Abstract
Some popular methods for polymorphism and mutation discovery involve ascertainment of novel bands by the examination of electrophoretic gel images. Although existing strategies for mapping bands work well for specific applications, such as DNA sequencing, these strategies are not well suited for novel band detection. Here, we describe a general strategy for band mapping that uses background banding patterns to facilitate lane calling and size calibration. We have implemented this strategy in GelBuddy, a user-friendly Java-based program for PC and Macintosh computers, which includes several utilities to assist discovery of mutations and polymorphisms. We demonstrate the use of GelBuddy in applications based on single-base mismatch cleavage of heteroduplexed PCR products. Use of software designed to facilitate novel band detection can significantly shorten the time needed for image analysis and data entry in a high-throughput setting. Furthermore, the interactive strategy implemented in GelBuddy has been successfully applied to DNA fingerprinting applications, such as AFLP. GelBuddy promises to make electrophoretic gel analysis a viable alternative to DNA resequencing for discovery of mutations and polymorphisms.
INTRODUCTION
The present genomic era was made possible by the automated determination of nucleotide sequences and restriction digest fingerprints from electrophoretic gel images. Systems to perform these tasks were developed soon after the introduction of large-format film scanners in the 1980s (1,2) and continue to facilitate what would otherwise be a rate-limiting step in sequence determination and assembly. Now a major challenge of genomics is the discovery of single-nucleotide differences, such as naturally occurring single-nucleotide polymorphisms (SNPs) and induced point mutations (3–5). Many methods to discover such differences are in use, among which are those that compare differences in physical characteristics of DNA fragments recognizable in images produced by gel electrophoresis and other technologies. However, recognition of subtle differences between otherwise identical patterns is potentially rate-limiting for these methods.
A popular strategy for SNP and mutation discovery is to observe novel fragments produced by cleavage of a mismatch between annealed DNA strands (6). Mismatch cleavage of heteroduplexed DNA allows discovery of single-base differences in pooled samples (7). This is an especially valuable feature for discovery of rare SNPs and induced mutations, which are difficult and costly to detect by sequencing (5). Software specifically designed for mapping novel fragments found in electrophoretic gel images should greatly increase the efficiency of mismatch cleavage analysis, making such a strategy more generally applicable.
Here, we describe a software tool that is designed for mismatch cleavage detection of single-nucleotide changes. GelBuddy is an interactive system that automates the most tedious aspects of the analysis. We introduce the use of background bands to track lanes and to compensate for differences in mobility between lanes. This information is used to map novel bands identified by the user, calculate fragment lengths and identify co-migrating bands for genotype characterization. GelBuddy can generate text reports or post results over the World Wide Web, and can be used as a stand-alone tool or as an adjunct to an integrated server-based workflow management system. The strategy used by GelBuddy is generally applicable to other types of electrophoretic images, such as those produced for DNA fingerprinting applications.
ALGORITHM AND METHODS
Lane tracking algorithm
Lane tracks are generated by detecting local maxima of intensity profiles formed by vertically integrating pixel values over either a set of horizontal sectors or the entire image. These data may be used to generate either segmented or straight lane tracks. To construct segmented lane tracks, the image is divided into horizontal sectors, features corresponding to lanes are detected in each sector, features in neighboring sectors are connected to form lane tracks, and incomplete lane tracks are extended to the bottom and top of the image by interpolation. To construct straight lane tracks, the entire image is treated as a single sector, using the detected peaks to determine the x-coordinate of each lane. In either case, missing lanes are added to fill in gaps, and artifactual lanes on the left and right sides of the image are eliminated to obtain the number of lanes specified by the user.
After forming each intensity profile, a sharp cutoff low-pass filter is applied to the intensity profile and local maxima are recorded. The cutoff frequency is proportional to the number of lanes specified by the user. Because this technique does not require the determination of an intensity threshold, it is relatively insensitive to variation in signal intensity between images and between lanes in a single image. However, this technique is sensitive to noise and filtration artifacts, which result in the detection of artifactual features at the left and right sides of the image. The lane tracks resulting from these features are removed at the end of the lane-tracking process.
