Summary
Understanding the contribution of gene-environment interactions (GxE) to complex trait variation can provide insights into disease mechanisms, explain sources of heritability, and improve genetic risk prediction. While large biobanks with genetic and deep phenotypic data hold promise for obtaining novel insights into GxE, our understanding of GxE architecture in complex traits remains limited. We introduce a method to estimate the proportion of trait variance explained by GxE (GxE heritability) and additive genetic effects (additive heritability) across the genome and within specific genomic annotations. We show that our method is accurate in simulations and computationally efficient for biobank-scale datasets.
We applied our method to common array SNPs (MAF ), fifty quantitative traits, and four environmental variables (smoking, sex, age, and statin usage) in unrelated white British individuals in the UK Biobank. We found 68 trait-E pairs with significant genome-wide GxE heritability () with a ratio of GxE to additive heritability of on average. Analyzing million imputed SNPs (MAF ), we documented an approximate increase in genome-wide GxE heritability compared to array SNPs. We partitioned GxE heritability across minor allele frequency (MAF) and local linkage disequilibrium (LD) values, revealing that, like additive allelic effects, GxE allelic effects tend to increase with decreasing MAF and LD. Analyzing GxE heritability near genes highly expressed in specific tissues, we find significant brain-specific enrichment for body mass index (BMI) and basal metabolic rate in the context of smoking and adipose-specific enrichment for waist-hip ratio (WHR) in the context of sex.
Keywords: gene-environment interaction, gene-context interaction, gene-drug interaction, scalable variance component analysis, genetic architecture of gene-environment interactions, complex traits, patitioning GxE heritability, noise heterogeneity, UK Biobank
Pazokitoroudi et al. introduced a scalable method to estimate trait variance explained by gene-environment interactions across the genome and within specific genomic annotations. Application of the method in the UK Biobank uncovered significant genome-wide GxE heritability and enrichment of GxE heritability within population and functional genomic annotations.
Introduction
Variation in a complex trait is modulated by an interplay between genetic and environmental factors. Characterizing the effects of gene-environment interactions (GxE) on complex trait variation has the potential to shed light on biological mechanisms underlying the trait,1,2,3 inform public health measures,4 identify sources of missing heritability,5 and improve the accuracy and portability of trait prediction.6,7 The growth of biobanks that collect genetic and deep phenotypic data (that span disease outcomes, clinical labs, lifestyle factors, and environmental exposures) across large numbers of individuals offers the possibility to gain novel insights into GxE.3,8 Nevertheless, characterizing GxE has proved challenging due, in part, to the small effect sizes of individual genetic variants.9,10
A potentially powerful methodological approach aims to quantify GxE effects aggregated across a set of variants without needing to pinpoint individual variants. In this approach, the proportion of trait variation explained by GxE (GxE heritability or ) is estimated by fitting a class of variance components models where the model parameters, i.e., the variance components, are informative of . Methods for estimating using this approach include GCTA-GxE,11 multitrait GREML (MV-GREML),5 random regression GREML (RR-GREML),5,12 and whole-genome reaction norm model (RNM) and its multitrait version (MRNM).13 All of these methods (except RNM) are able to account for differences in the noise or residual variance across environments (noise heterogeneity), which is important to mitigate biases in GxE heritability estimates.13,14 However, these methods work with discrete-valued environmental variables, with RNM and MRNM further restricted to fit bivariate and univariate environments, respectively. A more recent general framework, GxEMM,14 can be applied to both discrete and continuous environmental variables while modeling noise heterogeneity. However, none of these methods are practical for biobank-scale datasets with sample sizes in the hundreds of thousands and genetic variants in the millions. Two recent methods, GPLEMMA15 and MEMMA,16 attempt to scale GxE heritability estimation to large-scale datasets but do not model noise heterogeneity. A more recent method, MonsterLM,17 has been shown to be feasible for biobank-scale datasets and to produce unbiased estimates in many scenarios. However, MonsterLM requires SNPs to be filtered to common variants with low levels of linkage disequilibrium (LD), which may limit its application to discover GxE. As a result, current methods for estimating GxE heritability either do not scale to the biobank setting or are susceptible to biased estimates. Additional insights into the architecture of GxE can be gleaned if we can move beyond genome-wide estimates of GxE heritability and estimate GxE heritability across specific genomic annotations such as minor allele frequency (MAF), LD, and functional genomic annotations.
We propose a scalable and robust method, GENIE (gene-environment interaction estimator) that can estimate the proportion of trait variance explained by GxE and additive genetic effects (additive heritability). Using extensive simulations and real data analysis, we show that GENIE accurately estimates and provides calibrated tests of due to its ability to account for noise that is heterogeneous across environments. Importantly, GENIE is scalable: able to estimate GxE on datasets with hundreds of thousands of individuals, millions of SNPs, and tens of environmental variables in several hours. The ability of GENIE to be applied to large-scale datasets is important for power: we show that GENIE has adequate power to detect as low as 2% across a sample of unrelated individuals. Finally, GENIE is versatile: able to handle multiple environmental variables (discrete or continuous) and to estimate not only genome-wide but also partition across genomic annotations (both overlapping and non-overlapping).
To demonstrate its utility, we first applied GENIE to estimate the genome-wide on common SNPs ( SNPs with MAF ) and four environmental variables (smoking, sex, age, and statin usage) for fifty quantitative phenotypes measured across unrelated white British individuals in the UK Biobank (UKB). Second, we leveraged the scalability of GENIE to partition across common and low-frequency imputed SNPs ( with ) in UKB. We partitioned into genomic annotations based on the MAF and local LD score of each SNP to investigate the variation in GxE effects with population genetic features and to estimate genome-wide that includes the contribution of both common and low-frequency SNPs. Finally, we applied GENIE to assess whether shows tissue-specific enrichment by analyzing each of 53 tissue-specific gene sets identified from the GTEx dataset.18
Material and methods
Generalized GxE linear mixed model
Let denote a genotype matrix, denote a matrix of environmental variables, denote a matrix of fixed-effect covariates, and denote an N-vector of phenotypes. We assume the following linear mixed model:
| (Equation 1) |
Here, denotes an arbitrary distribution with mean and covariance , denotes l-th column of , and denotes row-wise Kronecker product. denotes the M-vector of SNP effect sizes, denotes the P-vector of fixed effects, denotes the M-vector of genetic effect sizes in the context of environment l (GxE effects) while denotes the N-vector of noise-by-environment effect sizes for environment l, and denotes the N-vector of noise. , , , and denote the residual variance, additive genetic, gene-by-environment, and noise-by-environment variance components, respectively. These variance components can then be transformed into the additive heritability or the proportion of variance explained by additive effects ( associated with ) and the GxE heritability or the proportion of variance explained by interactions of genetics with a given environment ( associated with ). The noise-by-environment matrix for environment l is obtained as the row-wise Kronecker product between the identity matrix and the environment vector so that the vector of environment-specific noise for each individual i (due to environment l) will be given by . In the simplest case of a binary environment that is coded as , the phenotype of an individual whose environmental variable is set to value 1 will have an additional contribution of noise () relative to an individual whose environment variable is set to 0. Further, all individuals whose environmental variable takes the value 1 will have an additional term that contributes to their phenotypic variance, quantified by , relative to individuals with environmental variable 0. This formulation generalizes to settings where the environment is coded as categorical (but with values different from ) and to continuous-valued environments. We now refer to the noise-by-environment (or heterogeneous noise) component as the NxE component and the variance as the NxE variance in the following sections.
Estimation in the GxE linear mixed model
We assume without loss of generality that is centered, and the columns of and are standardized. To estimate the variance components of our linear mixed model (LMM), we use a method-of-moments (MoM) estimator that searches for parameter values so that the population moments are close to the sample moments. Since , we derived the MoM estimates by equating the population covariance to the empirical covariance. For simplicity, we exclude the matrix of covariates from the model in the following derivation as the covariates can be efficiently projected out of the phenotype, genotypes, and interaction terms with minimal additional cost (Note S1).
For compactness, we denote , for , for , and . The population covariance is given by
| (Equation 2) |
where
and
Using as our estimate of the empirical covariance, we need to solve the following least squares problem to find the variance components.
| (Equation 3) |
The MoM estimator satisfies the following normal equations:
| (Equation 4) |
where is matrix with entries , and and are vectors with entries and , respectively, for .
The heritability associated with component i for a component that represents additive genetic or GxE effects (equivalently, the proportion of variance explained by component i) is defined as follows:
| (Equation 5) |
The aforementioned definition of heritability holds when the columns of each of the matrices have zero means and N is large. To explicitly ensure that the columns of GxE matrices also have zero means, a column consisting of all ones is included in the covariate matrix. Consequently, when the covariates are projected out of the GxE matrices (Note S1), it guarantees that all columns have zero means.
Computational challenges
Computing the coefficients of the system of linear Equation 4 presents computational challenges. The main computational bottleneck is the evaluation of the quantities for which requires . Therefore, the total time complexity for exact MoM is , imposing challenging memory or computation requirements for Biobank-scale data (N in the hundreds of thousands, M in the millions, and L in the hundreds or thousands).
Scalable estimation
Instead of computing the exact value of , GENIE uses a randomized estimator of the trace.19 This estimator uses the fact that for a given matrix , is an unbiased estimator of ( where is a random vector with mean zero and covariance ). Hence, we can estimate the values , as follows:
| (Equation 6) |
Here, are B independent random vectors with zero mean and covariance . In GENIE, we draw these random vectors independently from a standard normal distribution. Note that computing by using the above estimator involves matrix-vector multiplications, which are repeated B times. Therefore, the total running time is .
Moreover, we can leverage the structure of the genotype matrix, which only contains entries in . For a fixed genotype matrix , we can improve the per iteration time complexity of matrix-vector multiplication from to by using the Mailman algorithm.20 Solving the normal equations takes time so that for a small number of components (L), the overall time complexity of our algorithm is .
