Abstract
Previous work has shown that deletions of genomic segments at nucleotide (nt) positions +238 to +253, i.e., construct BH10-LD3, or nt positions +261 to +274, i.e., construct BH10-LD4, within the human immunodeficiency virus type 1 (HIV-1) dimerization initiation site (DIS) destroyed DIS secondary structure and dramatically reduced viral replication capacity. Surprisingly, two point mutations located within the viral peptide 2 (p2) and nucleocapsid (NC) protein termed MP2 and MNC, respectively, were able to compensate for this defect. Since the MP2 mutation involves an amino acid substitution near the cleavage site between p2 and NC, we investigated the effects of the above-mentioned deletions on the processing of Gag proteins. Immunoprecipitation assays performed with monoclonal antibodies against viral capsid (CA) (p24) protein showed that p2 was cleaved from CA with less efficiency in viruses that contained the LD3 and LD4 deletions than in wild-type viruses. The presence of the two compensatory mutations, MP2 and MNC, increased the efficiency of the cleavage of p2 from CA, but neither mutation alone had this effect or was sufficient to compensate for the observed impairment in infectiousness. A virus that contained both of the above-mentioned deletions within the DIS was also impaired in regard to processing and infectiousness, and it could likewise be compensated by the MP2 and MNC point mutations. These results suggest that the DIS region of HIV-1 RNA plays an important role in the processing of Gag proteins.
Human immunodeficiency virus type 1 (HIV-1) selectively encapsidates two identical copies of full-length plus-strand viral genomic RNA that exist as dimers within virions and are noncovalently linked across a segment at their 5′ ends (5). Encapsidation and dimerization are tightly associated events, as suggested by the colocalization of RNA signals responsible for both of these processes (5). An RNA segment between the primer binding site and the beginning of the gag gene has been identified as a major cis-acting element involved in viral RNA packaging and dimerization (1, 8, 17, 18, 21, 23, 25). Sequence analysis and nuclease accessibility mapping studies revealed the presence of four distinct stem-loop structures in this area, SL1, SL2, SL3, and SL4; of these, SL1, SL3, and SL4 are major motifs that determine selective encapsidation (3, 9, 16, 25, 26). The SL1 region of viral genomic RNA has been identified mainly because it contains palindromic loop sequences (GCGCGC), and it has been proposed to be the virus RNA dimerization initiation site (DIS) on the basis of cell-free experiments (2, 11, 19, 24, 29, 33).
Viral genomic RNA is packaged at the same time as HIV-1 structural proteins (Gag and Gag-Pol) begin to assemble at the cell membrane. Mutagenesis studies have shown that the viral nucleocapsid (NC) domain, a highly conserved feature of the Gag protein in retroviruses (except for spumaviruses), is required for packaging of viral RNA to occur. This is also supported by the finding that the NC protein can bind to viral RNA in the area of the packaging signals (i.e., SL1, SL3, and SL4) with high affinity in cell-free assays (1, 6, 7, 10, 12, 14, 31). This concept is also supported by experiments on viral SL1 deletion mutants for which packaging was restored to wild-type levels by a point mutation in the NC protein in which a Thr at amino acid position 24 was changed to Ile (22). However, the impaired infectiousness of the mutated viruses could be restored only if a second point mutation also occurred at the cleavage site between peptide 2 (p2) and NC, i.e., MP2, changing a Thr at amino acid position 12 to Ile (22). This involved one of eight amino acids within the cleavage sequence. Therefore, the SL1 motif may be involved in other aspects of viral replication in addition to its role in RNA dimerization and packaging of viral genomic RNA.
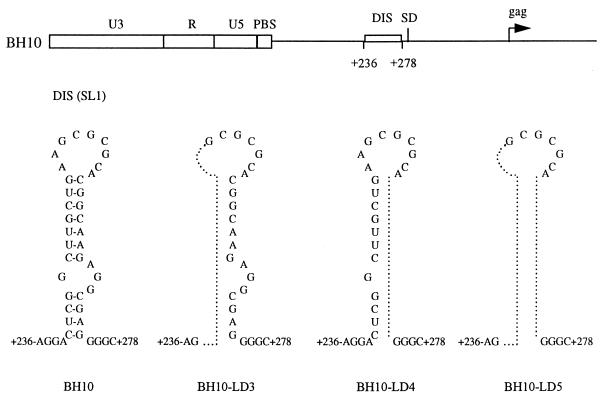
Previous work in our laboratory has shown that the second of the compensatory mutations referred to above, MP2, resulted in an altered amino acid sequence at the initial cleavage site in the Gag protein (22). This suggests that the deletion of the SL1 region may have resulted in abnormal processing of Gag proteins and that the MP2 point mutation may have corrected this deficit. To investigate this possibility, we studied the processing of Gag precursor proteins by radiolabeling transfected COS-7 cells and performing immunoprecipitation assays. Three deletion mutations involving various sequences within the DIS region were employed in these experiments: BH10-LD3 (nucleotides [nt] +238 to +253), BH10-LD4 (nt +261 to +274), and a larger deletion encompassing both of the above-mentioned alterations, i.e., BH10-LD5 (nt +238 to +253 and +261 to +274) (Fig. 1).
