Abstract
Background
Attention deficit hyperactivity disorder (ADHD) is a neurodevelopmental disorder influenced by genetic and environmental factors. This study examined the specific gene variants, dopamine transporter 1 (DAT1) rs6350, dopamine receptor D3 (DRD3) rs6280, dopamine receptor D2 (DRD2) rs6277, and catechol-O-methyltransferase (COMT) rs4633, in relation to ADHD among Pakistani children by exploring the potential gene-gene and gene-environment interactions.
Methods
A total of 100 cases of ADHD and 100 healthy children were recruited. The tetra-primer amplification refractory mutation system (ARMS) assays were designed for genotyping the selected variants in both groups, and their association with ADHD was determined in different genetic models. Gene-gene and gene-environmental interactions were determined by the multifactor dimensionality reduction (MDR) method.
Results
The DAT1 rs6350 SNV AA genotype showed a significantly increased risk for ADHD in the codominant and recessive models. Conversely, the AG genotype demonstrated a protective factor for ADHD in the codominant and overdominant models. The DRD3 rs6280 T allele exhibited a decreased risk for ADHD, and the TT genotype showed a reduced risk in the recessive and log-additive models. No association between the DRD2 rs6277 and COMT rs4633 SNVs with ADHD was found in our population. The MDR analysis of the best three-fold interaction model showed redundancy between DAT1 rs6350 and DRD3 rs6280; however, the risk was increased with the gender variable, which showed a weak synergistic interaction with these SNVs.
Conclusion
Genes associated with dopaminergic neurotransmission may contribute to the occurrence of ADHD. Furthermore, gene-gene and gene-environmental interactions may increase ADHD susceptibility.
Keywords: ADHD, Environment, Interaction, Gene, Variant
1. Introduction
Attention deficit hyperactivity disorder (ADHD) is a neurodevelopmental disorder that impacts around 2–7 % of children globally. It is characterized by difficulties in problem-solving, and attention, and hyperactivity, commonly observed in school-age children (Kian et al., 2022, Sayal et al., 2018). ADHD increases the likelihood of co-occurring mental disorders such as schizophrenia, autism spectrum disorder, major depression, substance abuse, academic and occupational struggles, accidents, and criminal behaviour. ADHD is influenced by a combination of genetic and environmental factors that impact the structure and functioning of brain networks related to cognition and behaviour (Cabana-Domínguez et al., 2023). Although ADHD, like other complex disorders, does not adhere to the typical Mendelian inheritance pattern (Al-Mubarak et al., 2020), twin, family, and adoption investigations have highlighted the substantial role of genetic factors, estimating ADHD's heritability at around 76–88 % (Faraone and Larsson, 2019, Faraone et al., 2005). Furthermore, the risk increases with a familial history of ADHD (Septier et al., 2019).
Evidence suggests that genes impacting the central dopamine system contribute to the development of ADHD and may interplay with environmental factors. Biomarker analysis has identified the involvement of the dopamine transporter (DAT), coded by the DAT1/SLC6A3 (solute carrier family six member 3) gene, and catechol-O-methyltransferase (COMT) in the etiopathogenesis of ADHD. The DAT controls dopaminergic firing in synapses, while the COMT influences dopamine metabolism, playing essential roles in monitoring behavioural functions contained by the prefrontal cortex (Maitra et al., 2022). Previous study showed an association with both alerting performance and executive attention in ADHD sib pairs with rs6350 SNV at the DAT1 locus (Konrad et al., 2010). Furthermore, homozygous genotypes of CC (rs4633) COMT were significantly associated with higher level of methylation involvement of the epigenetic variation of COMT loci in ADHD (Fageera et al., 2021). Another study found significantly increased risk of ADHD with DRD2 (odds ratio (OR) = 7.5) and DAT1 (OR = 6.6) gene variant risk alleles (Kopeckova et al., 2008).
The impact of gene variants on disease outcome may be synergistic or antagonistic, highlighting the significance of studying their combined effect in understanding the aetiology of the disease (Das et al., 2011). Intrinsic environmental factors, including socioeconomic status, family emotional climate, parental marital discord, and factors related to siblings, interact with ADHD's genetic vulnerability. The interplay between environmental factors and genetic susceptibility in ADHD is not fully understood, but investigating their combined effects is crucial for deeper insights and potential clinical applications (He and Li, 2022, Mooney et al., 2023). Therefore, the objectives of this study were to investigate the association between specific gene variants, DAT1 (rs6350), dopamine receptor D3 (DRD3) gene (rs6280), dopamine receptor D2 (DRD2) gene (rs6277), and COMT (rs4633), with ADHD among Pakistani children. Additionally, the study aimed to explore the potential gene-gene and gene-environment interactions among these genes.
2. Material and Methods
2.1. Clinical settings
One hundred cases of ADHD were identified and diagnosed on the criteria outlined in the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-V) (“Diagnostic and statistical manual of mental disorders: DSM-5™, 5th ed,” 2013). According to this criteria subjects with ADHD show a persistent pattern of inattention and/or hyperactivity-impulsivity that interferes with functioning or development. Based on the types of symptoms, three presentations of ADHD can occur: combined, predominantly inattentive, and predominantly hyperactive-impulsive presentation. Ethical approval was obtained from our institution, and informed consent was obtained from the parents or legal guardians of the participants. The level of hyperactivity was assessed by the Conners' Parents and Teachers Rating Scale, which is a 27-item Likert scale completed by parents (Conners et al., 1997). The Vanderbilt ADHD Diagnostic Rating Scale (VADRS) (Committee on Quality Improvement, 2000), a 55-item tool, was also employed for assessment. Table 1 displays the mean scores for Conner's Parent Rating Scale and VADRS. As controls, 100 healthy children were recruited. Sample size was taken from previous study reported on association of dopaminergic system genes in ADHD children (Kopeckova et al., 2008). Participants with neurological disorders or medical illnesses, known genetic conditions, mental retardation including fragile X syndrome, and those whose parents declined consent for participation were excluded from the study.
Table 1.
Mean score for Conner’s parent rating scale, and Vanderbilt assessment scale.
| Scale | Mean | Standard deviation | Standard error mean |
|---|---|---|---|
| Conner’s Parent Rating Scale | 125.090 | 37.4053 | 3.7405 |
| Vanderbilt Assessment Scale | |||
| Q1-9 | 20.45 | 6.478 | 0.648 |
| Q10-18 | 17.66 | 6.817 | 0.682 |
| Q19-26 | 14.41 | 7.171 | 0.724 |
| Q27-40 | 11.35 | 9.370 | 0.937 |
| Q41-47 | 3.03 | 4.367 | 0.439 |
| Reading | 1.28 | 0.726 | 0.073 |
| Mathematics | 1.40 | 0.778 | 0.078 |
| Written expression | 1.41 | 0.712 | 0.071 |
| Classroom Behavior | 1.67 | 0.995 | 0.100 |
| Following direction rules | 1.66 | 1.017 | 0.102 |
| Disrupting class | 1.52 | 0.979 | 0.098 |
| Assignment completion | 1.43 | 0.879 | 0.088 |
| Organizational skills | 1.75 | 1.132 | 0.113 |
2.2. Genes and SNVs selection
Four variants related to brain dopamine neurotransmission were selected for the present study. These variants are associated with four genes: COMT (rs4633; g.chr22:19962712) and DAT1 (rs6350; g.chr5:1443084), which play a role in regulating dopamine levels at synapses, as well as dopamine receptors D2 (rs6277; g.chr11:113412737) and D3 (rs6280; g.chr3:114171968). Their potential interaction is shown in Supplementary Fig. 1.
