Abstract
We have designed a novel, precise, and sensitive assay to measure unspliced (US) human immunodeficiency virus type 1 (HIV-1) mRNA in peripheral blood mononuclear cells of HIV-1-infected individuals by using real-time PCR and molecular beacons. Individuals were classified as either well suppressed (WS) or partially suppressed, based on longitudinal measurements of plasma HIV-1 RNA. The proportion of individuals with US mRNA undetectable over time was significantly higher among WS individuals; however, 30% of WS subjects still had detectable US mRNA after 24 months of effective antiviral therapy.
Effective antiviral therapy can suppress human immunodeficiency virus type 1 (HIV-1) replication to the extent that the level of plasma viremia falls below a detectable level for over 2 years (10, 11, 19). However, the quantitative definition of “undetectable” is limited by the sensitivity of currently available assays. Despite undetectable levels of HIV-1 RNA in plasma, replication-competent virus can reside latently within resting memory CD4+ T lymphocytes (4). This reservoir persists even after 2 years of seemingly effective antiviral therapy (5, 7, 30). If replication of HIV-1 is effectively inhibited, the reservoir of HIV-1 in resting memory CD4+ T lymphocytes should decay over time, with a half-life of ∼6 months (13). However, if there is persistent replication below the limits of detection of currently available assays, the reservoir will appear to persist longer due to ongoing replenishment.
Studies of untreated HIV-1-infected individuals demonstrate high levels of unspliced (US) and multiply spliced (MS) mRNA in peripheral blood mononuclear cells (PBMC) in roughly equimolar ratios (22–24, 28, 29). The overall amount of HIV-1 mRNA expression strongly correlates with disease progression (22–24, 29). Others report an association between a reduced MS-to-US mRNA ratio and a poor prognosis (8, 17). However, following treatment with effective antiviral therapy, there is a rapid decrease of MS mRNA compared to US mRNA in the first 48 h (28), with most individuals having undetectable MS mRNA by 2 weeks (1). Using a novel, rapid, and accurate real-time PCR assay, we therefore undertook a study to determine if the detection of US mRNA in PBMC of individuals on effective antiviral therapy for at least 2 years may serve as an indicator of persistent viral replication.
Total cellular RNA was extracted and reverse transcribed as previously described (28). To detect the input copy number of HIV-1 mRNA particles, a molecular beacon was used in combination with real-time PCR. The method of detection using molecular beacons (16, 27) and a fluorescence detector system (6, 9, 12, 26) has been previously described. Each 50-μl reaction mixture contained 2 μl of cDNA from the reverse transcription reaction, and the final concentration of each component was as follows: 1.0× Taqman buffer A (Perkin-Elmer, Norwalk, Conn.), 3.5 mM MgCl2, 0.4 pmol of molecular beacon per μl, 0.4 pmol of each primer per μl, and 1.25 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer). The primers used have been previously described (28). The molecular beacon was designed to recognize a conserved region within the HIV-1 long terminal repeat (5′-FAM-CGGGAGTACT CACCAGTCGCCGCCCCTCGCCCTCCCG-DABCYL-3′), where 6-carboxyfluorescein (FAM) serves as the reporter fluorochrome and 4-dimethylaminophenylazobenzoic acid (DABCYL) serves as the quencher (Fig. 1A). One cycle of denaturation (95°C for 10 min) was performed, followed by 45 cycles of amplification (95°C for 15 s, 55°C for 30 s, and 72°C for 30 s). PCR was carried out in a spectrofluorometric thermal cycler (ABI PRISM 7700; Applied Biosystems Inc.) that monitors changes in the fluorescence spectrum of each reaction tube during the annealing phase while simultaneously carrying out programmed temperature cycles.
