Abstract
Background:
Early detection of esophageal squamous cell carcinoma (ESCC) can considerably improve the prognosis of patients. Aberrant cell-free DNA (cfDNA) methylation signatures are a promising tool for detecting ESCC. However, available markers based on cell-free DNA methylation are still inadequate. This study aimed to identify ESCC-specific cfDNA methylation markers and evaluate the diagnostic performance in the early detection of ESCC.
Methods:
We performed whole-genome bisulfite sequencing (WGBS) for 24 ESCC tissues and their normal adjacent tissues. Based on the WGBS data, we identified 21,469,837 eligible CpG sites (CpGs). By integrating several methylation datasets, we identified several promising ESCC-specific cell-free DNA methylation markers. Finally, we developed a dual-marker panel based on methylated KCNA3 and OTOP2, and then, we evaluated its performance in our training and validation cohorts.
Results:
The ESCC diagnostic model constructed based on KCNA3 and OTOP2 had an AUC of 0.91 [95% CI: 0.85–0.95], and an optimal sensitivity and specificity of 84.91% and 94.32%, respectively, in the training cohort. In the independent validation cohort, the AUC was 0.88 [95% CI: 0.83–0.92], along with an optimal sensitivity of 81.5% and specificity of 92.9%. The model sensitivity for stage I–II ESCC was 78.4%, which was slightly lower than the sensitivity of the model (85.7%) for stage III–IV ESCC.
Conclusion:
The dual-target panel based on cfDNA showed excellent performance for detecting ESCC and might be an alternative strategy for screening ESCC.
Keywords: Esophageal squamous cell carcinoma, Cell-free DNA, DNA methylation landscape, Biomarker, Liquid biopsy, Early detection
Introduction
Esophageal cancer (ESCA) is one of the most common malignant tumors. It ranks seventh and sixth in tumor-related occurrence and death worldwide, respectively; approximately 604,100 new cases and 544,100 deaths related to ESCA occurred in 2020.[1–4] The number of new cases and deaths related to ESCA is expected to reach 957,000 and 880,000 as estimated in 2040, respectively.[3] Esophageal squamous cell carcinoma (ESCC) and adenocarcinoma (EAC) are the two major histological subtypes of ESCA. The most common (~90%) histological type of ESCA in China, Central Asia, Northern Iran, East Africa, and some other endemic areas is ESCC.[5,6] Patients with ESCC have a poor prognosis, with a five-year survival rate below 30%.[7,8] The poor prognosis is mainly attributed to the long asymptomatic period during the precancerous state, which can last for 5–10 years.[9] Detecting ESCC at an early stage is challenging due to its asymptomatic characteristics. However, the long precancerous period provides a critical window for the screening and prevention of ESCC. Endoscopy is the primary procedure performed for screening ESCC. It has different levels of success in detecting ESCC in high-risk populations.[10] Some researchers found that performing Lugol's Chromoendoscopy for high-risk populations in China significantly improved the early detection rate and decreased the incidence and mortality of ESCC.[11,12] However, considering that ESCC is prevalent in economically underdeveloped and medically under-resourced areas, endoscopic screening may not be feasible. Additionally, endoscopy is an expensive procedure for screening ESCC, considering that only one suitable case can be detected for curative resection for more than 100 endoscopic screenings, even in endemic areas.[11,13–15] Therefore, a robust, inexpensive, and highly accurate approach needs to be developed for screening ESCC.[10,16,17]
Liquid biopsy technology based on cell-free DNA (cfDNA) is an effective non-invasive tool for detecting tumors.[18,19] Aberrant DNA methylation is a hallmark of ESCC. It strongly influences the initiation and development of this disease.[20–22] The methylation pattern of cfDNA is usually the same as their originating cells or tissues, which suggests that detecting the ESCC-specific DNA methylation load on cfDNA might be an effective strategy.[23–25] Qiao et al[26] developed a diagnostic panel with 921 differentially methylated regions (DMRs) by analyzing methylation microarray data from public databases. This method had a sensitivity of 76.2% for detecting ESCA. However, too many markers used in this panel limited its application in clinical practice. The whole-genome bisulfite sequencing (WGBS) technology can be used to accurately determine the changes in methylation pattern at a single-nucleotide resolution across the whole genome.[27,28] Only a few studies have used WGBS data to identify ESCC methylation markers, although several promising markers have been reported.[28–31]
In this study, we performed WGBS for 24 paired tumors and normal adjacent tissues (NATs) obtained from Chinese patients with ESCC to elucidate the methylation patterns in ESCC at a genome-wide level. By integrating the methylation profiles from multiple datasets, we aimed to identify potential methylation markers that might be used for detecting ESCC. Finally, we developed and tested a blood-based diagnostic model for ESCC based on polymerase chain reaction (PCR) platform. We further evaluated the diagnostic performance of this classifier in an independent validation cohort.
Methods
Ethical approval
The protocols for this study were approved by the Ethics Committee of Shanghai Changhai Hospital (No. CHEC2023-018). All participants provided informed consent.
Sample collection
In this study, we collected 24 ESCC tissues and their NATs for WGBS. The clinical characteristics of the 24 patients are shown in Supplementary Table 1, http://links.lww.com/CM9/B701. Sanger sequencing was performed using 42 healthy plasma samples, 50 NATs, and 24 ESCC tissues to evaluate the methylation status of candidate markers. In total, 449 plasma samples consisting of the training set (n = 229) and validation set (n = 220) were collected from 118 individuals with ESCC, 105 healthy individuals, and 226 individuals with other diseases, respectively. The inclusion criteria for ESCC patients were as follows: (1) individuals were above 18 years; (2) the disease status was confirmed by endoscopy, imaging examination, or pathological biopsy. All tissue samples and plasma samples were collected from Changhai Hospital in Shanghai, China.
The samples from non-ESCC diseases were defined as the interfering group, and the inclusion criteria were as follows: (1) patients with benign diseases of the digestive system who underwent endoscopy (including esophagitis, gastritis, enteritis, appendicitis, gastric polyps, colorectal polyps, etc.); (2) untreated patients with other digestive system malignancies (including gastric cancer, colorectal cancer, liver cancer, pancreatic cancer, bile duct cancer, etc.); (3) patients with non-digestive system malignancies (including thyroid cancer, lung squamous cell carcinoma, cervical cancer, endometrial cancer, breast cancer, prostate cancer, etc.).
The following individuals or samples were excluded: (1) individuals with maligancies who showed signs of distant metastasis; (2) plasma samples that were not preserved in the required environment; and (3) the sample amount available was insufficient. The clinical features of the plasma samples are shown in Supplementary Table 2, http://links.lww.com/CM9/B701.
Whole-genome bisulfite sequencing and data preprocessing
About 200 ng of genomic DNA mixed with 1% unmethylated λ DNA was ultrasonically interrupted by Covaris LE220, and then, adapters were added to both ends of the fragmented DNA. The DNA fragments were treated with bisulfite using the DNA Methylation-Gold kit (Zymo, Genesee Scientific, San Diego, CA, USA). The bisulfite (BS)-treated single-stranded DNA was amplified by the KAPA HiFi Hot Start Uracil + Ready Mix (KAPA) and universal PCR primer reagents to prepare complete library.[32] The concentration and length of the DNA fragment were measured using a Qubit 3.0 fluorometer (Thermo Fisher Scientific, Waltham, USA) and an Agilent 2200 Bioanalyzer device (Agilent, Santa Clara, CA, USA), respectively. Then, PE150 sequencing was performed using an Illumina NovaSeq6000 S4 chip (Illumina, San Diego, CA, USA), and the sequencing depth was set at 30×.
