Abstract
The herpes simplex virus protein VP22 is a major phosphoprotein of infected cells. In this study, we identify two serine phosphorylation sites within VP22 and show that the N-terminal site is a substrate for casein kinase II, while the extreme C-terminal site is a substrate for another, as yet unidentified, cellular kinase. Furthermore, we show that a mutant of VP22 which has both sites altered is unable to incorporate phosphate in vivo, confirming that there are no other phosphorylation sites within VP22.
The herpes simplex virus type 1 structural protein VP22 is a major component of the virus tegument (6, 7, 10) and is highly phosphorylated during infection (5, 7). While the role of VP22 phosphorylation is yet to be defined, we have previously shown that there are at least two different forms of VP22 present within the infected cell, as judged by differential migration on sodium dodecyl sulfate (SDS)-polyacrylamide gels (4). The slower-migrating form of the protein (221) represents a heavily phosphorylated species of VP22, while the faster-migrating form of VP22 (222) represents the nonphosphorylated form of the protein and is present in infected cells in amounts approximately equivalent to those of 221 (4). Interestingly, it is the nonphosphorylated form of VP22 which is specifically incorporated into assembling virions, suggesting that the mechanism(s) involved in tegument and hence virus assembly in some way differentiates between these two species of VP22. Thus, the status of VP22 phosphorylation may determine the ultimate fate of the protein during infection.
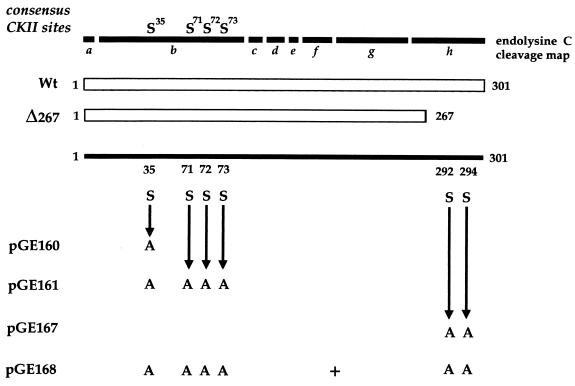
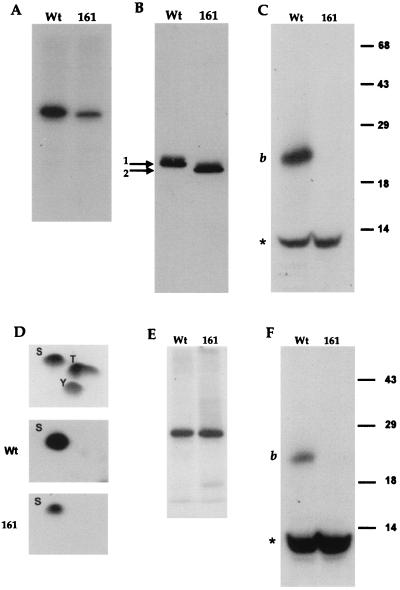
When VP22 is expressed by transient transfection, it is phosphorylated in the same manner as infected-cell VP22 (4), suggesting that the VP22 kinase(s) is of cellular origin. Previously, we have demonstrated that the majority of phosphate present on VP22 is located on serine residues within the N-terminal region of the 38-kDa protein and, more specifically, on a 20-kDa peptide produced by endolysine C cleavage of VP22 (4) (Fig. 1, peptide b). We have also shown that the cellular kinase casein kinase II (CKII) phosphorylates this region of VP22 in an in vitro phosphorylation assay. Within the N-terminal peptide b, there are four serine consensus CKII phosphorylation sites, one at residue 35 (S35) and a cluster of three at residues 71, 72, and 73 (S71, 72, 73) (Fig. 1). To determine whether these sites are utilized by CKII, we constructed two mutants of VP22 in the background of plasmid UL49ep (8), in which either S35 or S35 and S71, 72, 73 were changed to alanine residues by PCR mutagenesis, resulting in plasmids pGE160 and pGE161, respectively (Fig. 1). Plasmids expressing wild-type (WT) and mutant VP22 proteins were transfected into COS-1 cells, and high-salt extracts were prepared 40 h after transfection, as described previously (4). The proteins were immunoprecipitated with a polyclonal anti-VP22 antibody, AGV30 (2), and after extensive washing of the protein A Sepharose beads, 150 U of CKII (New England BioLabs) was added directly to the beads containing bound VP22 in the presence of [γ-32P]ATP (10 μCi), as described previously (4). Analysis of the resulting phosphoproteins by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) showed that while there was no difference between the levels of phosphate incorporated into WT and pGE160 proteins (data not shown), there was a fourfold drop in the level of 32P incorporated into pGE161 compared to that in WT protein (Fig. 2A). Both proteins were expressed at similar levels, as judged by Western blotting of the transfected-cell extracts (Fig. 2B). Moreover, while WT and pGE160 proteins migrated to the same position on SDS-polyacrylamide gels (data not shown), the pGE161 mutant of VP22 migrated faster than WT VP22 (Fig. 1B).