FIG. 1.
Schematic illustration of the BH10-LD3, BH10-LD4, and BH10-LD5 deletion mutant constructs, in which DNA sequences at nt positions +238 to +253, +261 to +274, and both +238 to +253 and +261 to +274, respectively, have been deleted. Sequences eliminated in each of the constructs are identified by a dotted line within the secondary structure of the DIS region. PBS, primer binding site; SD, splice donor.
Partial deletions in the DIS region delay cleavage between p2 and CA.
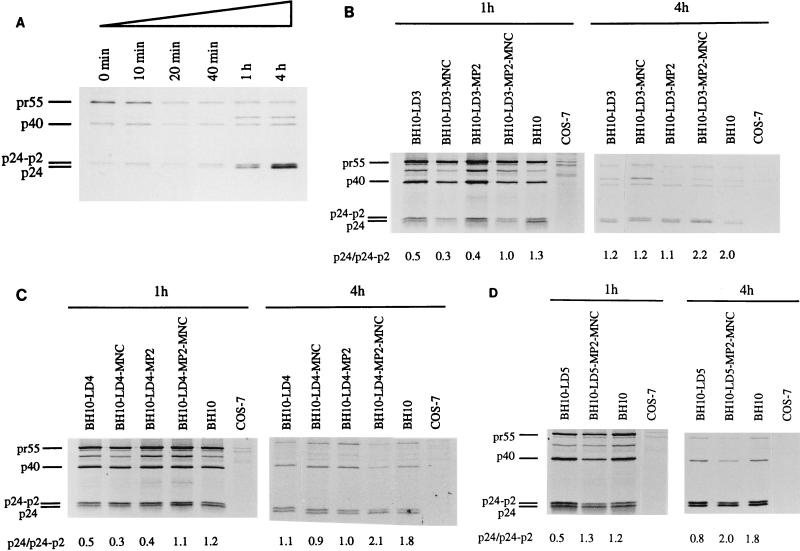
We have previously shown that the BH10-LD3 and BH10-LD4 deletion mutations, both of which eliminate select DIS sequences (Fig. 1), dramatically decreased viral replication capacity (22). To further explore the mechanisms responsible for this observation, we radiolabeled COS-7 cells that had been transfected with either wild-type BH10 DNA or DNA from the mutated BH10-LD3 or BH10-LD4 constructs, and we evaluated the processing of Gag proteins by immunoprecipitation of cell lysates with anti-capsid (anti-CA) immunoglobulin G monoclonal antibody. Time course experiments were performed in which cells transfected with wild-type BH10 DNA were lysed at 0, 10, 20, or 40 min or 1 or 4 h after radiolabeling with [35S]Met and [35S]Cys. Cleavage of pr55 is evident from the fact that decreasing levels of this protein were associated with increased amounts of CA (p24), as cells were cultured over longer periods (Fig. 2A). This process was also characterized by the presence of decreasing amounts of the CA-p2 protein as well as increasing amounts of CA over the time studied (Fig. 2A). Since CA (p24) is directly derived from the CA-p2 protein, the ratio between these two entities is an effective measure of the processing of Gag proteins. In the following studies, cells were lysed at 1 or 4 h after radiolabeling and the ratios of CA to CA-p2 were determined to evaluate Gag processing.
FIG. 2.
