y., Meng L., Han L., Jia Y., Zhao Y., Gao H., Kang R., Wang X., Tang D., Dai E. Lipid storage and lipophagy regulates ferroptosis. Biochem. Biophys. Res. Commun. 2019;508:997–1003. doi: 10.1016/j.bbrc.2018.12.039. [DOI] [PubMed] [Google Scholar]
- 19.Lin Z., Long F., Kang R., Klionsky D.J., Yang M., Tang D. The lipid basis of cell death and autophagy. Autophagy. 2023:1–20. doi: 10.1080/15548627.2023.2259732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Z., Liu J., Kang R., Yang M., Tang D. Lipid metabolism in ferroptosis. Adv Biol (Weinh) 2021;5 doi: 10.1002/adbi.202100396. [DOI] [PubMed] [Google Scholar]
- 21.Kagan V.E., Mao G., Qu F., Angeli J.P., Doll S., Croix C.S., Dar H.H., Liu B., Tyurin V.A., Ritov V.B., Kapralov A.A., Amoscato A.A., Jiang J., Anthonymuthu T., Mohammadyani D., Yang Q., Proneth B., Klein-Seetharaman J., Watkins S., Bahar I., Greenberger J., Mallampalli R.K., Stockwell B.R., Tyurina Y.Y., Conrad M., Bayır H. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 2017;13:81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doll S., Proneth B., Tyurina Y.Y., Panzilius E., Kobayashi S., Ingold I., Irmler M., Beckers J., Aichler M., Walch A., Prokisch H., Trumbach D., Mao G., Qu F., Bayir H., Fullekrug J., Scheel C.H., Wurst W., Schick J.A., Kagan V.E., Angeli J.P., Conrad M. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017;13:91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan H., Li X., Zhang X., Kang R., Tang D. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem. Biophys. Res. Commun. 2016;478:1338–1343. doi: 10.1016/j.bbrc.2016.08.124. [DOI] [PubMed] [Google Scholar]
- 24.Chen F., Kang R., Liu J., Tang D. The ACSL4 network regulates cell death and autophagy in diseases. Biology. 2023;12 doi: 10.3390/biology12060864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Z., Liu J., Long F., Kang R., Kroemer G., Tang D., Yang M. The lipid flippase SLC47A1 blocks metabolic vulnerability to ferroptosis. Nat. Commun. 2022;13:7965. doi: 10.1038/s41467-022-35707-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed A., Ware T., Li H., Fernando Bazan J., Cravatt B.F. TMEM164 is an acyltransferase that forms ferroptotic C20:4 ether phospholipids. Nat. Chem. Biol. 2023;19:378–388. doi: 10.1038/s41589-022-01253-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J., Liu Y., Wang Y., Li C., Xie Y., Klionsky D.J., Kang R., Tang D. TMEM164 is a new determinant of autophagy-dependent ferroptosis. Autophagy. 2023;19:945–956. doi: 10.1080/15548627.2022.2111635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magtanong L., Ko P.J., To M., Cao J.Y., Forcina G.C., Tarangelo A., Ward C.C., Cho K., Patti G.J., Nomura D.K., Olzmann J.A., Dixon S.J. Exogenous monounsaturated fatty acids promote a ferroptosis-resistant cell state. Cell Chem. Biol. 2019;26:420–432.e429. doi: 10.1016/j.chembiol.2018.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang D., Feng Y., Zandkarimi F., Wang H., Zhang Z., Kim J., Cai Y., Gu W., Stockwell B.R., Jiang X. Ferroptosis surveillance independent of GPX4 and differentially regulated by sex hormones. Cell. 2023;186:2748–2764 e2722. doi: 10.1016/j.cell.2023.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou Y., Li H., Graham E.T., Deik A.A., Eaton J.K., Wang W., Sandoval-Gomez G., Clish C.B., Doench J.G., Schreiber S.L. Cytochrome P450 oxidoreductase contributes to phospholipid peroxidation in ferroptosis. Nat. Chem. Biol. 2020;16:302–309. doi: 10.1038/s41589-020-0472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan B., Ai Y., Sun Q., Ma Y., Cao Y., Wang J., Zhang Z., Wang X. Membrane damage during ferroptosis is caused by oxidation of phospholipids catalyzed by the oxidoreductases POR and CYB5R1. Mol. Cell. 2021;81:355–369 e310. doi: 10.1016/j.molcel.2020.11.024. [DOI] [PubMed] [Google Scholar]
- 32.Liu L., Pang J., Qin D., Li R., Zou D., Chi K., Wu W., Rui H., Yu H., Zhu W., Liu K., Wu X., Wang J., Xu P., Song X., Cao Y., Wang J., Xu F., Xue L., Chen Y. Deubiquitinase OTUD5 as a novel protector against 4-HNE-triggered ferroptosis in myocardial ischemia/reperfusion injury. Adv. Sci. 2023;10 doi: 10.1002/advs.202301852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X., Huang J., Yu C., Liu J., Gao W., Li J., Song X., Zhou Z., Li C., Xie Y., Kroemer G., Liu J., Tang D., Kang R. A noncanonical function of EIF4E limits ALDH1B1 activity and increases susceptibility to ferroptosis. Nat. Commun. 2022;13:6318. doi: 10.1038/s41467-022-34096-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W., Dai J., Hou G., Liu H., Zheng S., Wang X., Lin Q., Zhang Y., Lu M., Gong Y., Xiang Z., Yu Y., Hu Y. SMURF2 predisposes cancer cell toward ferroptosis in GPX4-independent manners by promoting GSTP1 degradation. Mol. Cell. 2023;83:4352–4369 e4358. doi: 10.1016/j.molcel.2023.10.042. [DOI] [PubMed] [Google Scholar]
- 35.Xie Y., Kang R., Klionsky D.J., Tang D. GPX4 in cell death, autophagy, and disease. Autophagy. 2023:1–18. doi: 10.1080/15548627.2023.2218764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imai H., Hakkaku N., Iwamoto R., Suzuki J., Suzuki T., Tajima Y., Konishi K., Minami S., Ichinose S., Ishizaka K., Shioda S., Arata S., Nishimura M., Naito S., Nakagawa Y. Depletion of selenoprotein GPx4 in spermatocytes causes male infertility in mice. J. Biol. Chem. 2009;284:32522–32532. doi: 10.1074/jbc.M109.016139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao C., Liu X., Zhang Y., Lei G., Yan Y., Lee H., Koppula P., Wu S., Zhuang L., Fang B., Poyurovsky M.V., Olszewski K., Gan B. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593:586–590. doi: 10.1038/s41586-021-03539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seiler A., Schneider M., Forster H., Roth S., Wirth E.K., Culmsee C., Plesnila N., Kremmer E., Radmark O., Wurst W., Bornkamm G.W., Schweizer U., Conrad M. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metabol. 2008;8:237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., Cheah J.H., Clemons P.A., Shamji A.F., Clish C.B., Brown L.M., Girotti A.W., Cornish V.W., Schreiber S.L., Stockwell B.R. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viswanathan V.S., Ryan M.J., Dhruv H.D., Gill S., Eichhoff O.M., Seashore-Ludlow B., Kaffenberger S.D., Eaton J.K., Shimada K., Aguirre A.J., Viswanathan S.R., Chattopadhyay S., Tamayo P., Yang W.S., Rees M.G., Chen S., Boskovic Z.V., Javaid S., Huang C., Wu X., Tseng Y.Y., Roider E.M., Gao D., Cleary J.M., Wolpin B.M., Mesirov J.P., Haber D.A., Engelman J.A., Boehm J.S., Kotz J.D., Hon C.S., Chen Y., Hahn W.C., Levesque M.P., Doench J.G., Berens M.E., Shamji A.F., Clemons P.A., Stockwell B.R., Schreiber S.L. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Canli O., Alankus Y.B., Grootjans S., Vegi N., Hultner L., Hoppe P.S., Schroeder T., Vandenabeele P., Bornkamm G.W., Greten F.R. Glutathione peroxidase 4 prevents necroptosis in mouse erythroid precursors. Blood. 2016;127:139–148. doi: 10.1182/blood-2015-06-654194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang R., Zeng L., Zhu S., Xie Y., Liu J., Wen Q., Cao L., Xie M., Ran Q., Kroemer G., Wang H., Billiar T.R., Jiang J., Tang D. Lipid peroxidation drives gasdermin D-mediated pyroptosis in lethal polymicrobial sepsis. Cell Host Microbe. 2018;24:97–108 e104. doi: 10.1016/j.chom.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bersuker K., Hendricks J.M., Li Z., Magtanong L., Ford B., Tang P.H., Roberts M.A., Tong B., Maimone T.J., Zoncu R., Bassik M.C., Nomura D.K., Dixon S.J., Olzmann J.A. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doll S., Freitas F.P., Shah R., Aldrovandi M., da Silva M.C., Ingold I., Goya Grocin A., Xavier da Silva T.N., Panzilius E., Scheel C.H., Mourao A., Buday K., Sato M., Wanninger J., Vignane T., Mohana V., Rehberg M., Flatley A., Schepers A., Kurz A., White D., Sauer M., Sattler M., Tate E.W., Schmitz W., Schulze A., O'Donnell V., Proneth B., Popowicz G.M., Pratt D.A., Angeli J.P.F., Conrad M. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 45.Deshwal S., Onishi M., Tatsuta T., Bartsch T., Cors E., Ried K., Lemke K., Nolte H., Giavalisco P., Langer T. Mitochondria regulate intracellular coenzyme Q transport and ferroptotic resistance via STARD7. Nat. Cell Biol. 2023;25:246–257. doi: 10.1038/s41556-022-01071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mishima E., Nakamura T., Zheng J., Zhang W., Mourao A.S.D., Sennhenn P., Conrad M. DHODH inhibitors sensitize to ferroptosis by FSP1 inhibition. Nature. 2023;619:E9–E18. doi: 10.1038/s41586-023-06269-0. [DOI] [PubMed] [Google Scholar]
- 47.Jin D.Y., Chen X., Liu Y., Williams C.M., Pedersen L.C., Stafford D.W., Tie J.K. A genome-wide CRISPR-Cas9 knockout screen identifies FSP1 as the warfarin-resistant vitamin K reductase. Nat. Commun. 2023;14:828. doi: 10.1038/s41467-023-36446-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai E., Zhang W., Cong D., Kang R., Wang J., Tang D. AIFM2 blocks ferroptosis independent of ubiquinol metabolism. Biochem. Biophys. Res. Commun. 2020;523:966–971. doi: 10.1016/j.bbrc.2020.01.066. [DOI] [PubMed] [Google Scholar]
- 49.Muller F., Lim J.K.M., Bebber C.M., Seidel E., Tishina S., Dahlhaus A., Stroh J., Beck J., Yapici F.I., Nakayama K., Torres Fernandez L., Bragelmann J., Leprivier G., von Karstedt S. Elevated FSP1 protects KRAS-mutated cells from ferroptosis during tumor initiation. Cell Death Differ. 2023;30:442–456. doi: 10.1038/s41418-022-01096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bu C., Hu S., Yu J., Li N., Gu J., Sheng Z., Yan Z., Bu X. Fear stress promotes glioma progression through inhibition of ferroptosis by enhancing FSP1 stability. Clin. Transl. Oncol. 2023;25:1378–1388. doi: 10.1007/s12094-022-03032-1. [DOI] [PubMed] [Google Scholar]
- 51.Lee J.M., Hammaren H.M., Savitski M.M., Baek S.H. Control of protein stability by post-translational modifications. Nat. Commun. 2023;14:201. doi: 10.1038/s41467-023-35795-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dewson G., Eichhorn P.J.A., Komander D. Deubiquitinases in cancer. Nat. Rev. Cancer. 2023;23:842–862. doi: 10.1038/s41568-023-00633-y. [DOI] [PubMed] [Google Scholar]
- 53.Yu X., Li W., Liu H., Wang X., Coarfa C., Cheng C., Yu X., Zeng Z., Cao Y., Young K.H., Li Y. PD-L1 translocation to the plasma membrane enables tumor immune evasion through MIB2 ubiquitination. J. Clin. Invest. 2023;133 doi: 10.1172/JCI160456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He Y.M., Zhou X.M., Jiang S.Y., Zhang Z.B., Cao B.Y., Liu J.B., Zeng Y.Y., Zhao J., Mao X.L. TRIM25 activates AKT/mTOR by inhibiting PTEN via K63-linked polyubiquitination in non-small cell lung cancer. Acta Pharmacol. Sin. 2022;43:681–691. doi: 10.1038/s41401-021-00662-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakamura T., Hipp C., Santos Dias Mourão A., Borggräfe J., Aldrovandi M., Henkelmann B., Wanninger J., Mishima E., Lytton E., Emler D., Proneth B., Sattler M., Conrad M. Phase separation of FSP1 promotes ferroptosis. Nature. 2023;619:371–377. doi: 10.1038/s41586-023-06255-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gong J., Liu Y., Wang W., He R., Xia Q., Chen L., Zhao C., Gao Y., Shi Y., Bai Y., Liao Y., Zhang Q., Zhu F., Wang M., Li X., Qin R. TRIM21-Promoted FSP1 plasma membrane translocation confers ferroptosis resistance in human cancers. Adv. Sci. 2023;10 doi: 10.1002/advs.202302318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castelo-Soccio L., Kim H., Gadina M., Schwartzberg P.L., Laurence A., O'Shea J.J. Protein kinases: drug targets for immunological disorders. Nat. Rev. Immunol. 2023;23:787–806. doi: 10.1038/s41577-023-00877-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu K., Yan M., Liu T., Wang Z., Duan Y., Xia Y., Ji G., Shen Y., Wang L., Li L., Zheng P., Dong B., Wu Q., Xiao L., Yang X., Shen H., Wen T., Zhang J., Yi J., Deng Y., Qian X., Ma L., Fang J., Zhou Q., Lu Z., Xu D. Creatine kinase B suppresses ferroptosis by phosphorylating GPX4 through a moonlighting function. Nat. Cell Biol. 2023;25:714–725. doi: 10.1038/s41556-023-01133-9. [DOI] [PubMed] [Google Scholar]
- 59.Harvey K.F., Zhang X., Thomas D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 60.Wu J., Minikes A.M., Gao M., Bian H., Li Y., Stockwell B.R., Chen Z.N., Jiang X. Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signalling. Nature. 2019;572:402–406. doi: 10.1038/s41586-019-1426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang W.H., Ding C.C., Sun T., Rupprecht G., Lin C.C., Hsu D., Chi J.T. The Hippo pathway effector TAZ regulates ferroptosis in renal cell carcinoma. Cell Rep. 2019;28:2501–2508 e2504. doi: 10.1016/j.celrep.2019.07.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang W.H., Lin C.C., Wu J., Chao P.Y., Chen K., Chen P.H., Chi J.T. The Hippo pathway effector YAP promotes ferroptosis via the E3 ligase SKP2. Mol. Cancer Res. 2021;19:1005–1014. doi: 10.1158/1541-7786.MCR-20-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang H.L., Hu B.X., Li Z.L., Du T., Shan J.L., Ye Z.P., Peng X.D., Li X., Huang Y., Zhu X.Y., Chen Y.H., Feng G.K., Yang D., Deng R., Zhu X.F. PKCβII phosphorylates ACSL4 to amplify lipid peroxidation to induce ferroptosis. Nat. Cell Biol. 2022;24:88–98. doi: 10.1038/s41556-021-00818-3. [DOI] [PubMed] [Google Scholar]
