Summary
Frontotemporal dementia (FTD) is the second most common cause of early-onset dementia after Alzheimer disease (AD). Efforts in the field mainly focus on familial forms of disease (fFTDs), while studies of the genetic etiology of sporadic FTD (sFTD) have been less common. In the current work, we analyzed 4,685 sFTD cases and 15,308 controls looking for common genetic determinants for sFTD. We found a cluster of variants at the MAPT (rs199443; p = 2.5 × 10−12, OR = 1.27) and APOE (rs6857; p = 1.31 × 10−12, OR = 1.27) loci and a candidate locus on chromosome 3 (rs1009966; p = 2.41 × 10−8, OR = 1.16) in the intergenic region between RPSA and MOBP, contributing to increased risk for sFTD through effects on expression and/or splicing in brain cortex of functionally relevant in-cis genes at the MAPT and RPSA-MOBP loci. The association with the MAPT (H1c clade) and RPSA-MOBP loci may suggest common genetic pleiotropy across FTD and progressive supranuclear palsy (PSP) (MAPT and RPSA-MOBP loci) and across FTD, AD, Parkinson disease (PD), and cortico-basal degeneration (CBD) (MAPT locus). Our data also suggest population specificity of the risk signals, with MAPT and APOE loci associations mainly driven by Central/Nordic and Mediterranean Europeans, respectively. This study lays the foundations for future work aimed at further characterizing population-specific features of potential FTD-discriminant APOE haplotype(s) and the functional involvement and contribution of the MAPT H1c haplotype and RPSA-MOBP loci to pathogenesis of sporadic forms of FTD in brain cortex.
We identified associations at the MAPT, APOE, and MOBP loci with FTD, suggesting potential common genetic denominators across multiple neurological conditions (i.e., FTD, PSP, ALS, AD, PD, and CBD). These findings will aid in the further characterization of the common and disease-specific pathogenesis of, and interventions for, these disorders.
Introduction
The set of neuropathologies known as frontotemporal lobar degeneration (FTLD; MIM: 607485) make up the second most common cause of early-onset dementia after Alzheimer disease (AD; MIM: 104310).1,2 The most common presentation is frontotemporal dementia (FTD; MIM: 600274), which itself comprises heterogeneous clinical, pathological, and genetic features.
Clinically, the two major syndromes are the behavioral (bvFTD) and the language variants (primary progressive aphasias [PPAs]).3,4 The latter are further subdivided into semantic dementia (SD or semantic variant PPA [svPPA]) and progressive non-fluent aphasia (PNFA or nonfluent/agrammatic variant PPA [nfvPPA]).3,5 FTD can also overlap with motor-neuron disease (FTD-MND)6 and share clinical features with progressive supranuclear palsy (PSP; MIM: 601104) and the corticobasal syndrome (CBS; no MIM).7 Pathologically, Tau and TDP-43 are the most frequent protein aggregates that define the pathological subtypes of FTLD-tau and FTLD-TDP (≤45% and ≤50% of all affected individuals, respectively).8 Genetically, familial FTD (fFTD; ∼30% of all FTD-affected individuals) is predominantly linked to mutations in MAPT (MIM: 157140), GRN (MIM: 138945), and C9orf72 (MIM: 614260)9,10; of note, GFRA2 (MIM: 601956) and TMEM106B (MIM: 613413) (previously reported in a cohort with TDP-43 pathology11) were found to be associated with increased risk in a GRN mutation FTD cohort.12 Sporadic FTD (sFTD; ∼70% of all affected individuals) has been associated with genetic risk markers at the TMEM106B, DPP6 (MIM: 126141), UNC13A (MIM: 609894) and HLA-DQA2 (MIM: 613503) loci in cohorts with TDP-43 pathology11,13 and HLA-DRs (MIM: 142860) reported in an sFTD cohort encompassing all clinical subtypes.14
Efforts in the field have mainly focused on fFTDs, while fewer studies sought to determine the genetic etiology of apparently sFTDs (discussed in Eichler et al., 2010 and Ferrari et al., 201915,16). In the current work, we analyzed 4,685 sFTD cases and 15,308 controls to identify common genetic determinants contributing to increased risk of sFTD and assess their potential biological impact.
Subjects and methods
Study population
Individuals included in the study were clinically diagnosed with a variant of FTD, including the subtypes bvFTD, SD/svPPA, PNFA/nfvPPA, FTD-MND, and FTD-unspecified (i.e., if individuals were diagnosed with FTD but could not be assigned to a specific subtype; see introduction section for acronym definitions). Diagnoses were made according to international consensus criteria: Neary et al. (for FTD, until 2011), Rascovsky et al. (for bvFTD), Gorno-Tempini et al. (for PPA, svPPA and nfvPPA), and Strong et al. (for FTD-MND).3,4,5,17 Individuals with logopenic variant PPA were excluded because of its major association with AD.
A previous FTD cohort, divided into discovery (cohort I) and replication (cohort II) sets,14 was further elaborated by (1) accruing updated metadata leading to the exclusion of individuals that did not meet the diagnostic criteria detailed above, or for which updates were not provided, and (2) inclusion of additional sets of controls. A new cohort of samples (cohort III) was progressively collected between 2016 and 2019 by clinicians and research groups based in Europe (Belgium, France, Germany, Italy, the Netherlands, Norway, Slovenia, Spain, Sweden, and UK) and North America (USA and Canada). Each contributing site obtained written informed consent for the samples to be part of this genetic study (IRB approval #9811/001). All samples (cohorts I, II, and III) were progressively sent as extracted DNA from tissues (blood and/or brain) and stored at −80°C upon receipt (see also Ferrari et al., 201414). The bulk of the control samples used in this study (from France, Germany, Italy, the Netherlands, Spain, Sweden, UK, and USA) was available through a previous study,14 and additional control data were obtained from collaborators (at National Institutes of Health, USA and University College London, UK) and genotyped during the cohort III genotyping iterations (also including additional controls from Italy, Norway, and Slovenia). Overall, the control population at hand consisted of 16,821 samples (from France, Germany, Italy, the Netherlands, Spain, Sweden, UK, USA, Norway, and Slovenia), free of neurological illness at the time of sampling, and matched to cases based on population ancestry (see also QC—Samples below).
Cohorts
Before quality control checks (QCs), cohort I included 2,026 cases and 8,387 controls, cohort II 1,121 cases and 5,091 controls, and cohort III 2,504 cases and 3,343 controls. The three cohorts were independently QCed (see below), leaving 2,006 cases and 8,350 controls for cohort I, 1,090 cases and 5,067 controls for cohort II, and 2,315 cases and 3,230 controls for cohort III.
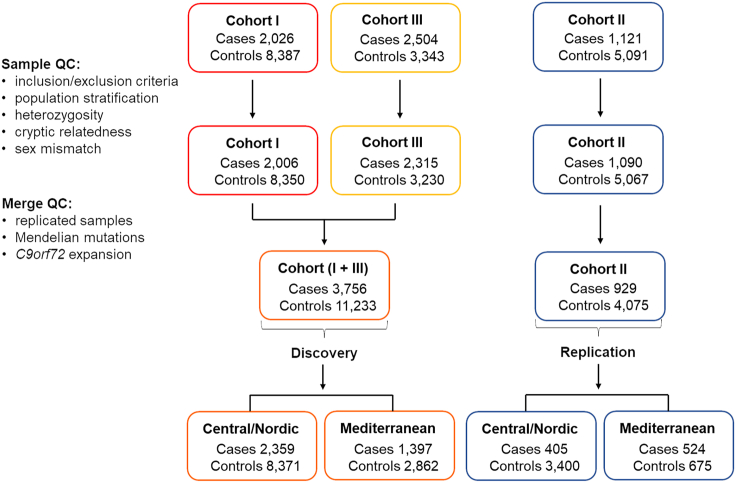
Cohorts I and III (run on genotyping chips) were combined into a new discovery cohort. Cohort II (run on NeuroX array14,18) was used as replication set. Duplicated samples and samples known to carry Mendelian mutations in neurodegenerative genes (APP [MIM: 104760], CHCHD10 [MIM: 615903], CHMP2B [MIM: 609512], FUS [MIM: 137070], GRN, HNRNPA1 [MIM: 164017], LRRK2 [MIM: 609007], MAPT, NOTCH3 [MIM: 600276], PSEN1 [MIM: 104311], PSEN2 [MIM: 600759], SERPINI1 [MIM: 602445], SORL1 [MIM: 602005], SQSTM1 [MIM: 601530], TMEM106B, TBK1 [MIM: 604834], VCP [MIM: 601023]), or a C9orf72 pathogenic expansion, were removed from the analyses, leading to the following final study cohorts (before sample QC): 3,756 cases and 11,233 controls for the discovery phase (cohorts I + III) and 929 cases and 4,075 controls for the replication phase (cohort II). A breakdown of samples (and FTD subtypes) is shown in Table S1. The three cohorts, the QC procedures, and the discovery and replication sets are shown in Figure 1. Of note, given the small number of samples for some of the FTD subtypes (SD, PNFA, and FTD-MND), subtype analysis was not informative, and therefore it is not reported.
