Abstract
The transforming proteins of the small DNA tumor viruses, simian virus 40 (SV40), adenovirus, and human papillomavirus (HPV) target a number of identical cellular regulators whose functional abrogation is required for transformation. However, while both adenovirus E1A and SV40 large T transforming properties also depend on the targeting of the transcriptional coactivator CBP/p300, no such interaction has been described for the HPV oncoprotein E6 or E7. Here, we demonstrate that the HPV-16 E6 protein, previously shown to facilitate the degradation of p53 in a complex with E6-associated protein (E6AP), also targets CBP/p300 in an interaction involving the C-terminal zinc finger of E6 and CBP residues 1808 to 1826. Furthermore, this interaction is limited to E6 proteins of high-risk HPVs associated with cervical cancer that have the capacity to repress p53-dependent transcription. An HPV-16 E6 mutant (L50G) that binds CBP/p300, but not E6AP, is still capable of down-regulating p53 transcriptional activity. Thus, HPV E6 proteins possess two distinct mechanisms by which to abrogate p53 function: the repression of p53 transcriptional activity by targeting the p53 coactivator CBP/p300, and the removal of cellular p53 protein through the proteosome degradation pathway.
The small DNA tumor viruses represented by simian virus 40 (SV40), adenoviruses, and human papillomaviruses (HPVs) have been the subject of intense study since their interactions with cellular targets provide insights into the processes involved in oncogenesis (16, 26, 40, 76). In the case of HPVs, certain types, such as HPV type 16 (HPV-16) and HPV-18, which are described as “high risk,” are associated with invasive cervical carcinoma, while other types, exemplified by HPV-6 and HPV-11, that are associated with benign lesions are described as “low risk” (9, 85). A comparison of functional differences between high-risk and low-risk HPV proteins is particularly useful in identifying important targets in tumorigenesis.
All three of these small DNA tumor viruses target regulators of the cell cycle in order to promote cell proliferation and a suitable environment for viral replication. Two of the proteins targeted for this purpose are the retinoblastoma gene product (pRB) and p53, both important inhibitors of cell cycle progression (77). By interacting with pRB, the adenovirus E1A protein (Ad E1A) (80), the SV40 large T antigen (SV40 TAg) (14), and the HPV E7 protein (17, 51) promote the dissociation of E2F from pRB (4, 10), thus inducing the expression of S-phase-specific genes (15). Targeting of the p53 protein by the adenovirus E1B 55-kDa protein (64), SV40 TAg (39, 46), and HPV E6 (79), appears to be equally important. This is illustrated by studies of retinal photoreceptor cell fate in transgenic mice (25, 59). In those cells in which only pRB has been deregulated (for example, by expression of the HPV-16 E7 protein), p53-dependent apoptosis is observed. Those cells expressing HPV E7 but lacking p53 gene expression, however, do not undergo apoptosis and may go on instead to form retinoblastomas (25). Identical results are observed when the absence of p53 gene expression is replaced by the coexpression of HPV-16 (16E6) (59). Together these results provide a convincing argument for the need to abolish p53 activity if pRB function has been abrogated and the virus is to avoid inducing host cell death. An important consequence of this viral strategy is that host cells in which p53 function has been abolished are compromised in the ability to mediate a response to the induction of DNA damage (19, 32, 68, 74). Subsequently, this may result in the accumulation of genetic changes that are associated with tumorigenesis.
The mechanism by which 16E6 down-regulates p53 activity has been shown to involve the active promotion of p53 degradation through the ubiquitin-dependent proteolytic pathway (28, 66, 70). 16E6 achieves this by forming a complex with E6-associated protein (E6AP), a cellular protein that acts as a ubiquitin ligase (65). The ability to form an E6-E6AP-p53 complex appears to be limited to high-risk E6 proteins (28, 79).
Another important cellular target of the Ad E1A and SV40 TAg proteins is the transcriptional coactivator CBP/p300 (2, 18, 47). Through the interaction with specific transcription factors, CBP/p300 regulates a variety of signal-modulated events (29). The mechanisms by which CBP/p300 activates gene expression include (i) the ability to modify histones and nonhistone transcription factors through intrinsic or associated acetyltransferase activity (6, 23, 58, 83) and (ii) bridging the gap between DNA-bound transcription factors and components of the general transcription machinery (55).
Increasing evidence suggests that there is also a role for CBP/p300 in the inhibition of cell cycle progression and cellular differentiation (20). This may explain, at least in part, why CBP/p300 is the target of SV40 and Ad E1A proteins. Recently published data have also demonstrated that CBP/p300 activates p53-dependent transcription (3, 24, 45, 67). Thus, part of the cell cycle-inhibitory properties of CBP/p300 may result from its involvement in p53-regulated events. Indeed, one mechanism by which SV40 and adenoviruses can abrogate p53 function is by targeting the p53 cofactor CBP/p300, and at least for Ad E1A, it has been shown that CBP-binding-deficient mutants are no longer capable of down-regulating p53-dependent transcription (24, 45, 67).
Interestingly, the down-regulation of p53-dependent transcription in vivo is not limited to SV40 TAg and Ad E1A but has also been demonstrated for high-risk HPV E6 proteins (49). However, to date no interaction with the transcriptional coactivator CBP/p300 has been described for the HPV E6 oncoprotein. It could be argued that the ability of high-risk HPV E6 proteins to degrade p53 through the E6AP pathway might be sufficient to explain the abrogation of p53 transcriptional activity. However, adenoviruses also possess the capacity to degrade p53 via the E1B 55-kDa protein (63, 69) and yet still target p53 transcriptional properties through an E1A-CBP/p300 interaction.
For both SV40 and adenoviruses, the targeting of CBP/p300 has been shown to be a prerequisite for transformation (18, 50, 81). In the light of this, and the ability of E6 to interact with multiple cellular targets (36), we wondered whether the 16E6 protein might also target the transcriptional coactivator CBP/p300.
Here we provide evidence that HPV E6 proteins can indeed interact with the transcriptional coactivator CBP/p300. In line with the observation that CBP/p300 is an important target for the transformation process, we also demonstrate that only E6 proteins from high-risk HPVs are capable of binding CBP/p300. Finally, we provide evidence that the ability to bind to CBP/p300 correlates with the down-regulation of p53 transcriptional activity in a manner similar to that of the Ad E1A protein.
MATERIALS AND METHODS
Plasmid constructs.
The vector used for the expression of glutathione S-transferase (GST) fusion proteins, unless otherwise stated, was pGEX-2TKP (a modified version of the Pharmacia pGEX-2TK vector containing a new polylinker), which was a gift from T. Kouzarides. Also a gift from T. Kouzarides were plasmids GST-CBP I (residues 461 to 662), GST-CBP II (residues 1621 to 1877), GST-P/CAF, GST-E1A [1–90] (residues 1 to 90 of Ad E1A), G5E1BCAT, pHK3NVP16, pHKnTCBP1VP16, pHKnCBP2VP16, and pHKGT. This last construct contains the DNA binding domain of GAL4 (residues 1 to 147) driven by an SV40 promoter and was used to create the GAL4-HPV E6 fusion proteins GAL-11E6 and GAL-16E6. Plasmids GST-11E6 and GST-16E6 were created by inserting HPV-11 and HPV-16 E6 sequences amplified by PCR into pGEX2TKP. The GST-6E6 and GST-18E6 constructs used to express the full-length HPV-6 and 18 E6, proteins respectively, were a kind gift from D. Pim (60). The GST-E6AP expression vector was kindly provided by P. Howley (28). The GST-CBP constructs described in Fig. 2A were created by cloning PCR-amplified fragments into pGEX2TKP. The maltose binding protein (MBP)-CBP fusion construct was created by cloning a CBP fragment (residues 1808 to 1852) from pGEX2TKP into the vector pMALP, a modified version of pMAL (New England Biolabs [NEB]) containing a new polylinker that was a gift from E. Manser. Also provided by E. Manser was the in vitro transcription and mammalian expression vector pXJ-FLAG (48). The original DNA for the 16E6 mutant L50G was a kind gift from T. Kanda (54) and was cloned via the BamHI and XhoI restriction sites into pXJ-FLAG. All the other constructs used for in vitro transcription reactions were cloned BamHI/HindIII into pXJ-FLAG.