Raw reads in the fastq format were generated using the BCL Convert (v3.10.5) software (https://support.illumina.com/sequencing/sequencing_software/bcl-convert.html). The base quality was first evaluated using FastQC v0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and then, the adapter sequences were removed using the Cutadapt (v4.1) software (https://cutadapt.readthedocs.io/en/v4.1/). Low-quality reads and bases, which are defined with more than 50% "N" base, or the length is less than 50 bp or with more than 50% low-quality bases (<Q20), were removed by the fastp[33] tool (v0.20.1, https://github.com/OpenGene/fastp/releases). The clean reads were aligned to the hg38 (p16) reference genome using the bowtie2 software (v2.4.4) (parameter default,https://bowtie-bio.sourceforge.net/bowtie2/index.shtml). The methylation levels of CpGs, defined as the proportion of methylated reads in the total number of reads (ranging from 0 to 1), were calculated by the bismark_methylation_extractor tool in BisMark (v0.22.3) (parameters: –paired-end–no_overlap–report–ignore 5– ignore_r2 5–comprehensive–bedGraph–counts–cytosine_report, https://www.bioinformatics.babraham.ac.uk/projects/bismark/). The BS-converted efficiency was estimated using λ DNA. The raw reads were submitted to the NCBI Sequence Read Archive (accession ID: PRJNA917325).
The quality control results of the WGBS data for 24 paired samples are shown in Supplementary Table 3, http://links.lww.com/CM9/B701. We found that the mapping rates of all samples were over 80%, the average effective sequencing depth was around 20×, and the average base quality was above Q30; these findings suggested that the WGBS data was acceptable. In the subsequent analysis, we retained only those CpGs that had at least five reads covered. If the methylation values were missing in more than 50% of the samples, the site was removed. The rest of the missing values were then filled in using the k-nearest neighbor algorithm (R package "bnstruct", https://github.com/sambofra/bnstruct).
Differential methylation analysis
We performed the non-parametric rank sum test to identify the differentially methylated CpGs (DMCs) between ESCC and NAT samples.[34] Significant DMCs were defined as fold change ≥1.5 and P-value <0.05. Differentially methylated regions (DMRs) were identified using the previously reported sliding window method,[35] with the following parameters—(1) the length of the DMR: between 50 bp and 300 bp, (2) DMCs required: at least two, and (3) distance between two adjacent DMCs: less than 50 bp. Based on the methylation status of DMCs on ESCC and NAT samples, we defined the DMR that was hypermethylated in ESCC as hyper-DMR and the hypermethylated DMR in NATs as hypo-DMR. The differentially methylated genes (DMGs) were determined based on the relative position of the DMRs and genes on the genome. We defined the genes that overlapped with hyper-DMRs as hyper-DMGs and overlapped with hypo-DMRs as hypo-DMGs. The DMRs were further divided into four categories based on their genomic locations on the genome, downstream (within 200 bp downstream of the gene), innergenic, intergenic, and upstream (within 2 kb upstream of the gene) regions. The DMGs that encoded proteins in the "upstream" and "downstream" groups were selected for the KEGG pathway enrichment analysis.
Preparation of public datasets
The WGBS dataset GSE149608 was downloaded from the Gene Expression Omnibus (GEO) database, which included 10 pairs of ESCC and NAT samples [Supplementary Table 4, http://links.lww.com/CM9/B701]. The methylation data of the other 33 types of cancer in The Cancer Genome Atlas (TCGA) were also retrieved from the NCI Genomic Data Commons (https://portal.gdc.cancer.gov/). The methylation data of the 33 types of cancer were first preprocessed, which yielded a total of 8968 samples, of which 710 were NATs and 8258 were primary cancer tissues. For the probes whose methylation values were missing data, K-nearest neighbor algorithm "bnstruct"[36] was used to fill in them.
Cell-free DNA in blood can be released from multiple tissues or organs, besides the ESCA tissue. Therefore, we further evaluated the methylation levels of the candidate markers in 32 cancer samples from TCGA (excluding 95 ESCC samples) to obtain ESCC-specific methylation markers. For each CpG probe, their average β values were calculated for each type of cancer.
Most cfDNA is released from blood cells. To exclude the interference of blood cell DNA, we evaluated the methylation levels of candidate markers in healthy blood samples using the GSE40279 dataset,[37] consisting of 656 whole blood samples of healthy individuals; the DNA methylation levels were assessed using the 450k platform. The average β values of the candidate probes for the 656 samples were calculated, and then the probes were sorted from smallest to largest according to this value. A smaller β value indicated a lower level of background methylation in blood cells.
DNA extraction and bisulfite conversion
The whole blood samples were centrifuged at 3000 r/min for 10 min. Only the plasma was collected from patients with ESCC, whereas the plasma and white blood cells were collected separately from healthy individuals. The genomic DNA of tissue samples, plasma, and white blood cells were extracted using the Nucleic Acid Extraction and Purification Kit (Wuhan Ammunition Life Science and Technology Co., Ltd, Wuhan, China) following the manufacturer's instructions.
Sodium bisulfite treatment of the purified DNA was performed using the Bisulfite Conversion Kit (Wuhan Ammunition Life Science and Technology Co., Ltd., Wuhan, China) following the protocols. For tissue and white blood samples, 1 μg of DNA was converted. For plasma samples, 50 μL of purified DNA was converted. After bisulfite treatment, 25 μL of the eluted DNA was either used immediately for PCR analysis or stored at -80°C until further use. The amplified products of BS-DNA were then sequenced by Sanger sequencing using specific primers (same as the PCR primers).
Cell lines and plasmids
The TE-1[38] and KLE[39] cell lines, obtained from Tongji Medical College, Huazhong University of Science and Technology, were used in this study. The cells were cultured in DMEM supplemented with 10% fetal bovine serum. Because the four candidate genes, including ZNF582, KCNA3, RAPGEFL1, and OTOP2, were hypermethylated and hypomethylated in TE-1 cells and KLE cells, as determined by Sanger sequencing, the two cells were selected as positive and negative controls, respectively. The fully methylated amplicon sequences of the four targets and the ACTB amplicon region after bisulfite conversion were artificially synthesized and cloned into the vector pUC57. The recombinant vectors were then inserted into Escherichia coli for the preservation and amplification. The recombinant plasmids extracted from E. coli were serially diluted to 105 copies/μL, 104 copies/μL, 103 copies/μL, and 102 copies/μL as templates for the standard curve analysis.
Methylation-specific PCR
We designed forward and reverse PCR primers for four candidate targets according to their genome sequences. The designed primers and minor groove binder (MGB) probes[40] are shown in Supplementary Table 5, http://links.lww.com/CM9/B701. The ACTB gene was used as an internal control to check whether each run was normal. Using synthetic recombinant plasmids, we estimated the parameters of these primers, including the amplification efficiency and primer specificity.