Figure 1.
QC pipeline for the three cohorts contributing to discovery and replication
The discovery and replication sets were further subdivided into Central/Nordic and Mediterranean Europeans (PCA-based genetically estimated ancestry) to assess potential population-specific disease risk loci.
Sample genotyping
Cohort I cases had been genotyped on either 660K or Omni-Express Illumina array chips.14 Cohort II cases had been genotyped on the NeuroX array.14,18 Cohort III cases were genotyped on NeuroChip.19 Samples were genotyped at the Laboratory of Neurogenetics of the National Institute on Aging, NIH or at the core facility at the Institute of Child Health, UCL (UCL Genomics). All arrays were run on the Illumina Infinium platform as per the manufacturer’s instructions. Control samples had been genotyped using a variety of array chips, including 330K, 550K, 660K, Omni-Express, NeuroX, and NeuroChip, and cohorts I and II were previously genotyped as per Ferrari et al., 2014;14 cohort III samples were genotyped within the current study (IRB approval #9811/001). Data were QCed following standard procedure using samples with genotyping call rate ≥95% and markers with GenTrain score ≥0.7.
QC—Samples
Sample-level QC was performed before carrying out separate imputation for each of the three cohorts. Genotypes were used to inform on population substructure via principal-component analysis (PCA). Linkage disequilibrium (LD)-pruned markers with a 95% genotyping rate (less than 5% missing), Hardy-Weinberg equilibrium exact test (HWE) p value ≥1 × 10−10 midp,20 and minor allele frequency (MAF) ≥0.01 were used to assess ancestry via PCA against HapMap Phase3 (hapmap3_r3_b36_fwd.consensus.qc.poly). This analysis allowed us to address population substructure and led to the exclusion of population outliers; briefly, PCA on the three different cohorts was performed, and overlap with European samples from HapMap was used to identify outliers (see details in Figure S1).
We also assessed samples’ potential contamination by evaluating individuals’ heterozygosity (removing samples with inbreeding coefficient estimates outside 4 standard deviations from the mean distribution) and cryptic relatedness (removing sample pairs with estimated identity by descent = PI_HAT greater than 0.125) using a set of LD-pruned high-quality SNPs (markers with a 95% genotyping rate, HWE ≥1 × 10−10 midp, and MAF ≥0.01). Finally, we excluded samples with a possible mismatch for sex by assessing the X chromosome heterozygosity (for cohorts II and III only, as for cohort I genotyping for the X chromosome was not available).
QC—Markers
Variant-level QC cleaning was performed before imputation for each of the three cohorts. Palindromic markers and markers with missing call rates exceeding 5% were removed. Finally, we assessed differences between case and control genotype data via non-random missingness, excluding markers with Bonferroni’s corrected p values <0.1 (indicating significant differential missingness between cases and controls).
Imputation
The three cohorts were imputed separately. Cohort I was converted from GRCh36/hg18 to GRCh37/hg19 using the LIFTOVER tool prior to imputation. We imputed markers through the Michigan server (https://imputationserver.sph.umich.edu/) using the following specifications and thresholds: Minimac4; HRC reference panel (GRCh37/hg19); imputation filter rsq 0.3; Eagle v.2.4 phasing; European (EUR) population. Multiallelic and palindromic variants were removed following imputation. Post-imputation dataset sizes were as follows: cohort I, ∼22 M markers; cohort II, ∼5 M markers; cohort III, ∼21 M markers. Imputation for cohort II resulted in a smaller number of imputed markers due to this cohort being genotyped on the NeuroX array, an exome chip not originally designed for imputation.14,18
Discovery cohort merge
Relatedness across the three cohorts was evaluated using genotyped-only communal markers (cohorts I ꓵ II = 7,480 genotyped communal markers, II ꓵ III = 10,328, I ꓵ III = 94,773). A set of LD-pruned high-quality SNPs (markers with a 95% genotyping rate, HWE ≥1 × 10−10 midp, and MAF ≥0.01) was used to evaluate cryptic relatedness (removing sample pairs with estimated identity by descent = PI_HAT greater than 0.125). Cohort I and III post-imputation were then combined using the overlapping markers as discovery cohort (3,756 cases and 11,233 controls; ∼18 M markers genotyped and imputed), while cohort II was used as replication cohort (929 cases and 4,075 controls; ∼5 M markers genotyped and imputed) (Figure S2).
Association analyses
We performed association analysis using markers with MAF >1% and HWE >10−4 midp through the PLINK case-control logistic regression association analysis with 20 principal components (PCs), sex, and study cohort (I or III) as covariates. The top markers were confirmed by running a similar association analysis in MAOS (https://dlin.web.unc.edu/software/maos/) using 20 PCs, sex, study cohort (I or III), and FTD subtypes as covariates.21 The same analytical pipeline was applied to the replication cohort. Markers of interest were meta-analyzed (discovery + replication) using METAL (run with STDERR scheme).22
Genome-wide significant markers were annotated using the Ensembl Variant Effect Predictor (VEP). Frequencies for 1000 Genomes (European samples) were used to further control for marker frequencies in the general population: a variation of >15% between our controls and the European general population as per 1000 Genomes was used as threshold to exclude variants from the current study. When single populations from 1000 Genomes were used, they were selected as follows: GBR = British in England and Scotland; CEU = Northern Europeans from Utah; IBR = Iberian populations in Spain; TSI = Tuscans from Italy.
The Bonferroni threshold for genome-wide significance was p ≤ 5 × 10−8.23 Variants with p values between 1 × 10−5 and 5 × 10−8 were reported as suggestive (only loci containing ≥20 markers in those p value ranges).
Heritability and genetic correlation analyses
We used LD score regression (LDSC)24 to derive an SNP-based heritability estimate (h2) for the discovery FTD summary statistics (3,756 cases and 11,233 controls). The analysis was performed using the pre-computed European SNP LD scores (https://data.broadinstitute.org/alkesgroup/LDSCORE/eur_w_ld_chr.tar.bz2).
Then, LDSC was employed for estimation of genetic correlation (rg) between FTD and publicly available genome-wide association study (GWAS) summary statistics of five neurodegenerative disorders: (1) clinical AD GWAS of 63,926 samples (AD)25; (2) AD clinical/proxy GWAS and related dementias (ADRD) of 487,511 samples26; (3) Parkinson Disease GWAS (PD; MIM: 168600) of 1,474,097 samples27; (4) amyotrophic lateral sclerosis GWAS (ALS; MIM: 612069) of 138,086 samples28; and (5) Lewy body dementia GWAS (LBD; MIM: 127750) of 6,618 samples.29
BUHMBOX (https://software.broadinstitute.org/mpg/buhmbox/) was run according to Han et al., 2016.30 SNPs associated with AD25 were extracted in the APOE (MIM: 107741) region and filtered to remove FTD SNPs with p < 0.05. The remaining SNPs were clumped (with r2 = 0.1 in 10,000-kb window), resulting in 64 SNPs for the analysis.
Locus analysis
Risk loci at chromosome 17 (MAPT region) and chromosome 19 (APOE region) were further characterized by extracting the genotypes of the following markers: rs17650901 (A:G) and rs242557 (G:A) on chromosome 17, where the A alleles tag the H1 and H1c clade, respectively,31,32 and rs429358 (C:T) and rs7412 (T:C) on chromosome 19 to assess the APOE alleles (C/C, C/C = Ɛ4/Ɛ4; T/T, T/T = Ɛ2/Ɛ2; and T/T, C/C = Ɛ3/Ɛ3). Additional controls for the APOE markers were downloaded from the 1000 Genomes Project using the Ensembl Genome Browser and obtained from an independent cohort of controls (731 Italian controls and 347 Central/Nordic European controls). Differences in Ɛ allele counts were assessed using Pearson’s χ2 test with Yates’ continuity correction.
Functional analysis
We used the full distribution of SNP p values and the top markers (post joint analysis) to further characterize biological and functional effects, including potential effects on expression and splicing, using the GTEx (https://gtexportal.org/) and Functional Mapping and Annotation (FUMA) (https://fuma.ctglab.nl/)33 platforms.
Software
All analyses were performed using R (R v.3.5.2), R studio (R v.3.6.2, studio v.1.2.1335), PLINK v.1.9, MAOS (http://dlin.web.unc.edu/files/2011/08/maos-1.2-linux.tar_.gz), and METAL (version for Windows, released March 25, 2011). LD score regression was run using LDSC v.1.0.1 (https://github.com/bulik/ldsc).