2.3. DNA extraction and ARMS PCR assay
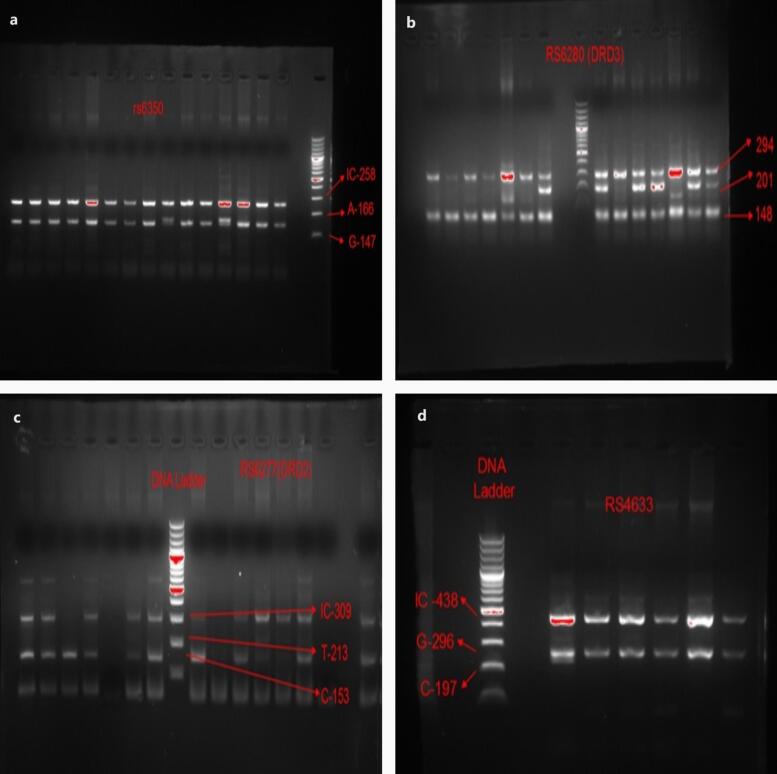
A 10 ml venous blood sample was collected following a standard procedure, and genomic DNA was extracted (Grimberg et al., 1989). The variants were genotyped by tetra-primer amplification refractory mutation system polymerase chain reaction (ARMS PCR) designed in our setting (Ye et al., 2001). The primers were designed using the PRIMER1 web tool (Collins and Ke, 2012), and their sequences were validated through the UCSC genome browser (Kent et al., 2002). The desired fragments were amplified using the 2720 thermocycler (Applied Biosystems) under optimized amplification conditions. PCR for SNVs was performed in a 20 μl reaction containing genomic DNA (2 μl), dNTPs (2 μl), Taq polymerase (0.6 μl), MgCl2 buffer (2 μl), and primers forward outer (0.5 μl), reverse outer (0.5 μl), forward inner (1 μl), and reverse inner (1 μl). PCR conditions were set as follows: 95 °C for 2 min, followed by 30 cycles at 95 °C for 30 s, annealing for 40 s, and extension at 72 °C for 50 s, with a subsequent final extension at 72 °C for 5 min. The PCR products were separated on a 2 % agarose gel (Fig. 1). The selected samples were amplified using forward and reverse outer primers for Sanger sequencing to validate the ARMS assays. The primer sequences for the ARMS assay and their corresponding annealing temperatures can be found in Table 2.
Fig. 1.
Agarose gel electrophoresis of newly designed tetra-primer ARMS PCR assays for variants a. DAT1 rs6350 (band size: 258, 166, and 147), b. DRD3 rs6280 (band size: 294, 201, and 148), c. DRD2 rs6277 (band size: 309, 213, and 153), and d. COMT rs4633 (band size: 438, 296, and 197).
Table 2.
Primer sequences and annealing temperature of gene variants.
| Genes/SNVs | Primers Sequences | Annealing Temperature |
|---|---|---|
| DRD2 (rs6277) |
IF-5″- CGT CCC ACC ACG GTC TCC ACA GCA CTA CC -3″ IR-5″-ATT CTT CTC TGG TTT GGC GGG GCT GGC A -3″ OF-5″-TTG TCC GGC TTT ACC CAG AGC CCT CTG CC-3″ OR-5″-ACG GCT CAT GGT CTT GAG GGA GGT CCG G-3″ |
60 °C |
| DRD3 (rs6280) |
IF-5″-CTG CCC CAC AGG TGT AGT TCA GGT GTC T -3″ IR-5″-CTA TGG CAT CTC TGA GCC AGC TGA TTG -3″ OF-5″-AGT AGT TGG TGG TAG TCT GCA GGG CCC -3″ OR-5″-CTA AGC AAC CAA GCC CCA AAG AGT CTG A-3″ |
69 °C |
| DAT 1 (rs6350) |
IF-5″- AGG CCC CAC CTG TGT GTC T TCA AGT GGT -3″ IR-5″-CTA TGG GCC AGC TGA TTG CAT CTC TGA -3″ OF-5″- TGA TTG CAT GGG C AGT TGG TGG TAG CC -3″ OR-5″- TGT GTC T TCA CAA GCC AGT TGG TGG TAG -3″ |
62 °C |
| COMT (rs4633) |
IF-5″- AGT TGG TGG TAG TGT GTC T GGG C AGT TGG -3″ IR-5″-CTA AGC TGA TTG GGG C AGT TGG TGA -3″ OF-5″- CAA GCC AGT TGG C AGT TGG TGG TAG CC -3″ OR-5″- T TCA AGT TGT GTC TGG TGG TAG TGG TAG -3″ |
64 °C |
IF: Inner forward, IR: Inner reverse, OF: Outer forward, OR: Outer reverse.
2.4. Statistical analysis
The data were analyzed using SPSS version 23. Categorical variables were presented as frequencies/percentages, and a two-sided Fisher exact test/Chi-square test (where applicable) was applied for analysis. Continuous variables were presented as means/standard deviations. To determine the association of independent variables with ADHD, bivariate and multivariate logistic regression analyses were performed. The associations were determined as OR and adjusted odds ratios (AOR) with 95 % confidence intervals (CI). Genotype and allele frequencies were calculated as frequencies and percentages. The Hardy-Weinberg equilibrium (HWE) was calculated to assess the SNV deviation. The association of single nucleotide variations (SNVs) with ADHD between cases and controls was determined under different genetic models by applying logistic regression using SNPStat software (Solé et al., 2006). The Benjamini and Hochberg method was applied using an online calculator (https://tools.carbocation.com/FDR) to account for multiple comparisons. Gene-gene interaction was analyzed using SHEsisPlus software (Yong and He, 2005), and gene-environmental interaction was studied using MDR v. 3.0.2. A p-value of less than 0.05 was considered significant.
