Abstract
One of the limitations of adenovirus vectors is the lack of machinery necessary for their integration into host chromosomes, resulting in short-term gene expression in dividing cells. We analyzed frequencies of integration and persistence of gene expression from integrated adenovirus vectors. Both E1-substituted and helper-dependent adenovirus vectors achieved similar integration efficiencies of ∼10−3 to 10−5 per cell, with the helper-dependent vector showing slightly higher efficiencies. In stable cell pools, gene expression of the integrated vector persisted for at least 50 cell divisions without selection. However, some stable cell clones showed changes in gene expression, which were accompanied by structural changes in the integrated vector DNA.
Recombinant adenovirus vectors are attracting increasing attention as in vivo gene transfer vehicles for human gene therapy (2, 31, 36). However, one of the limitations of E1-substituted adenovirus vectors currently used in most clinical gene therapy protocols is the relatively short-term expression of the transferred gene in vivo. E1-substituted vectors express viral antigens that induce a cytotoxic-T-lymphocyte-mediated immune response against the vector-transduced cells by the host, resulting in inflammation and short-term gene expression from the vector in vivo (3, 28, 32–35). To overcome this problem, we and others have recently developed a helper-dependent (gutless) adenovirus vector by removing all the viral coding sequences from adenovirus vector DNA (7, 11, 12, 15, 19). Such vectors are therefore expected to minimize the immune responses of the host that would cause rejection of the transduced cells (20, 24). However, even with this system, vector DNA is eventually lost in dividing cells because although adenoviruses exist as multiple episomal copies in the infected cell nuclei, they lack the machinery necessary for integration into host chromosomes. Furthermore, activation of T-helper cells and B cells in response to viral capsid proteins produces neutralizing antibodies that block the efficient readministration of vector (13, 37). Therefore, further improvement of an adenovirus vector that replicates or efficiently integrates into host chromosomes is required to obtain long-term expression, even in dividing cells. Although it is known that a wild-type adenovirus rarely integrates into the chromosomes of cells that are not permissive for viral DNA replication, there have not yet been any extensive investigations of how frequently replication-incompetent adenovirus vectors integrate into host chromosomes. In this study, we analyzed frequencies of integration of E1-substituted and helper-dependent adenovirus vectors and stability of gene expression from the integrated vectors.
Integration efficiencies of E1-substituted and helper-dependent adenovirus vectors in cell lines.
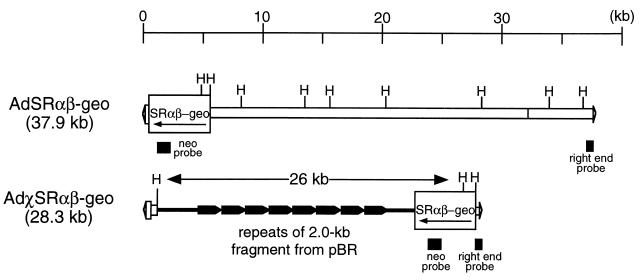
To compare the efficiencies of integration of the E1-substituted and helper-dependent adenovirus vectors, we rescued both types of vectors with the β-geo marker gene, a fusion of the E. coli β-galactosidase (β-Gal) gene and the neomycin phosphotransferase II gene (neo) (9) that is driven by the SRα promoter (27) (SRαβ-geo) (Fig. 1). The SRαβ-geo marker gene cassette (19) was subcloned into an adenovirus transfer plasmid, pXCX2 (26). AdSRαβ-geo, an E1-substituted adenovirus vector (ΔE1 vector), was rescued by cotransfection into the 293 cell line (Microbix, Toronto, Ontario, Canada) of the plasmid with pJM17 (18). The virus was then plaque isolated, propagated, and purified as described previously (10). The titer of the vector was 1.2 × 109 PFU/ml on 293 cells. A helper-dependent adenovirus vector DNA was constructed as follows. First, an AvrII-SmaI fragment of pFG140 (18) encompassing the junction of the ligated right end (452 bp) and left end (1,009 bp) of Ad5 was subcloned into an XbaI site of the charomid 9-22 vector (23). This adenovirus sequence contains two inverted terminal repeats and the packaging signal but does not encode any intact open reading frames of the parental human adenovirus type 5 (Ad5). Second, the SRαβ-geo cassette was subcloned into a SmaI site of the plasmid. The helper-dependent AdχSRαβ-geo vector (ΔAd vector) was rescued by cotransfection into 293 cells of the resultant plasmid with a helper-virus DNA–terminal protein complex. AdHprt, an adenovirus vector with E1 and E3 deleted and with a nonfunctional genomic sequence from the mouse Hprt locus, was used as a helper virus. The vector was propagated and purified as described previously (12, 19). After three rounds of purification by a CsCl density gradient, the titer of the vector was measured in situ, using 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) as a substrate, as previously described (17). The titer of the vector on the African green monkey cell line COS7 (American Type Culture Collection [ATCC], Rockville, Md.) was 2.4 × 109 β-Gal-transducing units (BTU)/ml, and that of the helper on 293 cells, as measured by a plaque assay, was 2.2 × 107 PFU/ml. Therefore, the vector stock contained 0.9% helper virus contamination.
