Abstract
The subgenomic mRNAs of the coronavirus mouse hepatitis virus (MHV) are composed of a leader sequence, identical to the 5′ 70 nucleotides of the genome, joined at distant downstream sites to a stretch of sequence that is identical to the 3′ end of the genome. The points of fusion occur at intergenic sequences (IGSs), loci on the genome that contain a tract of sequence homologous to the 3′ end of the leader RNA. We have constructed a mutant of MHV-A59 containing an extra IGS inserted into the genome immediately downstream of the 3′-most gene, that encoding the nucleocapsid (N) protein. We show that in cells infected with the mutant, there is synthesis of an additional leader-containing subgenomic RNA of the predicted size. Our study demonstrates that (i) an IGS can be a sufficient cis-acting element to dictate MHV transcription, (ii) the relative efficiency of an IGS must be influenced by factors other than the nucleotides immediately adjacent to the 5′AAUCUAAAC3′ core consensus sequence or its position relative to the 3′ end of the genome, (iii) a downstream IGS can exert a polar attenuating effect on upstream IGSs, and (iv) unknown factors prevent the insertion of large exogenous elements between the N gene and the 3′ untranslated region of MHV. These results confirm and extend conclusions previously derived from the analysis of defective interfering RNAs.
Coronaviruses are single-stranded, positive-sense RNA viruses possessing exceptionally large genomes (27 to 31 kb) (39). Coronavirus infection of cells results in a program of gene expression that includes, in addition to the replication of genomic RNA (gRNA), the synthesis of a set of smaller, subgenomic RNA (sgRNA) species of both polarities (44). The positive-sense sgRNAs, which serve as mRNAs for proteins encoded downstream, are composed of linked distant segments of the genome. Each contains a leader RNA (70 to 100 nucleotides [nt] long), identical to the 5′ end of the genome, joined at a downstream site to a stretch of sequence (the body of the sgRNA) that is identical to the 3′ end of the genome. The much less abundant negative-sense sgRNAs (34, 38) are complementary to their positive-sense counterparts, having 5′ oligo(U) tracts (8) and 3′ antileaders (37).
The mechanism that gives rise to the formation of sgRNAs is not well understood. It is generally agreed, from UV mapping studies (11, 40), that sgRNAs are not generated from the splicing of genome-length precursors. In an early model, that of leader-primed transcription, it was proposed that sgRNAs were generated by the pausing of the viral polymerase near the end of the leader sequence, followed by the detachment and reattachment of the leader RNA to an internal portion of the negative-strand antigenomic template, from whence it was subsequently elongated (19). More recent evidence supports models in which leader-to-body fusion is the result of quasicontinuous synthesis across two distant portions of a looped-out template, which are brought together via protein-RNA and protein-protein interactions (20, 47). The details of the strand transfer event, however, are unknown. Also unresolved are issues of whether the initial leader-to-body fusion event occurs during positive-strand or negative-strand RNA synthesis and whether the negative-strand sgRNA species are active templates or dead-end products (1, 12, 34–36, 38).
Common to all the various models of coronavirus transcription is a focus on what have been termed intergenic sequences (IGSs), loci on the genome that contain a short run of sequence identical or nearly identical to the 3′ end of the leader RNA (2, 17, 19, 44). These regions, which are often not truly intergenic since they overlap with the upstream gene, range from 9 to 18 nt in mouse hepatitis virus (MHV). The IGSs of MHV contain the core consensus sequence 5′AAUCUAAAC3′, and with rare exceptions, this 9-nt motif appears in the leader-body junctions of the sgRNAs. For MHV, early studies noted a rough correlation between the amount of leader and IGS homology and the relative efficiency of sgRNA synthesis from a given IGS. In addition, a rough correlation was seen between proximity to the 3′ end of the genome and the relative efficiency of sgRNA synthesis from a given IGS. However, for the sgRNAs of other coronaviruses, bovine coronavirus, porcine transmissible gastroenteritis virus (TGEV), and feline infectious peritonitis virus (FIPV), striking exceptions to one or both of these generalities have been demonstrated (5, 7, 9, 38).
Technical barriers, most notably that of genome size, have so far precluded the establishment of an infectious cDNA clone of any coronavirus. Thus, beyond the examination of IGSs in natural viral variants, the major strides that have been made in the study of transcription have been facilitated by the discovery and cloning of defective interfering (DI) RNAs of MHV (23, 25, 41) and, subsequently, of other coronaviruses (3, 28, 31). DI RNAs are deletion mutants, far smaller than gRNA and usually composed of several discontinuous regions of the genome. Their replication takes place only in the presence of a helper virus. Two of the most salient conclusions that have emerged from analyses of these are, first, that the insertion of an IGS is sufficient to generate the synthesis of sgRNA from a DI RNA and, second, that beyond the core consensus motif, the extent of homology between the leader and the IGS does not correlate with the efficiency of transcription, as reflected in the sgRNA/gRNA ratio (24, 42).