Standard errors of the estimates
We used a computationally efficient block jackknife21 to compute standard errors of the estimates, which does not require any assumptions on the distribution of the effect sizes. Each jackknife subsample was created by removing a block of the genotype matrix, and we approximated the true SE by the jackknife estimate. Specifically, if we partition the genotype into J non-overlapping blocks , , where is the heritability estimate based on (removing from ), and is the mean of estimates across J jackknife subsamples. The jackknife estimator was implemented efficiently in GENIE to compute the estimate in time . In our analysis, we used blocks defined over SNPs to compute the standard errors of the estimates.
Partitioning GxE heritability across the genome
Although the model defined in Equation 1 is beneficial in quantifying genome-wide GxE effects for a given E, it is interesting to identify and interpret the interaction of E with specific regions of the genome, such as SNPs with a particular range of minor allele frequencies or SNPs that lie within genes expressed specifically in a tissue. Following our previous work,21 the genotype component can be assigned to T (potentially overlapping) components with respect to a set of annotations (such as MAF/LD or functional annotations). Thus, we extend our model as follows:
| (Equation 7) |
Here, is the genotype of annotation t with SNPs, and refers to the effect sizes of SNPs in annotation t in the context of environment l. Analogously, refers to the variance component for SNPs in annotation t in the context of environment l while refers to the GxE heritability associated with annotation t in the context of environment l.
Given estimated GxE heritabilities under the above model, we define the enrichment of genetic effects in annotation t in the context of environment l (also termed GxE enrichment) as follows:
| (Equation 8) |
Estimating GxE in the UK Biobank
We applied GENIE to the UKB8 where we considered environmental variables such as smoking status, sex, age, and statin medication. The analyses utilized the UKB Resource under application 331277, with participants’ informed consents verified by the UKB.22 For every environmental variable, we applied GENIE to estimate additive heritability () and GxE heritability () across 50 quantitative phenotypes (in a model that included the environmental variable as a main effect and accounted for noise heterogeneity) (Table S2). In this study, we restricted our analysis to SNPs that were present in the UKB Axiom array used to genotype the UK Biobank. SNPs with greater than missingness and MAF smaller than were removed. Moreover, SNPs that failed the Hardy-Weinberg test at significance threshold were removed. We restricted our study to self-reported British white ancestry individuals who are degree relatives that are defined as pairs of individuals with kinship coefficient .8 Furthermore, we removed individuals who are outliers for genotype heterozygosity and/or missingness. Finally, we obtained a set of individuals and SNPs for real data analyses. No LD pruning or filtering was required by GENIE subsequently.
We included age, sex, , age sex, sex, and the top 20 genetic principal components (PCs) as covariates in our analysis for all traits. We always include the environmental variable as a covariate in these analyses. We used PCs precomputed by the UKB from a superset of individuals. Additional covariates were used for waist-to-hip ratio (adjusted for body mass index [BMI]) and diastolic/systolic blood pressure (adjusted for cholesterol-lowering medication, blood pressure medication, insulin, hormone replacement therapy, and oral contraceptives). We standardized environmental variables in our primary analyses. The standardized coding for binary environmental variables has an invariant property in the sense that the covariance matrix would be the same regardless of flipping the coding. We also considered the binary coding of environmental variables to be relevant. Statin usage is defined as a binary environmental variable based on C10AA (the American Therapeutic Chemical [ATC] code of statin), which corresponds to taking any subtype of statin medications. Smoking status is defined as a categorical variable with three possible values (never, previous, and current).
We considered an additional analysis of genotypes at high-quality imputed SNPs (with a hard call threshold of 0.2 and an INFO score ) with MAF in the unrelated white British individuals. We further restricted our analyses to SNPs that are under Hardy-Weinberg equilibrium () and are confidently imputed in more than of the individuals. Additionally, we excluded SNPs in the MHC region, resulting in a total of SNPs.
In our analysis of heritability partitioned based on MAF-LD annotations (primarily for the imputed SNPs), we divided SNPs into eight annotations based on quartiles of the LD scores (computed in-sample using GCTA) and two MAF bins (MAF and MAF ). In our analyses of heritability partitioned based on tissue-specific gene expression annotations, we used the annotations for the 53 tissue-specific genes generated by Finucane et al.18 using a matrix of normalized gene expression values from the Genotype-Tissue Expression (GTEx) database, which included samples from various tissues, including the focal tissue. The authors calculated a t statistic for each gene to determine its specific expression in the focal tissue and ranked all genes based on their t-statistics. They defined the top of genes with the highest t statistic as the set of specifically expressed genes for the focal tissue. To improve the accuracy of the gene set construction, 100-kb windows are added on either side of the transcribed region of each gene in the set of specifically expressed genes to generate a genome annotation that corresponds to the focal tissue.
Results
Calibration and power
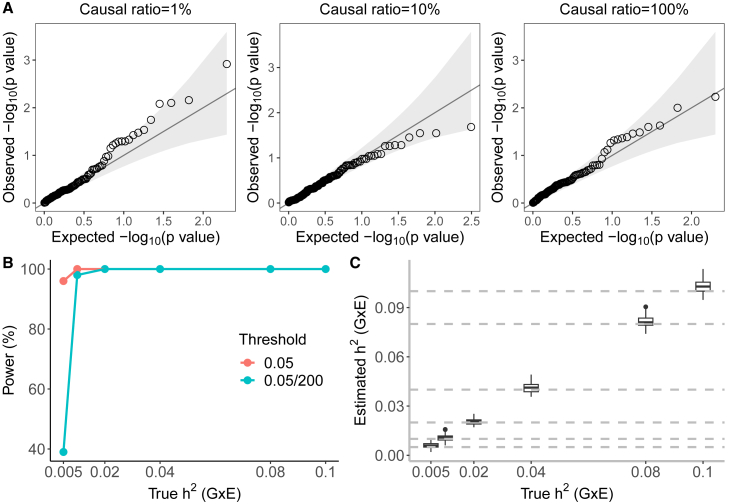
We assessed the false positive rate of tests of GxE heritability based on GENIE in simulations under different genetic architectures with no GxE heritability. For each architecture, we simulated 100 phenotype replicates across unrelated white British individuals in the UKB and SNPs with MAF genotyped on the UKB genotyping array. We chose statin usage in the UKB as the environmental variable. We varied the percentage of causal SNPs while fixing the additive heritability at . We ran GENIE with random vectors (see the following section on the choice of the number of random vectors).
Across all simulations, the false positive rate of rejecting the null hypothesis of no GxE heritability is controlled at levels 0.05 and (we consider this threshold, which controls for the number of trait-environmental variable [trait-E] pairs that we test in UKB): the average rejection at is and for and , respectively (Figure 1A).
Figure 1.
Calibration and power of GENIE in simulations ( unrelated individuals, SNPs)
(A) Q-Q plot of p values (of a test of the null hypothesis of zero GxE heritability) when GENIE is applied to phenotypes simulated in the absence of GxE effects. Each panel contains 100 replicates of phenotypes simulated with additive heritability and varying proportions of causal variants. The causal ratios are the same for the G and GxE components (), and the causal SNPs for the GxE component are independently sampled to those for the additive genetic component. Across all architectures, the mean of rejection at is and for and , respectively ( is not significantly different from the nominal rate of ).
(B) The power of GENIE across genetic architectures as a function of GxE heritability. We report power for p value thresholds of .
(C) The accuracy of estimates obtained by GENIE. Across all simulations, statin usage in UKB was used as the environmental variable.
To measure the power of GENIE to detect GxE heritability, we simulated phenotypes with a non-zero GxE heritability. Across genetic architectures, we varied the GxE heritability with no noise heterogeneity while fixing the additive heritability at 0.25 and the percentage of causal SNPs at (these are default values of additive heritability and causal ratio across our simulations unless otherwise specified). We also tested GENIE by varying the sample size from to . We simulated 100 replicates for every genetic architecture. Let be the estimate of and be the jackknife estimate of the standard error on the i-the replicate for . We computed the p value of a test of the null hypothesis of no on the i-th replicate from the Z score defined as for . We reported the percentage of replicates with p value as the power of GENIE on a given genetic architecture for a p value threshold of t.
GENIE has adequate power to detect GxE effects with in a sample of unrelated individuals at (Figure 1B). The power increases from around to as the sample size grows from to when at and remains almost for as the sample size reaches (Figure S2A). Additionally, GENIE yields unbiased estimates of GxE heritability (Figure 1C), and the SEs estimated by GENIE were concordant with the true SEs (Figure S3).
Next, we assessed the accuracy of GENIE in a setting with multiple environmental variables. We simulated phenotypes from a sub-sampled set of UKB genotypes, choosing a subset of individuals and SNPs on chromosome 1 of the UKB Axiom array. We considered a setting with environmental variables with , five environmental variables with , three environmental variables with , and two with . We generated 100 replicates of simulated phenotypes for each set of parameters. We find that GENIE obtains estimates of that are accurate across the environmental variables (Figure S1; Table S1).
Impact of randomization on GxE estimates
We investigated the impact of randomization on the estimates obtained by GENIE by comparing it to the exact MoM. Since exact MoM is computationally infeasible for large sample sizes, we choose to experiment on a small-scale dataset consisting of unrelated white British individuals and SNPs selected from the UK Biobank array SNPs on chromosome 1. We generated 100 replicates of phenotypes with no noise heterogeneity, , and varying with standardized smoking status as the environment variable. We ran GENIE using the G + GxE + NxE model with random vectors and compared the estimated G and GxE heritability with the results from GCTA-HE regression11 (exact MoM) on G and GxE GRM matrices. We see that exact MoM has a slightly higher statistical power than GENIE (with an increase in power of to across the values tested; Figure S4A). Further, the relative contribution of randomization to the SE of GENIE remains around despite the variation of power difference across simulations (Figure S4B).
Confirming that randomization makes a modest difference on the power of GENIE, we quantified the effect of the number of random vectors. We explored the choice of the number of random vectors in two ways. First, we quantified the contribution of randomization to the SE of the GxE estimator in GENIE. We simulated 100 phenotypes where . We compared the SE of GxE estimates with random vectors run 100 times over one of the replicates (the contribution of the randomization to the SE) to the SE of GxE estimates across 100 replicates to determine that, with , randomization contributes to about 30% of the total SE across various sample sizes (Figure S5). Second, we verified that our GxE estimates are highly correlated for the choice of random vectors vs. (Pearson’s correlation ; Figure S6). These results lead us to conclude that random vectors provide stable estimates, and we use this setting in our remaining analyses.