Results
Discovery association analyses
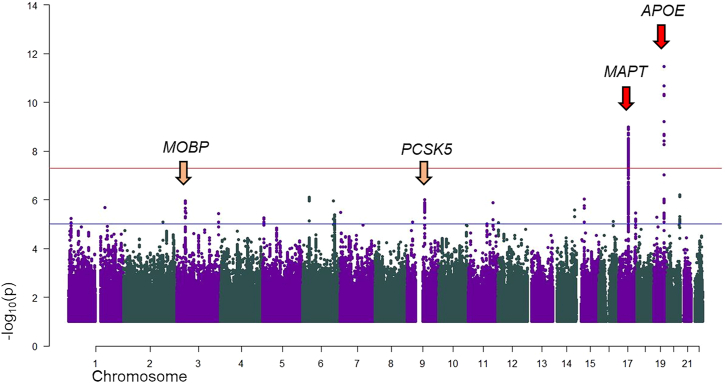
We performed GWAS analysis for the discovery cohort (3,756 cases and 11,233 controls). The genomic inflation factor (λ) was 1.035 (λ1000 = 1.006) (Figure S3). 1,886 markers, mapping to the MAPT (1,880 markers; chr17:43,572,419–44,862,347 [GRCh37/hg19]) and APOE (6 markers; chr19:45,387,596–45,396,144 [GRCh37/hg19]) loci, were genome-wide significant.
The SNPs with the lowest p values were rs199443 (MAPT locus; p = 1.03 × 10−9; β = 0.229 [C = risk allele, major allele]; SE = 0.037) and rs6857 (APOE locus; p = 6.2 × 10−10; β = 0.239 [T = risk allele, minor allele]; SE = 0.039) (Figure 2; Table 1; Table S2). After conditioning the regional analyses on the index SNPs, each association disappeared, suggesting there is a unique signal per locus (Figures S4A and S4B). To further support our discovery findings, we used another statistical method (MAOS; see subjects and methods), which allowed us to run five different association analyses, assessing each subtype, accounting for the same control sets, and meta-analyzing the outcomes. The genome-wide-significant hits shown above were confirmed and displayed improved statistics (Table S3).
Figure 2.
Discovery phase: Manhattan plot
Red arrows identify genome-wide-significant signals (chromosomes 17 and 19); yellow arrows identify suggestive towers (10−8 < p > 10−5) including at least 20 markers (chromosomes 3 and 9). The plot is cut at −log10(p) = 1. The gene symbols represent the locus and do not necessarily imply functional/biological relevance.
Table 1.
Top marker summary statistics at the significant loci
| rs number | Chr | Base pair | Locus | A1/A2 |
Discovery |
Replication |
Joint Analysis |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ImpScore (rsq)1,2 |
Minor allele frequency |
PLINK |
ImpScore (rsq) |
Minor allele frequency |
PLINK |
METAL |
|||||||||||||||||||
| Controls | Cases | 1000G EUR- | Beta | OR | SE | p value | Controls | Cases | Beta | OR | SE | p value | Beta | OR | SE | p value | I2 | HetPVal | |||||||
| rs199443 | 17 | 44,819,565 | MAPT | T/C∗ | 0.96, 0.96 | 0.224 | 0.187 | 0.223 | 0.229 | 1.257 | 0.037 | 1.03E-09 | genotyped | 0.220 | 0.210 | 0.267 | 1.306 | 0.077 | 5.34E-04 | 0.236 | 1.266 | 0.034 | 2.50E-12 | 0 | 6.53E-01 |
| rs6857 | 19 | 45,392,254 | APOE | T∗/C | 0.96, 0.98 | 0.151 | 0.179 | 0.165 | 0.239 | 1.270 | 0.039 | 6.22E-10 | 0.99 | 0.161 | 0.181 | 0.249 | 1.283 | 0.072 | 5.01E-04 | 0.241 | 1.273 | 0.034 | 1.31E-12 | 0 | 9.00E-01 |
| rs1009966 | 3 | 39,473,591 | MOBP | G/A∗ | 0.93, 0.97 | 0.475 | 0.433 | 0.460 | 0.144 | 1.155 | 0.030 | 1.12E-06 | 0.77 | 0.469 | 0.413 | 0.157 | 1.17 | 0.057 | 6.18E-03 | 0.147 | 1.158 | 0.026 | 2.36E-08 | 0 | 8.47E-01 |
We identified six suggestive signals comprising at least four markers with p < 1 × 10−5 and sought to take two of them (comprising at least 20 markers with p < 1 × 10−5) forward for replication and joint analysis to screen for their potential relevance (Table S4): chromosome 3 (top SNP rs13081054; myelin-associated oligodendrocyte basic protein [MOBP; MIM: 600948] locus) and chromosome 9 (top SNP rs76573513; PCSK5 [MIM: 600488] locus).
Replication and joint analyses
The following top markers in discovery analysis (n = 6 for the APOE locus; n = 1,880 for the MAPT locus; n = 29 for the MOBP locus) were present in the replication set (see also Tables S2 and S4).
In the replication set, the chromosome 17 and chromosome 19 top SNPs (rs199443 and rs6857, respectively) reached p value = 5.3 × 10−4 (β = 0.267; SE = 0.077) and p value = 5 × 10−4 (β = 0.249; SE = 0.072), respectively. Following joint analysis, each marker was genome-wide significant (p = 2.5 × 10−12; β = 0.236; SE = 0.034 for rs199443 [MAPT locus] and p = 1.31 × 10−12; β = 0.241; SE = 0.034 for rs6857 [APOE locus]), with the effect being in the same direction (Table 1; Table S2).
For the suggestive signals, only markers on chromosome 3 were available in the replication set (due to lower imputation coverage for the NeuroX exome-chip; see subjects and methods): rs1009966 was replicated and reached lowest p value (genome-wide significant) after joint analysis (p value = 2.36 × 10−8; β = 0.147; SE = 0.026), with the effect being in the same direction (Table S4).
MAPT and APOE loci
We further characterized the MAPT locus in the discovery cohort using rs17650901 (A:G) and rs242557 (G:A), where the A alleles tag the H1 and H1c clade, respectively. The frequencies of the A allele and the homozygous A/A genotype were significantly increased in cases compared to controls for rs17650901 (p = 1.9 × 10−9 and p = 7.7 × 10−9, respectively) and rs242557 (p = 5 × 10−3 and p = 8.8 × 10−3, respectively), suggesting association of the H1/H1 haplotype and H1c clade with sFTD (Table 2).
Table 2.
Haplotype analysis at the MAPT locus
| Marker | Genotype | Haplotype |
Cases |
Controls |
χ2A | χ2AA | ||
|---|---|---|---|---|---|---|---|---|
| Count | Freq | Count | Freq | |||||
| chr17:44039691A>G (rs17650901) | G/G | H2/H2 | 163 | 0.04 | 644 | 0.06 | 1.9 × 10−9 | 7.7 × 10−9 |
| G/A | H1/H2 | 1,221 | 0.33 | 4,098 | 0.36 | |||
| A/A | H1/H1 | 2,372 | 0.63 | 6,491 | 0.58 | |||
| chr17:44019712G>A (rs242557) | A/A | H1c/H1c | 509 | 0.14 | 1,338 | 0.12 | 5 × 10−3 | 8.8 × 10−3 |
| A/G | H1/H1c | 1,733 | 0.46 | 5,148 | 0.46 | |||
| G/G | H1/H1 | 1,514 | 0.40 | 4,747 | 0.42 | |||
The H1 and H2 haplotype distribution is shown for cases and controls. Count, number of subjects; freq, frequency of the haplotype.
The significant markers at chromosome 19 revealed a (genetically estimated) ancestry-related difference in allele frequencies (Table 3): whereas individuals with FTD showed similar frequencies for the chromosome 19 markers regardless of ancestry, Mediterranean European controls showed remarkably decreased frequencies compared to Central/Nordic European controls (Table 3). This trend was further confirmed in an independent cohort of 731 Italian and 347 Continental European controls (Table 3; additional characterization of the APOE locus [APOE Ɛ4 alleles] is included in the supplemental information [Note S1]).
Table 3.