To examine whether an IGS can be a sufficient cis-acting element to determine sgRNA synthesis in the context of the entire genome, we sought to insert an extra IGS into MHV by using targeted recombination, a procedure developed for the site-directed mutagenesis of this virus. Our choice for the site of insertion was the extreme 5′ boundary of the 3′ untranslated region (3′ UTR) of the viral genome. We have previously shown that this end of the 3′ UTR harbors a bulged stem-loop, essential for MHV replication, that begins one nt downstream from the nucleocapsid (N) gene stop codon (10). The discovery of this structure explained our previous inability to insert a new transcription module at a site now known to correspond to the loop of this element (27). In contrast, van der Most et al. had been able to add an IGS adjacent to the N gene stop codon in a DI RNA construct, and they observed transcript synthesis from this newly created leader-to-body fusion point (42). To determine whether the entire MHV genome would similarly tolerate an additional transcription unit, we designed a new IGS (designated IGS7/8) that was patterned to mimic both the sequence and the positioning of IGS6/7, the IGS between MHV genes 6 and 7 (i.e., the membrane protein [M] and N genes). IGS6/7 gives rise to the largest molar amount of sgRNA in MHV-infected cells (11, 21, 44).
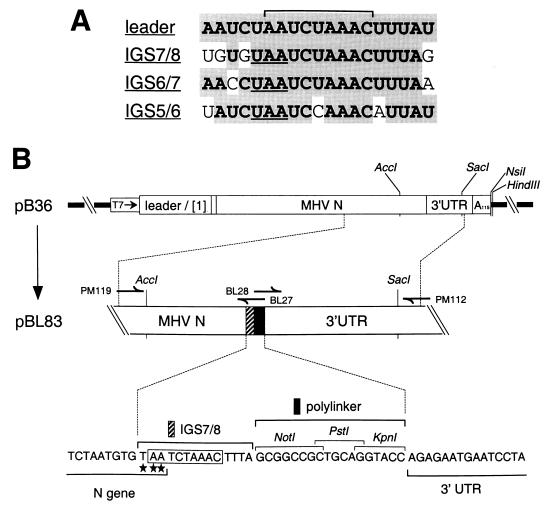
The sequence of IGS7/8 was not made entirely identical to that of IGS6/7 because we wished to maintain the overlap of the new IGS with the stop codon of the preceding (N) gene without introducing coding changes into the terminus of the N gene. A comparison of the resulting IGS7/8 with IGS6/7 and IGS5/6 is shown in Fig. 1A.
FIG. 1.
(A) Sequence homology among the 3′ end of the leader RNA, constructed IGS7/8, and naturally occurring IGS6/7 and IGS5/6 of MHV-A59. The bracket marks the nine-nt core consensus IGS motif. For each IGS, the stop codon of the upstream gene is underlined. (Note that the upstream nonhomologous C residue in IGS6/7 is a U residue in MHV-JHM.) (B) Construction of transcription vectors for the synthesis of MHV DI RNAs containing a new IGS (IGS7/8) downstream of the N gene. The parent vector, pB36, encodes a DI RNA consisting entirely of MHV components, except for a 48-nt linker that connects the 5′ 467 nt of the MHV genome (leader plus gene 1 fragment) to a 1,666-nt segment comprising the entire N gene and 3′ UTR (26). Plasmid pBL83 was derived from pB36 by the insertion of IGS7/8 (hatched rectangle), followed by an 18-bp polylinker region (solid rectangle), immediately downstream of (and partially overlapping with) the N gene stop codon. The PCR primers used in the construction of pBL83 are indicated above the schematic of the relevant segment of this plasmid, and the sequence of the insertion is shown beneath it. The MHV core consensus IGS sequence is boxed, and the N gene stop codon is marked with stars.