Noise heterogeneity
Previous studies have shown that accounting for noise heterogeneity (NxE component) is essential to avoid false positives and inflation in estimates of GxE effects.13,14,23 To demonstrate the importance of modeling NxE, we simulated phenotypes in the presence of NxE effect such that (we set to 0.04 when ). We ran GENIE, in turn, with and without the NxE component. Across all simulations, the model that does not account for the NxE component (G + GxE) yields statistically significant upward bias in its GxE estimates (relative bias ranges from to across genetic architectures) while the model that fits a noise heterogeneity component (G + GxE + NxE) achieves unbiased estimates of GxE (Figure S7).
Comparison with existing methods in simulations
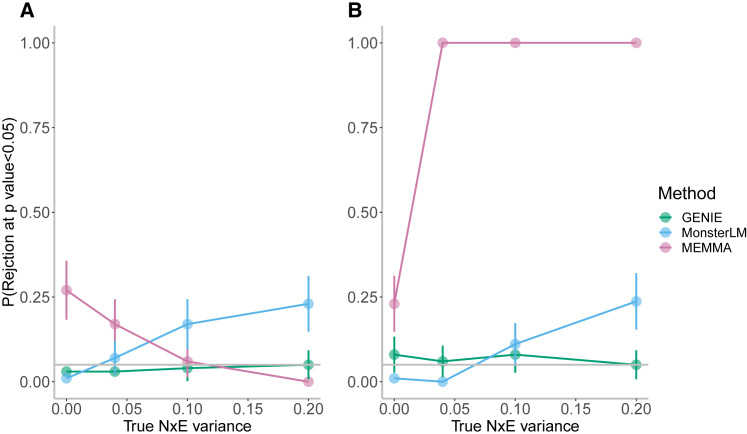
We compared the calibration of tests of GxE from GENIE with MEMMA16 and MonsterLM.17 GPLEMMA15 was excluded due to its focus on multiple environmental variables. We conducted the benchmark experiments on SNPs from a subset of unrelated white British individuals. To ensure a fair comparison with MonsterLM, which requires genotype QC steps, we filtered SNPs by removing those with high LD () and low MAF (MAF ), resulting in SNPs (we report results for GENIE and MEMMA on unfiltered SNPs in Figure S8). We then simulated phenotypes with both continuous (cystatin-C) and discrete (statin usage) environmental variables on the filtered SNPs. In simulations with no GxE or NxE effects, MEMMA had inflated false positive rates while GENIE and MonsterLM were calibrated (Figure 2). The inflated false positive rate for MEMMA in the absence of the NxE effect can be explained by a bias in their estimates of the SE of the variance components (Figure S9). Under scenarios with noise heterogeneity, GENIE remained calibrated while MonsterLM displayed inflation in its false positive rate with increasing NxE variance for both continuous and discrete environment variables. MEMMA showed elevated false positive rates with discrete environment variables, and lower but still inflated false positives with continuous environmental variables (Figure 2).
Figure 2.
Comparisons of false positive rates with existing methods with the presence of noise heterogeneity
False positive rates of tests for GxE heritability across GENIE, MEMMA, and MonsterLM using (A) continuous and (B) discrete environment exposures. We performed simulations with no GxE heritability but with varying magnitudes of the variance of the NxE effect. We computed the false positive rate as the fraction of rejections (p value of a test of the null hypothesis of zero GxE heritability ) over 100 replicates of phenotypes. The phenotypes were simulated from individuals and SNPs filtered from SNPs with the genotype QC steps in MonsterLM: SNPs that failed the Hardy-Weinberg test at the significance threshold were excluded, and highly correlated SNPs with LD and SNPs with MAF were removed. Error bars correspond to the estimated 95% CI of the rejection rate.
Robustness of GENIE in simulations
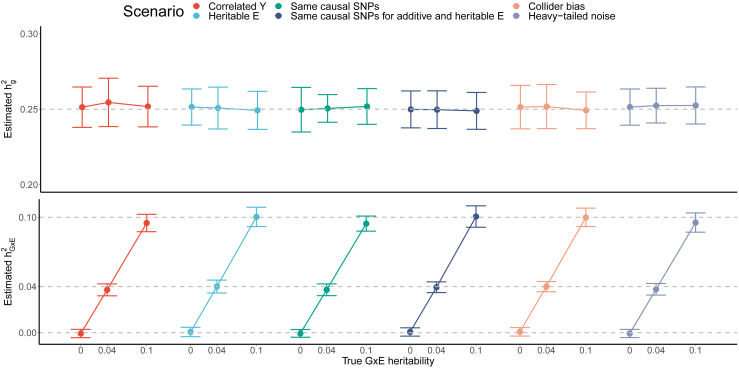
We tested the robustness of GENIE by varying the correlation between the phenotype (Y) and the environment (E), simulating heritable E, imposing that the causal SNPs are the same for the G and GxE components, simulating Y that has the same causal SNPs with the heritable E, and simulating a collider bias scenario. In addition, we also considered a scenario where the environment noise is drawn from a heavy-tailed distribution (see Note S3 for details). In these simulations, we use a continuous environmental exposure (to complement our previous set of simulations that used a discrete environmental exposure, i.e., statin usage). In scenarios where the environmental exposure is heritable, we simulated continuous environmental exposure with specific genetic architecture. In simulations where the environment exposure is not heritable, we use a continuous exposure measured in UKB (cystatin-C). In all simulations, we simulated phenotypes with NxE and varying GxE effects across individuals genotyped at SNPs for 100 replicates. The results summarized in Figure 3 indicate that GENIE obtains accurate estimates across these scenarios.
Figure 3.
Estimation of G and GxE heritability in six simulated scenarios
We investigated the performance of GENIE in estimating G and GxE heritability under six simulated scenarios. (1) Correlated Y: the phenotypes were correlated with the continuous environment exposure, with Pearson’s correlation ; (2) heritable E: the environment exposure E was simulated from the same set of genotype data as in the phenotype simulation, with an additive genetic heritability of 0.1; (3) same causal SNPs: additive genetic causal SNPs completely overlap with GxE causal SNPs; (4) same causal SNPs for additive and heritable E: additive genetic causal SNPs completely overlap with the causal SNPs explaining heritability in E, where E is the same as in scenario (2); (5) collider bias: the phenotype Y and environment exposure E are correlated through an unobserved confounder; we simulated a heritable environment variable with a genetic heritability of 0.1. The phenotypes were then generated to have a Pearson’s correlation with the heritable E. We assumed that the correlation was due to an unobserved confounder.17 (6) Heavy-tailed noise: we drew the environment noise component from the Student’s t-distribution with degrees of freedom = 4. In all scenarios, we simulated 100 replicates of phenotypes with NxE and varying magnitude of GxE effects across individuals genotyped at SNPs. The ground truth GxE heritability was 0, 0.04, and 0.1, with corresponding NxE variance of 0.04, 0.04, and 0.1. The additive genetic heritability was fixed at 0.25. The x and y axes denote the true GxE heritability and the estimated G and GxE heritability. Points and error bars represent the mean and estimated CI, respectively. Across all simulations where there is no GxE, the mean of P(rejection at ) are and for and , respectively ( is not significantly different from the nominal rate of ).
Computational efficiency
We evaluated the runtime of GENIE, MonsterLM, MEMMA, and GCTA(HE) (which implements an exact MoM estimator) with increasing sample size () for a fixed number of SNPs () and a single environmental variable. All methods were run on an Intel(R) Xeon(R) Gold 6140 CPU 2.30GHz, with 187GB RAM. Ten random vectors are used by GENIE and MEMMA. For GENIE, runtime measurements were obtained for the single component and eight MAF/LD components. All other methods fit a single G and GxE variance component. The runtime of GCTA(HE) includes the computation of the GRM matrix. Our comparison used the CPU implementation of MonsterLM, with runtime calculations excluding the preprocessing step for genotype filtering required by MonsterLM. GENIE is highly scalable and can estimate GxE on about individuals and roughly SNPs within an hour, with the eight-component model nearly as efficient as the single-component model (Figure S11).
Estimating GxE in the UKB
We applied GENIE to estimate additive heritability () and GxE heritability () for 50 quantitative phenotypes measured in UKB across unrelated white British individuals. These 50 phenotypes fall into eight broader phenotypic categories (blood biochemistry, kidney biomarkers, anthropometry, lipid metabolism biomarkers, blood pressure, liver biomarkers, lung, and glucose metabolism biomarkers) that have been analyzed in prior works.24,25,26 Following these studies, we applied a rank-based inverse normal transformation to all phenotypes. For certain phenotypes affected by medication usage (systolic/diastolic blood pressure, LDL direct, and total cholesterol), we adopted heuristic adjustments for medication variables.24,27 We then reevaluated the GxE heritability estimates using GENIE (see Note S4 for details). We considered, in turn, smoking status, sex, age, and statin usage as environmental variables. We included each environmental variable as a fixed effect in the relevant analyses. First, we explored the importance of modeling NxE in real data (building on our simulation results). We then analyzed, in turn, common SNPs genotyped on the UKB array (MAF ) and then common and low-frequency imputed SNPs (MAF ). For selected combinations of phenotypes and environmental variables, we also applied GENIE to partition GxE heritability across functional annotations to estimate GxE heritability in genes expressed in specific tissues.
We note that individuals with missing environmental or phenotype data were removed in the implementation of GENIE instead of being imputed by the mean value. We observed that the application of mean imputation to the phenotype results in underestimation of and while mean imputation of the environment variables affected the estimation of but not (Figure S12). We therefore recommend that users leave missing exposure and outcome data as it is when applying GENIE in their analysis based on the simulation results.