FIG. 1.
Structure of adenovirus vectors. H, HindIII sites used for Southern hybridization analysis of the structure of integrated vectors.
The human cell lines HeLa (ATCC), HT1080 (ATCC), and KB (ATCC), the African green monkey kidney cell lines CV-1 (ATCC) and Vero (provided by Harumi Kasamatsu), the baby hamster kidney (BHK) cell line (provided by Debi Nayak), the Chinese hamster ovary (CHO) cell line (provided by Owen N. Witte), and the mouse cell line NIH 3T3 (ATCC) were infected with the vectors at 10 PFU/cell for the ΔE1 vector or 10 BTU/cell for the ΔAd vector. At 48 h postinfection, the infected cells were diluted and replated onto 96-well plates at densities of 105, 104, and 103 cells/plate. To avoid low plating efficiencies at low cell densities, the total cell numbers in each plate were adjusted to 105 by adding uninfected cells. After 24 h, medium containing G418 was added. The final concentration of G418 was 500 μg/ml (HeLa, HT1080, BHK, and NIH 3T3 cells) or 1,000 μg/ml (KB, CHO, CV-1, and Vero cells). G418-containing medium was added again 1 week later, and the G418-resistant colonies were counted and integration efficiencies were calculated 4 weeks postinfection. The results are summarized in Table 1. In most of the cell lines, both vectors integrated efficiently at frequencies of 10−4 to 10−5, but we occasionally observed a higher efficiency of 10−3 in HT1080 and KB cells. CHO cells showed relatively high integration efficiencies, particularly with the ΔAd vector, which achieved levels above 1%. In the experiments with HeLa, KB, and CHO cell lines, in which both types of vectors were tested in parallel, integration frequencies were severalfold higher with the ΔAd vector than with the ΔE1 vector, but both vectors showed similar efficiencies in HT1080 cells.
TABLE 1.
Integration efficiency
| Cell line | Expt | Integration efficiency with vector
|
|
|---|---|---|---|
| ΔE1 (10 PFU/cell) | ΔAd (10 BTU/cell)a | ||
| Human | |||
| HeLa | 1 | 3.1 × 10−4 | 9.3 × 10−4 |
| 2 | 3.3 × 10−5 | 5.5 × 10−5 | |
| 3 | 7.0 × 10−5 | 2.6 × 10−4 | |
| 4 | 1.2 × 10−4 | NT | |
| 5 | 3.0 × 10−5 | NT | |
| HT1080 | 1 | 3.3 × 10−4 | 2.9 × 10−4 |
| 2 | 2.4 × 10−3 | 1.8 × 10−3 | |
| KB | 1 | 5.0 × 10−5 | 2.6 × 10−3 |
| 2 | 4.0 × 10−4 | 3.7 × 10−3 | |
| 3 | 5.0 × 10−5 | NT | |
| Nonhuman | |||
| CHO | 1 | 4.4 × 10−3 | 1.1 × 10−2 |
| 2 | 3.0 × 10−3 | 2.0 × 10−2 | |
| 3 | 2.8 × 10−2 | NT | |
| 4 | 3.0 × 10−4 | NT | |
| CV-1 | 1 | 4.4 × 10−5 | 1.0 × 10−4 |
| 2 | 2.3 × 10−4 | NT | |
| 3 | 2.8 × 10−4 | NT | |
| BHK | 1 | 7.4 × 10−4 | NT |
| 2 | 2.0 × 10−4 | NT | |
| Vero | 1 | 2.2 × 10−4 | NT |
| 2 | 6.0 × 10−5 | NT | |
| NIH 3T3 | 1 | 1.0 × 10−5 | NT |
NT, not tested.
The relation of the PFU titer of the ΔE1 vector to the BTU titer of the ΔAd vector was determined as follows. Infected HeLa cells were harvested 4 h postinfection, at which time viral DNA synthesis had not yet started, and total DNA was extracted. The DNA was digested with HindIII and subjected to Southern hybridization with a β-Gal fragment (nucleotides 118 to 581 of ECLACZ; GenBank no. V00296) as a probe. Based on the intensity of the signal measured by a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.), the copy number of the ΔAd vector at 10 BTU/cell is not as high as that of the ΔE1 vector at 10 PFU/cell but corresponds to the copy number at 6.4 PFU/cell. Therefore, the integration efficiency of the ΔAd vector in Table 1 might be an underestimate, compared to that of the ΔE1 vector.
