Abstract
Background
Recent studies have linked alterations in the gut microbiome and metabolic disruptions to the invasive behavior and metastasis of colorectal cancer (CRC), thus affecting patient prognosis. However, the specific relationship among gut microbiome, metabolite profiles, and mutated-RAS/BRAF metastatic colorectal cancer (M-mCRC) remains unclear. Furthermore, the potential mechanisms and prognostic implications of metabolic changes induced by gut microbiome alterations in patients with M-mCRC still need to be better understood.
Methods
We conducted Mendelian randomization (MR) to evaluate the causal relationship of genetically predicted 196 gut microbiome features and 1400 plasma metabolites/metabolite ratios on M-mCRC-specific survival. Additionally, we identified significant gut microbiome-metabolites/metabolite ratio associations based on M-mCRC. Metabolite information was annotated, and functional annotation and pathway enrichment analyses were performed on shared proteins corresponding to significant metabolite ratios, aiming to reveal potential mechanisms by which gut microbiome influences M-mCRC prognosis via modulation of human metabolism.
Results
We identified 11 gut microbiome features and 49 known metabolites/metabolite ratios correlated with M-mCRC-specific survival. Furthermore, we identified 17 gut microbiome-metabolite/metabolite ratio associations specific to M-mCRC, involving eight lipid metabolites and three bilirubin degradation products. The shared proteins corresponding to significant metabolite ratios were predominantly localized within the integral component of the membrane and exhibited enzymatic activities such as glucuronosyltransferase and UDP-glucuronosyltransferase, crucial in processes such as glucuronidation, bile secretion, and lipid metabolism. Moreover, these proteins were significantly enriched in pathways related to ascorbate and aldarate metabolism, pentose and glucuronate interconversions, steroid hormone biosynthesis, and bile secretion.
Conclusion
Our study offers novel insights into the potential mechanisms underlying the impact of the gut microbiome on the prognosis of M-mCRC. These findings serve as a meaningful reference for exploring potential therapeutic targets and strategies in the future.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00384-024-04686-9.
Keywords: Mendelian randomization(MR), Gut microbiome, Metabolites, Mutated-RAS/BRAF metastatic colorectal cancer(M-mCRC), Survival, Causal relationship
Introduction
Colorectal cancer (CRC) is a prevalent malignancy of the digestive system and ranks as the second leading cause of cancer-related deaths [1]. An estimated 50 to 60% of cases manifest distant metastasis upon diagnosis, resulting in a poor prognosis with a 5-year survival rate ranging from merely 10 to 14% [2]. Activating missense mutations in the RAS/BRAF gene occur in about 40% of CRC patients, leading to dysregulated activation of the mitogen-activated protein kinase (MAPK) signaling pathway. This prevents patients with metastatic CRC (mCRC) from benefiting from epidermal growth factor receptor (EGFR) inhibitors such as cetuximab. Studies have demonstrated that mCRC patients with mutant RAS exhibit a worse prognosis relative to those with wild-type RAS [3–5]. At present, therapeutic options for initially unresectable RAS/BRAF-mutated microsatellite stable (MSS)-type mCRC are limited, and although clinical research has explored targeted therapeutic strategies for mutated-RAS/BRAF mCRC (M-mCRC) and treatment approaches based on various potential signaling pathways and molecules, most have failed to translate into clinical practice. The current key to research on M-mCRC is to further investigate its biology, explore new selective drugs and combinations that directly target the mutation sites, and identify metabolic pathway inhibitors that indirectly target MAPK signaling. This necessitates the identification of additional prospective prognostic and predictive biomarkers as well as potential therapeutic targets, hoping to provide a basis for formulating individualized treatment regimens and ultimately improving patient survival outcomes.
Recent studies have revealed a significant correlation between alterations in the human gut microbiome and mCRC. For instance, Bertocchi et al. found that the gut microbiome contributed to tumor metastasis by disrupting the integrity of the gut vascular barrier [6]. Yuan et al. demonstrated that alterations in the gut microbiome could induce a remodeling of the hepatic immune microenvironment by regulating the proliferation of liver Kupffer cells, thereby promoting liver metastasis of CRC [7]. However, due to the influence of confounding factors such as geographic location, dietary habits, age characteristics, treatment regimens, and specimen DNA extraction methods across various studies, the causal relationship between the abundance of specific gut microbiomes and mCRC, along with its prognostic implications for patients, remains inconclusive. Metabolites are an important factor linking the gut microbiome to CRC. Yang et al. have identified alterations in the abundance of specific gut microbiomes and plasma metabolite concentrations in CRC patients compared to healthy individuals, establishing correlations between specific gut microbiomes and metabolite profiles during the development of CRC [8]. DT et al. have revealed that the metabolite formate derived from Fusobacterium nucleatum promoted CRC invasion by activating AhR signal transduction [9]. These findings suggest that gut microbiomes may impact CRC development and metastasis by modulating certain metabolite levels through their effects on metabolic pathways. Metabolic reprogramming is a common characteristic observed in malignant tumors, including CRC. Dysregulated lipid metabolism-induced metabolic reshaping can lead to the accumulation of lipid droplets, thereby influencing the progression and invasion of CRC. Furthermore, it may also induce chemotherapy resistance in CRC, thereby influencing patient prognosis [10–14]. Hutton et al. found that mutations in RAS and BRAF induce metabolic reprogramming by upregulating glucose uptake and enhancing the expression of glutamine metabolism proteins [15]. Furthermore, current clinical trials are exploring potential therapeutic strategies for M-mCRC by targeting the RAS through metabolic pathways, indicating the potential influence of metabolic dysregulation on the prognosis of M-mCRC. Presently, there have been studies exploring the risk and prognostic biomarkers of CRC based on microbiomics and metabolomics. S et al. examined the metagenomics and metabolomics of 616 patients with different stages of CRC. Their findings indicated that the abundance of certain species, including Fusobacterium nucleatum spp, phyla Firmicutes, Fusobacteria, and Bacteroidetes increased with the malignancy of CRC. Metabolomic analysis revealed notable increases in branched-chain amino acids and phenylalanine in colorectal mucosal adenocarcinoma (MA), while bile acids demonstrated significant elevation in cases of multiple adenomas and/or MA. Additionally, isovaleric acid (a branched-chain fatty acid generated via bacterial fermentation from leucine17) exhibited a progressive rise in accordance with tumor staging. Consequently, these findings underscore the identification of stage-specific potential intestinal microbiota and metabolites relevant to CRC [16]. E et al. identified two potential microbial taxa, Prevotella enoeca and Ruthenibacterium lactatiformans, which exhibited higher abundances in CRC subjects with BRAFV600E mutation and BRAF wild-type, respectively. These findings proposed the potential of these microbial taxa as discerning biomarkers for the BRAF status among CRC patients [17]. However, studies on the associations between gut microbiomes and metabolites in M-mCRC are limited, and the mechanisms through which specific gut microbiomes cause alterations in metabolic pathways remain unclear. Moreover, the effects of gut microbiome-induced metabolic dysregulation on the prognosis of M-mCRC patients remain inconclusive.
Mendelian randomization (MR) is a causal inference method based on genetic variations. It involves single nucleotide polymorphisms (SNPs) strongly associated with exposure factors as instrumental variables (IVs) to infer causal relationships with outcomes. MR offers a distinct advantage by leveraging the random distribution of genotypes to effectively minimize the influence of confounding factors in observational studies, thus ensuring the robustness of causal inference.
In this study, we conducted a two-sample MR to investigate the causal impacts of gut microbiome features and plasma metabolite/metabolite ratios on M-mCRC-specific survival. Subsequently, we estimated potential causal associations between significant gut microbiome features and metabolite/metabolite ratios. Finally, we annotated metabolite information and performed functional annotation as well as pathway enrichment analyses on shared proteins corresponding to significant metabolite ratios. Our objectives were as follows: (1) to identify specific gut microbiomes and metabolite molecules associated with M-mCRC-specific survival; (2) to identify gut microbiome-metabolite associations based on M-mCRC, thereby unveiling potential mechanisms through which gut microbiome influence the prognosis of M-mCRC; (3) to uncover potential therapeutic targets for M-mCRC, thus offering insights for further investigation into treatment strategies.
Methods
Study design
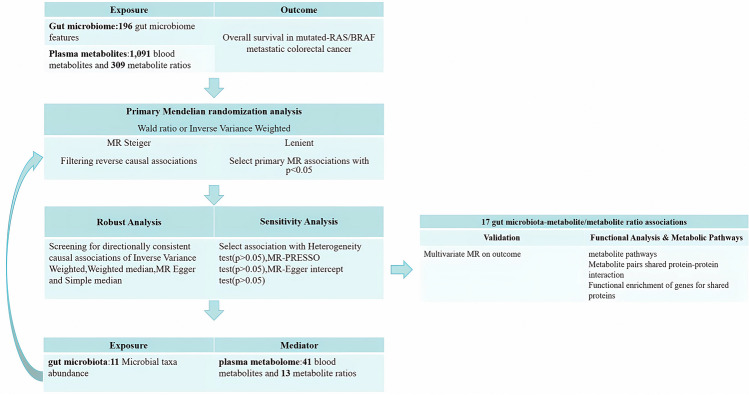
The study design schematic is shown in Fig. 1. First, we screened genetic variants representing the abundance of 196 gut microbiome features and genetic variants representing levels of 1091 plasma metabolites and 309 metabolite ratios. We then selected the overall survival (OS) of M-mCRC patients in the GWAS dataset as the outcome. Next, we performed MR to (1) estimate the causal effects of 196 gut microbiome features on the outcome and identify significant gut microbiome factors, (2) estimate the causal effects of 1400 plasma metabolite/metabolite ratios on the outcome and identify significant metabolite/metabolite ratios, and (3) estimate the causal effects between significant gut microbiome and metabolites/metabolite ratios. We further individually annotated the information on significant metabolites and the shared enzymes corresponding to metabolite ratios. At last, we analyzed the interactions among these proteins, identified genes for shared proteins, and performed functional enrichment analysis. Our study design followed the STROBE-MR statement [18] (Table S1).
Fig. 1.
Overview of study design
Data source
Summary statistics of gut microbiomes were extracted from the publicly available GWAS dataset of the MiBioGen Consortium, encompassing 18,340 subjects across 25 cohorts and revealing a total of 196 genetically controlled gut microbiome features [19]. Genetic variants associated with plasma metabolites were obtained from the Genome-wide association study(GWAS) conducted on 8299 European participants within the Canadian Longitudinal Study on Aging (CLSA) cohort, a large-scale longitudinal study encompassing biomedical data from over 50,000 Canadian participants [20]. The genome-wide genotyping data and circulating plasma metabolite information of 8299 European participants were obtained, and finally, a total of 1091 plasma metabolites and 309 metabolite ratios involving shared enzymes under genetic control were identified [21]. The outcome summary dataset was obtained from a publicly available GWAS study, which included genotype data of 694 M-mCRC patients in the COIN [22] and COIN-B [23] clinical trials. This study integrated clinically pathological factors associated with OS (defined as the time from diagnosis to the death of patients or trial completion) to formulate regression models for survival analyses. Cox proportional hazards regression was employed to evaluate the association between genetic instruments and M-mCRC-specific survival [24].
