Abstract
We have investigated the antigenicity of the C- and N-terminal halves of pIX of human adenovirus types 2 and 3 (Ad2 and Ad3) as well as their orientations in virions. We found that only the C-terminal halves of Ad2 pIX and Ad3 pIX reacted in a subgenus-specific manner by enzyme-linked immunosorbent assay and immunoblot analysis. Based on immunoelectron microscopy experiments, pIX in viral capsids appears to be positioned such that the C-terminal part of pIX constitutes the surface domain whereas the N terminus of the protein makes up the internal domain in icosahedral Ad capsids.
Human adenovirus (Ad) is composed of at least 11 different structural proteins (polypeptides II through IX, IIIa, μ, and tp) and a linear double-stranded DNA genome (31, 32, 36, 37). The fine structure of the complete Ad particle was determined by image reconstruction from cryoelectron microscopy and by X-ray crystallography and electron microscopy difference imaging (33). pIX is a 14.3-kDa minor component of the groups of nine hexons (GONs) that form the central region of each facet of the icosahedral capsid (6, 7). pIX functions as a cement stabilizing hexon-hexon interactions (19) and is essential for viral DNA packaging (20). A mutant of Ad serotype 5 (Ad5) containing a deletion in the gene for pIX produced virions that are more heat labile than those of the wild type (11). The mutant can grow only in 293 cells, which produce pIX coded for by their resident Ad5 sequences; however, the amount is too small to be incorporated into virions (21). Recently it was shown that pIX of Ad5, in addition to its structural contribution, exhibits transcriptional properties (27). We have also found that Ad2 pIX and Ad3 pIX are O-glycosylated and phosphorylated (1).
Antisera directed to virions or purified capsid proteins were used to elucidate the topographical organization of the structural proteins in studies based on the currently accepted model of the architecture of adenoviruses (6, 7, 18, 19). The pIX has not been characterized in as much detail as the hexon, fiber, or pentonbase with regard to its antigenicity (38). Polyclonal serum was produced against purified Ad2 pIX to study the immunological properties of pIX (6). On the basis of the results of immunodiffusion tests with Ad2 pIX-specific antibodies it was assumed that both type- and group-specific determinants exist on pIX. However, no information is available on whether specific regions of the protein are important for serotype or subgenus specificity or what may constitute a type-specific, as opposed to a cross-reactive, epitope. Furthermore, pIX is known to be buried within the capsid, although there is still the question of whether it is located on the inner or outer surface of the GON (6, 8). We used bacterially expressed recombinant pIX of Ad2 and Ad3 as a subgenus-specific antigen in enzyme-linked immunosorbent assay (ELISA) and immunoblot analysis (2). In this report we present data on the immunogenic regions and the orientation of pIX in virions, based on immunological analysis of the C- and N-terminal parts of pIX by ELISA, immunoblotting, immunogold electron microscopy, and neutralization assay.
Construction of plasmids, expression of pIX, and production of polyclonal rabbit sera.
The full-length gene encoding pIX was amplified by PCR with either an Ad2 genome or an Ad3 genome as a template, using primers as reported previously (2). A C-terminal fragment was prepared by PCR amplification by using the sense primers Ad2pIXC, 5′[3810]-TT aga tct ACC GCC CGC GGG ATT GTG A-3′ (with a BglII restriction site [underlined]), and Ad3pIXC, 5′[3691]-TT aga tct AAC ACC ATC CTT GGA ATG G-3′ (with a BglII restriction site [underlined]). The sequence encoding the N-terminal part of pIX was amplified by PCR by using the antisense primers Ad2pIXN, 5′[3672]-TTT aag ctt GGC GGC AGC AGT AGC-3′ (with a HindIII restriction site [underlined]), and Ad3pIXN, 5′[3771]-TTT aag ctt TTA GGC TGC AGC GGC TGA-3′ (with a HindIII restriction site [underlined]). The antisense primers for the C-terminal fragments and the sense primers for the N-terminal fragments were the same as those for the full-length pIX gene (2).
PCR-amplified BamHI-HindIII- and EcoRI-HindIII-restricted fragments of the pIX genes of Ad2 and Ad3, respectively, were cloned into appropriately digested pQE30 expression vector (QIAGEN, Hilden, Germany), generating Ad2pIX/ pQE30 and Ad3pIX/pQE30. In these constructs, pIX was fused with six histidine residues, enabling purification by metal chelate affinity chromatography. PCR fragments encoding only the N- and C-terminal halves of Ad2 pIX and Ad3 pIX were cleaved with the appropriate corresponding enzymes and cloned into pQE40 expression vector, generating Ad2pIXN/pQE40 or Ad2pIXC/pQE40 and Ad3pIXN/pQE40 or Ad3pIXC/pQE40. In these constructs, residues 2 to 70 and 71 to 140 of Ad2 pIX and residues 2 to 70 and 71 to 138 of Ad3 pIX were fused in frame to the C terminus of dihydrofolate reductase (DHFR), which has six histidine residues (His6 tag), at its N terminus. Polyclonal antisera against purified full-length pIX and against the C-terminal and N-terminal halves of Ad2 pIX and Ad3 pIX were produced by subcutaneous and intramuscular injections into rabbits (2, 25).