Ancestry-related difference at the APOE locus
|
Minor allele frequency |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Discovery | 1000G | Discovery Nordic/Central European | Independent Nordic/Central European cohort | Discovery Mediterranean | Independent Italian cohort | |||||||||
| Chr | rs number | Base pair | p value | OR | Allele | Cases | Controls | EUR | Cases | Controls | Controls | Cases | Controls | Controls |
| 19 | rs6857 | 45,392,254 | 6.22E-10 | 1.27 | T | 0.1793 | 0.1508 | 0.165 | 0.1855 | 0.1651 | 0.1686 | 0.1689 | 0.109 | 0.1204 |
| 19 | rs12972970 | 45,387,596 | 2.10E-09 | 1.28 | A | 0.1531 | 0.1283 | 0.1352 | 0.1598 | 0.1422 | 0.147 | 0.1417 | 0.0877 | 0.1026 |
| 19 | rs34342646 | 45,388,130 | 2.35E-09 | 1.276 | A | 0.1571 | 0.1321 | 0.1392 | 0.1636 | 0.1455 | 0.147 | 0.146 | 0.09294 | 0.1026 |
| 19 | rs71352238 | 45,394,336 | 3.87E-09 | 1.273 | C | 0.155 | 0.1304 | 0.1332 | 0.1624 | 0.1444 | 0.1527 | 0.1424 | 0.08945 | 0.1033 |
| 19 | rs2075650 | 45,395,619 | 5.29E-09 | 1.272 | G | 0.1523 | 0.128 | 0.1312 | 0.1598 | 0.1427 | 0.1455 | 0.1396 | 0.08491 | 0.1033 |
| 19 | rs11556505 | 45,396,144 | 5.50E-09 | 1.272 | T | 0.1519 | 0.1278 | 0.1312 | 0.1594 | 0.1426 | 0.1455 | 0.1392 | 0.08456 | 0.1033 |
The allele frequencies of the significant markers at the APOE locus are shown in different cohorts (the entire discovery cohort; the 1000 Genomes European cohort, the discovery cohort divided into Central European/Nordic and Mediterranean European populations, and independent samples of Central European/Nordic and Mediterranean European ancestry). 1000G, 1000 Genomes.
Central/Nordic and Mediterranean European independent analyses
To better understand the population-specific contribution to the discovery analysis results, we split the discovery cohort into Central/Nordic (2,359 cases and 8,371 controls) and Mediterranean Europeans (1,397 cases and 2,862 controls), based on genetically estimated ancestry (PCA based), and performed association analysis for these two sub-cohorts separately (Figures S5 and S6). The genomic inflation factors were λ = 1.0135 and λ = 1.0356, respectively.
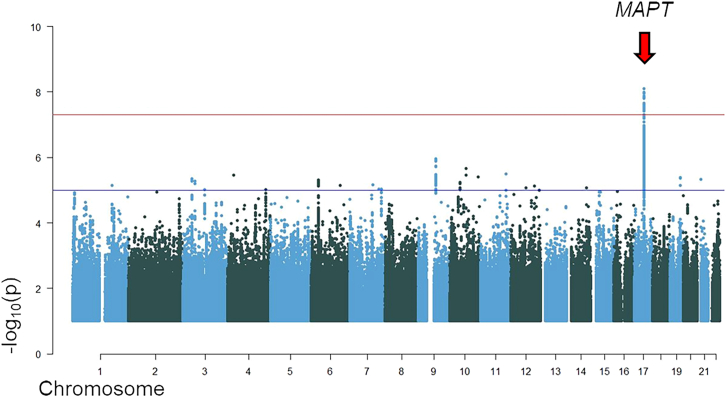
Association analysis for the Central/Nordic European cohort revealed one genome-wide-significant signal on chromosome 17 at the MAPT locus (rs199443; p = 1.08 × 10−8; β = 0.256 [C = risk allele, major allele]; SE = 0.045). In the replication set (also subdivided in Central/Nordic [405 cases and 3,400 controls] and Mediterranean Europeans [524 cases and 675 controls]), the chromosome 17 top SNP reached p value = 2.19 × 10−2 (β = 0.249; SE = 0.109), and joint analysis confirmed genome-wide-significant statistics (p = 8.02 × 10−10; β = 0.256; SE = 0.042), with the effect being in the same direction (Figure 3; Table 4). The APOE locus signal on chromosome 19 (rs6857) was not genome-wide significant (p value = 7.9 × 10−5) in the discovery Central/Nordic European cohort; it reached p value = 2.55 × 10−2 in the replication set and remained not significant (p = 6.8 × 10−6; β = 0.183; SE = 0.041) after joint analysis (Table 4).
Figure 3.
Manhattan plot for discovery-phase Central/Nordic European cohort
Red arrow indicates the genome-wide significant signal (chromosome 17). The plot is cut at −log10(p) = 1. The gene symbols represent the locus and do not necessarily imply functional/biological relevance.
Table 4.
Top marker summary statistics for the Central European/Nordic and Mediterranean European cohorts
|
Discovery |
Replication |
Joint Analysis |
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Minor allele frequency | PLINK | Minor allele frequency | PLINK | METAL | |||||||||||||||||||||
| Cohort | rs number | Chr | Base pair | Locus | A1/A2 | Controls | Cases | 1000G IBR | 1000G TSI | 1000G GBR | 1000G CEU | OR | Beta | SE | p value | Controls | Cases | OR | Beta | SE | p value | Beta | OR | SE | p value |
| Central European/Nordic | rs199443 | 17 | 44,819,565 | MAPT | T/C | 0.215 | 0.175 | - | - | 0.231 | 0.197 | 1.293 | 0.256 | 0.045 | 1.08E-08 | 0.211 | 0.179 | 1.283 | 0.249 | 0.109 | 2.19E-02 | 0.256 | 1.291 | 0.042 | 8.018E-10 |
| rs6857 | 19 | 45,392,254 | APOE | T/C | 0.165 | 0.186 | - | - | 0.170 | 0.197 | 1.193 | 0.176 | 0.045 | 7.90E-05 | 0.168 | 0.203 | 1.235 | 0.211 | 0.095 | 2.55E-02 | 0.183 | 1.201 | 0.041 | 6.799E-06 | |
| Mediterranean European | rs199443 | 17 | 44,819,565 | MAPT | T/C | 0.250 | 0.207 | 0.262 | 0.308 | - | - | 1.184 | 0.170 | 0.070 | 1.52E-02 | 0.265 | 0.234 | 1.142 | 0.133 | 0.108 | 2.19E-01 | 0.159 | 1.172 | 0.059 | 6.80E-03 |
| rs6857 | 19 | 45,392,254 | APOE | T/C | 0.109 | 0.169 | 0.164 | 0.117 | - | - | 1.457 | 0.376 | 0.082 | 4.00E-06 | 0.124 | 0.165 | 1.281 | 0.248 | 0.126 | 4.88E-02 | 0.338 | 1.402 | 0.068 | 7.67E-07 | |
Discovery, replication, and joint analysis statistics are shown for the top markers (smallest p value) at the significant loci. 1000G, 1000 Genomes MAF; GBR, British in England and Scotland; CEU, Northern Europeans from Utah; IBR, Iberian populations in Spain; TSI, Tuscans from Italy.
Association analysis for the Mediterranean cohort did not yield significant results (probably because of power issues due to the small Mediterranean cohort size). However, it is worth noting that, although the signal on chromosome 17 showed pjoint = 6.80 × 10−3, the signal on chromosome 19 was suggestive (pjoint = 7.67 × 10−7; β = 0.338; SE = 0.068) (Figure S7; Table 4).
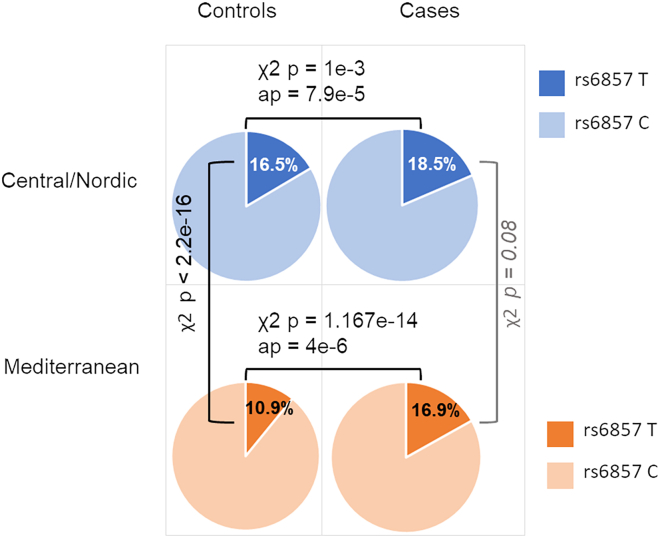
To further shed light on the APOE locus signal on chromosome 19 (rs6857) in the two cohorts, we performed χ2 test (cases vs. controls) for rs6857 in the Central/Nordic European and Mediterranean European cohorts (Figure 4): the unadjusted p value for the former group was 1 × 10−3, while for the latter it was 1.2 × 10−14 (and driven by the control frequencies).