- 64.Sun X., Ou Z., Xie M., Kang R., Fan Y., Niu X., Wang H., Cao L., Tang D. HSPB1 as a novel regulator of ferroptotic cancer cell death. Oncogene. 2015;34:5617–5625. doi: 10.1038/onc.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang C., Zhao Y., Wang L., Guo Z., Ma L., Yang R., Wu Y., Li X., Niu J., Chu Q., Fu Y., Li B. De novo pyrimidine biosynthetic complexes support cancer cell proliferation and ferroptosis defence. Nat. Cell Biol. 2023;25:836–847. doi: 10.1038/s41556-023-01146-4. [DOI] [PubMed] [Google Scholar]
- 66.Zhan M., Ding Y., Huang S., Liu Y., Xiao J., Yu H., Lu L., Wang X. Lysyl oxidase-like 3 restrains mitochondrial ferroptosis to promote liver cancer chemoresistance by stabilizing dihydroorotate dehydrogenase. Nat. Commun. 2023;14:3123. doi: 10.1038/s41467-023-38753-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen X., Tsvetkov A.S., Shen H.M., Isidoro C., Ktistakis N.T., Linkermann A., Koopman W.J.H., Simon H.U., Galluzzi L., Luo S., Xu D., Gu W., Peulen O., Cai Q., Rubinsztein D.C., Chi J.T., Zhang D.D., Li C., Toyokuni S., Liu J., Roh J.L., Dai E., Juhasz G., Liu W., Zhang J., Yang M., Liu J., Zhu L.Q., Zou W., Piacentini M., Ding W.X., Yue Z., Xie Y., Petersen M., Gewirtz D.A., Mandell M.A., Chu C.T., Sinha D., Eftekharpour E., Zhivotovsky B., Besteiro S., Gabrilovich D.I., Kim D.H., Kagan V.E., Bayir H., Chen G.C., Ayton S., Lunemann J.D., Komatsu M., Krautwald S., Loos B., Baehrecke E.H., Wang J., Lane J.D., Sadoshima J., Yang W.S., Gao M., Munz C., Thumm M., Kampmann M., Yu D., Lipinski M.M., Jones J.W., Jiang X., Zeh H.J., Kang R., Klionsky D.J., Kroemer G., Tang D. International consensus guidelines for the definition, detection, and interpretation of autophagy-dependent ferroptosis. Autophagy. 2024 doi: 10.1080/15548627.2024.2319901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J., Liu J., Xu Y., Wu R., Chen X., Song X., Zeh H., Kang R., Klionsky D.J., Wang X., Tang D. Tumor heterogeneity in autophagy-dependent ferroptosis. Autophagy. 2021;17:3361–3374. doi: 10.1080/15548627.2021.1872241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu H., Liu Q., Shan X., Gao W., Chen Q. ATM orchestrates ferritinophagy and ferroptosis by phosphorylating NCOA4. Autophagy. 2023;19:2062–2077. doi: 10.1080/15548627.2023.2170960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X., Song X., Li J., Zhang R., Yu C., Zhou Z., Liu J., Liao S., Klionsky D.J., Kroemer G., Liu J., Tang D., Kang R. Identification of HPCAL1 as a specific autophagy receptor involved in ferroptosis. Autophagy. 2023;19:54–74. doi: 10.1080/15548627.2022.2059170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song X., Zhu S., Chen P., Hou W., Wen Q., Liu J., Xie Y., Liu J., Klionsky D.J., Kroemer G., Lotze M.T., Zeh H.J., Kang R., Tang D. AMPK-mediated BECN1 phosphorylation promotes ferroptosis by directly blocking system X(c)(-) activity. Curr. Biol. : CB. 2018;28:2388–2399.e2385. doi: 10.1016/j.cub.2018.05.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Y., Wang Y., Liu J., Kang R., Tang D. Interplay between MTOR and GPX4 signaling modulates autophagy-dependent ferroptotic cancer cell death. Cancer Gene Ther. 2021;28:55–63. doi: 10.1038/s41417-020-0182-y. [DOI] [PubMed] [Google Scholar]
- 73.Jiang L., Kon N., Li T., Wang S.J., Su T., Hibshoosh H., Baer R., Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang S.J., Li D., Ou Y., Jiang L., Chen Y., Zhao Y., Gu W. Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep. 2016;17:366–373. doi: 10.1016/j.celrep.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen L., Cai Q., Yang R., Wang H., Ling H., Li T., Liu N., Wang Z., Sun J., Tao T., Shi Y., Cao Y., Wang X., Xiao D., Liu S., Tao Y. GINS4 suppresses ferroptosis by antagonizing p53 acetylation with Snail. Proc. Natl. Acad. Sci. U.S.A. 2023;120 doi: 10.1073/pnas.2219585120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Y., Ma Y., Li Q., Ling Y., Zhou Y., Chu K., Xue L., Tao S. STAT6 inhibits ferroptosis and alleviates acute lung injury via regulating P53/SLC7A11 pathway. Cell Death Dis. 2022;13:530. doi: 10.1038/s41419-022-04971-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen H., Lin X., Yi X., Liu X., Yu R., Fan W., Ling Y., Liu Y., Xie W. SIRT1-mediated p53 deacetylation inhibits ferroptosis and alleviates heat stress-induced lung epithelial cells injury. Int. J. Hyperther. 2022;39:977–986. doi: 10.1080/02656736.2022.2094476. [DOI] [PubMed] [Google Scholar]
- 78.Wang Y., Yu R., Wu L., Yang G. Hydrogen sulfide guards myoblasts from ferroptosis by inhibiting ALOX12 acetylation. Cell. Signal. 2021;78 doi: 10.1016/j.cellsig.2020.109870. [DOI] [PubMed] [Google Scholar]
- 79.Wang Y., Liu Y., Wang C., Kang R., Tang D., Liu J. EP300 promotes ferroptosis via HSPA5 acetylation in pancreatic cancer. Sci. Rep. 2023;13 doi: 10.1038/s41598-023-42136-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang Y., Yang L., Jiang S., Yang T., Lan J., Lei Y., Tan H., Pan K. HMGB1 mediates lipopolysaccharide-induced inflammation via interacting with GPX4 in colon cancer cells. Cancer Cell Int. 2020;20:205. doi: 10.1186/s12935-020-01289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H., Liu W., Zhang X., Wu F., Sun D., Wang Z. Ketamine suppresses proliferation and induces ferroptosis and apoptosis of breast cancer cells by targeting KAT5/GPX4 axis. Biochem. Biophys. Res. Commun. 2021;585:111–116. doi: 10.1016/j.bbrc.2021.11.029. [DOI] [PubMed] [Google Scholar]
- 82.Wang Y., Qiu S., Wang H., Cui J., Tian X., Miao Y., Zhang C., Cao L., Ma L., Xu X., Qiao Y., Zhang X. Transcriptional repression of ferritin light chain increases ferroptosis sensitivity in lung adenocarcinoma. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.719187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sui S., Zhang J., Xu S., Wang Q., Wang P., Pang D. Ferritinophagy is required for the induction of ferroptosis by the bromodomain protein BRD4 inhibitor (+)-JQ1 in cancer cells. Cell Death Dis. 2019;10:331. doi: 10.1038/s41419-019-1564-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang J., Long G., Hu K., Xiao D., Liu S., Xiao L., Zhou L., Tao Y. Targeting USP8 inhibits O-GlcNAcylation of SLC7A11 to promote ferroptosis of hepatocellular carcinoma via stabilization of OGT. Adv. Sci. 2023;10 doi: 10.1002/advs.202302953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu G., Murshed A., Li H., Ma J., Zhen N., Ding M., Zhu J., Mao S., Tang X., Liu L., Sun F., Jin L., Pan Q. O-GlcNAcylation enhances sensitivity to RSL3-induced ferroptosis via the YAP/TFRC pathway in liver cancer. Cell Death Dis. 2021;7:83. doi: 10.1038/s41420-021-00468-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang X., Liu M., Chu Y., Liu Y., Cao X., Zhang H., Huang Y., Gong A., Liao X., Wang D., Zhu H. O-GlcNAcylation of ZEB1 facilitated mesenchymal pancreatic cancer cell ferroptosis. Int. J. Biol. Sci. 2022;18:4135–4150. doi: 10.7150/ijbs.71520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun X., Ou Z., Chen R., Niu X., Chen D., Kang R., Tang D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173–184. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen P.H., Smith T.J., Wu J., Siesser P.F., Bisnett B.J., Khan F., Hogue M., Soderblom E., Tang F., Marks J.R., Major M.B., Swarts B.M., Boyce M., Chi J.T. Glycosylation of KEAP1 links nutrient sensing to redox stress signaling. EMBO J. 2017;36:2233–2250. doi: 10.15252/embj.201696113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen Y., Zhu G., Liu Y., Wu Q., Zhang X., Bian Z., Zhang Y., Pan Q., Sun F. O-GlcNAcylated c-Jun antagonizes ferroptosis via inhibiting GSH synthesis in liver cancer. Cell. Signal. 2019;63 doi: 10.1016/j.cellsig.2019.109384. [DOI] [PubMed] [Google Scholar]
- 90.Yu F., Zhang Q., Liu H., Liu J., Yang S., Luo X., Liu W., Zheng H., Liu Q., Cui Y., Chen G., Li Y., Huang X., Yan X., Zhou J., Chen Q. Dynamic O-GlcNAcylation coordinates ferritinophagy and mitophagy to activate ferroptosis. Cell Discov. 2022;8:40. doi: 10.1038/s41421-022-00390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gao C., Xiao F., Zhang L., Sun Y., Wang L., Liu X., Sun H., Xie Z., Liang Y., Xu Q., Wang L. SENP1 inhibition suppresses the growth of lung cancer cells through activation of A20-mediated ferroptosis. Ann. Transl. Med. 2022;10:224. doi: 10.21037/atm-21-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu X., Mao Y., Feng Z., Dai F., Gu T., Zheng J. SENP1 inhibits ferroptosis and promotes head and neck squamous cell carcinoma by regulating ACSL4 protein stability via SUMO1. Oncol. Rep. 2024;51:1–14. doi: 10.3892/or.2023.8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bai Y.T., Xiao F.J., Wang H., Ge R.L., Wang L.S. Hypoxia protects H9c2 cells against Ferroptosis through SENP1-mediated protein DeSUMOylation. Int. J. Med. Sci. 2021;18:1618–1627. doi: 10.7150/ijms.50804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peng B., Ling X., Huang T., Wan J. HSP70 via HIF-1 α SUMOylation inhibits ferroptosis inducing lung cancer recurrence after insufficient radiofrequency ablation. PLoS One. 2023;18 doi: 10.1371/journal.pone.0294263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen M., Jiang Y., Sun Y. KDM4A-mediated histone demethylation of SLC7A11 inhibits cell ferroptosis in osteosarcoma. Biochem. Biophys. Res. Commun. 2021;550:77–83. doi: 10.1016/j.bbrc.2021.02.137. [DOI] [PubMed] [Google Scholar]
- 96.Liu Y., Wang Y., Liu J., Kang R., Tang D. The circadian clock protects against ferroptosis-induced sterile inflammation. Biochem. Biophys. Res. Commun. 2020;525:620–625. doi: 10.1016/j.bbrc.2020.02.142. [DOI] [PubMed] [Google Scholar]
- 97.Feng S., Rao Z., Zhang J., She X., Chen Y., Wan K., Li H., Zhao C., Feng Y., Wang G., Hu J., Luo X. Inhibition of CARM1-mediated methylation of ACSL4 promotes ferroptosis in colorectal cancer. Adv. Sci. 2023 doi: 10.1002/advs.202303484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y., Yan S., Liu X., Deng F., Wang P., Yang L., Hu L., Huang K., He J. PRMT4 promotes ferroptosis to aggravate doxorubicin-induced cardiomyopathy via inhibition of the Nrf2/GPX4 pathway. Cell Death Differ. 2022;29:1982–1995. doi: 10.1038/s41418-022-00990-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Deng H., Jia S., Tang J., Rong F., Xu C., Chen X., Wang Z., Zhu C., Sun X., Liao Q., Liu W., Li W., Xiao W., Liu X. SET7 methylates the deubiquitinase OTUB1 at Lys (122) to impair its binding to E2 enzyme UBC13 and relieve its suppressive role on ferroptosis. J. Biol. Chem. 2023;299 doi: 10.1016/j.jbc.2023.103054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jiang H., Zhang X., Chen X., Aramsangtienchai P., Tong Z., Lin H. Protein lipidation: occurrence, mechanisms, biological functions, and enabling technologies. Chem. Rev. 2018;118:919–988. doi: 10.1021/acs.chemrev.6b00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Q., Li N., Deng L., Jiang X., Zhang Y., Lee L.T.O., Zhang H. ACSL1-induced ferroptosis and platinum resistance in ovarian cancer by increasing FSP1 N-myristylation and stability. Cell Death Dis. 2023;9:83. doi: 10.1038/s41420-023-01385-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Z., Wang Y., Shen N., Liu Y., Xu X., Zhu R., Jiang H., Wu X., Wei Y., Tang J. AMPKalpha1-mediated ZDHHC8 phosphorylation promotes the palmitoylation of SLC7A11 to facilitate ferroptosis resistance in glioblastoma. Cancer Lett. 2024 doi: 10.1016/j.canlet.2024.216619. [DOI] [PubMed] [Google Scholar]
- 103.Paulsen C.E., Carroll K.S. Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev. 2013;113:4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cui S., Ghai A., Deng Y., Li S., Zhang R., Egbulefu C., Liang G., Achilefu S., Ye J. Identification of hyperoxidized PRDX3 as a ferroptosis marker reveals ferroptotic damage in chronic liver diseases. Mol. Cell. 2023;83:3931–3939 e3935. doi: 10.1016/j.molcel.2023.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kotschi S., Jung A., Willemsen N., Ofoghi A., Proneth B., Conrad M., Bartelt A. NFE2L1-mediated proteasome function protects from ferroptosis. Mol. Metabol. 2022;57 doi: 10.1016/j.molmet.2022.101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Y., Wang C. Quantitative reactive cysteinome profiling reveals a functional link between ferroptosis and proteasome-mediated degradation. Cell Death Differ. 2023;30:125–136. doi: 10.1038/s41418-022-01050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen X., Yu C., Kang R., Kroemer G., Tang D. Cellular degradation systems in ferroptosis. Cell Death Differ. 2021;28:1135–1148. doi: 10.1038/s41418-020-00728-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu Y., Jiao H., Yue Y., He K., Jin Y., Zhang J., Zhang J., Wei Y., Luo H., Hao Z., Zhao X., Xia Q., Zhong Q., Zhang J. Ubiquitin ligase E3 HUWE1/MULE targets transferrin receptor for degradation and suppresses ferroptosis in acute liver injury. Cell Death Differ. 2022;29:1705–1718. doi: 10.1038/s41418-022-00957-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guo S., Chen Y., Xue X., Yang Y., Wang Y., Qiu S., Cui J., Zhang X., Ma L., Qiao Y., Wang J. TRIB2 desensitizes ferroptosis via βTrCP-mediated TFRC ubiquitiantion in liver cancer cells. Cell death discovery. 2021;7:196. doi: 10.1038/s41420-021-00574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Y., Liu Y., Liu J., Kang R., Tang D. NEDD4L-mediated LTF protein degradation limits ferroptosis. Biochem. Biophys. Res. Commun. 2020;531:581–587. doi: 10.1016/j.bbrc.2020.07.032. [DOI] [PubMed] [Google Scholar]