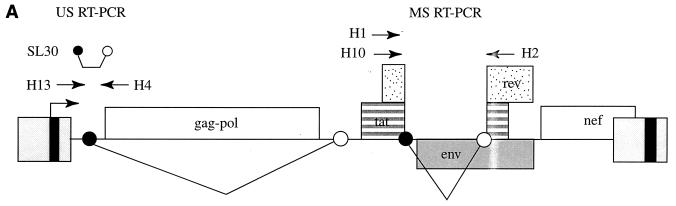
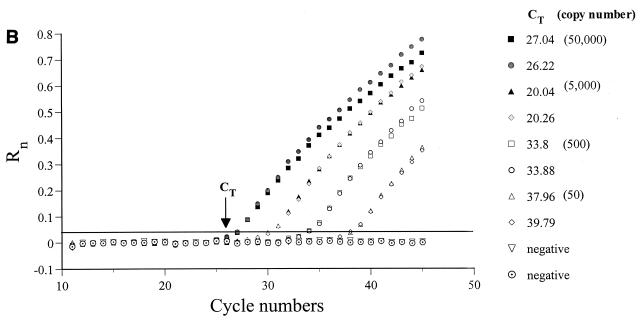
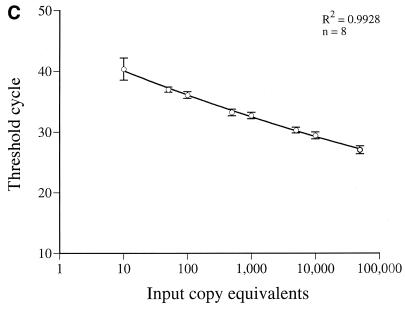
FIG. 1.
(A) Location of primers and beacon on the HIV-1 genome (adapted from reference 22). The primers and beacon shown correspond to the following positions on pNL4-3: H13, nucleotides (nt) 626 to 648; H4, nt 879 to 901; SL30, nt 641 to 865; H10, nt 6070 to 6093; H2, nt 8504 to 8526; and H1, nt 6080 to 6102. (B) Real-time PCR quantification of HIV-1 US mRNA. A reading of change in fluorescence (Rn) as a function of cycle number is demonstrated for a range of known input copy numbers of in vitro-generated HIV-1 US mRNA transcripts. The copy numbers range from 50,000 to 50 copy equivalents per reaction. The cycle number at which the mean fluorescence rises 10 standard deviations above the baseline is called the threshold cycle (CT). The CT is shown for duplicates of the standards used. (C) Relationship of known number of HIV-1 transcripts to the CT. The CT is directly proportional to the log of the input copy equivalents, as demonstrated by the standard curve generated.
For each run, a standard curve was generated from duplicate samples of purified in vitro-transcribed US mRNA transcripts (22) ranging from 50,000 to 10 copies (Fig. 1B and 1C). Each test specimen was reverse transcribed and amplified in duplicate. A third reaction mixture was processed and amplified, without the addition of reverse transcriptase, to control for potential DNA contamination. A sample was considered positive only if both replicates detected US mRNA. If one replicate was negative and the other positive, it was assumed that the concentration of US mRNA was at the lower limits of the assay and the result was reported as <50 copies/μg of RNA. Copy numbers were calculated by interpolation of the experimentally determined threshold cycle (CT) as previously described (6, 9, 12, 26). To control for the recovery of intact PBMC RNA and for the uniform efficiency of each reverse transcription reaction, a 189-bp glyceraldehyde-3-phosphate dehydrogenase (GAPDH) fragment was amplified with specific primers (28). GAPDH was quantified by using real-time PCR and a molecular beacon (5′-HEX-GCGTCGGCTCTCCAGAACATCATCCCTGCCTCGACGC-DABCYL-3′), where hexachlorofluorescein (HEX) served as the reporter fluorochrome. The PCR conditions were similar to those used for HIV-1 quantification, except that the hybridization temperature was 60°C. A standard curve was generated with 10-fold dilutions of total RNA quantified by spectrophotometry. The final results were expressed as the mean copy number of US mRNA per microgram of total RNA. For some individuals, MS mRNA was amplified by nested conventional PCR as previously described (28). The lower limit of sensitivity of this assay was 64 copies/μg of total RNA. All primer and beacon locations are shown in Fig. 1A.
