Abstract
Excessive weight (overweight and obesity) is a common disorder involving genetic and environmental factors, associated with cardiovascular diseases, type-2 diabetes, and others. NOTCH1 is critical for the maintenance of stem cells and adult tissues, being reported as a key player in metabolism and adipogenesis in animals. Thus, we test the hypothesis that NOTCH1 Single Nucleotide Polymorphisms (SNPs) are associated with excessive weight. Participants from the census-based cohort SABE (Saúde, Bem Estar e Envelhecimento—Health, Well-Being, and Aging), carried out in the city of São Paulo-Brazil, were stratified into cases and controls according to BMI. We filter the SNPs located at the start and end positions of NOTCH1 and 50 Kb on both sides. We selected SNPs with minor allelic frequency (MAF) greater than or equal to 0.01 and Hardy–Weinberg equilibrium (p > 0.05) and r2 ≥ 0.8. We performed an association study with genotypes and haplotypes, as well as in silico functional analysis of the identified SNPs. We observed an association of the SNP rs9411207 with the risk of excessive weight, under log-additive model, and the genotype distribution showed an increased frequency of homozygous TT (OR 1.50, CI 1.20–1.88; p = 0.0002). The haplotype GAT constructed from this and other SNPs in high Linkage Disequilibrium was more frequent in excessive-weight individuals (p = 0.003). In silico analyses suggested that these SNPs are likely to affect the transcription of NOTCH1 and other genes involved in adipogenesis and metabolism. This is the first study reporting association between NOTCH1 SNPs and the risk of excessive weight. Considering the possibility of NOTCH1 modulation, additional population studies are important to replicate these data and confirm the usefulness of risk genotypes for management strategies of excessive weight.
Keywords: Elderly, Excessive weight, Haplotype, NOTCH1 gene, Single nucleotide polymorphism
Subject terms: Biotechnology, Genetics
Introduction
According to the World Health Organization (WHO), the frequency of people with high Body Mass Index (BMI) is rising fast worldwide1,2. Overweight and obesity (excessive weight) are expected to affect 50–60% of the adult population by 20503. In tandem with this increase, the proportion of the population above age 65 grows, and consequently, the high prevalence of those who are both obese and elderly4. These conditions are associated with a higher risk of cardiovascular diseases, type 2 diabetes (T2D), and many other chronic conditions5.
Excessive weight is a multifactorial condition characterized by abnormal and/or excessive fat cells on specific depots in the body6,7. It is genetically complex, resulting from the interaction between environmental factors and at-risk genetic profile8. The genetic component for obesity accounts for 40% to 50% of the variability in body weight status, but it varies across BMI classes, being lower among individuals with normal weight (about 30%) and higher in individuals with obesity mild to severe (60–80%). Moreover, two-thirds of the heritability of BMI can be attributed to common DNA variants. According to Bouchard, obesity-promoting alleles exert minimal effects in individuals with normal weight but may have higher penetrance in individuals prone to obesity9. In this context, several genes and pathways have been identified in animal studies, as well in linkage and association studies10.
Notch signaling is a conserved pathway that regulates cell proliferation, differentiation, self-renewal potential, apoptosis, inflammatory response, and cell-fate decisions11,12. This complex pathway involves four Notch receptors encoded by NOTCH1-4 genes, and five ligands of the Jagged/Delta-like families (JAGGED1/2, DLL1/3/4)13. In the last decade, NOTCH1 has been highlighted as a key regulator of metabolism and a player in adipogenesis, browning of adipocytes, and resistance to high-fat diet-induced obesity, both in vitro and in mice experiments14–16. Recently, Yamaguchi et al.17 incidentally found that Notch1 heterozygous-deficient (N1+/−) mice can gain weight easily.
NOTCH1 gene (9q34) is composed of 34 exons and is expressed in stem cells and most adult tissues. Cryptic changes in this cytogenetic location cause syndromic obesity in childhood18–20. NOTCH1 variants have been identified in cardiovascular malformations21, health consequences of early-life stress conditions22, and cancer23. However, to the best of our knowledge, the association between NOTCH1 variants and excessive weight has not yet been explored. Thus, considering the above experimental evidence of the involvement of NOTCH1 in metabolism and adipogenesis, we hypothesize that genetic variations in this gene are associated with excessive weight phenotype. Therefore, our main goal is to verify whether genetic variations in NOTCH1 are associated with excessive weight and related traits.
Methods
Study cohort
All participants were unrelated and selected from the interdisciplinary project named SABE (Saúde, Bem Estar e Envelhecimento—Health, Well-Being, and Aging), which was coordinated by the Pan American Health Organization (PAHO/WHO) as a multicenter health survey and well-being of elder people in seven urban centers in the Caribbean and Latin America, including São Paulo—Brazil. The SABE project was approved by the Institutional Review Board of the University of São Paulo School of Public Health and CEP/CONEP (Brazilian local and national Ethical Committee Boards, CAAE: 47683115.4.0000.5421, Review: 3.600.782). All methods were performed in accordance with the Declaration of Helsinki. The collection contains all individual-level genomic data sets that agreed to participate and signed the written informed consent form. Since then, this cohort has been analyzed in genetic studies24.
Data collection
Data collection was carried out by trained staff and described elsewhere25. A standardized questionnaire (C10) focusing on medical history, lifestyle, and sociodemographic characteristics was collected from all individuals. The C10, proposed by the PAHO, was translated and adapted for use in the Brazilian cohort. We collected blood samples by venipuncture for biochemical and genomic analysis of all participants.
The following demographic and health variables were recorded: gender, age, systolic blood pressure (SBP) (mmHg), diastolic blood pressure (DBP) (mmHg), HDL cholesterol (mg/dL), LDL cholesterol (mg/dL), total cholesterol (TC) (mg/dL), fasting triglyceride (TG) (mg/dL), fasting plasma glucose (FPG) (mg/dL), glycated hemoglobin (Hb1Ac) (%), C-Reactive Protein (hsCRP) (mg/L), BMI (kg/m2) and Waist Circumference (WC) (cm).
The T2D was identified by the question “Has a doctor or nurse ever told you that you have diabetes mellitus, that is, high blood sugar levels?”, or use of medication, or FPG > 126 mg/dl or Hb1Ac > 6.5%. Hypertension is identified by the question “Has a doctor or nurse ever told you that you have hypertension, that is, high blood pressure? To the both above questions, the response alternatives were: yes, no, do not know, and did not answer; the final two were considered as missing. In relation to smoking, we also used the information reported as current smoking (yes/no) and ex-smokers (stop smoking five years or more).