- 111.Jiang L., Wang J., Wang K., Wang H., Wu Q., Yang C., Yu Y., Ni P., Zhong Y., Song Z., Xie E., Hu R., Min J., Wang F. RNF217 regulates iron homeostasis through its E3 ubiquitin ligase activity by modulating ferroportin degradation. Blood. 2021;138:689–705. doi: 10.1182/blood.2020008986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tang Z., Jiang W., Mao M., Zhao J., Chen J., Cheng N. Deubiquitinase USP35 modulates ferroptosis in lung cancer via targeting ferroportin. Clin. Transl. Med. 2021;11 doi: 10.1002/ctm2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Terzi E.M., Sviderskiy V.O., Alvarez S.W., Whiten G.C., Possemato R. Iron-sulfur cluster deficiency can be sensed by IRP2 and regulates iron homeostasis and sensitivity to ferroptosis independent of IRP1 and FBXL5. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abg4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu Q., Wang F., Chen Y., Cui H., Wu H. A regulatory module comprising G3BP1-FBXL5-IRP2 axis determines sodium arsenite-induced ferroptosis. J. Hazard Mater. 2023;465 doi: 10.1016/j.jhazmat.2023.133038. [DOI] [PubMed] [Google Scholar]
- 115.Lei S., Chen C., Han F., Deng J., Huang D., Qian L., Zhu M., Ma X., Lai M., Xu E., Zhang H. AMER1 deficiency promotes the distant metastasis of colorectal cancer by inhibiting SLC7A11- and FTL-mediated ferroptosis. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.113110. [DOI] [PubMed] [Google Scholar]
- 116.Huang F., Huang Z., Wei Q., Liu G., Pu J. E3 ubiquitin ligase HECTD3 is a tumor suppressor and mediates the polyubiquitination of SLC7A11 to promote ferroptosis in colon cancer. Exp. Cell Res. 2023;430 doi: 10.1016/j.yexcr.2023.113697. [DOI] [PubMed] [Google Scholar]
- 117.Liu R., Liu L., Bian Y., Zhang S., Wang Y., Chen H., Jiang X., Li G., Chen Q., Xue C., Li M., Liu L., Liu X., Ma S. The dual regulation effects of ESR1/nedd4l on SLC7A11 in breast cancer under ionizing radiation. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.772380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen Q., Zheng W., Guan J., Liu H., Dan Y., Zhu L., Song Y., Zhou Y., Zhao X., Zhang Y., Bai Y., Pan Y., Zhang J., Shao C. SOCS2-enhanced ubiquitination of SLC7A11 promotes ferroptosis and radiosensitization in hepatocellular carcinoma. Cell Death Differ. 2023;30:137–151. doi: 10.1038/s41418-022-01051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Y., Wang S., Zhang W. HRD1 functions as a tumor suppressor in ovarian cancer by facilitating ubiquitination-dependent SLC7A11 degradation. Cell Cycle. 2023;22:1116–1126. doi: 10.1080/15384101.2023.2178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhu Y., Zhang C., Huang M., Lin J., Fan X., Ni T. TRIM26 induces ferroptosis to inhibit hepatic stellate cell activation and mitigate liver fibrosis through mediating SLC7A11 ubiquitination. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.644901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang S., Zhang Q., Zhao M., Wang X., Zhang Y., Gan B., Zhang P. The deubiquitinase ZRANB1 is an E3 ubiquitin ligase for SLC7A11 and regulates ferroptotic resistance. J. Cell Biol. 2023;222 doi: 10.1083/jcb.202212072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tang J., Long G., Xiao D., Liu S., Xiao L., Zhou L., Tao Y. ATR-dependent ubiquitin-specific protease 20 phosphorylation confers oxaliplatin and ferroptosis resistance. MedComm. 2020;4(2023) doi: 10.1002/mco2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang B., Bao W., Zhang S., Chen B., Zhou X., Zhao J., Shi Z., Zhang T., Chen Z., Wang L., Zheng X., Chen G., Wang Y. LncRNA HEPFAL accelerates ferroptosis in hepatocellular carcinoma by regulating SLC7A11 ubiquitination. Cell Death Dis. 2022;13:734. doi: 10.1038/s41419-022-05173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li H., Wei Y., Wang J., Yao J., Zhang C., Yu C., Tang Y., Zhu D., Yang J., Zhou J. Long noncoding RNA LINC00578 inhibits ferroptosis in pancreatic cancer via regulating SLC7A11 ubiquitination. Oxid. Med. Cell. Longev. 2023;2023 doi: 10.1155/2023/1744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen C., Xie B., Li Z., Chen L., Chen Y., Zhou J., Ju S., Zhou Y., Zhang X., Zhuo W., Yang J., Mao M., Xu L., Wang L. Fascin enhances the vulnerability of breast cancer to erastin-induced ferroptosis. Cell Death Dis. 2022;13:150. doi: 10.1038/s41419-022-04579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhu Z., Zheng Y., He H., Yang L., Yang J., Li M., Dai W., Huang H. FBXO31 sensitizes cancer stem cells-like cells to cisplatin by promoting ferroptosis and facilitating proteasomal degradation of GPX4 in cholangiocarcinoma, Liver international. official journal of the International Association for the Study of the Liver. 2022;42:2871–2888. doi: 10.1111/liv.15462. [DOI] [PubMed] [Google Scholar]
- 127.Sun J., Lin X.M., Lu D.H., Wang M., Li K., Li S.R., Li Z.Q., Zhu C.J., Zhang Z.M., Yan C.Y., Pan M.H., Gong H.B., Feng J.C., Cao Y.F., Huang F., Sun W.Y., Kurihara H., Li Y.F., Duan W.J., Jiao G.L., Zhang L., He R.R. Midbrain dopamine oxidation links ubiquitination of glutathione peroxidase 4 to ferroptosis of dopaminergic neurons. J. Clin. Invest. 2023;133 doi: 10.1172/JCI165228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tang H., Jiang X., Hua Y., Li H., Zhu C., Hao X., Yi M., Li L. NEDD4L facilitates granulosa cell ferroptosis by promoting GPX4 ubiquitination and degradation. Endocrine connections. 2023;12 doi: 10.1530/ec-22-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cheng F., Dou J., Yang Y., Sun S., Chen R., Zhang Z., Wei H., Li J., Wu Z. Drug-induced lactate confers ferroptosis resistance via p38-SGK1-NEDD4L-dependent upregulation of GPX4 in NSCLC cells. Cell Death Dis. 2023;9:165. doi: 10.1038/s41420-023-01463-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sun X., Zhang Q., Lin X., Shu P., Gao X., Shen K. Imatinib induces ferroptosis in gastrointestinal stromal tumors by promoting STUB1-mediated GPX4 ubiquitination. Cell Death Dis. 2023;14:839. doi: 10.1038/s41419-023-06300-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sun X., Huang N., Li P., Dong X., Yang J., Zhang X., Zong W.X., Gao S., Xin H. TRIM21 ubiquitylates GPX4 and promotes ferroptosis to aggravate ischemia/reperfusion-induced acute kidney injury. Life Sci. 2023;321 doi: 10.1016/j.lfs.2023.121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li J., Liu J., Zhou Z., Wu R., Chen X., Yu C., Stockwell B., Kroemer G., Kang R., Tang D. Tumor-specific GPX4 degradation enhances ferroptosis-initiated antitumor immune response in mouse models of pancreatic cancer. Sci. Transl. Med. 2023;15 doi: 10.1126/scitranslmed.adg3049. [DOI] [PubMed] [Google Scholar]