HIV-1-infected individuals were enrolled in treatment protocols within the first 90 days after symptoms of primary HIV-1 infection. All individuals were receiving at least three antiretroviral drugs, including zidovudine (600 mg/day) and lamivudine (300 mg/day), with either ritonavir (1,200 mg/day), indinavir (2,400 mg/day), or ritonavir (800 or 1,200 mg/day) with saquinavir (800 or 1,200 mg/day). Individuals were classified as either well suppressed (WS) or partially suppressed (PS) in terms of viral load. With the Roche Ultrasensitive Amplicor HIV-1 Monitor assay (lower limit of detection, 50 HIV-1 RNA copies/ml; Roche Molecular Systems, Branchburg, N.J.) utilized to measure plasma viremia monthly or bimonthly, WS was defined as ≤1 episode of detectable virus over the duration of follow-up; PS was defined as >1 episode of detectable virus within the range of 50 to 400 copies per ml during the same period.
In order to quantify residual viral replication in individuals with sustained viral suppression, we compared WS and PS individuals. The characteristics of the 27 male individuals (WS, n = 20; PS, n = 7) are summarized in Table 1. All individuals studied had undetectable virus, as measured by the Roche Ultrasensitive Amplicor HIV-1 assay, for a median duration of 20 months. The mean baseline plasma HIV-1 RNA level was 596,000 copies/ml and the mean baseline CD4 cell count was 500 cells/μl, with no significant difference between WS and PS individuals for either parameter. The median time it took to achieve a plasma HIV-1 RNA level of <400 to 500 copies per ml was 45 days, with the median times for WS and PS groups being 20 and 63 days, respectively (P = 0.06, Mann-Whitney rank sum test).
TABLE 1.
Characteristics of individuals studied
| Characteristic | Total | WS | PS |
|---|---|---|---|
| No. of subjects | 27 | 20 | 7 |
| No. of subjects on the following drug combinationa (mean duration of follow-up): | |||
| AZT-3TC-IDV (34 mo) | 10 | 8 | 2 |
| AZT-3TC-RIT (28 mo) | 5 | 3 | 2 |
| AZT-3TC-SQV/RIT (22 mo) | 12 | 9 | 3 |
| Age (yr) | |||
| Mean ± SD | 33.3 ± 7.0 | 32.6 ± 6.5 | 35.2 ± 7.0 |
| Range | 24.2–53.9 | 26.7–53.9 | 24.2–51.2 |
| Baseline CD4 cell count/μl | |||
| Mean | 500 | 517 | 433 |
| Range | 47–1,224 | 197–1,224 | 47–838 |
| Baseline viral loadbc | |||
| Mean | 596 | 441 | 610 |
| Range | <0.5–4,760 | <0.5–4,760 | 14.8–2,730 |
| Time until viral loadb was <400 (days) | |||
| Median | 45 | 20 | 63 |
| Range | 0–182 | 0–182 | 33–132 |
| Duration of viral loadd at <50 (mo) | |||
| Median | 20 | 20 | 19 |
| Range | 6–34 | 9–34 | 6–25 |
| No. of episodes of viral loadd at 50–400 | |||
| Mean | <1 | 3 | |
| Range | 0–1 | 2–5 |
AZT, zidovudine; 3TC, lamivudine; RIT, ritonavir; SQV, saquinavir; and IDV, indinavir.
Viral load (branched DNA, copies per milliliter).
Values are in thousands.
Viral load (RT-PCR, copies per milliliter).
PBMC from these individuals were examined for the presence of US mRNA by using a novel real-time PCR assay together with molecular beacons. The threshold sensitivity was consistently 10 copy equivalents per reaction. The linear dynamic range is in excess of 5 log units (Fig. 1C). The linear relationship between CT and the log of the input copy number has been previously described (9, 12). Briefly, the CT is the cycle number at which the fluorescence rises 10 standard deviations above the baseline. The CT values decrease linearly as target quantity increases (Fig. 1C). The CT of duplicate samples of US RNA standards from eight experiments demonstrated a mean coefficient of variation of 1.4% (Fig. 1B). The CT of duplicate aliquots of eight samples of PBMC mRNA from infected individuals demonstrated a mean coefficient of variation of 0.65% (Fig. 1B). Based on studies of extraction and analysis of four replicate aliquots of the same PBMC samples from four individuals, the interassay variability of US mRNA measurements averaged 28%.