We prepared 50 μL of PCR solution with High-Affinity Hotstart Taq Polymerase (TIANGEN, Beijing, China). Then, 10 μL of template DNA, non-template control, and positive and negative controls were mixed together in every plate. PCR was performed using an ABI 7500 instrument (Thermo Fisher Scientific, Waltham, USA) under the following cycling conditions: 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 60°C for 30 s.
Statistical analysis
All statistical analyses and figures were made using the R software (version 4.0.5, www.r-project.org). The receiver operating characteristic (ROC) curve was plotted using the R package "pROC"[41] to assess the performance of candidate targets. Briefly, the cycle threshold (Ct) values of each target for all samples were used as the predictor variable, and the sample type was used as the response variable. They were then input to the "roc" method with a boot parameter equaling 100. The area under the ROC curve (AUC) and 95% confidence interval (CI) were calculated simultaneously. Each point on the ROC curve represented a sensitivity/specificity pair corresponding to a specific threshold. P-value less than 0.05 was selected as the significant threshold. We calculated the Youden index for each sensitivity/specificity pair. We selected the sensitivity/specificity corresponding to maximum value of this index as the optimal value for a given assay and the threshold as the optimal positive cut-off. The exact cut-off Ct value for each single target and the exact cut-off probability value for the logistic regression model are presented in Supplementary Table 6, http://links.lww.com/CM9/B701. The models for the combination of multiple targets were developed using logistic regression, implemented in the R software "glm" method. The probability of each sample predicted by the regression model was used as the input of the "roc" method to estimate the AUC values and the optimal sensitivity and specificity of the model.
Results
Flowchart of the test development
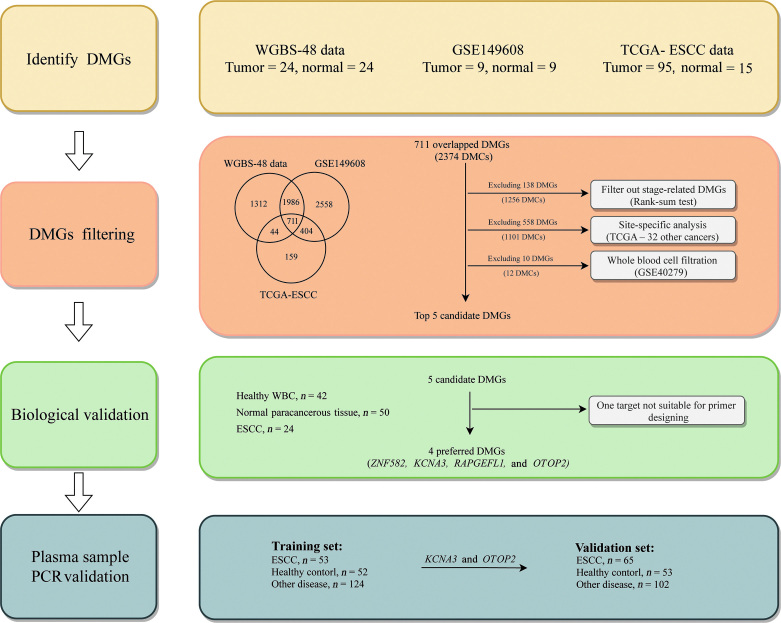
The ESCC detection test was developed based on four steps [Figure 1]. First, we performed WGBS for 24 pairs of ESCC tissues and their NATs to identify DMCs and DMRs across the whole genome. Then, we validated the identified DMCs and DMRs in an independent WGBS dataset and the TCGA-ESCC cohort. In the second step, we identified potential plasma methylation markers. The methylation levels of the DMCs shared by the three datasets were evaluated for 32 types of cancer in TCGA database to obtain ESCC-specific DMCs. For plasma markers, the level of methylation of ESCC-specific DMCs in blood cells was assessed using data from whole blood cells (WBC) of healthy individuals to exclude those with high methylation background in blood cells. In the third step, we performed Sanger sequencing for the most promising targets obtained in the second step to confirm their methylation status in ESCC tissues. In the final step, we established a panel based on two candidate markers and its performance, including the indicators of AUC, sensitivity, and specificity, was evaluated in plasma samples from the training and validation sets.
Figure 1.
The flowchart of this study. DMC: Differentially methylated CpG; DMG: Differentially methylated genes; DMR: Differentially methylated region; ESCC: Esophageal squamous cell carcinoma; PCR: Polymerase chain reaction; WBC: Whole blood cell; WGBS: Whole-genome bisulfite sequencing.
Identification of DMRs across the whole genome
We obtained 21,469,837 eligible CpGs, with an average of 17,762,493 per sample, by pretreating the WGBS data. Overall, abnormally methylated regions were not present at the chromosome level, such as large hypomethylated or hypermethylated blocks [Figure 2A]. Chr1 and chr2 had the most CpGs, and the number of CpGs in each chromosome was proportional to the chromosome size [Supplementary Figure 1, http://links.lww.com/CM9/B702]. Additionally, no significant differences in the number of CpGs were observed between the tumor and normal samples (P = 0.24). However, further analysis indicated that the DMCs in the normal samples exhibited higher methylation levels than those in tumor samples. The methylation values of DMCs in ESCC followed a bimodal distribution, one around 0.80 and the other around 0.65 [Supplementary Figure 2A, http://links.lww.com/CM9/B702].
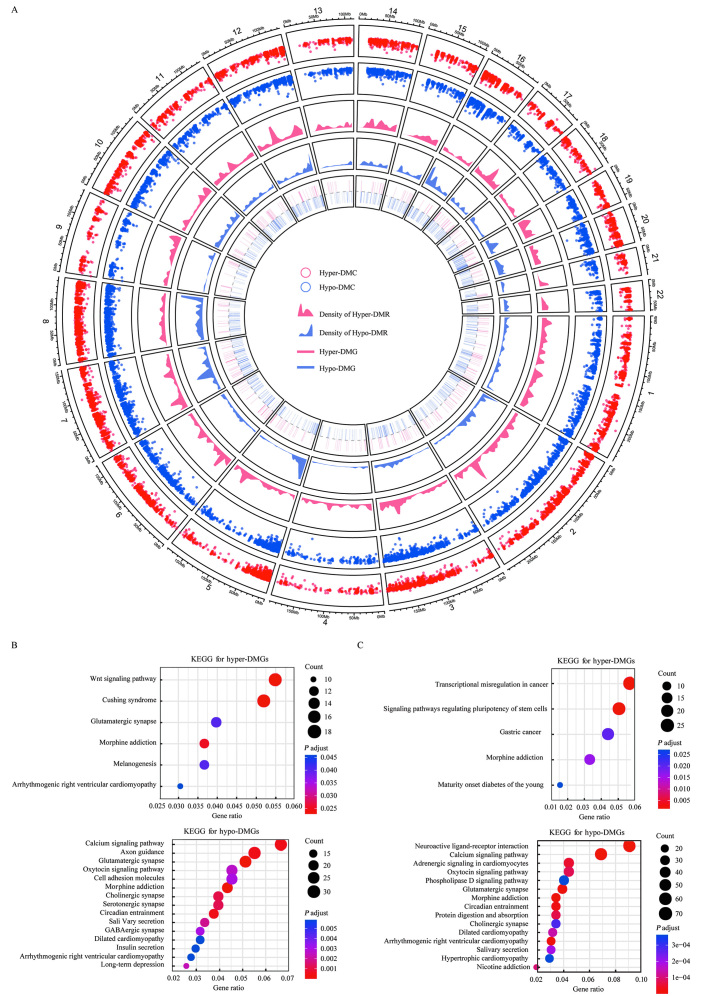
Figure 2.