Robustness of GENIE in the UKB
We first assessed the robustness of GENIE by estimating under three different models: G, G + GxE, and G + GxE + NxE, where each model is named by the set of variance components fitted jointly. The additive heritability estimates were highly correlated across the models (Pearson’s correlation for every pair of models), leading us to conclude that GENIE provides robust estimates of additive heritability across different models (Figure S13). We observed a significant difference in for a handful of trait-E pairs when estimated with G + GxE and G + GxE + NxE that include alcohol frequency intake and overall health with smoking status, sex, or age as the environmental variable. In previous work,21 we compared the additive estimates from RHE with S-LDSC,28 GRE,29 SumHer,30 and LDSC31 to find that RHE estimates of additive heritability for 22 complex traits are consistent with the existing methods. We additionally compared the additive heritability estimates from GENIE with those obtained using LDSC (run with in-sample LD scores estimated from a subset of unrelated white British individuals in UKB). The estimates of additive from LDSC were compared against those from GENIE with environmental exposures of smoking status, sex, age, and statin. The estimates across 50 traits were consistently correlated for the two methods, with Pearson’s correlations ranging from 0.87 to 0.93 (Figure S14).
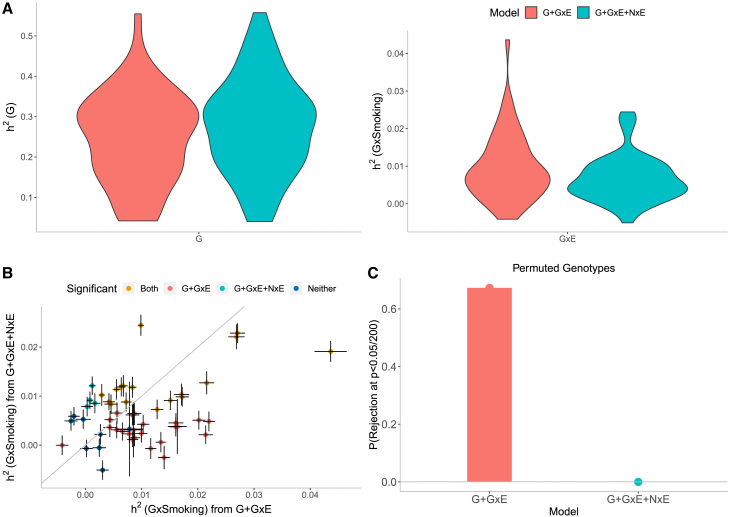
Our simulations in the previous section revealed the importance of modeling noise heterogeneity (Figure S7). To investigate the consequences of modeling NxE in real data, we fitted, in turn, models without and with NxE (in addition to G and GxE components). The number of trait-E pairs with significant () decreased from 135 under the G + GxE model to 68 under the G + GxE + NxE model: changing from 40 to 21 for smoking (Figure 4B), 27 to 28 for sex (Figure S15B), 28 to 12 for age (Figure S16B), and 40 to 7 for statin usage (Figure S17B). For traits with significant , the magnitudes of the estimates varied across the two models: the ratios of estimates under the G + GxE + NxE to the G + GxE model were on average (range: ), (), (), and () for smoking (Figure 4A), sex (Figure S15A), age (Figure S16A), and statin (Figure S17A), respectively. The magnitude of noise heterogeneity across trait-E pairs can be substantial: , , , and of the additive heritability on average for smoking, sex, age, and statin, respectively (Figures S18–S21). To further investigate the effect of modeling NxE, we performed permutation analyses by randomly shuffling the genotypes while preserving the trait-E relationship (a setting where there is expected to be no GxE by construction while the relationship between phenotype and E is preserved). We applied GENIE under the G + GxE and G + GxE + NxE models to each trait-E pair. The false positive rate of rejecting the null hypothesis of no GxE across the trait-E pairs is substantially inflated under the G + GxE model while being controlled under the G + GxE + NxE model (Figures 4C, S15C, S16C, and S17C for smoking, sex, age, and statin respectively). These results indicate that modeling NxE is critical to avoid spurious findings of GxE.
Figure 4.
Effect of noise heterogeneity (NxE) on estimates of heritability associated with GxSmoking across 50 quantitative phenotypes in UKB
Model G + GxE refers to a model with additive and gene-by-environment interaction components where the environmental variable is smoking status. Model G + GxE + NxE refers to a model with additive, gene-by-environment interaction, and noise heterogeneity (noise-by-environment interaction) components.
(A) We ran GENIE under G + GxE and G + GxE + NxE models to assess the effect of fitting an NxE component on the additive and GxE heritability estimates.
(B) Comparison of GxE heritability estimates obtained from GENIE under a G + GxE + NxE model (x axis) to a G + GxE model (y axis). Black error bars mark standard errors centered on the estimated GxE heritability. The color of the dots indicates whether estimates of GxE heritability are significant under each model.
(C) We performed permutation analyses by randomly shuffling the genotypes while preserving the trait-E relationship and applied GENIE in each setting under G + GxE and G + GxE + NxE models. We report the fraction of rejections P(p value of a test of the null hypothesis of zero GxE heritability that accounts for the number of phenotypes tested) over 50 UKB phenotypes.
Gene-by-smoking interaction
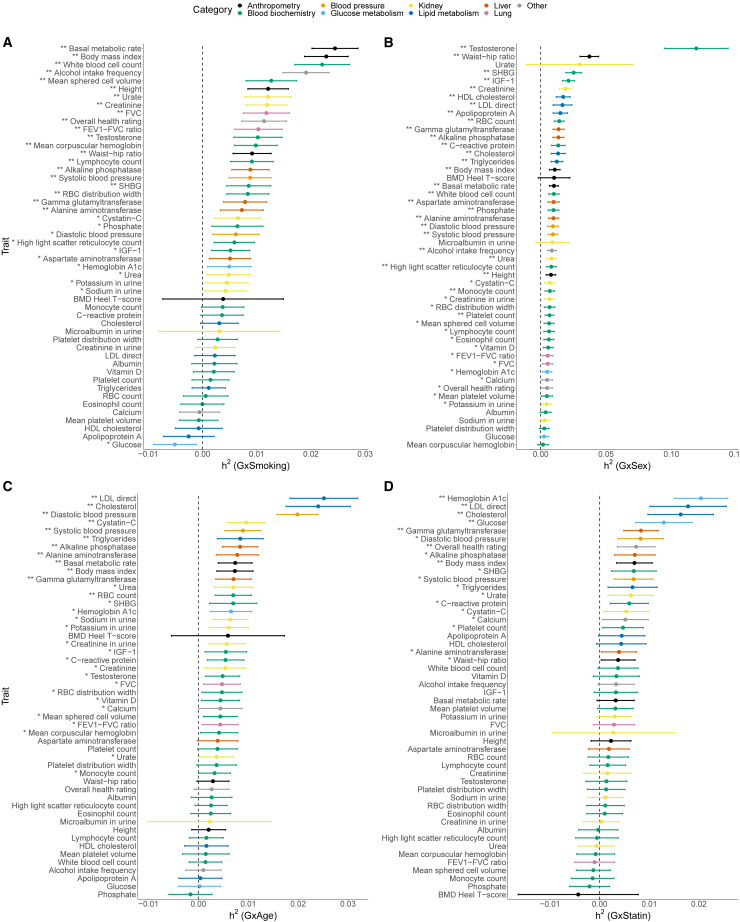
We applied GENIE to estimate the proportion of phenotypic variance explained by gene-by-smoking interactions () for 50 quantitative phenotypes. We find 21 traits showing statistically significant evidence for () with about of on average (Figures 5A and 6A). Two of the traits with the largest were basal metabolic rate and BMI with estimates of and , respectively (estimates remained significant when we used the binary coding of the smoking status variable obtained by merging the categories of never and previous; Figures S25 and S28C). Our estimates are consistent with a previous study that analyzed BMI and lifestyle factors in the UKB to find significant GxE for smoking behavior.5 The estimates for basal metabolic rate and BMI are about and of their respective estimates.
Figure 5.
Estimates of GxE heritability across phenotypes in UKB
Estimates of (A) GxSmoking, (B) GxSex, (C) GxAge, and (D) GxStatin heritability across 50 UKB phenotypes. We applied GENIE to unrelated white British individuals and array SNPs (MAF ). Our model includes the environmental variable as a fixed effect and accounts for noise heterogeneity. The environmental variable is standardized in these analyses. Error bars mark standard errors centered on the point estimates. The asterisk and double asterisk correspond to the nominal and , respectively.
Figure 6.
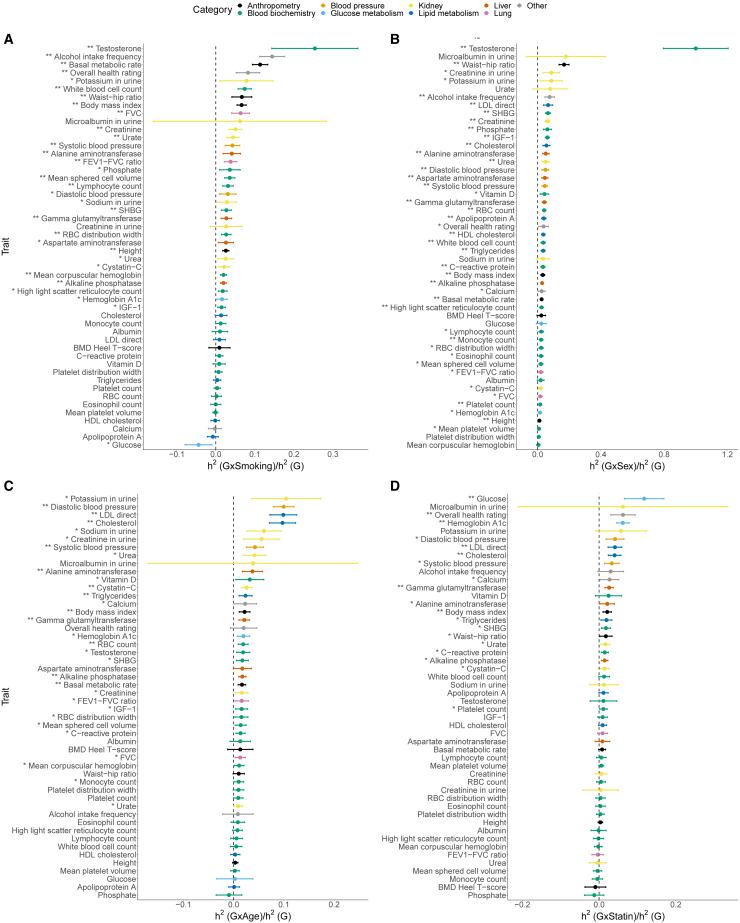
Estimates of the ratio of GxE to additive heritability across phenotypes in UKB
Estimates of the ratio of (A) GxSmoking, (B) GxSex, (C) GxAge, and (D) GxStatin to additive heritability across 50 UKB phenotypes. Error bars mark standard errors centered on the point estimates. The asterisk and double asterisk correspond to the nominal and , respectively.