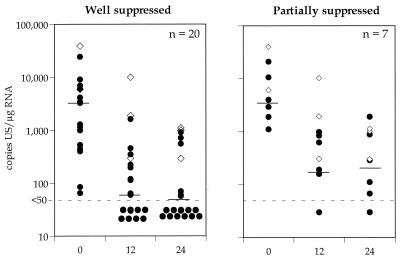
In this study, PBMC US mRNA levels were assessed before therapy and again at 12 and 24 months following the introduction of therapy. At each time point posttherapy the plasma viral load was <50 copies/ml. The results are summarized in Fig. 2. Levels of US mRNA in PBMC declined on treatment (median copy numbers [ranges] at 0, 12, and 24 months, respectively, were as follows: 3,446 [84 to 24,333], 60 [<50 to 1,621], and <50 [<50 to 720] for WS individuals and 3,487 [1,119 to 10,695], 193 [169 to 989], and 200 [<50 to 888] for PS individuals; P = 0.4, 0.03, and 0.11 [Mann-Whitney rank sum test]). Individuals who had persistently detectable virus in the periphery (n = 3) always had detectable US mRNA (Fig. 2).
FIG. 2.
Cross-sectional analysis of US mRNA copies per microgram of mRNA in WS and PS individuals. Levels of US mRNA in PBMC from WS and PS individuals (circles) and median values (black lines) are shown. Data from three nonsuppressed individuals (diamonds) are shown as positive controls. There is a significant difference between WS and PS individuals in the levels of US mRNA at 12 months (P = 0.03) and 24 months (P = 0.11). The lower limit of detection of US mRNA was 50 copies/μg of total RNA.
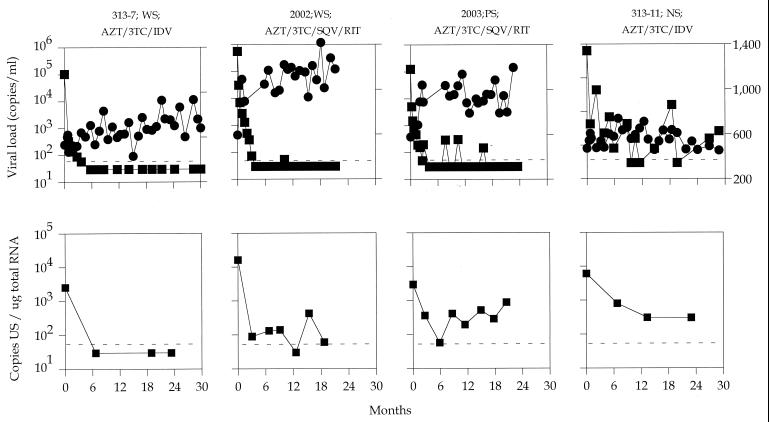
For some individuals, PBMC mRNA was assessed every 6 months. Longitudinal levels of CD4+ T cells, plasma HIV-1 RNA, and US mRNA over time in representative subjects are shown in Fig. 3. In all cases studied, US mRNA levels declined within the first 6 months of therapy. For most WS patients (14 of 20), US mRNA was undetectable after 24 months of therapy. However, for some patients from whom samples were taken more frequently, US mRNA was intermittently detectable (Fig. 3, patient 2002). For patients with partial suppression of plasma virus, US mRNA levels also declined initially, though afterward, US mRNA was persistently detectable (Fig. 3, patient 2003).
FIG. 3.
Sequential changes in plasma viral load (squares in top row), CD4 lymphocyte count (circles), and US mRNA level (squares in bottom row) after combination therapy. The infected individual’s designation (e.g., 2003), the treatment regimen (AZT, zidovudine; 3TC, lamivudine; RIT, ritonavir; SQV, saquinavir; IDV, indinavir), and the classification of suppression are listed at the top of each panel. The lower limits of detection of plasma viral load and US mRNA were 50 copies/ml and 50 copies/μg of total RNA, respectively. NS, not suppressed.