Integration efficiencies of the adenovirus vector that were assessed by G418 resistance could be affected by vector expression levels of the selectable marker gene, β-geo (25), and/or by the ability of vector DNA to integrate into chromosomes. To distinguish between these two possibilities, the infected HeLa, HT1080, KB, and CHO cells were plated into 100-mm dishes and subjected to G418 selection to obtain pools of stably transduced cells. Cell extracts were prepared from these cell pools, and the expression levels of the β-geo marker gene were compared. The levels of β-Gal activity showed no correlation to vector or cell types (data not shown), indicating that there are no significant differences in the levels of expression of SRαβ-geo once it integrates into a host chromosome, regardless of the vector backbones used. These results suggest that more efficient integration by the ΔAd vector is mainly due to the stronger ability of the ΔAd vector to integrate into cellular chromosomes. The higher integration efficiencies of the ΔAd vector can be attributed to the nature of completely nonviral genomic sequences or to the lack of leaky expression of viral genes, some of which inhibit normal cellular machinery, unlike those in the ΔE1 vector.
Although wild-type adenovirus does not integrate into the chromosomes of the permissive cells because its infection leads to lytic infection, it can integrate in nonpermissive cells (e.g., hamster cells infected with Ad12) (for a review, see reference 6). In addition, adenovirus mutants that code for a temperature-sensitive DNA-binding protein yield stable cell clones at nonpermissive temperatures (8). Chromosomal integration of an E1-substituted adenovirus vector in rat and simian cells has previously been reported (30). At a multiplicity of infection (MOI) of 200, the integration efficiencies were 0.4% for Rat2 cells and 0.75% for CV-1 cells. More recently, stable integration of E1-substituted adenovirus vector with the β-gal marker gene in mouse NIH 3T3, human A549, and primary human cells has been reported (38). Although the integration efficiency was 15% in these cell lines when ionizing radiation was used, the efficiency without radiation was not reported.
In our study, the ΔE1 and ΔAd vectors were used to investigate the frequencies of integration of adenovirus vectors in different cell types (Table 1). Human epidermoid cell lines such as HeLa and KB are known to be permissive for viral DNA replication of wild-type Ad5. BHK and Vero cells are semipermissive, and CV-1 and CHO cells are nonpermissive (16). Most of the cell lines infected by the ΔE1 vector showed similar integration efficiencies of ∼10−3 to 10−5 (Table 1), suggesting that factors determining cellular permissiveness do not affect the integration of adenovirus vectors. Under these conditions (MOI of 10), only up to 10 viral DNA molecules entered per cell, in contrast to approximately 106 DNA molecules per cell transferred by the calcium phosphate transfection method, in which 2.2 to 6.4% of DNA is internalized into the nucleus (21). Our results indicate that the ability of adenovirus DNA to integrate into host chromosomes seems extremely high compared to that of naked plasmid DNA, even though the end is protected by the terminal protein.
Sustained gene expression from integrated adenovirus vector.
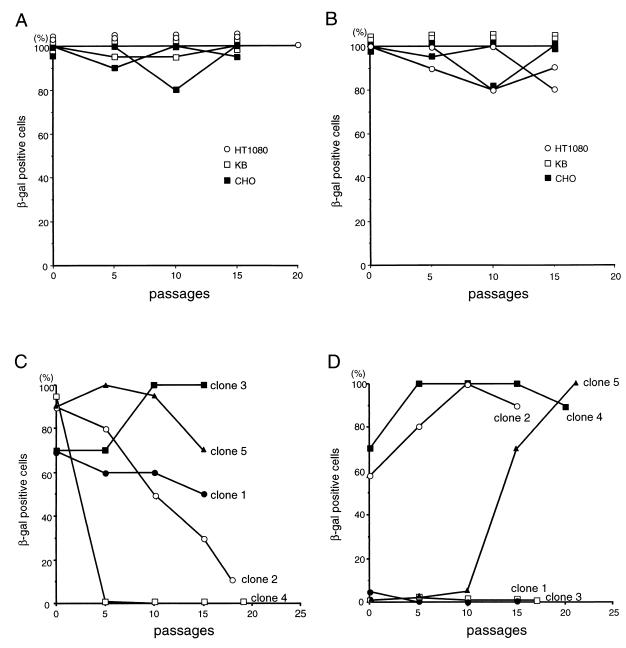