The weight and height were measured using a portable scale (Seca, Germany) with a capacity of 150 kg (sensitivity of 1 kg), and an anthropometer (Harpenden, England), respectively. The WC was measured midway between the lower margin of the last palpable rib and the top of the iliac crest using an inelastic measuring tape, to the nearest 0.1 cm after inhalation and exhalation. The BMI was calculated with the weight in kilograms divided by the square of height in meters (kg/m2). Measurement techniques were standardized according to literature and measurements were performed in triplicate using the mean values for the analysis. Using the WHO classification26, we split the individuals into groups according to the BMI values in excessive weight (≥ 25.0 kg/m2, including obesity) and normal weight (≤ 24.9 kg/m2). We excluded individuals with incomplete clinical or genetic data.
Next-generation sequencing data and tag SNPs selection
DNA extraction, whole-genome sequencing, and quality control of variants were followed as described elsewhere25. Genome sequence data were deposited in the public archive ABraOM—Arquivo Brasileiro Online de Mutações (Brazilian Online Mutations Archive, http://abraom.ib.usp.br). We used the individual European, African, Native American, and East Asian ancestries inferred by Naslavsky et al.25, as covariables in our models. In summary, Naslavsky et al. sequenced a total of N = 1200 individuals and conducted Kinship analyses, which led to the identification and exclusion of 29 closely related individuals. This process resulted in a final cohort comprising 1171 unrelated individuals. The majority of the cohort demonstrates admixture, with average global ancestry proportions of 72.6 ± 26.3% European, 17.8 ± 20.9% African, 6.7 ± 6.6% Native American, and 2.8 ± 16.2% East Asian. These proportions show partial correlation with self-declared race/ethnicities. Here, we calculated Fst using formula: Fst = (Ht − Hs)/Ht, considering the subpopulations White, Black, Mixed, Asian and Native-American for rs9411207, and the result was Fst = 0.1057, indicating moderate differentiation between populations.
We filtered SNPs located in the start and end positions of NOTCH1 plus 50-Kb on both sides, spanning Chr9:136,440,101 to 136,599,978 of the human reference sequence (GRCh38:NC_000009.12). After the exclusion of INDELs, firstly, we retained SNPs with Minor Allele Frequency (MAF) higher than or equal to 0.01, and in Hardy–Weinberg equilibrium (p > 0.05). Then, SNPs were selected using a tag SNP approach (pairwise tagging algorithm performed with a threshold of r2 ≥ 0.8) using HapMap genotype data (release 28) in Haploview 4.2 software27. Tag SNPs were included in the association analysis. The allele and genotype frequencies of associated SNPs were compared to the Allele Frequency Aggregator (ALFA) project from the National Center for Biotechnology Information (NCBI) database28.
Statistical analysis
Individuals' characteristics were compared using descriptive statistics. Categorical variables are presented as frequencies and percentages, N(%), and continuous variables are expressed as median and extreme values (minimum and maximum) for non-parametric data. The one-sample Kolmogorov–Smirnov test was used to test the normality of the distribution. Comparisons between excessive-weight and normal-weight individuals were carried out using the chi-square test and the non-parametric Mann–Whitney test.
The SNP-based association analysis was performed using the R package “SNPassoc”, under different genetic models (codominant, dominant, recessive, overdominant, and log-additive)29. Analysis was adjusted for age, gender, and ancestry (Model 1), and for all confounding variables (Model 2). Odds ratios (OR) and 95% confidence intervals (CI) were calculated by multinomial logistic regression. Haplotype blocks were defined based on Gabriel et al.30 and linkage disequilibrium (LD) plots were generated using Haploview 4.225. Haplotype frequencies were estimated by the Expectation—Maximization algorithm (EM algorithm) using the R statistical package “HaploStats”31.
We adopted the significance of p < 0.05 and Bonferroni correction for multiple test comparisons (p = 0.05/N of tag SNPs or N of Haplotypes tested) or when necessary. Statistical analysis was performed using SPSS version 27.0 (IBM, Armonk, NY, USA) and the computing environment R version 4.0.0 (R Development Core Team, 2020).
A power analysis was performed using the software G*Power version 3.1.9.2 to verify the rs9411207 association with excessive weight. The sample size was 1,024, and to perform the power analysis were considered: a significance level of 0.05, the OR of 1.5, statistical power of 90% and the expected squared coefficient of multiple correlations (R2) of 0.25 (moderate association).
In silico functional analysis
Functional annotation of the associated SNPs was obtained from the functional prediction websites: rVarBase31, HaploReg33, RegulomeDB34, and Gtex portal (Genotype-Tissue Expression)35. The rVarBase database (version 2.0 of rSNPBase) was used to describe the regulatory features of the SNP in the dimension of chromatin states, overlapping regulatory elements, and potential target genes32. HaploReg v4.133 and RegulomeDB34 were used to annotate the SNPs by systematic mining of comparative, regulatory, and epigenomic data, based on the Encyclopedia of DNA Elements (ENCODE) project. The RegulomeDB score was used to identify and compare potential regulatory variants; lower scores are associated with a wider range of data supporting functional importance. GTEx portal35 was used to determine the significant expression of quantitative trait loci (eQTL) for the associated SNPs. We also search for trait associations in GWAS Catalog.
Results
Study cohort
Of the total of individuals with phenotype and genotype data (N = 1024), 280 were normal weight (27.34%) and 744 were cases (72.65%), including 424 overweight (41.4%) and 320 obesity (31.25%). The median age was 71.31 years old (range 59 to 99 years old) and 64.28% were women. The clinical, anthropometric, and socio-demographic characteristics of the individuals, as well as the frequencies of European, African, Native American, and East Asian ancestries, are shown in Table 1. Cases showed increased values of DBP and prevalence of hypertension, even as FPG, Hb1Ac, hsCRP, TG levels, compared to normal weight (p < 0.001). Decreased levels of HDL cholesterol were observed in cases (p < 0.001). Excessive weight was less frequent in current smokers (p < 0.001) and ex-smokers (p < 0.05) individuals.
Table 1.
Sociodemographic, genetic ancestry, anthropometric, and clinical characterization of SABE cohort.