Gene-by-sex interaction
We find 28 traits with statistically significant () with observed to be on average (Figures 5B and 6B). Serum testosterone levels showed the largest of with the nearly as large as consistent with prior work showing differences in genetic associations32,33 and heritability34 across males and females. Beyond testosterone, we observe significant for several anthropometric traits, such as waist-hip-ratio (WHR) adjusted for BMI ( and ), and lipid measures (results consistent for binary encoding; Figures S26 and S28B) consistent with previous work documenting sex-specific differences in the genetic architecture of anthropometric traits.34,35,36,37,38,39 Consistent with prior GWAS that identified genetic variants with sex-dependent effects,40,41 our analyses of serum urate levels show substantial point estimates of , although these estimates are not statistically significant.
Gene-by-age interaction
We find 12 traits with statistically significant () with observed to be on average (Figures 5C and 6C). Lipid and blood pressure measures show some of the largest (about for LDL and total cholesterol and for diastolic blood pressure). Previous studies have found genetic variants in SORT1 to have age-dependent effects on LDL cholesterol42 and nominal evidence for age-dependent genetic effects on blood pressure regulation.43 We find that BMI shows evidence for significant while WHR does not, expanding on prior work that identified age-dependent genetic variants for BMI but not for WHR in genome-wide association studies (GWASs).36 Interestingly, we used a standardized encoding of age so that GxAge effects capture the interaction of genetic effects on the phenotype as a function of deviation from the mean age in UKB while previous studies typically focus on changes in genetic effects in bins of age. It is plausible that other codings of age, e.g., coding age to measure interactions as a function of older vs. younger individuals, could yield differing results.
Gene-by-statin interaction
We find seven traits that show statistically significant evidence for () with an average ratio of to across traits of (Figures 5D and 6D). We find that LDL and total cholesterol show significant ( and respectively) while HDL cholesterol with a point estimate of of does not (results consistent for binary encoding; Figures S27 and S28A). We observe the largest estimates of for HbA1c and blood glucose measurements ( and respectively), which are interesting in light of statin usage being shown to be associated with a small increase in risk for type 2 diabetes.44
GxE heritability estimates stratified by sex
Quantitative measurements like testosterone concentrations are strongly determined by sex, and therefore, one might be concerned with the possibility of collider bias in estimates on the whole population for these sex-determined traits. To address this issue, we repeated our previous analyses to estimate GxSmoking, GxAge, and GxStatin in females and males separately across the 50 traits. The results show that the sex-specific GxE heritability estimates are overall consistent with the results on all individuals (Pearson’s correlations ranging from 0.67 to 0.80). By comparing GxE heritability estimates between female and male individuals, we noted Pearson’s correlations of 0.50, 0.61, and 0.40 for GxSmoking, GxAge, and GxStatin, respectively (Figures S22–S24). In terms of the GxE heritability of testosterone specifically, we see that is no longer significant for testosterone in female and male individuals (Figure S22) while estimates of overlap with the previous results: and in females and males, respectively, and in the whole population. Hence, the attenuation of our estimates could be explained by the possibility of collider bias or a reduction in power. In general, the phenotypes that have the most significant GxE interactions are in the categories of anthropometry and blood biochemistry for GxSmoking, blood pressure and glucose metabolism for GxAge, and glucose metabolism and lipid metabolism for GxStatin in the sex-stratified analyses. In particular, GxSmoking estimates on BMI, basal metabolic rate, and white blood cell count remain significant for both males and females under . The differences in the GxE estimates between males and females could suggest the presence of sex-specific GxE interaction effects.
Comparison with existing methods on significant trait-E pairs
We compared GxE heritability estimates of MEMMA, MonsterLM, and GENIE on real UKB phenotypes. While the consistency of GxE estimates from methods based on different model assumptions can enhance our confidence in the results, such comparisons have inherent limitations—our simulations have revealed variations in false positive rates among different methods. With these caveats, we evaluated GxE heritability using MonsterLM and MEMMA on 68 significant trait-E pairs detected by GENIE (). We noted Pearson’s correlation between the point estimates of GENIE and MonsterLM and 0.24 between GENIE and MEMMA across the 68 trait-E pairs (Figure S10). The closer alignment between the point estimates by GENIE and MonsterLM can be attributed to the shared consideration of noise heterogeneity within both models.
Estimating GxE heritability from imputed SNPs
We applied GENIE to estimate , , , and attributable to imputed SNPs with MAF . Prior work has shown that analyzing common and low-frequency variants with a single variance component can result in biased estimates of additive heritability.45,46 A solution to this problem involves fitting multiple variance components obtained by partitioning SNPs based on their frequency and local LD scores (as quantified by the LD scores31 or the LDAK scores45).30,46,47,48 We follow this approach by partitioning SNPs into eight annotations based on quartiles of the LD scores and two MAF annotations (MAF and MAF ; material and methods).
We performed simulations to show that GENIE applied with SNPs partitioned based on MAF and LD scores can accurately estimate across varying MAF and LD-dependent genetic architectures while using a single component for all SNPs can lead to substantial biases (Note S2, Figure S29). We applied GENIE using MAF-LD partitions to jointly estimate and (Figures S30–S33). While estimates of from imputed SNPs are largely concordant with the estimates obtained from array SNPs, we identify nine trait-E pairs for which the estimates are significantly different (). In all these cases, estimates from imputed SNPs are higher than those from array SNPs. For example, we estimated for BMI , which is larger than our estimate based on array SNPs as well as a previous estimate of based on common HapMap3 SNPs.5 Across all trait-E pairs, we observed that the average ratio () is 1.17 (1.66, 1.23, 0.71, and 1.17, respectively, for GxSmoking, GxSex, GxAge, and GxStatin; Figure S34). Across trait-E pairs with significant , the average is on the imputed data compared to on array data while the ratio of is on the imputed data compared to on the array data (averaged across trait-E pairs, we estimated on imputed vs. on array data).
We explored the impact of fitting multiple variance components based on MAF and LD by applying GENIE to fit a single GxE and additive variance component using smoking status as the environmental variable. While ten traits showed significant in both analyses, five traits were exclusively significant in the MAF-LD model while one was exclusively significant in the single-component model. Restricting to traits with significant GxSmoking in both models, estimates in the MAF-LD model were about three times those from the single-component model on average (Figure S35). We also investigated whether MAF-LD partitioning affected estimates of obtained from array SNPs. We find that estimates are largely concordant whether obtained from a single component or an MAF-LD partitioned model (ratio of 0.99 on average) consistent with the array SNPs being relatively common (MAF ). Our analysis suggests that partitioning by MAF and LD is helpful for estimating from both common and low-frequency SNPs and the inclusion low-frequency SNPs can increase estimates of for specific traits.
Partitioning GxE heritability across MAF and LD annotations
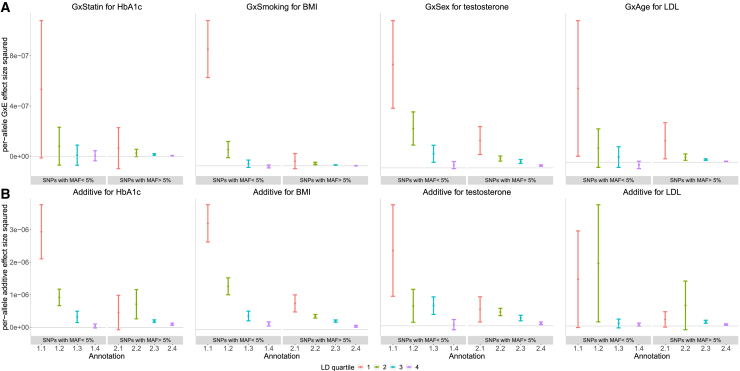
Previous studies have shown that the additive SNP effects increase with decreasing MAF and local levels of LD21,49,50,51 likely due to the effects of negative selection. Similar to previous analyses,15,17 we explored the MAF-LD dependence of SNP effects in the context of specific environmental factors. Our analyses in the preceding section, showing differences in the genome-wide estimates when partitioning by MAF and LD vs. fitting a single variance component, suggest that GxE effects are expected to vary by MAF and LD in a pattern that is distinct from what would be expected when fitting a single variance component, which assumes that the effect size at a SNP varies with its allele frequency f as while not varying with local LD (for a fixed value of the allele frequency f). To explore the MAF-LD dependence of GxE effects, we used GENIE to partition across MAF and LD annotations (while simultaneously partitioning additive heritability) of imputed SNPs divided into eight annotations based on quartiles of LD-scores and two MAF bins (low-frequency bins with MAF and high-frequency bins with MAF ). Within each of these eight bins, we defined the per-allele squared effect size as where is the GxE (or additive) heritability attributed to bin k, is the number of SNPs in bin k, and is the mean MAF in bin k.
For the sake of presentation, we selected one phenotype with high genome-wide GxE heritability for each of the four environmental variables analyzed (Figure 7; see Table S4 for results on all trait-E pairs). Across bins of MAF and LD, the magnitude of additive allelic effects tends to be larger than those of the GxE effects consistent with the genome-wide results. We observed that the per-allele squared GxE effect size tends to increase with lower MAF within a given quartile of LD score and to increase with lower bins of LD score for a fixed MAF bin (Figure 7A). These trends are analogous to the relationship observed for additive per-allele effect sizes (Figure 7B). Across the trait-E pairs, restricting to the lowest quartile of LD scores, low-frequency SNPs tend to have higher per-allele GxE effect sizes compared to high-frequency SNPs: the ratio of in low vs. high MAF bins is , , , and for HbA1c-statin, BMI-smoking, LDL-age, and testosterone-sex, respectively. In the highest quartile of LD scores, we found no statistically significant differences in across low and high MAF SNPs in any of the four trait-E pairs (we also plot the per-standardized genotype additive and GxE heritability, , in Figure S36).