Ad pIX amino acid sequences show high homology within subgenus and differences between subgenera.
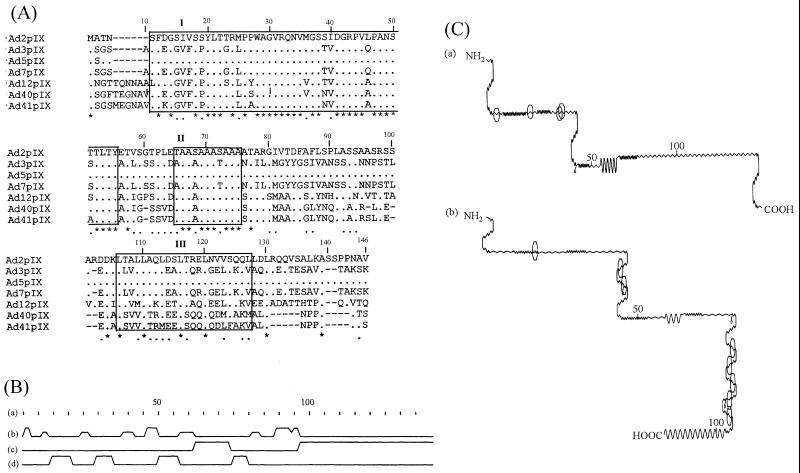
The nucleotide sequences of pIX genes of Ad2 (3) and Ad5 (28) (of subgenus C), Ad3 (16) and Ad7 (15) (of subgenus B), Ad12 (5) (of subgenus A), and Ad40 (12) and Ad41 (4) (of subgenus F) were translated and analyzed for amino acid sequence similarity. Pairwise alignment (30), secondary structure determinations (10), and amino acid similarity scoring were performed by using the global alignment program GAP from the Genetics Computer Group (13, 14). Multiple sequence alignment was accomplished with CLUSTAL V (22), by using a value of 10 for both fixed and floating gap penalties (Fig. 1A). Pairwise amino acid sequence alignments between serotypes confirmed that all those belonging to the same subgenus had over 95% identity over the entire length of pIX. However, the degrees of identity and similarity ranged between 50 and 65% for comparisons between serotypes belonging to different subgenera. The alignment revealed that the N-terminal half of the protein exhibited moderate levels of identity (67%) and similarity (74%), whereas the C-terminal half of the protein presented as a variable region (33% identity and 40% similarity). Three domains which showed high conservation of identical or similar amino acids were identified. These are domain I (residues 11 to 55), which contains two phosphorylation sites, domain II (residues 65 to 75), and domain III (residues 106 to 127), which contains a leucine zipper motive. Secondary structure predictions made by the Chou and Fasman method (10) are shown in Fig. 1B. The possible antigenic sites (23) of Ad2 pIX and Ad3 pIX have different locations, as indicated in Fig. 1C. While the few epitopes of Ad2 pIX are located in the N-terminal part, more antigenic sites were predicted to be present in the C-terminal part than in the N-terminal part of Ad3 pIX.
FIG. 1.
(A) Multiple amino acid sequence alignment of pIX from seven human Ad serotypes. Residue numbering includes the gaps (−) in the alignment. The asterisks and dots denote identical and conserved residues, respectively. Structural similarity is indicated by boxes (labelled I, II, and III). (B) Schematic diagram of the secondary structure derived from predictions made by the Chou and Fasman method (10) shows residue numbering according to the sequences of Ad2 (a), turns (b), alpha-helical regions (c), and β regions (d). (C) Schematic diagrams of the potential antigenic sites of Ad2pIX (a) and Ad3pIX (b) (23). The sites are represented by octagons.
Antigenicity of Ad2 pIX and Ad3 pIX.