A transcription vector encoding an MHV DI RNA containing IGS7/8 followed by a short polylinker was constructed by the simultaneous ligation of two PCR products into pB36 (26) (Fig. 1B). The first insert, generated by PCR with primers PM119 and BL27 (Table 1), contained IGS7/8 and the polylinker at its 3′ end. The second insert, generated by PCR with primers BL28 and PM112 (Table 1), contained the polylinker at its 5′ end. The former PCR product was restricted with AccI and PstI, the latter was restricted with PstI and SacI, and both were inserted in place of the AccI-SacI fragment of pB36 to generate pBL83 (Fig. 1B). DNA manipulations were performed by standard methods (32). All ligation junctions and sequences of inserts generated by PCR were confirmed by dideoxy sequencing according to the method of Sanger et al. (33) with modified T7 DNA polymerase (Sequenase; U.S. Biochemical).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence | Sense | Purpose |
|---|---|---|---|
| BL27 | 5′GGTACCTGCAGCGGCCGCTAAAGTTTAGATTACACATTAGAGTCATC3′ | − | Construction of pBL83 |
| BL28 | 5′AATTGCGGCCGCTGCAGGTACCAGAGAATGAATCCTATGT3′ | + | Construction of pBL83 |
| PM112 | 5′CCATGATCAACTTCATTC3′ | − | Construction of pBL83; RT-PCR analysis of viral mutants; complementary to nt 18 to 35 from the 3′ end of the genome |
| PM119 | 5′TAGTACTCTACCTGGTTT3′ | + | Construction of pBL83 |
| BL64 | 5′ATGAAGGTACCAATGTAAGCGTTGC3′ | + | Construction of pBL108 |
| BL65 | 5′ATTCAGGTACCTTACACATTAGAGT3′ | − | Construction of pBL108 |
| PM143 | 5′TATAAGAGTGATTGGCGTCCGTACGTAC3′ | + | RT-PCR analysis of viral mutants; identical to the 5′ end of the leader sequence |
| BL19 | 5′TTTTTTTTTTGTGATTCTTC3′ | − | RT-PCR analysis of viral mutants; complementary to nt 1 to 10 from the 3′ end of the genome plus 10 nt of the poly(A) tail |
Targeted recombination was employed to transduce IGS7/8 into the genome of Alb4, a thermolabile N gene deletion mutant of MHV-A59 (16) exactly as described previously (10, 26). Capped donor RNA transcribed from HindIII-truncated pBL83 with a T7 polymerase transcription kit (Ambion) was transfected into Alb4-infected mouse L2 cells, and recombinant progeny viruses were selected as those able to form large plaques at 39°C, in contrast to the tiny plaques formed by Alb4 (16). Two independent recombinants, Alb169 and Alb170, which resulted from separate transfections, were isolated and purified. Direct RNA sequencing (30) of total cytoplasmic RNA derived from Alb169- and Alb170-infected mouse 17 Cl1 cells demonstrated the repair of the Alb4 deletion and the incorporation of IGS7/8 and the adjacent polylinker into each mutant (data not shown).
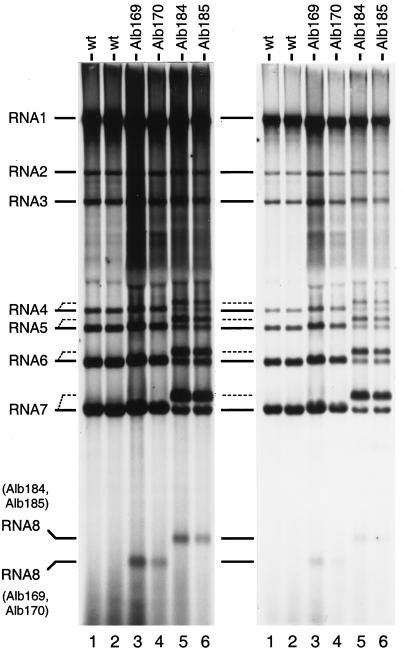
The metabolic labeling of RNA in cells infected with Alb169 and Alb170 revealed that each synthesized an extra RNA species (sgRNA8 [Fig. 2, lanes 3 and 4]) in addition to gRNA (RNA1) and sgRNA2 to sgRNA7, which were synthesized in common with cells infected with wild-type MHV (lanes 1 and 2). The mobility of sgRNA8 was consistent with its predicted size (394 nt plus polyadenylate). Although each of the other mutant RNAs (RNA1 to RNA7) was expected to be 29 nt larger than the wild-type counterpart, this was not clearly detectable at this level of resolution.
FIG. 2.
Analysis of MHV RNA synthesized in infected cells. Mouse 17 Cl1 cells infected with wild-type or recombinant virus were labeled with 123 μCi of [33P]orthophosphate per ml from 14.5 to 16.5 h postinfection in the presence of 20 μg of actinomycin D per ml, essentially as reported previously (10, 26). Purified cytoplasmic RNA was separated by electrophoresis through 1% agarose containing formaldehyde, and labeled RNA was visualized by fluorography. Lanes 1 and 2, wild-type (wt) MHV. Lanes 3 and 4, Alb169 and Alb170, independent recombinants containing inserted IGS7/8. Lanes 5 and 6, Alb184 and Alb185, independent recombinants containing inserted IGS7/8 and a duplication of domain III of the N gene. The left and right panels, respectively, show 72-h and 24-h exposures of the same gel. Broken lines indicate detectable mobility differences of RNA4 through RNA7 of Al6184 and Al6185.
To confirm that the additional RNA species had the leader-body structure of an authentic sgRNA, intracellular RNA was analyzed by reverse transcription followed by PCR (RT-PCR). RT was carried out with a primer complementary to the boundary between the 3′ UTR and the poly(A) tail, and this product was amplified by PCR under standard reaction conditions described previously (16), by using a pair of primers identical to the 5′ end of the leader sequence and complementary to a downstream segment of the 3′ UTR (Fig. 3). For Alb169 and Alb170, a PCR product corresponding to the predicted size (377 bp) derived from sgRNA8 was clearly detected (Fig. 3, lanes 4 and 5), and this product was absent from the wild-type control (lane 3). Also as expected, mRNA7 of Alb169 and Alb170 yielded a PCR product consistent with its predicted size of 1,756 bp (Fig. 3, lanes 4 and 5), detectably larger than the corresponding product from wild-type RNA7 (1,727 bp; lane 3). RT-PCR products originating from sgRNAs larger than mRNA7 were not seen, presumably because they were outcompeted by the smaller products and also because the standard reaction conditions used were not optimized for long RT-PCR.