| Variables | All individuals | Normal weight | Excessive weight | p-value |
|---|---|---|---|---|
| N | 1024 | 280 (27.34%) | 744 (72.65%) | – |
| Male/female (%) | 365 (35.71%)/657(64.28%) | 115 (41.21%)/164(58.78%) | 250 (33.64%)/493(66.35%) | 0.024 |
| Age (years) | 71.31 (59–99) | 74.36 (60–99) | 68.97 (59–98) | < 0.001 |
| Ancestry frequencies | ||||
| European | 72.53% | 72.08% | 72.69% | 0.79 |
| African | 17.89% | 16.13% | 18.55% | 0.04 |
| Asian | 2.80% | 5.26% | 1.88% | 0.22 |
| Native American | 6.75% | 6.49% | 6.85% | 0.82 |
| BMI (kg/m2) | 27.45 (15.13–53.61) | 22.83 (15.13–25) | 29.2 (25.01–53.61) | < 0.001 |
| WC (cm) | 94 (62–142) | 82 (64–99) | 98 (62–142) | < 0.001 |
| Hypertension (%) | 702 (68.55%) | 167 (59.64%) | 535 (71.9%) | < 0.001 |
| SBP (mmHg) | 138 (88–250) | 135 (88–242) | 138.5 (91–250) | 0.13 |
| DBP (mmHg) | 79 (49–125) | 76 (50–117) | 80 (49–125) | < 0.001 |
| HDL (mg/dl) | 48 (19–133) | 52 (20–124) | 46 (19–133) | < 0.001 |
| LDL (mg/dl) | 126 (28–299) | 126 (41–226) | 125 (28–299) | 0.89 |
| TC (mg/dl) | 204 (86–388) | 204 (89–321) | 204 (86–388) | 0.94 |
| TG (mg/dl) | 115 (25–1283) | 101 (25–436) | 121.5 (34–1283) | < 0.001 |
| T2D (%) | 259 (25.29%) | 53 (18.92%) | 206 (27.68%) | 0.004 |
| FPG (mg/dl) | 88 (80–102) | 85 (78–95) | 90 (82–105.8) | < 0.001 |
| Hb1Ac (%) | 5.8 (4.8–13.7) | 5.7 (4.9–13.6) | 5.8 (4.8–13.7) | < 0.001 |
| hsCRP (mg/l) | 2.34 (0.14–141) | 1.43 (0.14–61.5) | 2.83 (0.14–141) | < 0.001 |
| Ex/current smoker | 375 (36.62%)/121 (11.81%) | 110 (39.28%)/52 (18.57%) | 265 (35.61%)/69 (9.27%) | < 0.001 |
Results are presented as medians and extreme values (minimum and maximum). P-values indicate differences between normal weight (BMI < 25 kg/m2) and overweight/obesity (BMI ≥ 25 kg/m2) individuals according to Mann–Whitney test for non-parametric variables and the Chi-square test for sex variables and smoking status. Ancestry frequencies were expressed as percentage of medium values. BMI body mass index, WC Waist Circumference, SBP Systolic Blood Pressure, DBP Diastolic Blood Pressure, HDL high density lipoprotein cholesterol, LDL low density lipoprotein cholesterol, TC Total Cholesterol, TG Triglycerides, T2D Type 2 Diabetes, FPG Fasting Plasma Glucose, HbA1c glycated hemoglobin, hsCRP high-sensitivity C-reactive protein.
Association of individual SNPs with excessive weight
Of a total of 3816 SNPs in the NOTCH1 region and borders (SEC16A, C9orf163, and NALT1), 566 were common variants. Of these, 453 were in HWE and 161 tag SNPs were included in the association analysis (Supplemental Table 1).
We observed an association between the SNP rs9411207 and excessive weight, after adjustment for age, gender, and ancestry (Model 1). Using Akaike’s Information Criterion (AIC), the log-additive model for this SNP best fit the data (OR 1.49; 95% CI 1.21–1.85; p = 0.0002), surviving the Bonferroni correction [p ≤ 0.0003 (0.05/161)]. We also observed, in the log-additive model, a nominal association between excessive weight and the SNPs rs2229971 (OR 1.46; 95% CI 1.17–1.81; p = 0.0005), rs11574891 (OR 1.43; 95% CI 1.15–1.77; p = 0.0012), rs3125005 (OR 1.36; 95% CI 1.10–1.67; p = 0.0038) and rs3812604 (OR 1.35; 95% CI 1.10–1.65; p = 0.0041) (Supplemental Table 2).
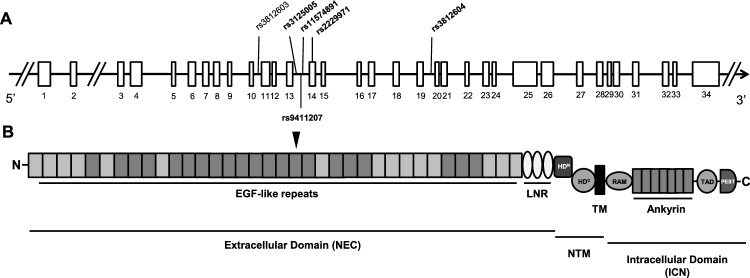
The SNPs rs9411207, rs11574891 and rs3125005 were located on intron 13; rs3812604 on intron 19. The rs2229971 (c.2265T>G; p.N775N) is a synonymous variant located on exon 14. All of these SNPs were found in the NOTCH1 gene, in the region corresponding Epidermal Growth Factor (EGF)-like domain, as shown in Fig. 1.
Figure 1.
Schematic diagram of the human NOTCH1 gene and its protein domain organization. (A) NOTCH1 gene and the positions of the investigated SNPs in our association study. Horizontal black arrows indicate the direction of transcription. The white bar represents the exonic region according to the National Center for Biotechnology Information (NCBI) and the University of California at Santa Cruz Browser (UCSC). (B) Domain organization of human NOTCH1. The NRR consists of the LNR and HD domains. HD N- and C-terminal portions of the heterodimerization domain, ANK ankyrin repeats, EGF repeats epidermal growth factor-like repeats, LNR LIN-12/Notch repeats, PEST proline/glutamic acid/serine/threonine rich domain, RAM RBP-Jκ-associated module, TM transmembrane domain, TAD transactivation domain, ICN intracellular NOTCH1, NEC N-terminal extracellular, NTM C-terminal transmembrane.
The genotypic distributions of the samples of individuals with normal weight and with excessive weight are in HWE. The worldwide frequency for the rs9411207 T allele is around 0.35 (ALFA project), and our cohort varied from 0.36 to 0.43 among the groups. Other variants analyzed also showed frequencies similar to the ALFA project (Supplemental Table 3). Genotype distribution for rs9411207 differed for ancestries, LDL and FPG. Among the marginally associated SNPs, genotype distribution also differed according to variables (Supplemental Table 4). Concerning anthropometry, the GG genotype of rs3125005 and rs3812604, and AA genotype of rs11574891 showed higher values for BMI (p < 0.0001, p < 0.0001, and p = 0.0186, respectively). The AA genotype of rs11574891 was also associated with WC (p = 0.0395). Concerning metabolic variables, the rs3125005 genotypes differed in LDL cholesterol levels; with exception of the rs11574891, all SNPs varied for FPG levels, and the rs2229971, rs3125005, and rs3812604 genotypes for Hb1Ac. Moreover, the rs3125005, and rs3812604 showed variation for hypertension (p = 0.0168) and SBP (p = 0.0307), respectively.