GelBuddy uses a simple heuristic to construct segmented lane tracks from the peaks detected in each sector. Each lane track begins as a peak in the center sector. The track is extended toward the bottom of the image by adding a peak in the next sector, only if there exists a peak, that is the mutual nearest neighbor of the current lane endpoint. Thus, peak A in sector k is connected to peak B in sector k + 1, only if B is the peak in sector k + 1 closest to A and A is the peak in sector k closest to B. The lane track is truncated if no mutual nearest neighbor exists. This process is repeated until the bottom of the image is reached and begins again at the center of the image to extend each lane track toward the top of the image. Truncated lanes are then extended one sector at a time using the positions of adjacent non-truncated lanes until the top and bottom of the image are reached.
At this point, the set of lane tracks constructed by the above procedure may contain gaps (caused by missing lanes in the source image) and artifactual lanes (caused by the detection of spurious peaks). Missing lanes are filled in by computing the mean distance between each pair of adjacent lanes and inserting new lane tracks between each pair of lanes, whose distance exceeds the overall mean lane separation by a factor of 3/4. A two-channel electropherogram is constructed for each lane by integrating intensity values within a 5-pixel wide horizontal window centered on the lane track, and the total intensity of each lane is calculated. Artifactual lanes at the left and right sides of the image are removed by repeatedly pruning either the leftmost or the rightmost lane (whichever is fainter) until the desired number of lanes is reached.
Algorithm for calibration curve construction
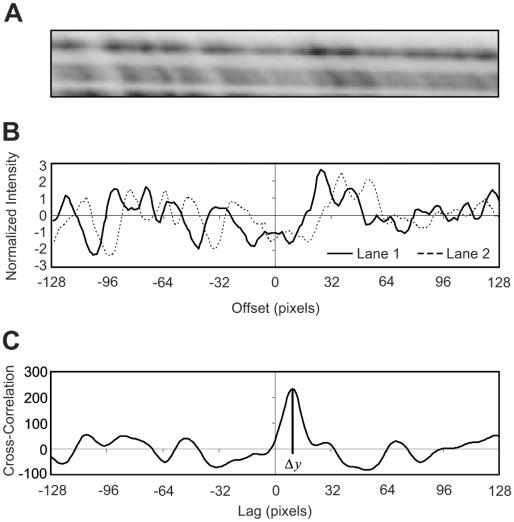
In its simplest form, the calibration algorithm constructs curves of equal fragment length by aligning signal patterns in adjacent lanes. The algorithm takes as input the previously constructed electropherogram for each lane and an initial position y0. To calculate yk+1 from yk, intensity values from a 128-pixel window centered on yk are extracted from the electropherograms for lanes k and k + 1. Each array is normalized to mean 0 and SD 1 to form the functions fk(y) and fk+1(y), and the cross-correlation function
is computed for each lag Δy. The lag at which the cross-correlation function is maximized is the amount by which the signal of lane k + 1 must be shifted to best match the signal of lane k (Figure 1).
Figure 1.
Determination of the cross-correlation function from raw image data for size calibration. The lag Δy maximizing the cross-correlation function determines the offset between lanes. (A) Raw image data, rotated 90° right. (B) Normalized intensity values for each lane. (C) Cross-correlation function. Image is from the Seattle TILLING Project.
Spurious correlation may be caused by reaction failure, detector saturation or by the presence of bands in lane k + 1 not present in lane k (or vice versa). We employ two techniques to reduce the effect of such events. First, a limit L is placed on the acceptable lag Δy. The cross-correlation is computed for each lag |Δy| ≤ L, and if the cross-correlation is greatest at either endpoint (±L), the alignment for lane k + 1 is discarded. Second, the procedure described above is repeated to compute optimal lag Δy′ (|Δy′| ≤ 2L), and cross-correlation between lanes k and k + 2. The alignment for lane k + 1 is discarded if the maximal cross-correlation between lanes k and k + 1 is less than half the maximal cross-correlation between lanes k and k + 2.