FIG. 3.
RT-PCR analysis of viral sgRNAs. RNA isolated from mock-, wild-type-, or mutant-infected cells was reverse transcribed with primer BL19 and then amplified by PCR with primers PM143 and PM112 (Table 1). In the schematic at the top, the leader and 3′ UTR are indicated by solid lines; the N gene and the IGS7/8 insert are represented by open and closed rectangles, respectively. PCR products were detected by electrophoresis through 1% agarose containing ethidium bromide. The predicted sizes (in base pairs) of PCR products are given to the right of the gel; sizes of DNA standards (std; lane 1) are given to the left.
In RT-PCRs identical to those whose results are shown in Fig. 3, the 377-bp bands obtained with Alb169 and Alb170 RNAs were isolated and entirely sequenced on both strands. This confirmed that both bands were of the predicted size and that both contained the expected leader-body junction. A portion of the sequence for the Alb169-derived fragment, encompassing the region of leader-to-body fusion, is shown in Fig. 4. As anticipated, the junction occurred at the core consensus motif 5′AAUCUAAAC3′, and there was no evidence of heterogeneity within the population of PCR fragment molecules. The sequence of the same fragment derived from Alb170 RNA was identical to that from Alb169 RNA (data not shown). These results indicated that sgRNA8 was, indeed, a leader-containing RNA species and not a nucleolytic degradation product. Thus, the IGS7/8 incorporated downstream of the N gene stop codon was sufficient to direct transcription on the MHV genome.
FIG. 4.
Sequence of the leader-body junction of sgRNA8. The 377-bp RT-PCR product obtained by using primers PM143 and PM112 with RNA isolated from Alb169-infected cells was sequenced with an Applied Biosystems model 377 DNA sequencer. Shown is a portion of the DNA sequence determined with a negative-sense primer corresponding to nt 86 to 103 downstream of the N gene stop codon. Above this is given the inferred (positive-sense) RNA sequence encompassing the leader-body junction; the nine-nt core consensus IGS motif is boxed.
To assess the transcriptional efficiency of the inserted IGS7/8, RNA bands in the first four lanes of the gel shown in Fig. 2 were quantitated by PhosphorImager scanning and normalized with respect to the molarity of gRNA in each sample (Table 2). These results showed that in cells infected with wild-type MHV-A59, the sgRNA/gRNA ratios averaged 9.7 for sgRNA7 and 3.9 for sgRNA6. These values agree well with the corresponding values of 7.6 and 2.9 reported by Jacobs et al. (11) when the data of these authors is normalized in the same manner and corrected for what are now known to be the true sizes of the RNA species. In contrast, the corresponding values of 50 and 16 reported by Leibowitz et al. (21) (similarly normalized and corrected) are considerably higher. This may reflect different conditions of infection, or it may be due to the reduced recovery of gRNA by these investigators. Notably, all three sets of data have a similar ratio of sgRNA7 to sgRNA6.
TABLE 2.
Relative molar amounts of sgRNAs synthesized in wild-type- and IGS7/8 mutant-infected cells
| Virus | RNA species | Intensitya (arbitrary units) | RNA lengthb (nt) | sgRNA/gRNA molar ratio |
|---|---|---|---|---|
| Wild type | gRNA | 22,380 | 31,528 | 1.00 |
| sgRNA6 | 7,380 | 2,641 | 3.94 | |
| sgRNA7 | 12,760 | 1,944 | 9.25 | |
| Wild type | gRNA | 22,200 | 31,528 | 1.00 |
| sgRNA6 | 7,330 | 2,641 | 3.94 | |
| sgRNA7 | 13,810 | 1,944 | 10.09 | |
| Alb169 | gRNA | 50,510 | 31,557 | 1.00 |
| sgRNA6 | 10,600 | 2,670 | 2.48 | |
| sgRNA7 | 13,160 | 1,973 | 4.17 | |
| sgRNA8 | 1,090 | 594 | 1.15 | |
| Alb170 | gRNA | 22,530 | 31,557 | 1.00 |
| sgRNA6 | 5,280 | 2,670 | 2.77 | |
| sgRNA7 | 7,480 | 1,973 | 5.31 | |
| sgRNA8 | 519 | 594 | 1.22 |
Radioactivity in RNA bands in the gel shown in Fig. 2 was quantitated by scanning with an Image Quant PhosphorImager (Molecular Dynamics). The subtracted background intensity for each band was determined from a rectangle of equal size drawn immediately above or below the rectangle for the band. Indicated RNA bands from two independent wild-type samples (lanes 1 and 2), Alb169 (lane 3), and Alb170 (lane 4) were quantitated.
Including a polyadenylate tail length of 200 nt.
