Abstract
The mobile transgene constructs of most human immunodeficiency virus (HIV)-based lentivirus vectors currently in use contain viral long terminal repeats, a 5′ untranslated region, gag sequences, and env sequences that include the Rev-responsive element (RRE). In this study, we examined the possibility of deleting HIV splice sites and gag and env sequences from an HIV type 1 recombinant vector established in our laboratory as part of our ongoing efforts to improve this vector system. Mutations in the major splice donor site (SD) markedly reduced viral RNA expression but had little effect on vector titer. Deletion of gag or env sequences, excluding RRE, led to a moderate reduction in vector titer. Interestingly, deletion of RRE slightly reduced viral RNA expression but markedly impaired vector function. Combined deletions of RRE, gag (except for the first 40 nucleotides), env, and the SD mutation resulted in a twofold increase in cytoplasmic viral RNA expression and a recovery of vector efficiency to ∼50% of the wild-type level. This increase in cytoplasmic RNA levels is likely to be due, at least in part, to effects of the TE671 host cells, a human rhabdomyosarcoma cell line used for vector production in our system, on the cytoplasmic distribution of spliced and unspliced viral RNA. These results show that optimal lentivirus vector function can be maintained in the absence of multiple essential viral elements.
Lentiviruses have advantages over traditional Moloney murine leukemia virus-derived vectors in transient and long-term transduction of various types of human cells (11, 23, 30, 31). To establish a safe lentivirus vector system, it is necessary to delete essential viral sequences without compromising vector functions. The human immunodeficiency virus (HIV) 5′ untranslated region (UTR) contains several essential regulatory elements, including the primer binding site for initiation of reverse transcription and a structure of four stem-loops as part of the packaging signal (Ψ) for genome packaging (13, 15, 28). Previous studies demonstrated that deletions in this region markedly reduced packaging efficiency and HIV replication (12, 25). Besides the four stem-loops in the 5′ UTR, other sequences have been identified throughout the HIV genome as additional packaging signals. These include 5′ gag, pol (13), 3′ env (13, 24), TAR (14, 17, 29), and the 5′ poly(A) hairpin motif in U5 (18, 39). Deletion of any of these elements from the vector system may diminish vector function. Another factor affecting vector function is the level of cytoplasmic viral RNA expression controlled by viral regulatory proteins and genetic elements (reviewed in references 19 and 20). This regulation is mediated by Rev, which counteracts the nuclear retention of viral RNA controlled by splice sites and cis-repressive sequences (CRS) (or inhibitory sequences [INS]) to allow nuclear export of Rev-responsive element (RRE)-containing RNAs (3, 7–9, 32, 35). Some of these CRS/INS elements have been mapped to reside within the splice donor site (SD) (6, 21, 36), p17 of gag (5, 37, 38), pol (16, 27, 33), and env (32). Deletion of these elements may affect viral RNA expression and thus vector function.
We have recently established an HIV type 1-based lentivirus vector system, HP/TV, which produces high vector titers, more than 106 infectious particles per ml upon transfection of human TE671 cells with three plasmids (pTV, pHP, and pHEFVSV-G), without detectable replication-competent virus (11). To further improve the safety and payload of this vector system, we attempted to delete more of the essential viral elements in pTV. Through mutation and deletion analyses, the contributions of these genetic elements to viral RNA expression and vector function were elucidated.
Mutations of the 5′ splice site down-regulate cytoplasmic RNA expression but have only moderate effects on vector titer.
