Abstract
Human immunodeficiency virus type 1 (HIV-1) amino acid substitutions observed during antiretroviral drug therapy may be caused by drug selection, non-drug-related evolution, or sampling error introduced by the sequencing process. We analyzed HIV-1 sequences from 371 untreated patients and from 178 patients receiving a single protease inhibitor. Amino acid substitution patterns during treatment were compared with inferred substitution patterns arising evolutionarily without treatment. Our results suggest that most treatment-associated amino acid substitutions are caused by selective drug pressure, including substitutions not previously associated with drug resistance.
To assess the statistical significance of sequence changes during anti-human immunodeficiency virus (HIV) therapy, sequences should ideally also be observed in a control group of patients not receiving drug treatment who have had samples obtained at baseline and after an equal period of follow-up. Generally, such data are not available. In this paper, we introduce a novel method for assessing the statistical significance of HIV-1 protease sequence changes during therapy with a protease inhibitor. We tested the method by using large sets of published subtype B protease sequences from untreated HIV-1-infected individuals and from patients receiving therapy with a single protease inhibitor.
Patients and sequences.
The patient group consisted of patients from whom published protease sequences were available before and after treatment. There were a total of 178 patients from eight different studies. Thirty patients had received indinavir, 44 had received ritonavir, 53 had received saquinavir (hard-gel formulation [Invirase]), and 51 had received nelfinavir. The number of patients per study, the drug treatment regimens, and the sequencing methods used in each study are shown in Table 1. Control sequences included the 178 baseline pretherapy sequences and sequences from 193 other, untreated individuals (371 control sequences in total [GenBank accession numbers given at the end of the text]).
TABLE 1.
Published studies with sequences before and after treatment with a single protease inhibitor
| Drug | Reference | No. of patients | Drug dosage | Sequencing methodology | GenBank accession no. |
|---|---|---|---|---|---|
| Indinavir | Condra et al. (2) | 13 | Dose finding | Multiple molecular clones from culture supernatant | U71606–U72026 |
| Ruiz et al. (13) | 11 | 2,400 mg daily (median 24 wk) | Population-based sequencing of cDNA derived from plasma HIV-1 RNA | Not in GenBank | |
| Zhang et al. (18) | 6 | 2,400 mg daily (median 72 wk) | Multiple molecular clones from plasma HIV-1 RNA | AF024721–AF025827 | |
| Ritonavir | Molla et al. (9) | 41 | Dose finding | Population-based sequencing of cDNA derived from plasma HIV-1 RNA | Not in GenBank |
| Schmit et al. (15) | 3 | 1,200 mg daily (median 48 wk) | Population-based sequencing of cDNA derived from plasma HIV-1 RNA | AJ002495–AJ002508 | |
| Saquinavir (Invirase) | Schapiro et al. (14) | 34 | 3,600–7,200 mg daily (median 26 wk) | Population-based sequencing of cDNA derived from plasma HIV-1 RNA | AF013841–AF013908 |
| Craig et al. (3) | 19 | Dose finding | Multiple molecular clones from plasma HIV-1 RNA | Not in GenBank | |
| Nelfinavir | Patick et al. (11) | 51 | Dose finding | Population-based sequencing of cDNA derived from plasma HIV-1 RNA | Not in GenBank |
For patients having more than one posttherapy sequence, only the final sequence was used. Only sequences obtained by dideoxynucleotide terminator cycle sequencing were included. When multiple clones were sequenced, the most common residue at each position was used. When a mixture was present in a sequence obtained by direct PCR sequencing, the residue associated with the greatest electrophoretic signal was used. Each control sequence was considered to belong to HIV-1 subtype B. For 85 isolates (including 48 from patients in South America, the Middle East, Asia, and Africa), the subtype had been confirmed by env and/or gag sequencing. In the remaining 286 cases, the isolates were considered to be subtype B based on the patients’ North American or European origin, phylogenetic analyses demonstrating clustering with known subtype B protease sequences, and comparison with reference subtypes (10).
To prevent the inadvertent inclusion of more than one sequence per individual and of laboratory contaminants, the nucleic acid sequences from the control and treated patients were examined for closely related pairs of sequences (see also reference 7). Neighbor-joining trees of sequences from the treated and control patients were derived (by using the PHYLIP programs [4]) and revealed several pairs of identical sequences. Only one sequence from each of the identical sequence pairs was included in the study. GenBank accession numbers and isolate names of the excluded sequences are given below. (The 193 sequence pairs from treated patients and the 371 control sequences from untreated persons represent the curated data set.)