Differential methylation analysis for ESCC and their NATs. (A) A circos plot showing hyper and hypo-events on different chromosomes. The circles from outer to inner indicate hyper-DMCs and hypo-DMCs, DMRs, and DMGs. Red and blue dots represent the methylation level of DMCs on ESCC and normal controls, respectively. Red and blue areas indicate the distribution density of DMRs on different chromosomes between ESCC and normal controls, respectively. Red and blue rectangles indicate the location of DMGs on different chromosomes between ESCC and normal controls, respectively. (B) The top 15 significantly enriched KEGG pathways associated with the upstream/downstream types of hyper-DMGs and hypo-DMGs. (C) The top 15 significantly enriched KEGG pathways associated with the innergenic hyper-DMGs and hypo-DMGs. DMC: Differentially methylated CpG; DMG: Differentially methylated genes; DMR: Differentially methylated region; ESCC: Esophageal squamous cell carcinoma; NATs: Normal adjacent tissues.
The differential methylation analysis yielded 1,554,374 DMCs [Supplementary Table 7, http://links.lww.com/CM9/B701], of which 1,455,679 (93.65%) DMCs were hypomethylated and 98,695 (6.35%) DMCs were hypermethylated. Based on the criteria described in this method, we obtained 14,530 hyper-DMRs and 158,978 hypo-DMRs, and more than half of the DMRs contained only two DMCs [Supplementary Table 8, http://links.lww.com/CM9/B701]. Hyper-DMRs were significantly more prevalent in the innergenic regions than in the hypo-DMRs (Supplementary Figure 2B, http://links.lww.com/CM9/B702). The differences between hyper-DMRs and hypo-DMRs in the distribution of chromosomes were not significant (P = 0.24, Supplementary Table 9, http://links.lww.com/CM9/B701). The average length of hyper-DMR was greater than that of hypo-DMR [Supplementary Figure 2C, http://links.lww.com/CM9/B702], and the average number of DMCs in hyper-DMR was higher than that in hypo-DMR [Supplementary Figure 2D, http://links.lww.com/CM9/B702].
All DMRs overlapped with 5083 genes, including 3590 hypo-DMGs and 1493 hyper-DMGs, of which the protein-coding and long non-coding (lncRNA) genes accounted for the largest proportions with 96.32% and 92.28%, respectively [Supplementary Table 10, http://links.lww.com/CM9/B701]. Since methylation occurred in gene bodies and the regulatory regions had an inconsistent effect on gene expression, we performed pathway enrichment analysis for the upstream/downstream types of DMGs and the innergenic DMGs separately. The results showed that hyper-DMGs of the two types were enriched in different pathways [Figure 2B], whereas hypo-DMGs shared some pathways between the types, such as the calcium signaling and oxygen signaling pathways [Figure 2C].
Identification of candidate markers for detecting ESCC
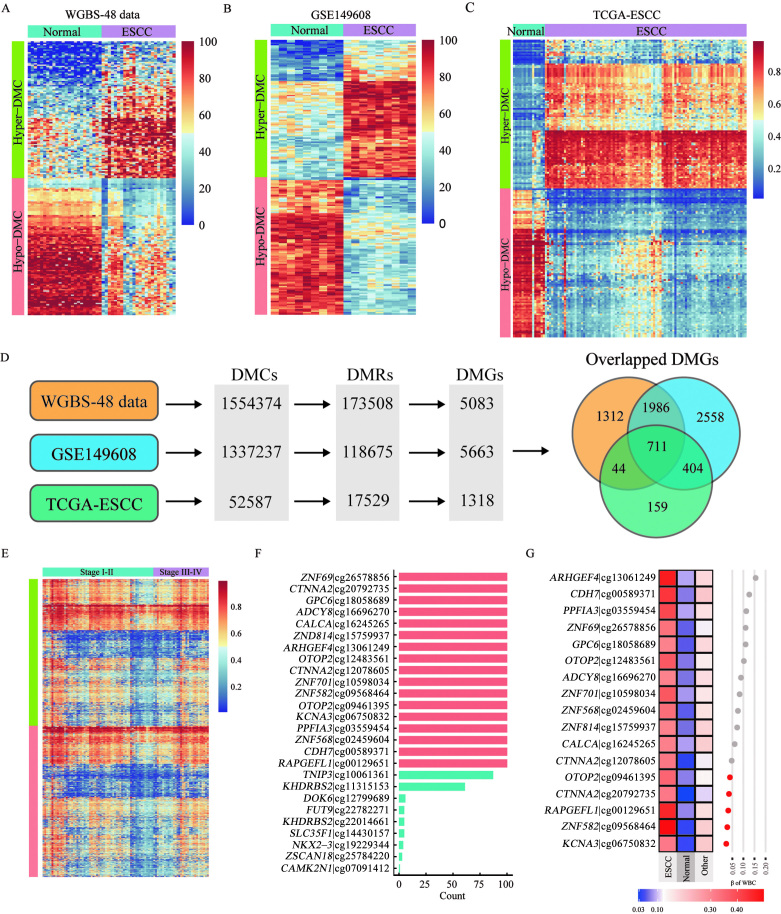
We obtained 1,554,374 DMCs from the in-house dataset [Figure 3A]. These DMCs were validated using the GSE149608 dataset. Using the same method, we identified 1,337,237 DMCs (57,632 hypo-DMCs and 1,279,605 hypo-DMCs) in this dataset [Figure 3B]; 775,008 DMCs were shared by both datasets. The results of the correlation analysis showed that the methylation levels of these shared DMCs were similar between the datasets [Supplementary Figure 3A–D, http://links.lww.com/CM9/B702]. Based on further analysis, we identified 118,675 DMRs in the GSE149608 dataset involving 5663 genes, including 2697 (53.06%) DMGs identified in the in-house dataset.
Figure 3.
Identification of candidate methylation markers for ESCC. (A–C): A heat map of the top 100 most significant hyper-DMCs and hypo-DMCs identified in the in-house dataset (A), the GSE149608 dataset (B), and the TCGA-ESCC dataset (C). (D): The DMCs, DMRs and DMGs were compared among the three datasets. (E) The methylation levels of candidate probes in stage I–II and stage III–IV ESCC. (F) The frequency of the probes in 100 time of the LASSO regressions (only 26 probes with non-zero coefficients are shown). (G) The methylation levels of 17 candidate probes in healthy WBC, 710 NATs, and 32 other types of cancer. DMC: Differentially methylated CpG; DMG: Differentially methylated genes; DMR: Differentially methylated region; ESCC: Esophageal squamous cell carcinoma; NATs: Normal adjacent tissues; TCGA: The Cancer Genome Atlas; WBC: Whole blood cells; WGBS: Whole-genome bisulfite sequencing.