Optionally, successive points of the calibration curve may be calculated by comparing each lane with a single template lane [typically the leftmost signal lane, f0(y)] instead of the signal from the previous lane [fk(y)]. The calculation proceeds as above, using the cross-correlation function
We found that by using the leftmost lane as a template to calculate all the offsets for each calibration curve, GelBuddy was usually able to construct calibration curves accurate to within the resolution of the gel. Using a single template across the entire width of the gel allows for greater precision but is sensitive to differences between the template lane and other sample lanes of the image.
Determining fragment length of novel bands
The locations of two size standards (typically 200 and 700 bp) at the left margin of the image are provided by the user. The number of calibration curves, and the initial position y0 of each calibration curve, is application-dependent (see Results). For each calibration curve, the two size standards are used to calculate a ‘virtual’ size standard (w, y0) for the leftmost lane, using an exponential formula empirically derived for use in the Perl program squint [(7); J. Henikoff, personal communication]. The remainder of the calibration curve is used to construct size standards (w, yk) for the remaining lanes.
To compute the fragment length of each novel band, GelBuddy first locates the nearest lane and the two virtual size standards in that lane closest to the y-coordinate of the novel band. From this information, the fragment length is computed using the formula described above.
GelBuddy implementation
GelBuddy was implemented using Java v1.4.2 and the Java Advanced Imaging Library v1.1.2 (Sun Microsystems, Santa Clara, CA), and has been successfully used in a production environment on computers running Windows (Microsoft, Redmond, WA) and Macintosh OS X (Apple, Cupertino, CA) operating systems. The TiledImage and DisplayJAI classes and ‘affine’ and ‘lookup’ operators provided by the Java Advanced Imaging Library were used to implement image display, magnification and dynamic range adjustment tools comparable in performance with those implemented in commercially available image editing software. GelBuddy may be downloaded from http://www.gelbuddy.org.
RESULTS
Image acquisition and inspection
GelBuddy allows the simultaneous analysis of two images of arbitrary size, limited by the amount of available virtual memory. We have applied GelBuddy to the LI-COR DNA analysis system, which produces images of differentially end-labeled, electrophoretically separated DNA molecules (8). The LI-COR analyzer is a vertical slab-gel system, that generates two 16-bit grayscale TIFF images, typically 1632 pixels wide and 4716 pixels tall, representing signals produced by IRDye 700 and IRDye 800 fluorophores. GelBuddy can read raw TIFF images or compressed 8-bit grayscale JPEG images compiled by an image archiving system (7).
The user begins an analysis session by selecting two image files. The aspect ratio of each image is adjusted to fit the display, and the IRDye 700 image is presented in black-on-white format with lowest-mobility fragments at the top of the image (Figure 2). Toolbar buttons allow the user to choose black-on-white or white-on-black display, alternate between the two images, adjust the dynamic range of a single image or synchronously adjust the magnification and aspect ratio of both images. Analysis of single-channel data may be accomplished by loading the same image into both channels.
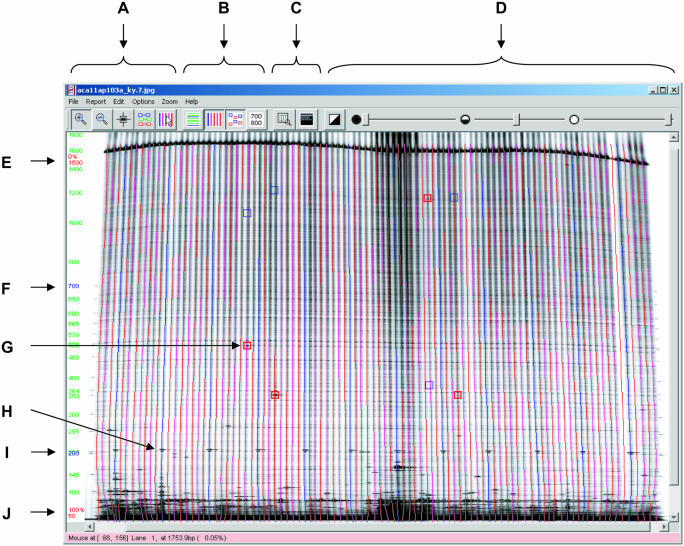
Figure 2.