To analyze the persistence of gene expression from integrated adenovirus vectors, the pools and clones of stably transduced cells, obtained as described above, were cultured without G418 selection. Expression of β-Gal in these cells was detected by X-Gal staining at every five passages (Fig. 2). In most of the stable cell pools, β-Gal expression did not diminish over at least 15 passages, which corresponds to approximately 50 cell divisions (Fig. 2A and B). There were no significant differences in the time-course profiles of marker gene expression between the ΔE1 and ΔAd vectors. For more quantitative analysis, the levels of β-Gal enzymatic activity were also measured in triplicate (with the luminescent β-Gal gene reporter system 2; Clontech, Palo Alto, Calif.) before and after the series of passages without G418 (Table 2). The amount of protein in each sample was standardized by the Bradford method (Bio-Rad protein assay; Bio-Rad, Hercules, Calif.). Each cell pool showed different patterns of stability of β-Gal expression from the integrated vectors. In HT1080 cells, the levels of β-Gal activity tended to decrease with both the ΔE1 (0.5- and 0.7-fold) and ΔAd (0.4- and 0.6-fold) vectors. In KB cells, the levels of β-Gal activity remained the same for both the ΔE1 (1.0- and 1.2-fold) and ΔAd (0.9- and 1.4-fold) vectors. In CHO cells, the levels of β-Gal activity from the integrated ΔE1 vector (0.7- and 0.5-fold), but not from the ΔAd vector (1.1- and 0.7-fold), tended to decrease. We also analyzed the persistence of gene expression of individual stable HeLa clones (Fig. 2C and D). In two of five HeLa cell clones with stably integrated ΔE1 vectors, the level of β-Gal activity decreased from high to low over time (clones 2 and 4) (Fig. 2C). Interestingly, in HeLa clone 5, which was transduced by the ΔAd vector, the percentage of β-Gal-positive cells increased dramatically over time (Fig. 2D). This observation is in contrast to the expression of viral genes from integrated wild-type adenovirus, which is often shut off by methylation (4–6).
FIG. 2.
β-Gal expression from the integrated vectors over passages of the cells infected with ΔE1 vector (A and C) and ΔAd vector (B and D) without G418 selection. The stable cell pools (A and B) and HeLa clones (C and D) were maintained without G418 selection and stained by X-Gal every five passages; the percentages of X-Gal-positive cells were determined by microscopic observation. Two independent pools were analyzed for each cell line.
TABLE 2.
β-Gal activity before and after passages without G418 selection
| Cell line or clone | Vector (no. of colonies pooled) | β-Gal activitya
|
Change (fold) | |
|---|---|---|---|---|
| Before passages | After passages | |||
| HT1080 | ΔE1 1 (80) | 191.1 | 90.3 | 0.5 |
| ΔE1 2 (37) | 155.4 | 109.8 | 0.7 | |
| ΔAd 1 (73) | 214.3 | 86.6 | 0.4 | |
| ΔAd 2 (48) | 90.9 | 53.3 | 0.6 | |
| KB | ΔE1 1 (9) | 47.3 | 46.7 | 1.0 |
| ΔE1 2 (59) | 58.6 | 70.9 | 1.2 | |
| ΔAd 1 (∼400) | 173.6 | 157.0 | 0.9 | |
| ΔAd 2 (∼240) | 170.7 | 244.0 | 1.4 | |
| CHO | ΔE1 1 (∼200) | 215.1 | 151.7 | 0.7 |
| ΔE1 2 (28) | 52.6 | 25.9 | 0.5 | |
| ΔAd 1 (∼200) | 250.6 | 272.1 | 1.1 | |
| ΔAd 2 (74) | 50.7 | 37.2 | 0.7 | |
| HeLa clones | ΔE1 1 | 43.7 | 9.2 | 0.2 |
| ΔE1 2 | 35.4 | 5.9 | 0.2 | |
| ΔE1 3 | 15.8 | 148.0 | 9.4 | |
| ΔE1 4 | 33.3 | 0.0 | 0.0 | |
| ΔE1 5 | 90.4 | 59.1 | 0.7 | |
| ΔAd 1 | 2.6 | 1.7 | 0.7 | |
| ΔAd 2 | 182.3 | 169.4 | 0.9 | |
| ΔAd 3 | 8.4 | 3.8 | 0.5 | |
| ΔAd 4 | 66.3 | 70.1 | 1.1 | |
| ΔAd 5 | 8.3 | 98.7 | 11.9 | |
Values are in units (105) of enzyme per microgram of protein.
Structure of the integrated vector.