3. Results
3.1. Characteristics and association of environmental and family factors with ADHD
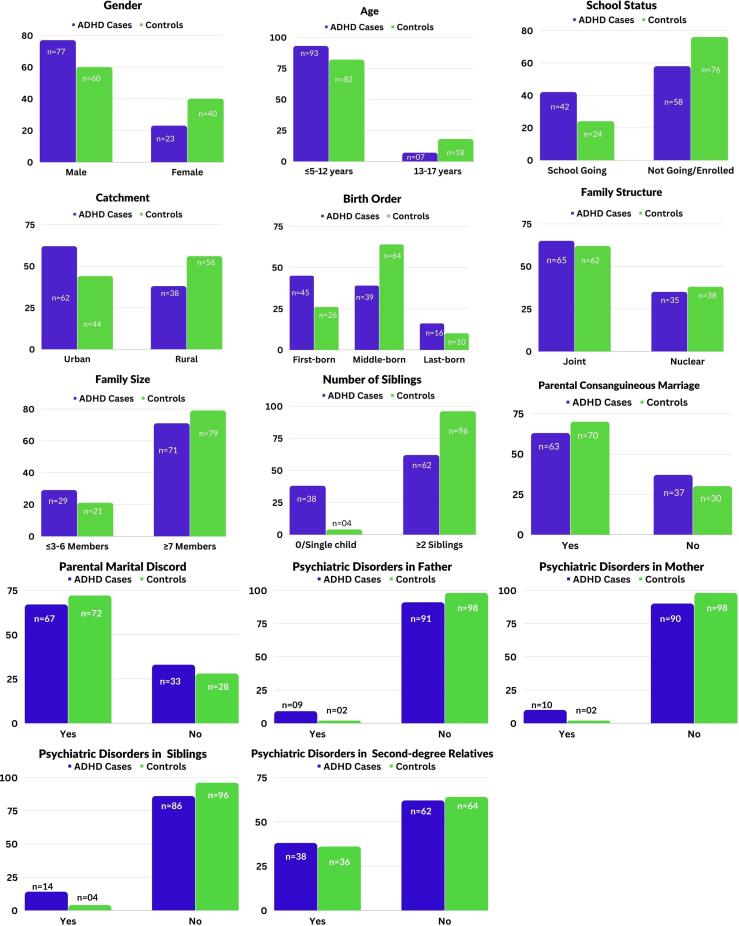
The mean age of children with ADHD was 8.42 ± 2.42, and the control was 8.92 ± 3.25 years. Fig. 2 presents the distribution of ADHD cases and controls based on personal factors, family factors, and family history of psychiatric disorders. Among the cases, the male-to-female ratio was 3.17:1. Most ADHD cases fell within the age range of ≤ 5 and 12 and predominantly resided in urban areas. Additionally, 42 % of ADHD cases attended school. Multiple logistic regression analyses were performed to consider potential confounding effects, adjusting for these variables (Table 3).
Fig. 2.
The distribution of ADHD cases and controls based on personal factors, family factors, and a family history of psychiatric disorders.
Table 3.
Association of environmental and family factors with attention deficit hyperactivity disorder (ADHD).
| Variables | Categories | Cases | Controls | OR (95 % CI) | P | AOR (95 % CI) | P |
|---|---|---|---|---|---|---|---|
| Personal Factors | |||||||
| Gender | Male | 77 | 60 | 2.232 (1.208–4.124) | 0.010 | 2.291 (1.186–4.424) | 0.014 |
| Female | 23 | 40 | Ref | ||||
| Age (Years) | ≤5–12 | 93 | 82 | 2.916 (1.16–7.334) | 0.023 | 3.44 (1.239–9.548) | 0.018 |
| 13–17 | 07 | 18 | Ref | ||||
| School Status | School going | 42 | 24 | 2.293 (1.25–4.207) | 0.007 | 2.238 (1.168–4.289) | 0.015 |
| Not going/enrolled | 58 | 76 | Ref | ||||
| Catchment | Urban | 62 | 44 | 2.077 (1.181–3.653) | 0.011 | 2.131 (1.158–3.923) | 0.015 |
| Rural | 38 | 56 | Ref | ||||
| Family factors | |||||||
| Birth order | First-born | 45 | 26 | 1.082 (0.429–2.731) | 0.868 | 0.572 (0.193–1.699) | 0.315 |
| Middle-born | 39 | 64 | 0.381 (0.157–0.923) | 0.032 | 0.63 (0.238–1.664) | 0.351 | |
| Last-born | 16 | 10 | Ref | ||||
| Family structure | Joint | 65 | 62 | 1.138 (0.64–2.025) | 0.66 | 1.487 (0.664–3.327) | 0.335 |
| Nuclear | 35 | 38 | Ref | ||||
| 1Family size (members) | ≤3-6 | 29 | 21 | 1.537 (0.805–2.933) | 0.193 | 1.572 (0.646–3.826) | 0.319 |
| ≥7 | 71 | 79 | Ref | ||||
| *Number of siblings | 0/Single child | 38 | 04 | 14.71 (5.002–43.256) | <0.0001 | 16.252 (4.696–56.24) | <0.0001 |
| ≥2 | 62 | 96 | Ref | ||||
| Parental consanguinity | Yes | 63 | 70 | 0.73 (0.405–1.316) | 0.295 | 0.908 (0.451–1.826) | 0.786 |
| No | 37 | 30 | Ref | ||||
| Parental marital discord | Yes | 67 | 72 | 1.267 (0.693–2.316) | 0.443 | 1.405 (0.711–2.773) | 0.328 |
| No | 33 | 28 | Ref | ||||
| Family history of psychiatric disorders | |||||||
| Father | Yes | 09 | 02 | 4.846 (1.020–23.028) | 0.047 | 3.121 (0.593–16.436) | 0.179 |
| No | 91 | 98 | Ref | ||||
| Mother | Yes | 10 | 02 | 5.444 (1.161–25.521) | 0.032 | 4.258 (0.849–21.352) | 0.078 |
| No | 90 | 98 | Ref | ||||
| Siblings | Yes | 14 | 04 | 3.907 (1.239–12.323) | 0.020 | 3.62 (1.07–12.251) | 0.039 |
| No | 86 | 96 | Ref | ||||
| Second-degree relatives | Yes | 38 | 36 | 1.090 (0.614–1.935) | 0.770 | 0.755 (0.402–1.417) | 0.382 |
| No | 62 | 64 | Ref | ||||
OR: Odds ratio, AOR: Adjusted odds ratio, CI: Confidence interval. Bold fonts indicate significant P value. AOR adjusted for factors related to personal, family and history of psychiatric disorders separately.
*No study subject had 1 sibling so that category excluded.
2 cases had family size ≤ 3; remaining had 4–6 family members.
Regarding personal factors, being a single child without siblings increased the risk of having ADHD by more than 16-fold (95 % CI: 4.696–56.24; P < 0.0001). Since no study participants had one sibling, this category was excluded. The protective effect of being a middle-born child against ADHD observed in the univariate model lost significance in the multivariate model. A history of psychiatric disorders among siblings was strongly associated with ADHD (P = 0.039). At the same time, the role of psychiatric history among fathers and mothers was also evident, but to a lesser extent.