We then assessed the proportion of individuals who had persistently undetectable levels of US mRNA during the follow-up period. The proportion of individuals with US mRNA undetectable over time in a Kaplan-Meir analysis (at 0, 6, 12, 18, and 24 months) was significantly higher among WS individuals (0, 0.2, 0.3, 0.6, and 0.7 for WS patients and 0, 0, 0, 0, and 0.2 for PS patients). However, 6 of 20 WS individuals had persistently detectable US mRNA after 24 months of undetectable plasma viremia. In order to determine the significance of detectable US mRNA, PBMC from 14 of 20 WS individuals and 2 of 7 PS individuals were assessed for the presence of MS mRNA after 12 to 24 months of therapy. No individuals had detectable MS mRNA.
The persistence of US mRNA in nearly all PS individuals suggests that the intermittent detection of low levels of RNA in plasma is biologically significant and most likely represents ongoing replication at the very lower limits of the sensitivity of the assay. We do not know the duration of these intermittent low-level rises in plasma virus, since for all PS individuals, plasma RNA was undetectable 1 month later. However, in contrast to the low-level fluctuation of plasma viremia, US mRNA remained persistently detectable for most of these individuals and may serve as a more sensitive indicator of ongoing viral replication.
Within the WS group, 30% of individuals had persistently detectable US mRNA. There are several possible explanations for this observation. Firstly, the definition of WS is dependent on sampling. Although in nearly all cases sampling was performed monthly or bimonthly, if low-level intermittent viremia was short-lived, then individuals could conceivably have been incorrectly classified as WS. Secondly, even in truly WS individuals, residual viral replication may occur at a level below the sensitivity of current plasma assays.
Studies of US and MS mRNA expression in cells infected in vitro (14, 15, 21) and in activated latently infected cells (1, 18, 20) demonstrate an early rise in MS mRNA levels followed within 5 to 24 h by a rapid rise in US mRNA levels. These findings suggest that levels of MS mRNA in PBMC are closely associated with the number of newly infected cells (1, 28). Following treatment, there is a rapid decline in MS mRNA levels within 48 h, but US mRNA persists (1, 28). Thus, the absence of MS mRNA in most WS individuals, even in the presence of US mRNA, is in agreement with these previous reports.
Given the absence of MS mRNA, what are the possible explanations for the persistence of US mRNA? The most likely explanation is that ongoing replication could continue within a site where antiviral therapy is ineffective. When cells from this “privileged” site enter the bloodstream, contact with effective antiviral drugs causes a rapid decline in the level of MS mRNA but a slower decline in the level of US mRNA, and therefore we are able to detect only persistently replenished reservoirs of US mRNA. Ineffective antiviral therapy could also result in the appearance of drug resistance. However, replication of resistant virus would be unlikely, as studies of similar patients on prolonged antiviral therapy with durable viral suppression do not demonstrate emergence of drug resistance in virus isolated from resting CD4+ T cells (7, 30). Virus isolated from resting CD4+ T cells would be representative of both reservoirs of infection, i.e., ongoing low-level productive infection as well as the so-called “latent reservoir.” In future studies, we plan to directly compare the quantity of virus isolated from this reservoir and the amount of detectable US mRNA. Alternatively, there may be ongoing replication of infected cells, such as T cells or macrophages, but with a very low level of viral expression. Another possibility is that the US mRNA represents preintegration complexes between the entry and integration stages in the viral life cycle (2, 3, 25, 31), but this is unlikely given the predicted short half-life of this form of HIV-1 RNA (2, 4). Finally, US RNA could represent viral RNA contained in particles bound to the surfaces of unfractionated PBMC. Our assay would be unable to differentiate between viral RNA and mRNA. However, in our studies of lymphoid tissue of patients with detectable US mRNA and undetectable MS mRNA, in situ hybridization with radiolabeled RNA probes revealed evidence of intracellular viral RNA, making bound particles an unlikely explanation for the persistence of US mRNA (data not shown).