Classification of amino acid substitution types.
The consensus of the 371 control amino acid sequences differed from the Los Alamos HIV Sequence Database subtype B consensus sequence at one position, residue 63 (L in Los Alamos, P in this data set) (5). 63L is more commonly considered to represent the wild-type amino acid at this position and thus was also used in this study. We define five types of possible amino acid pairs derived from the alignment of two sequences (Table 2). If the consensus residue at a given position is designated as C and all others by N (nonconsensus), then an aligned pair of amino acid residues could be CC (both sequences have the consensus residue), NN (both sequences contain the same nonconsensus residue), CN (a substitution from the consensus residue to a nonconsensus residue), NC (a substitution from a nonconsensus residue to the consensus residue), or NN′ (a substitution from the nonconsensus residue N to a different nonconsensus residue, N′).
TABLE 2.
Types of amino acid pairs comprising the alignment of two HIV-1 protease sequences obtained from an individual before and after treatment with a protease inhibitor
| Type | Description | Example |
|---|---|---|
| CC | Consensus residue, unchanged during therapy | L10L |
| NN | Nonconsensus residue, unchanged during therapy | I10I |
| CN | Consensus residue→nonconsensus residue | L10I |
| NC | Nonconsensus residue→consensus residue | I10L |
| NN′ | Nonconsensus residue→different nonconsensus residue | I10V |
Classification of positions according to the distribution of substitution types.
The distribution of amino acid substitution types was determined at each of the 99 protease sequence positions for the group of patients that had received a protease inhibitor (Table 3). Based on this distribution, each position was classified according to its pattern of substitution types: (i) conserved/invariant, all residue pairs are CC; (ii) polymorphic/invariant, all residue pairs are CC or NN; (iii) conserved/mutated, all residue pairs are CC or CN; (iv) polymorphic/mutated, all residue pairs are CC, NN, NN′, or CN (substitutions only occur away from the consensus); (v) polymorphic/revertant, all residue pairs are either CC, NN, or NC (substitutions only occur toward the consensus); or (vi) polymorphic/bidirectional, CN and NC occur.
TABLE 3.
Classification of HIV-1 protease amino acid positions according to the distribution of substitution types at that position in patients receiving a protease inhibitor
| Pattern | Description |
|---|---|
| I, conserved/invariant | All residue pairs are CC |
| II, polymorphic/invariant | Residue pairs are CC or NN |
| III, conserved/mutated | Residue pairs are CC or CN |
| IV, polymorphic/mutated | Residue pairs are CC, NN, CN, or NN′ |
| V, polymorphic/revertant | Residue pairs are CC, NN, or NC |
| VI, polymorphic/bidirectional | Residue pairs include CN and NC |
In our analysis, a single reversion to consensus (NC) was sufficient for classifying a position as polymorphic/bidirectional (pattern VI). An alternative approach would be to establish flexible cutoffs for the level of NC that distinguish a polymorphic/mutated (pattern IV) position from a polymorphic/bidirectional (pattern VI) position.
Sequence variability in HIV-1 subtype B protease from untreated patients.
It is estimated that HIV-1 variants with every possible amino acid substitution are produced daily in HIV-1-infected individuals (1). However, only those protease variants with enzymatic function approximating that of wild-type virus are likely to ever become the predominant variant in the plasma of an untreated person. To exclude technical sequencing errors and cases of circulating virus containing unusual variants, we examined polymorphisms that were present in at least two isolates from untreated individuals and that were present as the predominant clone whenever multiple clones were sequenced.
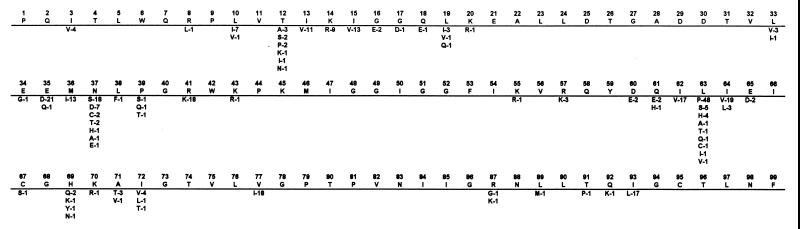
Figure 1 shows the polymorphisms present in the sequences from the 371 untreated individuals. There were 40 polymorphic positions. At 12 positions (residues 12, 13, 15, 35, 36, 37, 41, 62, 63, 64, 77, and 93), nonconsensus residues occurred with a frequency of >10%. At 15 positions, nonconsensus residues occurred with a frequency of 2 to 10%. At 13 positions, nonconsensus residues occurred in two or four sequences (0.7 to 1.4%). This distribution of polymorphisms is similar to that described in a previous publication (6).