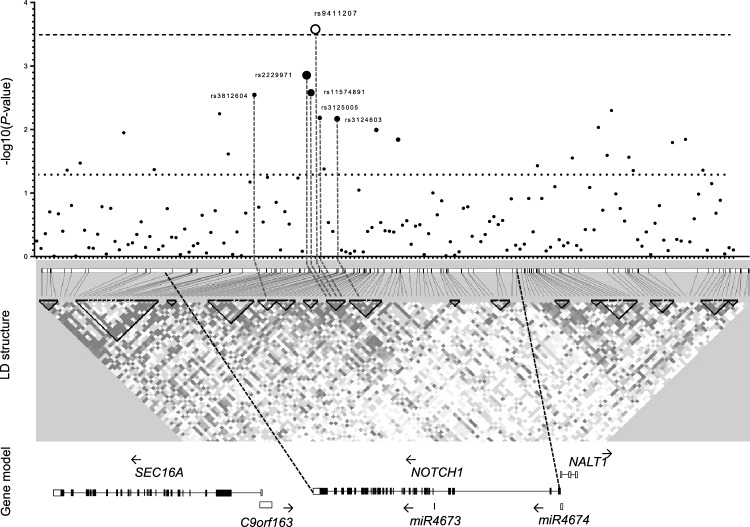
Considering that genotypes differed in relation to ancestry and the above clinical variables, we performed a multinomial logistic regression. As shown in Fig. 2, after adjusting all confounding variables in Model 2 (age, gender, ancestry, HDL, TG, hsCRP, Hb1Ac, and hypertension), the TT genotype of rs9411207 remained associated with excessive weight (OR 1.50, 95% CI 1.20–1.88; p = 0.0002) (Table 2). Genetic model analysis, along with p-values for all tagSNP are showed in Supplemental Table 5. In the Supplemental Table 6 we presented the corresponding OR, AIC and p-values for the selected SNPs.
Figure 2.
Regional association plot of NOTCH1 gene with excessive weight. –Log 10 (p-value) is shown in the upper panel, and the white circle represents the associated rs9411207, and the black circles are the marginally associated SNPs. The dimension of black circles is directly proportional to the LD (r2) with rs9411207. The dashed line represents the Bonferroni correction p-value and is the dotted line the nominal p-value. In the middle, the dashed vertical lines indicate the position of associated SNPs in the LD structure. LD blocks were shown in black triangles, representing high LD. In the lower panel, we showed the genomic structure. The LD map was created using HaploView software.
Table 2.
SNP association under the log-additive genetic model adjusted for confounding variables (Model 1and 2).
| SNP (consequence) | Genotypes | Normal weight N (%) | Excessive weight N (%) | OR (95% CI) M1 | p-value M1 | OR (95% CI) M2 | p-value M2 |
|---|---|---|---|---|---|---|---|
| rs9411207 (intronic) | CC | 116 (41.6%) | 244 (32.8%) | ||||
| CT | 124 (44.4%) | 353 (47.5%) | |||||
| TT | 39 (14%) | 146 (19.7%) | 1.49 (1.21–1.85) | 0.0002 | 1.50 (1.20–1.88) | 0.0002 | |
| rs2229971 (synonymous) | AA | 127 (45.5%) | 266 (35.8%) | ||||
| AG | 114 (40.9%) | 347 (46.8%) | |||||
| GG | 38 (13.6%) | 129 (17.4%) | 1.46 (1.17–1.81) | 0.0005 | 1.44 (1.15–1.81) | 0.0013 | |
| rs11574891 (intronic) | GG | 137 (48.9%) | 304 (40.9%) | ||||
| GA | 123 (43.9%) | 335 (45.0%) | |||||
| AA | 20 (7.1%) | 105 (14.1%) | 1.43 (1.15–1.77) | 0.0012 | 1.41 (1.12–1.77) | 0.0026 | |
| rs3125005 (intronic) | AA | 46 (16.5%) | 93 (12.5%) | ||||
| AG | 132 (47.3%) | 318 (42.8%) | |||||
| GG | 101 (36.2%) | 332 (44.7%) | 1.36 (1.10–1.67) | 0.0038 | 1.35 (1.09–1.69) | 0.0065 | |
| rs3812604 (intronic) | AA | 82 (29.4%) | 168 (22.6%) | ||||
| GA | 133 (47.7%) | 352 (47.4%) | |||||
| GG | 64 (22.9%) | 222 (29.9%) | 1.35 (1.10–1.65) | 0.0041 | 1.38 (1.12–1.71) | 0.0028 | |
| rs3124603 (intronic) | TT | 109 (38.92%) | 249 (33.46%) | ||||
| TC | 122 (43.57%) | 346 (46.50%) | |||||
| CC | 49 (17.50%) | 149 (20.02%) | 1.32 (1.07–1.63) | 0.01 | 1.36 (1.09–1.69) | 0.0068 |
CI Confidence Interval, OR Odds Ratio. Model 1 (M1): Adjusted for age, gender and ancestry; Model 2 (M2): Adjusted for age, gender, ancestry, HDL, triglycerides, hsCRP and HbA1c.
We also included the rs3124603, which has a significant increase in Model 2, it has a moderate LD with rs9411207. In this last model, the inclusion of the “smoke” variable did not alter the association for rs9411207 (OR 1.51, 95% CI 1.20–1.89; p = 0.0003) (Supplemental Table 7).
LD structure and haplotype analysis
The rs9411207 showed high LD with rs2229971 (D′ = 0.93 and r2 = 0.79) and rs11574891 (D′ = 0.99 and r2 = 0.74). We investigated the effect of the combined association of these SNPs in haplotype analysis. Considering eight possible haplotypes containing these SNPs, seven were identified (Supplemental Table 8). Of these, three had a frequency above 5% and the GAT haplotype, encompassing risk alleles, was more frequent in excessive (32.30%) than normal weight (25.66%) (OR 1.42; 95% CI 1.14–1.78, p = 0.003) (Table 3).
Table 3.
Haplotype analysis of NOTCH1 tag SNPs.
| No. | rs2229971 | rs11574891 | rs9411207 | Normal weight | Excessive weight | Odds Ratio (95% CI) | p-value |
|---|---|---|---|---|---|---|---|
| 1 | A | G | C | 62.45% | 54.97% | 1 | – |
| 2 | G | G | T | 7.14% | 6.79% | 1.08 (0.76–1.54) | 0.80 |
| 3 | G | A | T | 25.66% | 32.30% | 1.42 (1.14–1.78) | 0.003 |
In silico functional analysis
The rVarBase predicted that the rs9411207, rs2229971, rs11574891, rs3125005, rs3812604, and rs3124603 were located in a region which might regulate distally and RNA–protein bound elements. This database suggests that these SNPs interact with NOTCH1, AGPAT2, C9orf163, NALT1, and the microRNAs miR4673 and miR4674, both originating from the NOTCH1 gene sequence. These SNPs are located in the chromatin interactive region with a predominantly strong transcription function in various cell types, including adipose-derived mesenchymal stem cells and adipose nuclei (Table 4). In addition, the rs9411207 has a RegulomeDB score of 3a, and the SNPs rs11574891 and rs3125005 of 2b, predict their role as likely to affect gene expression level. Moreover, the region encompassing these SNPs is a binding site for RFX1, a transcriptional regulator. The HaploReg database further highlighted these SNPs as enhancing histone marks in the brain and muscular tissues. Of these, only the SNP rs2229971 is likely to influence histone marks in fat adipose nuclei. Using the GTEx portal, we found significant eQTL activity for the SNP rs3125005 related to CARD9 in the pancreas tissue and SEC16A in the stomach, for the SNP rs3812604 related to SDCCAG3 in the subcutaneous adipose tissue, and for rs3124603 related to NALT1 in Whole Blood and other tissues. Furthermore, GWAS Catalog showed association between the T allele of rs9411207 and NOTCH1 protein level measurement (beta = 0.107 unit increase; CI 0.08–0.134, p = 8 × 10–15).