Using GelBuddy to detect polymorphisms in 8-fold pooled Arabidopsis DNA. (A) Mode selection tools (zoom in, zoom out, edit signals, edit groups and edit lanes). (B) Detail selection tools (show calibration curves, show lanes, show signals and select channel). (C) Window tools (resize image to fit window and view log window). (D) Image adjustment tools (invert image, adjust black level, adjust gamma and adjust white level). (E) Adjustable upper limit marker. (F) Adjustable 700 bp calibration marker. (G) Recorded signals (foreground channel signals in red, background channel signals in blue). (H) Lane tracks. Blue lanes coincide with 200 bp marker DNA added to every eighth lane. (I) Adjustable 200 bp calibration marker. (J) Adjustable lower limit marker. Image is from the Seattle TILLING Project.
Tracking, verification and correction of lanes
Lane tracks are generated when the user clicks on the ‘Show Lanes’ button, specifies the number of lanes in the image (typically 96), whether straight or segmented lane tracks are to be constructed, and whether a single-channel image or a summed composite image is to be used. Within a few seconds, GelBuddy superimposes a set of lane tracks on the gel image (colored vertical lines in Figure 2).
Our strategy for visual identification of lane-tracking errors relies on a 200 bp DNA marker added to the sample in every eighth lane. The tracks of lanes expected to contain the marker are colored blue; other lane tracks are alternately colored red and magenta. Missing and superfluous lane tracks (the most common lane-tracking errors) are easily detected by failure of the blue lane tracks to coincide with the 200 bp markers.
Several interactive tools are provided to allow correction of lane-tracking errors. Most errors can be corrected by deleting incorrect lane tracks and interpolating new lanes from the remaining correct lane tracks. GelBuddy also allows the user to drag and drop individual lane-tracking points or entire lane tracks if necessary.
Size calibration
The user clicks on the ‘Show Calibration Information’ button to immediately display the calibration ladder (Figure 3). At the left margin of the image, two adjustable calibration points (200 and 700 bp) are displayed in blue and a 100–2000 bp calibration ladder is displayed in green. The adjustable calibration points are initialized to fixed positions relative to the top and bottom of the image, and the positions of the remaining calibration points are interpolated from the two adjustable points. The user drags the 200 and 700 bp markers to match DNA ladder bands or other image features. As the user adjusts the markers, GelBuddy rapidly recalculates and updates the calibration ladder and superimposed calibration curves. Mobility limit points may be adjusted by the user in a similar manner.
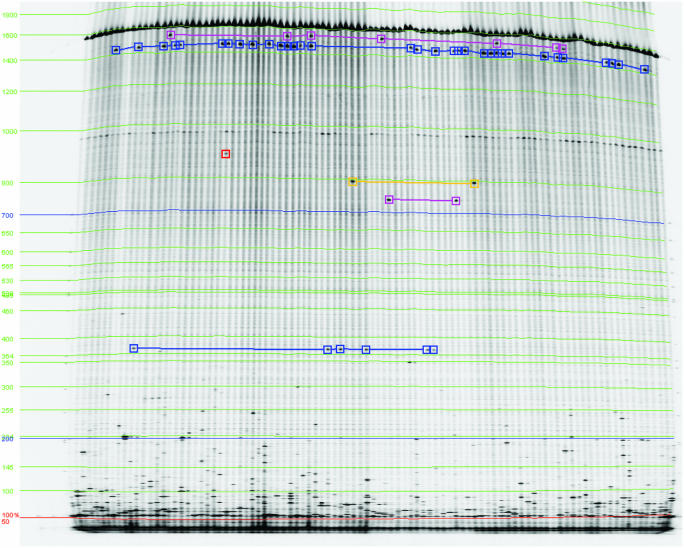
Figure 3.