Figure 4.
Risk allele frequencies of top marker (rs6857) at the chromosome 19 locus in the Central/Nordic and Mediterranean European discovery cohorts
Uncorrected p values calculated via χ2 are reported; association p values (ap) are reported for the case-control comparisons.
In summary, taken together, these data suggest that, in our extended cohort, the signal at the MAPT locus appeared to be mainly driven by the Central/Nordic European cohort and that at the APOE locus by the Mediterranean European cohort.
Functional analysis
We found no meaningful impacted biological pathways using the full distribution of SNP p values (Table S5). The assessment of potential effects on expression and splicing (e/sQTL) exerted by the risk markers highlighted in the current study revealed the following (Table S6): (1) there was no significant effect on expression or splicing of in-cis genes for the APOE locus marker (rs6857); (2) the MAPT locus marker (rs199443) revealed effects on both expression and splicing, in cis, in brain cortex, affecting genes involved in transcription regulation (e.g., KANSL1 antisense RNA 1 [KANSL1-AS1; MIM: 612452]), protein trafficking (e.g., ADP ribosylation factor-like GTPase 17A [ARL17A]), and signal transduction (e.g., corticotropin-releasing hormone receptor 1 [CRHR1; MIM: 122561]), as well as MAPT itself; and (3) the top marker on chromosome 3 (rs1009966) revealed effects on expression in the cerebellum and splicing in brain cortex of the ribosomal protein SA gene (RPSA; MIM: 150370); the RPSA protein is involved in stabilizing ribosomal subunits and in the signal transduction as a cell surface receptor for laminin.34
Heritability and genetic correlation analyses
Heritability estimation with LDSC regression for FTD summary statistics returned h2 = 0.118 (se = 0.02) (Table S7A). Genetic correlation of FTD with five other traits related to neurodegenerative diseases revealed positive significant correlation with all of them (p < 0.05), with the exception of Alzheimer disease-related dementia (ADRD) GWAS26 (see Table S7B). The largest genetic correlations were with LBD (rg = 0.91), ALS (rg = 0.71), and AD (rg = 0.55), and the overall results of this analysis indicated substantial shared genetic liability or potential misdiagnoses of FTD with all of the following conditions: LBD, ALS, AD, and PD. BUHMBOX was run to compare the APOE region (chr19:44.4–46.5 Mb) between AD and FTD, resulting in p = 0.006 and Mendelian randomization (MR) p = 5.8 × 10−6.
Cross-check of markers across different neurodegenerative diseases
We sought to verify the statistics of key markers previously identified in other neurodegenerative conditions (etiologically close to FTD) in our current dataset (Table S8). The TMEM106B and GFRA2 markers11,12 showed negligible association in our dataset (likely because of the current study design, i.e., clinical cohort excluding individuals with GRN mutations). Some of the significant ALS markers as shown in van Rheenen et al., 2021 and Nicolas et al., 201828,35 reached p = 1.07 × 10−3 and p = 1.6 × 10−4 for the UNC13A (MIM: 609894) and MOBP loci markers (rs12973192 [proxy] and rs631312, respectively) with similar effects sizes (OR = 1.1), while the C9orf72 marker (rs3849943, highly significant in ALS) showed negligible association in our dataset (probably because of the current study design, i.e., individuals with C9orf72 expansion were excluded from the study). Most historically established AD markers—including rs6656401 and rs679515 (CR1; MIM: 120620), rs6733839 (BIN1; MIM: 601248), rs9331896 and rs11787077 (CLU; MIM: 185430), and rs6605556 (HLA)26,36,37—showed negligible association. PICALM (rs3851179; MIM: 603025) showed p = 6.5 × 10−3 and, interestingly, MAPT (rs199515) p = 1.1 × 10−9.26,36
In addition, for the APOE locus, we assessed several markers that were extensively reported as being genome-wide significant in AD (rs4420638, rs439401, and rs7412)37,38: one of these markers showed significant p value levels, though displaying smaller effect size in the current cohort (rs4420638; p = 9.6 × 10−8 with OR = 3.95 in AD and 1.2 in FTD), while the other two did not (rs439401; p = 8.1 × 10−1 and rs7412; p = 1.9 × 10−4). Conversely, we also sought to verify the MOBP hit reported as a novel potential FTD locus in the current work (rs1009966) in AD datasets: it showed negligible p values in two recent AD GWASs, i.e., p = 3.4 × 10−1 and 7.5 × 10−1.25,26 A previously reported hit at the HLA locus14 reached suggestive significance (rs9268877; p = 9.57 × 10−7). Furthermore, whereas several previously reported PSP and CBD risk variants39,40,41 showed negligible association, some PSP risk markers reached p values in the range of 10−3 (rs1411478 [STX6; MIM: 603944] and rs11568563 [SLCO1A2; MIM: 602883]) and 10−4 (rs1768208 [MOBP]) in our dataset. Finally, the MAPT locus risk variants previously reported in PD, AD, PSP, and CBD26,39,40,42 all resulted genome-wide significant in our dataset.
Discussion
In the current work, we analyzed 4,685 sFTD cases and 15,308 controls, looking for common genetic determinants contributing to increased risk of sFTD. Compared to the previous work,14 we here increased sample size and improved the cohort (updated diagnoses) and provided insights highlighting a cluster of variants at the MAPT (rs199443; chromosome 17) and APOE (rs6857; chromosome 19) loci and a candidate locus on chromosome 3 (rs1009966 in the intergenic region between RPSA and MOBP) contributing to increased risk for sFTD by potentially mediating effects on expression and splicing of functionally relevant in-cis genes at the MAPT43 and RPSA-MOBP loci.
Further analysis of the MAPT locus suggested an increase of the H1/H1 haplotype and of the H1c clade in individuals with sFTD. This signature was previously reported in two small FTD cohorts from France and the UK,44,45 and it also appears to be consistent across different neurodegenerative conditions, as the H1 haplotype was shown to be associated with AD,46,47 CBD, PSP,48,49,50 and PD.51 This, and the fact that the MAPT locus risk variants previously reported in PD, AD, PSP, and CBD26,39,40,42 all resulted genome-wide significant in our dataset (and in complete and perfect LD with each other), supports the notion that the MAPT locus may be a common denominator at the crossroad of multiple etiologically close conditions such as FTD, AD,26 PSP, CBD,50 and PD.52
The association with the RPSA-MOBP locus appears of particular interest considering its involvement in PSP39 and ALS.28 Our reported marker with the lowest p value after joint analysis (rs1009966; p = 2.36 × 10−8, OR = 1.16) is in complete (D’ = 1) although not perfect (R2 = 0.3) LD in the European population (LDlink [https://ldlink.nih.gov/?tab=home] CEU 1000 Genomes, GRCh37) with the PSP (rs1768208) and ALS (rs631312) genome-wide-significant markers, suggesting potential pleiotropy at this locus across a subset of individuals with FTD, PSP, and ALS.
The signal at the APOE locus appeared to be population specific and driven by the control frequencies. More specifically, although the frequencies of the markers at the APOE locus were relatively similar in the Central/Nordic European (18.5%) and Mediterranean European (16.9%) cases making up the study cohort, they were significantly less frequent in Mediterranean (10.9%) compared to Central/Nordic European (16.5%) controls. The frequency patterns observed in our Central/Nordic vs. Mediterranean European controls were further supported by the 1000 Genomes cohort (IBR + TSI vs. CEU + GBR general populations), an independent cohort of controls (Italian and Central/Nordic Europeans), and a population-specific study analyzing APOE frequencies in the Treviso longevity (TRELONG) longitudinal study.53 All this taken together may suggest there being a genuine variation in the genetic architecture of these two (close yet different) European population groups at this locus.54,55 The link to the APOE locus could lead to different explanations/interpretations: (1) it may reflect a presence of individuals in our cohort with behavioral variant AD that mimics FTD56,57; (2) it may underpin comorbidities at play, given that some individuals with FTD coming to autopsy can show concurrent AD or vascular changes in addition to changes attributed directly to FTD58,59,60; (3) it may represent a genuine association pertaining to a population-specific subset of individuals with FTD. In relation to this latter point, it is worth noting that, per study design, we excluded all samples diagnosed with the logopenic variant in the current work (thus reducing potential diagnosis bias), and we verified that many of the historical AD risk loci (e.g., CR1, BIN1, CLU) reached negligible p values in our cohort, while the MOBP locus hit reported for FTD in the current work shows negligible p values in two recent AD GWASs. Moreover, a literature survey supports the notion of the involvement of the APOE locus in some forms of FTD.52,61,62,63,64
Previous work14 reported association with the HLA locus; in the current analyses, although genome-wide-significant levels are not reached, we show rs9268863 as the top SNP at the HLA locus (p = 7.86 × 10−7, OR = 1.2; A = risk allele) in the discovery analysis, with rs9268877 and rs9268863 being in complete (D’ = 1) and perfect (R2 = 1) LD in the European population (LDlink [https://ldlink.nih.gov/?tab=home] CEU 1000 Genomes, GRCh37). Clearly, despite other work supporting the notion of an involvement of immune system processes in the pathogenesis of sFTD,65 additional studies are needed to shed light on the level and type of involvement of the HLA locus in sFTD.