Two significant observations were clear from the quantitation of viral RNA species synthesized in cells infected with the mutants Alb169 and Alb170. First, the sgRNA/gRNA ratio for sgRNA8 averaged 1.2 (Table 2), indicating that IGS7/8 was markedly less efficient in driving transcription than the highly similar IGS6/7, assuming that sgRNA8 has a stability comparable to that of the other sgRNAs. This argues that proximity to the 3′ end of the genome is not the primary determinant of IGS efficiency, in accord with the transcription patterns observed with the coronaviruses porcine TGEV (7, 38) and FIPV (5) and also with a set of MHV DI RNA constructs (45). It additionally suggests that, besides the core consensus sequence and its immediately adjacent nucleotides, there must be other factors making a substantial contribution to the efficiency of a given IGS. Although we cannot rule out the possibility that there is a strong effect exerted by the second and fourth nucleotides upstream of the core consensus IGS motif (which differ between IGS7/8 and IGS6/7 [Fig. 1A]), this seems unlikely in light of the results of van der Most et al. (42), who observed similarly low sgRNA/gRNA ratios for IGSs inserted adjacent to the N gene stop codon in a DI RNA construct.
A second conclusion emerging from the data shown in Table 2 is that the inserted IGS7/8 in Alb169 and Alb170 exerted a polar influence on upstream IGSs. Thus, for the mutants, the molar amounts of sgRNA7 and sgRNA6 were reduced by roughly one-half and one-third, respectively, relative to wild-type levels. A similar effect has been observed with DI RNAs of MHV (13, 45) and bovine coronavirus (18) into which multiple IGSs had been engineered, although it should be noted that this phenomenon was not seen with one MHV DI RNA (1). In one case (45), the inhibitory effect of a downstream MHV IGS was found to act over a distance of 761 nt. Our results show that the same type of attenuation occurs in the complete MHV genome and that it can persist over a span of at least 2,076 nt (the distance between IGS7/8 and IGS5/6).
To determine whether IGS7/8 could have utility in the expression of foreign genetic material in MHV, we initially inserted the gene encoding the green fluorescent protein (GFP) or luciferase into the polylinker site of pBL83. However, in multiple targeted recombination trials, donor RNAs containing either of these heterologous reporter genes never gave rise to progeny viruses. Among a number of explanations that might have accounted for this failure, one was that there possibly exists an RNA element in the 3′ portion of the N gene that is essential for MHV replication. Evidence from deletion mapping studies of a number of MHV DI RNAs suggests that the minimal cis-acting sequence at the 3′ end of the MHV genome that can support replication extends beyond the 3′ UTR into the distal portion of the N gene (15, 22, 43), encompassing the region that encodes domain III of the N protein (29). Therefore, it was possible that N gene domain III could be separated from the 3′ UTR by a short insert such as the 29-nt fragment containing IGS7/8, but an insertion as large as the GFP or luciferase gene might not be tolerated.
To test this possibility, two vectors were constructed in which a duplication of N gene domain III (the 3′ 141 nt of the N gene flanked by PCR-generated KpnI sites) was placed immediately upstream of the 3′ UTR (Fig. 5). In one vector, pBL108, the duplicated segment was inserted into the original IGS7/8 polylinker of pBL83. In the other, pBL110, the duplicated segment was inserted following the GFP gene of pBL86, which in turn, had been generated from pBL83 by the transfer of the GFP gene (as a 732-bp NotI-NotI fragment from Green Lantern-1 vector [Life Technologies]) into the NotI site of the polylinker.
FIG. 5.
Construction of transcription vectors for the synthesis of MHV DI RNAs containing IGS7/8 as well as a duplication of the 3′ portion of the N gene inserted immediately upstream of the 3′ UTR. The region encompassing nt 1225 to 1365 of the N gene (corresponding to domain III of the N protein [NIII] [29]), flanked by primer-created KpnI sites, was amplified by PCR with primers BL64 and BL65, with pB36 as the template. This domain III-containing fragment was inserted into the KpnI site of the polylinker (solid rectangle) following inserted IGS7/8 (hatched rectangle) of either pBL83 or pBL86 to generate plasmids pBL108 and pBL110, respectively.
Targeted recombination experiments were carried out with donor RNA from pBL108, the vector containing duplicated domain III without an exogenous reporter gene. This resulted in the selection of two independent recombinants, Alb184 and Alb185, which formed wild-type-sized plaques at the nonpermissive temperature for the Alb4 parent. Following the plaque purification of these mutants, direct RNA sequencing demonstrated that in each the Alb4 deletion had been repaired and that each harbored IGS7/8 and duplicated N gene domain III (data not shown).
In contrast, despite repeated attempts, no recombinants were obtained with donor RNA from pBL110, the vector containing duplicated domain III and a GFP reporter gene. Consistent with this, we were unable to detect the replication of pBL110 RNA by the metabolic labeling of MHV-infected cells that were transfected with this pseudo-DI RNA. Synthetic RNA from pBL86, the parent of pBL110, was also incapable of replicating as an MHV DI RNA (data not shown). These results implied that it was the presence of the GFP gene adjacent to the 3′ UTR, rather than the absence of a hypothetical cis-acting RNA element in domain III of the N gene, that was somehow lethal to the virus. Moreover, since it had been previously shown that most of gene 4 can be replaced by the GFP gene (6), this suggested that it was the location of the insertion, not the gene itself, that was inhibitory to MHV replication.