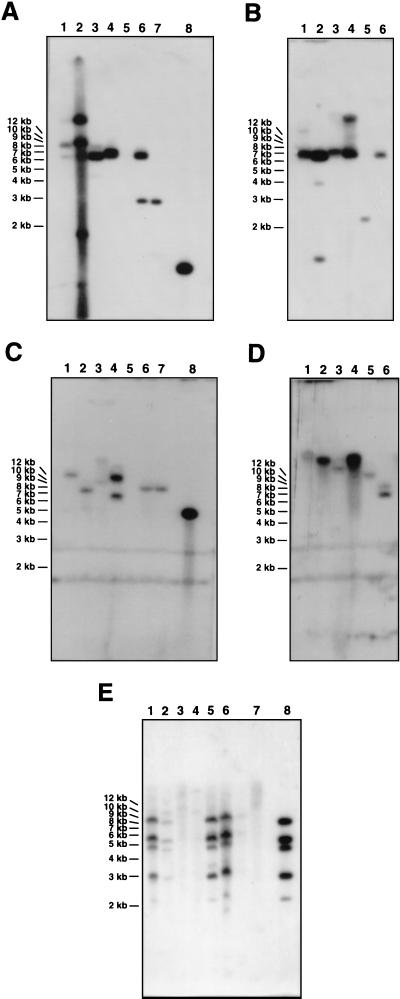
The structure of adenovirus DNA integrated into host chromosomes is known to vary, depending on the cell line and the type of virus. Rat embryonic cells transformed by Ad5 usually contain only the left end of the genome, whereas cell lines transformed by temperature-sensitive adenovirus mutants at semi- or nonpermissive temperatures often contain multiple copies of all or most of the adenovirus genome (6). To analyze the structure of integrated adenovirus vectors, genomic DNA was extracted from stably transduced HeLa cell clones, digested with HindIII, and subjected to Southern hybridization. Fragments of the neo gene, the right end of Ad5 (nucleotides 35032 to 35780 of ADRCOMPGEN; GenBank no. M73260 and M29978), and full-length Ad5 DNA were used as probes (Fig. 1). The right-end probe hybridized with the DNA fragment between the rightmost HindIII site in the adenovirus genome and a HindIII site to the right of the integration site, which produced a band representing a size unique to the integration site. In ΔE1 stable clones, the right-end probe hybridized with one band in three of five clones, indicating a single-copy integration of the right end of the vector. However, clones 2 and 5 had three and two bands, respectively, indicating multiple integrations of the right end in these clones (Fig. 3A). Similarly, the neo probe with ΔE1 integrants produced a band representing a size unique to a left-end integration junction. Contrary to the results with the right-end probe, clones 1, 2, and 5 showed single-copy integration, while clones 3 and 4 showed two integrants each (Fig. 3C). Clone 1 had one additional faint band hybridizing with the right-end probe. This faint band might indicate that in some cells the integrated vector is rearranged. Finally, the full-length adenovirus probe should produce eight bands specific for the full-length vector, with two additional bands corresponding to the junctions of integration sites at both ends of the vector (Fig. 1). All of the clones showed the bands common to AdSRαβ-geo viral DNA (Fig. 3E, lanes 1, 2, 5, and 6). These results indicate that all of the HeLa ΔE1 stable clones had at least one copy of full-length vector integrated into their chromosomes, which was often accompanied by additional integration events. In the case of the stable ΔAd clones, all the integrants showed a single band with both the right-end and the neo probes (Figs. 3B and D), suggesting that most of the stable cell clones had a single-copy integration. Clone 2 had two additional faint bands hybridizing with the right-end probe, suggesting rearrangement of integrated vectors in some cells. The neo probe should produce a 26-kb internal fragment encompassing the SRαβ-geo cassette and the stuffer sequence consisting of repeats of a 2-kb fragment from pBR (Fig. 1). However, at least two clones (clones 3 and 5) (Fig. 3D) showed fragments smaller than those of other clones. Therefore, in these clones, an internal fragment was deleted before or after integration. The deletions might be associated with the nature of the repetitive sequences of the fragment. In CV-1 cell clones, ΔE1 vector showed patterns of integration similar to those in HeLa cells (i.e., a single full-length copy with an additional end sequence). On the other hand, ΔAd vector tended to integrate at multiple sites in many clones but, unlike in HeLa cells, no internal deletion was detected by Southern analysis (data not shown). It is not clear why the ΔE1 and ΔAd vectors show distinct patterns of integration.
FIG. 3.