The previously mentioned 2697 DMGs were then validated based on the TCGA-ESCC cohort. Using the same approach, we identified 52,587 DMCs [Figure 3C] (36,685 hyper-DMCs and 15,902 hypo-DMCs) between ESCC tissues and their NATs, which consisted of 17,529 DMRs involving 1318 DMGs. By integrating the three datasets, we obtained 2374 overlapped DMCs (711 DMGs) [Figure 3D]. To filter the DMCs that did not show different methylation levels at different stages of ESCC, we compared the methylation values of the 2374 DMCs between stage I–II and III–IV ESCC. We selected the top 50% of the probes, corresponding to 1118 probes (573 DMGs), with the smallest delta β values between stages I–II and III–IV, for the next analysis [Figure 3E]. Next, we identified the ESCC-specific hyper-DMCs using the data on the other 32 types of cancer in TCGA database. Using 710 NATs and the 32 types of cancer tissues (n = 8163) as controls, we compressed the 1118 probes using LASSO algorithm. We found that 17 probes (15 genes) appeared in all 100 times of LASSO regression. These probes were selected for the subsequent analysis [Figure 3F].
We expected these 17 probes to have the lowest methylation levels in blood cells, considering that they were plasma cfDNA methylation markers. Therefore, the methylation levels of the 17 candidate probes were evaluated in healthy blood cells. The top five probes with minimal methylation values in blood cells were cg06750832 (KCNA3), cg09568464 (ZNF582), cg00129651 (RAPGEFL1), cg20792735 (CTNNA2), and cg09461395 (OTOP2) [Figure 3G], and thus, they were considered to be the most promising markers.
Validating the methylation status of candidate genes by Sanger sequencing
We found that four of the five DMCs, including cg06750832 (KCNA3), cg09568464 (ZNF582), cg00129651 (RAPGEFL1), and cg09461395 (OTOP2), met the requirements of methylation specific PCR (MSP) primer design. Therefore, we performed Sanger sequencing for the four targets to verify their methylation status in healthy blood cells (n = 42), NATs (n = 50), and ESCC tissues (n = 24). The four genes were prevalently hypermethylated in ESCC samples but hypomethylated in NATs and healthy blood cells [Supplementary Table 11, http://links.lww.com/CM9/B701]. Compared to the other three genes, OTOP2 showed lower methylation levels in healthy individuals and NATs [Supplementary Figure 4A–D, http://links.lww.com/CM9/B702], which matched the results obtained by analyzing theTCGA-ESCC dataset. Similar findings were also observed for KCNA3, as demonstrated by the higher proportion of methylated CpGs in ESCC tissues compared to that in the other three genes. Additionally, more than 90% of the CpGs in the four genes exhibited hypermethylation in the ESCC samples [Supplementary Figure 5A–D, http://links.lww.com/CM9/B702].
Performance of four candidate genes for detecting ESCC in the training set
The preliminary results suggested that ZNF582, KCNA3, RAPGEFL1, and OTOP2 might be the most promising plasma cfDNA methylation markers for ESCC. Therefore, we performed PCR-based assays using the four genes. In the standard curve experiments, we found that the amplification efficiency of the primers designed for these four targets was 102.44, 103.39, 111.02, and 100.11, respectively. As these values were high [Supplementary Figure 6A–D, http://links.lww.com/CM9/B702], the performance of the four assays was considered to be satisfactory.
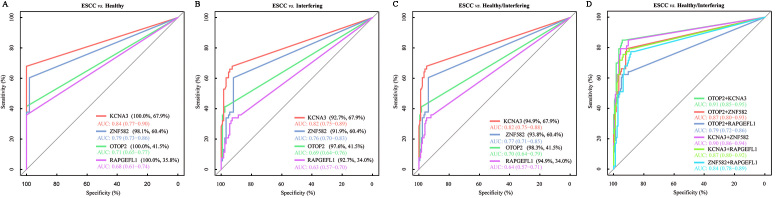
The training set for this study included plasma samples from 52 healthy individuals, 124 individuals with non-esophageal lesions (interfering disease), and 53 patients with ESCC. The ROC curves suggested that KCNA3 had the highest AUC value (0.84 [95% CI: 0.77–0.90]), followed by ZNF582 (0.79 [95% CI: 0.73–0.86]), RAPGEFL1 (0.71 [95% CI: 0.65–0.77]), and OTOP2 (0.68 [95% CI: 0.61–0.74]) [Figure 4A]. The optimal specificity for KCNA3, RAPGEFL1, and OTOP2 was 100% at the maximum value of the Youden index, but it was slightly lower for ZNF582 (98.1%). The optimal sensitivities of the four targets were 67.9%, 60.4%, 41.5%, and 35.8%, respectively. When the interfering disease was used as a control, the AUC values for all targets and the optimal specificities decreased [Figure 4B]. When the healthy condition and interfering disease together were used as the control, the AUC values of the four markers were lower than those for the healthy control but higher than those for the interfering disease used as a control [Figure 4C]. No single target had a sensitivity value exceeding 70% for detecting ESCC, although they all showed high specificity. Therefore, we improved the sensitivity of ESCC detection by combining the four markers. When the healthy condition and interfering disease together were used as the control, we obtained higher AUC values for all combinations of any two targets than that for any single target [Figure 4D]. Among the six combinations (any two of the four targets), the panel of OTOP2 and KCNA3 had the highest AUC value, with optimal sensitivity and specificity of 84.91% and 94.32%, respectively. Thus, the performance of this panel was further assessed using the validation set.
Figure 4.
The ROC curve analysis was conducted to evaluate the performance of the four targets in the training set. (A) ROC curves for the four targets when the healthy condition was used as the control. (B) ROC curves for the four targets when the interfering disease was used as the control. (C) ROC curves for the four targets when the healthy condition and interfering disease were used as the control. (D) ROC curves for the six combinations of the four targets. AUC: Area under the curve; ROC curves: Receiver operating characteristic curves.