Figure 7.
Per-allele squared GxE and additive effect sizes as a function of MAF and LD
(A) The squared per-allele GxE effect size for four selected pairs of trait and environment (trait-E pairs).
(B) The squared per-allele additive effect size for the same trait-E pairs. The x axis corresponds to MAF-LD annotations where annotation includes SNPs in MAF bin i and LD quartile j where MAF bin 1 and MAF bin 2 correspond to SNPs with MAF and MAF , respectively, while the first quartile of LD scores correspond to SNPs with the lowest LD scores respectively). The y axis shows the per-allele GxE (or additive) effect size squared defined as where is the GxE (or additive) heritability attributed to bin k, is the number of SNPs in bin k, and is the mean MAF in bin k. Error bars mark standard errors centered on the estimated effect sizes.
Partitioning GxE heritability across tissue-specific genes
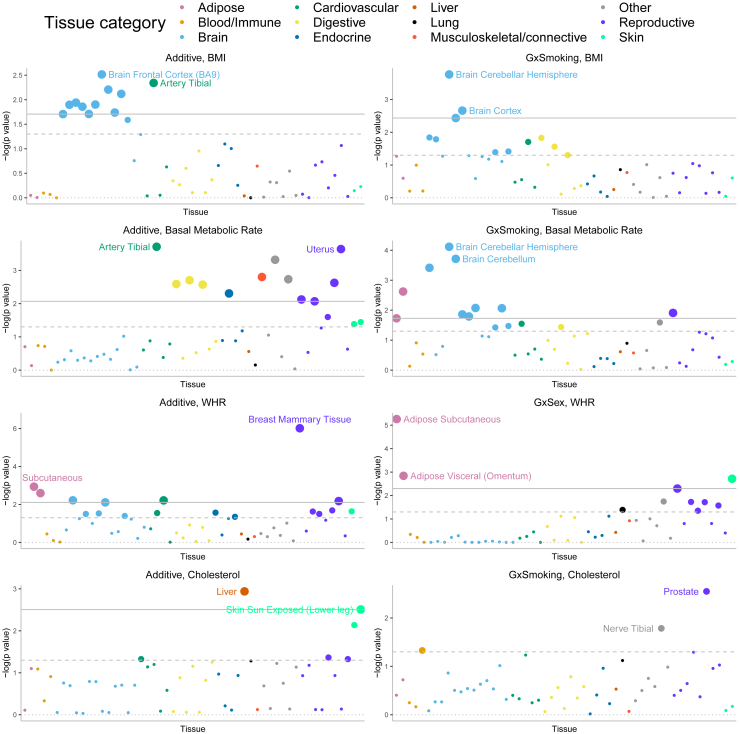
The ability of GENIE to simultaneously estimate multiple, potentially overlapping, additive and GxE variance components enables us to explore how is localized across the genome. Specifically, we set to answer the question of whether is enriched in genes specifically expressed in a given tissue as a means to identify tissues that are relevant to a trait in a specific environmental context.
We applied GENIE to estimate and across each of 53 sets of genomic annotations defined as regions around genes that are highly expressed in a specific tissue in the GTEx dataset18 (Table S3). For each of the four environmental variables, we analyzed only traits with genome-wide significant based on our prior analyses of the array SNPs. For every set of tissue-specific genes, we followed prior work18 by jointly modeling the tissue-specific gene annotation as well as 28 genomic annotations that are part of the baseline LDSC annotations that include genic regions, enhancer regions, and conserved regions.28 Specifically, our model has 29 additive variance components and 29 GxE variance components and estimates the additive and GxE heritability that can be attributed to genes specifically expressed in a tissue while controlling for the effects of the background annotations. A positive represents a positive contribution of genetic effects in a tissue to additive heritability.18 Analogously, a positive represents a positive contribution of genetic effects in this tissue to trait heritability in the context of the specific environment. We test estimates of to answer whether a tissue of interest is enriched for GxE (additive) heritability conditional on the remaining genomic annotations included in the model.
We first verified that our approach is able to detect previously reported enrichments for additive effects such as brain-specific enrichment for BMI and adipose-specific enrichment for WHR (Figure 8).18 Across 68 trait-E pairs with significant genome-wide GxE that we tested, we observed significant enrichment of (FDR ) for at least one tissue in five trait-E pairs (we plot four of these pairs in Figure 8 since the results from the fifth LDL-age are highly correlated with cholesterol-age). Across these trait-E pairs, we documented differential patterns of enrichments for GxE effects compared to additive effects. BMI exhibits brain-specific enrichment of and while WHR exhibits enrichment of and in adipose and breast tissue (in addition to the enrichment of in the uterus and cardiovascular tissues). The adipose-tissue-specific enrichment of in WHR is notable in light of known instances of genes associated with WHR in adipose tissue in a sex-dependent manner. ADAMTS9, a gene involved in insulin sensitivity,35 is specifically expressed in adipose tissue and has been shown to be located near GWAS hits for WHR that are specific to females.35,36,52 The transcription factor, KLF14, is located near a sex-dependent GWAS variant for WHR, type 2 diabetes, and multiple other metabolic and anthropometric traits.53 Further, the expression level of this gene is associated with the GWAS variant in adipose but not with other tissues.53 We also found instances where tissues that are enriched for are distinct from those that are enriched for . We observed that the enrichment of for basal metabolic rate in brain and adipose tissues is distinct from the tissues that are enriched in for the same trait (cardiovascular and digestive tissues) (Figure 8). Finally, we find suggestive evidence that the liver is the most enriched tissue for in HbA1c () as well as for in testosterone (), although neither enrichment is significant at FDR of 0.10. These enrichments recapitulate known biology: the liver-specific enrichment of GxStatin effects for HbA1c reflect the tissues in which the target of statins (HMG-CoA-reductase) is expressed54 while the liver-specific enrichment of GxSex for testosterone is consistent with previous findings implicating CYP3A7, a gene involved in testosterone metabolism that is specifically expressed in the liver and lies within a locus that contains one of the strongest GWAS signals for serum testosterone in females.32
Figure 8.
Partitioning GxE heritability across 53 tissue-specific genes
We plot where p is the corresponding p value of the tissue-specific GxE enrichment defined as . For every tissue-specific annotation, we use GENIE to test whether this annotation is significantly enriched for per-SNP heritability, conditional on 28 functional annotations that are part of the baseline LDSC annotations. The dashed and solid lines correspond to the nominal and FDR threshold, respectively. We have labeled two tissues with the most significant p values for each figure.
Discussion
We have described GENIE, a method that can jointly estimate the proportion of variation in a complex trait that can be attributed to GxE and additive genetic effects. GENIE can also partition GxE heritability across the genome with respect to annotations such as functional and tissue-specific annotations or annotations defined based on the MAF and local LD score of each SNP to localize signals of GxE. GENIE provides well-calibrated tests for the existence of a GxE effect and has high power to detect GxE effects while being scalable to large datasets.
Our simulations and real data analysis results confirm the importance of including noise heterogeneity in GxE models. Simulations comparing the calibration of GENIE to MEMMA and MonsterLM suggest that modeling NxE does not introduce biases in scenarios without noise heterogeneity. Furthermore, it aids in controlling false positive rates when noise heterogeneity exists. In UKB data analyses, we observed that about half of trait-E pairs with significant under the G + GxE model are no longer significant under the G + GxE + NxE model. Consistent with this observation, we estimated a substantial contribution of noise heterogeneity to trait variation. While our results demonstrated the importance of integrating noise heterogeneity for a more reliable and accurate estimation of GxE heritability, alternative methods—adjusting the phenotype values of individuals in different quantile bins of the environment variable separately as proposed in Di Scipio et al.17—can prove effective under moderate levels of noise heterogeneity.