Table 4.
In silico functional analysis of NOTCH1 tag SNPs.
| SNP | Chromatin state | Variant interacting gene | Regulome Db | Enhance histone marks | Gtex (eQTL) |
|---|---|---|---|---|---|
| rs9411207 | ↑ (ADMSC; AN) | NOTCH1; AGPAT2; C9orf163; NALT1; MIR4673; MIR4674 | 3a | BRN, MUS and 5 tissues | – |
| rs2229971 | Enhancer (AN) and ↑ (ADMSC) | NOTCH1; AGPAT2; C9orf163; NALT1; MIR4673; MIR4674 | 4 | AN, BRN, MUS and 8 tissues | – |
| rs11574891 | ↑ (ADMSC; AN) | NOTCH1; AGPAT2; C9orf163; NALT1; MIR4673; MIR4674 | 2b | BRN, MUS and 5 tissues | – |
| rs3125005 | ↑ (ADMSC; AN) | NOTCH1; AGPAT2; C9orf163; NALT1; MIR4673; MIR4674 | 2b | BRN, MUS and 5 tissues | Pancreas, Stomach and WB |
| rs3812604 | ↑ (AN) and ↓ (ADMSC) | C9orf163; NOTCH1 | 4 | 2 tissues | Adipose Subcutaneous |
| rs3124603 | ↑ (AN) and ↓ (ADMSC) | NOTCH1; NALT1; MIR4673; MIR4674 | 2b | BRN and 5 tissues | WB and 3 tissues |
Data are derived from rvarBase (https://rv.psych.ac.cn), RegulomeDB (https://www.regulomedb.org), HaploReg v4 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) and Gtex portal (https://www.gtexportal.org). ADMSC Adipose Derived Mesenchymal Stem Cell Cultured Cells, AN Adipose Nuclei, MUS skeletal muscle, WB whole blood. ↑ means strong transcription and ↓ means weak transcription.
Discussion
The evolutionarily conserved Notch pathway has emerged as a regulator of metabolism in both in vitro and animal studies14–16,36. Based on this, we evaluated the genetic association of SNPs in the NOTCH1 gene and borders with excessive weight in a Brazilian elderly cohort. We showed an independent association between the rs9411207 and the risk of excessive weight conferred by the TT genotype, which increased 1.5-fold compared to other genotypes (Table 2). This SNP is in high LD with the rs2229971 and rs11574891, and the haplotype GAT, constituted by these three SNPs, might be a risk factor for excessive weight. The rs9411207 is associated with NOTCH1 protein levels. Using in silico functional analysis, we found that these SNPs were located in a region involved in the transcriptional regulation of NOTCH1 and other genes. Moreover, the rs2229971 was located in an enhancer region.
Synonymous variants might affect nucleic acid stability, the secondary conformation of RNA, and also the structure, function, and protein levels37. In turn, SNPs located in intronic regions might have a strong functional impact by mechanisms such as alterations in the stability of mRNA, activation of cryptic splice sites, and/or loss of regulatory repressor elements38,39. Intronic SNPs have been associated with several diseases, including obesity, T2D, and other disorders, e.g. variants in FTO40–42 and TCF7L223,43,44. Although intronic variants in NOTCH1 are found associated with cardiac developmental defects45, to date, we have not identified any studies of the association between genetic variants in NOTCH1 and excessive weight or related traits.
The possibility of LD with other real functional variants or non-synonymous variants associated with the phenotype should be considered. We did not identify LD with non-synonymous variants, however, the rs9411207 and the rs11574891 are in high LD with the synonymous rs2229971, which has been associated with bicuspid aortic valve46 and cancer47,48. In line with this, both haplotype studies and in silico functional analysis provide crucial information to enhance our comprehension of the interaction between genetic variations and the traits of diseases, as well as the region responsible for regulating gene expression49. Thus, it is plausible that one of these variants is in a regulatory region and that the risk allele boosts or decreases the transcription rate50, deregulating the Notch1 signaling pathway.
In this context, according to in silico functional analysis, the rs9411207 interacts with the chromatin of the NOTCH1 and other genes. The rVarBase suggests that this region interacts with AGPAT2, which triggers the synthesis of triglycerides inside the adipocyte51 and regulates adipogenesis52. In vitro and animal models, studies showed that AGPAT2 is essential for postnatal development and maintenance of white and brown adipose tissue (WAT and BAT, respectively), along with insulin signaling53–55. Also, AGPAT2 mutations cause human lipodystrophy (OMIM 603100), a condition in which individuals, although thin, have metabolic syndrome, similar to that found in common obesity52.
Moreover, we also verified that these SNPs might interact with the long noncoding RNA gene NALT1 and the microRNAs miR4673 and miR4674, all involved in the NOTCH1 expression by different mechanisms. While overexpression of NALT1 is associated with up-regulation of the Notch1 signaling pathway56, the miR4673 and miR4674 might be involved in the inhibition or degradation of NOTCH157. Besides, the miR4673 is involved in oxaguanine-DNA repair and inflammation58 and, the miR4674 in the regulation of γ-synuclein, an adipocyte-neuron gene with increased activity in obesity and control of body lipid metabolism59,60, and angiogenesis61.
The SNPs rs11574891 and rs3125005 showed a score of 2b in Regulome DB, indicating their potential to affect gene expression levels. Also, these SNPs seem to be in a binding site for RFX1, a transcription factor important to adipogenesis and implicated in Alstron syndrome (OMIM 203800), which is considered a human model for obesity and other metabolic disorders62,63. It is important to note that the rs3125005, rs3812604, and rs3124603 are in eQTL region. The GG genotype for rs3125005 increases the CARD9 expression in the pancreas and SEC16A in the stomach; and the GG genotype for rs3812604 increases the SDCCAG3 in subcutaneous adipose tissue. The CARD9 plays a role in multiple metabolic diseases, such as obesity, insulin resistance, and atherosclerosis64. The SEC16A is a RAB10 effector required for insulin-stimulated GLUT4 trafficking in adipocytes65. SNPs in SEC16B, a SEC16A ortholog, are consistently associated with obesity risk in different populations66.