3.2. Characteristics of ADHD cases
Among the 100 ADHD cases, the most frequent was type 3 combined (ADHD-C), accounting for 55 %, followed by type 2 hyperactive-impulsive (ADHD-HI) at 27 % and type 1 predominantly inattentive (ADHD-I) at 18 %. When examining the distribution of ADHD types by gender, ADHD-C was present in 42 males and 13 females, ADHD-HI in 17 males and ten females, and ADHD-I in 18 males.
3.3. Genotype/allele distribution and association with ADHD
The genotype distribution of all SNVs in the controls was found to be in concordance with HWE. Table 4 presents the genotype frequencies and the association of SNVs with ADHD. For the DAT1 rs6350 SNV, the AA genotype showed an increased risk for ADHD in both the codominant (AA vs. GG: OR = 5.90, 95 % CI = 1.69–20.65; P* < 0.0001) and recessive (AA vs. GG-AG: OR = 9.12, 95 % CI = 2.63–31.60; P* < 0.0001) models. Conversely, the AG genotype demonstrated a protective factor for ADHD in the codominant (AG vs. GG: OR = 0.02, 95 % CI = 0.00–0.17; P* < 0.0001) and overdominant (AG vs. GG-AA: OR = 0.02, 95 % CI = 0.00–0.14; P* < 0.0001) models. The DRD3 rs6280 T allele exhibited a decreased risk for ADHD (T vs C: OR = 0.57, 95 % CI = 0.38–0.86; P* = 0.03), and the TT genotype showed a reduced risk in the recessive (TT vs CC-CT: OR = 0.33, 95 % CI = 0.15–0.74; P* = 0.0235) and log-additive (OR = 0.60, 95 % CI = 0.41–0.90; P* = 0.044) models. Although the association was significant in the codominant model, the significance was lost after false discovery rate correction using the Benjamini-Hochberg method. No association between the DRD2 rs6277 and COMT rs4633 SNVs and ADHD was found in our population.
Table 4.
Allele and genotype frequencies and association of SNVs with ADHD.
| SNV/Genetic model/ HWE (p) | Allele/ Genotype |
Case n (%) |
Control n (%) |
OR (95 %CI) | P | P* |
|---|---|---|---|---|---|---|
| rs6277 | C | 178 | 179 | 1.00 | 0.87 | 0.87 |
| T | 22 | 21 | 1.05 (0.56–1.98) | |||
| Codominant | CC | 81 | 80 | 1.00 | 0.52 | 0.8 |
| CT | 16 | 19 | 0.83 (0.40–1.73) | |||
| TT | 03 | 01 | 2.96 (0.30–29.09) | |||
| Dominant | CC | 81 | 80 | 1.00 | 0.86 | 0.95 |
| CT-TT | 19 | 20 | 0.94 (0.47–1.89) | |||
| Recessive | CC-CT | 97 | 99 | 1.00 | 0.3 | 0.025 |
| TT | 03 | 01 | 3.06 (0.31–29.95) | |||
| Overdominant | CC-TT | 84 | 81 | 1.00 | 0.58 | 0.725 |
| CT | 16 | 19 | 0.81 (0.39–1.69) | |||
| Log-additive | − | − | − | 1.05 (0.57–1.92) | 0.88 | 0.926 |
| HWE (p) | − | 0.09 | 1 | − | − | |
| rs6280 | C | 136 | 110 | 1.00 | 0.007 | 0.03 |
| T | 64 | 90 | 0.57 (0.38–0.86) | |||
| Codominant | CC | 46 (46) | 35 (35) | 1.00 | 0.016 | 0.053 |
| CT | 44 (44) | 40 (40) | 0.84 (0.45–1.55) | |||
| TT | 10 (10) | 25 (25) | 0.30 (0.13–0.72) | |||
| Dominant | CC | 46 (46) | 35 (35) | 1.00 | 0.11 | 0.275 |
| CT-TT | 54 (54) | 65 (65) | 0.63 (0.36–1.12) | |||
| Recessive | CC-CT | 90 (90) | 75 (75) | 1.00 | 0.0047 | 0.0235 |
| TT | 10 (10) | 25 (25) | 0.33 (0.15–0.74) | |||
| Overdominant | CC-TT | 56 (56) | 60 (60) | 1.00 | 0.57 | 0.759 |
| CT | 44 (44) | 40 (40) | 1.18 (0.67–2.07) | |||
| Log-additive | − | − | − | 0.60 (0.41–0.90) | 0.011 | 0.044 |
| HWE (p) | −- | 1 | 0.068 | − | − | |
| rs6350 | G | 155 | 159 | 1.00 | 0.63 | 0.835 |
| A | 45 | 41 | 1.13 (0.69–1.82) | |||
| Codominant | GG | 77 (77) | 62 (62) | 1.00 | <0.0001 | <0.0001 |
| AG | 1 (1) | 35 (35) | 0.02 (0.00–0.17) | |||
| AA | 22 (22) | 3 (3) | 5.90 (1.69–20.65) | |||
| Dominant | GG | 77 (77) | 62 (62) | 1.00 | 0.021 | 0.06 |
| AG-AA | 23 (23) | 38 (38) | 0.49 (0.26–0.90) | |||
| Recessive | GG-AG | 78 (78) | 97 (97) | 1.00 | <0.0001 | <0.0001 |
| AA | 22 (22) | 3 (3) | 9.12 (2.63–31.60) | |||
| Overdominant | GG-AA | 99 (99) | 65 (65) | 1.00 | <0.0001 | <0.0001 |
| AG | 1 (1) | 35 (35) | 0.02 (0.00–0.14) | |||
| Log-additive | − | − | − | 1.08 (0.73–1.61) | 0.69 | 0.811 |
| HWE (p) | − | <0.0001 | 0.76 | − | − | |
| rs4633 | G | 182 | 188 | 1.00 | 0.26 | 0.51 |
| C | 18 | 12 | 1.55 (0.73–3.31) | |||
| Codominant | GG | 86 | 89 | 1.00 | 0.37 | 0.62 |
| GC | 10 | 10 | 1.03 (0.41–2.61) | |||
| CC | 04 | 01 | 4.14 (0.45–37.74) | |||
| Dominant | GG | 86 | 89 | 1.00 | 0.52 | 0.74 |
| GC-CC | 14 | 11 | 1.32 (0.57–3.06) | |||
| Recessive | GG-GC | 96 | 99 | 1.00 | 0.16 | 0.35 |
| CC | 04 | 01 | 4.12 (0.45–37.53) | |||
| Overdominant | GG-CC | 90 | 90 | 1.00 | 1 | 1 |
| GC | 10 | 10 | 1.00 (0.40–2.52) | |||
| Log-additive | −- | − | − | 1.41 (0.72–2.78) | 0.31 | 0.56 |
| HWE (p) | − | 0.0026 | 0.3 | − | − |
HWE: Hardy-Weinberg equilibrium. P*: Benjamini-Hochberg adjusted P value. Bold fonts indicate significant P value.
3.4. Binary logistic regression model for gene-gene interaction among ADHD cases and controls
The binary logistic regression model, or two gene-gene interaction, showed significant interactions between DRD2 rs6277 and COMT rs4633 (P*=0.044), DRD3 rs6280 and COMT rs4633 (P*=0.006) and DAT1 rs6350 and COMT rs4633 (P*=0.044) (Table 5).