FIG. 1.
Phosphorylation mutants of VP22. At the top is the endolysine C cleavage map of the 301-amino-acid VP22 open reading frame (ORF), with consensus serine CKII sites denoted. Each of the cleavage products is labelled a to h. The regions of the ORF expressed from plasmids pUL49ep (Wt) and Δ267 and the serine (S)-to-alanine (A) substitutions present in plasmids pGE160, pGE161, pGE167, and pGE168 are shown. These substitutions were introduced by PCR mutagenesis into the parental plasmid pUL49ep.
FIG. 2.
Identification of two phosphorylation sites within VP22. Plasmids expressing WT or pGE161 (161) proteins were transfected into COS-1 cells, and high-salt extracts were made. (A) WT and pGE161 protein-containing extracts were immunoprecipitated with antibody AGV30, and in vitro phosphorylation was performed with CKII. The radiolabelled samples were analyzed by SDS-PAGE on a 10% acrylamide gel. (B) The same extracts used in panel A were analyzed by Western blotting using antibody AGV30. To observe the shift in mobility of pGE161, a 10-cm-long gel was utilized. (C) The radiolabelled samples from panel A were transferred to nitrocellulose, excised, and cleaved with endolysine C. The cleaved samples were analyzed by SDS-PAGE on a 15% acrylamide gel. The positions of cleavage peptide b and a 10-kDa radiolabelled peptide (∗) are shown. The positions of molecular mass markers (in kilodaltons) are indicated to the right of the gel. (D) The radiolabelled samples from panel A were transferred to a polyvinylidene difluoride membrane, subjected to acid hydrolysis, and analyzed by two-dimensional chromatography. The relative migration of cold markers is shown in the top panel. S, serine; T, threonine; Y, tyrosine. (E) Extracts containing WT and pGE161 proteins were treated as in panel A but in the absence of added CKII. (F) Radiolabelled samples from panel E were treated with endolysine C as described above for panel C.
To further analyze the CKII in vitro-labelled VP22, we subjected both the WT and pGE161 proteins to cleavage with the protease endolysine C. Cleavage of the WT protein resulted in the 20-kDa labelled peptide observed previously (4) (Fig. 2C, band b). Strikingly, the cleavage profile of the pGE161 protein demonstrated that the 20-kDa peptide was no longer phosphorylated in this mutant of VP22 (Fig. 2C, lane 161), confirming that we had correctly identified and mutated the phosphorylation site within the N-terminal 20-kDa peptide. Moreover, in both the WT and pGE161 proteins, there was an additional 10-kDa peptide which was efficiently labelled in both proteins (Fig. 2C, bands labelled with the asterisk). Phosphoamino acid analysis was also done on the same labelled proteins, as described previously (4), and demonstrated that both WT and pGE161 proteins were labelled solely on the serine residues (Fig. 2D), implying that the second 10-kDa phosphopeptide also contained phosphorylated serines. To determine whether this additional site was also a substrate for exogenous CKII, both WT and pGE161 extracts were immunoprecipitated as described above, and [γ-32P]ATP was added to the protein A Sepharose beads, but in this case without the addition of CKII. Interestingly, both WT and pGE161 proteins were phosphorylated in the absence of CKII, but in this assay the efficiency of incorporation was equal for the two proteins (Fig. 2E). Endolysine C cleavage demonstrated that the 10-kDa peptide was the major phosphopeptide generated in this assay and was present in both proteins (Fig. 2F). Thus, these results indicate that VP22 coprecipitates a cellular kinase, which we have termed VAK (VP22-associated kinase) and which phosphorylates VP22 within the 10-kDa peptide. Furthermore, it appears that the in vitro phosphorylation assays of immunoprecipitated VP22, done in the presence of exogenous CKII measured both CKII activity on the 20-kDa peptide and VAK activity on the 10-kDa peptide.