The role of the Notch pathway in adipogenesis is not fully understood. There is a crosstalk between Notch1 and Wnt signaling pathways, which in turn negatively regulates adipogenesis15,67. Several studies claim to have a positive role of NOTCH1 for the adipocyte differentiation process68–71, energy metabolism, and adipocyte browning14,72–74. Inhibition or deletion of Notch1 reduces WAT mass and increases expression of BAT-signature genes, promoting the formation of beige adipocytes7. Furthermore, it increases energy expenditure, improves insulin sensitivity, and protects mice from obesity induced by a high-fat diet75. Conversely, activation of Notch1 signaling in adipocytes is sufficient to promote a whitening phenotype in perivascular adipose tissue35.
More recently, Wan et al. (2021)76 showed that adipogenesis promoted by period circadian regulator 3 (PER3) was mediated by Notch1 pathway inhibition, and Yamaguchi et al.17 verified that haploinsufficiency of Notch1 promotes fat accumulation and adipogenesis. Yamaguchi et al. discussed that the difference between the investigations could be due to the timing of activation and dose effects on Notch1 signaling downstream transcription factors, as well as differences in the study protocols (e.g., pharmacological or genetic interference, and time course). Further analysis will be required to gain insight into the underlying mechanisms.
Our study has limitations. Older adults are particularly susceptible to sarcopenic obesity, which involves a decrease in muscle and bone mass, meanwhile an increase in fat mass. Thus, the BMI values apparently remain stable and might be overestimated for the elderly with sarcopenic obesity. We did not evaluate important environmental factors such as physical activity and diet. Despite these limitations, the strength of the present study includes the median age of our population, which exceeds the age of onset obesity and common comorbidities, thus minimizing a typical bias in the selection of the control group77. Moreover, we evaluated a multiethnic population, which might increase the potential for the identification of new genes and variants78.
Together with previous experimental findings on Notch1 signaling in adipocytes, adipogenesis, and metabolism, our results of the association study and functional in silico SNP analyses provide insights into human excessive weight phenotype. Thus, considering the advances in the knowledge of synthetic and natural NOTCH1 modulators on cancer therapies79, as well as a better refinement of overweight-related phenotypes, the validation of these results in other populations is important and might contribute to precision medicine.
Conclusion
In summary, our data suggest that the T allele and TT genotype of rs9411207, as well as GAT haplotype in the NOTCH1 gene, are associated with an increased risk of excessive weight in the Brazilian population. Although the exact mechanism accounting for their influence on excessive weight remains to be determined, our data suggest that these NOTCH1 genetic variants might affect the transcription of relevant genes for adipogenic pathways and corroborate the possibility of abnormal NOTCH1 activity, opening a new perspective of the investigation of overweight and obesity-related traits.
Supplementary Information
Author contributions
E.C.S.B. wrote the manuscript text, participated in the conceptualization, methodology, formal analysis and investigation. M.S.N., Y.A.O.D and M.Z. participated in the methodology, validation, investigation and data curation. I.S.F., J.W., L.B. and L.H.S.P. participated in the methodology and formal analysis. I.S.F., L.B. and V.P.S. participated in the writing—manuscript text. M.N., M.O.S., G.L.Y., J.W., P.S., F.P., S.L.V.V.Z., Y.A.O.D., M.R.P.B. and M.Z. participated in the writing—review and editing. F.I.V.E. participated in the conceptualization, methodology, formal analysis, investigation, writing - manuscript text, writing—review & editing, and supervision.
Funding
Fundação de Amparo à Pesquisa do Espírito Santo - FAPES to ECSB as a PhD scholarship recipient (PROCAP 2016, n. 0123/2016, SIAFEM: 73772542/16), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - CAPES to ECSB (PDSE Scholarship to Study Abroad Grant No. 19/2016), INCT/FAPESP (Grant No. 50931-3/2014), FAPESP/CEPID (Grant No. 08028-1/2013) and CNPq-Casadinho-Procad (Grant No. 552672/2011-4).
Data availability
The datasets generated and analyzed during the current study are available in the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under EGA Study accession number EGAS00001005052. EGA's website, https://ega-archive.org, has additional information. To be examined and approved by the designated Data Access Committee, all requests must be submitted through EGA (DAC) as previously described.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-65771-1.
References
- 1.Kalish, V. B. Obesity in older adults. Prim. Care43(1), 137–144 (2016). 10.1016/j.pop.2015.10.002 [DOI] [PubMed] [Google Scholar]
- 2.Sarma, S., Sockalingam, S. & Dash, S. Obesity as a multisystem disease: Trends in obesity rates and obesity-related complications. Diabetes Obes. Metab.23(S1), 3–16 (2021). 10.1111/dom.14290 [DOI] [PubMed] [Google Scholar]
- 3.Agha, M. & Agha, R. The rising prevalence of obesity: Part A: Impact on public health. Int. J. Surg. Oncol.2(7), e17 (2017). 10.1097/IJ9.0000000000000017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhana, K. et al. Obesity in older adults and life expectancy with and without cardiovascular disease. Int. J. Obes.40(10), 1535–1540 (2016). 10.1038/ijo.2016.94 [DOI] [PubMed] [Google Scholar]
- 5.Powell-Wiley, T. M. et al. Obesity and cardiovascular disease: A scientific statement from the American heart association. Circulation143, E984–E1010 (2021). 10.1161/CIR.0000000000000973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuko, O.-O. & Saito, M. Brown fat as a regulator of systemic metabolism beyond thermogenesis. Diabetes Metab. J.45, 840–852 (2021). 10.4093/dmj.2020.0291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu, J. et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell150(2), 366–376 (2012). 10.1016/j.cell.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh, S. & Bouchard, C. Convergence between biological, behavioural and genetic determinants of obesity. Nat. Rev. Genet.18(12), 731–749 (2017). 10.1038/nrg.2017.72 [DOI] [PubMed] [Google Scholar]
- 9.Bouchard, C. Genetics of obesity: What we have learned over decades of research. Obesity29(5), 802–820 (2021). 10.1002/oby.23116 [DOI] [PubMed] [Google Scholar]
- 10.Goodarzi, M. O. Genetics of obesity: What genetic association studies have taught us about the biology of obesity and its complications. Lancet Diabetes Endocrinol.6(3), 223–236 (2018). 10.1016/S2213-8587(17)30200-0 [DOI] [PubMed] [Google Scholar]
- 11.Perdigoto, C. N. & Bardin, A. J. Sending the right signal: Notch and stem cells. Biochim. Biophys. Acta Gen. Subj.1830(2), 2307–2322 (2013). 10.1016/j.bbagen.2012.08.009 [DOI] [PubMed] [Google Scholar]