Performance of KCNA3 and OTOP2 for detecting ESCC in the validation set
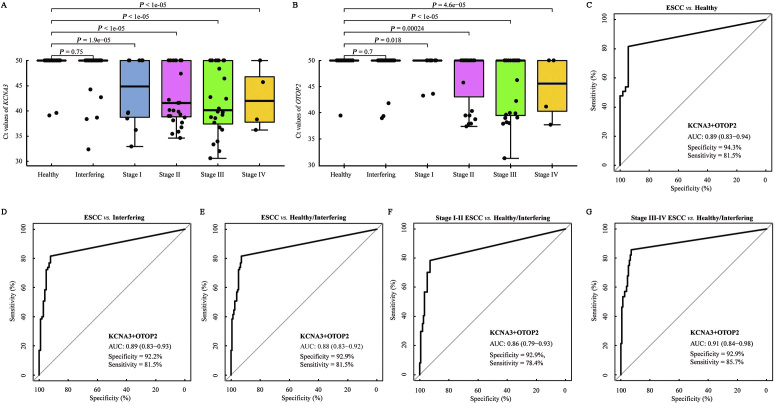
The validation set consisted of 220 plasma samples collected from 53 healthy individuals, 102 non-esophageal cancer patients, and 65 patients with ESCC. The results of the PCR in the validation set are presented in Supplementary Table 12, http://links.lww.com/CM9/B701. We first evaluated the Ct values of KCNA3 and OTOP2 at different stages of ESCC. The Ct values of both targets were not significantly different between the data on healthy individuals and interfering diseases, but they were significantly lower in all stages of ESCC [Figure 5A,B]. The results of the ROC curve analysis showed that the combination of KCNA3 and OTOP2 had an AUC of 0.89 (95% CI: 0.83–0.94) for the difference between patients with ESCC and healthy individuals. The optimal sensitivity was 81.5% and the specificity was 94.3% [Figure 5C]. When the interfering disease was used as the control, the AUC for KCNA3/OTOP2 was 0.89 (95% CI: 0.83–0.93), and the optimal specificity decreased to 92.2% [Figure 5D]. When the healthy condition and interfering disease were used together as the control, the AUC for KCNA3/OTOP2 was 0.88 (0.83–0.92), with the optimal specificity of 92.9% [Figure 5E]. The optimal sensitivity was 78.4% [Figure 5F] for stage I–II ESCC, whereas, it was 85.7% for stage III–IV ESCC [Figure 5G], which was higher than the sensitivity for early-stage ESCC, although the difference was not statistically significant (P >0.05). Additionally, the differences in KCNA3/OTOP2 for the detection of ESCC stratified by age and gender [Supplementary Table 13, http://links.lww.com/CM9/B701] were not significant.
Figure 5.
The performance of methylated KCNA3 and OTOP2 for detecting ESCC in the validation set. (A) The Ct values of KCNA3 in different types of plasma samples. (B) The Ct values of OTOP2 in different types of plasma samples. (C) The ROC curve shows the performance of methylated KCNA3/OTOP2, indicating the difference between ESCC and the healthy condition. (D) The ROC curve shows the performance of methylated KCNA3/OTOP2, indicating the difference between ESCC and interfering diseases. (E) The ROC curve shows the performance of methylated KCNA3/OTOP2, indicating the difference between ESCC and healthy condition/interfering diseases. (F) The ROC curve shows the performance of methylated KCNA3/OTOP2, indicating the difference between stage I–II ESCC and healthy condition/interfering disease. (G) The ROC curve shows the performance of methylated KCNA3/OTOP2, indicating the difference between stage III–IV ESCC and healthy condition/interfering disease. AUC: Area under the curve; Ct: Cycle threshold; ESCC: Esophageal squamous cell carcinoma.
Discussion
The lack of appropriate screening methods or tools is the main problem in areas with a high prevalence of ESCC.[10] Endoscopy is the gold standard for the diagnosis of ESCC, but its usage has limitations of invasiveness, inconvenience, and less cost-effectiveness.[42] As endoscopy is invasive and extremely uncomfortable for the patient, it would reduce the compliance of high-risk populations. In addition, the effectiveness of endoscopy highly depends on the experience of the endoscopists and the infrastructure of endoscopy units, which is not applicable in mass screening, especially for resource-limited areas. Hence, blood-based liquid biopsy assays need to be developed as an alternative approach. In this study, we identified five novel promising markers by integrating multiple datasets, including WGBS-48 data (in-house data), GSE149608, methylation data in TCGA, and GSE40279. Based on further analysis, we found that a panel consisting of methylated KCNA3 and OTOP2 showed excellent performance for the detection of ESCC in the training and independent plasma validation cohorts. Our results indicated that the dual-target panel might be an effective tool for detecting ESCC.
We aimed to develop one panel with the best detection capacity using the fewest targets. In this study, we incorporated three datasets to ensure that the biomarkers could adequately account for the tumor heterogeneity linked to ESCC. First, we performed WGBS for 24 ESCC tissues and their NATs, which allowed us to identify more candidate DMCs. Then, we validated the candidate DMCs in two independent datasets, GSE149608 and TCGA-ESCC, and obtained 2374 DMCs shared by the three datasets. To obtain the stage-insensitive CpGs, we only retained those DMCs that did not show significant differences between stage I–II and III–IV ESCCs. The ESCC-specific hyper-DMCs were identified using the other 32 types of cancer in TCGA datasets via LASSO regression. Finally, we evaluated the methylation levels of ESCC-specific hyper-DMCs in 656 healthy blood cell samples to eliminate the CpGs that showed high methylation levels in blood cells. Based on this method, we obtained the top five DMCs with the lowest blood cell methylation for the subsequent analysis.
In the biological validation stage, the candidate marker CTNNA2 was excluded due to the inaccessible primers and probes. The methylation status of the other four candidate targets was confirmed by Sanger sequencing. The ESCC tissues showed the highest methylated CpG ratios, and healthy individuals showed the lowest methylated CpG ratios. Although all single markers could be used to differentiate between ESCC and healthy controls, their sensitivity did not exceed 70%. Therefore, we developed a diagnostic panel combining multiple markers to enhance their performance for detecting ESCC. Based on the amount of plasma used and the ease of interpreting the results of MSP in clinical practice, we focused on analyzing the two-by-two combination of the four candidate markers to obtain the optimal combination and their optimal cut-off value in the training set. We simultaneously evaluated the performance of the three-marker panels and the four-marker panel, and they all had higher AUC and sensitivity than the two-marker panels but exhibited lower specificities. Therefore, in a trade-off between sensitivity and specificity, we selected the two-marker panel consisting of KCNA3 and OTOP2, which had the highest AUC and sensitivity, as the best dual-marker combination and evaluated its diagnostic performance using an independent validation set. To minimize bias, the analysts conducting the sequencing and classification analysis were unaware of the clinical information.
Although several blood-based markers are widely used in clinical practice, including squamous cell carcinoma antigen (SCC-Ag), carcinoembryonic antigen (CEA), and cytokeratin-19 fragment (CYFRA21-1), they do not serve as independent diagnostic markers for ESCC.[42] The sensitivity of our panel for detecting ESCC was 81.5%, which was higher than the sensitivity of the three traditional serum biomarkers.[43] Our findings showed that ESCC can be effectively detected with fewer DNA methylation markers. Early detection of ESCC can significantly improve the prognosis of patients. Therefore, strategies to diagnose ESCC in the early stage are clinically valuable. In this study, the panel of KCNA3 and OTOP2 showed a sensitivity of 78.4% in the validation cohort for stage I–II ESCC and a high specificity of 92.9%; these values were better than those of the panels reported in other studies. For example, the panel of 921 DMRs proposed by Qiao et al[26] showed a sensitivity of 66.7% for stage I-II ESCC. Another panel with five DNA methylation markers could detect stage I–II ESCC detection with a sensitivity of 53.6%.[44]
The better diagnostic performance of the dual-target panel suggested that it can be used for screening large-scale populations. Individuals diagnosis with a positive condition need to be further confirmed by endoscopy. To simulate real-world scenarios, we included several interfering samples as benign lesions and various other malignancies in this study. The dual-target panel showed a specificity of 92.2% for interfering samples in the independent validation set. Additionally, compared to endoscopy, the panel was cheaper, minimally invasive, less dependent on operator expertise, and more reproducible. Thus, the two methylation markers are more suitable for routine clinical assessments and population studies.