Southern analysis of integrated vector DNA. DNA from HeLa cell clones infected with the ΔE1 (A, C, and E) or ΔAd (B, D, and E) vector was digested with HindIII and subjected to Southern hybridization with the right-end (A and B), neo (C and D), and full-length Ad5 DNA (E) probes. (A and C) Lanes 1 to 3, ΔE1 clones 1 to 3, respectively; lane 4, clone 4 before the passages without selection; lane 5, clone 4 after the passages; lane 6, clone 5 before the passages; lane 7, clone 5 after the passages; lane 8, AdSRαβ-geo DNA. (B and D) Lanes 1 to 4, ΔAd clones 1 to 4, respectively; lane 5, clone 5 before the passages without selection; lane 6, clone 5 after the passages. (E) Lane 1, ΔE1 clone 5; lane 2, ΔE1 clone 3; lane 3, ΔAd clone 3; lane 4, ΔAd clone 4, lane 5, ΔE1 clone 1; lane 6, ΔE1 clone 2; lane 7, uninfected HeLa; lane 8, AdSRαβ-geo DNA.
We also examined the changes in the structure of integrated vector in three HeLa clones before and after passages without selection. In ΔE1 clone 4, the integrated vector signal disappeared, as indicated by both probes after 15 passages, consistent with the decrease in levels of β-Gal-positive cells from 95 to 0% (Fig. 3A and C, lanes 4 and 5). In ΔE1 clone 5, which had a moderate decrease in β-Gal-positive cells (from 90 to 70%), one of the two fragments hybridizing with the right-end probe disappeared (Fig. 3A, lanes 6 and 7), while the only band that hybridized with the neo probe remained unaltered before and after passages without selection (Fig. 3C, lanes 6 and 7). On the other hand, in the ΔAd stable clone 5, which had increased activity (from 1 to 100%), the integrated vector showed a rearranged pattern after 15 passages (Fig. 3B and D, lanes 5 and 6). This rearrangement might account for the increased β-Gal activity. Rearrangement of integrated viral DNA was also documented in hamster cells with stably integrated Ad12 (14).
In summary, although adenovirus vectors integrate into host chromosomes relatively efficiently, unlike retroviral integration, most of the stable clones have an extra fragment(s) of the vector or deleted vector. Gene expression from the integrated vector is relatively stable. However, integrated vectors sometimes become further rearranged, resulting in an altered level of gene expression. Adenovirus vectors can infect a variety of cell types at very high efficiencies. Our results suggest that it might be possible to use an adenovirus vector to establish stable cell clones in cells which are refractory to other gene delivery methods. Considering the somewhat unstable nature of integrated adenovirus vector DNA, selection strategies for the integrated DNA might be needed to obtain long-term expression. It is known that gene expression from retroviral vectors is shut off in some primary cell cultures. Therefore, it would be of interest to analyze the integration frequencies of adenovirus vectors and the longevity of gene expression, especially in primary cultures such as cultures of hematopoietic cells, which are one of the main targets for human gene therapy. On the other hand, because one of the advantages of an adenovirus vector for human gene therapy is its rare chromosomal integration, thereby circumventing potential insertional mutagenesis of cancer-related genes, it would be important to determine how frequently an adenovirus vector integrates in vivo. There are reports suggesting that stable integration of adenovirus vectors into the host chromosome occurs after in vivo gene transfer in animals (1, 22). Efficient production in transgenic mice by adenovirus gene transfer into fertilized eggs was also reported (29). Therefore, considering the high MOIs usually used for in vivo gene transfer, elucidation of adenovirus integration in vivo is a very important issue for evaluating the safety of adenovirus vectors for human gene therapy.
Acknowledgments
A.H. and S.S. contributed equally to this work.
We thank Harumi Kasamatsu, Debi Nayak, and Owen N. Witte for providing cell lines, Frank L. Graham for providing the pXCX2 and pJM17 plasmids, Izumu Saito for providing the charomid vector, Arnie Berk for critical discussion and for reading the manuscript, Wendy Aft for preparation of the manuscript, Amit Oberai for generating preliminary data, and Lena Kim for technical support.
This work was supported by NIH grant AI-42214.