FIG. 1.
Sequence polymorphisms present in published HIV-1 protease sequences obtained from 371 individuals not receiving treatment with a protease inhibitor. The uppermost sequence contains the consensus residue at that position among the 371 sequences and is identical to the subtype B consensus sequence (5). Amino acid residues beneath the consensus occurred in sequences from at least two individuals. The number following the residue is the percentage of sequences with that residue.
Pairwise differences between sequences of HIV-1 protease from patients not receiving protease inhibitors.
To assess the effect of naturally occurring protease sequence variation on the distribution of genetic distances between HIV-1 isolates from untreated patients, we calculated all possible pairwise nucleotide and amino acid distances between the control sequences (Fig. 2). Nucleotide distances were measured by using the Kimura two-parameter method (4). Amino acid distances were measured as the number of amino acid differences between two sequences.
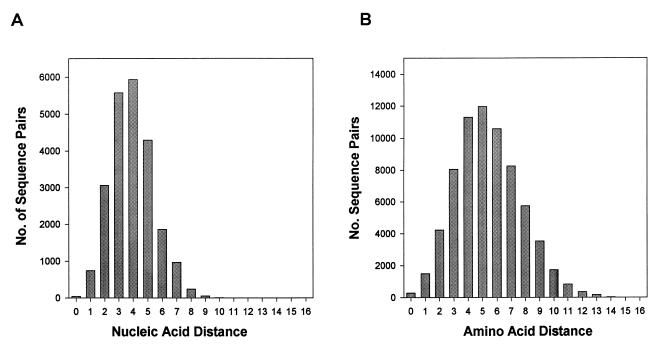
FIG. 2.
Distribution of pairwise genetic distances among the control patient sequences. (A) Distribution of uncorrected nucleic acid sequence distances among the 214 control patients from whom nucleic acid sequences were obtained. (B) Distribution of pairwise amino acid sequence distances (number of amino acid differences) among all 371 control patients.
For the 371 control patients in whom amino acid sequences were available, the mean pairwise sequence distance was 5.4 amino acids (range, 0 to 16 amino acid substitutions). For the 214 patients in whom nucleic acid sequences were available, the mean pairwise sequence distance was 4.4% (range, 0.3 to 11.3%). Uncorrected nucleic acid sequence distances of <1% were found in 46 (0.2%) sequence pairs, and sequence distances of 1 to 2% were found in 745 (3.3%) sequence pairs. Among the 46 sequence pairs with a distance of <1%, seven pairs included BRU and eight pairs included sequences determined in the same laboratory. Although the distribution of pairwise sequence differences is several-fold lower for pol than for env (7, 12, 16, 17), HIV-1 isolates with pairwise genetic distances of <1 to 2% in their protease genes should be examined for the possibility of laboratory contamination or epidemiologic linkage.
Estimation of amino acid substitution rates in HIV-1 isolates from untreated individuals.
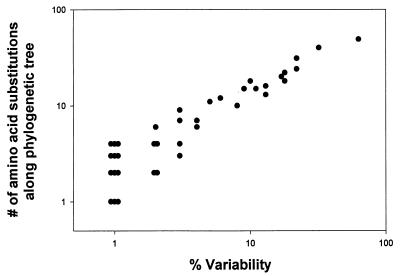
Sequence variability at a given position may indicate the tendency of this position to mutate. Alternatively, it may represent sequence changes that are ancestral but not necessarily frequent. To distinguish between these possibilities, we correlated sequence variability with inferred substitution rates at the 40 variable positions. Neighbor-joining and maximum-parsimony phylogenetic trees of the control sequences were created, and the estimated numbers of inferred amino acid substitutions along the branches of each tree were calculated (4, 8). Figure 3 shows that percent variability and inferred amino acid substitution rate were highly correlated (r2 = 0.91; P < 0.001). This analysis suggests that the sequence variability at a given position is a reliable indicator of the tendency of this position to mutate.
FIG. 3.