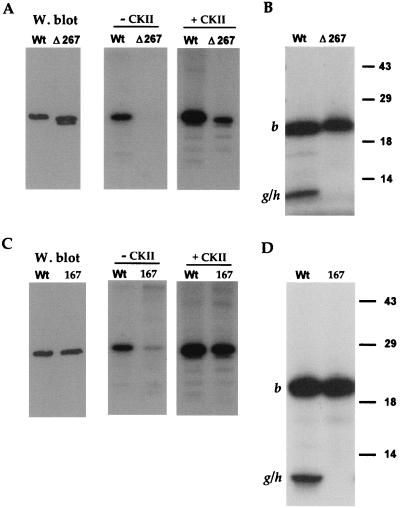
Analysis of the endolysine C cleavage map of VP22 reveals that there are no potential candidate peptides for this second 10-kDa phosphopeptide (Fig. 1). However, the C terminus of VP22 consists of two colinear 5.6-kDa peptides (Fig. 1, fragments g and h), which could conceivably account for the 10-kDa peptide if the lysine between fragments g and h were not accessible to the protease. To determine whether the additional phosphorylation site was present on either of these two peptides, we first made use of a previously constructed VP22 mutant called Δ267 (4). The Δ267 mutant lacks the C-terminal 34 residues which contain four serines at residues 277, 280, 292, and 294 (Fig. 1). COS-1 cells were transfected with WT and Δ267 protein-expressing plasmids, high-salt extracts were prepared as described above, and Western blotting was performed to determine the relative expression levels of these two proteins (Fig. 3A). These extracts were then immunoprecipitated in duplicate with antibody AGV30, and in vitro kinase assays were performed either in the presence or absence of added CKII. Both WT and Δ267 proteins were labelled in the presence of CKII, although Δ267 protein was labelled fivefold less efficiently than WT protein (Fig. 3A). Strikingly, while WT protein was labelled in the absence of CKII (Fig. 3A), as observed before, there was no detectable phosphorylation of Δ267, indicating that either it did not interact with VAK or it did not function as a substrate (Fig. 3A). Consistent with this interpretation, endolysine C cleavage of the CKII labelled proteins revealed that while the 20-kDa peptide was present in both the WT and Δ267 proteins (Fig. 3B, band b), the 10-kDa phosphopeptide was absent from the Δ267 profile (Fig. 3B). While it is possible that the g-h fusion peptide of Δ267 was not detectable on this gel because of its reduced size, the simplest interpretation of these results is that the smaller phosphopeptide from the WT protein represents phosphorylation within the extreme C terminus of VP22, that is, in peptide h (Fig. 1, endolysine C cleavage map).
FIG. 3.
VP22 possesses a C-terminal phosphorylation site. (A) Extracts were made from COS-1 cells transfected with plasmids expressing either WT or Δ267 protein. These extracts were analyzed by Western blotting using antibody AGV30 (W. blot) and by immunoprecipitation with AGV30 followed by in vitro phosphorylation in the absence (− CKII) or presence (+ CKII) of CKII. The doublet in lane Δ267 of the Western blot represents full-length Δ267 (upper band) and a slightly smaller breakdown product (lower band). (B) The samples from the in vitro CKII phosphorylation in panel A were transferred to nitrocellulose and subjected to cleavage with endolysine C. The resulting peptides were analyzed by SDS-PAGE on a 15% acrylamide gel. The positions of cleavage peptide b and the partial cleavage product encompassing cleavage peptides g and h (g/h) are indicated. (C) Extracts were made from COS-1 cells transfected with plasmids expressing either WT or pGE167 (167) protein. These extracts were analyzed by Western blotting using antibody AGV30 (W. blot) and by immunoprecipitation with AGV30 followed by in vitro phosphorylation in the absence (− CKII) or presence (+ CKII) of CKII. (D) The samples from the in vitro CKII phosphorylation in panel A were transferred to nitrocellulose and subjected to cleavage with endolysine C. The resulting peptides were analyzed by SDS-PAGE on a 15% acrylamide gel. The positions of cleavage peptide b and the partial cleavage product encompassing cleavage peptides g and h (g/h) are indicated.