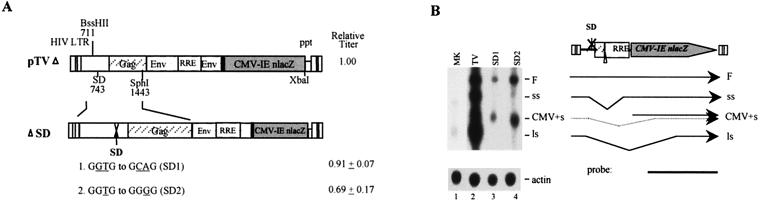
We first mutated the consensus SD sequence in pTV by PCR site-specific mutagenesis (primers are listed in Table 1). Mutations of SD from GGTG to GCAG (SD1) or to GGGG (SD2) resulted in a moderate decrease (10 to 30%) in vector titer (Fig. 1A). To determine the mutational effects of SD on pTV RNA expression, TE671 cells were transfected with 20 μg of pTV, 2 μg of pCEP4tat, 4 μg of pCMVrev, and 0.2 μg of pXGH5, which encodes human growth hormone, as a transfection control. Forty hours posttransfection, the cells were lysed and cytoplasmic poly(A)+ RNA was harvested for Northern analysis, as previously described (10). As shown in Fig. 1B, wild-type pTV produced four different sizes of RNA: full length, short intron spliced, a cytomegalovirus (CMV) promotor-driven nlacZ transcript which comigrated with a spliced RNA species, and large intron spliced. The SD mutations (SD1 and SD2), as predicted, abrogated splicing. However, minute amounts of short intron-spliced RNA, due to the use of a cryptic 5′ splice site as previously reported (36), were detected (Fig. 1B, lanes 3 and 4). It is noteworthy that mutations of SD reduced expression of cytoplasmic full-length RNA by more than 70% compared with wild-type vector, possibly due to decreased stability of the unspliced RNA or to pretermination of transcription. It has been reported that the 5′ SD imposes a suppressive effect on activation of the polyadenylation site in the 5′ long terminal repeat (1, 2), and thus mutations of SD may activate 5′ polyadenylation. The decline in genomic RNA expression (70%), however, did not correlate with the reduction in vector titer (10 to 30%). This could be explained by the following two possibilities. First, the amount of full-length RNA is always in excess of the viral packaging requirement and thus is not a determining factor of vector titer. Second, the elimination of spliced RNA by SD mutation abrogates its interference in genomic RNA packaging and thus results in indirect enhancement of packaging of full-length RNA. In any event, our results demonstrated that SD itself is not directly involved in HIV packaging and can be mutated in the vector system.
TABLE 1.
PCR mutagenesis primers used for construction of pTV mutants
| Primer | Mutant | Mutation or deletion site | Primer sequencea |
|---|---|---|---|
| 1 | SD1 | GGTG→GCAG | 733-5′-ggcggcgactgcagagtacgccaa-3′-756 |
| 2 | SD2 | GGTG→GGGG | 733-5′-ggcggcgactggggagtacgccaa-3′-756 |
| 3 | gag/env.dl.1 | Δ (7431–7610) | 7628-5′-ctctggtctgaagatct/ttgaccttcagtactc-3′-7414 |
| 4 | gag/env.dl.2 | Δ (1511–7610) | 7628-5′-ctctggtctgaagatct/actagtagttcctgctatg-3′-1494 |
| 5 | gag/env.dl.3 | Δ (1278–7610) | 7628-5′-ctctggtctgaagatct/gccttctcttctactact-3′-1260 |
| 6 | gag/env.dl.4 | Δ (1045–7610) | 7628-5′-ctctggtctgaagatct/gaggactgctattgtatt-3′-1027 |
| 7 | gag/env.dl.5 | Δ (830–7610) | 7628-5′-ctctggtctgaagatct/ctaattctcccccgctt-3′-812 |
| 8 | dl.RRE | Δ (7759–7992) | 8006-5′-aaccccaaatcccc/attcccactgctcttttt-3′-7741 |
The numbering system corresponds to pNL4-3.
FIG. 1.
Analyses of SD mutations. (A) Schematic diagram of SD mutations and relative vector titers. For virus production, plasmids pTV, pHP, pHEF VSV-G, and pXGH were used to cotransfect TE671 cells. Culture supernatants containing viral vectors were harvested at 48 h posttransfection and used to determine viral titer or to determine transfection efficiency via radioimmunoassay measurement of human growth hormone. The relative vector titer of each mutant was normalized against pTV, which was arbitrarily set at 1.00. The titer of pTV was consistently 2 × 105 to 10 × 105 CFU/ml. The relative titer represents an average of 4 to 7 independent experiments ± standard error. (B) Northern analysis of poly(A)+ cytoplasmic RNA of SD mutants (SD1 and SD2) in transfected TE671 cells. The four major RNA spices are abbreviated as follows: F, full-length unspliced RNA; ss, short intron-spliced RNA (from 5′ SD to SA in HIV env); CMV+s, CMV promoter-driven nlacZ transcript plus a spliced RNA population with 5′ SD and a cryptic splice site in the CMV promoter; ls, long intron-spliced RNA (from 5′ SD to a cryptic SA in the nlacZ gene). A lacZ probe was used for hybridization, as indicated.