Selection of genetic variants
We extracted eligible genetic variants as IVs from the GWAS summary datasets of gut microbiome and plasma metabolite/metabolite ratios through the following process. First, we selected SNPs strongly associated with each exposure at the genome-wide locus significance level (p < 1 × 10−5). Second, we eliminated (1) SNPs with multiple alleles (> 2) or those located on chromosome 23, (2) SNPs with minor allele frequency (MAF) < 0.01, and (3) SNPs with linkage disequilibrium (LD) based on the criteria of r2 < 0.01 and window size > 10,000 kb with reference to the 1000 Genomes European Project. Third, we calculated F-statistic using the formula F = R2 × (N − 2)/(1 − R2) [25] to evaluate the efficacy of each genetic variant. A higher F-statistic indicated that the IV could better explain the variation in the dependent variable. Fourth, we determined R2 of the linear regression fit model using the formula R2 = 2 × EAF × (1 − EAF) × beta2/(2 × EAF × (1 − EAF) × beta2) + 2 × EAF × (1 − EAF) × SE × N × beta2 [26, 27] to quantify the degree of variance in the dependent variable explained by genetic variations. At last, we retained genetic variants with F-statistic > 10 as IVs to reduce the impact of weakly predictive IVs on MR analyses.
MR analysis
We performed preliminary MR analyses to evaluate the causal effects of 196 gut microbiome features and 1400 plasma metabolite/metabolite ratios on the outcome, respectively. For exposures with only one IV, we employed the Wald ratio to estimate causal effects, while the inverse variance weighted (IVW) method was used for exposures with multiple IVs. IVW combines the effects of multiple IVs on outcomes to achieve an accurate estimation of causal effects through least squares estimation, thereby improving the accuracy of the estimation. We identified exposures that reached statistical significance (p < 0.05) and conducted supplementary MR analyses using MR-Egger, weighted median, and simple median methods. Additionally, we employed the MR Steiger test to determine the directionality of causality, mitigating the impact of reverse causal associations on MR [28]. Similarly, we evaluated the causal effect between significant gut microbiomes and plasma metabolite/metabolite ratios using the same method. Finally, we utilized multivariate MR to evaluate the independent contribution of significant gut microbiomes-metabolite/metabolite ratios to the outcome [29].
Sensitivity analysis
To ensure the robustness of our findings, a series of sensitivity analyses were conducted for each statistically significant causal association (IVW p < 0.05). First, Cochran’s Q statistic was calculated by summing the squared residuals of each IV on the outcome estimates. Subsequently, a chi-square test was performed to determine the presence of variations in the impact of different IVs on the outcome. Additionally, to assess the heterogeneity of the IVs’ effects, I2 was calculated using the formula I2 = (Q-Q_df)/Q, with an I2 value of 25% or more indicating moderate to high heterogeneity [30, 31]. Moreover, we employed several approaches to mitigate the influence of IVs causally linking outcomes from confounding pathways other than exposure factors and identify and correct potential horizontal pleiotropy (HP). First, we applied the MR-Egger method to build regression models incorporating an intercept term (θ0) and test the significance of the Egger regression slopes to identify HP [32]. Second, we employed MR-PRESSO to (1) compute regression residuals of IVs on outcome estimates to identify HP, (2) detect and eliminate outlier IVs, and (3) generate adjusted causal estimates, thereby providing more robust MR results [33]. Third, we employed the leave-one-out method to assess the consistency of the MR results by excluding each IV and recalculating the causal effect to determine whether the causal association between exposure and outcome was significantly influenced by any of the IVs. At last, we generated scatter plots and funnel plots to visually present the estimates and their corresponding confidence intervals for each IV, with the aim of detecting potential anomalies or outliers. All statistical analyses were conducted using the TwoSampleMR and MR-PRESSO packages in R version 4.2.0 [33–35].
Functional and metabolic pathway analysis
To investigate the potential pathways through which the gut microbiomes may impact the prognosis of M-mCRC, we further analyzed significant metabolites and shared proteins corresponding to metabolite ratios. Leveraging the Human Metabolome Database (HMDB), which provides comprehensive information on small molecule metabolites present in the human body [36], we pursued the following steps. First, we retrieved metabolite information from HMDB and annotated metabolite super and sub-pathways. Second, building upon the work of J et al., who identified 309 pairs of metabolite enzymes or transporter proteins using HMDB, we acquired information about metabolite ratios, including their shared enzymes or transporter proteins, protein types, and corresponding genes [21]. Third, we utilized the Database for Annotation, Visualization, and Integrated Discovery (DAVID) tools to annotate the cellular components (CC), biological processes (BP), and molecular functions (MF) of the shared genes, followed by pathway enrichment analysis, thereby revealing the gene functions and their roles in biological processes [37]. At last, we employed the STRING database to construct protein–protein interaction (PPI) networks of shared proteins with a minimum required interaction score of 0.4 [38].
Results
Association between the gut microbiome and M-mCRC-specific survival
We investigated the causal correlation between 196 gut microbiome features and the outcome at five taxonomic levels: phylum, class, order, family, and genus. We actually analyzed 187 microbiome features as IVs for 9 were not present in the outcome GWAS summary dataset. We identified 14 significant associations (p < 0.05) (Table S2). Further details of the IVs involved in these 14 relevant microbiomes are provided in Table S3. Notably, all IVs exhibited F-statistics greater than 10, indicating their capacity to explain the causal association between gut microbiomes and outcome. However, the results of MR Steiger did not support a unidirectional causal association of the genus Eubacteriumeligens, Ruminococcus1, and class Alphaproteobacteria with the outcome (Table S3). Consequently, our analysis concluded that ten gut microbiome features, including the genus Eubacteriumhallii (hazard ratio [HR] = 5.34, 95% confidence interval [CI] = 1.33–21.51, p = 0.018) and Eubacteriumnodatum (HR = 1.79, CI = 1.13–2.84, p = 0.013), were associated with worse M-mCRC-specific survival. However, the genus Butyrivibrio (HR = 0.56, CI = 0.34–0.93, p = 0.025) significantly improved patient survival (Table 1). Sensitivity analyses, as presented in Table S4 and Supplementary Figs. 1–8, confirmed the robustness of the significant causal associations (I2 < 25%, Cochran’s Q > 0.05). Both the MR-Egger intercept term and the MR-PRESSO-based test did not detect any outlier IVs or significant HP (Egger intercept p > 0.05, MR-PRESSO Global Test p > 0.05). Due to the limited IVs for the family Oxalobacteraceae, genus Catenibacterium, and Oxalobacter, corresponding sensitivity analyses were not performed.
Table 1.
MR results of causal links between gut microbiome and outcome
| Classification | Bacterial traits | Nsnp | Methods | HR (95% CI) | p-value |
|---|---|---|---|---|---|
| Family | Oxalobacteraceae.id.2966 | 1 | Wald ratio | 3.45 (1.03, 11.59) | 0.0454 |
| Veillonellaceae.id.2172 | 4 | Inverse variance weighted (fixed effects) | 8.04 (2.53, 25.56) | 0.0004 | |
| Inverse variance weighted (multiplicative random effects) | 8.04 (2.73, 23.70) | 0.0002 | |||
| Weighted median | 9.76 (2.07, 46.07) | 0.0040 | |||
| MR Egger | 0.58 (0.00, 504.24) | 0.8907 | |||
| Simple median | 12.27 (2.72, 55.26) | 0.0011 | |||
| Genus | Eubacteriumhalliigroup.id.11338 | 3 | Inverse variance weighted (fixed effects) | 5.34 (1.33, 21.51) | 0.0184 |
| Inverse variance weighted (multiplicative random effects) | 5.34 (1.28, 22.23) | 0.0213 | |||
| Weighted median | 3.03 (0.46, 20.01) | 0.2486 | |||
| MR Egger | 0.04 (0.00, 724.06) | 0.8603 | |||
| Simple median | 3.17 (0.45, 22.23) | 0.2457 | |||
| Eubacteriumnodatumgroup.id.11297 | 5 | Inverse variance weighted (fixed effects) | 1.79 (1.13, 2.84) | 0.0137 | |
| Inverse variance weighted (multiplicative random effects) | 1.79 (1.03, 3.11) | 0.0391 | |||
| Weighted median | 1.54 (0.81, 2.95) | 0.1890 | |||
| MR Egger | 10.36 (1.17, 91.48) | 0.1261 | |||
| Simple median | 1.56 (0.79, 3.10) | 0.1996 | |||
| Butyrivibrio.id.1993 | 5 | Inverse variance weighted (fixed effects) | 0.56 (0.34, 0.93) | 0.0252 | |
| Inverse variance weighted (multiplicative random effects) | 0.56 (0.36, 0.88) | 0.0112 | |||
| Weighted median | 0.61 (0.30, 1.25) | 0.1758 | |||
| MR Egger | 1.34 (0.13, 13.81) | 0.8218 | |||
| Simple median | 0.61 (0.31, 1.20) | 0.1521 | |||
| Catenibacterium.id.2153 | 1 | Wald ratio | 3.92 (1.43, 10.76) | 0.0079 | |
| Faecalibacterium.id.2057 | 2 | Inverse variance weighted (fixed effects) | 6.04 (1.02, 35.67) | 0.0472 | |
| Inverse variance weighted (multiplicative random effects) | 6.04 (4.03, 9.05) | 0.0000 | |||
| Olsenella.id.822 | 3 | Inverse variance weighted (fixed effects) | 2.31 (1.22, 4.37) | 0.0102 | |
| Inverse variance weighted (multiplicative random effects) | 2.31 (1.64, 3.25) | 0.0000 | |||
| Weighted median | 2.42 (1.08, 5.43) | 0.0323 | |||
| MR Egger | 3.80 (0.41, 35.01) | 0.4483 | |||
| Simple median | 2.53 (1.09, 5.85) | 0.0305 | |||
| Oxalobacter.id.2978 | 1 | Wald ratio | 3.30 (1.02, 10.61) | 0.0454 | |
| Ruminiclostridium5.id.11355 | 4 | Inverse variance weighted (fixed effects) | 5.64 (1.68, 19.01) | 0.0052 | |
| Inverse variance weighted (multiplicative random effects) | 5.64 (3.51, 9.08) | 0.0000 | |||
| Weighted median | 6.18 (1.41, 27.02) | 0.0156 | |||
| MR Egger | 7.59 (0.06, 895.66) | 0.4926 | |||
| Simple median | 6.26 (1.47, 26.57) | 0.0130 | |||
| Phylum | Proteobacteria.id.2375 | 3 | Inverse variance weighted (fixed effects) | 8.29 (2.03, 33.92) | 0.0033 |
| Inverse variance weighted (multiplicative random effects) | 8.29 (4.06, 16.93) | 0.0000 | |||
| Weighted median | 8.97 (1.55, 51.84) | 0.0143 | |||
| MR Egger | 80.47 (0.12, 556.72) | 0.4139 | |||
| Simple median | 11.29 (1.59, 79.99) | 0.0152 |
Association between plasma metabolite/metabolite ratio and M-mCRC-specific survival
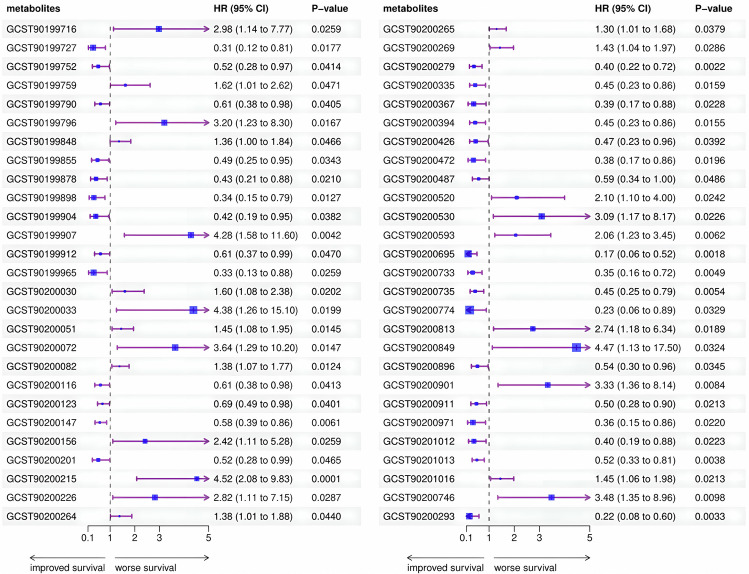
We selected eligible genetic variants as IVs for 1091 plasma metabolites and 309 metabolite ratios and performed preliminary MR analyses. After excluding metabolites lacking IV-related information in the outcome dataset, we conducted an analysis involving 1376 metabolite-outcome associations. From this analysis, we identified 54 metabolites/metabolite ratios, of which 49 were known to have a causal association with the outcome. The remaining metabolites, X-11850, X-11880, X-13007, X-17335, and X-21834, were unknown. Among the identified associations, 20 plasma metabolites and eight metabolite ratios were positively associated with M-mCRC-specific survival (HR < 1, p < 0.05), while 16 plasma metabolites and five metabolite ratios were negatively associated with M-mCRC-specific survival (HR > 1, p < 0.05). Notably, 18 metabolites were involved in lipid metabolic pathways and 6 in amino acid metabolic pathways, highlighting the potential influence of lipids and amino acids on the prognosis of M-mCRC. Additionally, succinylcarnitine may impact energy metabolism by participating in the tricarboxylic acid cycle (TCA), whereas inosine and 2′-o-methyluridine may affect nucleotide metabolism by participating in purine and pyrimidine metabolic pathways. These findings suggest a potential role of energy and nucleotide metabolism in the progression of M-mCRC (Table S6 and Fig. 2). Supplementary Table S5 presents correlation results for all 1376 metabolites. Tables S7–8 show IVs for all 54 significant metabolites/metabolite ratios, with F-statistic > 10, indicating robust causal associations without the potential for reverse causal associations or heterogeneity.