Table 5.
Binary model of gene-gene interaction.
| SNV set | Case interaction | Control interaction | Difference | P | P* |
|---|---|---|---|---|---|
| rs6277,rs6280 | −0.028 | −0.036 | 0.007 | 0.726 | 0.871 |
| rs6277,rs6350 | −0.01 | −0.024 | 0.014 | 0.311 | 0.467 |
| rs6277,rs4633 | −0.056 | −0.003 | −0.053 | 0.022 | 0.044 |
| rs6280,rs6350 | −0.024 | −0.031 | 0.007 | 0.893 | 0.893 |
| rs6280,rs4633 | −0.015 | −0.091 | 0.075 | 0.001 | 0.006 |
| rs6350,rs4633 | −0.007 | −0.052 | 0.045 | 0.017 | 0.044 |
P*: Benjamini-Hochberg adjusted P value. Bold fonts indicate significant P value.
3.5. Multifactor dimensionality reduction (MDR), the best model for two-fold and three-fold gene-gene and gene-environmental interactions
Multifactor dimensionality reduction (MDR) was used to analyze the best two-fold and three-fold interaction models for gene-gene and gene-environmental variable interactions. Table 6 displays the MDR models selected as the best models based on the highest average testing accuracy and cross-validation (CV) consistency. Among the two-fold interaction models, DAT1 rs6350 and gender exhibited an average testing accuracy of 71.5 % and a CV consistency of 9/10. The three-fold interaction model, consisting of DAT1 rs6350, DRD3 rs6280, and gender, demonstrated an average testing accuracy of 68 % and a CV consistency 7/10.
Table 6.
Best models for single variable, two-fold and three-fold gene-environmental interactions selected by MDR.
| Model | Single variable | Two-fold interaction | Three-fold interaction |
|---|---|---|---|
| Best interaction models | rs6350 | rs6350, Gender | rs6350, rs6280, Gender |
| CV consistency | 10/10 | 9/10 | 7/10 |
| Training Accuracy | 0.67 | 0.735 | 0.7589 |
| Testing Accuracy | 0.67 | 0.715 | 0.68 |
| Sensitivity | 0.99 | 0.85 | 0.86 |
| Specificity | 0.35 | 0.62 | 0.65 |
| Odds Ratio (95 % Confidence Interval) | 53.307 (7.12–398.76) | 9.2456 (4.67–18.27) | 11.4082 (5.67–22.939) |
| Χ2: | 39.159 | 46.6477 | 54.4199 |
| Significance test (P) | P < 0.0001 | P < 0.0001 | P < 0.0001 |
| Precision | 0.6037 | 0.6911 | 0.7107 |
| Kappa | 0.34 | 0.47 | 0.51 |
| F-Measure: | 0.75 | 0.7623 | 0.7783 |
CV: Cross validation. P value based on 1000 permutation test. Bold fonts indicate significant P value.
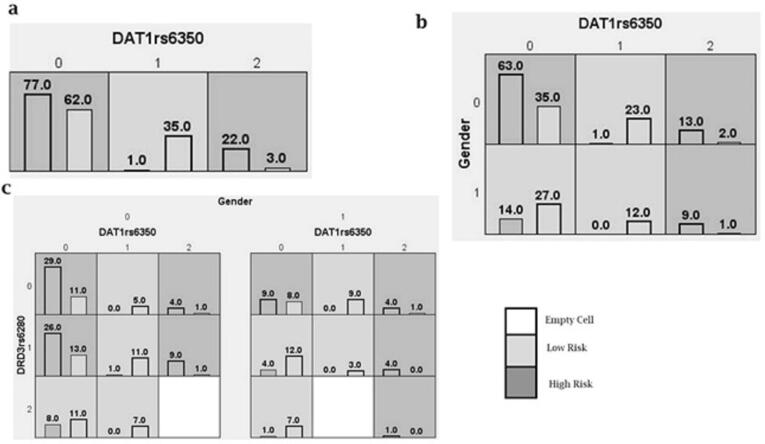
Fig. 3 displays the visual representation of the MDR models. Each cell in the diagram represents the distribution of genotypes among cases and controls. Cells shaded in dark grey indicate a 'high-risk' category identified by MDR, while other shades represent 'low-risk' cells. According to the twofold interaction MDR model, males with both common and rare homozygous genotypes have a higher risk than females, where risk is associated only with the rare homozygous genotype. On the other hand, the threefold interaction MDR model suggests that males carrying one or two common alleles for DRD3 rs6280 have an increased risk of developing ADHD if they also possess homozygous alleles for DAT1 rs6350. The risk is also amplified when rare homozygous alleles of DAT1 rs6350 are combined with one or two common alleles of DRD3 rs6280. Among females, the risk is primarily observed in those with the combination of common homozygous alleles for DAT1 rs6350 and DRD3 rs6280, as well as the rare homozygous allele of DAT1 rs6350 in combination with DRD3 rs6280.
Fig. 3.
The best (a) one locus, (b) two, and (c) three loci models selected by multifactor dimensionality reduction (MDR). Each cell represents the genotype distribution (“0” for common allele homozygous, “1” for heterozygous, and “2” for rare allele homozygous). Among the numbers across the y-axis, “0” codes for “Male” and “1” for “Female”. The left bar in each cell represents the number of cases, and the right bar shows the number of controls. Dark grey cells show “high risk” combinations, light grey cells show “low risk” combinations and empty cells are blank.