- 133.Wang Z., Xia Y., Wang Y., Zhu R., Li H., Liu Y., Shen N. The E3 ligase TRIM26 suppresses ferroptosis through catalyzing K63-linked ubiquitination of GPX4 in glioma. Cell Death Dis. 2023;14:695. doi: 10.1038/s41419-023-06222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang J., Xie H., Yao J., Jin W., Pan H., Pan Z., Xie D., Xie D. TRIM59 promotes steatosis and ferroptosis in non-alcoholic fatty liver disease via enhancing GPX4 ubiquitination. Hum. Cell. 2023;36:209–222. doi: 10.1007/s13577-022-00820-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dong K., Wei R., Jin T., Zhang M., Shen J., Xiang H., Shan B., Yuan J., Li Y. HOIP modulates the stability of GPx4 by linear ubiquitination. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2214227119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zheng Z., Shang X., Sun K., Hou Y., Zhang X., Xu J., Liu H., Ruan Z., Hou L., Guo Z., Wang G., Xu F., Guo F. P21 resists ferroptosis in osteoarthritic chondrocytes by regulating GPX4 protein stability. Free Radic. Biol. Med. 2024 doi: 10.1016/j.freeradbiomed.2023.12.047. [DOI] [PubMed] [Google Scholar]
- 137.Yang L., Chen X., Yang Q., Chen J., Huang Q., Yao L., Yan D., Wu J., Zhang P., Tang D., Zhong N., Liu J. Broad spectrum deubiquitinase inhibition induces both apoptosis and ferroptosis in cancer cells. Front. Oncol. 2020;10:949. doi: 10.3389/fonc.2020.00949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li D., Wang Y., Dong C., Chen T., Dong A., Ren J., Li W., Shu G., Yang J., Shen W., Qin L., Hu L., Zhou J. CST1 inhibits ferroptosis and promotes gastric cancer metastasis by regulating GPX4 protein stability via OTUB1. Oncogene. 2023;42:83–98. doi: 10.1038/s41388-022-02537-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chu L.K., Cao X., Wan L., Diao Q., Zhu Y., Kan Y., Ye L.L., Mao Y.M., Dong X.Q., Xiong Q.W., Fu M.C., Zhang T., Zhou H.T., Cai S.Z., Ma Z.R., Hsu S.W., Wu R., Chen C.H., Yan X.M., Liu J. Autophagy of OTUD5 destabilizes GPX4 to confer ferroptosis-dependent kidney injury. Nat. Commun. 2023;14:8393. doi: 10.1038/s41467-023-44228-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yao L., Yan D., Jiang B., Xue Q., Chen X., Huang Q., Qi L., Tang D., Chen X., Liu J. Plumbagin is a novel GPX4 protein degrader that induces apoptosis in hepatocellular carcinoma cells. Free Radic. Biol. Med. 2023;203:1–10. doi: 10.1016/j.freeradbiomed.2023.03.263. [DOI] [PubMed] [Google Scholar]
- 141.Han L., Bai L., Fang X., Liu J., Kang R., Zhou D., Tang D., Dai E. SMG9 drives ferroptosis by directly inhibiting GPX4 degradation. Biochem. Biophys. Res. Commun. 2021;567:92–98. doi: 10.1016/j.bbrc.2021.06.038. [DOI] [PubMed] [Google Scholar]
- 142.Liu K., Liu J., Zou B., Li C., Zeh H.J., Kang R., Kroemer G., Huang J., Tang D. Trypsin-mediated sensitization to ferroptosis increases the severity of pancreatitis in mice. Cell Mol Gastroenterol Hepatol. 2022;13:483–500. doi: 10.1016/j.jcmgh.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yuan J., Lv T., Yang J., Wu Z., Yan L., Yang J., Shi Y. HDLBP-stabilized lncFAL inhibits ferroptosis vulnerability by diminishing Trim69-dependent FSP1 degradation in hepatocellular carcinoma. Redox Biol. 2022;58 doi: 10.1016/j.redox.2022.102546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ma Y., Huang L., Zhang Z., Yang P., Chen Q., Zeng X., Tan F., Wang C., Ruan X., Liao X. CD36 promotes tubular ferroptosis by regulating the ubiquitination of FSP1 in acute kidney injury. Genes & diseases. 2024;11:449–463. doi: 10.1016/j.gendis.2022.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu M.R., Shi C., Song Q.Y., Kang M.J., Jiang X., Liu H., Pei D.S. Sorafenib induces ferroptosis by promoting TRIM54-mediated FSP1 ubiquitination and degradation in hepatocellular carcinoma. Hepatology communications. 2023;7 doi: 10.1097/hc9.0000000000000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jin Z.L., Gao W.Y., Guo F., Liao S.J., Hu M.Z., Yu T., Yu S.Z., Shi Q. Ring finger protein 146-mediated long-chain fatty-acid-coenzyme a ligase 4 ubiquitination regulates ferroptosis-induced neuronal damage in ischemic stroke. Neuroscience. 2023;529:148–161. doi: 10.1016/j.neuroscience.2023.08.007. [DOI] [PubMed] [Google Scholar]
- 147.Qiu M., Yan W., Liu M. YAP facilitates nedd4l-mediated ubiquitination and degradation of ACSL4 to alleviate ferroptosis in myocardial ischemia-reperfusion injury. Can. J. Cardiol. 2023;39:1712–1727. doi: 10.1016/j.cjca.2023.07.030. [DOI] [PubMed] [Google Scholar]
- 148.Nguyen K.T., Mun S.H., Yang J., Lee J., Seok O.H., Kim E., Kim D., An S.Y., Seo D.Y., Suh J.Y., Lee Y., Hwang C.S. The MARCHF6 E3 ubiquitin ligase acts as an NADPH sensor for the regulation of ferroptosis. Nat. Cell Biol. 2022;24:1239–1251. doi: 10.1038/s41556-022-00973-1. [DOI] [PubMed] [Google Scholar]
- 149.Chen C., Yang Y., Guo Y., He J., Chen Z., Qiu S., Zhang Y., Ding H., Pan J., Pan Y. CYP1B1 inhibits ferroptosis and induces anti-PD-1 resistance by degrading ACSL4 in colorectal cancer. Cell Death Dis. 2023;14:271. doi: 10.1038/s41419-023-05803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yang Y., Luo M., Zhang K., Zhang J., Gao T., Connell D.O., Yao F., Mu C., Cai B., Shang Y., Chen W. Nedd4 ubiquitylates VDAC2/3 to suppress erastin-induced ferroptosis in melanoma. Nat. Commun. 2020;11:433. doi: 10.1038/s41467-020-14324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huang G., Xiang Z., Wu H., He Q., Dou R., Lin Z., Yang C., Huang S., Song J., Di Z., Wang S., Xiong B. The lncRNA BDNF-AS/WDR5/FBXW7 axis mediates ferroptosis in gastric cancer peritoneal metastasis by regulating VDAC3 ubiquitination. Int. J. Biol. Sci. 2022;18:1415–1433. doi: 10.7150/ijbs.69454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Feng L., Zhao K., Sun L., Yin X., Zhang J., Liu C., Li B. SLC7A11 regulated by NRF2 modulates esophageal squamous cell carcinoma radiosensitivity by inhibiting ferroptosis. J. Transl. Med. 2021;19:367. doi: 10.1186/s12967-021-03042-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Anandhan A., Dodson M., Shakya A., Chen J., Liu P., Wei Y., Tan H., Wang Q., Jiang Z., Yang K., Garcia J.G., Chambers S.K., Chapman E., Ooi A., Yang-Hartwich Y., Stockwell B.R., Zhang D.D. NRF2 controls iron homeostasis and ferroptosis through HERC2 and VAMP8. Sci. Adv. 2023;9 doi: 10.1126/sciadv.ade9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li J., Lu K., Sun F., Tan S., Zhang X., Sheng W., Hao W., Liu M., Lv W., Han W. Panaxydol attenuates ferroptosis against LPS-induced acute lung injury in mice by Keap1-Nrf2/HO-1 pathway. J. Transl. Med. 2021;19:96. doi: 10.1186/s12967-021-02745-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang H., Huang Q., Xia J., Cheng S., Pei D., Zhang X., Shu X. The E3 ligase MIB1 promotes proteasomal degradation of NRF2 and sensitizes lung cancer cells to ferroptosis. Mol. Cancer Res. : MCR. 2022;20:253–264. doi: 10.1158/1541-7786.Mcr-21-0342. [DOI] [PubMed] [Google Scholar]