Fig. 2.
Association of genetically predicted plasma metabolite/metabolite ratios with M-mCRC-specific survival
Association between the gut microbiome and plasma metabolite/metabolite ratio based on M-mCRC
We analyzed the 539 pairs causal correlation between eleven gut microbiomes and 49 plasma metabolite/metabolite ratios revealed in the aforementioned studies and identified 17 significant associations between gut microbiomes and metabolite/metabolite ratios, including eight products related to lipid metabolism pathways, three products related to bilirubin degradation, and one additional compound. The genus Eubacteriumhallii (OR = 1.18, CI = 1.01–1.38, p = 0.0324) and Oxalobacter (OR = 1.19, CI = 1.07–1.32, p = 0.0012) were associated with increased lipid metabolites concentration, while genus Butyrivibrio (OR = 0.90, CI = 0.84–0.97, p = 0.0079), Catenibacterium (OR = 0.85, CI = 0.73–0.98, p = 0.0233), and Faecalibacterium (OR = 0.79, CI = 0.67–0.94, p = 0.0080) and phylum Proteobacteria (OR = 0.80, CI = 0.68–0.95, p = 0.0123) exhibited negative effects on lipid metabolites. Moreover, genus Ruminiclostridium was negatively correlated with two lipid metabolites, sphingolipids (OR = 0.81, CI = 0.67–0.98, p = 0.0278) and ceramides (OR = 0.78, CI = 0.63–0.97, p = 0.0226) (Table 2). Further analysis of 8 lipid metabolites revealed associations with M-mCRC-specific survival. For instance, 10-undecenoate (a medium-chain fatty acid) (HR = 2.98, CI = 1.14–7.77, p = 0.0259); sphingomyelin (d18:2/23:0, d18:1/23:1, d17:1/24:1) (HR = 4.38, CI = 1.26–15.15, p = 0.0199); and 1-palmitoyl-2-dihomo-linolenoyl-GPC (a phosphatidylcholine) (HR = 1.45, CI = 1.08–1.95, p = 0.0145) were associated with worse M-mCRC-specific survival, while 1-oleoyl-GPC (HR = 0.31, CI = 0.12–0.81, p = 0.0177); 2-palmitoleoyl-GPC (both lysophospholipid) (HR = 0.61, CI = 0.38–0.98, p = 0.0405); 5alpha-androstan-3beta, 17beta-diol monosulfate (an Androgenic Steroid) (HR = 0.49, CI = 0.25–0.95, p = 0.0343); ceramide (d18:1/14:0, d16:1/16:0) (HR = 0.61, CI = 0.38–0.98, p = 0.0413); and stearoyl sphingomyelin (d18:1/18:0) (both Sphingomyelins) (HR = 0.45, CI = 0.23–0.86, p = 0.0159) were associated with improved survival. These findings suggested that gut microbiomes could impact the prognosis of M-mCRC by participating in lipid metabolism. Notably, genus Olsenella was positively correlated with the concentrations of the three bilirubin degradation products (OR > 1, p < 0.05), and higher Olsenella abundance and bilirubin degradation products were both correlated with poor M-mCRC-specific survival (HR > 1, p < 0.05), suggesting a potential association between genus Olsenella and poor survival of M-mCRC by promoting bilirubin degradation (Table S6). Table S9 displays the correlation results for the 539 association pairs. Tables S10–11 and Supplementary Figs. 9–25 provide information on IVs, Steiger test results, and sensitivity analysis results for the 17 significant gut microbiome-metabolite associations.
Table 2.
MR results of causal links between gut microbiome and metabolites/metabolite ratios based on M-mCRC
| Gut microbiome | ID | Metabolites/Metabolite ratios | Super_pathway | Sub_pathway | Nsnp | Methods | OR(95%CI) | p−value |
|---|---|---|---|---|---|---|---|---|
| genus.Eubacteriumhalliigroup.id.11338 | GCST90199716 | 10-undecenoate (11:1n1) levels | Lipid | Medium Chain Fatty Acid | 12 | Inverse variance weighted (fixed effects) | 1.18(1.01,1.38) | 0.0324 |
| 12 | Inverse variance weighted (multiplicative random effects) | 1.18(1.01,1.38) | 0.0321 | |||||
| 12 | Weighted median | 1.20(0.97,1.49) | 0.0953 | |||||
| 12 | MR Egger | 1.09(0.77,1.52) | 0.6442 | |||||
| 12 | Simple median | 1.24(1.02,1.51) | 0.0329 | |||||
| GCST90200901 | Ornithine to phosphate ratio | Amino Acid / Energy | / | 12 | Inverse variance weighted (fixed effects) | 1.30(1.11,1.50) | 0.0007 | |
| 12 | Inverse variance weighted (multiplicative random effects) | 1.30(1.12,1.50) | 0.0005 | |||||
| 12 | Weighted median | 1.16(0.94,1.43) | 0.1709 | |||||
| 12 | MR Egger | 1.34(0.97,1.85) | 0.1100 | |||||
| 12 | Simple median | 1.16(0.94,1.42) | 0.1652 | |||||
| genus.Butyrivibrio.id.1993 | GCST90199727 | 1-oleoyl-GPC (18:1) levels | Lipid | Lysophospholipid | 13 | Inverse variance weighted (fixed effects) | 0.90(0.84,0.97) | 0.0079 |
| 13 | Inverse variance weighted (multiplicative random effects) | 0.90(0.84,0.98) | 0.0110 | |||||
| 13 | Weighted median | 0.93(0.83,1.03) | 0.1819 | |||||
| 13 | MR Egger | 0.98(0.69,1.39) | 0.9046 | |||||
| 13 | Simple median | 0.93(0.84,1.03) | 0.1707 | |||||
| GCST90200971 | Leucine to phosphate ratio | Amino Acid / Energy | / | 13 | Inverse variance weighted (fixed effects) | 0.92(0.86,0.99) | 0.0221 | |
| 13 | Inverse variance weighted (multiplicative random effects) | 0.92(0.87,0.98) | 0.0089 | |||||
| 13 | Weighted median | 0.92(0.84,1.01) | 0.0873 | |||||
| 13 | MR Egger | 0.71(0.53,0.96) | 0.0476 | |||||
| 13 | Simple median | 0.92(0.83,1.02) | 0.1000 | |||||
| GCST90201012 | 3-methyl-2-oxovalerate to 3-methyl-2-oxobutyrate ratio | Amino Acid / Amino Acid | / | 13 | Inverse variance weighted (fixed effects) | 0.91(0.85,0.98) | 0.0151 | |
| 13 | Inverse variance weighted (multiplicative random effects) | 0.91(0.85,0.98) | 0.0110 | |||||
| 13 | Weighted median | 0.94(0.85,1.05) | 0.2593 | |||||
| 13 | MR Egger | 0.90(0.65,1.24) | 0.5329 | |||||
| 13 | Simple median | 0.94(0.85,1.04) | 0.2243 | |||||
| GCST90201016 | Bilirubin (Z,Z) to etiocholanolone glucuronide ratio | Cofactors and Vitamins / Lipid | / | 13 | Inverse variance weighted (fixed effects) | 0.87(0.81,0.94) | 0.0004 | |
| 13 | Inverse variance weighted (multiplicative random effects) | 0.87(0.80,0.94) | 0.0009 | |||||
| 13 | Weighted median | 0.87(0.78,0.97) | 0.0097 | |||||
| 13 | MR Egger | 0.73(0.51,1.04) | 0.1122 | |||||
| 13 | Simple median | 0.87(0.78,0.97) | 0.0110 | |||||
| genus.Catenibacterium.id.2153 | GCST90199790 | 2-palmitoleoyl-GPC (16:1) levels | Lipid | Lysophospholipid | 4 | Inverse variance weighted (fixed effects) | 0.85(0.73,0.98) | 0.0233 |
| 4 | Inverse variance weighted (multiplicative random effects) | 0.85(0.78,0.92) | 0.0001 | |||||
| 4 | Weighted median | 0.86(0.73,1.02) | 0.0768 | |||||
| 4 | MR Egger | 1.03(0.17,6.31) | 0.9760 | |||||
| 4 | Simple median | 0.86(0.73,1.01) | 0.0669 | |||||
| genus.Oxalobacter.id.2978 | GCST90199855 | 5alpha-androstan-3beta,17beta-diol monosulfate (2) levels | Lipid | Androgenic Steroids | 8 | Inverse variance weighted (fixed effects) | 1.19(1.07,1.32) | 0.0012 |
| 8 | Inverse variance weighted (multiplicative random effects) | 1.19(1.05,1.34) | 0.0051 | |||||
| 8 | Weighted median | 1.12(0.97,1.29) | 0.1267 | |||||
| 8 | MR Egger | 1.38(0.78,2.46) | 0.3102 | |||||
| 8 | Simple median | 1.13(0.98,1.30) | 0.0891 | |||||
| genus.Ruminiclostridium5.id.11355 | GCST90200033 | Sphingomyelin (d18:2/23:0, d18:1/23:1, d17:1/24:1) levels | Lipid | Sphingomyelins | 6 | Inverse variance weighted (fixed effects) | 0.81(0.65,1.01) | 0.0670 |