ELISA was performed as described previously (25). Wells were coated with 17 ng of pIXC- or pIXN-DHFR fusion proteins/well and 10 ng of full-length pIX antigen/well. Optical densities (OD) at 492 nm were measured at a dilution of 1:800 in duplicate samples in each assay. Ad2 pIX-specific antibodies reacted strongly with homologous full-length pIX (OD, 2.11) and moderately with heterologous full-length pIX (OD, 0.73) (Table 1). At the same serum dilution, Ad3 pIX-specific antibodies reacted to homologous and to heterologous pIX with absorbance values of 2.32 and 1.07, respectively. Anti-Ad2 pIX serum reacted weakly with the homologous C-terminal half of pIX (OD, 0.317) and did not react with the heterologous C-terminal half of pIX (OD, 0.017). This serum showed absorbance values of 0.480 and 0.064 in reactions with the homologous and the heterologous N-terminal halves of pIX, respectively. In contrast with this, anti-Ad3 pIX serum had an absorbance value of 0.61 in reaction with the homologous C-terminal half of the Ad3 pIX but did not react with Ad2pIXC. Polyclonal rabbit serum directed against Ad3 pIX also reacted with the N-terminal halves of Ad3 pIX (OD, 0.307) and Ad2 pIX (OD, 0.365). None of the experimental results shown in Tables 1 and 2 is due to a possible cross-reactivity with DHFR because either the antigen used lacked DHFR or the antibodies used were raised against a recombinant protein which contains no DHFR sequences. In order to find out whether the reactivity of our antisera with the recombinant proteins was not partly caused by the His6 tag, we tested the antisera against purified virus samples by immunoblotting. Anti-Ad2 pIX and anti-Ad3 pIX sera reacted well with the corresponding homologous pIX. The Ad3pIXN- and Ad3pIXC-specific sera weakly recognized Ad3 pIX (not shown). Additionally, Ad2 pIX-specific antiserum did not recognize Ad3pIXC, and Ad3 pIX-specific antiserum did not bind to the Ad2pIXC under the conditions chosen, as mentioned above. The weak binding indicates that any reaction with the His6 tag is very weak and can be ignored.
TABLE 1.
Reactivities of anti-pIX polyclonal rabbit sera directed against the homologous and heterologous antigens of Ad2 and Ad3, respectively
| Antigen | OD for reaction witha:
|
|
|---|---|---|
| Anti-Ad2 pIX | Anti-Ad3 pIX | |
| Ad2 pIX | 2.11 ± 0.33 | 1.07 ± 0.13 |
| Ad2pIXC | 0.317 ± 0.027 | 0 |
| Ad2pIXN | 0.48 ± 0.13 | 0.365 ± 0.054 |
| Ad3 pIX | 0.73 ± 0.13 | 2.32 ± 0.28 |
| Ad3pIXC | 0.017 ± 0.015 | 0.61 ± 0.15 |
| Ad3pIXN | 0.064 ± 0.015 | 0.307 ± 0.062 |
The means and standard deviations of two or three independent experiments are displayed. The values of the corresponding preimmune sera, multiplied by a factor of 1.5, were subtracted from the values for the antisera.
TABLE 2.
Reactivities of anti-pIXC and anti-pIXN polyclonal rabbit sera directed against the homologous and heterologous antigens of Ad2 and Ad3, respectively
| Serum | OD for reaction witha:
|
|
|---|---|---|
| Ad2 pIX | Ad3 pIX | |
| Anti-Ad2pIXC | 0.195 ± 0.059 | 0.032 ± 0.018 |
| Anti-Ad2pIXN | 0.84 ± 0.29 | 0.092 ± 0.052 |
| Anti-Ad3pIXC | 0.071 ± 0.014 | 2.27 ± 0.10 |
| Anti-Ad3pIXN | 0.505 ± 0.075 | 1.82 ± 0.36 |
The means and standard deviations of two or three independent experiments are displayed. The values of the corresponding preimmune sera, multiplied by a factor of 1.5, were subtracted from the values for the antisera.
One of the main aims of our study was to analyze the orientation of pIX in the virion by immunoelectron microscopy by using antisera directed to the terminal halves of pIX. To get this information, it was also necessary to test the reactivities of these antisera in the ELISA. Ad3pIXC- and Ad3pIXN-specific antibodies strongly detected full-length Ad3 pIX, with OD of 2.27 and 1.82, respectively (Table 2). Ad2pIXC- and Ad2pIXN-specific antibodies reacted weakly or moderately with full-length Ad2 pIX, with OD of 0.195 and 0.84, respectively. These results indicated that more or stronger immunogenic sites are situated in the N-terminal part than in the C-terminal part of Ad2 pIX and that the reactivities of these antisera are weaker than those of anti-Ad3pIXC and -Ad3pIXN sera.