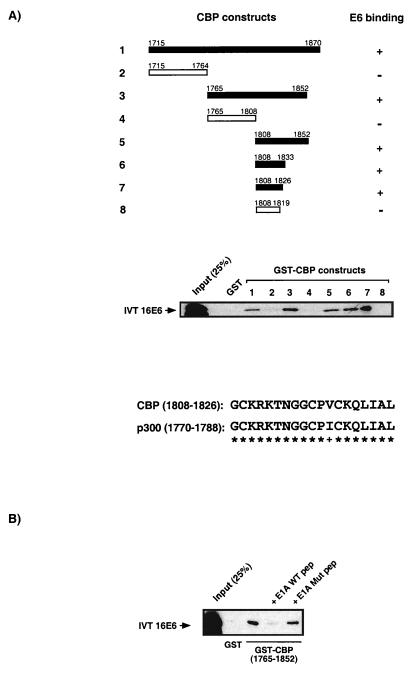
FIG. 2.
Identification of an HPV-16 E6 binding site on CBP/p300. (A) GST-CBP fusion constructs used in micro-affinity column experiments to define CBP sequences capable of binding 16E6 are shown schematically. A 19-amino-acid sequence of CBP (residues 1808 to 1826; lane 7) and larger fragments containing this sequence are able to bind 16E6. Deletion into these sequences abolishes E6 binding (lane 8). Also shown is an alignment of this 19-residue binding site of CBP and the corresponding p300 sequence. Eighteen asterisks represent the conservation of 18-amino-acid residues in that sequence, while + represents the single conservative change. (B) 16E6 and Ad E1A bind the same 19-amino-acid motif in CBP. The interaction between 16E6 and GST-CBP (1765 to 1852) can be disrupted by the presence of a peptide (pep) consisting of Ad E1A sequences previously shown to bind the CBP TRAM (57a). This observation is specific because a peptide containing mutant Ad E1A sequences fails to prevent the E6-CBP interaction. WT, wild type; Mut, mutant.
Expression of recombinant bacterial fusion proteins.
GST fusion and MBP fusion proteins were expressed in Escherichia coli, extracted with lysis buffer (50 mM Tris-HCl [pH 8.0], 0.5 mM EDTA, 5 mM dithiothreitol [DTT], 15% glycerol, 1 mg of lysozyme per ml, 1 mM phenylmethylsulfonyl fluoride), and, after sonication and centrifugation, stored at −70°C.
Partial purification of CBP/p300 from HeLa nuclear extract.
HeLa nuclear extract was diluted 1:3 with 20 mM morpholineethanesulfonic acid buffer (pH 6.1)–10 mM NaF–0.1% Triton X-100 before being passed over a 0.2-ml SP Sepharose ion-exchange column (Pharmacia) equilibrated with the same buffer. Elution of CBP/p300 using a step gradient of increasing [KCl] was maximal in the 300 and 400 mM KCl fractions (as determined by Western blot analysis using an anti-p300 antibody [data not shown]). These two fractions were pooled and then diluted 1:7 with binding buffer (20 mM Tris-HCl [pH 8.0], 0.5 mM EDTA, 0.5 mM DTT, 20% glycerol, 01% Nonidet P-40 [NP-40]) to bring the final salt concentration down to 50 mM KCl. The partially purified CBP/p300 was then passed over GST fusion protein micro-affinity columns as described below.
Detection of protein-protein interactions by using micro-affinity columns.
Bacterial lysate containing GST fusion protein was incubated with glutathione Sepharose beads (Pharmacia) for 30 min at 4°C in 1× NENT buffer (100 mM NaCl, 1 mM EDTA, 0.5% NP-40, Tris-HCl [pH 8.0]). After spinning down and washing with 1 ml 1× NENT, the beads were loaded into a yellow Gilson pipette tip containing a glass bead (BDH catalog no. 332134Y) to create a 25-μl GST micro-column. For MBP micro-columns, a similar approach was taken in which amylose resin (NEB) was used in place of glutathione-Sepharose beads. These columns were then used to detect interactions with in vitro-translated (IVT) and radiolabelled proteins, bacterially expressed fusion proteins, or partially purified nuclear CBP/p300.
For IVT proteins, expression and incorporation of [35S]methionine was performed by using TNT kits (Promega) according to the manufacturer’s recommendations. After a 1-h incubation at 30°C, 40 μl of a 50-μl IVT reaction was diluted with 360 μl of IPD buffer (50 mM KCl, 40 mM HEPES [pH 7.5], 5 mM 2-β-mercaptoethanol, 0.1% Tween 20, 0.5% milk) before being passed over the GST micro-column. After washing the column twice with 200 μl wash buffer (IPD buffer containing 150 mM KCl), proteins were eluted from the column by adding 25 μl of 2× sodium dodecyl sulfate (SDS) loading dye, heating to 95°C for 5 min, chasing with 25 μl water, and spinning in a micro-centrifuge. Samples were then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), and after staining and drying of the gel, proteins were detected via exposure to autoradiographic film.
To detect the interaction between two bacterially expressed recombinant proteins, GST or MBP fusion proteins were passed over MBP or GST fusion micro-affinity columns, respectively. After purification of the target recombinant fusion protein on glutathione-Sepharose beads or amylose resin, the proteins were eluted in recombinant binding buffer (RBB; 25 mM HEPES [pH 7.6], 50 mM KCl, 12.5 mM MgCl2, 10% glycerol, 0.1% NP-40) containing either 10 mM reduced glutathione (for GST proteins) or 20 mM maltose (for MBP fusion proteins). After passage of the recombinant target protein in RBB over the micro-affinity column and washing (using RBB containing 150 mM KCl), the samples were eluted as described above for IVT proteins and run on SDS-gels. Samples were then transferred onto polyvinylidene difluoride (PVDF) membranes, and GST or MBP fusion proteins were detected by using the appropriate antibodies by Western blot analysis (see below). A similar analysis was performed for the partially purified nuclear CBP/p300 proteins. In this case, the binding buffer consisted of 20 mM Tris-HCl (pH 8.0), 50 mM KCl, 0.5 mM EDTA, 0.5 mM DTT, 20% glycerol, and 0.1% NP-40. Detection of CBP/p300 was by Western blot analysis using antibodies specific for these proteins.
Western blot analysis.