| 6 | Inverse variance weighted (multiplicative random effects) | 0.81(0.67,0.98) | 0.0278 | |||||
| 6 | Weighted median | 0.87(0.66,1.15) | 0.3329 | |||||
| 6 | MR Egger | 0.95(0.37,2.42) | 0.9129 | |||||
| 6 | Simple median | 0.84(0.63,1.12) | 0.2261 | |||||
| GCST90200116 | Ceramide (d18:1/14:0, d16:1/16:0) levels | Lipid | Ceramides | 6 | Inverse variance weighted (fixed effects) | 0.78(0.59,1.02) | 0.0683 | |
| 6 | Inverse variance weighted (multiplicative random effects) | 0.78(0.63,0.97) | 0.0226 | |||||
| 6 | Weighted median | 0.73(0.52,1.03) | 0.0760 | |||||
| 6 | MR Egger | 1.49(0.48,4.60) | 0.5301 | |||||
| 6 | Simple median | 0.71(0.50,1.02) | 0.0658 | |||||
| phylum.Proteobacteria.id.2375 | GCST90200051 | 1-palmitoyl-2-dihomo-linolenoyl-GPC (16:0/20:3n3 or 6) levels | Lipid | Phosphatidylcholine (PC) | 7 | Inverse variance weighted (fixed effects) | 0.80(0.65,1.00) | 0.0506 |
| 7 | Inverse variance weighted (multiplicative random effects) | 0.80(0.68,0.95) | 0.0123 | |||||
| 7 | Weighted median | 0.78(0.59,1.04) | 0.0928 | |||||
| 7 | MR Egger | 0.54(0.30,0.99) | 0.1032 | |||||
| 7 | Simple median | 0.79(0.59,1.06) | 0.1124 | |||||
| GCST90200226 | 5-hydroxy-2-methylpyridine sulfate levels | Xenobiotics | Chemical | 7 | Inverse variance weighted (fixed effects) | 0.74(0.58,0.94) | 0.0134 | |
| 7 | Inverse variance weighted (multiplicative random effects) | 0.74(0.59,0.92) | 0.0064 | |||||
| 7 | Weighted median | 0.76(0.55,1.06) | 0.1047 | |||||
| 7 | MR Egger | 0.69(0.36,1.34) | 0.3201 | |||||
| 7 | Simple median | 0.76(0.54,1.06) | 0.1078 | |||||
| genus.Olsenella.id.822 | GCST90200264 | Bilirubin degradation product, C17H20N2O5 (2) levels | Partially Characterized Molecules | Partially Characterized Molecules | 8 | Inverse variance weighted (fixed effects) | 1.14(1.03,1.26) | 0.0130 |
| 8 | Inverse variance weighted (multiplicative random effects) | 1.14(1.03,1.26) | 0.0136 | |||||
| 8 | Weighted median | 1.08(0.94,1.23) | 0.2745 | |||||
| 8 | MR Egger | 1.04(0.67,1.60) | 0.8750 | |||||
| 8 | Simple median | 1.08(0.95,1.24) | 0.2449 | |||||
| GCST90200265 | Bilirubin degradation product, C16H18N2O5 (4) levels | Partially Characterized Molecules | Partially Characterized Molecules | 8 | Inverse variance weighted (fixed effects) | 1.12(1.01,1.24) | 0.0270 | |
| 8 | Inverse variance weighted (multiplicative random effects) | 1.12(1.02,1.23) | 0.0169 | |||||
| 8 | Weighted median | 1.15(1.00,1.31) | 0.0473 | |||||
| 8 | MR Egger | 1.26(0.84,1.90) | 0.3010 | |||||
| 8 | Simple median | 1.15(1.00,1.31) | 0.0421 | |||||
| GCST90200269 | Bilirubin degradation product, C17H20N2O5 (1) levels | Partially Characterized Molecules | Partially Characterized Molecules | 8 | Inverse variance weighted (fixed effects) | 1.13(1.02,1.25) | 0.0168 | |
| 8 | Inverse variance weighted (multiplicative random effects) | 1.13(1.02,1.25) | 0.0159 | |||||
| 8 | Weighted median | 1.07(0.94,1.23) | 0.2988 | |||||
| 8 | MR Egger | 1.05(0.68,1.61) | 0.8388 | |||||
| 8 | Simple median | 1.08(0.95,1.24) | 0.2352 | |||||
| genus.Faecalibacterium.id.2057 | GCST90200335 | Stearoyl sphingomyelin (d18:1/18:0) levels | Lipid | Sphingomyelins | 7 | Inverse variance weighted (fixed effects) | 0.79(0.67,0.94) | 0.0080 |
| 7 | Inverse variance weighted (multiplicative random effects) | 0.79(0.63,0.99) | 0.0415 | |||||
| 7 | Weighted median | 0.87(0.68,1.11) | 0.2571 | |||||
| 7 | MR Egger | 0.97(0.66,1.44) | 0.8984 | |||||
| 7 | Simple median | 0.72(0.54,0.97) | 0.0313 | |||||
| GCST90201012 | 3-methyl-2-oxovalerate to 3-methyl-2-oxobutyrate ratio | Amino Acid / Amino Acid | / | 7 | Inverse variance weighted (fixed effects) | 1.32(1.11,1.57) | 0.0019 | |
| 7 | Inverse variance weighted (multiplicative random effects) | 1.32(1.07,1.63) | 0.0099 | |||||
| 7 | Weighted median | 1.38(1.07,1.77) | 0.0123 | |||||
| 7 | MR Egger | 1.27(0.84,1.93) | 0.3047 | |||||
| 7 | Simple median | 1.37(1.03,1.81) | 0.0307 |
Independent effects of gut microbiome-metabolite/metabolite ratios on M-mCRC-specific survival
We performed multivariate MR to estimate the independent effects of the aforementioned 17 pairs of gut microbiomes and metabolite/metabolite ratios on M-mCRC-specific survival. As shown in Table 3, the genus Ruminiclostridium and Olsenella and the ornithine-to-phosphate ratio were significantly associated with worse M-mCRC-specific survival (β > 0, p < 0.05). Conversely, only stearoyl sphingomyelin (β = − 1.54, CI = − 2.82 to − 0.25, p = 0.033) was positively associated with M-mCRC-specific survival. Notably, after removing the effect of the gut microbiome, the effects of ceramide (d18:1/14:0, d16:1/16:0), 1-palmitoyl-2-dihomo-linolenoyl-GPC, and 5-hydroxy-2-methylpyridine sulfate on outcome were inconsistent with the original effects, although not statistically significant. We hypothesized that this inconsistency might be attributed to the inhibition of these metabolites by the gut microbiome. Although the residual independent impacts of gut microbiome and metabolites on the outcome did not attain statistical significance, they consistently exhibited the same trend as the initial effect.
Table 3.
Multivariate MR estimates of the effect of gut microbiome-metabolite/metabolite ratio on M-mCRC-specific survival
| Exposure | Metabolites/metabolite ratio | Method | β (95% CI) | P | Cochran Q | PHeterogeneity | Egger intercept | Pintercept |
|---|---|---|---|---|---|---|---|---|
| IVW | 0.32 (− 0.81, 1.44) | 0.583 | 8.488 | 0.387 | 8.104 | 0.324 | ||
| 0.87 (0.07, 1.67) | 0.033 | |||||||
| genus.Eubacteriumhalliigroup.id.11338/GCST90200901 | Ornithine to phosphate ratio | MR Egger | 0.71 (− 1.48, 2.90) | 0.526 | 8.280 | 0.309 | ||
| 0.82(-0.06,1.69) | 0.068 | |||||||
| Weighted median | 1.05 (− 0.71, 2.81) | 0.242 | ||||||
| 0.84 (− 0.25, 1.93) | 0.131 | |||||||
| IVW | 0.31 (− 0.93, 1.54) | 0.629 | 8.745 | 0.272 | 8.327 | 0.215 | ||
| 0.87 (− 0.06, 1.81) | 0.067 | |||||||
| genus.Eubacteriumhalliigroup.id.11338/GCST90199716 | 10-undecenoate (11:1n1) levels | MR Egger | 0.47 (− 1.79, 2.73) | 0.683 | 8.699 | 0.191 | ||
| 0.89 (− 0.14, 1.93) | 0.091 | |||||||
| Weighted median | 0.69 (− 1.05, 2.42) | 0.44 | ||||||
| 1.10 (− 0.18, 2.38) | 0.093 | |||||||
| IVW | − 0.29 (− 0.94, 0.35) | 0.374 | 18.043 | 0.021 | 17.825 | 0.013 | ||
| 0.06 (− 1.39, 1.50) | 0.94 | |||||||
| genus.Butyrivibrio.id.1993/GCST90199727 | 1-oleoyl-GPC (18:1) levels | MR Egger | − 0.43 (− 1.84, 0.98) | 0.551 | 17.924 | 0.012 | ||
| 0.02 (− 1.56, 1.60) | 0.982 | |||||||
| Weighted median | − 0.40 (− 1.06, 0.27) | 0.239 | ||||||
| − 0.80 (− 2.28, 0.67) | 0.285 | |||||||
| IVW | − 0.29 (− 0.83, 0.24) | 0.282 | 15.112 | 0.088 | 14.755 | 0.064 | ||
| − 0.46 (− 1.78, 0.85) | 0.489 | |||||||
| genus.Butyrivibrio.id.1993/GCST90200971 | Leucine to phosphate ratio | MR Egger | 0.07 (− 0.93, 1.07) | 0.888 | 13.843 | 0.086 | ||