After accounting for noise heterogeneity, we observe significant genome-wide across more than a quarter of the trait-E pairs analyzed. Our finding has implications for understanding trait heritability by moving beyond the definition of narrow-sense heritability that only includes additive genetic effects. Based on our analyses, it is conceivable that approaches that can jointly model the hundreds of environmental variables measured in biobank-scale datasets will further increase estimates of . Additionally, our recovery of additional from low-frequency SNPs ( MAF ) point to traits where an understanding of GxE effects can benefit from whole-exome and whole-genome studies. Our analyses of common and low-frequency SNPs lead us to recommend that SNPs should be partitioned based on MAF and LD when estimating GxE heritability (while such partitioning does not qualitatively affect results for common SNPs). Further, our results point to traits where GxE has the potential to improve genome-wide polygenic scores (GPSs) of complex traits (since quantifies the maximum predictive accuracy that is achievable by a linear predictor based on GxE effects). In the context of sex as an environmental variable, sex-specific GPS has been shown to provide improved accuracy over agnostic scores.34,39,55,56 GxE has also been recently proposed as a possible explanation for why GPS may not generalize beyond the cohort on which these predictors were trained6 so that modeling GxE in relevant traits could improve their transferability. Our finding that allelic effects for GxE increase with decreasing MAF and LD analogous to the relationship observed for additive allelic effects motivates an evolutionary understanding of these trends and can inform what we expect to learn from studies of rare genetic variation. Finally, our identification of sets of genes that are enriched for GxE can offer clues on trait-relevant tissues and pathways and has the potential to inform functional genomic studies.57,58
We discuss the limitations of our work as well as directions for future research. First, GENIE does not explicitly model G-E correlations.13 While such correlations can lead to biases in estimates of GxE in the fixed-effect setting,59 it has been shown that, in the polygenic setting, the GxE variance component estimates remain unbiased when G-E correlations are independent of the polygenic GxE effects.14 Further, our simulations suggest that GENIE is robust in the presence of G-E correlations. Nevertheless, there are plausible settings, where such correlations can lead to false positive or biased estimates of GxE, e.g., where the phenotype directly affects the environmental variable. Developing scalable methods that are accurate in these settings is an important direction for future work. Second, estimates of GxE heritability are sensitive to the scale on which traits and environmental variables are measured and how environmental variables are encoded. In this work, we analyze quantile-normalized traits (following prior studies) and encode discrete environmental variables using a univariate parameterization (either as a 0–1 vector for each environmental variable or as a standardized version). It might be preferable to work with traits measured on their original scale and to encode each level of discrete environmental variables by a separate 0–1 covariate (leading to k environmental covariates for a k-valued environmental variable). While such choices would necessarily be guided by domain knowledge and interpretability, GENIE supports easy-to-use and rapid exploration of the consequences of these choices and can aid in assessing the robustness of these choices (we have explored a limited space of these choices here). Third, the environmental variable relevant for GxE may not be measured directly or accurately, so the environmental variable that is measured in a dataset is best viewed as a proxy for the relevant latent environmental covariate. It is essential to acknowledge that the missingness patterns of phenotypes in biobanks frequently display structure that is more intricate than random missingness.60,61 Consequently, removing individuals with missing data on Es can potentially affect GxE and other heritability estimates. One approach to tackle this complexity involves accurate imputation of missing data while mitigating the introduction of additional biases as observed in the mean imputation simulations (Figure S12). We view this as an important direction for future work. Fourth, the model underlying GENIE is not applicable to binary traits (either with or without ascertainment). GENIE can be extended to be applicable to binary traits (e.g., disease status) along the lines proposed in the context of additive62,63 and GxE estimation.14
Apart from the constraints inherent to the GENIE model, we stress the need for cautious interpretations of the results of this study due to several limitations. While GENIE can model the impact of heterogeneous noise resulting from observed environmental variables by introducing NxE components, it is important to note that the heterogeneous noise may also arise due to non-observed environmental variables. Several recent works have tried to test for GxE when the environmental variables are not observed.10,64 These issues along with the possibility of reverse causality, i.e., where the phenotype affects the environmental variable, warrant caution in any causal interpretation of our results (although it might be possible to overcome some of these limitations in specific analyses such as GxSex). Moreover, while the primary focus of our work is on the methodological aspects of GxE heritability estimation, our application of GENIE to medication-sensitive traits highlights the complexities arising in this setting that warrant care in interpreting the results. To explore these issues, we repeated our previous analyses after performing heuristic adjustments of phenotypes for relevant medications. Our additional analyses of GxE estimates on measurements adjusted for medication usage suggest that, while most of our results are robust to these issues (e.g., GxE for systolic and diastolic blood pressure, GxStatin on HbA1c), some are less so (e.g., GxAge on LDL and cholesterol) (see Note S4 for details). Finally, while analyses in this work were based on a cohort of self-identified white British individuals, it is valuable to investigate GxE effects using GENIE across a broader range of populations for stronger and more comprehensive results.
Data and code availability
GENIE software is an open-source software freely available at https://github.com/sriramlab/GENIE. The software requires , cmake, and make to compile the code on a Linux machine. Please see the documentation in the GitHub repository for further information.
Acknowledgments
This research was conducted using the UK Biobank Resource under application 331277. We thank the participants of UK Biobank for making this work possible. This work was funded by NIH grants R35GM125055 (A.P. and S.S.), HG006399 (S.S.), and NSF grant CAREER-1943497 (A.P. and S.S.).
The authors would like to thank Alkes Price and Arbel Harpak for their feedback on the manuscript. The authors would also like to acknowledge the stimulating discussions at the UCLA Computational Genomics Summer Institute (supported by NIH grants GM135043 and GM112625) and the 2018 Bertinoro workshop in Statistical and Computational Genomics that enabled this work.
Declaration of interests
The authors declare no competing interests.
Published: June 11, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.05.015.
Contributor Information
Ali Pazokitoroudi, Email: alipazoki@cs.ucla.edu.
Sriram Sankararaman, Email: sriram@cs.ucla.edu.
Web resources
GCTA (v1.94.1), https://yanglab.westlake.edu.cn/software/gcta
LDSC (v1.0.1), https://github.com/bulik/ldsc
LEMMA (v1.0.4), https://github.com/mkerin/LEMMA
MonsterLM (v0.1.1), https://github.com/GMELab/MonsterLM
PLINK (v1.90), https://www.cog-genomics.org/plink
Supplemental information
References
- 1.Yang J., Loos R.J., Powell J.E., Medland S.E., Speliotes E.K., Chasman D.I., Rose L.M., Thorleifsson G., Steinthorsdottir V., Mägi R., et al. FTO genotype is associated with phenotypic variability of body mass index. Nature. 2012;490:267–272. doi: 10.1038/nature11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gagneur J., Stegle O., Zhu C., Jakob P., Tekkedil M.M., Aiyar R.S., Schuon A.-K., Pe’er D., Steinmetz L.M. Genotype-environment interactions reveal causal pathways that mediate genetic effects on phenotype. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Virolainen S.J., VonHandorf A., Viel K.C.M.F., Weirauch M.T., Kottyan L.C. Gene-environment interactions and their impact on human health. Genes Immun. 2023;24:1–11. doi: 10.1038/s41435-022-00192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khoury M.J., Wagener D.K. Epidemiological evaluation of the use of genetics to improve the predictive value of disease risk factors. Am. J. Hum. Genet. 1995;56:835–844. [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson M.R., English G., Moser G., Lloyd-Jones L.R., Triplett M.A., Zhu Z., Nolte I.M., van Vliet-Ostaptchouk J.V., Snieder H., et al. LifeLines Cohort Study Genotype–covariate interaction effects and the heritability of adult body mass index. Nat. Genet. 2017;49:1174–1181. doi: 10.1038/ng.3912. [DOI] [PubMed] [Google Scholar]
- 6.Mostafavi H., Harpak A., Agarwal I., Conley D., Pritchard J.K., Przeworski M. Variable prediction accuracy of polygenic scores within an ancestry group. Elife. 2020;9 doi: 10.7554/eLife.48376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laville V., Majarian T., Sung Y.J., Schwander K., Feitosa M.F., Chasman D.I., Bentley A.R., Rotimi C.N., Cupples L.A., de Vries P.S., et al. Gene-lifestyle interactions in the genomics of human complex traits. Eur. J. Hum. Genet. 2022;30:730–739. doi: 10.1038/s41431-022-01045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore R., Casale F.P., Jan Bonder M., Horta D., BIOS Consortium. Franke L., Barroso I., Stegle O. A linear mixed-model approach to study multivariate gene–environment interactions. Nat. Genet. 2019;51:180–186. doi: 10.1038/s41588-018-0271-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young A.I., Wauthier F.L., Donnelly P. Identifying loci affecting trait variability and detecting interactions in genome-wide association studies. Nat. Genet. 2018;50:1608–1614. doi: 10.1038/s41588-018-0225-6. [DOI] [PubMed] [Google Scholar]
- 11.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S.H., van der Werf J.H.J. MTG2: an efficient algorithm for multivariate linear mixed model analysis based on genomic information. Bioinformatics. 2016;32:1420–1422. doi: 10.1093/bioinformatics/btw012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni G., Van Der Werf J., Zhou X., Hyppönen E., Wray N.R., Lee S.H. Genotype-covariate correlation and interaction disentangled by a whole-genome multivariate reaction norm model. Nat. Commun. 2019;10:2239. doi: 10.1038/s41467-019-10128-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahl A., Nguyen K., Cai N., Gandal M.J., Flint J., Zaitlen N. A robust method uncovers significant context-specific heritability in diverse complex traits. Am. J. Hum. Genet. 2020;106:71–91. doi: 10.1016/j.ajhg.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerin M., Marchini J. Inferring Gene-by-Environment Interactions with a Bayesian Whole-Genome Regression Model. Am. J. Hum. Genet. 2020;107:698–713. doi: 10.1016/j.ajhg.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerin M., Marchini J. A non-linear regression method for estimation of gene–environment heritability. Bioinformatics. 2020;36:5632–5639. doi: 10.1093/bioinformatics/btaa1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Scipio M., Khan M., Mao S., Chong M., Judge C., Pathan N., Perrot N., Nelson W., Lali R., Di S., et al. A versatile, fast and unbiased method for estimation of gene-by-environment interaction effects on biobank-scale datasets. Nat. Commun. 2023;14:5196. doi: 10.1038/s41467-023-40913-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finucane H.K., Reshef Y.A., Anttila V., Slowikowski K., Gusev A., Byrnes A., Gazal S., Loh P.-R., Lareau C., Shoresh N., et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 2018;50:621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchinson M. A stochastic estimator of the trace of the influence matrix for laplacian smoothing splines. Commun. Stat. Simulat. Comput. 1989;18:1059–1076. [Google Scholar]