Processing of Gag polyproteins in mutated and wild-type viruses as studied by gel analysis of samples that had been subjected to radiolabeling and immunoprecipitation. COS-7 cells were transfected with wild-type BH10 or with mutated viral DNA (BH10-LD3, BH10-LD4, and BH10-LD5), and they were radiolabeled with [35S]Met and [35S]Cys for 1 h thereafter. Cells were lysed at either 1 or 4 h after radiolabeling, and viral proteins in cell lysates were immunoprecipitated with immunoglobulin G monoclonal antibodies against the CA protein. (A) Time course experiments after the radiolabeling of COS-7 cells that had been transfected with wild-type BH10 DNA. The cells were lysed at 0, 10, 20, or 40 min or at 1 or 4 h after radiolabeling. (B to D) Results of studies performed with mutated BH10-LD3, BH10-LD4, and BH10-LD5, respectively. Mock-transfected COS-7 cells served as negative controls and underwent the radiolabeling and immunoprecipitation protocols described above. The relative density ratios of the CA (p24) and CA-p2 bands are shown at the bottoms of the gels.
Studies that compared the mutant (BH10-LD3 and BH10-LD4) and wild-type (BH10) viruses showed that the relative densities of the CA and CA-p2 bands, quantified by molecular imaging (with an apparatus from Bio-Rad, Mississauga, Ontario, Canada), differed in each case and that the ratios of CA to CA-p2 were significantly lower for both of the mutant viruses than they were for the wild type at each time point (Fig. 2B and C). Therefore, the CA-p2 protein was processed to a lesser extent in the mutant viruses, indicating that Gag processing had been delayed with each of the BH10-LD3 and BH10-LD4 constructs.
Roles of the MP2 and MNC point mutations.
The MP2 and MNC point mutations can restore wild-type replication kinetics to viruses containing the LD3 and LD4 deletions (22). To determine whether these mutations might also correct the diminished processing efficiency of CA-p2, we performed radiolabeling and immunoprecipitation experiments with the viruses BH10-LD3-MP2-MNC and BH10-LD4-MP2-MNC, into which these point mutations had been inserted by site-directed mutagenesis. Figure 2B and C show that similar ratios of CA to CA-p2 existed among the wild-type BH10 virus and each of the two deletion-mutated viruses, into which the MP2 and MNC substitutions had been inserted, at both 1 and 4 h after radiolabeling. In contrast, neither the MP2 nor the MNC mutation restored processing to wild-type levels (Fig. 2B and C); this is consistent with previous observations that neither mutation alone could restore wild-type replication levels to the BH10-LD3 and BH10-LD4 viruses. Future studies will assess whether the MP2 and MNC mutations can affect other changes in the psi (ψ) region of viral RNA.
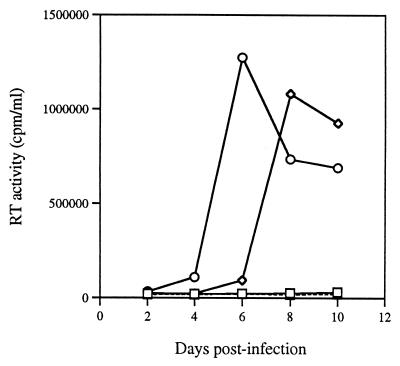
We also generated a larger deletion, BH10-LD5, in which all of the stem sequences of the DIS had been deleted. The results of studies in which viruses harvested from transfected COS-7 cells were used to infect MT-2 cells showed that very low levels of reverse transcriptase were present (Fig. 3) and that a loss of viral infectiousness had occurred. Furthermore, Fig. 2D shows that the cleavage of p2 from CA-p2 in BH10-LD5 viruses was delayed at both 1 and 4 h compared with p2 cleavage in wild-type BH10.
FIG. 3.
Compensation of defects caused by the LD5 deletion by the MP2 and MNC point mutations. MT-2 cells were infected with equivalent amounts of mutant (BH10-LD5 [□] and BH10-LD5-MP2-MNC [◊]) or wild-type (BH10 [○]) viruses on the basis of CA protein levels, and virus production was monitored by levels of reverse transcriptase (RT) generated thereafter. Mock-infected cells (▵) exposed to a heat-inactivated wild-type virus served as a negative control.
Site-directed mutagenesis was used to generate the construct BH10-LD5-MP2-MNC, and infectivity assays in MT-2 cells showed that its replication capacity was delayed by only 1 to 2 days compared to that of the wild-type BH10 (Fig. 3). Consistently, the processing of CA-p2 in BH10-LD5-MP2-MNC proceeded normally (Fig. 2D).
Abnormal ultrastructure of virus particles containing the LD5 deletion.