| − 0.24 (− 1.67, 1.19) | 0.744 | |||||||
| Weighted median | − 0.62 (− 1.27, 0.03) | 0.062 | ||||||
| − 1.01 (− 2.38, 0.36) | 0.149 | |||||||
| IVW | − 0.26 (− 0.77, 0.25) | 0.32 | 15.440 | 0.117 | 14.758 | 0.098 | ||
| − 0.78 (− 1.90, 0.35) | 0.177 | |||||||
| genus.Butyrivibrio.id.1993/GCST90201012 | 3-methyl-2-oxovalerate to 3-methyl-2-oxobutyrate ratio | MR Egger | − 0.77 (− 1.92, 0.38) | 0.191 | 13.986 | 0.123 | ||
| − 0.90 (− 2.06, 0.26) | 0.128 | |||||||
| Weighted median | − 0.49 (− 1.15, 0.18) | 0.149 | ||||||
| − 0.13 (− 1.46, 1.21) | 0.852 | |||||||
| IVW | − 0.20 (− 0.77, 0.36) | 0.479 | 14.087 | 0.080 | 13.577 | 0.059 | ||
| 0.68 (− 0.50, 1.86) | 0.258 | |||||||
| genus.Butyrivibrio.id.1993/GCST90201016 | Bilirubin (Z,Z) to etiocholanolone glucuronide ratio | MR Egger | 0.48 (− 0.72, 1.67) | 0.432 | 11.495 | 0.118 | ||
| 0.50 (− 0.67, 1.67) | 0.404 | |||||||
| Weighted median | − 0.39 (− 1.06, 0.28) | 0.254 | ||||||
| 0.79 (− 0.61, 2.19) | 0.268 | |||||||
| IVW | 0.60 (− 0.26, 1.46) | 0.174 | 5.514 | 0.239 | 5.162 | 0.160 | ||
| − 0.36 (− 1.30, 0.59) | 0.458 | |||||||
| genus.Catenibacterium.id.2153/GCST90199790 | 2-palmitoleoyl-GPC (16:1) levels | MR Egger | 0.91 (− 0.58, 2.41) | 0.231 | 5.028 | 0.170 | ||
| − 0.18 (− 1.41, 1.05) | 0.775 | |||||||
| Weighted median | 0.76 (− 0.66, 2.19) | 0.293 | ||||||
| − 0.30 (− 1.40, 0.81) | 0.599 | |||||||
| IVW | 0.21 (− 0.56, 0.98) | 0.59 | 3.821 | 0.576 | 3.684 | 0.451 | ||
| − 0.70 (− 1.50, 0.10) | 0.085 | |||||||
| genus.Oxalobacter.id.2978/GCST90199855 | 5alpha-androstan-3beta, 17beta-diol monosulfate (2) levels | MR Egger | 0.58 (− 0.43, 1.58) | 0.26 | 2.592 | 0.628 | ||
| − 0.76 (− 1.56, 0.05) | 0.065 | |||||||
| Weighted median | 0.02 (− 1.16, 1.20) | 0.972 | ||||||
| − 0.44 (− 1.56, 0.67) | 0.436 | |||||||
| IVW | 1.49 (0.48, 2.50) | 0.004 | 10.692 | 0.297 | 9.831 | 0.277 | ||
| − 0.62 (− 1.61, 0.38) | 0.225 | |||||||
| genus.Ruminiclostridium5.id.11355/GCST90200033 | Sphingomyelin (d18:2/23:0, d18:1/23:1, d17:1/24:1) levels | MR Egger | 1.45 (− 0.32, 3.23) | 0.108 | 10.689 | 0.220 | ||
| − 0.62 (− 1.68, 0.44) | 0.252 | |||||||
| Weighted median | 1.25 (− 0.07, 2.57) | 0.063 | ||||||
| 0.29 (− 1.33, 1.91) | 0.727 | |||||||
| IVW | 1.41 (0.49, 2.33) | 0.003 | 3.026 | 0.990 | 2.846 | 0.985 | ||
| 0.30 (− 0.345, 0.96) | 0.36 | |||||||
| genus.Ruminiclostridium5.id.11355/GCST90200116 | Ceramide (d18:1/14:0, d16:1/16:0) levels | MR Egger | 1.43 (− 0.04, 2.89) | 0.056 | 3.025 | 0.981 | ||
| 0.31 (− 0.36, 0.97) | 0.365 | |||||||
| Weighted median | 1.19 (− 0.05, 2.43) | 0.06 | ||||||
| 0.34 (− 0.56, 1.24) | 0.46 | |||||||
| IVW | 0.49 (− 0.50, 1.48) | 0.332 | 14.295 | 0.353 | 14.154 | 0.291 | ||
| − 0.22 (− 0.86, 0.43) | 0.51 | |||||||
| phylum.Proteobacteria.id.2375/ − GCST90200051 | 1-palmitoyl-2-dihomo-linolenoyl-GPC (16:0/20:3n3 or 6) levels | MR Egger | 1.13 (− 0.26, 2.52) | 0.111 | 12.626 | 0.397 | ||
| − 0.29 (− 0.93, 0.35) | 0.378 | |||||||
| Weighted median | 0.47 (− 1.11, 2.05) | 0.561 | ||||||
| − 0.08 (− 0.95, 0.79) | 0.856 | |||||||
| IVW | 0.13 (− 1.34, 1.60) | 0.858 | 13.631 | 0.058 | 13.621 | 0.034 | ||
| − 0.05 (− 1.29, 1.19) | 0.937 | |||||||
| phylum.Proteobacteria.id.2375/GCST90200226 | 5-hydroxy-2-methylpyridine sulfate levels | MR Egger | − 3.89 (− 8.11, 0.34) | 0.071 | 8.336 | 0.215 | ||
| − 1.61 (− 3.48, 0.27) | 0.094 | |||||||
| Weighted median | − 0.04 (− 1.75, 1.66) | 0.961 | ||||||
| − 1.18 (− 2.95, 0.59) | 0.19 | |||||||
| IVW | 0.99 (0.20, 1.78) | 0.014 | 1.645 | 0.649 | 1.421 | 0.491 | ||
| 0.32 (− 0.67, 1.31) | 0.525 | |||||||
| genus.Olsenella.id.822/GCST90200264 | Bilirubin degradation product, C17H20N2O5 (2) levels | MR Egger | 1.70 (− 0.36, 3.76) | 0.107 | 1.113 | 0.573 | ||
| − 0.43 (− 2.68, 1.82) | 0.706 | |||||||
| Weighted median | 0.94 (− 0.30, 2.19) | 0.137 | ||||||
| 0.23 (− 1.26, 1.73) | 0.76 | |||||||
| IVW | 0.68 (0.06, 1.30) | 0.031 | 2.851 | 0.827 | 2.675 | 0.750 | ||
| − 0.01 (− 0.76, 0.75) | 0.983 | |||||||
| genus.Olsenella.id.822/GCST90200265 | Bilirubin degradation product, C16H18N2O5 (4) levels | MR Egger | 1.22 (0.18, 2.26) | 0.021 | 1.255 | 0.940 | ||
| − 0.11 (− 0.88, 0.66) | 0.778 | |||||||
| Weighted median | 0.39 (− 0.52, 1.30) | 0.403 | ||||||
| 0.10 (− 0.92, 1.11) | 0.851 | |||||||
| IVW | 0.10 (0.20, 1.79) | 0.014 | 2.013 | 0.733 | 1.763 | 0.623 | ||
| 0.19 (− 0.69, 1.06) | 0.68 | |||||||
| genus.Olsenella.id.822/GCST90200269 | Bilirubin degradation product, C17H20N2O5 (1) levels | MR Egger | 1.29 (− 0.56, 3.15) | 0.171 | 1.891 | 0.595 | ||
| − 0.16 (− 2.31, 1.98) | 0.881 | |||||||
| Weighted median | 0.93 (− 0.26, 2.11) | 0.125 | ||||||
| − 0.22 (− 1.56, 1.12) | 0.746 | |||||||
| IVW | 1.23 (− 0.64, 3.09) | 0.197 | 6.759 | 0.149 | 6.359 | 0.095 | ||
| − 0.56 (− 1.86, 0.74) | 0.4 | |||||||
| genus.Faecalibacterium.id.2057/GCST90200335 | Stearoyl sphingomyelin (d18:1/18:0) levels | MR Egger | − 2.03 (− 5.07, 1.01) | 0.191 | 1.109 | 0.775 | ||
| − 1.54 (− 2.82, − 0.25) | 0.019 | |||||||
| Weighted median | 1.52 (− 0.54, 3.58) | 0.149 | ||||||
| − 0.16 (− 1.77, 1.45) | 0.846 | |||||||
| IVW | 1.26 (− 0.56, 3.08) | 0.174 | 3.403 | 0.334 | 3.145 | 0.208 | ||
| − 0.76 (− 1.88, 0.35) | 0.179 | |||||||
| genus.Faecalibacterium.id.2057/GCST90201012 | 3-methyl-2-oxovalerate to 3-methyl-2-oxobutyrate ratio | MR Egger | 0.08 (− 3.81, 3.98) | 0.967 | 2.749 | 0.253 | ||
| − 0.56 (− 1.92, 0.81) | 0.425 | |||||||
| Weighted median | 2.45 (− 0.56, 5.45) | 0.11 | ||||||
| − 0.72 (− 2.20, 0.77) | 0.343 |
Analysis of the functional roles and metabolic pathways associated with shared proteins corresponding to significant metabolite ratios
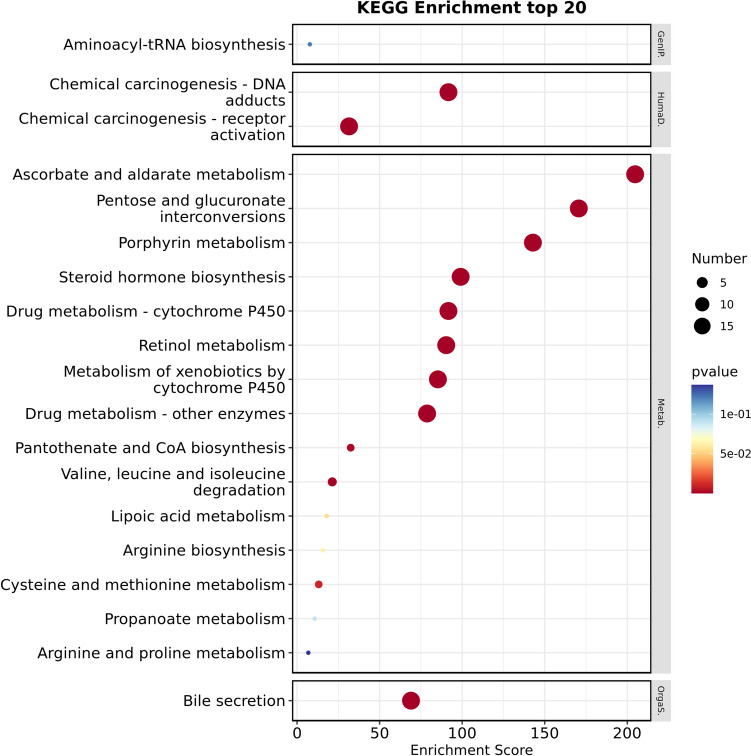
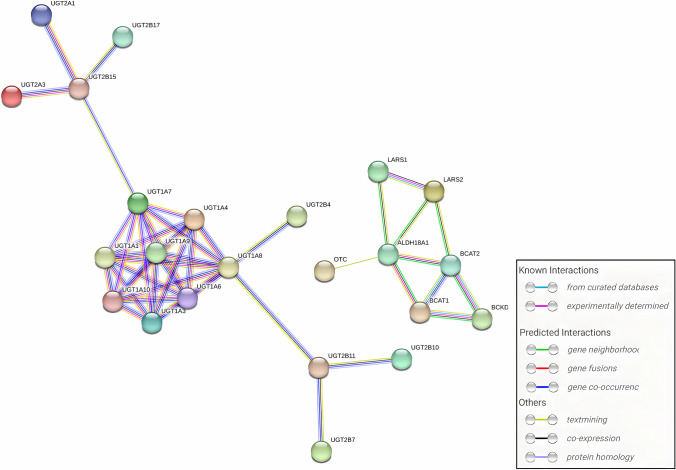
We obtained shared proteins and genes for shared proteins of significant metabolite ratios and performed functional annotation of the shared genes using the DAVID tool (Table 4). The shared proteins were mainly localized in the endoplasmic reticulum membrane, with a lesser presence in the intracellular membrane-bounded organelle and integral components of membranes. They demonstrated diverse enzyme activities, including glucuronosyltransferase and UDP-glycosyltransferase, involved in metabolic processes such as glucuronidation, steroid and lipid metabolism, and branched-chain amino acid metabolism. Moreover, they exhibited significant enrichment in pathways such as ascorbate and aldarate metabolism, pentose and glucuronate interconversions, porphyrin metabolism, steroid hormone biosynthesis, and bile secretion (Figs. 3–4 and Tables S12–13). These findings suggest that the gut microbiome may affect M-mCRC-specific survival by regulating the enzymatic activities of organelle membrane proteins involved in glucuronidation and the metabolism of a variety of substances such as steroids, lipids, and amino acids. The PPI network constructed based on STRING revealed interactions between genes encoding shared proteins (Fig. 5).
Table 4.