Proteins analyzed by SDS-PAGE on 0.75-mm-thick gels were blotted onto PVDF membranes (NEN) overnight. The membranes were blocked with 5% (wt/vol) nonfat dry milk in TBST (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% Tween 20). A 1-h incubation at room temperature with the first monoclonal antibody was followed by washing with TSBT. The membrane was then incubated for 30 min with a horseradish peroxidase-coupled second antibody (1:4,000; DAKO) before washing in TSBT. Proteins were visualized with hyperfilm in the presence luminol (Amersham) for 10 to 60 s, depending on signal intensity. The commercially available monoclonal antibodies used in this study included the anti-p300 antibody p300 Ab-1 (Calbiochem), the anti-CBP/p300 antibody NM11 (Pharmingen), the anti-GST antibody B14 (Santa Cruz Biotechnology), and the anti-MBP antibody (NEB).
In vitro competition assays using recombinant proteins.
For analysis of the disruption of in vitro binding of p53 and CBP by 16E6 protein, His-tagged p53 in bacterial lysate was first bound to nickel-nitrilotriacetic acid agarose beads (Qiagen) before incubation for 30 min with 3 μg of purified MBP-CBP II and a various amount of purified GST or GST-16E6 (0.5, 5, or 10 μg) in IPD buffer. After the beads were washed twice in IPD buffer containing an additional 200 mM KCl, proteins were eluted by heating the beads to 95°C in 2× SDS loading dye for 5 min. Samples were then run on an 10% SDS-polyacrylamide, gel and the proteins were subjected to Western blot analysis using an anti-MBP antibody (NEB).
Mammalian two hybrid-experiments.
To study protein-protein interactions in vivo, we made use of a GAL4-VP16 chloramphenicol acetyltransferase (CAT) reporter system described previously (5). Full-length 11E6 and 16E6 sequences were fused to the DNA binding domain of GAL4, resulting in the constructs pGAL4-11E6 and pGAL4-16E6, respectively. U2-OS cells were cotransfected with 1 μg pGAL4-11E6 or pGAL4-16E6 and 4 μg pG5E1BCAT (a CAT reporter vector containing multiple GAL4 DNA binding sites). Cotransfected together with these plasmids was either pHK3NVP16 (the activation domain of VP16, residues 415 to 490, driven by the SV40 promoter), 2 μg of pHKnTCBP1VP16 (expressing CBP residues 461 to 662 in frame with the VP16 activation domain), or 2 μg of pHKnCBP2VP16 (expressing CBP residues 1621 to 1877 in frame with the VP16 activation domain); 48 h after transfection, the cells were harvested and CAT assays were performed as described below.
In vitro and in vivo p53 degradation assays.
E6-mediated degradation of p53 was assayed by previously described methods (54, 65, 57a). For in vitro degradation of p53, 12.5 μl of IVT E6 protein was mixed with 2 μl of IVT 35S-labelled p53 in a total volume of 25 μl of assay buffer (25 mM Tris-HCl [pH 7.5], 100 mM NaCl, 3 mM DTT). The sample was then incubated at room temperature for 30, 90, or 180 min. At the indicated time points, the reaction was stopped by adding 2× SDS loading dye and boiling for 5 min. The samples were then analyzed by SDS-PAGE and autoradiography. In vivo degradation assays were performed as previously described (57a). U2-OS cells were transfected with 0.1 μg of a p53 expression vector and 1 μg of different E6-expressing vectors; 24 h after transfection, cells were shifted to medium containing 25 μg of cycloheximide per ml. Cells were harvested 0, 1, and 3 h after cycloheximide treatment as previously described (57a). Cell lysates were subjected to SDS-PAGE and analyzed by Western blotting using the anti-p53 antibody DO-1 (Santa Cruz).
Transfections and CAT assays.
U2-OS cells were plated onto 10-cm-diameter culture dishes and transfected at 50 to 70% confluency, using Lipofectin reagent (GIBCO-BRL). For the p53 transcription studies, 1 μg of the p53 reporter PG13CAT or the control vector MG15CAT was cotransfected with 2 to 5 μg of expression vectors for HPV E6 proteins, Ad E1A, or full-length CBP. Expression plasmids used in transfections included those based on the expression vector pXJ-FLAG (for E6 constructs), the Rous sarcoma virus-driven Ad5 12S E1A expression plasmid pBJ9Ω (a gift from H. Land), and plasmid pRc/RSV-mCBP.HA.RK (a gift from R. Goodman). CAT assays have been described elsewhere (56), and the data presented, unless otherwise stated, represent between three and eight experiments using at least two independent DNA preparations.
RESULTS
The 16E6 protein interacts with full-length nuclear CBP/p300.
To determine whether the 16E6 protein could interact with CBP/p300, we partially purified these transcriptional coactivators from HeLa nuclear extract (see Materials and Methods) and then passed the fraction enriched for CBP/p300 over an E6 affinity column. Western blot analysis using the monoclonal antibodies p300 Ab-1 (Fig. 1A) and NM11 (data not shown) detected a specific interaction between CBP/p300 and GST-16E6. No interaction was detected for the control GST column, even though a greater amount of protein was used. Also shown in Fig. 1A is the interaction between full-length nuclear CBP/p300 and GST-P/CAF and GST-YY1, both of which have previously been shown to interact with CBP/p300 (43, 57, 83). These data provide the first evidence that a papillomavirus oncoprotein can associate with the transcriptional coactivator CBP/p300, although they do not provide information about the nature of the interaction.
FIG. 1.
16E6 interacts with the transcriptional coactivator CBP/p300. (A) Equal amounts of partially purified full-length (FL) CBP/p300 from HeLa nuclear extract were passed over GST, GST-16E6, GST-P/CAF, and GST-YY1 micro-affinity columns. After SDS-PAGE and transfer to PVDF membranes, Western blot analysis detected the presence of CBP/p300. The positions of the molecular weight markers are also indicated. (B) GST micro-affinity columns were used to detect the interaction of IVT radiolabelled 16E6 with GST-CBP II (residues 1621 to 1877). No interaction was detected for the control GST column or the GST-CBP I (residues 461 to 662) column. The lower panel shows the Coomassie blue-stained SDS-gel of GST and GST fusion proteins eluted from the micro-affinity columns and also shows the molecular weight marker ladder. (C) Comparison of the 16E6-CBP II interaction with known E1A-CBP II and 16E6-E6AP interactions in GST micro-affinity column assays. (D) Demonstration of a direct interaction between 16E6 and CBP using two recombinant bacterially expressed proteins. GST or GST-E6 was passed over a column containing MBP-CBP (residues 1808 to 1852) fusion protein. Bound GST-fusion protein was detected by Western blot analysis using an anti-GST antibody. The MBP-CBP fusion protein was also passed over a GST or GST-E6 column, and the interaction detected with an anti-MBP antibody.
Both Ad E1A and SV40 TAg bind the CBP II domain of CBP (residues 1621 to 1877; also referred to as the C/H3 domain), which represents a hot spot for transcription factor interactions (30). We tested whether 16E6 was also able to bind to this region of CBP, using a micro-affinity column containing GST-CBP (1621 to 1877). In Fig. 1B it can be seen that IVT radiolabelled 16E6 does indeed bind to the GST-CBP II domain but not to GST or to GST-CBP I (461 to 662), another region of CBP that binds multiple cellular transcription factors (30).
To gain an insight into the relative strength of the E6-CBP II association, we compared this protein-protein interaction with two previously described interactions, namely, that of E1A and the CBP II domain (2, 5, 47), and the binding of HPV E6 to the cellular factor E6AP (28). As can be seen from the results presented in Fig. 1C, the association of HPV-16 E6 with the CBP II domain is similar in strength to those seen with the two previously documented interactions. Nevertheless, it should be noted that while 16E6 and Ad E1A bind the CBP II domain at comparable levels, E1A binds full-length nuclear CBP/p300 with a much higher affinity (data not shown). This is most likely due to the fact that E1A can bind multiple sites on CBP/p300 in addition to the CBP II domain (37).