Relationship between percentage of variability and inferred number of amino acid substitutions arising evolutionarily among subtype B HIV-1 isolates from untreated individuals. The percentage of isolates with a nonconsensus residue is plotted on the x axis. The inferred number of amino acid substitutions was derived from a neighbor-joining tree by using the programs PHYLIP and MacClade (4, 8). For this analysis, the subtype A isolate U455 (5) was used as an outgroup. Nearly identical results were obtained if a maximum-parsimony tree was used instead of the neighbor-joining tree.
Comparison of the patterns of amino acid substitutions in HIV-1 isolates from patients receiving protease inhibitors with those obtained from untreated patients.
Table 4 contains a summary of those substitutions that occurred at least twice during protease inhibitor therapy. There were means of 5.8 amino acid sequence substitutions in the 30 sequence pairs from patients receiving indinavir, 3.8 substitutions in the 44 sequence pairs from patients receiving ritonavir, 2.7 substitutions in the 53 sequence pairs from patients receiving saquinavir, and 2.7 substitutions in the 51 sequence pairs from patients receiving nelfinavir. The distribution of sequence patterns included 40 pattern I positions (conserved/invariant), 3 pattern II positions (polymorphic/invariant), 19 positions with pattern III substitutions (conserved/mutated), 12 positions with pattern IV substitutions (polymorphic/mutated), 2 positions demonstrating only reversion to consensus (pattern V), and 23 positions with bidirectional substitutions (pattern VI, polymorphic/bidirectional).
TABLE 4.
Patterns of amino acid substitutions during treatment with a single protease inhibitora
| Residue | % Variability | Mutation(s) | No. of patient isolates with mutation (no. of patients)
|
||||
|---|---|---|---|---|---|---|---|
| Consensus→nonconsensus
| |||||||
| Treated (178) | Ritonavir (44) | Indinavir (30) | Saquinavir (Invirase) (53) | Nelfinavir (51) | |||
| Pattern III (conserved/mutated) | |||||||
| 82 | 0 | V82A/F/T/S/I | 61 | 40 | 18 | 3 | 0 |
| 54 | 0 | I54V/L | 30 | 18 | 11 | 1 | 0 |
| 46 | 0 | M46I/L | 31 | 7 | 15 | 2 | 7 |
| 90 | 0 | L90M | 27 | 1 | 8 | 16 | 2 |
| 30 | 0 | D30N | 24 | 0 | 0 | 0 | 24 |
| 88 | 0 | N88D/S/T | 12 | 0 | 1 | 1 | 10 |
| 24 | 0 | L24I | 11 | 1 | 10 | 0 | 0 |
| 48 | 0 | G48V | 10 | 0 | 2 | 8 | 0 |
| 84 | 0 | I84V | 9 | 4 | 2 | 3 | 0 |
| 73 | 0 | G73S | 9 | 0 | 5 | 3 | 1 |
| 58 | 0 | Q58E | 5 | 2 | 1 | 1 | 1 |
| 75 | 0 | V75I | 3 | 1 | 0 | 0 | 2 |
| 32 | 0 | V32I | 2 | 0 | 2 | 0 | 0 |
| 66 | 0 | I66V | 2 | 0 | 2 | 0 | 0 |
| Pattern IV (polymorphic/mutated) | |||||||
| 60 | 2 | D60E/N/Y | 9 | 1 | 0 | 5 | 3 |
| 57 | 4 | R57K | 5 | 0 | 1 | 1 | 3 |
| 34 | 1 | E34K/D/Q | 4 | 3 | 0 | 0 | 1 |
| 89 | 1 | L89M/I | 3 | 0 | 3 | 0 | 0 |
| 33 | 4 | L33F | 3 | 3 | 0 | 0 | 0 |
| Pattern VI (polymorphic/bidirectional) | |||||||
| 71 | 4 | A71V/T | 40 | 10 | 13 | 9 | 8 |