Identifying co-migrating bands to determine genotypes of unpooled DNA. Calibration curves are displayed in green and blue. Five sets of co-migrating bands are indicated by connected boxes of various colors and one rare polymorphism is marked in red. Image is from the Seattle TILLING Project.
Recording and grouping novel bands
The user records visually identified novel bands corresponding to cleavage fragments by clicking on the image. GelBuddy records the channel number, lane number, and inferred fragment length of the band and groups co-migrating bands. Because correct grouping is critical for genotype characterization applications, GelBuddy provides an ‘edit groups’ mode to permit the user to override automatic signal grouping.
Reporting and posting results
GelBuddy can present results in an XML-based machine-readable format or as text. The text report contains the names of the source images, lane tracks, the list of recorded signals, a list of grouped signals (corresponding to co-migrating bands) and a list of genotypes (sets of lanes with identical co-migrating bands). The machine-readable report omits the list of genotypes but includes lane-tracking data and option settings, and may be saved locally and subsequently reloaded to resume an analysis session or posted directly to a web server for further analysis and archival storage.
Application to mutation detection for reverse genetics
We first applied GelBuddy to a reverse-genetic strategy called TILLING (7). TILLING combines genome-wide chemical mutagenesis with efficient SNP discovery to generate an allelic series of point mutations. High-throughput TILLING consists of a series of steps, beginning with PCR amplification of pooled DNAs using end-labeled primers. PCR products are heated and cooled to form heteroduplexes and novel fragments are generated by cleavage with CEL I or other mismatch-sensitive endonucleases (9). Fragments are separated by electrophoresis, allowing visualization of labeled bands that differ in mobility (Figure 2).
The generality of chemical mutagenesis makes TILLING applicable to a wide variety of organisms. However, because chemically induced mutations are relatively rare [1 in 105–106 bp (10)], thousands of individuals must be tested in order to find an allelic series of mutations for a single gene. Mutation discovery by heteroduplex mismatch cleavage of pooled samples is remarkably efficient: using 8-fold pooling, a 96-lane slab gel can detect single-base changes in a 1.5 kb region of the genomic DNA of 768 individuals, interrogating over one million base pairs of sequence in a single pass. Each mutation in the amplified region of the sample results in a novel band whose mobility indicates the position of the polymorphism. Faint signals may be visually distinguished from background bands by comparing adjacent lanes.
Sporadic mispriming products and other artifacts result in bands that may be mistaken for cleavage products. To reduce false positives, TILLING employs differentially labeled forward and reverse primers, which result in differentially labeled cleavage fragments: one fragment appears as a band in the IRDye 700 image and the other as a band in the IRDye 800 image. Analyzing both images during a single session allows the user to distinguish true signals from artifacts by flipping between the two images: the pattern of true cleavage fragments is inverted in the IRDye 800 image. GelBuddy automatically pairs the two fragments produced by mismatch cleavage and calculates the sum of their lengths in base pairs during report generation.
Once the user identifies novel bands corresponding to a mutation, these bands must be mapped to a lane number and inferred fragment length in order to determine the location of the mutation and the sample (or pool) in which the mutation was found. Prior to implementation of GelBuddy, TILLING laboratories used the Perl program squint to record manually compiled mutation data (7). To automate the data entry process, this program was adapted to accept data posted by GelBuddy, using the Perl module XML::Simple to parse GelBuddy's output (S. Kwong, personal communication). The remainder of the database system, which compiles customer orders, coordinates activity in the TILLING laboratory and records experimental results, required no modification.