The ability to quantify residual replication in individuals with undetectable viremia is critical to determining future management strategies for infected individuals. Current estimates of the half-life of the latent reservoir suggest that 7 to 10 years of continuous truly effective therapy will be necessary to eliminate the latent reservoir (13). However, if therapeutic strategies, such as activation, vaccination, or even cessation of therapy (13), are designed to facilitate the decay of the latent pool, then it is critical to have a simple, accurate test to identify individuals with residual viral replication. Intensification of therapy may be prudent for individuals with any evidence of residual replication. The detection of US mRNA in PBMC may prove to be a useful marker in identifying these individuals and may provide a virologic marker of response to intensified therapies in individuals with undetectable plasma HIV-1 RNA.
Acknowledgments
S.R.L. is supported by a CJ Martin fellowship from the National Health and Medical Research Council of Australia. This work was supported by NIH grants MOI-RR00102, AI41534, and IP30AI42848-01; an unrestricted grant from Glaxo-Wellcome; the Concerned Parents for AIDS Research (CPAR); and the Columbia/Rockefeller Center for AIDS Research (CFAR).
We thank Wen Chen for assistance in preparation of the table and figures and Olga Ford and Rosemary Schluger for clinical assistance.
REFERENCES
- 1.Bagnarelli P, Valenza A, Menzo S, Sampaolesi R, Varaldo P E, Butini L, Montroni M, Perno C-F, Aquaro S, Mathez D, Leibowitch J, Balotta C, Clementi M. Dynamics and modulation of human immunodeficiency virus type 1 transcripts in vitro and in vivo. J Virol. 1996;70:7603–7613. doi: 10.1128/jvi.70.11.7603-7613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukrinsky M, Stanwick T, Dempsey M, Stevenson M. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science. 1991;254:423–427. doi: 10.1126/science.1925601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun T W, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano R F. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1:1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 4.Chun T-W, Carruth L, Finzi D, Shen X, DiGiuseppe J, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T, Kuo Y-H, Brookmeyer R, Zeiger M, Barditch-Crovo P, Siliciano R. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 5.Chun T-W, Stuyer L, Mizell S, Ehler L, Mican J, Baseler M, Lloyd A, Nowak M, Fauci A. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fink L, Seeger W, Ermert L, Hanze J, Stahl U, Grimminger F, Kummer W, Bohle R. Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med. 1998;4:1329–1333. doi: 10.1038/3327. [DOI] [PubMed] [Google Scholar]
- 7.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 8.Furtado M R, Kingsley L A, Wolinsky S M. Changes in the viral mRNA expression pattern correlate with a rapid rate of CD4+ T-cell number decline in human immunodeficiency virus type 1-infected individuals. J Virol. 1995;69:2092–2100. doi: 10.1128/jvi.69.4.2092-2100.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson U E, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 10.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 11.Hammer S M, Squires K E, Hughes M D, Grimes J M, Demeter L M, Currier J S, Eron J J, Jr, Feinberg J E, Balfour H H, Jr, Deyton L R, Chodakewitz J A, Fischl M A. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 12.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 13.Ho D D. Toward HIV eradication or remission: the tasks ahead. Science. 1998;280:1866–1867. doi: 10.1126/science.280.5371.1866. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, Byrn R, Groopman J, Baltimore D. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J Virol. 1989;63:3708–3713. doi: 10.1128/jvi.63.9.3708-3713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klotman M, Kim S, Buchbinder A, DeRossi A, Baltimore D, Wong-Staal F. Kinetics of expression of multiply spliced RNA in early human immunodeficiency virus type 1 infection of lymphocytes and monocytes. Proc Natl Acad Sci USA. 1991;88:5011–5015. doi: 10.1073/pnas.88.11.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kostrikis L G, Tyagi S, Mhlanga M M, Ho D D, Kramer F R. Spectral genotyping of human alleles. Science. 1998;279:1228–1229. doi: 10.1126/science.279.5354.1228. [DOI] [PubMed] [Google Scholar]