In an attempt to identify the phosphorylated serine(s) within the h peptide more precisely, we initially constructed a VP22 mutant in which serine residues 292 and 294 were changed to alanine residues, resulting in plasmid pGE167 (Fig. 1). Both WT and pGE167 plasmids were transfected into COS-1 cells, extracts were prepared as described above, and the relative expression of the two proteins was assessed by Western blotting (Fig. 3C). After immunoprecipitation, in vitro kinase assays in the presence or absence of exogenous CKII were performed. Incubation with CKII resulted in phosphorylation of the mutated VP22 which was less than twofold reduced in comparison to WT (Fig. 3C, + CKII gel). However, in the absence of added CKII, phosphorylation of the pGE167 protein was 10-fold lower than that observed for WT protein (Fig. 3C). Endolysine C cleavage of the two CKII phosphorylated proteins revealed that while the 20-kDa phosphopeptide was present in both proteins (Fig. 3D, band b), the 10-kDa phosphopeptide was absent from the profile obtained for pGE167 (Fig. 3D). Therefore, mutation of the two extreme C-terminal serine residues resulted in the total loss of C-terminal phosphorylation on VP22.
To ensure that we had now identified all the potential phosphorylation sites of VP22, we constructed a mutant of VP22 containing both the CKII mutation and the C-terminal mutation (pGE168 [Fig. 1]). Plasmids expressing WT and pGE168 proteins were transfected into COS-1 cells, and extracts were prepared as described above. Western blotting of these extracts demonstrated that both forms of VP22 were expressed at approximately similar levels (Fig. 4A). To compare the relative efficiency of phosphorylation of these proteins, the same extracts were immunoprecipitated with antibody AGV30 and phosphorylated in vitro in either the presence or absence of CKII. This demonstrated that while WT protein was efficiently phosphorylated due to CKII and/or VAK (Fig. 4A, − CKII and + CKII gels, Wt lanes) incorporation of phosphate into the double mutant was entirely abolished in both assays (Fig. 4A, − CKII and + CKII gels).
FIG. 4.
A VP22 mutant with both the N- and C-terminal sites altered is phosphorylation negative. (A) Extracts were made from COS-1 cells transfected with plasmids expressing either WT or pGE168 protein. These extracts were analyzed by Western blotting using antibody AGV30 (W. blot) and by immunoprecipitation with AGV30 followed by in vitro phosphorylation in the absence (− CKII) or presence (+ CKII) of CKII. (B) COS-1 cells transfected with plasmids expressing either WT (Wt) or pGE168 (168) proteins or mock transfected (M) were labelled in vivo with [32P]orthophosphate. Extracts were made and analyzed by Western blotting with AGV30 (W. blot), and immunoprecipitation with AGV30 was followed by SDS-PAGE and autoradiography. The positions of radiolabelled protein which coprecipitates with both WT and pGE168 proteins (●) and heavily radiolabelled material at the top of the gel which coprecipitates with both WT and pGE168 proteins (◂) are indicated.