Application to SNP detection
We have also used GelBuddy to characterize naturally occurring variation. EcoTilling uses the single-base mismatch discovery method developed for TILLING, but instead of being pooled, samples are combined with reference DNA from a standard accession (11). Because naturally occurring polymorphisms are more frequent than those produced by a single round of chemical mutagenesis, EcoTilling generates many more cleavage fragments: typically a hundred or more per gel (Figure 3) versus 1–10 for the mutation discovery gels described above.
Once such naturally occurring polymorphisms are found, it is desirable to group individuals by genotype and sequence one individual of each genotype. This does not require that the size of each cleavage fragment be determined precisely—indeed, in genotyping applications calculating molecular length from mobility can be avoided entirely (12). However, to identify individuals with identical genotypes, it is necessary to correctly match co-migrating bands. We have found that smiling and other gel imperfections in our gel images cause co-migrating bands to appear at somewhat different vertical positions in the gel image; as a result, automatically classifying bands by vertical position alone is unreliable. Using the pattern of background bands present in each lane to construct per-lane calibration information allows GelBuddy to accurately identify co-migrating bands, eliminating what was once a tedious and error-prone step. We have used this strategy to infer common haplotypes among different Arabidopsis accessions (11).
Several implementation details facilitate the analysis of TILLING and EcoTilling gel images. Segmented lane tracks are constructed by default, as we have found that the lanes of a TILLING image typically converge toward the top of the image, and segmented lane tracks are necessary for accurate band mapping. GelBuddy allows construction of calibration curves only at the position of the lower (200 bp) and upper (700 bp) size standards. This simplification is feasible because TILLING applications do not require highly accurate fragment length calculation and because TILLING images contain a consistent, primer-specific pattern of background bands in each lane. Calculating lane offsets using adjacent lanes instead of a single template lane allows us to analyze images in which the leftmost and rightmost lanes are poorly defined. Also, percentage mobility is reported for compatibility with squint. The range of percentage mobility is determined by two mobility limit points (‘0%’, initially set to 1500 bp and ‘100%’ initially set to 50 bp), which can be adjusted by the user by dragging red markers at the left margin of the image.
Analysis of DNA fingerprinting images
Although we developed GelBuddy for TILLING and EcoTilling applications, the basic algorithms should be applicable to lane calling and calibration of AFLP (13) and other electrophoretic DNA fingerprinting images. Because these images lack the consistent pattern of background bands present in TILLING images, constructing de-smiling curves at the positions of the lower and upper size standards usually fails. To remedy this problem, we added the ability to create several calibration curves and to manually adjust the initial position of each curve by dragging markers at the left margin of the image. By interactively adjusting the vertical position of these markers and inspecting the alignment of the superimposed calibration ladder with the image, a user can rapidly line up co-migrating bands across the gel image. In this manner, we were able to generate useful calibration curves for the 12 AFLP images that we tested. These images cover a wide range of signal strengths, lane counts and running conditions.
We also found that in 11 of the 12 images analyzed, using the ‘straight lanes’ option provided more accurate lane-tracking results than the ‘segmented lanes’ option, a result we attribute to the lack of lane curvature, lack of background bands and scarcity of signal bands near the top of these images. We also found that the exponential formula used for inferring fragment length from relative mobility often led to increasing calibration inaccuracies with increasing fragment length.
A typical example of an AFLP gel image after GelBuddy lane calling and interactive calibration is shown in Figure 4. Despite the complexity of the AFLP fingerprint pattern, where most bands are represented in different sets of lanes, GelBuddy is nevertheless able to draw curves of equal fragment length throughout the gel (Figure 4, left). A closeup view of the same gel at 1:1 pixel aspect ratio shows that calibration curves are drawn with sufficient accuracy to allow base pair resolution (Figure 4, right). Thus, the interactive features of GelBuddy can be generally applied to fingerprinting applications, even those that generate complex banding patterns.