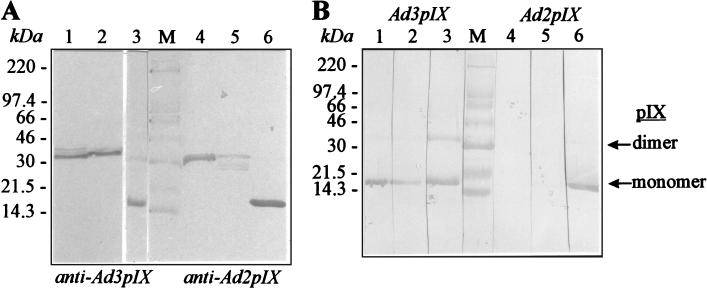
For further refinement immunoblotting analysis (2) was performed (Fig. 2A and B). The C- and N-terminal halves of Ad3 pIX were recognized by full-length Ad3 pIX-specific antibodies (Fig. 2A, lanes 1 and 2), and the C- and N-terminal halves of Ad2 pIX were recognized by full-length Ad2 pIX-specific antibodies (Fig. 2A, lanes 4 and 5). The N-terminal half of Ad2 pIX reacted weakly with Ad2 pIX-specific antibodies. Protein IX-specific antibodies bound strongly to the homologous full-length pIX (Fig. 2A and B, lanes 3 and 6). Ad2 pIX-specific antibodies reacted weakly with Ad3 pIX and Ad3pIXN, but they did not recognize Ad3pIXC (not shown). Ad3 pIX-specific antibodies bound to Ad2 pIX and very weakly to Ad2pIXN but did not recognize Ad2pIXC (not shown). Finally, the reactivities of antisera directed to the terminal parts of pIX were analyzed by using the homologous complete pIX as an antigen. Anti-Ad3pIXC and anti-Ad3pIXN sera bound to the complete Ad3 pIX (Fig. 2B, lanes 1 and 2). As a result these antisera could be used in an electron microscopy study performed for the detection of pIX on the virion. In contrast, anti-Ad2pIXC and anti-Ad2pIXN sera did not detect or only weakly detected Ad2 pIX antigen (Fig. 2B, lanes 4 and 5). All the data obtained from immunoblot analysis support the results of antigen analysis by ELISA.
FIG. 2.
Reactivities of antisera to recombinant full-length pIX and partially deleted pIX demonstrated by immunoblotting. (A) Analysis of reactivities of recombinant full-length pIX and C- and N-terminal halves of pIX with full-length pIX-specific polyclonal rabbit sera. Proteins Ad3pIXC, Ad3pIXN, and Ad3 pIX were electrotransferred to nitrocellulose after denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis and incubated with anti-Ad3 pIX as a primary antibody (lanes 1, 2 and 3); Ad2pIXC, Ad2pIXN, and Ad2 pIX were incubated with anti-Ad2 pIX (lanes 4, 5, and 6). Sera were diluted at 1:500. Horseradish peroxidase-conjugated secondary antibody was used with 3,3′-5,5′-tetramethylbenzidine (Boehringer, Mannheim, Germany) as a substrate. (B) Analysis of full-length recombinant Ad3 pIX (lanes 1 to 3) and Ad2 pIX (lanes 4 to 6). The antibodies used were as follows: lane 1, anti-Ad3pIXC; lane 2, anti-Ad3pIXN; lane 3, anti-Ad3 pIX; lane 4, anti-Ad2pIXC; lane 5, anti-Ad2pIXN; lane 6, anti-Ad2 pIX. Lane M, molecular weight markers. The positions of markers are given on the left.
Computer-based analysis of the potential antigenic sites of Ad2 and Ad3 pIX predicted that while the few antigenic determinants of Ad2 pIX are located in the N-terminal half of the protein, more antigenic sites are located in the C-terminal half of Ad3 pIX. Experimental results obtained from ELISA, immunoblot analysis, and predictions led to the conclusion that the major antigenic sites of Ad2 pIX differ from those of Ad3 pIX. One reason for the weak reactivities of anti-Ad2pIXN and -Ad2pIXC sera could be that one major epitope is located in the middle of Ad2 pIX, which was destroyed by construction of the expression vectors for Ad2pIXC and Ad2pIXN. The biological variability of the animals could be an additional factor. Our results on the cross-reactivity of antibodies showed that only the epitopes in the C-terminal part of pIX, and not those in the N-terminal half, are subgenus specific. This observation corresponds with the greater degrees of difference in the amino acid sequences of the C-terminal parts of pIX and the greater degrees of similarity in the N-terminal parts of pIX of Ad serotypes belonging to different subgenera. Further information on the amino acid residues which are involved in antigenic epitopes is required for precise epitope mapping. This could be achieved by construction of a series of mutants carrying deletions in the regions of pIX indicated in the present study, by use of monoclonal antibodies or by means of polyclonal antibodies and overlapping synthetic peptides which could be useful as antigens and for production of antibodies (26, 34).
Virus neutralization by anti-pIX sera.