- 156.Meng C., Zhan J., Chen D., Shao G., Zhang H., Gu W., Luo J. The deubiquitinase USP11 regulates cell proliferation and ferroptotic cell death via stabilization of NRF2 USP11 deubiquitinates and stabilizes NRF2. Oncogene. 2021;40:1706–1720. doi: 10.1038/s41388-021-01660-5. [DOI] [PubMed] [Google Scholar]
- 157.Wei J., Wang L., Zhang Y., Sun T., Zhang C., Hu Z., Zhou L., Liu X., Wan J., Ma L. TRIM25 promotes temozolomide resistance in glioma by regulating oxidative stress and ferroptotic cell death via the ubiquitination of keap1. Oncogene. 2023;42:2103–2112. doi: 10.1038/s41388-023-02717-3. [DOI] [PubMed] [Google Scholar]
- 158.Huang D., Li Q., Sun X., Sun X., Tang Y., Qu Y., Liu D., Yu T., Li G., Tong T., Zhang Y. CRL4(DCAF8) dependent opposing stability control over the chromatin remodeler LSH orchestrates epigenetic dynamics in ferroptosis. Cell Death Differ. 2021;28:1593–1609. doi: 10.1038/s41418-020-00689-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yan B., Guo J., Wang Z., Ning J., Wang H., Shu L., Hu K., Chen L., Shi Y., Zhang L., Liu S., Tao Y., Xiao D. The ubiquitin-specific protease 5 mediated deubiquitination of LSH links metabolic regulation of ferroptosis to hepatocellular carcinoma progression. MedComm. 2020;4:e337. doi: 10.1002/mco2.337. 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Duan J., Huang D., Liu C., Lv Y., Zhang L., Chang F., Zeng X., Li L., Wang W., Shao G. USP11-mediated LSH deubiquitination inhibits ferroptosis in colorectal cancer through epigenetic activation of CYP24A1. Cell Death Dis. 2023;14:402. doi: 10.1038/s41419-023-05915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li K., Chen B., Xu A., Shen J., Li K., Hao K., Hao R., Yang W., Jiang W., Zheng Y., Ge F., Wang Z. TRIM7 modulates NCOA4-mediated ferritinophagy and ferroptosis in glioblastoma cells. Redox Biol. 2022;56 doi: 10.1016/j.redox.2022.102451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang Z., Guo M., Li Y., Shen M., Kong D., Shao J., Ding H., Tan S., Chen A., Zhang F., Zheng S. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy. 2020;16:1482–1505. doi: 10.1080/15548627.2019.1687985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rong Y., Fan J., Ji C., Wang Z., Ge X., Wang J., Ye W., Yin G., Cai W., Liu W. USP11 regulates autophagy-dependent ferroptosis after spinal cord ischemia-reperfusion injury by deubiquitinating Beclin 1. Cell Death Differ. 2022;29:1164–1175. doi: 10.1038/s41418-021-00907-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chen F., Cai X., Kang R., Liu J., Tang D. Autophagy-dependent ferroptosis in cancer. Antioxidants Redox Signal. 2023;39:79–101. doi: 10.1089/ars.2022.0202. [DOI] [PubMed] [Google Scholar]
- 165.Hou W., Xie Y., Song X., Sun X., Lotze M.T., Zeh H.J., 3rd, Kang R., Tang D. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12:1425–1428. doi: 10.1080/15548627.2016.1187366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gao M., Monian P., Pan Q., Zhang W., Xiang J., Jiang X. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26:1021–1032. doi: 10.1038/cr.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wu Y., Kram H., Gempt J., Liesche-Starnecker F., Wu W., Schlegel J. ALDH1-Mediated autophagy sensitizes glioblastoma cells to ferroptosis. Cells. 2022;11 doi: 10.3390/cells11244015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu L., Zheng B., Luo M., Du J., Yang F., Huang C., Ma Z., Li C., Guo D., Peng H. Suppression of USP8 sensitizes cells to ferroptosis via SQSTM1/p62-mediated ferritinophagy. Protein Cell. 2023;14:230–234. doi: 10.1093/procel/pwac004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xiong Q., Li X., Li W., Chen G., Xiao H., Li P., Wu C. WDR45 mutation impairs the autophagic degradation of transferrin receptor and promotes ferroptosis. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.645831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen X., Li J., Kang R., Klionsky D.J., Tang D. Ferroptosis: machinery and regulation. Autophagy. 2021;17:2054–2081. doi: 10.1080/15548627.2020.1810918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wu Z., Geng Y., Lu X., Shi Y., Wu G., Zhang M., Shan B., Pan H., Yuan J. Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc. Natl. Acad. Sci. U.S.A. 2019;116:2996–3005. doi: 10.1073/pnas.1819728116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yu S., Li Z., Zhang Q., Wang R., Zhao Z., Ding W., Wang F., Sun C., Tang J., Wang X., Zhang H., Huang R., Wu Q., Jiang J., Zhao X. GPX4 degradation via chaperone-mediated autophagy contributes to antimony-triggered neuronal ferroptosis. Ecotoxicol. Environ. Saf. 2022;234 doi: 10.1016/j.ecoenv.2022.113413. [DOI] [PubMed] [Google Scholar]
- 173.Xue Q., Yan D., Chen X., Li X., Kang R., Klionsky D.J., Kroemer G., Chen X., Tang D., Liu J. Copper-dependent autophagic degradation of GPX4 drives ferroptosis. Autophagy. 2023:1–15. doi: 10.1080/15548627.2023.2165323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Xue Q., Kang R., Klionsky D.J., Tang D., Liu J., Chen X. Copper metabolism in cell death and autophagy. Autophagy. 2023;19:2175–2195. doi: 10.1080/15548627.2023.2200554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bi Y., Liu S., Qin X., Abudureyimu M., Wang L., Zou R., Ajoolabady A., Zhang W., Peng H., Ren J., Zhang Y. FUNDC1 interacts with GPx4 to govern hepatic ferroptosis and fibrotic injury through a mitophagy-dependent manner. J. Adv. Res. 2024;55:45–60. doi: 10.1016/j.jare.2023.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Thayyullathil F., Cheratta A.R., Alakkal A., Subburayan K., Pallichankandy S., Hannun Y.A., Galadari S. Acid sphingomyelinase-dependent autophagic degradation of GPX4 is critical for the execution of ferroptosis. Cell Death Dis. 2021;12:26. doi: 10.1038/s41419-020-03297-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lee Y. Roles of circadian clocks in cancer pathogenesis and treatment. Exp. Mol. Med. 2021;53:1529–1538. doi: 10.1038/s12276-021-00681-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yang M., Chen P., Liu J., Zhu S., Kroemer G., Klionsky D.J., Lotze M.T., Zeh H.J., Kang R., Tang D. Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aaw2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Liu J., Li J., Kang R., Tang D. Cell type-specific induction of ferroptosis to boost antitumor immunity. OncoImmunology. 2023;12 doi: 10.1080/2162402X.2023.2282252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang C., Zheng C., Wang H., Shui S., Jin H., Liu G., Xu F., Liu Z., Zhang L., Sun D., Xu P. Dual degradation mechanism of GPX4 degrader in induction of ferroptosis exerting anti-resistant tumor effect. Eur. J. Med. Chem. 2023;247 doi: 10.1016/j.ejmech.2022.115072. [DOI] [PubMed] [Google Scholar]