Last, we analyzed the in vivo phosphorylation levels of WT and pGE168 proteins. COS-1 cells, which had been mock transfected or transfected with either WT or pGE168 plasmid 24 h previously, were incubated in the presence of [32P]orthophosphate (50 μCi/ml) for 16 h. High-salt extracts were prepared as described above and used in either Western blotting (Fig. 4B) or immunoprecipitation with antibody AGV30 (Fig. 4B, In vivo 32P). While phosphate was efficiently incorporated into the WT VP22 protein (Fig. 4B, In vivo 32P), as we have observed previously (4), there was no detectable phosphate present in the pGE168 protein (Fig. 4B, In vivo 32P), in spite of the protein being expressed at the same level as WT (Fig. 4B, W. blot). Thus, the pGE168 mutant of VP22 is constitutively expressed as the 222 form of the protein (i.e., the nonphosphorylated form), confirming that we have identified all potential VP22 phosphorylation sites.
The results presented here extend our previous observations on the phosphorylation of VP22 (4), where we demonstrated that CKII utilized the N terminus of VP22 as a substrate. We have now located the CKII phosphorylation site to a cluster of three serines present at residues 71, 72, and 73 of VP22 and have shown that it is the phosphorylation of this site which causes the mobility shift from the 222 form of the protein to the 221 form. Moreover, an additional mutant of VP22 which has the cluster of three serines (residues 71, 72, and 73) mutated but still has serine at residue 35 behaves identically to the protein expressed by pGE161 (data not shown), confirming that Ser35 is not used as a phosphorylation site in VP22. In addition, we have located a C-terminal phosphorylation site which is not utilized by CKII but by an alternative cellular kinase and which has little or no effect on the mobility of VP22. While we have shown that both these sites are substrates for cellular kinases, we cannot rule out the possibility that they may also be substrates for viral kinases during infection.
The phosphorylation and/or dephosphorylation of VP22 may control its function by directing the protein to specific subcellular compartments or by enabling interactions with individual cellular or viral components. Interestingly, it has recently been suggested that phosphorylation may be involved in the dissociation of VP22 from the tegument upon virus entry into the cell (9). Moreover, phosphorylation of VP22 may play a role in one or more of the properties we have previously described for VP22, such as its interaction with VP16 (1), its ability to spread between cells (2), or its capacity to reorganize microtubules (3). Further studies on the mutants we describe here may help determine the importance of VP22 phosphorylation throughout the virus replication cycle and, more specifically, its role in virus maturation.
Acknowledgments
We thank John McLauchlan for plasmids pUL49ep and Δ267.
This work was funded by Marie Curie Cancer Care.
REFERENCES
- 1.Elliott G, Mouzakitis G, O’Hare P. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J Virol. 1995;69:7932–7941. doi: 10.1128/jvi.69.12.7932-7941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elliott G, O’Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 3.Elliott G, O’Hare P. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J Virol. 1998;72:6448–6455. doi: 10.1128/jvi.72.8.6448-6455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elliott G, O’Reilly D, O’Hare P. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology. 1996;226:140–145. doi: 10.1006/viro.1996.0638. [DOI] [PubMed] [Google Scholar]
- 5.Elliott G D, Meredith D M. The herpes simplex virus type 1 tegument protein VP22 is encoded by gene UL49. J Gen Virol. 1992;73:723–726. doi: 10.1099/0022-1317-73-3-723. [DOI] [PubMed] [Google Scholar]
- 6.Gibson W, Roizman B. Proteins specified by herpes simplex virus. X. Staining and radiolabeling properties of B capsid and virion proteins in polyacrylamide gels. J Virol. 1974;13:155–165. doi: 10.1128/jvi.13.1.155-165.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knopf K W, Kaerner H C. Virus-specific basic phosphoproteins associated with herpes simplex virus type 1 (HSV-1) particles and the chromatin of HSV-1 infected cells. J Gen Virol. 1980;46:405–414. doi: 10.1099/0022-1317-46-2-405. [DOI] [PubMed] [Google Scholar]
- 8.Leslie J, Rixon F J, McLauchlan J. Overexpression of the herpes simplex virus type 1 tegument protein VP22 increases its incorporation into virus particles. Virology. 1996;220:60–68. doi: 10.1006/viro.1996.0286. [DOI] [PubMed] [Google Scholar]
- 9.Morrison E E, Wang Y, Meredith D. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J Virol. 1998;72:7108–7114. doi: 10.1128/jvi.72.9.7108-7114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spear P G, Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972;9:143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]