Information of significant metabolite ratios
| ID | Metabolite ratios | Super pathways | Gene for shared protein | Protein names | Protein type |
|---|---|---|---|---|---|
| GCST90200901 | Ornithine/phosphate | Amino acid/energy | ALDH18A1;OTC |
Delta-1-pyrroline-5-carboxylate synthase; Ornithine carbamoyltransferase |
Enzyme; enzyme |
| GCST90200971 | Leucine/phosphate | Amino acid/energy | LARS2;LARS |
Probable leucine–tRNA ligase; Leucine–tRNA ligase |
Enzyme; enzyme |
| GCST90201016 | Bilirubin (Z,Z)/etiocholanolone glucuronide | Cofactors and vitamins/lipid |
UGT2B28;UGT2B4;UGT1A4;UGT2B10; UGT2B7;UGT2B15;UGT2A1;UGT1A1; UGT1A9;UGT1A8;UGT1A3;UGT1A10; UGT2B17;UGT1A6;UGT1A5;UGT2B11; UGT1A7; UGT2A3 |
UDP-glucuronosyltransferase 2B28; 2B4; 1–4;2B10;2B7;2B15;2A1;1–1;1–9;1–8; 1–3;1–10;2B17;1–6;1–5;2B11;1–7;2A3 |
Enzyme; enzyme; Enzyme; Enzyme; enzyme; Enzyme; Enzyme; enzyme; Enzyme; Enzyme; enzyme; Enzyme; Enzyme; enzyme; Enzyme; Enzyme; enzyme; Enzyme |
| GCST90201012 | 3-Methyl-2-oxovalerate/3-methyl-2-oxobutyrate | Amino acid/amino acid | BCKDHB;BCAT1;BCAT2 |
2-oxoisovalerate dehydrogenase subunit beta; Branched-chain-amino-acid aminotransferase; Branched-chain-amino-acid aminotransferase |
Enzyme; enzyme; Enzyme |
Fig. 3.
BP, CC and MF annotation analysis of gene for shared protein of significant metabolite ratios
Fig. 4.
KEGG pathways annotation analysis of gene for shared protein of significant metabolite ratios. Classification level: Genetic Information Processing (GenIP), Human Diseases (HumaD), Metabolism (Metab)
Fig. 5.
PPI network of interactions between shared protein genes
Discussion
In this study, MR analyses were employed to evaluate the potential causal effects of genetically predicted gut microbiome and blood metabolites on M-mCRC-specific survival and identify the gut microbiome-metabolite/metabolite ratios associations based on M-mCRC. Moreover, functional annotation and pathway enrichment analyses of shared proteins corresponding to significant metabolite ratios were performed to reveal the potential mechanisms underlying the impacts of the gut microbiome on M-mCRC-specific survival through modulation of human metabolism. In total, we identified significant correlations between eleven gut microbiome features and 49 known metabolites/metabolite ratios with M-mCRC-specific survival. Moreover, correlation analyses of significant gut microbiome and metabolites/metabolite ratios revealed 17 potential associations for gut microbiome-metabolites-M-mCRC-specific survival pathways.
Our findings unveiled a negative correlation between genus Eubacteriumhallii and M-mCRC-specific survival via positively regulating the ornithine to phosphate ratio (O/P). δ-1-pyrroline-5-carboxylate synthase (P5CS) and ornithine carbamoyltransferase (OTC) were shared proteins for the O/P. MR results indicated an association between elevated O/P and worse M-mCRC-specific survival. OTC catalyzes the urea cycle, while P5CS is primarily involved in synthesizing δ-1-pyrroline5-carboxylate (P5C), an intermediate product in the conversion of proline, glutamate, and ornithine. This process interconnects the TCA cycle, urea cycle, and proline metabolism, impacting cell growth, redox homeostasis, ATP production, and immune regulation [39–41]. Previous studies have demonstrated elevated P5CS expression in various malignancies, including hepatocellular carcinoma, where it plays a pivotal role in tumor metabolic remodeling and progression, correlating with poor patient prognosis [42, 43]. Additionally, the long isoform of P5CS (P5CS.long) encoded by selective splicing of the RNA transcript is significantly up-regulated by the oncogene p53 during apoptosis of DLD-1, a type of CRC cells [44]. Despite limited literature on the correlation of genus Eubacteriumhallii with CRC, we hypothesized that the gut microbiome may trigger upregulation of P5CS, thereby impacting cellular functions and dysregulating various metabolic pathways, including proline metabolism. This could lead to remodeling of the tumor microenvironment, affecting the efficacy of anti-cancer treatments and ultimately impacting the prognosis of patients with M-mCRC.
Our study highlighted the genus Butyrivibrio as the sole microbiome associated with improved M-mCRC-specific survival. Butyrivibrio, a bacterium known for producing butyrate, a short-chain fatty acid, has been linked to a reduced risk of CRC [45]. Butyrate plays a crucial role in preserving the integrity of the colonic epithelial cell barrier. Moreover, it inhibits angiogenesis and metastasis of CRC cells by suppressing Sp1 activation and down-regulating Neuropilin-1 (NRP-1) expression [46, 47]. Thus, butyrate may act as a protective factor against CRC by impeding its development and progression through multiple pathways. Our study further revealed that Butyrivibrio and its byproduct, butyrate, potentially correlate with improved prognoses of M-mCRC by modulating human metabolism. Elevated abundance of Butyrivibrio exhibited a negative correlation with three metabolite ratios, indicating a potential suppression of enzymes corresponding to the metabolite ratios, including leucine-tRNA ligase (LRS), branched-chain-amino-acid aminotransferase (BCAT1, BCAT2), and various UDP-glucuronosyltransferases. Studies have demonstrated that the LRS may activate the mammalian target of the rapamycin family (mTOR) by binding to RagD GTPases. Dysregulated activation of the mTOR signaling pathway has been implicated in the development and progression of CRC [48–50]. Furthermore, researchers have identified mTORC1 inhibitors targeting LRS with specific cytotoxicity against mTORC1-overexpressing CRC cells as potential therapeutic targets for CRC [51]. In addition, clinical studies have linked hypermethylated BCAT1 after radical CRC surgery to a higher recurrence rate. Methylated BCAT1 in human plasma may serve as a potential diagnostic and prognostic indicator for CRC [52, 53]. However, the specific interactions of Butyrivibrio and its product butyrate with LRS, mTOR, and BCAT are currently unknown, necessitating further investigation to elucidate their potential associations. In conclusion, our study sheds light on the impact of the butyrate-producing bacterium Butyrivibrio on M-mCRC-specific survival and identifies potential functional targets and metabolic pathways worthy of further exploration.
Our study revealed an intriguing finding regarding another butyrate-producing bacterium, the genus Faecalibacterium, which exhibited an opposite effect. Several studies have highlighted its role in immune system modulation and preservation of intestinal barrier integrity [54]. Elevated abundance of Faecalibacterium has been associated with enhanced immune therapeutic responses in various cancers, including non-small cell lung carcinoma (NSCLC), hepatocellular carcinoma, and melanoma [55–59]. For instance, Dikeocha et al. constructed an Azoxymethane (AOM)-induced CRC model in rats and observed that Faecalibacterium could suppress lipid peroxidation and CRC cell proliferation in colonic tissues, indicating potential anti-tumor properties [60]. Therefore, caution should be warranted in interpreting the relationship between Faecalibacterium and poor prognostic implications in M-mCRC, as identified in our study.
Our study also unveiled a potential correlation between eight lipid metabolites, potentially influenced by the abundance of the gut microbiome, and M-mCRC-specific survival. We found that genus Butyrivibrio and phylum Proteobacteria were correlated with reduced levels of 1-oleoyl-GPC (18:1) and 1-palmitoyl-2-dihomo-linolenoyl-GPC (16:0/20:3n3 or 6), respectively. This suggests the involvement of phosphatidylcholine (PC) and its hydrolysis product, lysophosphatidylcholine (LPC), in the gut microbiome’s influence on the host. Studies have demonstrated that an elevated abundance of Proteobacteria is associated with metabolic disturbances in the human body, closely linked to inflammatory bowel disease, and may serve as a potential diagnostic indicator for dysbiosis and disease susceptibility [61–63]. Georg et al. identified a significant increase in Proteobacteria abundance in CRC patients by comparing fecal genomes between the CRC and control groups [64]. Further investigation is required to understand the pathogenic mechanisms of Proteobacteria, and our study sheds light on its potential association with lipid metabolism dysregulation. Several studies have demonstrated the correlation between overexpression of lysophosphatidylcholine acyltransferase 1 (LPCAT1), a crucial enzyme influencing PC metabolism and remodeling, with the progression of various malignancies, including CRC [65–67]. Moreover, Kitamura et al. observed elevated levels of lysophospholipids in CRC tissues compared to normal tissues, with LPC, a primary constituent of oxidized low-density lipoprotein, implicated in endothelial dysfunction and potentially contributing to CRC development and progression [68]. Melissa et al. demonstrated that butyric inhibited LPC-induced epithelial barrier dysfunction by enhancing nitric oxide (NO) production derived from neuronal nitric oxide synthase (nNOS) and reducing reactive oxygen species (ROS) generation [69]. Thus, we hypothesize that Butyrivibrio may suppress LPC-induced endothelial damage, thereby inhibiting cancer cell invasion and metastasis and ultimately improving the prognosis of M-mCRC.
Moreover, our study revealed a potential role of sphingomyelins (SM) metabolism in mediating gut microbiome’s influence on the prognosis of M-mCRC. MR analysis unveiled a negative association of the abundance of genus Faecalibacterium and Ruminiclostridium5 with specific SM metabolites, including stearoyl sphingomyelin (d18:1/18:0) and sphingomyelins (d18:2/23:0, d18:1/23:1, and d17:1/24:1). Additionally, Ruminiclostridium5 was found to inhibit the levels of SM hydrolysis product ceramides. SM serves as a critical component of cellular membranes, contributing to cellular signaling pathways. Its biosynthesis can modulate the proliferation and apoptosis of cancer cells [70]. SM can be hydrolyzed by sphingomyelinase (SMase) to generate anti-proliferative molecules such as ceramide and sphingosine, as well as pro-proliferative molecules such as sphingosine-1-phosphate [71, 72]. Reduced SMase activity may contribute to aberrant SM metabolism, consequently facilitating the progression of CRC. Notably, Wu et al. found that microbial metabolite butyrate can enhance the activity of acidic SMase in human CRC HT29 cells, promoting the conversion of SM into ceramide [73], suggesting a potential correlation between the gut microbiome and SM metabolism. However, the impact of butyrate-mediated upregulation of acidic SMase expression on tumor development and progression remains to be further studied. Our study revealed a potential correlation between the abundance of butyrate-producing bacterium Faecalibacterium and SM metabolites, suggesting that gut microbiome could potentially impact the progression of CRC by acting on the cell membrane, modulating SM metabolism, and inducing aberrant signal transduction pathways. Furthermore, obesity is regarded as a risk factor for CRC. It is postulated that obesity-related dysbiosis of the gut microbiota may increase the risk of cancer [74]. Obesity-associated dysregulation of lipid metabolism has been observed to disrupt intestinal barrier integrity, consequently leading to the induction of metabolic endotoxemia which synergizes with existing adipose tissue inflammation. Intestinal inflammation and toxin-induced DNA damage in intestinal cells are considered potential mechanisms through which dysbiosis of the gut microbiome contributes to the process of carcinogenesis [75, 76]. J et al. identified alterations in the gut microbiome profile among CRC patients with obesity compared to those without obesity. Specifically, there was a reduction in butyrate-producing bacteria and an increase in opportunistic pathogens such as Prevotella, Fusobacterium nucleatum, Enterobacteriaceae, and Escherichia coli. These changes may contribute to elevated levels of pro-inflammatory cytokine IL-1β, harmful bacterial metabolite trimethylamine N-oxide (TMAO), and increased intestinal permeability observed in these patients. Thus, it is proposed that the obesity-associated gut microbiome could potentially play a role in the pathogenesis of CRC [77]. Our study provided additional insights into the potential correlation between the gut microbiome and the risk and prognosis of CRC through its impact on lipid metabolism. Nonetheless, further studies are required to validate these observations.