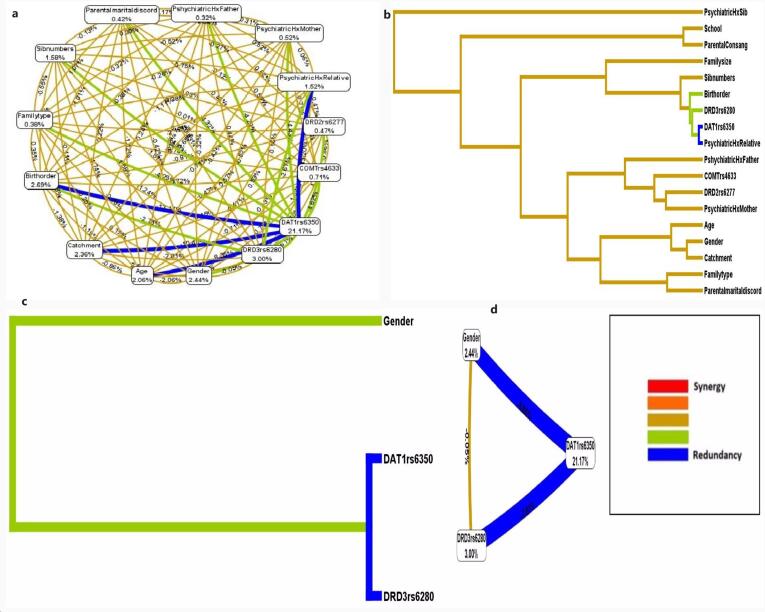
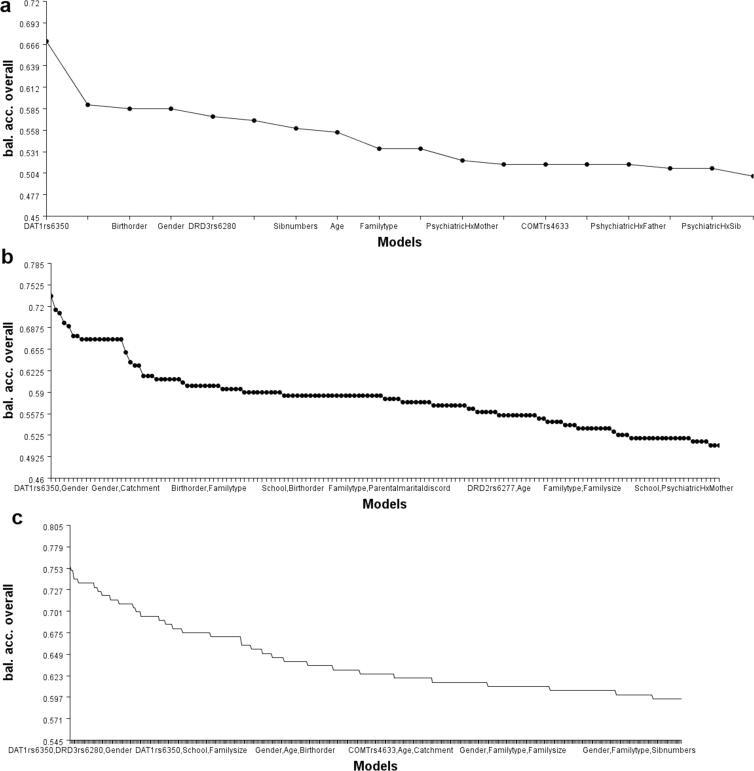
Fig. 4 shows the dendrogram, circle graph, and the interaction between genes and environmental variables. The MDR analysis of the best three-fold interaction model showed redundancy between DAT1 rs6350 and DRD3 rs6280; however, the risk was increased with the gender variable, which showed a weak synergistic interaction with these SNVs. The dendrogram of the combined MDR attribute network showed the synergistic interaction of multiple variables, potentially increasing the ADHD risk due to possible gene-environmental interactions. Fig. 5 displays the accuracy line chart of various models.
Fig. 4.
(a) Dendogram of the combined MDR attribute network showing interaction among genes and environmental factors; (b) Dendogram of the best three-fold interaction model selected by MDR (c) Entropy-based interaction network of genetic and environmental variables (d) Circle graph showing three-fold interaction model. The dendrogram shows a spectrum of colours that represent a continuum from synergy to redundancy, with red representing a relatively high degree of synergy (positive information gain), tan lines representing synergistic interaction, green lines showing weak synergistic interaction, and blue representing redundancy (negative information gain).
Fig. 5.
Line chart showing (a) single variable models, (b) two-fold interaction models, and (c) three-fold interaction models were chosen based on overall higher balanced accuracy to lower balanced accuracy.
4. Discussion
ADHD is a complex condition with polygenic inheritance influenced by social and environmental factors. Pharmacological and genetic investigations provide evidence supporting the role of dopamine neurotransmission in ADHD (RibaséS et al., 2012). It is worth noting that there is a lack of published data on the genetic basis and environmental interactions of ADHD, particularly in Pakistani children. Therefore, our study aimed to explore the influence of four specific genes associated with dopaminergic neurotransmission (DAT1, DRD2, DRD3, and COMT) on the susceptibility to ADHD by investigating gene-gene and gene-environment interactions.
The DAT1 gene codes for the DAT; it controls synaptic activity by transporting dopamine back into presynaptic dopaminergic neurons hence limiting the duration of synaptic activity. Neuroimaging studies have provided evidence of DAT's role in ADHD. Additionally, methylphenidate, a commonly used treatment for ADHD, down-regulates DAT activity and increases dopamine levels. Conversely, its withdrawal increases DAT activity beyond the initial level (Shang et al., 2011). Our study found that the DAT1 rs6350 AA genotype is associated with an increased risk of ADHD, whereas the rs6350 AG appears to have a protective effect. In agreement with that, Konard K et al. (Konrad et al., 2010) showed a rs6350 association with ADHD in alerting performance and executive attention. However, previous studies have not reported the influence of intrinsic environmental and genetic factors. Although the small sample size is a limitation of the present study, it is sufficient to provide over 80 % power at the OR of 5.9 found with the DAT1 rs6350 AA risk genotype at a 95 % confidence level in our study. Further studies with large sample size and investigation into other genetic variants could reveal additional findings and associations within our population.
The DRD3 gene and its specific variant, Ser-9-Gly (rs6280), have been widely investigated in ADHD. In vitro settings have revealed that the C allele of this variant exhibits greater affinity for dopamine and demonstrates more pronounced cAMP-MAPK signalling than the T allele (Jeanneteau et al., 2006). In our study, we noted a protective effect of the T allele in relation to ADHD development. Children with the TT genotype exhibited a reduced risk of developing ADHD compared to those with the CC genotype. Contrary to our findings, Fageera et al. (Fageera et al., 2018) observed a link between rs6280 and ADHD. They reported an increased T allele transmission from parents to the affected child, suggesting a potential linkage between this genetic locus and ADHD. Notably, the risk allele showed a higher occurrence of ADHD, hyperactivity, and impulsivity. In contrast, a meta-analysis reported a negative association between rs6280 and ADHD (Gizer, Ficks, & Waldman, 2009).
The DRD2 variants have been linked with childhood aggression, oppositional defiance, conduct problems, and ADHD. The synonymous SNV rs6277T allele (957C > T; Pro319Pro) has shown a reduction in translation and mRNA stability, resulting in a decrease of up to 50 % in protein synthesis compared to the C allele. Furthermore, a significant association has been found between the T allele and genotypes CT and TT with defiant and oppositional problems, attention and hyperactivity (Della Torre et al., 2018). In contrast to these findings, our study suggests no association between rs6277 and ADHD in Pakistani children. The Pakistani population demonstrates genetic variations and notable distinctions in susceptibility to disorders and disease progression (Khidri et al., 2019). Moreover, variances in family environment, behaviours, culture, ethnicities, genetic characteristics, and geographical locations may have varying effects as risk factors for ADHD in Pakistani children.
The COMT enzyme inactivates catecholamines, including the neurotransmitters dopamine, epinephrine, and norepinephrine. The COMT rs4860 (Val158Met) functions in the breakdown of extraneuronal dopamine and is linked to prefrontal-mediated cognition, attention, and social behaviour. Studies have linked COMT gene mutations to reduced grey matter volume and cortical abnormalities in children with ADHD. The Val158 version of COMT lowers dopamine levels, enhancing self-regulation, cognitive flexibility, and emotion processing. At the same time, carriers of the Met allele have increased dopamine availability, leading to cognitive rigidity and hyperactivity. Met-carriers also exhibit weaker white matter connections, potentially contributing to ADHD symptoms. However, research investigating the relationship between the Val158Met COMT variant and ADHD has produced conflicting results, likely due to gene-environmental interactions and the influence of early-life environmental adversity on dopaminergic states in the prefrontal cortex (Abraham et al., 2020, Cahill et al., 2022).