The metabolic labeling of cells infected with Alb184 and Alb185, the mutants containing IGS7/8 and duplicated N gene domain III, revealed that, as expected, they produced an extra sgRNA species (Fig. 2, lanes 5 and 6). In this case, the new sgRNA8 was 147 nt larger than that of Alb169 and Alb170 because of the duplicated domain III segment. The sizes of all other RNA species of Alb184 and Alb185 were 176 nt larger than their wild-type counterparts, and for sgRNA4 through sgRNA7 this resulted in clearly detectable mobility differences (Fig. 2). Interestingly, in addition to these more slowly migrating species, a set of minor RNA bands having the same mobilities as wild-type sgRNAs was also observed in cells infected with Alb184 and Alb185. Since the inocula for the RNA labeling experiment had been passage 1 virus from purified plaques, this suggested that duplicated domain III was extremely unstable in the MHV genome. Homologous recombination between the duplication and the intact N gene would have been expected to yield a revertant identical to wild-type MHV except for a KpnI site immediately 3′ to the N gene stop codon (Fig. 1B and 5). Because of the instability of Alb184 and Alb185, we could not quantitate the sgRNA/gRNA ratios for these mutants, since the observed gRNAs were a mixture of mutant and revertant gRNAs, the latter being produced continuously during the course of the infection. However, it was clear that the presence of the domain III duplication, while not significantly enhancing the transcription of sgRNA8, did not significantly inhibit it either. Thus, the new transcriptional unit would permit an insertion of 147 nt, although the insertion of a larger amount of heterologous material was apparently not allowed.
RT-PCR was used to confirm the leader-body structure of subgenomic RNA8 produced by Alb184 and Alb185. A product having the expected size of 551 bp was amplified from RNA isolated from cells infected with passage 2 of each of these viruses (Fig. 6, lanes 5 and 8). This fragment persisted faintly in passage 3 (Fig. 6, lanes 6 and 9) but was undetectable by passage 4 (lanes 7 and 10). The same RT-PCR product was not obtained from the RNA of three successive passages of wild-type-infected cells (Fig. 6, lanes 2 to 4). In RT-PCRs identical to those whose results are shown in Fig. 6, the 551-bp bands obtained with passage 2 Alb184 and Alb185 RNAs were isolated and completely sequenced on both strands. This confirmed the predicted sizes and leader-body junction sequences of both bands, and the leader-body junctions were seen to be homogeneous (data not shown).
FIG. 6.
RT-PCR analysis of viral sgRNAs. RNA isolated from cells infected with different passages of wild-type or mutant virus was reverse transcribed with primer BL19 and then amplified by PCR with primers PM143 and BL19 (Table 1). In the schematic at the top, the leader and 3′ UTR are indicated by solid lines; the N gene (or N gene domain III fragment) and the IGS7/8 insert are represented by open and closed rectangles, respectively. PCR products were detected by electrophoresis through 1% agarose containing ethidium bromide. The predicted sizes (in base pairs) of PCR products are given to the right of the gel; sizes of DNA standards (std; lane 1) are given to the left.
In accord with the results of metabolic labeling, two sgRNA7 species were detected for passage 2 Alb184 and Alb185 (Fig. 6, lanes 5 and 8). The larger one, consistent with the predicted size of 1,930 bp, was generated from sgRNA7 of the original mutant; the smaller one, predicted to be 1,760 bp in length, was generated from sgRNA7 of the revertant that was formed by recombination between the two copies of N gene domain III. This latter fragment was almost the same size as the corresponding 1,754-bp RT-PCR product obtained from wild-type-infected cells (Fig. 6, lanes 2 to 4). By passage 4 of Alb184 and Alb185, the RT-PCR product from the revertant predominated and that from the original mutant was no longer detectable. Thus, in the absence of any pressure to maintain duplicated domain III, mutants containing this segment were swiftly outcompeted by the revertant. Engineered duplications of this sort may provide a model for the study of homologous recombination in MHV.
In summary, our results permit the following conclusions. First, an inserted IGS can be sufficient to dictate transcription in the context of the entire MHV genome; this is congruous with conclusions drawn from a number of DI RNA studies (12, 13, 18, 24, 42, 45, 47). Second, IGS efficiency can be influenced by factors other than the sequence immediately adjacent to the core consensus nucleotides or the position of the IGS relative to the 3′ end of the genome. This conclusion runs counter to the general trends that have been noted for the IGSs of MHV, but it is in accord with the observation that the smallest transcripts of porcine TGEV (7, 38) and FIPV (5), both originating downstream of the N gene, are produced in much smaller molar amounts than the next larger transcripts. It also agrees well with a previous MHV DI RNA study (42), in which an IGS was inserted into almost exactly the same position, between the N gene and the 3′ UTR, as our IGS7/8. Third, a downstream IGS can exert a polar effect on the efficiency of upstream IGSs. This effect decreases over distance (the attenuation by IGS7/8 of IGS6/7 was greater than that of IGS5/6), but it is more long-ranging in the MHV genome than had been seen previously in work on DI RNAs (13, 18, 45). Finally, we have found that unknown factors prevent the insertion of large exogenous elements between the N gene and the 3′ UTR. This is perplexing, since porcine TGEV and FIPV each have one or two small genes situated in the analogous position downstream of the N gene (4, 14). It may reflect a limitation on the overall size of the MHV genome, since the previous insertion of the GFP gene in place of most of gene 4 created a net increase in genome size of 560 nt (6); in the present case, we were attempting to add at least 732 nt to the total genome. This would not, however, also explain the inability of the corresponding donor DI RNAs to replicate. Alternatively, it is possible that the inserted reporter genes interfere with the correct folding of cis-acting RNA elements in the 3′ UTR, such as the essential bulged stem-loop that we have described adjacent to the N gene stop codon (10) or other downstream secondary structures (46). We may obtain a clearer understanding of this issue when the sequence and structural requirements of the 3′ UTR are more completely elucidated.