Furthermore, our MR results revealed a positive correlation between the abundance of genus Olsenella and three bilirubin degradation products. Additionally, the butyrate-producing bacterium Butyrivibrio exhibited a negative correlation with the ratio of Bilirubin (Z, Z) to etiocholanolone glucuronide, indicating a potential suppression of multiple UDP-glucuronosyltransferases (UGTs) corresponding to this ratio. Moreover, these metabolite/metabolite ratios were all associated with worse M-mCRC-specific survival. In hepatic cells, indirect bilirubin (IBI) can be converted to direct bilirubin (DBIL) by binding to glucuronic acid catalyzed by UGT1A1. Elevated levels of total bilirubin (TBIL) and DBIL have been found to be associated with poor survival in patients with NSCLC and CRC. Yang et al. revealed that elevated preoperative levels of TBIL and DBIL were correlated with worse OS in patients with mCRC. They further suggested that DBIL might be an independent prognostic biomarker for mCRC [78, 79]. The gut microbiome could metabolize DBIL into bilirubin derivatives, including urobilin, which can subsequently undergo reabsorption via the hepatic portal vein, thereby modulating signal transmission along the liver-gut axis [80, 81]. We have identified three bilirubin derivatives associated with the prognosis of M-mCRC as potential risk factors and hypothesized that specific gut microbiome could catalyze bilirubin degradation, which in turn affects the gut-liver axis, thereby influencing the metabolism of bilirubin and bile acids. Therefore, further investigation into the interplay of the gut microbiome with bilirubin and its degradation products is crucial for understanding how alterations in bilirubin and bile acid metabolism mediated by gut microbiome contribute to the poor prognosis of M-mCRC. In addition, members of the UGT1A, UGT2A, and UGT2B families predominantly catalyze glucuronidation. Previous investigations have shown that extensive UGT polymorphisms in UGT1A and UGT2B genes were associated with the risk of various malignancies [82, 83]. UGT1A1 gene polymorphisms are common in CRC and have been linked to diminished response rates to irinotecan-based chemotherapy regimens and adverse prognostic outcomes in patients with mCRC [84, 85]. Our study revealed that polymorphisms in several UGTs, including UGT1A1, might be influenced by specific gut microbiomes, suggesting that various potential UGT polymorphisms might affect bilirubin metabolism in M-mCRC patients, thereby potentially inhibiting chemotherapy efficacy and contributing to poor prognoses. Further research is warranted to validate and explore the impact of these polymorphisms in UGTs on M-mCRC.
Despite these significant findings, our study is still subject to several limitations. Firstly, the limited SNPs associated with gut microbiome features lead to a scarcity of IVs, potentially compromising the accuracy of exposure-outcome effect estimates. Secondly, there are limited GWAS studies of survival data in M-mCRC patients. This prevents us from validating our findings regarding gut microbiome and metabolite/metabolite ratios across diverse population cohorts. Third, although mutations in both RAS and BRAF genes can lead to abnormal activation of the MAPK signaling pathway, there are currently no GWAS specifically targeting mCRC patients with individual RAS or BRAF mutations. As a result, we cannot further analyze whether there are clinical prognostic differences in mCRC patients with RAS and BRAF mutations due to changes in the gut microbiome and metabolites. Therefore, we advocate for future genomic studies focusing on mCRC patients with individual RAS and BRAF mutations to estimate the correlation between genetic variants and mCRC-specific survival. This will facilitate comprehensive research involving multi-omics approaches, including microbiomics, proteomics, and metabolomics, in relation to mCRC risk, prognosis, and therapeutic targets. Such research could be helpful for more detailed pathogenic mechanisms and therapeutic strategies, ultimately improving the overall prognosis for these patients.
In conclusion, our study identified associations of gut microbiome and metabolites with the prognosis of M-mCRC. These findings hold significant promise in guiding the exploration of potential therapeutic targets and intervention strategies. Further studies on the microbiome and metabolomics of M-mCRC are warranted.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We appreciate all the volunteers and patients who participated in the GWAS study. We are grateful to the MiBioGen consortium study for releasing the gut microbiota GWAS summary statistics, J Brent Richards et al. and Jeremy P. Cheadle et al., for releasing the latest GWAS summary data on plasma metabolites and the GWAS summary data on M-mCRC-specific survival.
Abbreviations
- CRC
Colorectal cancer
- MAPK
Mitogen-activated protein kinase
- mCRC
Metastatic colorectal cancer
- EGFR
Epidermal growth factor receptor
- MSS
Microsatellite stable
- M-mCRC
Mutated-RAS/BRAF metastatic colorectal cancer
- MR
Mendelian randomization
- SNP
Single nucleotide polymorphism
- GWAS
Genome-wide association study
- IV
Instrumental variable
- OS
Overall survival
- CLSA
Canadian Longitudinal Study on Aging
- MAF
Minor allele frequency
- LD
Linkage disequilibrium
- IVW
Inverse variance weighted
- HP
Horizontal pleiotropy
- HMDB
Human Metabolome Database
- TCA
Tricarboxylic acid cycle
- O/P
Ornithine to phosphate ratio
- P5C
δ-1-Pyrroline5-carboxylate
- P5CS
δ-1-Pyrroline-5-carboxylate synthase
- OTC
Ornithine carbamoyltransferase
- NRP-1
Neuropilin-1
- LRS
Leucine–tRNA ligase
- PC
Phosphatidylcholine
- LPC
Lysophosphatidylcholine
- SM
Sphingomyelins
- IBI
Indirect bilirubin
- DBIL
Direct bilirubin
- TBIL
Total bilirubin
Author contribution
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Yaoxian Xiang, Dong Yan, Chan Zhang and Jing Wang. The first draft of the manuscript was written by Yaoxian Xiang and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by The Capital Health Research and Development of Special Projects (2022–2-7083).
Data availability
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Declarations
Ethics approval and consent to participate
Ethical approval was waived by the local Ethics Committee of Beijing Luhe Hospital Affiliated to Capital Medical University in view of the retrospective nature of the study and all the procedures being performed were part of the routine care. All GWAS used in this study were previously approved by respective institutional review boards (IRBs). No new IRB approval was required. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, Ferlay J, Soerjomataram I et al (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries [J]. CA: a cancer J Clin 68(6):394–424 [DOI] [PubMed] [Google Scholar]
- 2.Nitsche U, Maak M, Schuster T et al (2011) Prediction of prognosis is not improved by the seventh and latest edition of the TNM classification for colorectal cancer in a single-center collective [J]. Annals Surg 254(5):793–800 [DOI] [PubMed] [Google Scholar]
- 3.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F et al (2007) Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies [J]. Can Res 67(6):2643–2648 [DOI] [PubMed] [Google Scholar]
- 4.Li Vre A, Bachet JB, Boige V et al (2008) KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab [J]. J Clin Oncol : official J Am Soc Clin Oncol 26(3):374–379 [DOI] [PubMed] [Google Scholar]
- 5.Dienstmann R, Connor K, Byrne AT (2020) Precision therapy in RAS mutant colorectal cancer [J]. Gastroenterology 158(4):806–811 [DOI] [PubMed] [Google Scholar]
- 6.Spadoni I, Zagato E, Bertocchi A et al (2015) A gut-vascular barrier controls the systemic dissemination of bacteria [J]. Science (New York, NY) 350(6262):830–834 [DOI] [PubMed] [Google Scholar]
- 7.Sun L, Zhu Z, Jia X et al (2022) The difference of human gut microbiome in colorectal cancer with and without metastases [J]. Front Oncol 2022(12):982744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Misra BB, Liang L et al (2019) Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer [J]. Theranostics 9(14):4101–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ternes D, Tsenkova M (2022) The gut microbial metabolite formate exacerbates colorectal cancer progression [J]. Nat Metabol 4(4):458–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pavlova NN, Thompson CB (2016) The emerging hallmarks of cancer metabolism [J]. Cell Metab 23(1):27–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu B, Ackerman D, Sanchez DJ et al (2015) HIF2α-dependent lipid storage promotes endoplasmic reticulum homeostasis in clear-cell renal cell carcinoma [J]. Cancer Discov 5(6):652–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bozza PT, Viola JP (2010) Lipid droplets in inflammation and cancer [J]. Prostaglandins Leukot Essent Fatty Acids 82(4–6):243–250 [DOI] [PubMed] [Google Scholar]
- 13.Accioly MT, Pacheco P, Maya-Monteiro CM et al (2008) Lipid bodies are reservoirs of cyclooxygenase-2 and sites of prostaglandin-E2 synthesis in colon cancer cells [J]. Can Res 68(6):1732–1740 [DOI] [PubMed] [Google Scholar]
- 14.Cotte AK, Aires V, Fredon M et al (2018) Lysophosphatidylcholine acyltransferase 2-mediated lipid droplet production supports colorectal cancer chemoresistance [J]. Nat Commun 9(1):322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutton JE, Wang X, Zimmerman LJ et al (2016) Oncogenic KRAS and BRAF drive metabolic reprogramming in colorectal cancer [J]. Mole Cellul Proteomics : MCP 15(9):2924–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yachida S, Mizutani S, Shiroma H et al (2019) Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer [J]. Nat Med 25(6):968–976 [DOI] [PubMed] [Google Scholar]
- 17.Trivieri N, Pracella R, Cariglia MG et al (2020) BRAF(V600E) mutation impinges on gut microbial markers defining novel biomarkers for serrated colorectal cancer effective therapies [J]. J Exp Clin Cancer Res 39(1):285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skrivankova VW, Richmond RC, Woolf BAR et al (2021) Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE-MR statement [J]. JAMA 326(16):1614–1621 [DOI] [PubMed] [Google Scholar]
- 19.Kurilshikov A, Medina-Gomez C, Bacigalupe R et al (2021) Large-scale association analyses identify host factors influencing human gut microbiome composition [J]. Nat Genet 53(2):156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raina P, Wolfson C, Kirkland S et al (2019) Cohort profile: the Canadian Longitudinal Study on Aging (CLSA) [J]. Int J Epidemiol 48(6):1752–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Lu T, Pettersson-Kymmer U et al (2023) Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases [J]. Nat Genet 55(1):44–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maughan TS, Adams RA, Smith CG et al (2011) Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial [J]. Lancet (London, England) 377(9783):2103–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wasan H, Meade AM, Adams R et al (2014) Intermittent chemotherapy plus either intermittent or continuous cetuximab for first-line treatment of patients with KRAS wild-type advanced colorectal cancer (COIN-B): a randomised phase 2 trial [J]. Lancet Oncol 15(6):631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wills C, Watts K, Maughan TS et al (2023) Germline variation in RASAL2 may predict survival in patients with RAS-activated colorectal cancer [J]. Genes Chromosom Cancer 62(6):332–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess S, Thompson SG (2011) Avoiding bias from weak instruments in Mendelian randomization studies [J]. Int J Epidemiol 40(3):755–764 [DOI] [PubMed] [Google Scholar]