- 12.Pakvasa, M. et al. Notch signaling: Its essential roles in bone and craniofacial development. Genes Dis.8(1), 8–24 (2021). 10.1016/j.gendis.2020.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuno, K. Notch signaling. Dev. Growth Differ.62(1), 3–3 (2020). 10.1111/dgd.12642 [DOI] [PubMed] [Google Scholar]
- 14.Bi, P. et al. Notch signaling regulates adipose browning and energy metabolism. Nat. Med.20(8), 911–918 (2014). 10.1038/nm.3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, M. C., Logan, H. & Newman, J. J. Distinct roles for Notch1 and Notch3 in human adipose-derived stem/stromal cell adipogenesis. Mol. Biol. Rep.47(11), 8439–8450 (2020). 10.1007/s11033-020-05884-8 [DOI] [PubMed] [Google Scholar]
- 16.Bartolome, A., Zhu, C., Sussel, L. & Pajvani, U. B. Notch signaling dynamically regulates adult β cell proliferation and maturity. J. Clin. Invest.129(1), 268–280 (2019). 10.1172/JCI98098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi, K. et al. Notch1 haploinsufficiency in mice accelerates adipogenesis. Sci. Rep.11, 1 (2021). 10.1038/s41598-021-96017-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cormier-Daire, V. et al. Cryptic terminal deletion of chromosome 9q34: A novel cause of syndromic obesity in childhood?. J. Med. Genet.40(4), 300–303 (2003). 10.1136/jmg.40.4.300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gawlik-Kuklinska, K. et al. A girl with duplication 9q34 syndrome. Am. J. Med. Genet. A143A(17), 2019–2023 (2007). 10.1002/ajmg.a.31847 [DOI] [PubMed] [Google Scholar]
- 20.D’Angelo, C. S. et al. Chromosomal microarray analysis in the genetic evaluation of 279 patients with syndromic obesity. Mol. Cytogenet.11(1), 1–18 (2018). 10.1186/s13039-018-0363-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pileggi, S. et al. Sequencing of NOTCH1 gene in an Italian population with bicuspid aortic valve: Preliminary results from the GISSI OUTLIERS VAR study. Gene715, 143970 (2019). 10.1016/j.gene.2019.143970 [DOI] [PubMed] [Google Scholar]
- 22.Steine, I. M. et al. Implication of NOTCH1 gene in susceptibility to anxiety and depression among sexual abuse victims. Transl. Psychiatry6(12), e977–e977 (2016). 10.1038/tp.2016.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva Barcelos, E. C. et al. NOTCH1-mutated chronic lymphocytic leukemia displays high endoplasmic reticulum stress response with druggable potential. Front. Oncol.13, 1218989, 1–14. 10.3389/fonc.2023.1218989 (2023). [DOI] [PMC free article] [PubMed]
- 24.Bride, L. et al. TCF7L2 rs7903146 polymorphism association with diabetes and obesity in an elderly cohort from Brazil. Peer J.9, e11349 (2021). [DOI] [PMC free article] [PubMed]
- 25.Naslavsky, M. S. et al. Whole-genome sequencing of 1,171 elderly admixed individuals from Brazil. Nat. Commun.13(1), 1–11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WHO. Obesity and Overweight. (2021). https://www.who.int/en/news-room/fact-sheets/detail/obesity-and-overweight. Accessed 29 Jul 2021.
- 27.Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics21(2), 263–265 (2005). 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 28.Phan, L. et al. ALFA: Allele Frequency Aggregator. (National Center for Biotechnology Information, U.S. National Library of Medicine, 2020). www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa.
- 29.González, J. R. et al. SNPassoc: An R package to perform whole genome association studies. Bioinformatics23(5), 644–645 (2007). 10.1093/bioinformatics/btm025 [DOI] [PubMed] [Google Scholar]
- 30.Gabriel, S. B. et al. The structure of haplotype blocks in the human genome. Science296(5576), 2225–2229 (2002). 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- 31.Schaid, D. J., Rowland, C. M., Tines, D. E., Jacobson, R. M. & Poland, G. A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet.70(2), 425–434 (2002). 10.1086/338688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo, L., Du, Y., Qu, S. & Wang, J. rVarBase: An updated database for regulatory features of human variants. Nucleic Acids Res.44(D1), D888–D893 (2016). 10.1093/nar/gkv1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward, L. D. & Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res.44(D1), D877–D881 (2016). 10.1093/nar/gkv1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyle, A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res.22(9), 1790–1797 (2012). 10.1101/gr.137323.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ardlie, K. G. et al. The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science348(6235), 648–660 (2015). 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boucher, J. M. et al. Pathological conversion of mouse perivascular adipose tissue by notch activation. Arterioscler. Thromb. Vasc. Biol.40, 2227–2243 (2020). 10.1161/ATVBAHA.120.314731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robert, F. & Pelletier, J. Exploring the impact of single-nucleotide polymorphisms on translation. Front. Genet.9, 507 (2018). 10.3389/fgene.2018.00507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooper, D. N. Functional intronic polymorphisms: Buried treasure awaiting discovery within our genes. Hum. Genomics4(5), 1–5 (2010). 10.1186/1479-7364-4-5-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaul, O. How introns enhance gene expression. Int. J. Biochem. Cell Biol.91, 145–155 (2017). 10.1016/j.biocel.2017.06.016 [DOI] [PubMed] [Google Scholar]
- 40.Frayling, T. M. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science316(5826), 889–894 (2007). 10.1126/science.1141634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tm, F. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science316(5826), 889–894 (2007). 10.1126/science.1141634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao, L., Fang, Y., Campbell, M. & Southerland, W. M. Population differentiation in allele frequencies of obesity-associated SNPs. BMC Genomics18(1), 1–16 (2017). 10.1186/s12864-017-4262-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Struan, G. et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet.38(3), 320–323 (2006). 10.1038/ng1732 [DOI] [PubMed] [Google Scholar]
- 44.Locke, A. E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature518(7538), 197–206 (2015). 10.1038/nature14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helle, E. et al. Loss of function, missense, and intronic variants in NOTCH1 confer different risks for left ventricular outflow tract obstructive heart defects in two European cohorts. Genet. Epidemiol.43(2), 215–226 (2019). 10.1002/gepi.22176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foffa, I. et al. Sequencing of NOTCH1, GATA5, TGFBR1 and TGFBR2 genes in familial cases of bicuspid aortic valve. BMC Med. Genet.14(1), 1–8 (2013). 10.1186/1471-2350-14-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, W. et al. Cigarette smoke enhances initiation and progression of lung cancer by mutating Notch1/2 and dysregulating downstream signaling molecules. Oncotarget8(70), 115128–115139 (2017). 10.18632/oncotarget.22924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jyk, C. et al. Case report: Exome sequencing reveals recurrent RETSAT mutations and a loss-of-function POLDIP2 mutation in a rare undifferentiated tongue sarcoma. F1000 Res.7, 499 (2018). 10.12688/f1000research.14383.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nianjun, L., Kui, Z. & Hongyu, Z. Haplotype-association analysis. Adv. Genet.60, 335–405 (2008). 10.1016/S0065-2660(07)00414-2 [DOI] [PubMed] [Google Scholar]