Figure 1D demonstrates that the interaction between 16E6 and the CBP II domain can occur directly, since binding can be detected using only purified, recombinant proteins. The interaction of GST-16E6 with an MBP-CBP affinity column was detected by Western blot analysis using anti-GST antibodies, while in the reciprocal experiment MBP-CBP binding to a GST-E6 column was detected with an anti-MBP antibody. Together, these results provide evidence that the HPV-16 E6 protein can associate with full-length nuclear CBP/p300 via the CBP II domain in an interaction that is most likely direct.
Characterization of the HPV-16 E6-CBP interaction.
To determine the E6 binding site within the CBP II domain, we used a number of GST-CBP constructs in micro-affinity column assays with IVT radiolabelled full-length HPV-16 E6 protein (Fig. 2A). We were able to identify a 19-amino-acid region of CBP (1808 to 1826) that was capable of binding full-length E6 (lane 7). Deletion into this sequence abolished binding to the E6 protein (lane 8). It can be seen from Fig. 2A that the 19-amino-acid sequence is virtually identical in both CBP and p300, with only one conservative change present, and a comparable level of conservation is also observed for CBP/p300 proteins from other species that are not shown here.
Interestingly, we have recently demonstrated that this same 19-amino-acid sequence of CBP (1808 to 1826), which we have termed a transcriptional adapter motif, or TRAM, binds numerous cellular factors and is also targeted by the Ad E1A protein (57a). Confirmation that 16E6 and Ad E1A both bind the CBP TRAM is demonstrated in Fig. 2B. An E1A peptide that can bind the CBP TRAM is capable of inhibiting the 16E6-CBP interaction, while a mutant version of the E1A peptide that is incapable of binding the CBP TRAM (57a) lacks this capacity.
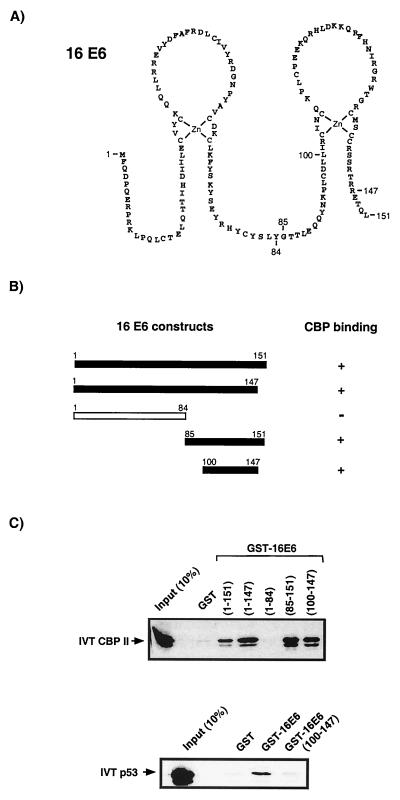
To identify the CBP binding region within 16E6, a similar binding assay was performed. As can be seen from Fig. 3, removal of the C-terminal residues 148 to 151, which have been implicated in the binding of another E6-interacting protein, hDLG (34, 44), had no effect on CBP binding. Dissection of the 16E6 protein into N-terminal (1 to 84) and C-terminal (85 to 151) halves demonstrated that while the N terminus of E6 does not bind CBP, the C-terminal half of the protein maintained the ability to bind CBP. Further analysis demonstrated that a smaller C-terminal region (amino acids 100 to 147) within the proposed second zinc finger structure of 16E6 also maintained the ability to bind CBP. This same region of 16E6 does not, however, bind IVT radiolabelled p53 protein (Fig. 3C), demonstrating that these two properties, namely, CBP and p53 binding, are separable.
FIG. 3.
Mapping of a 16E6 region involved in the interaction with CBP. (A) Amino acid sequence of the 16E6 protein. The two zinc finger structures are hypothetical, since they have not been based on spectroscopic methods but are supported by the strict conservation of eight cysteine residues in all HPV E6 proteins, as well as by the stoichiometric binding of zinc ions by E6 molecules (7, 21). Indicated are the numbers of the amino acid residues which mark the start or end points of 16E6 fragments used in interaction studies. (B) Schematic representation of GST-E6 fusion constructs used in micro-affinity column assays. (C) Interaction experiments define a region between 16E6 residues 100 to 147 as sufficient for the binding of CBP. This same region is not able to bind p53, however, indicating that p53 and CBP binding are dependent on different 16E6 domains.
In the context of a GST fusion protein (E6 amino acids 100 to 142), the cysteine residues (C103, C139, and C140) could be substituted with glycine residues without affecting the ability to bind CBP (data not shown). This finding suggests that specific sequences within the second zinc finger of E6 are involved in the interaction with CBP and that, at least in this context, an intact zinc finger structure is not necessary. However, we have not tested these mutations in the context of full-length E6 proteins alone and therefore cannot rule out the possibility that an intact zinc finger structure is necessary to present the E6 residues contacting CBP under these conditions.
E6 proteins of high-risk but not low-risk HPV types bind CBP/p300.
It has been suggested that functional differences between the E6 and E7 proteins of different HPV types represent a cardinal factor in their ability to transform cells and are reflected by their classification as either high-risk or low-risk types (8, 77, 85). In the case of other DNA tumor virus proteins, such as the Ad E1A protein and the SV40 TAg, interaction with the transcriptional coactivator CBP/p300 has been shown to be absolutely required for their transforming capabilities (18, 50, 81). If CBP/p300 is considered an important target in the transformation processes of other DNA tumor viruses, we postulated that an ability to target CBP/p300 might also be an important factor in distinguishing E6 proteins of high-risk from low-risk HPVs. Consequently, we compared the abilities the E6 proteins from two high-risk types (HPV-16 and HPV-18) to bind CBP with those from two low-risk types (HPV-6 and HPV-11).
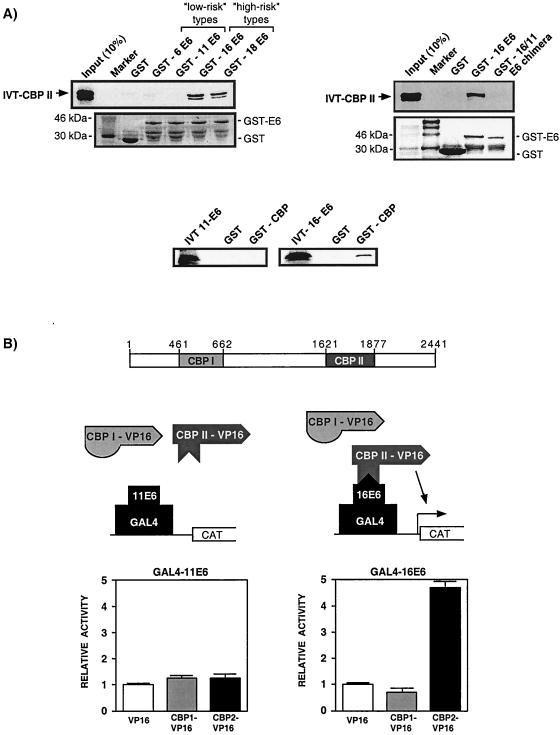
In fact, Fig. 4A clearly demonstrates that only the GST-E6 proteins of the high-risk types (HPV-16 and HPV-18) bind to IVT CBP II, while those of the low-risk types (HPV-6 and HPV-11) fail to bind CBP. This observation is reproducible, as can be seen from the inability of IVT 11E6 protein to bind to a GST-CBP affinity column. Also shown in Fig. 4A is the inability of a chimeric 16/11 E6 protein to bind CBP II. In this E6 protein, all amino acids are from HPV-16 except those from positions 107 to 135, which represent HPV-11 sequences. This substitution mutation of 29 amino acids contained within a region of HPV-16 implicated in Fig. 3 to bind CBP again suggests a fundamental difference in the ability of E6 proteins from high-risk and low-risk types to bind CBP in vitro.