In summary, we report that the MAPT (pointing to the H1c clade), APOE, and RPSA-MOBP loci contribute to increased genetic risk of sFTD pathogenesis. Notably, the involvement of the MAPT locus in sFTD, and its association also with AD, PSP, PD, and CBD, strongly suggests potential common genetic pleiotropy for these neurological conditions at this locus. Moreover, our results pointing at the RPSA-MOBP locus shed light on an additional potential genetic overlap between FTD and PSP and between FTD and ALS. It is worth noting that the diagnosis of neurodegenerative diseases is challenging due to subtle overlap of clinical presentations62; therefore, future studies will need to be powered enough to allow for further assessment of pleiotropy vs. potential misdiagnosis (e.g., through structural equation modeling66). Moreover, an interesting way forward will be to not only continue addressing the genetic etiology of familial vs. sporadic FTD separately, but also broaden approaches by carefully designing studies to test for modifiers and the polygenic nature of Mendelian cases, provided adequate power.
Our study, however, gives grounds to the inference that FTD may be part of a wider spectrum of genetically mediated neurodegenerative conditions67 spanning from ALS to AD phenotypes and encompassing PSP, PD, and CBD. The current study also lays the foundations for future work aimed at further characterizing population-specific features of potential FTD-discriminant APOE haplotype(s) and the functional involvement and contribution of the MAPT H1c haplotype and RPSA-MOBP loci to pathogenesis in brain cortex of sporadic forms of FTD.
Data and code availability
The accession number for the summary statistics (available for download from UCL Research Data Repository) is https://doi.org/10.5522/04/25600692.v1. Underlying participant-level data are available to potential collaborators, where individual-study data-access consent and pre-approved ethics are obtained to permit such data sharing: directly contact the site PIs (Note S2) to obtain both permission and consent to access their samples’ data.
Acknowledgments
This work was funded by the Alzheimer’s Society (grant number 284 to R.F.). Additional funding info and acknowledgments can be found in the supplemental information.
Author contributions
C.M.—whole-data analyses, data generation, writing of the paper; D.A.K.—whole-data analyses, data generation, revision of the paper; R.F.—funding, project design, genotyping, analyses, data generation, initial drafting of the paper; G.L.—analysis support; B.C.—curation of metadata, revision of the paper; V.S.—curation of metadata, revision of the paper; E.J.—genotyping, revision of the paper; M.M.X.T.—genotyping, provision of control samples, revision of the paper; M.A.N.—previous analyses, data generation, project design, revision of the paper; P.M.—funding, project design, provision of control samples, revision of the paper; A.B.S.—funding, project design, revision of the paper; J.H.—funding, project design, revision of the paper; V.E.-P.—project design, supervision, writing of the paper; D.A., G.F., E.S., S.B.-C., S.W.S.—control collection and characterization, revision of the paper; M.K., G.K.M.—genotyping, revision of the paper; all other authors— sample collection and characterization, revision of the paper.
Declaration of interests
O.A.A. has received speakers’ honoraria from Janssen, Lundbeck, and Sunovion and is a consultant to Cortechs.ai. C.C. received research support from GSK and EISAI. The funders of the study had no role in the collection, analysis, or interpretation of data; in the writing of the report; or in the decision to submit the paper for publication. C.C. is a member of the advisory board of Vivid Genomics and Circular Genomics. M.A.N. and H.L.L. hold part of a competitive contract awarded to Data Tecnica International LLC by the National Institutes of Health to support open science research. M.A.N. currently serves on the scientific advisory board for Character Bio Inc. and Neuron23 Inc. I.R.M. receives license royalties for patent related to PGRN therapy and is a member of the scientific advisory committee for Prevail Therapeutics. H.R.M. is employed by UCL. In the last 12 months he reports paid consultancy from Roche, Aprinoia, AI Therapeutics, and Amylyx; lecture fees/honoraria from BMJ, Kyowa Kirin, and Movement Disorders Society; and research grants from Parkinson’s UK, Cure Parkinson’s Trust, PSP Association, Medical Research Council, and the Michael J. Fox Foundation. H.R.M. is a co-applicant on a patent application related to C9ORF72—Method for diagnosing a neurodegenerative disease (PCT/GB2012/052140). R.P. has received honoraria for advisory boards and speaker engagements from Roche, EISAI, Eli Lilly, Biogen, Janssen-Cilag, Astra Zeneca, Schwabe, Grifols, Novo Nordisk, and Tabuk. R.S.-V. served in advisory board meetings for Wave Life Sciences, Ionis, and Novo Nordisk; has received personal fees for participating in educational activities from Janssen, Roche Diagnostics, and Neuraxpharm; and has received funding to her institution for research projects from Biogen and Sage Pharmaceuticals. S.W.S. received research support from Cerevel Therapeutics and is a member of the scientific advisory board of the Lewy Body Dementia Association and the Multiple System Atrophy Coalition. J.S.Y. serves on the scientific advisory board for the Epstein Family Alzheimer’s Research Collaboration. J.H. does consulting and gives talks for Eli-Lilly, Roche, and Eisai and is on the Ceracuity advisory board.
Published: June 17, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.05.017.
Contributor Information
Claudia Manzoni, Email: c.manzoni@ucl.ac.uk.
Valentina Escott-Price, Email: escottpricev@cardiff.ac.uk.
Supplemental information
References
- 1.Rabinovici G.D., Miller B.L. Frontotemporal lobar degeneration: epidemiology, pathophysiology, diagnosis and management. CNS Drugs. 2010;24:375–398. doi: 10.2165/11533100-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ratnavalli E., Brayne C., Dawson K., Hodges J.R. The prevalence of frontotemporal dementia. Neurology. 2002;58:1615–1621. doi: 10.1212/wnl.58.11.1615. [DOI] [PubMed] [Google Scholar]
- 3.Neary D., Snowden J.S., Gustafson L., Passant U., Stuss D., Black S., Freedman M., Kertesz A., Robert P.H., Albert M., et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 4.Rascovsky K., Hodges J.R., Knopman D., Mendez M.F., Kramer J.H., Neuhaus J., van Swieten J.C., Seelaar H., Dopper E.G.P., Onyike C.U., et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134:2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorno-Tempini M.L., Hillis A.E., Weintraub S., Kertesz A., Mendez M., Cappa S.F., Ogar J.M., Rohrer J.D., Black S., Boeve B.F., et al. Classification of primary progressive aphasia and its variants. Neurology. 2011;76:1006–1014. doi: 10.1212/WNL.0b013e31821103e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rohrer J.D., Warren J.D. Phenotypic signatures of genetic frontotemporal dementia. Curr. Opin. Neurol. 2011;24:542–549. doi: 10.1097/WCO.0b013e32834cd442. [DOI] [PubMed] [Google Scholar]