In this study, we also identified the markers that could be used to specifically detect ESCC, as reflected in the process of the identification candidate makers and the inclusion of interfering diseases in the training and validation sets. However, the two markers showed high methylation levels in TCGA-EACs. Although ESCC and EAC have biological differences, we found a significant overlap in their DNA methylation signatures. We also showed that DNA methylation sites were more indicative of the host organ epithelium than the biology of the underlying tumor,[44] as reported in several other studies.[26,45] This epigenetic similarity suggested that our dual-marker panel might also have excellent diagnostic performance for EAC. However, further studies are needed to confirm this speculation.
Our study had certain limitations. First, the sample sizes of the training and validation cohorts in this study were small. Thus, the dual-target panel needs to be evaluated in a larger clinical study. To address this issue, a national multicenter clinical trial has been initiated (NCT05680077). Second, the ability of this panel to diagnose asymptomatic ESCC patients should be assessed more extensively because the patients included in this study exhibited symptoms that are inconsistent with the situation that more asymptomatic patients are present in the population. Third, multiple datasets from different sources were used in this study, which might have introduced bias while identifying methylation markers. Specifically, we verified the candidate DMCs obtained from the WGBS data in the TCGA-ESCC dataset. This strategy caused us to miss many promising DMCs, especially those DMCs associated with WGBS that were not covered by the Illumina 450K BeadChip.
To summarize, genome-wide methylation profiling of ESCC provides valuable epigenetic information that can be used for developing novel methylation markers. Our findings suggested that cfDNA methylation signatures can be used for accurately diagnosing ESCC, and they might be used for screening high-risk populations. The dual-target diagnostic model constructed in this study also showed that liquid biopsy techniques based on cfDNA methylation might be further developed for application in the clinical setting.
Acknowledgments
None.
Funding
This research was supported by the Science and Technology Commission of Shanghai Municipality (No. 21Y31900100).
Conflicts of interest
None.
Data sharing
The raw reads have been submitted to NCBI Sequence Read Archive with the accession number PRJNA917325. All data associated with this study are present in the paper or the Supplementary Materials. Further inquiries can be directed to the corresponding authors.
Supplementary Material
Footnotes
Yan Bian, Ye Gao, Chaojing Lu, and Bo Tian contributed equally to this work.
How to cite this article: Bian Y, Gao Y, Lu CJ, Tian B, Xin L, Lin H, Zhang YH, Zhang X, Zhou SW, Wan KK, Zhou J, Li ZS, Chen HZ, Wang LW. Genome-wide methylation profiling identified methylated KCNA3 and OTOP2 as promising diagnostic markers for esophageal squamous cell carcinoma. Chin Med J 2024;137:1724–1735. doi: 10.1097/CM9.0000000000002832
References
- 1.Sung H Ferlay J Siegel RL Laversanne M Soerjomataram I Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71: 209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Qiu H, Cao S, Xu R. Cancer incidence, mortality, and burden in China: A time‐trend analysis and comparison with the United States and United Kingdom based on the global epidemiological data released in 2020. Cancer Commun (Lond) 2021;41: 1037–1048. doi: 10.1002/cac2.12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan E Soerjomataram I Rumgay H Coleman HG Thrift AP Vignat J, et al. The global landscape of esophageal squamous cell carcinoma and esophageal adenocarcinoma incidence and mortality in 2020 and projections to 2040: New estimates from GLOBOCAN 2020. Gastroenterology 2022;163: 649–658.e2. doi: 10.1053/j.gastro.2022.05.054. [DOI] [PubMed] [Google Scholar]
- 4.Xia C Dong X Li H Cao M Sun D He S, et al. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin Med J 2022;135: 584–590. doi: 10.1097/CM9.0000000000002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology 2018;154: 360–373. doi: 10.1053/j.gastro.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smyth EC Lagergren J Fitzgerald RC Lordick F Shah MA Lagergren P, et al. Oesophageal cancer. Nat Rev Dis Primers 2017;3: 17048. doi: 10.1038/nrdp.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng H Chen W Zheng R Zhang S Ji JS Zou X, et al. Changing cancer survival in China during 2003–15: A pooled analysis of 17 population-based cancer registries. Lancet Glob Health 2018;6: e555–e567. doi: 10.1016/S2214-109X(18)30127-X. [DOI] [PubMed] [Google Scholar]
- 8.Lagergren J, Mattson F. Diverging trends in recent population-based survival rates in oesophageal and gastric cancer. PLoS One 2012;7: e41352. doi: 10.1371/journal.pone.0041352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang GQ Abnet CC Shen Q Lewin KJ Sun XD Roth MJ, et al. Histological precursors of oesophageal squamous cell carcinoma: Results from a 13 year prospective follow up study in a high risk population. Gut 2005;54: 187–192. doi: 10.1136/gut.2004.046631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Codipilly DC Qin Y Dawsey SM Kisiel J Topazian M Ahlquist D, et al. Screening for esophageal squamous cell carcinoma: Recent advances. Gastrointest Endosc 2018;88: 413–426. doi: 10.1016/j.gie.2018.04.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen R Liu Y Song G Li B Zhao D Hua Z, et al. Effectiveness of one-time endoscopic screening programme in prevention of upper gastrointestinal cancer in China: A multicentre population-based cohort study. Gut 2020;70: 251–260. doi: 10.1136/gutjnl-2019-320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei WQ Chen ZF He YT Feng H Hou J Lin DM, et al. Long-term follow-up of a community assignment, one-time endoscopic screening study of esophageal cancer in China. J Clin Oncol 2015;33: 1951–1957. doi: 10.1200/JCO.2014.58.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z Guo C He Y Chen Y Ji P Fang Z, et al. A clinical model predicting the risk of esophageal high-grade lesions in opportunistic screening: A multicenter real-world study in China. Gastrointest Endosc 2020;91: 1253–1260.e3. doi: 10.1016/j.gie.2019.12.038. [DOI] [PubMed] [Google Scholar]
- 14.He Z Liu Z Liu M Guo C Xu R Li F, et al. Efficacy of endoscopic screening for esophageal cancer in China (ESECC): Design and preliminary results of a population-based randomised controlled trial. Gut 2019;68: 198–206. doi: 10.1136/gutjnl-2017-315520. [DOI] [PubMed] [Google Scholar]
- 15.Xin L Gao Y Cheng Z Wang T Lin H Pang Y, et al. Utilization and quality assessment of digestive endoscopy in China: Results from 5-year consecutive nationwide surveys. Chin Med J 2022;135: 2003–2010. doi: 10.1097/CM9.0000000000002366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Y Xin L Feng YD Yao B Lin H Sun C, et al. Feasibility and accuracy of artificial intelligence-assisted sponge cytology for community-based esophageal squamous cell carcinoma screening in China. Am J Gastroenterol 2021;116: 2207–2215. doi: 10.14309/ajg.0000000000001499. [DOI] [PubMed] [Google Scholar]