FIG. 4.
The E6-CBP/p300 interaction is specific for E6 proteins of high-risk HPVs. (A) Micro-affinity column experiments using either GST fusion or IVT E6 proteins demonstrate that only E6 proteins from the high-risk HPV types 16 and 18, but not the low-risk HPV types 6 and 11, are capable of interacting with CBP. Replacing the 16E6 sequences (residues 107 to 135) that have been implicated in CBP binding with those from 11E6 results in a chimeric protein (GST-16/11) that can no longer bind CBP. (B) Mammalian two-hybrid experiments (described in Materials and Methods), shown schematically, indicate that the distinction between E6 proteins of high-risk and low-risk HPVs extends to the in vivo interaction with the CBP II domain. Activation of the G5E1BCAT reporter is seen only after cotransfection of GAL4-16E6 and CBP II-VP16, not for those experiments in which GAL4-11E6 or CBP I-VP16 proteins were expressed.
This difference in CBP binding is also observed in vivo, as demonstrated by the mammalian two-hybrid assay presented in Fig. 4B. Transient cotransfection experiments were performed with U2-OS cells in which a CAT reporter construct, driven by multiple GAL4 binding sites (G5E1BCAT), was introduced along with either an expression vector for full-length 11E6 fused to the DNA binding domain of GAL4 (GAL4-11E6) or a similar construct containing HPV-16 sequences (GAL4-16E6). Activation of transcription was then determined for those cells containing these two plasmids in conjunction with either the expression vector for the VP16 activation domain alone, VP16 fused to the CBP I domain, or VP16 fused to the CBP II domain. The level of CAT activity obtained with cells cotransfected with the VP16 activation domain was set to 1 and the CAT activity of cells receiving either CBP I-VP16 or CBP II-VP16 was then compared to this.
As can be seen from the results in Fig. 4B, cells containing GAL4-16E6 could be activated by CBP II-VP16, while those containing the GAL4-11E6 expression vector could not. This effect was specific for the 16E6-CBP II interaction, since VP16 sequences fused to the CBP I domain failed to activate GAL4-16E6. Taken together, these results strongly suggest that there are functional differences between high-risk and low-risk proteins with respect to their ability to bind CBP/p300 both in vitro and in vivo.
The down-regulation of p53 transcriptional activity by HPV-16 E6 correlates with CBP binding.
One of the main functions proposed for E6 proteins of high-risk HPVs is the targeting of p53 in order to suppress apoptosis of the host cell (27, 53, 77). In the last few years, many lines of evidence have suggested that one way in which this might be achieved is by stimulating the degradation of p53 through the ubiquitination pathway (65, 66). Evidence has been provided both in vitro (28) and in vivo (70) that this activity is dependent on the interaction of E6 with a cellular factor termed E6AP which then acts as a ubiquitin ligase (65). The ability of E6 proteins to interact with E6AP has been shown to be limited to those of high-risk HPV types (79). It has also been reported previously that E6 proteins of high-risk but not low-risk types are able to down-regulate p53 transcriptional activity (49). One explanation for these observations is that down-regulation of p53-dependent transcription results from the E6AP-dependent degradation of p53.
Recently, it was also shown that p53-dependent transcription can be activated by CBP/p300 and that this activation can be abrogated by wild-type E1A but not a CBP-binding deficient mutant of E1A (3, 24, 45, 67). The results presented here have demonstrated that, like E1A, E6 proteins of high-risk HPVs can also target CBP/p300. We therefore examined whether the down-regulation of p53 transcriptional activity by E6 proteins could be achieved through the binding of CBP/p300 in a manner analogous to E1A.
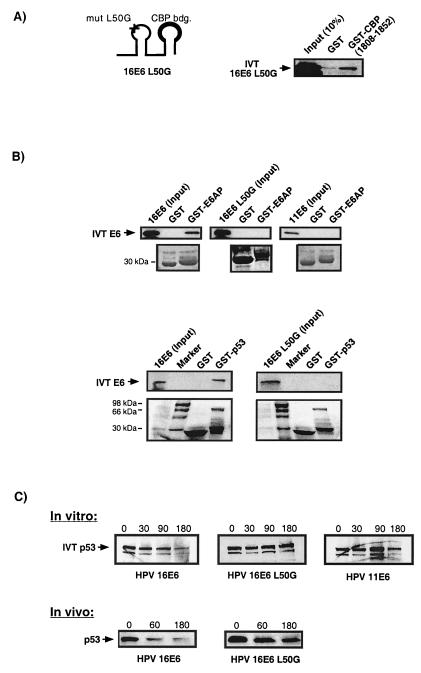
To answer this question, we required a 16E6 mutant that was deficient in targeting p53 for degradation through the E6AP pathway yet was still capable of binding CBP/p300. We assessed a number of existing 16E6 mutants before finding one with the desired properties. The 16E6 mutant L50G contains a point mutation in the first zinc finger of the E6 protein and has previously been shown to be p53 degradation deficient (54). Figure 5A demonstrates that this mutant is still able to interact with CBP in binding assays. However, when we tested this mutant for its ability to bind either E6AP or p53 in a similar assay, we found that it was deficient in this capacity (Fig. 5B). Furthermore, results in Fig. 5C confirm that this mutant, like the low-risk 11E6 protein, is unable to degrade p53 in standard in vitro and in vivo degradation assays (see Materials and Methods).
FIG. 5.
The 16E6 mutant L50G binds CBP but is unable to interact with E6AP or p53 and cannot degrade p53 in vitro or in vivo. (A) Schematic representation of the 16E6 mutant (mut) L50G showing the position of the amino acid exchange in the first zinc finger (marked by +) and the identified CBP interaction (binding [bdg.]) domain within the second zinc finger (bold line). GST micro-affinity column experiments using IVT 16E6 L50G protein demonstrate the ability of this mutant to interact with GST-CBP. (B) Similar in vitro micro-affinity column experiments show that unlike the wild-type 16E6 protein but similar to 11E6, the 16E6 mutant L50G is unable to interact with either GST-E6AP or GST-p53. The lower panels indicate the loading of GST, GST-E6AP, and GST-p53 proteins on the micro-affinity columns. (C) The upper panel shows p53 degradation assays using IVT 35S-labelled p53 mixed with various IVT E6 proteins. The numbered columns indicate the levels of p53 protein after various incubation times (0, 30, 90, and 180 min) at room temperature. The lower panel shows an in vivo degradation assay in which cellular levels of p53 are detected by Western blot analysis after cotransfection of U2-OS cells with p53 and 16E6 constructs. The numbers represent the incubation period (in minutes) in media containing cycloheximide before harvesting of the cellular proteins (see Materials and Methods). While the transfection of wild-type 16E6 leads to an observed decrease in cellular p53 levels after 60 min, the 16E6 mutant L50G is abrogated in this capacity.