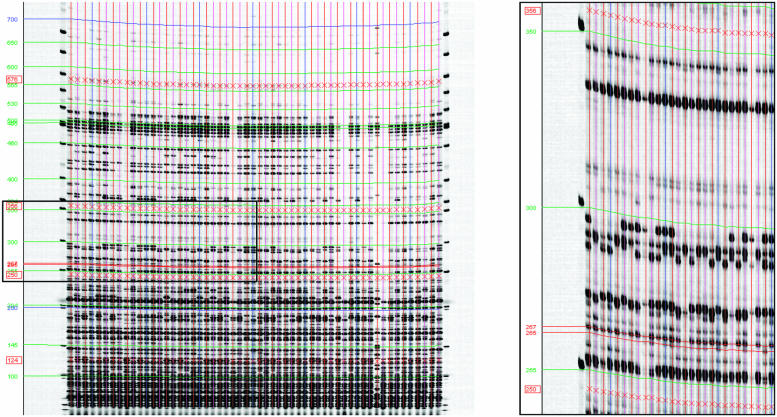
Figure 4.
GelBuddy lane calling and calibration of a typical AFLP gel image. Red ‘X’ marks indicate four calibration curves constructed by GelBuddy. Red, green and blue curves across width of gel indicate positions of equal mobility, interpolated from the calibration curves. Left: gel image of the full region of interest across all 56 lanes, where the straight lanes option was chosen. Right: an expansion of the boxed region of the left image displayed at a 1:1 pixel aspect ratio shows accurate calibration of two highly polymorphic fragments that are two bases apart in the 250–300 bp range. The AFLP LI-COR gel analyzer image of sorghum DNA was kindly provided by Patricia Klein (Texas A&M University).
DISCUSSION
We have described a strategy for the characterization of novel bands in electrophoretic images that combines automated lane detection, calibration and band mapping with visual identification of image features. By using the information present in the background bands of these images, lane tracks and calibration curves can be constructed rapidly by relatively simple algorithms, without the detection of individual background bands and without requiring the presence of dedicated marker lanes. We have also shown the applicability of this strategy to general fingerprinting applications, such as AFLP. Furthermore, we have presented a tool implementing this strategy and demonstrated its utility in the discovery of induced mutations and naturally occurring polymorphisms.
GelBuddy is a practical PC and Macintosh tool used on a daily basis in high-throughput TILLING laboratories. Further applications for the detection of mismatch cleavage products are envisioned. For example, there is a growing demand for technologies that can economically discover rare SNPs, which have been implicated in common human diseases (14). Mismatch cleavage is a highly suitable strategy for the discovery of rare heterozygous SNPs in individual and pooled DNA samples. Used as a prescreening method, it can greatly reduce the amount of resequencing that needs to be done. As mismatch cleavage strategies are further developed and applied to different gel and capillary platforms, GelBuddy can be adapted to serve as a general tool for SNP and mutation discovery.
GelBuddy takes advantage of the visual pattern recognition abilities of experienced users to identify novel bands and calibration standards. In principle, these tasks could also be automated, but the user would still need to visually review and possibly correct the results. Since only two calibration points must be adjusted and only a few signals must be recorded, little time would be saved by automated detection of these features in a TILLING image. However, if EcoTilling becomes widely used for the detection of rare polymorphisms, a fully automated gel reading tool will be desirable. The algorithms and user interface elements described in this paper provide much of the functionality such a tool will require.