- 7.Murley A.G., Coyle-Gilchrist I., Rouse M.A., Jones P.S., Li W., Wiggins J., Lansdall C., Rodríguez P.V., Wilcox A., Tsvetanov K.A., et al. Redefining the multidimensional clinical phenotypes of frontotemporal lobar degeneration syndromes. Brain. 2020;143:1555–1571. doi: 10.1093/brain/awaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halliday G., Bigio E.H., Cairns N.J., Neumann M., Mackenzie I.R.A., Mann D.M.A. Mechanisms of disease in frontotemporal lobar degeneration: gain of function versus loss of function effects. Acta Neuropathol. 2012;124:373–382. doi: 10.1007/s00401-012-1030-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Zee J., Gijselinck I., Dillen L., Van Langenhove T., Theuns J., Engelborghs S., Philtjens S., Vandenbulcke M., Sleegers K., Sieben A., et al. A pan-European study of the C9orf72 repeat associated with FTLD: geographic prevalence, genomic instability, and intermediate repeats. Hum. Mutat. 2013;34:363–373. doi: 10.1002/humu.22244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Deerlin V.M., Sleiman P.M.A., Martinez-Lage M., Chen-Plotkin A., Wang L.S., Graff-Radford N.R., Dickson D.W., Rademakers R., Boeve B.F., Grossman M., et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat. Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pottier C., Zhou X., Perkerson R.B., 3rd, Baker M., Jenkins G.D., Serie D.J., Ghidoni R., Benussi L., Binetti G., López de Munain A., et al. Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: a genome-wide association study. Lancet Neurol. 2018;17:548–558. doi: 10.1016/S1474-4422(18)30126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pottier C., Ren Y., Perkerson R.B., 3rd, Baker M., Jenkins G.D., van Blitterswijk M., DeJesus-Hernandez M., van Rooij J.G.J., Murray M.E., Christopher E., et al. Genome-wide analyses as part of the international FTLD-TDP whole-genome sequencing consortium reveals novel disease risk factors and increases support for immune dysfunction in FTLD. Acta Neuropathol. 2019;137:879–899. doi: 10.1007/s00401-019-01962-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrari R., Hernandez D.G., Nalls M.A., Rohrer J.D., Ramasamy A., Kwok J.B.J., Dobson-Stone C., Brooks W.S., Schofield P.R., Halliday G.M., et al. Frontotemporal dementia and its subtypes: a genome-wide association study. Lancet Neurol. 2014;13:686–699. doi: 10.1016/S1474-4422(14)70065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichler E.E., Flint J., Gibson G., Kong A., Leal S.M., Moore J.H., Nadeau J.H. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrari R., Manzoni C., Hardy J. Genetics and molecular mechanisms of frontotemporal lobar degeneration: an update and future avenues. Neurobiol. Aging. 2019;78:98–110. doi: 10.1016/j.neurobiolaging.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Strong M.J., Abrahams S., Goldstein L.H., Woolley S., McLaughlin P., Snowden J., Mioshi E., Roberts-South A., Benatar M., HortobáGyi T., et al. Amyotrophic lateral sclerosis - frontotemporal spectrum disorder (ALS-FTSD): Revised diagnostic criteria. Amyotroph. Lateral Scler. Frontotemporal Degener. 2017;18:153–174. doi: 10.1080/21678421.2016.1267768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nalls M.A., Bras J., Hernandez D.G., Keller M.F., Majounie E., Renton A.E., Saad M., Jansen I., Guerreiro R., Lubbe S., et al. NeuroX, a fast and efficient genotyping platform for investigation of neurodegenerative diseases. Neurobiol. Aging. 2015;36 doi: 10.1016/j.neurobiolaging.2014.07.028. 1605 e7-e1605.e1.605E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blauwendraat C., Faghri F., Pihlstrom L., Geiger J.T., Elbaz A., Lesage S., Corvol J.C., May P., Nicolas A., Abramzon Y., et al. NeuroChip, an updated version of the NeuroX genotyping platform to rapidly screen for variants associated with neurological diseases. Neurobiol. Aging. 2017;57:247.e9–247.e13. doi: 10.1016/j.neurobiolaging.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graffelman J., Moreno V. The mid p-value in exact tests for Hardy-Weinberg equilibrium. Stat. Appl. Genet. Mol. Biol. 2013;12:433–448. doi: 10.1515/sagmb-2012-0039. [DOI] [PubMed] [Google Scholar]
- 21.Lin D.Y., Sullivan P.F. Meta-analysis of genome-wide association studies with overlapping subjects. Am. J. Hum. Genet. 2009;85:862–872. doi: 10.1016/j.ajhg.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., ReproGen Consortium. Psychiatric Genomics Consortium. Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3. Duncan L., et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunkle B.W., Grenier-Boley B., Sims R., Bis J.C., Damotte V., Naj A.C., Boland A., Vronskaya M., van der Lee S.J., Amlie-Wolf A., et al. Author Correction: Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019;51:1423–1424. doi: 10.1038/s41588-019-0495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellenguez C., Küçükali F., Jansen I.E., Kleineidam L., Moreno-Grau S., Amin N., Naj A.C., Campos-Martin R., Grenier-Boley B., Andrade V., et al. New insights into the genetic etiology of Alzheimer's disease and related dementias. Nat. Genet. 2022;54:412–436. doi: 10.1038/s41588-022-01024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nalls M.A., Blauwendraat C., Vallerga C.L., Heilbron K., Bandres-Ciga S., Chang D., Tan M., Kia D.A., Noyce A.J., Xue A., et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Rheenen W., van der Spek R.A.A., Bakker M.K., van Vugt J.J.F.A., Hop P.J., Zwamborn R.A.J., de Klein N., Westra H.J., Bakker O.B., Deelen P., et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat. Genet. 2021;53:1636–1648. doi: 10.1038/s41588-021-00973-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chia R., Sabir M.S., Bandres-Ciga S., Saez-Atienzar S., Reynolds R.H., Gustavsson E., Walton R.L., Ahmed S., Viollet C., Ding J., et al. Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat. Genet. 2021;53:294–303. doi: 10.1038/s41588-021-00785-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han B., Pouget J.G., Slowikowski K., Stahl E., Lee C.H., Diogo D., Hu X., Park Y.R., Kim E., Gregersen P.K., et al. A method to decipher pleiotropy by detecting underlying heterogeneity driven by hidden subgroups applied to autoimmune and neuropsychiatric diseases. Nat. Genet. 2016;48:803–810. doi: 10.1038/ng.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caffrey T.M., Joachim C., Paracchini S., Esiri M.M., Wade-Martins R. Haplotype-specific expression of exon 10 at the human MAPT locus. Hum. Mol. Genet. 2006;15:3529–3537. doi: 10.1093/hmg/ddl429. [DOI] [PubMed] [Google Scholar]
- 32.Valenca G.T., Srivastava G.P., Oliveira-Filho J., White C.C., Yu L., Schneider J.A., Buchman A.S., Shulman J.M., Bennett D.A., De Jager P.L. The Role of MAPT Haplotype H2 and Isoform 1N/4R in Parkinsonism of Older Adults. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiGiacomo V., Meruelo D. Looking into laminin receptor: critical discussion regarding the non-integrin 37/67-kDa laminin receptor/RPSA protein. Biol. Rev. Camb. Philos. Soc. 2016;91:288–310. doi: 10.1111/brv.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicolas A., Kenna K.P., Renton A.E., Ticozzi N., Faghri F., Chia R., Dominov J.A., Kenna B.J., Nalls M.A., Keagle P., et al. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron. 2018;97:1267–1288. doi: 10.1016/j.neuron.2018.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naj A.C., Jun G., Beecham G.W., Wang L.S., Vardarajan B.N., Buros J., Gallins P.J., Buxbaum J.D., Jarvik G.P., Crane P.K., et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Rojas I., Moreno-Grau S., Tesi N., Grenier-Boley B., Andrade V., Jansen I.E., Pedersen N.L., Stringa N., Zettergren A., Hernández I., et al. Common variants in Alzheimer's disease and risk stratification by polygenic risk scores. Nat. Commun. 2021;12:3417. doi: 10.1038/s41467-021-22491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Höglinger G.U., Melhem N.M., Dickson D.W., Sleiman P.M., Wang L.S., Klei L., Rademakers R., de Silva R., Litvan I., Riley D.E., et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat. Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kouri N., Ross O.A., Dombroski B., Younkin C.S., Serie D.J., Soto-Ortolaza A., Baker M., Finch N.C.A., Yoon H., Kim J., et al. Genome-wide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy. Nat. Commun. 2015;6:7247. doi: 10.1038/ncomms8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez-Contreras M.Y., Kouri N., Cook C.N., Serie D.J., Heckman M.G., Finch N.A., Caselli R.J., Uitti R.J., Wszolek Z.K., Graff-Radford N., et al. Replication of progressive supranuclear palsy genome-wide association study identifies SLCO1A2 and DUSP10 as new susceptibility loci. Mol. Neurodegener. 2018;13:37. doi: 10.1186/s13024-018-0267-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang D., Nalls M.A., Hallgrímsdóttir I.B., Hunkapiller J., van der Brug M., Cai F., International Parkinson's Disease Genomics Consortium. 23andMe Research Team. Kerchner G.A., Ayalon G., et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson's disease risk loci. Nat. Genet. 2017;49:1511–1516. doi: 10.1038/ng.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reus L.M., Pasaniuc B., Posthuma D., Boltz T., International FTD-Genomics Consortium. Pijnenburg Y.A.L., Ophoff R.A. Gene Expression Imputation Across Multiple Tissue Types Provides Insight Into the Genetic Architecture of Frontotemporal Dementia and Its Clinical Subtypes. Biol. Psychiatry. 2021;89:825–835. doi: 10.1016/j.biopsych.2020.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughes A., Mann D., Pickering-Brown S. Tau haplotype frequency in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Exp. Neurol. 2003;181:12–16. doi: 10.1016/s0014-4886(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 45.Verpillat P., Camuzat A., Hannequin D., Thomas-Anterion C., Puel M., Belliard S., Dubois B., Didic M., Michel B.F., Lacomblez L., et al. Association between the extended tau haplotype and frontotemporal dementia. Arch. Neurol. 2002;59:935–939. doi: 10.1001/archneur.59.6.935. [DOI] [PubMed] [Google Scholar]