16E6 can inhibit the interaction between p53 and CBP.
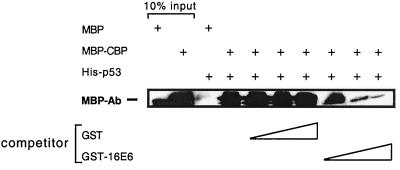
The work presented here and in our previous study (57a) indicates that 16E6 and p53 bind to the same motif (TRAM) in the CBP II domain. E1A, which also binds the CBP TRAM, can displace CBP from p53 (57a). We therefore tested whether the 16E6 protein could also displace CBP from p53. In Fig. 6 it can be seen that increasing concentrations of 16E6 do indeed inhibit the interaction between CBP and p53 in vitro, but not similar levels of GST protein alone. In this particular experiment, all three proteins used (His-p53, MBP-CBP II, and GST-16E6) are recombinant, bacterially expressed proteins, suggesting that this effect is direct and due to competitive binding between E6 and p53 for the CBP TRAM.
FIG. 6.
16E6 protein can disrupt a p53-CBP interaction in vitro. Bacterially expressed His-p53 can interact directly with bacterially expressed MBP-CBP II. While this interaction is not disrupted by the presence of purified GST protein, it is disrupted by increasing amounts of purified GST-16E6 in a concentration-dependent manner. MBP-Ab, antibody specific to MBP.
The repression of p53 transcriptional activity by 16E6 correlates with CBP binding and not p53 degradation.
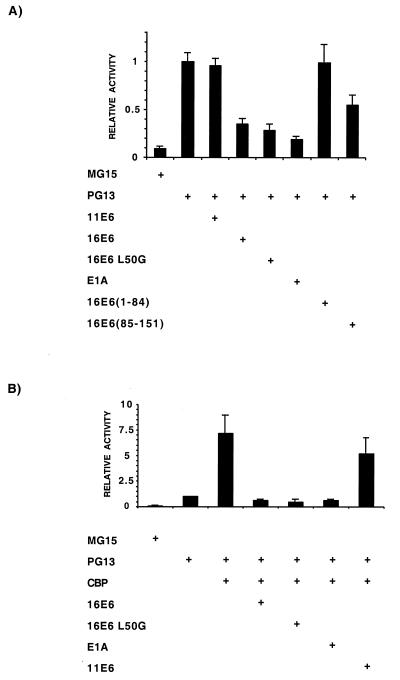
We next carried out a series of experiments in which we assessed the ability of the wild-type and L50G mutant HPV-16 E6 proteins to down-regulate p53-dependent transcription. U2-OS cells were transfected with the p53-responsive CAT reporter PG13CAT or the control vector MG15CAT, which contains mutated p53 binding sites. Cotransfected with PG13CAT were various expression plasmids coding for E6 proteins or Ad5 12S E1A. It can be seen from Fig. 7A that the PG13CAT vector is stimulated by endogenous p53 in U2-OS cells in a manner dependent on intact p53 binding sites. No effect on the level of transcriptional activity obtained with PG13CAT is seen upon the introduction of an expression vector containing full-length HPV-11 E6 sequences. By contrast, the expression of wild-type 16E6 protein results in a significant reduction in p53-dependent transcription. Consistent with our earlier analysis of the CBP binding domain within 16E6, the N-terminal 84 amino acids, which do not bind CBP, fail to repress p53 activity, while the C-terminal half of 16E6, which contains the CBP binding domain, can repress p53-dependent transcription, albeit slightly less efficiently than the full-length protein. Also shown for comparison is the level of repression of p53 activity obtained upon the introduction of the Ad5 12S E1A protein. Significantly, the 16E6 L50G mutant results in a similar level of transcriptional repression as wild-type 16E6. Thus, in this respect the 16E6 L50G mutant does not behave like a protein of a low-risk type but rather like wild-type 16E6. These data are consistent with the idea that by targeting CBP/p300, an E6 protein from a high-risk type can repress p53 transcriptional activity. Furthermore, the use of the 16E6 L50G mutant suggests that this ability is independent of E6AP-mediated degradation.
FIG. 7.
HPV-16 E6 targets the ability of CBP to activate p53-dependent transcription. (A) U2-OS cells were transfected with the p53-responsive CAT reporter (PG13) or a control vector with mutated p53 binding sites (MG15). Cotransfection of expression vectors for viral proteins show that HPV proteins able to interact with CBP can down-regulate p53 transactivation to a level comparable with Ad E1A. Those that fail to interact with CBP also fail to down-regulate p53-dependent transcription. (B) Overexpression of full-length CBP in experiments similar to those described above show that HPV proteins able to interact with CBP, including the HPV-16 L50G mutant, abolish the CBP-dependent superactivation of p53-dependent transcription seen with full-length CBP alone.
We wished to provide further evidence that the repression of p53 transcriptional activity by wild-type 16E6, 16E6 L50G, and E1A was due to the targeting of CBP/p300. Therefore, we overexpressed full-length CBP in a similar set of transfection experiments and examined whether the observed superactivation of p53-dependent transcription could be abrogated by these proteins. Figure 7B demonstrates that the cotransfection of full-length CBP into U2-OS cells stimulates p53-dependent transcription by approximately sevenfold. Like E1A, both wild-type 16E6 and the 16E6 L50G mutant abolish this CBP-induced superactivation of p53-dependent transcription. This is in contrast to 11E6, which is severely abrogated in this capacity.
In summary, the results presented in Fig. 5 to 7 provide evidence that E6 proteins from high-risk HPV types possess an alternative mechanism by which to down-regulate p53 activity. That is, by targeting CBP/p300, these E6 proteins can displace p53 from the CBP II domain and consequently abrogate p53-dependent transcription in a manner similar to Ad E1A.
DISCUSSION
In the last few years, study of the transformation processes that are instigated by SV40 and adenoviruses has led to the discovery of a number of important cellular regulators as viral targets. This knowledge has often resulted in the confirmation of identical targets for the high-risk HPVs (17, 79). While both SV40 and adenovirus proteins have recently been shown to bind to the transcriptional adapter CBP/p300, no such interaction has been described for HPV oncoproteins. One possible explanation for this is that all previously known HPV functions that mirror Ad E1A and SV40 TAg properties have been found to be present in the HPV E7 protein. To date no interaction between HPV E7 and CBP/p300 has been described. Moreover, the recent finding that HPV E7 proteins bind to and abrogate the function of p21Cip1 in a p53-independent manner (19a, 31) has suggested that E7 might not need to affect p53 activation by targeting CBP/p300, since it can directly affect the function of this important downstream inhibitor of cell cycle progression.
We set about identifying whether the HPV E6 oncoprotein could interact with CBP/p300 for three reasons. First, we believed that since CBP/p300 represented an indispensable target for transformation by Ad E1A and SV40 TAg, it was likely that at least one of the HPV oncoproteins would also target CBP/p300. Second, both adenoviruses and SV40 abrogate p53 activity in more than one way: by affecting p53 directly and also indirectly through an interaction with the transcriptional cofactor CBP/p300. Third, since HPV E6 proteins of high-risk types had previously been shown to down-regulate p53 transcriptional activity, we felt that they might also bring about this effect through the targeting of the coactivator CBP/p300.