Most of gag and the entire env sequences, except RRE, are dispensable for optimal lentivirus vector function.
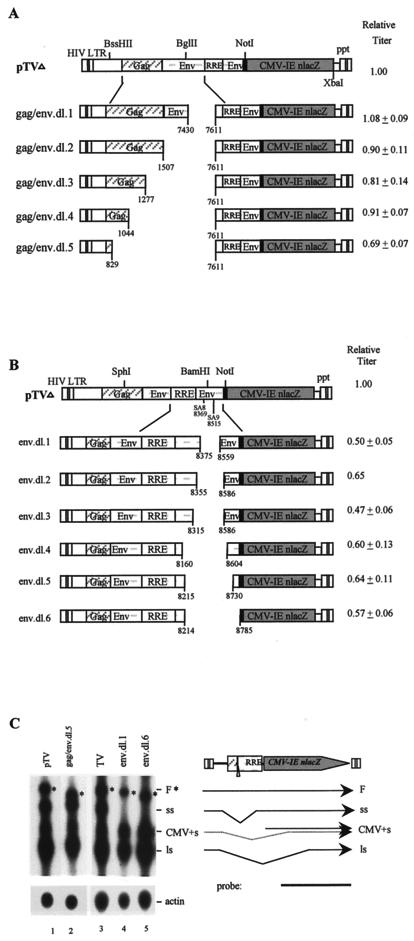
gag and env sequences contain potential packaging signals, splice sites, and CRS/INS elements. To examine their effects on vector functions, a series of deletions in gag and env were generated and tested. An array of deletions of 180, 361, 591, 824, and 1039 bp, in gag and env (upstream of RRE) was made by PCR site-specific mutagenesis (Table 1; Fig. 2A). Deletions of env 5′ to RRE and most of gag 3′ to the first 255-nucleotide (nt) coding sequence in pTV had no significant effects on vector titer (dl.1 to dl.4 [Fig. 2A]). Deletion of gag up to the first 40 nt led to a 30% decrease in vector titer (dl.5 [Fig. 2A]). Northern analyses showed that these mutants expressed cytoplasmic RNA at levels similar to that of wild-type pTV (not shown). Therefore, deletions of 5′ env and 3′ gag did not significantly affect RNA expression and vector titer. Deletion of gag 5′ coding sequences (dl.5) had a mild effect on vector titer (Fig. 2A), although it did not affect viral RNA expression (Fig. 2C), suggesting a role for the gag 5′ 255 nt in RNA packaging. These results are consistent with previous observations that the first 21 to 653 nt of gag are important for RNA packaging (4, 13, 26, 28, 34).
FIG. 2.
Analyses of 5′ gag and env deletions. (A) Schematic diagram of 5′ gag and 5′ env mutants and relative vector titers. The relative titer represents the mean of four to six experiments and is normalized to that of pTV. (B) Schematic diagram of 3′ env mutants and relative vector titers. The relative titer represents an average of four to nine experiments (except for env.dl.2) and is normalized against pTV, which was set at 1.00. (C) Northern analyses of gag and env deletion mutants. The full-length RNA is denoted by asterisks.
To further dissect the effects of env sequences on vector function, six more deletions (dl.1 to dl.6) in env 3′ to RRE, which contains two 3′ splice acceptor sites (SA), were generated by Bal31 exonuclease digestion (Fig. 2B). Deletion of 3′ env SA resulted in an overall ∼40% reduction in vector titer (Fig. 2B), and Northern analyses showed reduction in full-length genomic RNA and abrogation of the short intron-spliced RNA species (Fig. 2C, lanes 4 and 5). Therefore, the sequences of env SA appear to have moderate effects on full-length RNA expression as well as on vector function.
Deletion of RRE markedly diminishes vector titer.