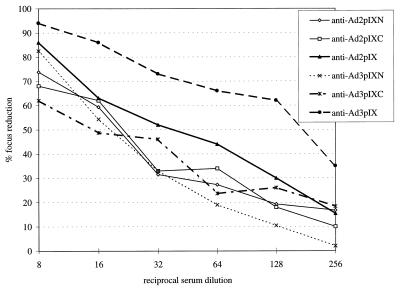
In order to find out whether Ad pIX-, AdpIXN-, and AdpIXC-specific antisera are able to prevent virus infection in vitro, neutralization tests (Fig. 3) were carried out by methods similar to those described previously (25, 29). The neutralizing effect of the antiserum was determined in relation to a virus control (titer, 50% neutralization of the virus at reciprocal serum dilution). Ad3 pIX-specific antiserum showed a neutralization titer of approximately 200 against the homologous virus. The neutralization titer of Ad2 pIX-specific antiserum was approximately 40, and this was lower than that of Ad3 pIX-specific antiserum. The neutralization titers of Ad2pIXC-, Ad2pIXN-, Ad3pIXC- and Ad3pIXN-specific sera against the homologous virus were in the range from 25 to 15. This means that the neutralizing capacities of anti-pIXN and anti-pIXC sera are very low and not significant. Cross-reactivity of the antisera was also tested (data not shown). None of the anti-Ad pIX-specific sera displayed a significant neutralizing activity against the heterologous virus.
FIG. 3.
Neutralization of the homologous Ad by polyclonal antisera directed to the full-length Ad2 and Ad3 pIX and to the terminal halves of each pIX. The result for each focus reduction assay is the average of three independent determinations of duplicate samples. Standard deviations were within the range from 5 to 20%.
Orientation of pIX within the virion.
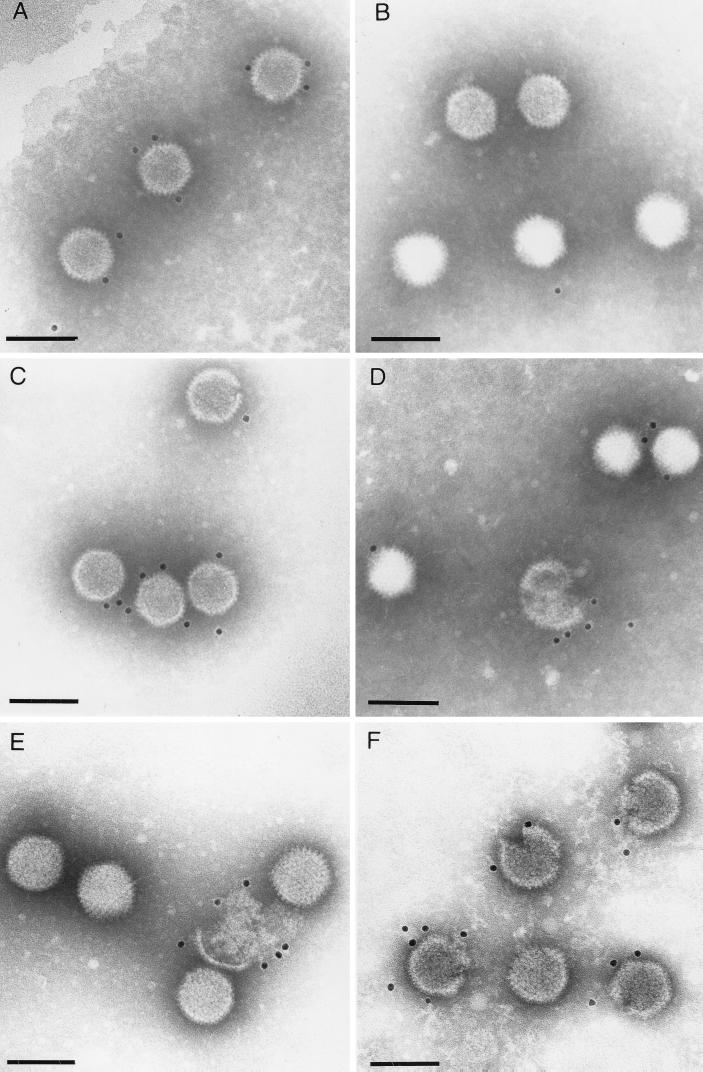
In order to examine the external locations of the antigenic regions and the orientation of pIX (in terms of the N and C termini) in virus particles, immunoelectron microscopy was performed with purified virions and pIX-specific antisera. Viruses were purified by two CsCl density gradient centrifugations as described previously (24, 25). For immunogold electron microscopy, purified virions were adsorbed to Formvar-coated, carbon-stabilized, and glow discharge-treated copper grids. After adsorption, the suspension was removed and the grids were incubated for 1 h with appropriately diluted antibodies. After several washing steps the grids were incubated with gold-tagged protein A (PAG10) or antispecies immunoglobulin G (GAR10, BioCell International, Cardiff, United Kingdom) for 1 h at room temperature. The grids were stained with 2% phosphotungstic acid (pH 7.4) and examined by transmission electron microscopy (EM 400T; Philips, Eindhoven, The Netherlands). Intact Ad3 virions (Fig. 4A) as well as disrupted and not quite intact Ad3 virus particles (Fig. 4F) were labeled by using full-length Ad3 pIX-specific antibodies and gold-tagged protein A. Whereas no reaction was detected when anti-Ad3pIXN serum was incubated with Ad3 (Fig. 4B), anti-Ad3pIXC serum reacted with the intact Ad3 virus particles (Fig. 4C). No cross-reactivity between Ad2 and Ad3 pIX antisera was observed in reciprocal immunoelectron microscopy experiments (data not shown). By using full-length Ad2 pIX-specific antibodies, the pIX could be strongly labeled with gold particles in morphologically incomplete Ad2 virus particles but reacted weakly in intact virions (Fig. 4D and E, respectively). Thus, the weak reactivity of the Ad2 pIX antiserum with the complete virus particles along with its stronger reactivity with the disintegrated and incomplete virus particles indicated that either only a few of the antigenic determinants of Ad2 pIX are accessible at the capsid surface of intact virions or the antibodies have only a weak avidity. Labeling of complete Ad2 virions by Ad2pIXC- and Ad2pIXN-specific antibodies and gold-tagged protein A was not detected (not shown).
FIG. 4.