Here, for the first time, we have provided evidence that the E6 proteins of high-risk HPVs can bind CBP/p300, thus demonstrating an Ad E1A and SV40 TAg-like property for HPV E6. One reason why the E6-CBP/p300 interaction might not have been previously detected is that unlike E1A, which demonstrates an extremely high affinity for this transcriptional coactivator and binds multiple domains, 16E6 binds the CBP II domain with only moderate affinity. Coupled with this is the fact that any attempts to analyze HPV E6-CBP/p300 interactions using IVT full-length CBP/p300 would, from our experience, have failed to detect such an interaction. The inability of in vitro-expressed full-length CBP/p300 to interact with proteins that bind only the CBP II domain is not limited to E6. Similar observations are also seen for other proteins, such as c-Fos and YY1 (84). We interpret these results to imply that access to the CBP II domain, in the context of full-length CBP/p300, is probably dependent on posttranslational modification. For example, access to the CBP II domain might be dependent on phosphorylation, because CBP/p300 phosphorylation is a dynamic process regulated in a cell cycle-dependent manner (1, 33, 82). By partially purifying full-length CBP/p300 from HeLa cell nuclear extract, we have been able to overcome these obstacles and demonstrate the ability of 16E6 to bind full-length CBP/p300 as do other transcription factors that bind only the CBP II domain.
The 16E6 protein, although only 151 amino acids in length, has been shown to have multiple properties and interact with a plethora of cellular proteins (36, 53). These include, in addition to E6AP, (i) a calcium-binding protein (E6-BP) that may play a role in epithelial differentiation (11), (ii) the Bak protein, a regulator of the apoptosis pathway (71), (iii) the focal adhesion protein paxillin (75), (iv) the human homologue of the Drosophila discs large tumor suppressor protein, hDLG (34, 44), and (v) IRF-3, a transcriptional activator possibly involved in antiviral cellular responses (62). Interestingly, HPV E6 proteins have also been shown to stimulate telomerase activity (35), thus potentially lengthening the life span of the host cell in a manner that is likely to contribute to the transformation process (78). In light of the results presented here, we can now add the transcriptional coactivator CBP/p300 to this impressive list of interacting cellular proteins. One advantage of targeting an important transcriptional coactivator such as CBP/p300 is that it may result in an ability to control multiple signaling pathways. That is because, in addition to p53, CBP/p300 acts as a cofactor for many other cellular regulators. Such a strategy appears to have been adopted by the Ad E1A protein (57a) and could also apply to high-risk HPV E6 proteins.
Comparisons between the E6 and E7 proteins of high-risk and low-risk HPVs have indicated functional distinctions based on differential affinities for cellular target proteins. For example, differences have been observed for the binding of E7 to pRB and p21Cip1 (31, 52) and for E6 binding to p53 and E6AP (28, 79). The capacity to target negative regulators of cell growth almost certainly represents a critical factor in the ability of these proteins to transform cells. Our observation that CBP/p300 binding is limited to E6 proteins of high-risk HPVs suggests that this transcriptional coactivator may also represent an important target for the transformation process. Such an idea is consistent with the finding that CBP/p300-binding-deficient mutants of Ad E1A are no longer capable of transforming cells. This finding, together with the fact that all three small DNA tumor viruses target CBP/p300, adds weight to the idea that CBP/p300 may act as a tumor suppressor protein.
One important role of CBP/p300 in cell cycle inhibition is in the facilitation of p53-dependent gene expression. Previous observations have indicated that p53 transcriptional activity can be abrogated by E6 proteins of high-risk HPVs (49). One potential mechanism to explain this would be the promotion of p53 degradation through the E6-E6AP pathway. However, the results presented here in Fig. 5 and 7 show that an HPV-16 mutant (L50G) that can bind CBP/p300 but fails to bind E6AP and degrade p53 in vitro and in vivo is still capable of repressing p53 transcriptional activity. This finding suggests that for this particular property the correlation is with CBP/p300 binding and not with the promotion of p53 degradation through the E6AP pathway. However, the previous analysis that demonstrated the inability of the 16E6 mutant L50G, in conjunction with HPV-16 E7, to transform human embryonic kidney cells (54) suggests that E6AP-dependent degradation of p53 is still likely to be a prerequisite for the induction of cellular transformation. We therefore propose that the E6 proteins of high-risk HPVs target p53 in two ways. The first, which may be a more immediate response, is the abrogation of p53 transcriptional activity by binding to the cofactor CBP/p300. The second would consist of the removal of p53 protein through E6AP-dependent degradation. Together, these complementary functions of E6 could facilitate the effective elimination of cellular p53 activity. Support for such a proposal comes from a number of previous studies that utilize HPV E6 mutants and demonstrate an independence of different aspects of transcriptional regulation from the ability to promote p53 degradation (12, 13, 42, 73).
It is still not certain at this time exactly how E6 abrogates the CBP/p300-dependent activation of p53. However, it is interesting that all three DNA tumor virus proteins (Ad E1A, SV40 TAg, and 16E6) bind the same region of CBP, which is also the p53 binding site within the CBP II domain. Although we have not yet determined whether SV40 TAg binds the 19-amino-acid CBP TRAM present within CBP II, we have established that this is the binding site for p53, 16E6, and Ad E1A (this study and reference 57a). This finding, together with the fact that both 16E6 and the Ad E1A proteins can displace p53 from the CBP TRAM in vitro, suggests that inhibition of a p53-CBP TRAM interaction could represent an important part of the mechanism by which p53 function is abrogated. Other potential p53 binding sites within CBP/p300 have been described, however (22, 24), which makes it difficult to predict the outcome of such displacement.
An analysis of the primary amino acid sequence within the region of 16E6 (residues 100 to 142) implicated in Fig. 3 in binding CBP/p300 has not revealed any obvious similarities to those in Ad E1A or p53 that have previously been shown to bind the CBP TRAM (57a). However, there are some similarities to the bipartite CBP/p300 binding domain of SV40 previously described (18), although the significance of these observations, if any, has yet to be established.
CBP/p300 has been shown to activate transcription by at least two mechanisms: one involving acetylation of histone and nonhistone proteins, and one involving the bridging of DNA-bound transcription factors to components of the basal transcription machinery. HPV E6 proteins could potentially abrogate both of these mechanisms of activation by binding to the CBP II domain. Recent reports have indicated that as a consequence of p53 acetylation, CBP/p300 can indirectly increase the affinity of p53 for its cognate DNA binding sites (23). If HPV E6 (as well as Ad E1A and SV40 TAg) prevented this acetylation by interacting with CBP/p300, then the affinity of p53 for target promoters might be reduced. This represents a distinct possibility, since a number of reports have demonstrated that the presence of E6 proteins from high-risk HPVs significantly reduces p53 DNA binding activity (41, 72, 73). The CBP/p300-associated acetyltransferase, P/CAF, also binds to the CBP II region and has been shown to be displaced by Ad E1A (83). P/CAF promotes cell cycle inhibition and cellular differentiation processes through its acetyltransferase properties (61, 83). Thus, by binding to the CBP II domain, HPV E6 might also down-regulate these pathways in addition to its effect on p53. HPV E6 proteins might also target the bridging mechanism for CBP/p300-dependent activation. CBP/p300 was recently shown to activate CREB-dependent transcription by recruiting RNA helicase A, a component of an RNA polymerase II complex to a promoter containing a functional CRE site (55). Since RNA helicase A has also been shown to bind the CBP II domain, 16E6, along with Ad E1A and SV40 TAg, might also inhibit this particular mechanism of CBP/p300-dependent activation by binding to the CBP II domain. Similar possibilities exist for the disruption of CBP/p300-TFIIB interactions (38). Future studies should permit an analysis of these potential mechanisms.