| 63 | 63 | L63P/A/H/S/Q | 32 | 10 | 6 | 7 | 9 |
| 36 | 14 | M36I/L/V | 24 | 8 | 6 | 3 | 7 |
| 10 | 10 | L10I/V/H/T/Y | 23 | 4 | 13 | 6 | 0 |
| 20 | 1 | K20R/I/M | 19 | 8 | 6 | 2 | 3 |
| 77 | 18 | V77I | 16 | 2 | 3 | 3 | 8 |
| 13 | 11 | I13V | 15 | 3 | 2 | 6 | 4 |
| 93 | 18 | I93L | 13 | 3 | 4 | 2 | 4 |
| 62 | 17 | I62V | 13 | 1 | 4 | 3 | 5 |
| 74 | 1 | T74A/S | 10 | 1 | 0 | 7 | 2 |
| 15 | 13 | I15V | 10 | 4 | 2 | 1 | 3 |
| 35 | 22 | E35D | 9 | 4 | 1 | 0 | 4 |
| 64 | 22 | I64V/M | 9 | 1 | 0 | 4 | 4 |
| 72 | 7 | I72V/L/M/T | 8 | 3 | 1 | 3 | 1 |
| 37 | 34 | N37S/C/D/E | 6 | 1 | 1 | 1 | 3 |
| 41 | 19 | R41K | 5 | 0 | 0 | 3 | 2 |
| 14 | 9 | K14R | 4 | 2 | 0 | 2 | 0 |
| 19 | 6 | L19I | 3 | 0 | 1 | 1 | 1 |
| —b | — | — | — | — | <0.0001 | <0.001 | |
| — | — | — | — | — | <0.0001 | <0.001 | |
| — | — | — | — | — | <0.0001 | <0.001 | |
| — | — | — | — | — | <0.0001 | <0.001 | |
| — | — | — | — | — | <0.0001 | <0.001 | |
| — | — | — | — | — | <0.0001 | <0.001 | |
| — | — | — | — | — | <0.0001 | <0.001 | |
| — | — | — | — | — | <0.0001 | <0.001 | |
| — | — | — | — | — | <0.0001 | <0.001 | |
| — | — | — | — | — | <0.0001 | <0.001 | |
| — | — | — | — | — | <0.0001 | <0.001 | |
| — | — | — | — | — | 0.01 | NS | |
| — | — | — | — | — | 0.07 | NS | |
| — | — | — | — | — | 0.07 | NS | |
| — | — | — | — | — | 0.0002 | 0.008 | |
| — | — | — | — | — | 0.03 | NS | |
| — | — | — | — | — | 0.001 | 0.04 | |
| — | — | — | — | — | 0.01 | NS | |
| — | — | — | — | — | 0.2 | NS | |
| 1 | 0 | 0 | 1 | 0 | 0.0001 | 0.004 | |
| 3 | 1 | 0 | 0 | 2 | 0.0005 | 0.02 | |
| 4 | 1 | 1 | 1 | 1 | 0.0001 | 0.004 | |
| 1 | 0 | 0 | 1 | 0 | 0.0001 | 0.004 | |
| 1 | 0 | 0 | 0 | 1 | 0.0001 | 0.004 | |
| 3 | 1 | 1 | 0 | 1 | 0.03 | NS | |
| 3 | 0 | 1 | 1 | 1 | 0.003 | 0.1 | |
| 2 | 0 | 0 | 1 | 1 | 0.005 | NS | |
| 1 | 1 | 0 | 0 | 0 | 0.07 | NS | |
| 1 | 0 | 0 | 1 | 0 | 0.0001 | 0.004 | |
| 1 | 0 | 0 | 1 | 0 | 0.04 | NS | |
| 1 | 0 | 1 | 0 | 0 | 0.2 | NS | |
| 9 | 2 | 4 | 3 | 0 | 0.5 | NS | |
| 1 | 0 | 1 | 0 | 0 | 0.01 | NS | |
| 4 | 2 | 1 | 1 | 0 | 0.6 | NS | |
| 2 | 0 | 1 | 1 | 0 | 0.5 | NS | |
| 3 | 0 | 1 | 1 | 1 | 0.5 | NS | |
| 1 | 0 | 0 | 1 | 0 | 0.6 | NS | |
The uncorrected P value is obtained by comparing CN − NC in the treated patients to the empirical distribution of this statistic when the permutation method is applied to the control sequences. The corrected P value is obtained by multiplying the P value by 37, since 37 positions were examined (Bonferroni correction). L5I/F, G40T, K45R, F53L/I, and I85V are pattern III mutations that each occurred in only one patient. R8Q, Q18R, K55R, E65D, H69P/Q/Y, K70R, and Q92K are pattern IV mutations that each occurred in only one patient. V50I and I98N are pattern V mutations that each occurred in only 1 patient. T12A/K/E and A/S/I12T, G16E and E16G, L38F and F38L, P39Q and T39P, and Q61H and E61Q are pattern VI mutations that each occurred in only 1 patient. NS, not significant.