Acknowledgments
The authors thank the other members of the Seattle Tilling Project (Elisabeth Bowers, Chris Burtner, Christine Codomo, Luca Comai, Jennifer Cooper, Thomas Donn, Elizabeth Greene, Aaron Holm, Samson Kwong, Robert Laport, Brad Till and Kim Young) for TILLING and EcoTILLING images and for helpful suggestions and enthusiastic adoption of GelBuddy during its earliest stages of development. The authors also thank Patricia Klein and Jeff Harford for providing AFLP gel images and Samson Kwong and Jorja Henikoff for modifying the Perl program squint to accept GelBuddy input. This work was funded by a grant from the National Science Foundation (BDI0234960). Funding to pay the Open Access publication charges for this article was provided by the National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Elder J.K., Green D.K., Southern E.M. Automatic reading of DNA sequencing gel autoradiographs using a large format digital scanner. Nucleic Acids Res. 1986;14:417–424. doi: 10.1093/nar/14.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sulston J., Mallett F., Durbin R., Horsnell T. Image analysis of restriction enzyme fingerprint autoradiograms. Comput. Appl. Biosci. 1989;5:101–106. doi: 10.1093/bioinformatics/5.2.101. [DOI] [PubMed] [Google Scholar]
- 3.Henikoff S., Comai L. Single-nucleotide mutations for plant functional genomics. Ann. Rev. Plant Biol. 2003;54:375–401. doi: 10.1146/annurev.arplant.54.031902.135009. [DOI] [PubMed] [Google Scholar]
- 4.Nagy A., Perrimon N., Sandmeyer S., Plasterk R. Tailoring the genome: the power of genetic approaches. Nature Genetics. 2003;33(Suppl.):276–284. doi: 10.1038/ng1115. [DOI] [PubMed] [Google Scholar]
- 5.Nickerson D.A., Kolker N., Taylor S.L., Rieder M.J. Sequence-based detection of single nucleotide polymorphism. Methods Mol. Biol. 2001;175:29–35. doi: 10.1385/1-59259-235-X:029. [DOI] [PubMed] [Google Scholar]
- 6.Oleykowski C.A., Bronson Mullins C.R., Godwin A.K., Yeung A.T. Mutation detection using a novel plant endonuclease. Nucleic Acids Res. 1998;26:4597–4602. doi: 10.1093/nar/26.20.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colbert T., Till B.J., Tompa R., Reynolds S., Steine M.N., Yeung A.T., McCallum C.M., Comai L., Henikoff S. High-throughput screening for induced point mutations. Plant Physiol. 2001;126:480–484. doi: 10.1104/pp.126.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Middendorf L.R., Bruce J.C., Bruce R.C., Eckles R.D., Grone D.L., Roemer S.C., Sloniker G.D., Steffens D.L., Sutter S.L., Brumbaugh J.A. Continuous on-line DNA sequencing using a versatile infrared laser scanner/electrophoresis apparatus. Electrophoresis. 1992;13:487–494. doi: 10.1002/elps.11501301103. [DOI] [PubMed] [Google Scholar]
- 9.Till B.J., Burtner C., Comai L., Henikoff S. Mismatch cleavage by single-strand specific nucleases. Nucleic Acids Res. 2004;32:2632–2641. doi: 10.1093/nar/gkh599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greene E.A., Codomo C.A., Taylor N.E., Henikoff J.G., Till B.J., Reynolds S.H., Enns L.C., Burtner C., Johnson J.E., Odden A.R., et al. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics. 2003;164:731–740. doi: 10.1093/genetics/164.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comai L., Young K., Till B.J., Reynolds S.H., Greene E.A., Codomo C.A., Enns L.C., Johnson J.E., Burtner C., Odden A.R., et al. Efficient discovery of DNA polymorphisms in natural populations by Ecotilling. Plant J. 2004;37:778–786. doi: 10.1111/j.0960-7412.2003.01999.x. [DOI] [PubMed] [Google Scholar]
- 12.Sulston J., Mallett F., Staden R., Durbin R., Horsnell T., Coulson A. Software for genome mapping by fingerprinting techniques. Comput. Appl. Biosci. 1988;4:125–132. doi: 10.1093/bioinformatics/4.1.125. [DOI] [PubMed] [Google Scholar]
- 13.Vos P., Hogers R., Bleeker M., Reijans M., van de Lee T., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M., et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen J.C., Kiss R.S., Pertsemlidis A., Marcel Y.L., McPherson R., Hobbs H.H. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]