- 17.Michael N L, Mo T, Merzouki A, O’Shaughnessy M, Oster C, Burke D S, Redfield R R, Birx D L, Cassol S A. Human immunodeficiency virus type 1 cellular RNA load and splicing patterns predict disease progression in a longitudinally studied cohort. J Virol. 1995;69:1868–1877. doi: 10.1128/jvi.69.3.1868-1877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michael N L, Morrow P, Mosca J, Vahey M, Burke D S, Redfield R R. Induction of human immunodeficiency virus type 1 expression in chronically infected cells is associated primarily with a shift in RNA splicing patterns. J Virol. 1991;65:1291–1303. doi: 10.1128/jvi.65.3.1291-1303.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perelson A, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho D. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 20.Pomerantz R J, Trono D, Feinberg M B, Baltimore D. Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral RNA expression: a molecular model for latency. Cell. 1990;61:1271–1276. doi: 10.1016/0092-8674(90)90691-7. [DOI] [PubMed] [Google Scholar]
- 21.Ranki A, Lagerstedt A, Ovod V, Aavik E, Krohn K. Expression kinetics and subcellular localization of HIV-1 regulatory proteins Nef, Tat and Rev in acutely and chronically infected lymphoid cell lines. Arch Virol. 1994;139:365–378. doi: 10.1007/BF01310798. [DOI] [PubMed] [Google Scholar]
- 22.Saksela K, Stevens C, Rubinstein P, Baltimore D. Human immunodeficiency virus type 1 mRNA expression in peripheral blood cells predicts disease progression independently of the numbers of CD4+ T lymphocytes. Proc Natl Acad Sci USA. 1994;91:1104–1108. doi: 10.1073/pnas.91.3.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saksela K, Stevens C E, Rubinstein P, Taylor P E, Baltimore D. HIV-1 messenger RNA in peripheral blood mononuclear cells as an early marker of risk for progression to AIDS. Ann Intern Med. 1995;123:641–648. doi: 10.7326/0003-4819-123-9-199511010-00001. [DOI] [PubMed] [Google Scholar]
- 24.Saltarelli M J, Hadziyannis E, Hart C E, Harrison J V, Felber B K, Spira T J, Pavlakis G N. Analysis of human immunodeficiency virus type 1 mRNA splicing patterns during disease progression in peripheral blood mononuclear cells from infected individuals. AIDS Res Hum Retroviruses. 1996;12:1443–1456. doi: 10.1089/aid.1996.12.1443. [DOI] [PubMed] [Google Scholar]
- 25.Stevenson M, Stanwick T, Dempsey M, Lamonica C. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suryanarayana K, Wiltrout T A, Vasquez G M, Hirsch V M, Lifson J D. Plasma SIV RNA viral load determination by real-time quantification of product generation in reverse transcriptase-polymerase chain reaction. AIDS Res Hum Retroviruses. 1998;14:183–189. doi: 10.1089/aid.1998.14.183. [DOI] [PubMed] [Google Scholar]
- 27.Tyagi S, Kramer F R. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 28.Vesanen M, Markowitz M, Cao Y, Ho D D, Saksela K. Human immunodeficiency virus type-1 mRNA splicing pattern in infected persons is determined by the proportion of newly infected cells. Virology. 1997;236:104–109. doi: 10.1006/viro.1997.8718. [DOI] [PubMed] [Google Scholar]
- 29.Vesanen M, Stevens C E, Taylor P E, Rubinstein P, Saksela K. Stability in controlling viral replication identifies long-term nonprogressors as a distinct subgroup among human immunodeficiency virus type 1-infected persons. J Virol. 1996;70:9035–9040. doi: 10.1128/jvi.70.12.9035-9040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong J, Hezareh M, Gunthard H, Havlir D, Ignacio C, Spina C, Richman D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1294. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 31.Zack J, Arrigo S, Weitsman S, Go A, Haislip A, Chen I. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]