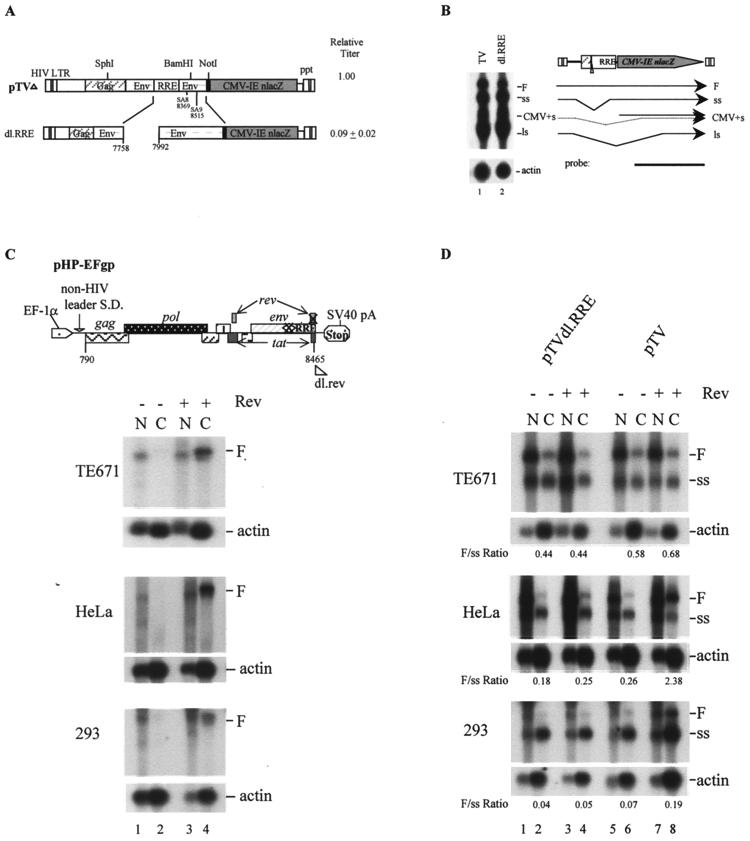
To determine the effects of RRE on lentivirus vector function, precise RRE deletion was made by PCR mutagenesis (Table 1; Fig. 3A). RRE deletion alone led to a marked reduction in vector titer (>90%). Interestingly, Northern analyses indicated that the RRE deletion had only moderate effects on vector RNA expression in TE671 cells (Fig. 3B, lane 2 versus lane 1). This is inconsistent with previous observations that removal of RRE from HIV usually abolishes cytoplasmic expression of full-length viral RNA (24, 34). It is possible that the recombinant vector backbone might contribute to this phenotype. Alternatively, the specific cell type (TE671) that we used for production of vectors might be different from those used by others. To clarify these issues, three different cell lines—TE671, HeLa, and 293—were transfected with different vector constructs—pTV, dl.RRE, or pHP-EFgp (a pHP mutant helper construct with rev deletion [Fig. 3C])—in the presence or absence of a rev plasmid. Cytoplasmic and nuclear RNAs were harvested and analyzed by Northern blotting. The results showed that pHP-EFgp did not transport unspliced RNA into the cytoplasm of transfected cells in the absence of rev (Fig. 3C, lane 2). When rev was present, full-length, unspliced RNA species were detected in the cytoplasm of all three cell types, suggesting that a similar Rev phenotype can be seen with a less-truncated HIV vector construct (Fig. 3C, lane 4). Interestingly, when RNA of the wild-type pTV construct was examined, differences in RNA partition ratio were observed in different cell types (Fig. 3D). In the presence of Rev, HeLa cells exhibited the highest ratio of cytoplasmic unspliced RNA to spliced RNA (2.38), while 293 cells exhibited the lowest (0.19) (Fig. 3D, lane 8). However, both HeLa and 293 cells showed a loss of full-length RNA in the cytoplasm when rev was absent (Fig. 3D, lane 6). TE671 cells, in contrast, did not exhibit an obvious rev effect when transfected with either pTV or dl.RRE (TE671 [Fig. 3D], 0.58 versus 0.68 for pTV and 0.44 versus 0.44 for pTVdl.RRE). Therefore, the specific RNA partition phenotype of the RRE mutant in TE671 is due to both the cell type and the specific pTV construct that we used.
FIG. 3.