—, not applicable.
A permutation method was used to assess the statistical significance of high levels of CN substitutions (away from the consensus) relative to NC substitutions (toward the consensus) at a particular position. For each of the 178 treatment sequence pairs, we determined the substitution type at each position. We then randomly picked an equal number of sequence pairs from the control group with the same distances as those observed in the treated group. Thus the random sequence pairs differed overall as much as the treatment sequence pairs, but variability occurred in how the substitutions were distributed among the substitution types.
The statistical significance of the observed treatment counts was assessed by repeating the random selection 10,000 times and deriving the empirical distribution of the aggregate counts of substitution patterns at each sequence position. The counts observed in the random pairings of control group sequences were ordered by value. The uncorrected P value is the tail probability of the count for the treatment group data relative to this empirical distribution. For example, the count of 13 CN/2 NC mutations at position 93 ranked tied 29 to 54 in the empirical distribution (28 random pairings had higher counts, 9,946 had lower counts), giving a conservative tail probability of 54/10,001 = 0.0054. To assess the significance of CN substitutions at sites with <1% variability (pattern III positions in Table 4), these positions were assumed to have a nonconsensus residue in 1 of 371 (0.0027) untreated individuals (nonconsensus residues at these positions were actually present in 0 to 1 of 371 sequences).
Of the 37 positions undergoing at least 2 substitutions during protease inhibitor therapy, 36 had an increased rate of CN substitutions relative to NC substitutions (residue 64 gives a tie [Table 4]). The statistical significance of changes at any one position is weakened by the necessary adjustment for multiple comparisons. However, the large number of CN substitutions compared with NC substitutions suggests that most treatment-associated amino acid substitutions are caused by selective drug pressure.
CN substitutions predominated at polymorphic positions as well as at highly conserved positions. Many of the CN substitutions occurred at positions previously shown to confer drug resistance by experimental methods. However, several positions with high rates of CN mutations, such as I13V, Q58E, G73S, and T74A/S (and possibly V75I), have not been previously associated with drug resistance. Given enough sequence data from patients receiving protease inhibitors, the statistical association between mutations such as these and protease inhibitor treatment should be reexamined.
Our analysis included protease sequences obtained in several different studies, including dose-finding studies in which the drugs were often used at suboptimal dosages. Therefore, we made no attempt to compare the sequence changes observed with different protease inhibitors. Studies in which sufficient numbers of sequenced protease isolates are obtained from patients receiving optimal protease inhibitor therapy are needed to compare sequence changes associated with different treatments.
Nucleotide sequence accession number.
The accession numbers of the baseline and follow-up sequences from patients receiving protease inhibitor therapy are given in Table 1. Accession numbers for the HIV-1 protease sequences from 195 additional untreated patients include the following: D10112, K02007, K02013, L02317, L08463 to L08464, M17449, M17451, M26727, M38429, M93258, M96155, U12738, U12745 to U12756, U19411 to U19415, U19417 to U19431, U19436, U19441, U19446 to U19449, U19457 to U19458, U21122, U21135, U26546, U31385 to U31395, U31399 to U31406, U31408, U31412, U34603, U37270, U43096, U43141, AF004394, AF005495, AF009369 to AF009375, AF009379 to AF009381, AF025722, AF025724, AF025726, AF025731, AF025732, AF025734, AF025736, AF025744, AF027708, AF027710, AF027715 to AF027716, AF027718, AF027720, AF040579, AF040584, AF040591, AF040596, AF040603, AF040608, AF040611, AF042100 to AF042101, AF042103 to AF042105, AF047306, AF047317, AF013857, AF078556 to AF078606, and AJ006287. Accession numbers of sequences excluded from analysis include AJ002496, AJ002497, AJ002499, AJ002500, U19452, U31409, AF025738, AF042100, AF078577, AF078578, and AF078669. Alignments of the sequences analyzed in this paper can be accessed at the web address in reference 16.
Acknowledgments
Robert Shafer was supported in part by NIH grant AI27666.
We thank Dale Kempf and Akhter Molla for providing the complete amino acid sequences of the isolates published in reference 9. We also thank David Katzenstein for critical review of the manuscript.