Although previous genetic studies have linked ADHD to the dopaminergic synaptic pathway genes, the results have been conflicting across various populations. A recent study conducted on Chinese children with ADHD children compared to control subjects found the significant association of DRD2 gene variants rs6277 and rs6275, and the SLC6A3 gene variant rs2652511 with ADHD out of 13 SNVs related to dopaminergic synaptic pathway examined (Zhong et al., 2024). Previous studies have reported the association of DRD2, DRD4 and COMT SNVs with ADHD in Finnish (Nyman et al., 2007), East Indian (Ghosh et al., 2013), and Chinese Han populations (Wang et al., 2019). A familial study involving ADHD children, along with their parents and siblings from Israel and Europe, found a link between the DRD2 gene and ADHD (Lasky‐Su et al., 2008). However, genome wide association studies conducted on ADHD populations in the USA and Australia did not find any such association (Middeldorp et al., 2016).
ADHD is unique among psychiatric conditions due to its association with specific environmental factors. A recent study reported that children from more chaotic home environments displayed higher initial ADHD symptoms and a slower regression in severity. Sensitivity analyses revealed that a substantial portion of the association between household chaos and ADHD symptoms could be attributed to a gene-environment relationship (Agnew-Blais et al., 2022). In the present study, we found the synergistic effect of various intrinsic environmental factors and potential gene interactions, suggesting possible gene-gene and gene-environmental interactions in ADHD. Assessing these risk factors in genetic studies and considering their influence during genetic analysis can contribute to a comprehensive understanding of the genetic contributions and environmental exposures associated with ADHD. Further research investigating these aspects in different populations may offer valuable insights.
Ethical Approval
The study was approved by the Research Ethics Committee of Liaquat University of Medical and Health Science, Jamshoro, Pakistan and Departmental review board of the Institute of Biotechnology and Genetic Engineering, University of Sindh, Jamshoro, Pakistan.
Authors' contributions
MAA and AMW conceived the research idea for the study. MAA and HAN designed the study. MAA, AHR, AM conducted the experiments. MAA and FFK performed statistical analysis and drafted the manuscript along with AMW and HAN. All authors contributed important intellectual contents during revision.
Funding
This study was supported by the Liaquat University of Medical and Health Sciences, Jamshoro and University of Sindh, Jamshoro. The funding sources had no role in the design of the study.
CRediT authorship contribution statement
Moin Ahmed Ansari: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. Habib Ahmed Naqvi: Resources, Supervision, Validation, Writing – review & editing. Feriha Fatima Khidri: Formal analysis, Investigation, Methodology, Software, Validation, Writing – original draft, Writing – review & editing, Visualization. Aatir Hanif Rajput: Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – review & editing. Ambar Mahmood: Data curation, Investigation, Methodology, Software, Visualization, Writing – review & editing. Ali Muhammad Waryah: Conceptualization, Funding acquisition, Project administration, Resources, Software, Supervision, Validation, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2024.104045.
Contributor Information
Moin Ahmed Ansari, Email: Moin.Ansari@lumhs.edu.pk.
Habib Ahmed Naqvi, Email: habib.naqvi@usindh.edu.pk.
Feriha Fatima Khidri, Email: feriha.fatima@lumhs.edu.pk.
Aatir Hanif Rajput, Email: aatir@lumhs.edu.pk.
Ambar Mahmood, Email: amber.mahmood@lumhs.edu.pk.
Ali Muhammad Waryah, Email: aliwaryah@lumhs.edu.pk.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
The data and materials may be obtained on request from the corresponding authors.
References
- Diagnostic and statistical manual of mental disorders: DSM-5™, 5th ed, American Psychiatric Publishing, Inc. xliv, 947-xliv, 947 (2013).
- Abraham E., Scott M.A., Blair C. Catechol-O-methyltransferase Val158Met genotype and early-life family adversity interactively affect attention-deficit hyperactivity symptoms across childhood. Front. Genet. 2020;11:724. doi: 10.3389/fgene.2020.00724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnew-Blais J.C., Wertz J., Arseneault L., Belsky D.W., Danese A., Pingault J.-B., Moffitt T.E. Mother's and children's ADHD genetic risk, household chaos and children's ADHD symptoms: A gene–environment correlation study. J. Child Psychol. Psychiatry. 2022;63(10):1153–1163. doi: 10.1111/jcpp.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mubarak B.R., Omar A., Baz B., Al-Abdulaziz B., Magrashi A.I., Al-Yemni E., Al-Tassan N.A. Whole exome sequencing in ADHD trios from single and multi-incident families implicates new candidate genes and highlights polygenic transmission. Eur. J. Hum. Genet. 2020;28(8):1098–1110. doi: 10.1038/s41431-020-0619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabana-Domínguez J., Antón-Galindo E., Fernàndez-Castillo N., Singgih E.L., O’Leary A., Norton W.H.G., Cormand B. The translational genetics of ADHD and related phenotypes in model organisms. Neurosci. Biobehav. Rev. 2023;144 doi: 10.1016/j.neubiorev.2022.104949. [DOI] [PubMed] [Google Scholar]
- Cahill S., Chandola T., Hager R. Genetic Variants Associated With Resilience in Human and Animal Studies. Front. Psych. 2022;13 doi: 10.3389/fpsyt.2022.840120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A., Ke X. Primer1: primer design web service for tetra-primer ARMS-PCR. Open Bioinformat. J. 2012;6:55–58. [Google Scholar]
- Committee on Quality Improvement, S. o. A.-D. H. D. (2000). Clinical practice guideline: diagnosis and evaluation of the child with attention-deficit/hyperactivity disorder. Pediatrics, 105(5), 1158-1170. [DOI] [PubMed]
- Conners C.K., Sitarenios G., Parker J.D., Epstein J.N. Conners' parent rating scale–revised. J. Abnorm. Child Psychol. 1997 doi: 10.1023/a:1022602400621. [DOI] [PubMed] [Google Scholar]
- Das M., Bhowmik A.D., Bhaduri N., Sarkar K., Ghosh P., Sinha S., Mukhopadhyay K. Role of gene–gene/gene–environment interaction in the etiology of eastern Indian ADHD probands. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2011;35(2):577–587. doi: 10.1016/j.pnpbp.2010.12.027. [DOI] [PubMed] [Google Scholar]
- Della Torre O.H., Paes L.A., Henriques T.B., de Mello M.P., Celeri E.H.R.V., Dalgalarrondo P., Santos-Júnior A.d. Dopamine D2 receptor gene polymorphisms and externalizing behaviors in children and adolescents. BMC Med. Genet. 2018;19(1):1–9. doi: 10.1186/s12881-018-0586-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fageera W., Sengupta S.M., Labbe A., Grizenko N., Joober R. DRD3 Gene and ADHD: A Pharmaco-Behavioural Genetic Study. NeuroMol. Med. 2018;20(4):515–524. doi: 10.1007/s12017-018-8504-z. [DOI] [PubMed] [Google Scholar]