- 50.Christina, M., Alejandra, C. & Luis, D. Bicodon bias can determine the role of synonymous SNPs in human diseases. BMC Genom.18, 1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garg, A. Lipodystrophies: Genetic and acquired body fat disorders. J. Clin. Endocrinol. Metab.96(11), 3313–3325 (2011). 10.1210/jc.2011-1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subauste, A. R. et al. Alterations in lipid signaling underlie lipodystrophy secondary to AGPAT2 mutations. Diabetes61(11), 2922–2931 (2012). 10.2337/db12-0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cortés, V. A. et al. Molecular mechanisms of hepatic steatosis and insulin resistance in the AGPAT2-deficient mouse model of congenital generalized lipodystrophy. Cell Metab.9(2), 165–176 (2009). 10.1016/j.cmet.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lila, G.-H. et al. Decreased caveolae in AGPAT2 lacking adipocytes is independent of changes in cholesterol or sphingolipid levels: A whole cell and plasma membrane lipidomic analysis of adipogenesis. Biochim. Biophys. Acta. Mol. Basis Dis.1867, 9 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Kelly, C. et al. AGPAT2 is essential for postnatal development and maintenance of white and brown adipose tissue. Mol. Metab.5(7), 491–505 (2016). 10.1016/j.molmet.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piao, H. Y., Guo, S., Wang, Y. & Zhang, J. Long noncoding RNA NALT1-induced gastric cancer invasion and metastasis via NOTCH signaling pathway. World J. Gastroenterol.25(44), 6508–6526 (2019). 10.3748/wjg.v25.i44.6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hammond, S. M. An overview of microRNAs. Adv. Drug Deliv. Rev.87, 3–14 (2015). 10.1016/j.addr.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang, H. et al. MiR-4673 modulates paclitaxel-induced oxidative stress and loss of mitochondrial membrane potential by targeting 8-oxoguanine-DNA glycosylase-1. Cell. Physiol. Biochem.42(3), 889–900 (2017). 10.1159/000478644 [DOI] [PubMed] [Google Scholar]
- 59.Millership, S., Ninkina, N., Rochford, J. J. & Buchman, V. L. γ-Synuclein is a novel player in the control of body lipid metabolism. Adipocyte2(4), 276 (2013). 10.4161/adip.25162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oort, P. J. et al. Gamma-synuclein is an adipocyte-neuron gene coordinately expressed with leptin and increased in human obesity. J. Nutr.138(5), 841–848 (2008). 10.1093/jn/138.5.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Icli, B. et al. MiR-4674 regulates angiogenesis in tissue injury by targeting p38K signaling in endothelial cells. Am. J. Physiol. Cell Physiol.318(3), C524–C535 (2020). 10.1152/ajpcell.00542.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Purvis, T. L. et al. Transcriptional regulation of the Alström syndrome gene ALMS1 by members of the RFX family and Sp1. Gene460(1–2), 20 (2010). 10.1016/j.gene.2010.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hearn, T. ALMS1 and Alström syndrome: A recessive form of metabolic, neurosensory and cardiac deficits. J. Mol. Med.97(1), 1 (2019). 10.1007/s00109-018-1714-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian, C., Tuo, Y.-L., Lu, Y., Xu, C.-R. & Xiang, M. The role of CARD9 in metabolic diseases*. Curr. Med. Sci.40, 2 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Bruno, J., Brumfield, A., Chaudhary, N., Iaea, D. & McGraw, T. E. SEC16A is a RAB10 effector required for insulin-stimulated GLUT4 trafficking in adipocytes. J. Cell Biol.214(1), 61–76 (2016). 10.1083/jcb.201509052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sahibdeen, V. et al. Genetic variants in SEC16B are associated with body composition in black South Africans. Nutr. Diabetes8(1), 1–10 (2018). 10.1038/s41387-018-0050-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sarah, R. et al. Inhibition of adipogenesis by Wnt signaling. Science289(5481), 950–953 (2000). 10.1126/science.289.5481.950 [DOI] [PubMed] [Google Scholar]
- 68.Garcés, C. et al. Notch-1 controls the expression of fatty acid-activated transcription factors and is required for adipogenesis. J. Biol. Chem.272(47), 29729–29734 (1997). 10.1074/jbc.272.47.29729 [DOI] [PubMed] [Google Scholar]
- 69.Ba, K. et al. Jagged-1-mediated activation of notch signalling induces adipogenesis of adipose-derived stem cells. Cell Prolif.45(6), 538–544 (2012). 10.1111/j.1365-2184.2012.00850.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ross, D. A., Rao, P. K. & Kadesch, T. Dual roles for the notch target gene hes-1 in the differentiation of 3T3-L1 preadipocytes. Mol. Cell. Biol.24(8), 3505–3513 (2004). 10.1128/MCB.24.8.3505-3513.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Urs, S. et al. Effect of soluble Jagged1-mediated inhibition of Notch signaling on proliferation and differentiation of an adipocyte progenitor cell model. Adipocyte1(1), 46–57 (2012). 10.4161/adip.19186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gridley, T. & Kajimura, S. Lightening up a notch: Notch regulation of energy metabolism. Nat. Med.20(8), 811–812 (2014). 10.1038/nm.3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pasut, A. et al. Notch signaling rescues loss of satellite cells lacking Pax7 and promotes brown adipogenic differentiation. Cell Rep.16(2), 333–343 (2016). 10.1016/j.celrep.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shan, T., Liu, J., Wu, W., Xu, Z. & Wang, Y. Roles of notch signaling in adipocyte progenitor cells and mature adipocytes. J. Cell. Physiol.232(6), 1258–1261 (2017). 10.1002/jcp.25697 [DOI] [PubMed] [Google Scholar]
- 75.Bi, P. et al. Inhibition of Notch signaling promotes browning of white adipose tissue and ameliorates obesity. Nat. Med.20(8), 911–918 (2014). 10.1038/nm.3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wan, X. et al. hPER3 promotes adipogenesis via hHSP90AA1-mediated inhibition of Notch1 pathway. Cell Death Dis.12(301), 1–15 (2021). [DOI] [PMC free article] [PubMed]
- 77.Paolo, G. et al. Centenarians as super-controls to assess the biological relevance of genetic risk factors for common age-related diseases: A proof of principle on type 2 diabetes. Aging5(5), 373–385 (2013). 10.18632/aging.100562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Halder, I. & Shriver, M. D. Measuring and using admixture to study the genetics of complex diseases. Hum. Genomics1(1), 52–62 (2003). 10.1186/1479-7364-1-1-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gharaibeh, L., Elmadany, N., Alwosaibai, K. & Alshaer, W. Notch1 in cancer therapy: Possible clinical implications and challenges. Mol. Pharmacol.98(5), 559–576 (2020). 10.1124/molpharm.120.000006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available in the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under EGA Study accession number EGAS00001005052. EGA's website, https://ega-archive.org, has additional information. To be examined and approved by the designated Data Access Committee, all requests must be submitted through EGA (DAC) as previously described.