- 20.Liberty E., Zucker S.W. The mailman algorithm: A note on matrix–vector multiplication. Inf. Process. Lett. 2009;109:179–182. [Google Scholar]
- 21.Pazokitoroudi A., Wu Y., Burch K.S., Hou K., Zhou A., Pasaniuc B., Sankararaman S. Efficient variance components analysis across millions of genomes. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-17576-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M., et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sul J.H., Bilow M., Yang W.-Y., Kostem E., Furlotte N., He D., Eskin E. Accounting for population structure in gene-by-environment interactions in genome-wide association studies using mixed models. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1005849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinnott-Armstrong N., Tanigawa Y., Amar D., Mars N., Benner C., Aguirre M., Venkataraman G.R., Wainberg M., Ollila H.M., Kiiskinen T., et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 2021;53:185–194. doi: 10.1038/s41588-020-00757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pazokitoroudi A., Chiu A.M., Burch K.S., Pasaniuc B., Sankararaman S. Quantifying the contribution of dominance deviation effects to complex trait variation in biobank-scale data. Am. J. Hum. Genet. 2021;108:799–808. doi: 10.1016/j.ajhg.2021.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei X., Robles C.R., Pazokitoroudi A., Ganna A., Gusev A., Durvasula A., Gazal S., Loh P.-R., Reich D., Sankararaman S. The lingering effects of Neanderthal introgression on human complex traits. Elife. 2023;12 doi: 10.7554/eLife.80757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warren H.R., Evangelou E., Cabrera C.P., Gao H., Ren M., Mifsud B., Ntalla I., Surendran P., Liu C., Cook J.P., et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat. Genet. 2017;49:403–415. doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.-R., Anttila V., Xu H., Zang C., Farh K., et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou K., Burch K.S., Majumdar A., Shi H., Mancuso N., Wu Y., Sankararaman S., Pasaniuc B. Accurate estimation of SNP-heritability from biobank-scale data irrespective of genetic architecture. Nat. Genet. 2019;51:1244–1251. doi: 10.1038/s41588-019-0465-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Speed D., Balding D.J. Sumher better estimates the SNP heritability of complex traits from summary statistics. Nat. Genet. 2019;51:277–284. doi: 10.1038/s41588-018-0279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulik-Sullivan B.K., Loh P.-R., Finucane H.K., Ripke S., Yang J., Schizophrenia Working Group of the Psychiatric Genomics Consortium. Patterson N., Daly M.J., Price A.L., Neale B.M., et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinnott-Armstrong N., Naqvi S., Rivas M., Pritchard J.K. GWAS of three molecular traits highlights core genes and pathways alongside a highly polygenic background. Elife. 2021;10 doi: 10.7554/eLife.58615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruth K.S., Day F.R., Tyrrell J., Thompson D.J., Wood A.R., Mahajan A., Beaumont R.N., Wittemans L., Martin S., Busch A.S., et al. Using human genetics to understand the disease impacts of testosterone in men and women. Nat. Med. 2020;26:252–258. doi: 10.1038/s41591-020-0751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu C., Ming M.J., Cole J.M., Edge M.D., Kirkpatrick M., Harpak A. Amplification is the primary mode of gene-by-sex interaction in complex human traits. Cell Genom. 2023;3 doi: 10.1016/j.xgen.2023.100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Randall J.C., Winkler T.W., Kutalik Z., Berndt S.I., Jackson A.U., Monda K.L., Kilpeläinen T.O., Esko T., Mägi R., Li S., et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkler T.W., Justice A.E., Graff M., Barata L., Feitosa M.F., Chu S., Czajkowski J., Esko T., Fall T., Kilpeläinen T.O., et al. The influence of age and sex on genetic associations with adult body size and shape: a large-scale genome-wide interaction study. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pulit S.L., Stoneman C., Morris A.P., Wood A.R., Glastonbury C.A., Tyrrell J., Yengo L., Ferreira T., Marouli E., Ji Y., et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019;28:166–174. doi: 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rask-Andersen M., Karlsson T., Ek W.E., Johansson Å. Genome-wide association study of body fat distribution identifies adiposity loci and sex-specific genetic effects. Nat. Commun. 2019;10:339. doi: 10.1038/s41467-018-08000-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernabeu E., Canela-Xandri O., Rawlik K., Talenti A., Prendergast J., Tenesa A. Sex differences in genetic architecture in the UK Biobank. Nat. Genet. 2021;53:1283–1289. doi: 10.1038/s41588-021-00912-0. [DOI] [PubMed] [Google Scholar]
- 40.Döring A., Gieger C., Mehta D., Gohlke H., Prokisch H., Coassin S., Fischer G., Henke K., Klopp N., Kronenberg F., et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat. Genet. 2008;40:430–436. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- 41.Kolz M., Johnson T., Sanna S., Teumer A., Vitart V., Perola M., Mangino M., Albrecht E., Wallace C., Farrall M., et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirts B.H., Hasstedt S.J., Hopkins P.N., Hunt S.C. Evaluation of the gene–age interactions in HDL cholesterol, LDL cholesterol, and triglyceride levels: the impact of the SORT1 polymorphism on ldl cholesterol levels is age dependent. Atherosclerosis. 2011;217:139–141. doi: 10.1016/j.atherosclerosis.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simino J., Shi G., Bis J.C., Chasman D.I., Ehret G.B., Gu X., Guo X., Hwang S.-J., Sijbrands E., Smith A.V., et al. Gene-age interactions in blood pressure regulation: a large-scale investigation with the CHARGE, Global BPgen, and ICBP consortia. Am. J. Hum. Genet. 2014;95:24–38. doi: 10.1016/j.ajhg.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sattar N., Preiss D., Murray H.M., Welsh P., Buckley B.M., de Craen A.J.M., Seshasai S.R.K., McMurray J.J., Freeman D.J., Jukema J.W., et al. Statins and risk of incident diabetes: a collaborative meta-analysis of randomised statin trials. Lancet. 2010;375:735–742. doi: 10.1016/S0140-6736(09)61965-6. [DOI] [PubMed] [Google Scholar]
- 45.Speed D., Hemani G., Johnson M.R., Balding D.J. Improved heritability estimation from genome-wide snps. Am. J. Hum. Genet. 2012;91:1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans L.M., Tahmasbi R., Vrieze S.I., Abecasis G.R., Das S., Gazal S., Bjelland D.W., de Candia T.R., Haplotype Reference Consortium. Goddard M.E., et al. Comparison of methods that use whole genome data to estimate the heritability and genetic architecture of complex traits. Nat. Genet. 2018;50:737–745. doi: 10.1038/s41588-018-0108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Speed D., Cai N., UCLEB Consortium. Johnson M.R., Nejentsev S., Balding D.J., et al. Reevaluation of SNP heritability in complex human traits. Nat. Genet. 2017;49:986–992. doi: 10.1038/ng.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gazal S., Loh P.-R., Finucane H.K., Ganna A., Schoech A., Sunyaev S., Price A.L. Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat. Genet. 2018;50:1600–1607. doi: 10.1038/s41588-018-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gazal S., Finucane H.K., Furlotte N.A., Loh P.-R., Palamara P.F., Liu X., Schoech A., Bulik-Sullivan B., Neale B.M., Gusev A., Price A.L. Linkage disequilibrium–dependent architecture of human complex traits shows action of negative selection. Nat. Genet. 2017;49:1421–1427. doi: 10.1038/ng.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schoech A.P., Jordan D.M., Loh P.-R., Gazal S., O’Connor L.J., Balick D.J., Palamara P.F., Finucane H.K., Sunyaev S.R., Price A.L. Quantification of frequency-dependent genetic architectures in 25 UK Biobank traits reveals action of negative selection. Nat. Commun. 2019;10:790. doi: 10.1038/s41467-019-08424-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng J., De Vlaming R., Wu Y., Robinson M.R., Lloyd-Jones L.R., Yengo L., Yap C.X., Xue A., Sidorenko J., McRae A.F., et al. Signatures of negative selection in the genetic architecture of human complex traits. Nat. Genet. 2018;50:746–753. doi: 10.1038/s41588-018-0101-4. [DOI] [PubMed] [Google Scholar]
- 52.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Mägi R., Strawbridge R.J., Pers T.H., Fischer K., Justice A.E., et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Small K.S., Todorčević M., Civelek M., El-Sayed Moustafa J.S., Wang X., Simon M.M., Fernandez-Tajes J., Mahajan A., Horikoshi M., Hugill A., et al. Regulatory variants at KLF14 influence type 2 diabetes risk via a female-specific effect on adipocyte size and body composition. Nat. Genet. 2018;50:572–580. doi: 10.1038/s41588-018-0088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stancu C., Sima A. Statins: mechanism of action and effects. J. Cell Mol. Med. 2001;5:378–387. doi: 10.1111/j.1582-4934.2001.tb00172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rawlik K., Canela-Xandri O., Tenesa A. Evidence for sex-specific genetic architectures across a spectrum of human complex traits. Genome Biol. 2016;17:166. doi: 10.1186/s13059-016-1025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flynn E., Tanigawa Y., Rodriguez F., Altman R.B., Sinnott-Armstrong N., Rivas M.A. Sex-specific genetic effects across biomarkers. Eur. J. Hum. Genet. 2021;29:154–163. doi: 10.1038/s41431-020-00712-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dixit A., Parnas O., Li B., Chen J., Fulco C.P., Jerby-Arnon L., Marjanovic N.D., Dionne D., Burks T., Raychowdhury R., et al. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Findley A.S., Monziani A., Richards A.L., Rhodes K., Ward M.C., Kalita C.A., Alazizi A., Pazokitoroudi A., Sankararaman S., Wen X., et al. Functional dynamic genetic effects on gene regulation are specific to particular cell types and environmental conditions. Elife. 2021;10 doi: 10.7554/eLife.67077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dudbridge F., Fletcher O. Gene-environment dependence creates spurious gene-environment interaction. Am. J. Hum. Genet. 2014;95:301–307. doi: 10.1016/j.ajhg.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mitra R., McGough S.F., Chakraborti T., Holmes C., Copping R., Hagenbuch N., Biedermann S., Noonan J., Lehmann B., Shenvi A., et al. Learning from data with Structured Missingness. Nat. Mach. Intell. 2023;5:13–23. [Google Scholar]
- 61.An U., Pazokitoroudi A., Alvarez M., Huang L., Bacanu S., Schork A.J., Kendler K., Pajukanta P., Flint J., Zaitlen N., et al. Deep learning-based phenotype imputation on population-scale Biobank data increases genetic discoveries. Nat. Genet. 2023;55:2269–2276. doi: 10.1038/s41588-023-01558-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Golan D., Lander E.S., Rosset S. Measuring missing heritability: inferring the contribution of common variants. Proc. Natl. Acad. Sci. USA. 2014;111:E5272–E5281. doi: 10.1073/pnas.1419064111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weissbrod O., Flint J., Rosset S. Estimating snp-based heritability and genetic correlation in case-control studies directly and with summary statistics. Am. J. Hum. Genet. 2018;103:89–99. doi: 10.1016/j.ajhg.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marderstein A.R., Davenport E.R., Kulm S., Van Hout C.V., Elemento O., Clark A.G. Leveraging phenotypic variability to identify genetic interactions in human phenotypes. Am. J. Hum. Genet. 2021;108:49–67. doi: 10.1016/j.ajhg.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GENIE software is an open-source software freely available at https://github.com/sriramlab/GENIE. The software requires , cmake, and make to compile the code on a Linux machine. Please see the documentation in the GitHub repository for further information.