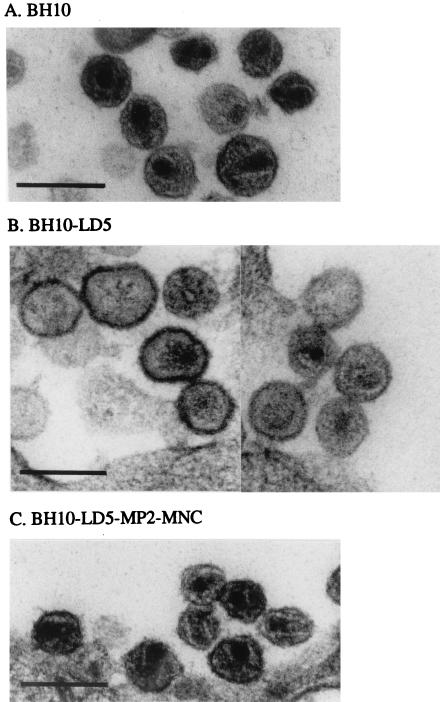
Gag processing is normally accompanied by the maturation of virus particles either during or following budding from the cell membrane, and this is characterized by the formation of a conical core. To investigate the maturation and ultrastructure of virus particles containing the LD5 deletion, COS-7 cells transfected by BH10-LD5 or wild-type BH10 were fixed with fresh 2.5% glutaraldehyde in phosphate-buffered saline, and thin sections were evaluated by electron microscopy. In the case of cells transfected with BH10, ≈70% of the virus particles contained a condensed conical core, indicating that proper maturation had occurred (Fig. 4A). In contrast, ≈75% of the viruses from transfections with BH10-LD5 were immature and possessed abnormal structures (Fig. 4B; see the electron-dense material under the cellular envelope in the absence of a condensed conical core as well as improperly condensed cores). Studies performed with the BH10-LD5-MP2-MNC virus revealed that these defects had been corrected (Fig. 4C).
FIG. 4.
Electron microscopy of COS-7 cells transfected with wild-type BH10 DNA, mutated BH10-LD5 DNA, or mutated BH10-LD5 DNA that also contained the MP2 and MNC substitutions, i.e., BH10-LD5-MP2-MNC. One hundred virus particles were scored for each micrograph. Bars, 0.2 μm.
The HIV-1 DIS structure was first identified on the basis of a highly conserved six-base palindrome (GCGCGC) within its loop portion, and on this basis a kissing-loop model for viral RNA dimer initiation has been proposed. In this model, the two RNA genomic strands interact through base-pairing between loop palindromes and the two stems can then form an interstrand duplex. Cell-free studies have shown that disruption of these palindromic sequences abrogates RNA dimerization (2, 11, 19, 24, 29, 33). However, data obtained through in vivo approaches have been equivocal; some studies have shown that mutagenesis of the DIS decreased the thermal stability of RNA dimers (10, 20), while others have reported an absence of effect in this regard (4, 32). In one case, viruses containing a modified DIS reverted to repair the palindromic nature of the loop (4). In contrast, the DIS is known to be involved in the encapsidation of viral genomic RNA, as shown through mutagenesis studies (4, 20, 22, 26, 28). This study further demonstrates the effects of deletions of DIS sequences on Gag polyprotein processing.
The HIV-1 gag gene is translated as a polyprotein that is cleaved by protease to yield final products that include a matrix (MA), a CA, p2, a NC, peptide 1 (p1), and peptide 6 (p6). The initial cleavage event occurs between the C terminus of p2 and the N terminus of NC to yield a MA-CA-p2 intermediate that is subsequently cleaved to generate MA and CA-p2. In contrast, cleavage of p2 from CA is a relatively late event in viral maturation (27). These various cleavages occur sequentially and are tightly associated with virion maturation; hence, the cleavage intermediates may regulate both this ordered polyprotein processing and virion morphogenesis.
NC-p1-p6, once it is cleaved from the Gag polyprotein, is believed to bind to genomic RNA to form the inner ribonucleoprotein core. Cleavage between MA and CA-p2 releases CA-p2 from the membrane, and the final removal of p2 from CA has been shown to be critical to the formation of a normal cone-shaped core (34). We have demonstrated that deletions of the DIS region resulted in the accumulation of an intermediate CA-p2 product as well as in aberrant virion maturation and diminished viral infectivity. This research provides support for the notion that cleavage of p2 from CA is required to form a well-condensed conical core within mature virus particles.