- 26.Papadimitriou N, Dimou N, Tsilidis KK et al (2020) Physical activity and risks of breast and colorectal cancer: a Mendelian randomisation analysis [J]. Nat Commun 11(1):597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shim H, Chasman DI, Smith JD et al (2015) A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians [J]. PLoS ONE 10(4):e0120758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hemani G, Tilling K, Davey SG (2017) Correction: orienting the causal relationship between imprecisely measured traits using GWAS summary data [J]. PLoS Genet 13(12):e1007149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter AR, Sanderson E, Hammerton G et al (2021) Mendelian randomisation for mediation analysis: current methods and challenges for implementation [J]. Eur J Epidemiol 36(5):465–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greco MF, Minelli C, Sheehan NA et al (2015) Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome [J]. Stat Med 34(21):2926–2940 [DOI] [PubMed] [Google Scholar]
- 31.Lu Y, Xu Z, Georgakis MK et al (2021) Smoking and heart failure: a Mendelian randomization and mediation analysis [J]. ESC heart failure 8(3):1954–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowden J, Davey Smith G, Burgess S (2015) Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression [J]. Int J Epidemiol 44(2):512–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verbanck M, Chen CY (2018) Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases [J]. Nat Gen 50(5):693–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hemani G, Zheng J (2018) The MR-base platform supports systematic causal inference across the human phenome [J]. Elife 7:e34408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R_Core_Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna,Austria. [M]. Available online: https://www.R-project.org/
- 36.Wishart DS, Feunang YD, Marcu A et al (2018) HMDB 4.0: the human metabolome database for 2018 [J]. Nucl Acids Res 46(D1):D608–D617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dennis G, Sherman BT, Hosack DA et al (2003) DAVID: database for annotation, visualization, and integrated discovery [J]. Gen Biol 4(5):P3 [PubMed] [Google Scholar]
- 38.Szklarczyk D, Gable AL, Lyon D et al (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets [J]. Nucleic Acids Res 47(D1):D607–D613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phang JM (2019) Proline metabolism in cell regulation and cancer biology: recent advances and hypotheses [J]. Antioxid Redox Signal 30(4):635–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuo CL, Chou HY, Chiu YC et al (2020) Mitochondrial oxidative stress by Lon-PYCR1 maintains an immunosuppressive tumor microenvironment that promotes cancer progression and metastasis [J]. Cancer Lett 474:138–150 [DOI] [PubMed] [Google Scholar]
- 41.Hata A, Tsuzuki T, Shimada K et al (1986) Isolation and characterization of the human ornithine transcarbamylase gene: structure of the 5’-end region [J]. J Biochem 100(3):717–725 [DOI] [PubMed] [Google Scholar]
- 42.Ding Z, Ericksen RE, Escande-Beillard N et al (2020) Metabolic pathway analyses identify proline biosynthesis pathway as a promoter of liver tumorigenesis [J]. J Hepatol 72(4):725–735 [DOI] [PubMed] [Google Scholar]
- 43.Nilsson R, Jain M, Madhusudhan N et al (2014) Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer [J]. Nat Commun 5:3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu CA, Khalil S, Zhaorigetu S et al (2008) Human Delta1-pyrroline-5-carboxylate synthase: function and regulation [J]. Amino Acids 35(4):665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donohoe DR, Holley D, Collins LB et al (2014) A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner [J]. Cancer Discov 4(12):1387–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cobo ER, Kissoon-Singh V, Moreau F et al (2017) MUC2 mucin and butyrate contribute to the synthesis of the antimicrobial peptide cathelicidin in response to entamoeba histolytica- and dextran sodium sulfate-induced colitis [J]. Infect Immun 85(3):10–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu DC, Waby JS, Chirakkal H et al (2010) Butyrate suppresses expression of neuropilin I in colorectal cell lines through inhibition of Sp1 transactivation [J]. Mol Cancer 9:276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han JM, Jeong SJ, Park MC et al (2012) Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway [J]. Cell 149(2):410–424 [DOI] [PubMed] [Google Scholar]
- 49.Bonfils G, Jaquenoud M, Bontron S et al (2012) Leucyl-tRNA synthetase controls TORC1 via the EGO complex [J]. Mol Cell 46(1):105–110 [DOI] [PubMed] [Google Scholar]
- 50.Bordonaro M, Lazarova DL (2015) Hypothesis: cell signalling influences age-related risk of colorectal cancer [J]. J Cell Mol Med 19(1):74–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoon S, Kim JH, Kim SE et al (2016) Discovery of leucyladenylate sulfamates as novel leucyl-trna synthetase (LRS)-targeted mammalian target of rapamycin complex 1 (mTORC1) inhibitors [J]. J Med Chem 59(22):10322–10328 [DOI] [PubMed] [Google Scholar]
- 52.Xu K, Yu AR, Pan SB et al (2023) Diagnostic value of methylated branched chain amino acid transaminase 1/IKAROS family zinc finger 1 for colorectal cancer [J]. World J Gastroenterol 29(36):5240–5253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pedersen SK, Symonds EL (2023) Detection of methylated BCAT1 and IKZF1 after curative-intent treatment as a prognostic indicator for colorectal cancer recurrence [J]. Cancer Med 12(2):1319–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miquel S, Mart NR, Rossi O et al (2013) Faecalibacterium prausnitzii and human intestinal health [J]. Curr Opin Microbiol 16(3):255–261 [DOI] [PubMed] [Google Scholar]
- 55.Chaput N, Lepage P, Coutzac C et al (2017) Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab [J]. Ann Oncol : official j Euro Soc Med Oncol 28(6):1368–1379 [DOI] [PubMed] [Google Scholar]
- 56.Limeta A, Ji B, Levin M et al (2020) Meta-analysis of the gut microbiota in predicting response to cancer immunotherapy in metastatic melanoma [J]. JCI insight 5(23). 10.1172/jci.insight.140940 [DOI] [PMC free article] [PubMed]
- 57.Spencer CN, Mcquade JL (2021) Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response [J]. Science 374(6575):1632–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li L, Ye J (2020) Characterization of gut microbiota in patients with primary hepatocellular carcinoma received immune checkpoint inhibitors: a Chinese population-based study [J]. Medicine 99(37):e21788 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Newsome RC, Gharaibeh RZ, Pierce CM et al (2022) Interaction of bacterial genera associated with therapeutic response to immune checkpoint PD-1 blockade in a United States cohort [J]. Gen Med 14(1):35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dikeocha IJ, Al-Kabsi AM, Chiu HT, Alshawsh MA (2022) Faecalibacterium prausnitzii ameliorates colorectal tumorigenesis and suppresses proliferation of HCT116 colorectal cancer cells. Biomedicines 10(5):1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shin NR, Whon TW, Bae JW (2015) Proteobacteria: microbial signature of dysbiosis in gut microbiota [J]. Trends Biotechnol 33(9):496–503 [DOI] [PubMed] [Google Scholar]
- 62.Frank DN, St Amand AL, Feldman RA et al (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases [J]. Proc Natl Acad Sci USA 104(34):13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sartor RB (2008) Microbial influences in inflammatory bowel diseases [J]. Gastroenterology 134(2):577–594 [DOI] [PubMed] [Google Scholar]
- 64.Zeller G, Tap J, Voigt AY et al (2014) Potential of fecal microbiota for early-stage detection of colorectal cancer [J]. Mol Syst Biol 10(11):766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abdelzaher E, Mostafa MF (2015) Lysophosphatidylcholine acyltransferase 1 (LPCAT1) upregulation in breast carcinoma contributes to tumor progression and predicts early tumor recurrence [J]. Tumour Biol :J Intl Soc Oncodev Biol Med 36(7):5473–5483 [DOI] [PubMed] [Google Scholar]
- 66.Morita Y, Sakaguchi T, Ikegami K et al (2013) Lysophosphatidylcholine acyltransferase 1 altered phospholipid composition and regulated hepatoma progression [J]. J Hepatol 59(2):292–299 [DOI] [PubMed] [Google Scholar]
- 67.Uehara T, Kikuchi H, Miyazaki S et al (2016) Overexpression of lysophosphatidylcholine acyltransferase 1 and concomitant lipid alterations in gastric cancer [J]. Ann Surg Oncol 23(2):S206–S213 [DOI] [PubMed] [Google Scholar]
- 68.Kitamura C, Sonoda H, Nozawa H et al (2019) The component changes of lysophospholipid mediators in colorectal cancer [J]. Tumour Biol J Intl Soc Oncodev Biol Med 41(5):1010428319848616 [DOI] [PubMed] [Google Scholar]
- 69.Dias MTS, Aguilar EC, Campos GP et al (2023) Butyrate inhibits LPC-induced endothelial dysfunction by regulating nNOS-produced NO and ROS production [J]. Nitric Oxide: Biol Chem 138–139:42–50 [DOI] [PubMed] [Google Scholar]
- 70.Lewis AC, Wallington-Beddoe CT, Powell JA et al (2018) Targeting sphingolipid metabolism as an approach for combination therapies in haematological malignancies [J]. Cell Death Discov 4:72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hannun YA, Linardic CM (1993) Sphingolipid breakdown products: anti-proliferative and tumor-suppressor lipids [J]. Biochem Biophys Acta 1154(3–4):223–236 [DOI] [PubMed] [Google Scholar]
- 72.Spiegel S, Milstien S (2003) Sphingosine-1-phosphate: an enigmatic signalling lipid [J]. Nat Rev Mol Cell Biol 4(5):397–407 [DOI] [PubMed] [Google Scholar]
- 73.Wu J, Cheng Y, Nsson J, B A, et al (2005) Acid sphingomyelinase is induced by butyrate but does not initiate the anticancer effect of butyrate in HT29 and HepG2 cells [J]. J lipid Res 46(9):1944–1952 [DOI] [PubMed] [Google Scholar]
- 74.Elinav E, Nowarski R, Thaiss CA et al (2013) Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms [J]. Nat Rev Cancer 13(11):759–771 [DOI] [PubMed] [Google Scholar]
- 75.Wu S, Rhee KJ, Albesiano E et al (2009) A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses [J]. Nat Med 15(9):1016–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Font-Burgada J, Sun B, Karin M (2016) Obesity and cancer: the oil that feeds the flame [J]. Cell Metab 23(1):48–62 [DOI] [PubMed] [Google Scholar]
- 77.Sánchez-Alcoholado L, Ordóñez R, Otero A et al (2020) Gut microbiota-mediated inflammation and gut permeability in patients with obesity and colorectal cancer [J]. Intl J Mol Sci 21(18):6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang L, Ge LY, Yu T, Liang Y, Yin Y, Chen H (2018) The prognostic impact of serum bilirubin in stage IV colorectal cancer patients. J Clin Lab Anal 32(2):e22272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li N, Xu M, Cai MY et al (2015) Elevated serum bilirubin levels are associated with improved survival in patients with curatively resected non-small-cell lung cancer [J]. Cancer Epidemiol 39(5):763–768 [DOI] [PubMed] [Google Scholar]
- 80.Vítek L, Majer F, Muchová L et al (2006) Identification of bilirubin reduction products formed by Clostridium perfringens isolated from human neonatal fecal flora [J] J Chromatog B. Anal Technol Biomed Life Scie 833(2):149–157 [DOI] [PubMed] [Google Scholar]
- 81.Nakamura T, Sato K, Akiba M et al (2006) Urobilinogen, as a bile pigment metabolite, has an antioxidant function [J]. J Oleo Sci 55(4):191–197 [Google Scholar]
- 82.Hu DG, Meech R, Mckinnon RA et al (2014) Transcriptional regulation of human UDP-glucuronosyltransferase genes [J]. Drug Metab Rev 46(4):421–458 [DOI] [PubMed] [Google Scholar]
- 83.Hu DG, Mackenzie PI, Mckinnon RA et al (2016) Genetic polymorphisms of human UDP-glucuronosyltransferase (UGT) genes and cancer risk [J]. Drug Metab Rev 48(1):47–69 [DOI] [PubMed] [Google Scholar]
- 84.Yu QQ, Qiu H, Zhang MS et al (2016) Predictive effects of bilirubin on response of colorectal cancer to irinotecan-based chemotherapy [J]. World J Gastroenterol 22(16):4250–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu C, Tang X, Qu Y et al (2016) UGT1A1 gene polymorphism is associated with toxicity and clinical efficacy of irinotecan-based chemotherapy in patients with advanced colorectal cancer [J]. Cancer Chemother Pharmacol 78(1):119–130 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.