Localization of pIX on capsid surfaces of Ad particles by immunogold electron microscopy performed by using anti-Ad3 pIX serum (A and F), anti-Ad3pIXN serum (B), anti-Ad3pIXC serum (C), and anti-Ad2 pIX serum (D and E). In all panels bars indicate 100 nm.
The C-terminal part of pIX constitutes the surface domain.
The locations of polypeptides in the Ad particles were initially determined by identification of specific morphological features observed by electron microscopy and by disruption of the virion (33, 35). Further studies utilized chemical cross-linking to provide information on the connections of polypeptides in virions (9, 17). The location of pIX within the virus capsid has been explored by various techniques, including enzymatic iodination (18) and immunological methods (6). The results of those studies were consistent in demonstrating that pIX was buried within the capsid but differed as to whether it was located on the inner or outer surface of the GON. pIX should be situated on the inner surface of the GON, because it appears to be inaccessible to antibody when positioned in the Ad2 virion (6). Antibodies against Ad2 pIX showed no reactivity with the intact virion and a low-level response to GON (8). Another study (7) showed that Ad2 pIX trimers are buried within the GON. The outer monomers are buried within the capsid edge formed by interlocking GON and would be exposed to solution only upon capsid dissociation. In the study of the GON, stoichiometric determinations of pIX (36) and a combination of electron microscopy and X-ray crystallography data (19) revealed that there are 12 copies of pIX per GON and 240 copies per virion, inserted as trimers. These studies also determined that four trimers of pIX are embedded in the large cavities in the upper surface of the GON, presumably to stabilize the GON assembly.
In the present immunoelectron microscopical study, we found a weak reactivity of anti-Ad2 pIX serum with intact Ad2 virions and a stronger reactivity of anti-Ad3 pIX serum with intact Ad3 virions. We also demonstrated that anti-Ad3 pIX serum has a moderate neutralizing activity and anti-Ad2 pIX serum has a weak neutralizing activity, indicating the presence of neutralizing epitopes on pIX. At this point, it is unknown why the Ad2 and Ad3 pIX differ with respect to surface exposure of their antigenic sites and how the anti-pIX sera neutralize the virion. By using Ad3pIXC- and Ad3pIXN-specific antisera and Ad3 virions in the immunogold electron microscopy, we could demonstrate that only the C terminus-specific antibodies bound to intact virions. From these results, we propose that the C-terminal sequences of the pIX constitute the surface domain and that the N-terminal sequences of pIX build up the internal domain of the protein in the capsid. This assignment of the orientation is also partly supported by enzymatic iodination studies of pIX (18). Enzymatic iodination of intact virions will reveal only external tyrosine residues of the proteins accessible to lactoperoxidase. There is only the unique tyrosine residue at position 13 of the N terminus of Ad2 pIX which should be located in the inside part of the intact virion according to the result of the present study. The fact that this tyrosine residue is accessible to enzymatic iodination (6) in purified pIX also supports the present finding, since in purified protein the N terminus of pIX should be partly or completely exposed in the solution. In addition, it has been proposed that pIX can be divided into N-terminal and C-terminal domains that are joined by an alanine-rich spacer, composed of 13 residues in Ad2 and 11 residues in Ad3, which is suggestive of the existence of at least two functional domains (16).