- 17.Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet 2017;390: 2383–2396. doi: 10.1016/S0140-6736(17)31462-9. [DOI] [PubMed] [Google Scholar]
- 18.Song P Wu LR Yan YH Zhang JX Chu T Kwong LN, et al. Limitations and opportunities of technologies for the analysis of cell-free DNA in cancer diagnostics. Nat Biomed Eng 2022;6: 232–245. doi: 10.1038/s41551-021-00837-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Pol Y, Mouliere F. Toward the early detection of cancer by decoding the epigenetic and environmental fingerprints of cell-free DNA. Cancer Cell 2019;36: 350–368. doi: 10.1016/j.ccell.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Xi Y Lin Y Guo W Wang X Zhao H Miao C, et al. Multi-omic characterization of genome-wide abnormal DNA methylation reveals diagnostic and prognostic markers for esophageal squamous-cell carcinoma. Signal Transduct Target Ther 2022;7: 53. doi: 10.1038/s41392-022-00873-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talukdar FR Soares Lima SC Khoueiry R Laskar RS Cuenin C Sorroche BP, et al. Genome-wide DNA methylation profiling of esophageal squamous cell carcinoma from global high-incidence regions identifies crucial genes and potential cancer markers. Cancer Res 2021;81: 2612–2624. doi: 10.1158/0008-5472.CAN-20-3445. [DOI] [PubMed] [Google Scholar]
- 22.Lin L, Cheng X, Yin D. Aberrant DNA methylation in esophageal squamous cell carcinoma: Biological and clinical implications. Front Oncol 2020;10: 549850. doi: 10.3389/fonc.2020.549850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein EA Richards D Cohn A Tummala M Lapham R Cosgrove D, et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann Oncol 2021;32: 1167–1177. doi: 10.1016/j.annonc.2021.05.806. [DOI] [PubMed] [Google Scholar]
- 24.Luo H, Wei W, Ye Z, Zheng J, Xu RH. Liquid biopsy of methylation biomarkers in cell-free DNA. Trends Mol Med 2021;27: 482–500. doi: 10.1016/j.molmed.2020.12.011. [DOI] [PubMed] [Google Scholar]
- 25.Liu MC Oxnard GR Klein EA Swanton C Seiden MV, CCGA Consortium . Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol 2020;31: 745–759. doi: 10.1016/j.annonc.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiao G Zhuang W Dong B Li C Xu J Wang G, et al. Discovery and validation of methylation signatures in circulating cell-free DNA for early detection of esophageal cancer: A case-control study. BMC Med 2021;19: 243. doi: 10.1186/s12916-021-02109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Habibi E Brinkman AB Arand J Kroeze LI Kerstens HH Matarese F, et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 2013;13: 360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Yu Q Xia N Zhao Y Jin H Chen R Ye F, et al. Genome-wide methylation profiling identify hypermethylated HOXL subclass genes as potential markers for esophageal squamous cell carcinoma detection. BMC Med Genomics 2022;15: 247. doi: 10.1186/s12920-022-01401-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, DeFina SM, Bajpai M, Yan Q, Yang L, Zhou Z. DNA methylation markers in esophageal cancer: An emerging tool for cancer surveillance and treatment. Am J Cancer Res 2021;11: 5644–5658. eCollection 2021. [PMC free article] [PubMed] [Google Scholar]
- 30.Salta S Macedo-Silva C Miranda-Gonçalves V Lopes N Gigliano D Guimarães R, et al. A DNA methylation-based test for esophageal cancer detection. Biomark Res 2020;8: 68. doi: 10.1186/s40364-020-00248-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng X, Xue H, Lü L, Shi P, Wang J, Wang J. Accumulated promoter methylation as a potential biomarker for esophageal cancer. Oncotarget 2017;8: 679–691. doi: 10.18632/oncotarget.13510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong T, Borgard H, Zhang Z, Chen S, Gao Z, Deng Y. Analysis and performance assessment of the whole genome bisulfite sequencing data workflow: Currently available tools and a practical guide to advance DNA methylation studies. Small Methods 2022;6: e2101251. doi: 10.1002/smtd.202101251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S, Zhou Y, Chen Y, Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018;34: i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Ge X, Peng F, Li W, Li JJ. Exaggerated false positives by popular differential expression methods when analyzing human population samples. Genome Biol 2022;23: 79. doi: 10.1186/s13059-022-02648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li R Qu B Wan K Lu C Li T Zhou F, et al. Identification of two methylated fragments of an SDC2 CpG island using a sliding window technique for early detection of colorectal cancer. FEBS Open Bio 2021;11: 1941–1952. doi: 10.1002/2211-5463.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franzin A, Sambo F, Di Camillo B. bnstruct: An R package for Bayesian Network structure learning in the presence of missing data. Bioinformatics 2017;33: 1250–1252. doi: 10.1093/bioinformatics/btw807. [DOI] [PubMed] [Google Scholar]
- 37.Hannum G Guinney J Zhao L Zhang L Hughes G Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013;49: 359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishihira T Kasai M Mori S Watanabe T Kuriya Y Suda M, et al. Characteristics of two cell lines (TE-1 and TE-2) derived from human squamous cell carcinoma of the esophagus. Gan 1979;70: 575–584. [PubMed] [Google Scholar]
- 39.Richardson GS Dickersin GR Atkins L MacLaughlin DT Raam S Merk LP, et al. KLE: A cell line with defective estrogen receptor derived from undifferentiated endometrial cancer. Gynecol Oncol 1984;17: 213–230. doi: 10.1016/0090-8258(84)90080-5. [DOI] [PubMed] [Google Scholar]
- 40.Dragan AI Pavlovic R McGivney JB Casas-Finet JR Bishop ES Strouse RJ, et al. SYBR Green I: Fluorescence properties and interaction with DNA. J Fluoresc 2012;22: 1189–1199. doi: 10.1007/s10895-012-1059-8. [DOI] [PubMed] [Google Scholar]
- 41.Robin X Turck N Hainard A Tiberti N Lisacek F Sanchez JC, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12: 77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma K Kalra A Tsai HL Okello S Cheng Y Meltzer SJ, et al. Accurate nonendoscopic detection of esophageal squamous cell carcinoma using methylated DNA biomarkers. Gastroenterology 2022;163: 507–509.e2. doi: 10.1053/j.gastro.2022.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng Q Zhang L Tu M Yin X Cai L Zhang S, et al. Development of a panel of autoantibody against NSG1 with CEA, CYFRA21-1, and SCC-Ag for the diagnosis of esophageal squamous cell carcinoma. Clin Chim Acta 2021;520: 126–132. doi: 10.1016/j.cca.2021.06.013. [DOI] [PubMed] [Google Scholar]
- 44.Qin Y Wu CW Taylor WR Sawas T Burger KN Mahoney DW, et al. Discovery, validation, and application of novel methylated DNA markers for detection of esophageal cancer in plasma. Clin Cancer Res 2019;25: 7396–7404. doi: 10.1158/1078-0432.CCR-19-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hlady RA Tiedemann RL Puszyk W Zendejas I Roberts LR Choi JH, et al. Epigenetic signatures of alcohol abuse and hepatitis infection during human hepatocarcinogenesis. Oncotarget 2014;5: 9425–9443. doi: 10.18632/oncotarget.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]