The regulation of Gag polyprotein processing is mainly attributable to amino acids located at cleavage junctions and to the conformation of individual Gag proteins that serve as substrates for these reactions. For instance, the rate of cleavage between the p2 and NC proteins is approximately 10-fold higher than that at other sites on pr55Gag, accounting for the fact that NC is the first protein to be released from the Gag precursor (30). Our research raises the possibility that noncoding viral RNA leader sequences may also play a role in the processing of Gag proteins.
The mechanisms whereby deletions within the DIS might affect Gag processing remain unclear. The defective encapsidation of viral genomic RNA that results from the aforesaid deletions might play a role. Studies performed with baculovirus-infected insect cells that expressed Gag showed that immature virus-like particles can be generated without the packaging of viral RNAs (13). However, the proper assembly and maturation of virus particles may indeed require the normal packaging of viral genomic RNA. In support of this notion, cell-free experiments have shown that the assembly of helically hollow cylinders by CA-NC polyproteins required the presence of viral RNA and that efficient cleavage of NC-p1-p6 depended on the binding of these polyproteins to viral RNA molecules (15). Since the BH10-LD3 deletion results in diminished viral RNA packaging (22), a lack of viral genomic RNA in virus particles may contribute to the Gag processing defect in this situation.
In summary, we have provided evidence that the DIS, located within the noncoding leader region of viral RNA, can participate in the processing of Gag polyproteins, in addition to having a well-described role in dimerization and encapsidation of viral genomic RNA.
Acknowledgments
This work was supported by grants to Mark A. Wainberg from the Medical Research Council of Canada.
REFERENCES
- 1.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awang G, Sen D. Mode of dimerization of HIV-1 genomic RNA. Biochemistry. 1993;32:11453–11457. doi: 10.1021/bi00093a024. [DOI] [PubMed] [Google Scholar]
- 3.Baudin G, Marquet R, Isel C, Darlix J L, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout B, van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 6.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 7.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clavel F, Orenstein J M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J Virol. 1990;64:5230–5234. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clever J L, Wong M L, Parslow T G. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wikde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 14.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross I, Hohenberg H, Huckhagel C, Krausslich H-G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 16.Harrison G P, Lever A M L. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi T, Shioda T, Iwakura Y, Shibuta H. RNA packaging signal of human immunodeficiency virus type 1. Virology. 1992;188:590–599. doi: 10.1016/0042-6822(92)90513-o. [DOI] [PubMed] [Google Scholar]
- 18.Kim H J, Lee K, O’Rear J J. A short sequence upstream of the 5′ major splice site is important for encapsidation of HIV-1 genomic RNA. Virology. 1994;198:336–340. doi: 10.1006/viro.1994.1037. [DOI] [PubMed] [Google Scholar]
- 19.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 20.Laughrea M, Jetté L, Mak J, Kleiman L, Liang C, Wainberg M A. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J Virol. 1997;71:3397–3406. doi: 10.1128/jvi.71.5.3397-3406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang C, Rong L, Laughrea M, Kleiman L, Wainberg M A. Compensatory point mutations in the human immunodeficiency virus type 1 Gag region that are distal from deletion mutations in the dimerization initiation site can restore viral replication. J Virol. 1998;72:6629–6636. doi: 10.1128/jvi.72.8.6629-6636.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luban J, Goff S P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J Virol. 1994;68:3784–3793. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marquet R, Paillart J C, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus type 1 RNA involves sequences located upstream of the splice donor site. Nucleic Acids Res. 1994;22:145–151. doi: 10.1093/nar/22.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McBride M S, Panganiban A T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mergener K, Facke M, Welker R, Brinkmann V, Gelderblom H R, Krausslich H-G. Analysis of HIV particle formation using transient expression of subviral constructs in mammalian cells. Virology. 1992;186:25–39. doi: 10.1016/0042-6822(92)90058-w. [DOI] [PubMed] [Google Scholar]
- 28.Paillart J-C, Berthoux L, Ottmann M, Darlix J-L, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paillart J C, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- 30.Partin K, Kräusslich H-G, Ehrlich L, Wimmer E, Carter C. Mutational analysis of a native substrate of the human immunodeficiency virus type 1 proteinase. J Virol. 1990;64:3938–3947. doi: 10.1128/jvi.64.8.3938-3947.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poon D T K, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakuragi J-I, Panganiban A T. Human immunodeficiency virus type 1 RNA outside the primary encapsidation and dimer linkage region affects RNA dimer stability in vivo. J Virol. 1997;71:3250–3254. doi: 10.1128/jvi.71.4.3250-3254.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skripkin E, Paillart J C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Kräusslich H-G. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]