REFERENCES
- 1.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 2.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Craig C, Race E, Sheldon J, Whittaker L, Gilbert S, Moffatt A, Rose J, Dissanayeke S, Chirn G W, Duncan I B, Cammack N. HIV protease genotype and viral sensitivity to HIV protease inhibitors following saquinavir therapy. AIDS. 1998;12:1611–1618. doi: 10.1097/00002030-199813000-00007. [DOI] [PubMed] [Google Scholar]
- 4.Felsenstein J. PHYLIP (Phylogenetic Inference Package). Version 3.5. Seattle: University of Washington; 1993. [Google Scholar]
- 5.Korber B T, Hahn B, Foley B, Mellors J W, Leitner T, Myers G, McCutchan F E, Kuiken C L. Human retroviruses and AIDS: a compilation and analysis of nucleic and amino acid sequences. Los Alamos, N. Mex: Theoretical Biology and Biophysics, Los Alamos National Laboratory; 1997. [Google Scholar]
- 6.Kozal M J, Shah N, Shen N P, Yang R, Fucini R, Merigan T C, Richman D D, Morris D, Hubbell E R, Chee M, Gingeras T R. Extensive polymorphisms observed in HIV-1 clade-B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 7.Learn G H, Jr, Korber B T M, Foley B, Hahn B H, Wolinsky S M, Mullins J I. Maintaining the integrity of human immunodeficiency virus sequence databases. J Virol. 1997;70:5720–5730. doi: 10.1128/jvi.70.8.5720-5730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maddison W P, Maddison D R. MacClade, version 3.01. Sunderland, Mass: Sinauer and Associates; 1992. [Google Scholar]
- 9.Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper P J, Mo H M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G R, Ho D D, Boucher C A, Leonard J M, Norbeck D W, Kempf D J. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 10.National Center for Biotechnology Information. 1998. HIV-1 Subtyping Tool [Online.] www.ncbi.nlm.nih.gov/retroviruses/HIV1. [6 May 1999, last date accessed.]
- 11.Patick A K, Duran M, Cao Y, Shugarts D, Keller M R, Mazabel E, Knowles M, Chapman S, Kuritzkes D R, Markowitz M. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob Agents Chemother. 1998;42:2637–2644. doi: 10.1128/aac.42.10.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quinones-Mateu M E, Holguin A, Dopazo J, Najera I, Domingo E. Point mutation frequencies in the pol gene of human immunodeficiency virus type 1 are two- to threefold lower than those of env. AIDS Res Hum Retroviruses. 1996;12:1117–1128. doi: 10.1089/aid.1996.12.1117. [DOI] [PubMed] [Google Scholar]
- 13.Ruiz L, Nijhuis M, Boucher C, Puig T, Bonjoch A, Martinez-Picado J, Marfil S, de Jong D, Burger D, Arno A, Balague M, Clotet B. Efficacy of adding indinavir to previous reverse transcriptase nucleoside analogues in relation to genotypic and phenotypic resistance development in advanced HIV-1-infected patients. J Acquired Immune Defic Syndr Hum Retrovirol. 1998;19:19–28. doi: 10.1097/00042560-199809010-00003. [DOI] [PubMed] [Google Scholar]
- 14.Schapiro J M, Winters M A, Stewart F, Efron B, Norris J, Kozal M J, Merigan T C. The effect of high-dose saquinavir on viral load and CD4+ T-cell counts in HIV-infected patients. Ann Intern Med. 1996;124:1039–1050. doi: 10.7326/0003-4819-124-12-199606150-00003. [DOI] [PubMed] [Google Scholar]
- 15.Schmit J-C, Ruiz L, Clotet B, Raventos A, Tor J, Leonard J, Desmyter J, De Clercq E, Vandamme A M. Resistance related mutations in the HIV-1 protease gene of patients treated for 1 year with the protease inhibitor ritonavir (ABT-538) AIDS. 1996;10:995–999. doi: 10.1097/00002030-199610090-00010. [DOI] [PubMed] [Google Scholar]
- 16.Shafer R W, Stevenson D, Chan B. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 1999;27:348–352. doi: 10.1093/nar/27.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shafer R W, Chuang T K, Hsu P, Bodley White C, Katzenstein D A. Sequence and drug susceptibility of subtype C protease from human immunodeficiency virus type 1 seroconverters in Zimbabwe. AIDS Res Hum Retroviruses. 1999;15:65–69. doi: 10.1089/088922299311727. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y-M, Imamichi H, Imamichi T, Lane H C, Falloon J, Vasudevachari M B, Salzman N P. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]