Effects of RRE deletion on vector titer and RNA expression in different human cell types. (A) Schematic diagram of RRE deletion and relative vector titers. (B) Northern analysis of cytoplasmic RNA of the RRE deletion mutant in TE671 cells. (C) Northern analyses of full-length pHP-EFgp RNA in the presence or absence of Rev in TE671, HeLa, and 293 cells. Cells were transfected with pHP-EFgp, a pHP mutant helper construct with rev deletion, in the presence (+) or absence (−) of a rev plasmid. Cytoplasmic (C) and nuclear (N) RNA was harvested and hybridized with an env probe from nt 8133 to 8465 of pNL4-3. (D) Northern analyses of nuclear and cytoplasmic RNA of pTV and pTVdl.RRE in TE671, HeLa, and 293 cells, in the presence and absence of Rev. A single HIV env probe was used; it hybridized to both full-length (F) and spliced (ss) RNA. The ratios of full-length to spliced RNA in the cytoplasm, which were determined by quantification of RNA bands with a Fuji BAS1000 phosphorimager, are indicated.
Despite the presence of similar amounts of cytoplasmic full-length RNA in TE671 cells transfected with wild-type pTV and the dl.RRE mutant, the titer of the RRE mutant was less than 10% of that of wild-type pTV, suggesting an alternative effect of RRE on vector function independent of the known RRE effect on cytoplasmic viral RNA expression.
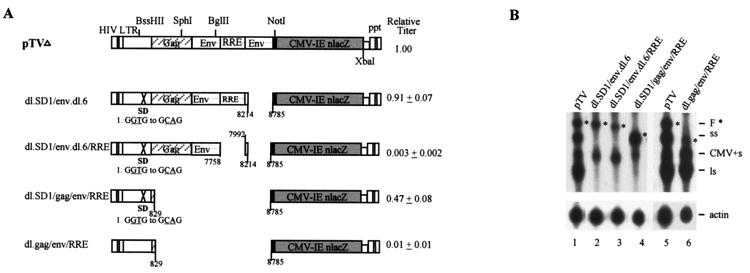
The combination of SD, SA, RRE and gag-env CRS mutations restores vector titer to 50% of the wild-type level.
The above results suggest that most of the HIV regulatory elements in pTV, except for RRE, have only moderate effects on vector function. To examine whether these regulatory elements could be deleted together, we combined different mutations and analyzed their effects. The combination of SD-SA mutation generates a vector with a near-wild-type level of titer (dl.SD1/env.dl.6) (Fig. 4A), which apparently repaired the defect of the env.dl.6 (SA) mutation (Fig. 2B). When RRE mutation was included, the resulting dl.SD1/env.dl.6/RRE mutant exhibited a vector titer close to zero (0.3% of wild-type level) (Fig. 4A). Interestingly, further removal of gag and env sequences resulted in a mutant, dl.SD1/gag/env/RRE, which showed a vector titer close to 50% of the wild-type level. This restored vector function diminished to about 1% of the wild-type level when the SD mutation was reverted to the wild type (dl.gag/env/RRE [Fig. 4A]), again suggesting a negative effect of splice site (SD-SA) disparity.
FIG. 4.
Analyses of combinations of RRE, SD, SA gag, and env mutations. (A) Schematic diagram of combinations of mutants and their relative vector titers. Relative vector titers are averages of four to nine experiments and are normalized against pTV, which was set at 1.00. (B) Northern analysis of cytoplasmic RNA of different combinations of mutants, as indicated.
Northern analyses demonstrated that when both SD and SA were deleted (dl.SD1/env.dl.6) (Fig. 4B, lane 2), full-length viral RNA expression was reduced by about 50% but the vector titer was maintained at 90% of the wild-type level. Further deletion of RRE from this mutant did not reduce RNA expression to a great extent (dl.SD1/env.dl.6/RRE) (Fig. 4B, lane 3), but vector function was markedly diminished (∼0.3% of the wild-type level [Fig. 4A]). This result corroborated the observed effects of the pTV dl.RRE mutant (Fig. 3). Surprisingly, when most of the gag (except for the first 40 nt) and the entire env CRS/INS, including RRE, were deleted, a twofold increase in full-length viral RNA expression, compared to wild-type vector (dl.SD1/gag/env/RRE) (Fig. 4B, lane 4), with an accompanying increase in vector titer (∼50% of the wild-type level [Fig. 4A]), was observed. When the mutated 5′ SD was reverted to generate dl.gag/env/RRE, expression of full-length viral RNA was again suppressed to about 40% of the wild-type level (Fig. 4B, lane 6) and vector function was again abolished (∼1% of the wild-type level [Fig. 4A]). Therefore, only when all of the CRS/INS regulatory elements, including SD, most of gag, the entire env, and RRE, are deleted can optimal vector function be restored.