- 46.Pastor P., Moreno F., Clarimón J., Ruiz A., Combarros O., Calero M., López de Munain A., Bullido M.J., de Pancorbo M.M., Carro E., et al. MAPT H1 Haplotype is Associated with Late-Onset Alzheimer's Disease Risk in APOEϵ4 Noncarriers: Results from the Dementia Genetics Spanish Consortium. J. Alzheimers Dis. 2016;49:343–352. doi: 10.3233/JAD-150555. [DOI] [PubMed] [Google Scholar]
- 47.Sánchez-Juan P., Moreno S., de Rojas I., Hernández I., Valero S., Alegret M., Montrreal L., García González P., Lage C., López-García S., et al. The MAPT H1 Haplotype Is a Risk Factor for Alzheimer's Disease in APOE epsilon4 Non-carriers. Front. Aging Neurosci. 2019;11:327. doi: 10.3389/fnagi.2019.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Houlden H., Baker M., Morris H.R., MacDonald N., Pickering–Brown S., Adamson J., Lees A., Rossor M., Quinn N., Kertesz A., et al. Corticobasal degeneration and progressive supranuclear palsy share a common tau haplotype. Neurology. 2001;56:1702–1706. doi: 10.1212/wnl.56.12.1702. [DOI] [PubMed] [Google Scholar]
- 49.Pittman A.M., Myers A.J., Abou-Sleiman P., Fung H.C., Kaleem M., Marlowe L., Duckworth J., Leung D., Williams D., Kilford L., et al. Linkage disequilibrium fine mapping and haplotype association analysis of the tau gene in progressive supranuclear palsy and corticobasal degeneration. J. Med. Genet. 2005;42:837–846. doi: 10.1136/jmg.2005.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yokoyama J.S., Karch C.M., Fan C.C., Bonham L.W., Kouri N., Ross O.A., Rademakers R., Kim J., Wang Y., Höglinger G.U., et al. Shared genetic risk between corticobasal degeneration, progressive supranuclear palsy, and frontotemporal dementia. Acta Neuropathol. 2017;133:825–837. doi: 10.1007/s00401-017-1693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Healy D.G., Abou-Sleiman P.M., Lees A.J., Casas J.P., Quinn N., Bhatia K., Hingorani A.D., Wood N.W. Tau gene and Parkinson's disease: a case-control study and meta-analysis. J. Neurol. Neurosurg. Psychiatry. 2004;75:962–965. doi: 10.1136/jnnp.2003.026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferrari R., Wang Y., Vandrovcova J., Guelfi S., Witeolar A., Karch C.M., Schork A.J., Fan C.C., Brewer J.B., International FTD-Genomics Consortium IFGC Genetic architecture of sporadic frontotemporal dementia and overlap with Alzheimer's and Parkinson's diseases. J. Neurol. Neurosurg. Psychiatry. 2017;88:152–164. doi: 10.1136/jnnp-2016-314411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gustafson D.R., Mazzuco S., Ongaro F., Antuono P., Forloni G., Albani D., Gajo G.B., Durante E., Caberlotto L., Zanardo A., et al. Body mass index, cognition, disability, APOE genotype, and mortality: the "Treviso Longeva" Study. Am. J. Geriatr. Psychiatry. 2012;20:594–602. doi: 10.1097/JGP.0b013e31823031a4. [DOI] [PubMed] [Google Scholar]
- 54.Corbo R.M., Scacchi R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE∗4 a 'thrifty' allele? Ann. Hum. Genet. 1999;63:301–310. doi: 10.1046/j.1469-1809.1999.6340301.x. [DOI] [PubMed] [Google Scholar]
- 55.Moreno-Grau S., de Rojas I., Hernández I., Quintela I., Montrreal L., Alegret M., Hernández-Olasagarre B., Madrid L., González-Perez A., Maroñas O., et al. Genome-wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with Alzheimer's disease and three causality networks: The GR@ACE project. Alzheimers Dement. 2019;15:1333–1347. doi: 10.1016/j.jalz.2019.06.4950. [DOI] [PubMed] [Google Scholar]
- 56.Ossenkoppele R., Pijnenburg Y.A.L., Perry D.C., Cohn-Sheehy B.I., Scheltens N.M.E., Vogel J.W., Kramer J.H., van der Vlies A.E., La Joie R., Rosen H.J., et al. The behavioural/dysexecutive variant of Alzheimer's disease: clinical, neuroimaging and pathological features. Brain. 2015;138:2732–2749. doi: 10.1093/brain/awv191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ossenkoppele R., Singleton E.H., Groot C., Dijkstra A.A., Eikelboom W.S., Seeley W.W., Miller B., Laforce R.J., Scheltens P., Papma J.M., et al. Research Criteria for the Behavioral Variant of Alzheimer Disease: A Systematic Review and Meta-analysis. JAMA Neurol. 2022;79:48–60. doi: 10.1001/jamaneurol.2021.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joshua Stevenson-Hoare A.-K.S., Sandor C., Hardy J., Escott-Price V. New cases of dementia are rising in elderly populations in Wales, UK. J. Neurol. Sci. 2023 doi: 10.1016/j.jns.2023.120715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naasan G., Rabinovici G.D., Ghosh P., Elofson J.D., Miller B.L., Coppola G., Karydas A., Fong J., Perry D., Lee S.E., et al. Amyloid in dementia associated with familial FTLD: not an innocent bystander. Neurocase. 2016;22:76–83. doi: 10.1080/13554794.2015.1046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rabinovici G.D., Carrillo M.C., Forman M., DeSanti S., Miller D.S., Kozauer N., Petersen R.C., Randolph C., Knopman D.S., Smith E.E., et al. Multiple comorbid neuropathologies in the setting of Alzheimer's disease neuropathology and implications for drug development. Alzheimers Dement. 2017;3:83–91. doi: 10.1016/j.trci.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferrari R., Grassi M., Salvi E., Borroni B., Palluzzi F., Pepe D., D'Avila F., Padovani A., Archetti S., Rainero I., et al. A genome-wide screening and SNPs-to-genes approach to identify novel genetic risk factors associated with frontotemporal dementia. Neurobiol. Aging. 2015;36 doi: 10.1016/j.neurobiolaging.2015.06.005. 2904 e13-e2904.e2.904E26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ingannato A., Bagnoli S., Bessi V., Ferrari C., Mazzeo S., Sorbi S., Nacmias B. Intermediate alleles of HTT: A new pathway in longevity. J. Neurol. Sci. 2022;438 doi: 10.1016/j.jns.2022.120274. [DOI] [PubMed] [Google Scholar]
- 63.Mishra A., Ferrari R., Heutink P., Hardy J., Pijnenburg Y., Posthuma D., International FTD-Genomics Consortium Gene-based association studies report genetic links for clinical subtypes of frontotemporal dementia. Brain. 2017;140:1437–1446. doi: 10.1093/brain/awx066. [DOI] [PubMed] [Google Scholar]
- 64.Rubino E., Vacca A., Govone F., De Martino P., Pinessi L., Rainero I. Apolipoprotein E polymorphisms in frontotemporal lobar degeneration: a meta-analysis. Alzheimers Dement. 2013;9:706–713. doi: 10.1016/j.jalz.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 65.Malpetti M., Cope T.E., Street D., Jones P.S., Hezemans F.H., Mak E., Tsvetanov K.A., Rittman T., Bevan-Jones W.R., Patterson K., et al. Microglial activation in the frontal cortex predicts cognitive decline in frontotemporal dementia. Brain. 2023 doi: 10.1093/brain/awad078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grotzinger A.D., Rhemtulla M., de Vlaming R., Ritchie S.J., Mallard T.T., Hill W.D., Ip H.F., Marioni R.E., McIntosh A.M., Deary I.J., et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat. Hum. Behav. 2019;3:513–525. doi: 10.1038/s41562-019-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koretsky M.J., Alvarado C., Makarious M.B., Vitale D., Levine K., Bandres-Ciga S., Dadu A., Scholz S.W., Sargent L., Faghri F., et al. Genetic risk factor clustering within and across neurodegenerative diseases. Brain. 2023 doi: 10.1093/brain/awad161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the summary statistics (available for download from UCL Research Data Repository) is https://doi.org/10.5522/04/25600692.v1. Underlying participant-level data are available to potential collaborators, where individual-study data-access consent and pre-approved ethics are obtained to permit such data sharing: directly contact the site PIs (Note S2) to obtain both permission and consent to access their samples’ data.