REFERENCES
- 1.Brown G R, Thiele D L, Silva M, Beutler B. Adenoviral vectors given intravenously to immunocompromised mice yield stable transduction of the colonic epithelium. Gastroenterology. 1997;112:1586–1594. doi: 10.1016/s0016-5085(97)70040-4. [DOI] [PubMed] [Google Scholar]
- 2.Crystal R G. Transfer of genes to humans: early lessons and obstacles to success. Science. 1995;270:404–410. doi: 10.1126/science.270.5235.404. [DOI] [PubMed] [Google Scholar]
- 3.Dai Y, Schwarz E M, Gu D, Zhang W W, Sarvetnick N, Verma I M. Cellular and humoral immune responses to adenoviral vectors containing factor IX gene: tolerization of factor IX and vector antigens allows for long-term expression. Proc Natl Acad Sci USA. 1995;92:1401–1405. doi: 10.1073/pnas.92.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doerfler W. Abortive infection and malignant transformation by adenoviruses: integration of viral DNA and control of viral gene expression by specific patterns of DNA methylation. Adv Virus Res. 1991;39:89–128. doi: 10.1016/s0065-3527(08)60793-9. [DOI] [PubMed] [Google Scholar]
- 5.Doerfler W. DNA methylation and its functional significance: studies on the adenovirus system. Curr Top Microbiol Immunol. 1984;108:79–98. doi: 10.1007/978-3-642-69370-0_6. [DOI] [PubMed] [Google Scholar]
- 6.Doerfler W. Uptake, fixation, and expression of foreign DNA in mammalian cells: the organization of integrated adenovirus DNA sequences. Curr Top Microbiol Immunol. 1982;101:127–194. doi: 10.1007/978-3-642-68654-2_6. [DOI] [PubMed] [Google Scholar]
- 7.Fisher K J, Choi H, Burda J, Chen S, Wilson J M. Recombinant adenovirus deleted of all viral genes for gene therapy of cystic fibrosis. Virology. 1996;217:11–22. doi: 10.1006/viro.1996.0088. [DOI] [PubMed] [Google Scholar]
- 8.Fisher P B, Babiss L E, Weinstein I B, Ginsberg H S. Analysis of type 5 adenovirus transformation with a cloned rat embryo cell line (CREF) Proc Natl Acad Sci USA. 1982;79:3527–3531. doi: 10.1073/pnas.79.11.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 10.Graham F L, Prevec L. Methods for construction of adenovirus vectors. Mol Biotechnol. 1995;3:207–220. doi: 10.1007/BF02789331. [DOI] [PubMed] [Google Scholar]
- 11.Haecker S E, Stedman H H, Balice-Gordon R J, Smith D B, Greelish J P, Mitchell M A, Wells A, Sweeney H L, Wilson J M. In vivo expression of full-length human dystrophin from adenoviral vectors deleted of all viral genes. Hum Gene Ther. 1996;7:1907–1914. doi: 10.1089/hum.1996.7.15-1907. [DOI] [PubMed] [Google Scholar]
- 12.Kochanek S, Clemens P R, Mitani K, Chen H-H, Chan S, Caskey C T. A new adenoviral vector: replacement of all viral coding sequences with 28 kb of DNA independently expressing both full-length dystrophin and β-galactosidase. Proc Natl Acad Sci USA. 1996;93:5731–5736. doi: 10.1073/pnas.93.12.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozarsky K F, McKinley D R, Austin L L, Raper S E, Stratford-Perricaudet L D, Wilson J M. In vivo correction of low density lipoprotein receptor deficiency in the Watanabe heritable hyperlipidemic rabbit with recombinant adenoviruses. J Biol Chem. 1994;269:13695–13702. [PubMed] [Google Scholar]
- 14.Kuhlmann I, Doerfler W. Loss of viral genomes from hamster tumor cells and nonrandom alterations in patterns of methylation of integrated adenovirus type 12 DNA. J Virol. 1983;47:631–636. doi: 10.1128/jvi.47.3.631-636.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar-Singh R, Chamberlain J S. Encapsidated adenovirus minichromosomes allow delivery and expression of a 14 kb dystrophin cDNA to muscle cells. Hum Mol Genet. 1996;5:913–921. doi: 10.1093/hmg/5.7.913. [DOI] [PubMed] [Google Scholar]
- 16.Lucher L A. Abortive adenovirus infection and host range determinants. Curr Top Microbiol Immunol. 1995;199:119–152. doi: 10.1007/978-3-642-79496-4_8. [DOI] [PubMed] [Google Scholar]
- 17.MacGregor G R, Mogg A E, Burke J F, Caskey C T. Histochemical staining of clonal mammalian cell lines expressing E. coli β-galactosidase indicates heterogeneous expression of the bacterial gene. Somatic Cell Mol Genet. 1987;13:253–265. doi: 10.1007/BF01535207. [DOI] [PubMed] [Google Scholar]
- 18.McGrory W J, Bautista D S, Graham F J. A simple technique for the rescue of early region I mutants into infectious human adenovirus type 5. Virology. 1988;163:614–617. doi: 10.1016/0042-6822(88)90302-9. [DOI] [PubMed] [Google Scholar]