In summary, we have demonstrated that deletion of essential HIV regulatory elements, including splice sites, gag, env, and RRE, from the HP/TV vector system is possible without significant loss of vector titer. Together with further deletion of long terminal repeat elements in pTV from a separate study (22), we have reduced the HIV sequences in pTV to less than 550 bp. This has effectively increased the lentivirus vector payload to more than 9 kb, because the HIV genome is approximately 9,700 nt. These modifications have effectively reduced the sequence homology between pHP and pTV and thus have greatly improved the safety of this vector system. Future modifications of remaining lentivirus sequences will require replacement of native viral essential elements with alternative functional elements while necessary functions for efficient vector production and gene transduction are preserved.
Acknowledgments
We thank Martin Stoltzfus and Maurice Swanson for critical comments and Gang Guo and Edward Mason for technical assistance.
Lung-Ji Chang is a Markey Scholar of the Lucille P. Markey Charitable Trust, and this work was supported by grants from the National Health and Research Development Program (NHRDP) of Canada and the National Institutes of Health (HL-59412) of the United States.
REFERENCES
- 1.Ashe M P, Griffin P, James W, Proudfoot N J. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 1995;9:3008–3025. doi: 10.1101/gad.9.23.3008. [DOI] [PubMed] [Google Scholar]
- 2.Ashe M P, Pearson L H, Proudfoot N J. The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J. 1997;16:5752–5763. doi: 10.1093/emboj/16.18.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barksdale S K, Baker C C. The human immunodeficiency virus type 1 Rev protein and the Rev-responsive element counteract the effect of an inhibitory 5′ splice site in a 3′ untranslated region. Mol Cell Biol. 1995;15:2962–2971. doi: 10.1128/mcb.15.6.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz R D, Hammarskjold M L, Helga-Maria C, Rekosh D, Goff S P. 5′ regions of HIV-1 RNAs are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology. 1995;212:718–723. doi: 10.1006/viro.1995.1530. [DOI] [PubMed] [Google Scholar]
- 5.Berthold E, Maldarelli F. cis-acting elements in human immunodeficiency virus type 1 RNAs direct viral transcripts to distinct intranuclear locations. J Virol. 1996;70:4667–4682. doi: 10.1128/jvi.70.7.4667-4682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borg K T, Favaro J P, Arrigo S J. Involvement of human immunodeficiency virus type-1 splice sites in the cytoplasmic accumulation of viral RNA. Virology. 1997;236:95–103. doi: 10.1006/viro.1997.8726. [DOI] [PubMed] [Google Scholar]
- 7.Brighty D W, Rosenberg M. A cis-acting repressive sequence that overlaps the Rev-responsive element of human immunodeficiency virus type 1 regulates nuclear retention of env mRNAs independently of known splice signals. Proc Natl Acad Sci USA. 1994;91:8314–8318. doi: 10.1073/pnas.91.18.8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang D D, Sharp P A. Messenger RNA transport and HIV rev regulation. Science. 1990;249:614–615. doi: 10.1126/science.2143313. [DOI] [PubMed] [Google Scholar]
- 9.Chang D D, Sharp P A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 10.Chang L-J, Zhang C. Infection and replication of Tat− human immunodeficiency viruses: genetic analyses of LTR and tat mutations in primary and long-term human lymphoid cells. Virology. 1995;211:157–169. doi: 10.1006/viro.1995.1388. [DOI] [PubMed] [Google Scholar]
- 11.Chang, L.-J., V. Urlacher, T. Iwakuma, Y. Cui, and J. Zucali. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther., in press. [DOI] [PubMed]