Acknowledgments
We are grateful to Tim Moran and Matthew Shudt of the Molecular Genetics Core Facility of the Wadsworth Center for the synthesis of oligonucleotides and automated DNA sequencing.
This work was supported in part by Public Health Service grant AI 39544 from the National Institutes of Health.
REFERENCES
- 1.An S, Makino S. Characterizations of coronavirus cis-acting RNA elements and the transcription step affecting its transcription efficiency. Virology. 1998;243:198–207. doi: 10.1006/viro.1998.9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Budzilowicz C J, Wilczynski S P, Weiss S R. Three intergenic regions of coronavirus mouse hepatitis virus strain A59 genome RNA contain a common nucleotide sequence that is homologous to the 3′ end of the viral mRNA leader sequence. J Virol. 1985;53:834–840. doi: 10.1128/jvi.53.3.834-840.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang R-Y, Hofmann M A, Sethna P B, Brian D A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Groot R J, Andeweg A C, Horzinek M C, Spaan W J M. Sequence analysis of the 3′-end of the feline coronavirus FIPV 79-1146 genome: comparison with the genome of porcine coronavirus TGEV reveals large insertions. Virology. 1988;167:370–376. doi: 10.1016/0042-6822(88)90097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Groot R J, ter Haar R J, Horzinek M C, van der Zeijst B A M. Intracellular RNAs of the feline infectious peritonitis coronavirus strain 79-1146. J Gen Virol. 1987;68:995–1002. doi: 10.1099/0022-1317-68-4-995. [DOI] [PubMed] [Google Scholar]
- 6.Fischer F, Stegen C F, Koetzner C A, Masters P S. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J Virol. 1997;71:5148–5160. doi: 10.1128/jvi.71.7.5148-5160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiscox J A, Cavanagh D, Britton P. Quantification of individual subgenomic mRNA species during replication of the coronavirus transmissible gastroenteritis virus. Virus Res. 1995;36:119–130. doi: 10.1016/0168-1702(94)00108-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann M A, Brian D A. The 5′ end of coronavirus minus-strand RNAs contains a short poly(U) tract. J Virol. 1991;65:6331–6333. doi: 10.1128/jvi.65.11.6331-6333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofmann M A, Chang R-Y, Ku S, Brian D A. Leader-mRNA junction sequences are unique for each subgenomic mRNA species in the bovine coronavirus and remain so throughout persistent infection. Virology. 1993;196:163–171. doi: 10.1006/viro.1993.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsue B, Masters P S. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs L, Spaan W J M, Horzinek M C, van der Zeijst B A M. Synthesis of subgenomic mRNAs of mouse hepatitis virus is initiated independently: evidence from UV transcription mapping. J Virol. 1981;39:401–406. doi: 10.1128/jvi.39.2.401-406.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong Y S, Makino S. Mechanism of coronavirus transcription: duration of primary transcription initiation activity and effects of subgenomic RNA transcription on RNA replication. J Virol. 1992;66:3339–3346. doi: 10.1128/jvi.66.6.3339-3346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joo M, Makino S. The effect of two closely inserted transcription consensus sequences on coronavirus transcription. J Virol. 1995;69:272–280. doi: 10.1128/jvi.69.1.272-280.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapke P A, Brian D A. Sequence analysis of the porcine transmissible gastroenteritis coronavirus nucleocapsid protein gene. Virology. 1986;151:41–49. doi: 10.1016/0042-6822(86)90102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y-N, Jeong Y S, Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koetzner C A, Parker M M, Ricard C S, Sturman L S, Masters P S. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J Virol. 1992;66:1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konings D A M, Bredenbeek P J, Noten J F H, Hogeweg P, Spaan W J M. Differential premature termination of transcription as a proposed mechanism for the regulation of coronavirus gene expression. Nucleic Acids Res. 1988;16:10849–10860. doi: 10.1093/nar/16.22.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishnan R, Chang R-Y, Brian D A. Tandem placement of a coronavirus promoter results in enhanced mRNA synthesis from the downstream-most initiation site. Virology. 1996;218:400–405. doi: 10.1006/viro.1996.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai M M C. Coronavirus: organization, replication and expression of genome. Annu Rev Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- 20.Lai M M C, Liao C-L, Lin Y-J, Zhang X. Coronavirus: how a large RNA viral genome is replicated and transcribed. Infect Agents Dis. 1994;3:98–105. [PubMed] [Google Scholar]