- 181.Wang H., Wang C., Li B., Zheng C., Liu G., Liu Z., Zhang L., Xu P. Discovery of ML210-Based glutathione peroxidase 4 (GPX4) degrader inducing ferroptosis of human cancer cells. Eur. J. Med. Chem. 2023;254 doi: 10.1016/j.ejmech.2023.115343. [DOI] [PubMed] [Google Scholar]
- 182.Zheng C., Wang C., Sun D., Wang H., Li B., Liu G., Liu Z., Zhang L., Xu P. Structure-activity relationship study of RSL3-based GPX4 degraders and its potential noncovalent optimization. Eur. J. Med. Chem. 2023;255 doi: 10.1016/j.ejmech.2023.115393. [DOI] [PubMed] [Google Scholar]
- 183.Ding Y., Chen X., Liu C., Ge W., Wang Q., Hao X., Wang M., Chen Y., Zhang Q. Identification of a small molecule as inducer of ferroptosis and apoptosis through ubiquitination of GPX4 in triple negative breast cancer cells. J. Hematol. Oncol. 2021;14:19. doi: 10.1186/s13045-020-01016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhang W., Jiang B., Liu Y., Xu L., Wan M. Bufotalin induces ferroptosis in non-small cell lung cancer cells by facilitating the ubiquitination and degradation of GPX4. Free Radic. Biol. Med. 2022;180:75–84. doi: 10.1016/j.freeradbiomed.2022.01.009. [DOI] [PubMed] [Google Scholar]
- 185.Zhou C., Yu T., Zhu R., Lu J., Ouyang X., Zhang Z., Chen Q., Li J., Cui J., Jiang F., Jin K.Y., Sarapultsev A., Li F., Zhang G., Luo S., Hu D. Timosaponin AIII promotes non-small-cell lung cancer ferroptosis through targeting and facilitating HSP90 mediated GPX4 ubiquitination and degradation. Int. J. Biol. Sci. 2023;19:1471–1489. doi: 10.7150/ijbs.77979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wu J., Liu C., Wang T., Liu H., Wei B. Deubiquitinase inhibitor PR-619 potentiates colon cancer immunotherapy by inducing ferroptosis. Immunology. 2023;170:439–451. doi: 10.1111/imm.13683. [DOI] [PubMed] [Google Scholar]
- 187.Wang S.A., Wu Y.C., Yang F.M., Hsu F.L., Zhang K., Hung J.J. NCI677397 targeting USP24-mediated induction of lipid peroxidation induces ferroptosis in drug-resistant cancer cells. Mol. Oncol. 2023 doi: 10.1002/1878-0261.13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sun Y., Berleth N., Wu W., Schlutermann D., Deitersen J., Stuhldreier F., Berning L., Friedrich A., Akgun S., Mendiburo M.J., Wesselborg S., Conrad M., Berndt C., Stork B. Fin56-induced ferroptosis is supported by autophagy-mediated GPX4 degradation and functions synergistically with mTOR inhibition to kill bladder cancer cells. Cell Death Dis. 2021;12:1028. doi: 10.1038/s41419-021-04306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Shimada K., Skouta R., Kaplan A., Yang W.S., Hayano M., Dixon S.J., Brown L.M., Valenzuela C.A., Wolpaw A.J., Stockwell B.R. Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat. Chem. Biol. 2016;12:497–503. doi: 10.1038/nchembio.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gryzik M., Asperti M., Denardo A., Arosio P., Poli M. NCOA4-mediated ferritinophagy promotes ferroptosis induced by erastin, but not by RSL3 in HeLa cells. Biochim. Biophys. Acta Mol. Cell Res. 2021;1868 doi: 10.1016/j.bbamcr.2020.118913. [DOI] [PubMed] [Google Scholar]
- 191.Xue Q., Yan D., Chen X., Li X., Kang R., Klionsky D.J., Kroemer G., Chen X., Tang D., Liu J. Copper-dependent autophagic degradation of GPX4 drives ferroptosis. Autophagy. 2023;19:1982–1996. doi: 10.1080/15548627.2023.2165323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chen Y., Li N., Wang H., Wang N., Peng H., Wang J., Li Y., Liu M., Li H., Zhang Y., Wang Z. Amentoflavone suppresses cell proliferation and induces cell death through triggering autophagy-dependent ferroptosis in human glioma. Life Sci. 2020;247 doi: 10.1016/j.lfs.2020.117425. [DOI] [PubMed] [Google Scholar]
- 193.Tang X., Ding H., Liang M., Chen X., Yan Y., Wan N., Chen Q., Zhang J., Cao J. Curcumin induces ferroptosis in non-small-cell lung cancer via activating autophagy. Thorac Cancer. 2021;12:1219–1230. doi: 10.1111/1759-7714.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhou L., Yang C., Zhong W., Wang Q., Zhang D., Zhang J., Xie S., Xu M. Chrysin induces autophagy-dependent ferroptosis to increase chemosensitivity to gemcitabine by targeting CBR1 in pancreatic cancer cells. Biochem. Pharmacol. 2021;193 doi: 10.1016/j.bcp.2021.114813. [DOI] [PubMed] [Google Scholar]
- 195.Shi H., Xiong L., Yan G., Du S., Liu J., Shi Y. Susceptibility of cervical cancer to dihydroartemisinin-induced ferritinophagy-dependent ferroptosis. Front. Mol. Biosci. 2023;10 doi: 10.3389/fmolb.2023.1156062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Xu X., Mao C., Zhang C., Zhang M., Gong J., Wang X. Salvianolic acid B inhibits ferroptosis and apoptosis during myocardial ischemia/reperfusion injury via decreasing the ubiquitin-proteasome degradation of GPX4 and the ROS-JNK/MAPK pathways. Molecules. 2023;28 doi: 10.3390/molecules28104117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chen J., Ou Z., Gao T., Yang Y., Shu A., Xu H., Chen Y., Lv Z. Ginkgolide B alleviates oxidative stress and ferroptosis by inhibiting GPX4 ubiquitination to improve diabetic nephropathy. Biomed. Pharmacother. 2022;156 doi: 10.1016/j.biopha.2022.113953. [DOI] [PubMed] [Google Scholar]
- 198.Zhao Y., Lu J., Mao A., Zhang R., Guan S. Autophagy inhibition plays a protective role in ferroptosis induced by alcohol via the p62-keap1-nrf2 pathway. J. Agric. Food Chem. 2021;69:9671–9683. doi: 10.1021/acs.jafc.1c03751. [DOI] [PubMed] [Google Scholar]
- 199.Tian Y., Lu J., Hao X., Li H., Zhang G., Liu X., Li X., Zhao C., Kuang W., Chen D., Zhu M. FTH1 inhibits ferroptosis through ferritinophagy in the 6-OHDA model of Parkinson's disease. Neurotherapeutics. 2020;17:1796–1812. doi: 10.1007/s13311-020-00929-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.