- 12.Clavel F, Orenstein J M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J Virol. 1990;64:5230–5234. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clever J L, Eckstein D A, Parslow T G. Genetic dissociation of the encapsidation and reverse transcription functions in the 5′ R region of human immunodeficiency virus type 1. J Virol. 1999;73:101–109. doi: 10.1128/jvi.73.1.101-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cochrane A W, Jones K S, Beidas S, Dillon P J, Skalka A M, Rosen C A. Identification and characterization of intragenic sequences which repress human immunodeficiency virus structural gene expression. J Virol. 1991;65:5305–5313. doi: 10.1128/jvi.65.10.5305-5313.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das A T, Klaver B, Berkhout B. The 5′ and 3′ TAR elements of human immunodeficiency virus exert effects at several points in the virus life cycle. J Virol. 1998;72:9217–9223. doi: 10.1128/jvi.72.11.9217-9223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das A T, Klaver B, Klasens B I, van Wamel J L, Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J Virol. 1997;71:2346–2356. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emerman M, Malim M H. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 20.Frankel A D, Young J A. HIV-1: fifteen proteins and an RNA. Annu Rev Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Hammarskjold M L, Li H, Rekosh D, Prasad S. Human immunodeficiency virus env expression becomes Rev-independent if the env region is not defined as an intron. J Virol. 1994;68:951–958. doi: 10.1128/jvi.68.2.951-958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwakuma, T., Y. Cui, and L.-J. Chang. Improved self-inactivating lentiviral vectors with U5 and U3 modifications. Submitted for publication. [DOI] [PubMed]
- 23.Kafri T, Blomer U, Peterson D A, Gage F H, Verma I M. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat Genet. 1997;17:314–317. doi: 10.1038/ng1197-314. [DOI] [PubMed] [Google Scholar]
- 24.Kaye J F, Richardson J H, Lever A M L. cis-acting sequences involved in human immunodeficiency virus type 1 RNA packaging. J Virol. 1995;69:6588–6592. doi: 10.1128/jvi.69.10.6588-6592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luban J, Goff S P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J Virol. 1994;68:3784–3793. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maldarelli F, Martin M A, Strebel K. Identification of posttranscriptionally active inhibitory sequences in human immunodeficiency virus type 1 RNA: novel level of gene regulation. J Virol. 1991;65:5732–5743. doi: 10.1128/jvi.65.11.5732-5743.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride M S, Panganiban A T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBride M S, Schwartz M D, Panganiban A T. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naldini L, Blomer U, Gage F H, Trono D, Verma I M. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 32.Nasioulas G, Zolotukhin A S, Tabernero C, Solomin L, Cunningham C P, Pavlakis G N, Felber B K. Elements distinct from human immunodeficiency virus type 1 splice sites are responsible for the Rev dependence of env mRNA. J Virol. 1994;68:2986–2993. doi: 10.1128/jvi.68.5.2986-2993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen H S, Cochrane A W, Rosen C. Interaction of cellular factors with intragenic cis-acting repressive sequences within the HIV genome. Virology. 1992;191:709–715. doi: 10.1016/0042-6822(92)90246-l. [DOI] [PubMed] [Google Scholar]
- 34.Parolin C, Dorfman T, Palu G, Gottlinger H, Sodroski J. Analysis in human immunodeficiency virus type 1 vectors of cis-acting sequences that affect gene transfer into human lymphocytes. J Virol. 1994;68:3888–3895. doi: 10.1128/jvi.68.6.3888-3895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powell D M, Amaral M C, Wu J Y, Maniatis T, Greene W C. HIV Rev-dependent binding of SF2/ASF to the Rev response element: possible role in Rev-mediated inhibition of HIV RNA splicing. Proc Natl Acad Sci USA. 1997;94:973–978. doi: 10.1073/pnas.94.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell D F, Martin M A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber B K, Pavlakis G N. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J Virol. 1992;66:7176–7182. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz S, Felber B K, Pavlakis G N. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J Virol. 1992;66:150–159. doi: 10.1128/jvi.66.1.150-159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vicenzi E, Dimitrov D S, Engelman A, Migone T S, Purcell D F, Leonard J, Englund G, Martin M A. An integration-defective U5 deletion mutant of human immunodeficiency virus type 1 reverts by eliminating additional long terminal repeat sequences. J Virol. 1994;68:7879–7890. doi: 10.1128/jvi.68.12.7879-7890.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]