- 21.Leibowitz J L, Wilhelmsen K C, Bond C W. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-A59 and MHV-JHM. Virology. 1981;114:39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin Y-J, Lai M M C. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makino S, Fujioka N, Fujiwara K. Structure of the intracellular defective viral RNAs of defective interfering particles of mouse hepatitis virus. J Virol. 1985;54:329–336. doi: 10.1128/jvi.54.2.329-336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makino S, Joo M, Makino J K. A system for the study of coronavirus mRNA synthesis: a regulated expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991;65:6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino S, Shieh C-K, Keck J G, Lai M M C. Defective interfering particles of murine coronavirus: mechanism of synthesis of defective viral RNAs. Virology. 1988;163:104–111. doi: 10.1016/0042-6822(88)90237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masters P S, Koetzner C A, Kerr C A, Heo Y. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J Virol. 1994;68:328–337. doi: 10.1128/jvi.68.1.328-337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masters P S, Peng D, Fischer F. Mutagenesis of the genome of mouse hepatitis virus by targeted RNA recombination. In: Talbot P J, Levy G A, editors. Advances in experimental medicine and biology. 380. Corona- and related viruses. New York, N.Y: Plenum Press; 1995. pp. 543–549. [DOI] [PubMed] [Google Scholar]
- 28.Méndez A, Smerdou C, Izeta A, Gebauer F, Enjuanes L. Molecular characterization of transmissible gastroenteritis coronavirus defective interfering genomes: packaging and heterogeneity. Virology. 1996;217:495–507. doi: 10.1006/viro.1996.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker M M, Masters P S. Sequence comparison of the N genes of five strains of the coronavirus mouse hepatitis virus suggests a three domain structure for the nucleocapsid protein. Virology. 1990;179:463–468. doi: 10.1016/0042-6822(90)90316-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng D, Koetzner C A, Masters P S. Analysis of second-site revertants of a murine coronavirus nucleocapsid protein deletion mutant and construction of nucleocapsid protein mutants by targeted RNA recombination. J Virol. 1995;69:3449–3457. doi: 10.1128/jvi.69.6.3449-3457.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Penzes Z, Tibbles K, Shaw K, Britton P, Brown T D K, Cavanagh D. Characterization of a replicating and packaged defective RNA of avian coronavirus infectious bronchitis virus. Virology. 1994;203:286–293. doi: 10.1006/viro.1994.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawicki S G, Sawicki D L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaad M C, Baric R S. Genetics of mouse hepatitis virus transcription: evidence that subgenomic negative strands are functional templates. J Virol. 1994;68:8169–8179. doi: 10.1128/jvi.68.12.8169-8179.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sethna P B, Brian D A. Coronavirus genomic and subgenomic minus-strand RNAs copartition in membrane-protected replication complexes. J Virol. 1997;71:7744–7749. doi: 10.1128/jvi.71.10.7744-7749.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sethna P B, Hofmann M A, Brian D A. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J Virol. 1991;65:320–325. doi: 10.1128/jvi.65.1.320-325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sethna P B, Hung S-L, Brian D A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc Natl Acad Sci USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siddell S G. The Coronaviridae: an introduction. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 1–10. [Google Scholar]
- 40.Stern D F, Sefton B M. Synthesis of coronavirus mRNAs: kinetics of inactivation of infectious bronchitis virus RNA synthesis by UV light. J Virol. 1982;42:755–759. doi: 10.1128/jvi.42.2.755-759.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Most R G, Bredenbeek P J, Spaan W J M. A domain at the 3′ end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J Virol. 1991;65:3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Most R G, de Groot R J, Spaan W J M. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J Virol. 1994;68:3656–3666. doi: 10.1128/jvi.68.6.3656-3666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Most R G, Luytjes W, Rutjes S, Spaan W J M. Translation but not the encoded sequence is essential for the efficient propagation of defective interfering RNAs of the coronavirus mouse hepatitis virus. J Virol. 1995;69:3744–3751. doi: 10.1128/jvi.69.6.3744-3751.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Most R G, Spaan W J M. Coronavirus replication, transcription, and RNA recombination. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 11–31. [Google Scholar]
- 45.van Marle G, Luytjes W, van der Most R G, van der Straaten T, Spaan W J M. Regulation of coronavirus mRNA transcription. J Virol. 1995;69:7851–7856. doi: 10.1128/jvi.69.12.7851-7856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams G D, Chang R-Y, Brian D A. Evidence for a pseudoknot in the 3′ untranslated region of the bovine coronavirus genome. In: Talbot P J, Levy G A, editors. Advances in experimental medicine and biology. 380. Corona- and related viruses. New York, N.Y: Plenum Press; 1995. pp. 511–514. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Liao C-L, Lai M M C. Coronavirus leader RNA regulates and initiates subgenomic mRNA transcription both in trans and in cis. J Virol. 1994;68:4738–4746. doi: 10.1128/jvi.68.8.4738-4746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]