- 19.Mitani K, Graham F L, Caskey C T, Kochanek S. Rescue, propagation, and partial purification of a helper virus-dependent adenovirus vector. Proc Natl Acad Sci USA. 1995;92:3854–3858. doi: 10.1073/pnas.92.9.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morsy M A, Gu M, Motzel S, Zhao J, Lin J, Su Q, Allen H, Franlin L, Parks R J, Graham F L, Kochanek S, Bett A J, Caskey C T. An adenoviral vector deleted for all viral coding sequences results in enhanced safety and extended expression of a leptin transgene. Proc Natl Acad Sci USA. 1998;95:7866–7871. doi: 10.1073/pnas.95.14.7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orrantia E, Chang P L. Intracellular distribution of DNA internalized through calcium phosphate precipitation. Exp Cell Res. 1990;190:170–174. doi: 10.1016/0014-4827(90)90181-9. [DOI] [PubMed] [Google Scholar]
- 22.Overturf K, Al-Dhalimy M, Ou C N, Finegold M, Tanguay R, Lieber A, Kay M, Grompe M. Adenovirus-mediated gene therapy in a mouse model of hereditary tyrosinemia type I. Hum Gene Ther. 1997;8:513–521. doi: 10.1089/hum.1997.8.5-513. [DOI] [PubMed] [Google Scholar]
- 23.Saito I, Stark G R. Charomids: cosmid vectors for efficient cloning and mapping of large or small restriction fragments. Proc Natl Acad Sci USA. 1986;83:8664–8668. doi: 10.1073/pnas.83.22.8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiedner G, Morral N, Parks R J, Wu Y, Koopmans S C, Langston C, Graham F L, Beaudet A L, Kochanek S. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat Genet. 1998;18:180–183. doi: 10.1038/ng0298-180. [DOI] [PubMed] [Google Scholar]
- 25.Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 26.Spessot R, Inchley K, Hupel T M, Bacchetti S. Cloning of the herpes simplex virus ICP4 gene in an adenovirus vector: effects on adenovirus gene expression and replication. Virology. 1989;168:378–387. doi: 10.1016/0042-6822(89)90279-1. [DOI] [PubMed] [Google Scholar]
- 27.Takebe Y, Seiki M, Fujisawa J-I, Hoy P, Yokota K, Arai K-I, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tripathy S K, Black H B, Goldwasser E, Leiden J M. Immune responses to transgene-encoded proteins limit the stability of gene expression after injection of replication-defective adenovirus vectors. Nat Med. 1996;2:545–550. doi: 10.1038/nm0596-545. [DOI] [PubMed] [Google Scholar]
- 29.Tsukui T, Kanegae Y, Saito I, Toyoda Y. Transgenesis by adenovirus-mediated gene transfer into mouse zona-free eggs. Nat Biotechnol. 1996;14:982–985. doi: 10.1038/nbt0896-982. [DOI] [PubMed] [Google Scholar]
- 30.Van Doren K, Hanahan D, Gluzman Y. Infection of eucaryotic cells by helper-independent recombinant adenoviruses: early region 1 is not obligatory for integration of viral DNA. J Virol. 1984;50:606–614. doi: 10.1128/jvi.50.2.606-614.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verma I M, Somia N. Gene therapy—promises, problems and prospects. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, Haecker S E, Su Q, Wilson J M. Immunology of gene therapy with adenoviral vectors in mouse skeletal muscle. Hum Mol Genet. 1996;5:1703–1712. doi: 10.1093/hmg/5.11.1703. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Nunes F A, Berencsi K, Furth E E, Gonczol E, Wilson J M. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc Natl Acad Sci USA. 1994;91:4407–4411. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y, Wilson J M. Clearance of adenovirus-infected hepatocytes by MHC class I-restricted CD4+ CTLs in vivo. J Immunol. 1995;155:2564–2570. [PubMed] [Google Scholar]
- 35.Yang Y, Xiang Z, Ertl H C, Wilson J M. Upregulation of class I major histocompatibility complex antigens by interferon-γ is necessary for T-cell-mediated elimination of recombinant adenovirus-infected hepatocytes in vivo. Proc Natl Acad Sci USA. 1995;92:7257–7261. doi: 10.1073/pnas.92.16.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeh P, Perricaudet M. Advances in adenoviral vectors: from genetic engineering to their biology. FASEB J. 1997;11:615–623. doi: 10.1096/fasebj.11.8.9240963. [DOI] [PubMed] [Google Scholar]
- 37.Yei S, Mittereder N, Tang K, Sullivan C O, Trapnell B C. Adenovirus-mediated gene transfer for cystic fibrosis: quantitative evaluation of repeated in vivo vector administration to the lung. Gene Ther. 1994;1:192–200. [PubMed] [Google Scholar]
- 38.Zeng M, Cerniglia G J, Eck S L, Stevens C W. High-efficiency stable gene transfer of adenovirus into mammalian cells using ionizing radiation. Hum Gene Ther. 1997;8:1025–1032. doi: 10.1089/hum.1997.8.9-1025. [DOI] [PubMed] [Google Scholar]