- Fageera W., Chaumette B., Fortier M.-È., Grizenko N., Labbe A., Sengupta S.M., Joober R. Association between COMT methylation and response to treatment in children with ADHD. J. Psychiatr. Res. 2021;135:86–93. doi: 10.1016/j.jpsychires.2021.01.008. [DOI] [PubMed] [Google Scholar]
- Faraone S.V., Larsson H. Genetics of attention deficit hyperactivity disorder. Mol. Psychiatry. 2019;24(4):562–575. doi: 10.1038/s41380-018-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone S.V., Perlis R.H., Doyle A.E., Smoller J.W., Goralnick J.J., Holmgren M.A., Sklar P. Molecular genetics of attention-deficit/hyperactivity disorder. Biol. Psychiatry. 2005;57(11):1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Ghosh, P., Sarkar, K., Bhaduri, N., Ray, A., Sarkar, K., Sinha, S., & Mukhopadhyay, K. (2013). Catecholaminergic gene variants: contribution in ADHD and associated comorbid attributes in the eastern Indian probands. BioMed Res. Int. [DOI] [PMC free article] [PubMed]
- Gizer I.R., Ficks C., Waldman I.D. Candidate gene studies of ADHD: a meta-analytic review. Hum. Genet. 2009;126(1):51–90. doi: 10.1007/s00439-009-0694-x. [DOI] [PubMed] [Google Scholar]
- Grimberg J., Nawoschik S., Belluscio L., McKee R., Turck A., Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17(20):8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Li J.J. A gene-environment interaction study of polygenic scores and maltreatment on childhood ADHD. Res. Child Adolescent Psychopathol. 2022;50(3):309–319. doi: 10.1007/s10802-021-00873-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanneteau F., Funalot B., Jankovic J., Deng H., Lagarde J.-P., Lucotte G., Sokoloff P. A functional variant of the dopamine D3 receptor is associated with risk and age-at-onset of essential tremor. Proc. Natl. Acad. Sci. 2006;103(28):10753–10758. doi: 10.1073/pnas.0508189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khidri F.F., Waryah Y.M., Ali F.K., Shaikh H., Ujjan I.D., Waryah A.M. MTHFR and F5 genetic variations have association with preeclampsia in Pakistani patients: a case control study. BMC Med. Genet. 2019;20(1):163. doi: 10.1186/s12881-019-0905-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kian N., Samieefar N., Rezaei N. Prenatal risk factors and genetic causes of ADHD in children. World J. Pediatr. 2022;18(5):308–319. doi: 10.1007/s12519-022-00524-6. [DOI] [PubMed] [Google Scholar]
- Konrad K., Dempfle A., Friedel S., Heiser P., Holtkamp K., Walitza S., Herpertz-Dahlmann B. Familiality and molecular genetics of attention networks in ADHD. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010;153B(1):148–158. doi: 10.1002/ajmg.b.30967. [DOI] [PubMed] [Google Scholar]
- Kopeckova M., Paclt I., Petrasek J., Pacltová D., Malíková M., Zagatova V. Some ADHD polymorphisms (in genes DAT1, DRD2, DRD3, DBH, 5-HTT) in case-control study of 100 subjects 6–10 age. Neuroendocrinol. Lett. 2008;29(2):246–251. [PubMed] [Google Scholar]
- Lasky-Su J., Neale B.M., Franke B., Anney R.J., Zhou K., Maller J.B., Buitelaar J. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147(8):1345–1354. doi: 10.1002/ajmg.b.30867. [DOI] [PubMed] [Google Scholar]
- Maitra S., Chatterjee M., Roychowdhury A., Panda C.K., Sinha S., Mukhopadhyay K. Specific dopaminergic genetic variants influence impulsivity, cognitive deficit, and disease severity of Indian ADHD probands. Mol. Biol. Rep. 2022;49(8):7315–7325. doi: 10.1007/s11033-022-07521-y. [DOI] [PubMed] [Google Scholar]
- Middeldorp C.M., Hammerschlag A.R., Ouwens K.G., Groen-Blokhuis M.M., Pourcain B.S., Greven C.U., Nolte I.M. A genome-wide association meta-analysis of attention-deficit/hyperactivity disorder symptoms in population-based pediatric cohorts. J. Am. Acad. Child Adolesc. Psychiatry. 2016;55(10) doi: 10.1016/j.jaac.2016.05.025. 896–905. e896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney, M.A., Ryabinin, P., Morton, H., Selah, K., Gonoud, R., Kozlowski, M., and Nigg, J.T. 2023. Joint polygenic and environmental risks for childhood attention-deficit/hyperactivity disorder (ADHD) and ADHD symptom dimensions. JCPP Adv., n/a(n/a), e12152. DOI: 10.1002/jcv2.12152. [DOI] [PMC free article] [PubMed]
- Nyman E.S., Ogdie M.N., Loukola A., Varilo T., Taanila A., Hurtig T., Jaervelin M.-R. ADHD candidate gene study in a population-based birth cohort: association with DBH and DRD2. J. Am. Acad. Child Adolesc. Psychiatry. 2007;46(12):1614–1621. doi: 10.1097/chi.0b013e3181579682. [DOI] [PubMed] [Google Scholar]
- RibaséS M., Ramos-Quiroga J.A., HerváS A., Sánchez-Mora C., Bosch R., Bielsa A., Cormand B. Candidate system analysis in ADHD: Evaluation of nine genes involved in dopaminergic neurotransmission identifies association with DRD1. World J. Biol. Psychiatry. 2012;13(4):281–292. doi: 10.3109/15622975.2011.584905. [DOI] [PubMed] [Google Scholar]
- Sayal K., Prasad V., Daley D., Ford T., Coghill D. ADHD in children and young people: prevalence, care pathways, and service provision. Lancet Psychiatry. 2018;5(2):175–186. doi: 10.1016/S2215-0366(17)30167-0. [DOI] [PubMed] [Google Scholar]
- Septier M., Peyre H., Amsellem F., Beggiato A., Maruani A., Poumeyreau M., Delorme R. Increased risk of ADHD in families with ASD. Eur. Child Adolesc. Psychiatry. 2019;28(2):281–288. doi: 10.1007/s00787-018-1206-0. [DOI] [PubMed] [Google Scholar]
- Shang C.-Y., Gau S.-S.-F., Liu C.-M., Hwu H.-G. Association between the dopamine transporter gene and the inattentive subtype of attention deficit hyperactivity disorder in Taiwan. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2011;35(2):421–428. doi: 10.1016/j.pnpbp.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Solé X., Guinó E., Valls J., Iniesta R., Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- Wang Y., Hu D., Chen W., Xue H., Du Y. Prenatal tobacco exposure modulated the association of genetic variants with diagnosed ADHD and its symptom domain in children: a community based case–control study. Sci. Rep. 2019;9(1):4274. doi: 10.1038/s41598-019-40850-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S., Dhillon S., Ke X., Collins A.R., Day I.N. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001;29(17):e88–e. doi: 10.1093/nar/29.17.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong Y., He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- Zhong L., He H., Zhang J., Gao X., Yin F., Zuo P., Song R. Gene interaction of dopaminergic synaptic pathway genes in attention-deficit hyperactivity disorder: a case-control study in Chinese children. Mol. Neurobiol. 2024;61(1):42–54. doi: 10.1007/s12035-023-03523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and materials may be obtained on request from the corresponding authors.