In summary, we found that only the C-terminal halves of Ad2 and Ad3 pIX reacted in a subgenus-specific manner and constitute the surface domains in icosahedral Ad capsids. Based on the present study, it will be interesting in future to study the binding properties of pIX with hexon and other structural proteins in the adenovirus virion.
Acknowledgments
This work was partially supported by Deutsche Forschungsgemeinschaft (SFB 286/YE1, Se 700/1-3) and also by MIKROGEN, Munich. A.A. is a recipient of grants from the Konrad Adenauer Stiftung.
We thank Lutz Gürtler and Robert Jack for critical reviewing of the manuscript and Maria Liebermann, Margret Wolfgramm, Millie Heitmann, and Christel Möller for excellent technical assistance.
REFERENCES
- 1.Akalu A. Antigenic characterization and posttranslational modification analysis of the protein pIX of human adenovirus serotypes 2 and 3. Thesis. Greifswald, Germany: University of Greifswald; 1997. [Google Scholar]
- 2.Akalu A, Seidel W, Liebermann H, Bauer U, Döhner L. Rapid identification of subgenera of human adenovirus by serological and PCR assays. J Virol Methods. 1998;71:187–196. doi: 10.1016/s0166-0934(97)00213-9. [DOI] [PubMed] [Google Scholar]
- 3.Aleström P, Akusjärvi G, Perricaudet M, Mathews M B, Klessig D F, Pettersson U. The gene for polypeptide IX of adenovirus type 2 and its unspliced messenger RNA. Cell. 1980;19:671–681. doi: 10.1016/s0092-8674(80)80044-4. [DOI] [PubMed] [Google Scholar]
- 4.Allard A, Wadell G. The E1B transcription map of the enteric adenovirus type 41. Virology. 1992;188:319–330. doi: 10.1016/0042-6822(92)90761-d. [DOI] [PubMed] [Google Scholar]
- 5.Bos J L, Polder L J, Bernards R, Schrier P I, van der Elsen P J, van der Eb A J, van Ormondt H. The 2.2 kb E1b mRNA of human Ad12 and Ad5 codes for two tumor antigens starting at different AUG triplets. Cell. 1981;27:121–131. doi: 10.1016/0092-8674(81)90366-4. [DOI] [PubMed] [Google Scholar]
- 6.Boulanger P, Lemay P, Blair G E, Russell W C. Characterization of adenovirus protein IX. J Gen Virol. 1979;44:783–800. doi: 10.1099/0022-1317-44-3-783. [DOI] [PubMed] [Google Scholar]
- 7.Burnett R M. The structure of the adenovirus capsid. II. The packaging symmetry of hexon and its implications for viral architecture. J Mol Biol. 1985;185:125–143. doi: 10.1016/0022-2836(85)90187-1. [DOI] [PubMed] [Google Scholar]
- 8.Cepko C L, Changelian P S, Sharp P A. Immunoprecipitation with two-dimensional pools as a hybridoma screening technique: production and characterization of monoclonal antibodies against adenovirus 2 proteins. Virology. 1981;110:385–401. doi: 10.1016/0042-6822(81)90069-6. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee P J, Vayda M E, Flint S J. Interactions among the three adenovirus core proteins. J Virol. 1985;55:379–386. doi: 10.1128/jvi.55.2.379-386.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou P Y, Fasman G D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 11.Colby W W, Shenk T. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J Virol. 1981;39:977–980. doi: 10.1128/jvi.39.3.977-980.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davison A J, Tellford E A R, Watson M S, McBride K, Mautner V. The DNA sequence of adenovirus type 40. J Mol Biol. 1993;234:1308–1316. doi: 10.1006/jmbi.1993.1687. [DOI] [PubMed] [Google Scholar]
- 13.Dayhoff M O, Barker W C, Hunt L T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- 14.Devereux J, Hereby P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dijkema R, Maat J, Dekker B M M, van Ormondt H, Boyer H W. The gene for polypeptide IX of human adenovirus type 7. Gene. 1981;13:375–385. doi: 10.1016/0378-1119(81)90017-2. [DOI] [PubMed] [Google Scholar]
- 16.Engler J A. The nucleotide sequence of the polypeptide IX gene of human adenovirus type 3. Gene. 1981;13:379–386. doi: 10.1016/0378-1119(81)90018-4. [DOI] [PubMed] [Google Scholar]
- 17.Everitt E, Sundquist B, Pettersson U, Philipson L. Structural proteins of adenoviruses. X. Isolation and topography of low molecular weight antigens from the virion of adenovirus type 2. Virology. 1973;52:130–147. doi: 10.1016/0042-6822(73)90404-2. [DOI] [PubMed] [Google Scholar]
- 18.Everitt E, Lutter L, Phillipson L. Structural proteins of adenoviruses. XII. Location and neighbor relationships among proteins of adenovirion type 2 as revealed by enzymatic iodination, immunoprecipitation and chemical cross-linking. Virology. 1975;67:197–208. doi: 10.1016/0042-6822(75)90417-1. [DOI] [PubMed] [Google Scholar]
- 19.Furcinitti P S, van Oostrum J, Burnett R M. Adenovirus polypeptide IX revealed as capsid cement by difference images from electron microscopy and crystallography. EMBO J. 1989;8:3563–3570. doi: 10.1002/j.1460-2075.1989.tb08528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh-Choudhury G, Haj-Ahmed Y, Graham F L. Protein IX, a minor component of the human adenovirus capsid, is essential for the packaging of full length genomes. EMBO J. 1987;6:1733–1739. doi: 10.1002/j.1460-2075.1987.tb02425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 22.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 23.Jameson B A, Wolf H. The antigenic index: a novel algorithm for predicting antigenic determinants. Comput Appl Biosci. 1988;4:181–186. doi: 10.1093/bioinformatics/4.1.181. [DOI] [PubMed] [Google Scholar]
- 24.Liebermann H, Mentel R. Quantification of adenovirus particles. J Virol Methods. 1994;50:281–292. doi: 10.1016/0166-0934(94)90184-8. [DOI] [PubMed] [Google Scholar]
- 25.Liebermann H, Mentel R, Döhner L, Modrow S, Seidel W. Inhibition of cell adhesion to the virus by synthetic peptides of fiber knob of human adenovirus serotypes 2 and 3 and virus neutralization by anti-peptide antibodies. Virus Res. 1996;45:111–121. doi: 10.1016/S0168-1702(96)01369-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liebermann H, Mentel R, Bauer U, Pring-Akerblom P, Dölling R, Modrow S, Seidel W. Receptor binding sites and antigenic epitopes on the fiber knob of human adenovirus serotype 3. J Virol. 1998;72:9121–9130. doi: 10.1128/jvi.72.11.9121-9130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lutz P, Rosa-Calatrava M, Kedinger C. The product of the adenovirus intermediate gene IX is a transcriptional activator. J Virol. 1997;71:5102–5109. doi: 10.1128/jvi.71.7.5102-5109.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maat J, van Bevern C P, van Ormondt H. The nucleotide sequence of Ad5 early region E1: the region between map positions 8.0 (HindIII site) and 11.8 (SmaI site) Gene. 1980;10:27–38. doi: 10.1016/0378-1119(80)90140-7. [DOI] [PubMed] [Google Scholar]
- 29.Mentel R, Matthes E, Janta-Lipinski M, Wegner U. Fluorescent focus reduction assay for the screening of anti-adenoviral agents. J Virol Methods. 1996;59:99–104. doi: 10.1016/0166-0934(96)02026-5. [DOI] [PubMed] [Google Scholar]
- 30.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequences of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 31.Pettersson U. Structural and nonstructural adenovirus proteins. In: Ginsberg H S, editor. The adenoviruses. New York, N.Y: Plenum Publishing Corp.; 1984. pp. 205–270. [Google Scholar]
- 32.Russell W C, Laver W G, Sanderson P J. Internal components of adenovirus. Nature. 1968;219:1127–1130. doi: 10.1038/2191127a0. [DOI] [PubMed] [Google Scholar]
- 33.Stewart P L, Fuller S D, Burnett R M. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 1993;12:2589–2599. doi: 10.1002/j.1460-2075.1993.tb05919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toogood C I A, Crompton J, Hay R T. Antipeptide antisera define neutralizing epitopes on the adenovirus hexon. J Gen Virol. 1992;73:1429–1435. doi: 10.1099/0022-1317-73-6-1429. [DOI] [PubMed] [Google Scholar]
- 35.Valentine R C, Pereira H G. Antigens and structure of the adenovirus. J Mol Biol. 1965;13:13–20. doi: 10.1002/rmv.375. [DOI] [PubMed] [Google Scholar]
- 36.van Oostrum J, Burnett R M. Molecular composition of the adenovirus type 2 virion. J Virol. 1985;56:439–448. doi: 10.1128/jvi.56.2.439-448.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wadell G, Doerfler W, Russell W C, Kitchingman G R. Adenoviruses. In: Webster R G, Granoff A, editors. Encyclopedia of virology. Vol. 2. London, United Kingdom: Academic Press; 1994. pp. 1–23. [Google Scholar]
- 38.Wohlfart C E G, Svensson U K, Everitt E. Interaction between HeLa cells and adenovirus type 2 virions neutralized by different antisera. J Virol. 1985;56:896–903. doi: 10.1128/jvi.56.3.896-903.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]