Finally, both Ad E1A and SV40 TAg proteins depend on an interaction with CBP/p300 for their cellular transformation properties. Here, we have provided evidence that an ability to bind CBP/p300 is limited to E6 proteins of high-risk HPVs (Fig. 4). Taken together, these observations suggest a possible role for the E6-CBP/p300 interaction in HPV-mediated oncogenesis. Future studies should be able to assess the ability of CBP-binding-deficient mutants of 16E6 to contribute towards cellular transformation. This in turn should provide an insight into the potential clinical relevance of the 16E6-CBP/p300 interaction described here.
ACKNOWLEDGMENTS
We thank T. Kouzarides, R. Goodman, T. Kanda, E. Manser, E. Androphy, and D. Pim for plasmid constructs, Choon-Heng Koh for technical assistance, and Benjamin Li and Ed Manser for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Ait-Si-Ali S, Ramirez S, Barre F X, Dkhissi F, Magnaghi-Jaulin L, Girault J A, Robin P, Knibiehler M, Pritchard L L, Ducommun B, Trouche D, Harel-Bellan A. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- 2.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adapter proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 3.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 4.Bagchi S, Raychaudhuri P, Nevins J R. Adenovirus E1A proteins can dissociate heteromeric complexes involving the E2F transcription factor: a novel mechanism for E1A trans-activation. Cell. 1990;62:659–669. doi: 10.1016/0092-8674(90)90112-r. [DOI] [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 7.Barbosa M S, Lowy D R, Schiller J T. Papillomavirus polypeptides E6 and E7 are zinc-binding proteins. J Virol. 1989;63:1404–1407. doi: 10.1128/jvi.63.3.1404-1407.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbosa M S, Vass W C, Lowy D R, Schiller J T. In vitro biological activities of the E6 and E7 genes vary among human papillomaviruses of different oncogenic potential. J Virol. 1991;65:292–298. doi: 10.1128/jvi.65.1.292-298.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosch F X, Manos M M, Munoz N, Sherman M, Jansen A M, Peto J, Schiffman M H, Moreno V, Kurman R, Shah K V, Alihonou E, Bayo S, Mokhtar H C, Chicareon S, Daudt A, Delosrios E, Ghadirian P, Kitinya J N, Koulibaly M, Ngelangel C, Tintore L M P, Riosdalenz J L, Sarjadi-Schneider A, Tafur L, Tayssie A R, Rolon P A, Torroella M, Tapia A V, Wabinga H R, Zatonski W, Sylla B, Vizcaino P, Magnin D, Kaldore J, Greer C, Wheeler C. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International Biological Study on Cervical Cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 10.Chellappan S, Kraus V B, Kroger B, Munger K, Howley P M, Phelps W C, Nevins J R. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc Natl Acad Sci USA. 1992;89:4549–4553. doi: 10.1073/pnas.89.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J J, Reid C E, Band V, Androphy E J. Interaction of papillomavirus E6 oncoproteins with a putative calcium- binding protein. Science. 1995;269:529–531. doi: 10.1126/science.7624774. [DOI] [PubMed] [Google Scholar]
- 12.Crook T, Fisher C, Masterson P J, Vousden K H. Modulation of transcriptional regulatory properties of p53 by HPV E6. Oncogene. 1994;9:1225–1230. [PubMed] [Google Scholar]
- 13.Crook T, Tidy J A, Vousden K H. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell. 1991;67:547–556. doi: 10.1016/0092-8674(91)90529-8. [DOI] [PubMed] [Google Scholar]
- 14.DeCaprio J A, Ludlow J W, Figge J, Shew J Y, Huang C M, Lee W H, Marsilio E, Paucha E, Livingston D M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 15.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyson N, Harlow E. Adenovirus E1A targets key regulators of cell proliferation. Cancer Surv. 1992;12:161–195. [PubMed] [Google Scholar]
- 17.Dyson N, Howley P M, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 18.Eckner R, Ludlow J W, Lill N L, Oldread E, Arany Z, Modjtahedi N, DeCaprio J A, Livingston D M, Morgan J A. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol. 1996;16:3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster S A, Demers G W, Etscheid B G, Galloway D A. The ability of human papillomavirus E6 proteins to target p53 for degradation in vivo correlates with their ability to abrogate actinomycin D-induced growth arrest. J Virol. 1994;68:5698–5705. doi: 10.1128/jvi.68.9.5698-5705.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Funk J O, Waga S, Harry J, Espling E, Stillman B, Galloway D A. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giles R H, Peters D J, Breuning M H. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 21.Grossman S R, Laimins L A. E6 protein of human papillomavirus type 18 binds zinc. Oncogene. 1989;4:1089–1093. [PubMed] [Google Scholar]
- 22.Grossman S R, Perez M, Kung A L, Joseph M, Mansur C, Xiao Z X, Kumar S, Howley P M, Livingston D M. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 23.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 24.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 25.Howes K A, Ransom N, Papermaster D S, Lasudry J G, Albert D M, Windle J J. Apoptosis or retinoblastoma: alternative fates of photoreceptors expressing the HPV-16 E7 gene in the presence or absence of p53. Genes Dev. 1994;8:1300–1310. doi: 10.1101/gad.8.11.1300. [DOI] [PubMed] [Google Scholar]
- 26.Howley P M, Munger K, Romanczuk H, Scheffner M, Huibregtse J M. Cellular targets of the oncoproteins encoded by the cancer associated human papillomaviruses. Proc Princess Takamatsu Symp. 1991;22:239–248. [PubMed] [Google Scholar]
- 27.Howley P M, Scheffner M, Huibregtse J, Munger K. Oncoproteins encoded by the cancer-associated human papillomaviruses target the products of the retinoblastoma and p53 tumor suppressor genes. Cold Spring Harbor Symp Quant Biol. 1991;56:149–155. doi: 10.1101/sqb.1991.056.01.019. [DOI] [PubMed] [Google Scholar]
- 28.Huibregtse J M, Scheffner M, Howley P M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janknecht R, Hunter T. Transcription. A growing coactivator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 30.Janknecht R, Hunter T. Versatile molecular glue. Curr Biol. 1996;6:951–954. doi: 10.1016/s0960-9822(02)00636-x. [DOI] [PubMed] [Google Scholar]
- 31.Jones D L, Alani R M, Munger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kessis T D, Slebos R J, Nelson W G, Kastan M B, Plunkett B S, Han S M, Lorincz A T, Hedrick L, Cho K R. Human papillomavirus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proc Natl Acad Sci USA. 1993;90:3988–3992. doi: 10.1073/pnas.90.9.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitabayashi I, Eckner R, Arany Z, Chiu R, Gachelin G, Livingston D M, Yokoyama K K. Phosphorylation of the adenovirus E1A-associated 300 kDa protein in response to retinoic acid and E1A during the differentiation of F9 cells. EMBO J. 1995;14:3496–3509. doi: 10.1002/j.1460-2075.1995.tb07356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klingelhutz A J, Foster S A, McDougall J K. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 36.Kubbutat M H, Vousden K H. New HPV E6 binding proteins: dangerous liaisons? Trends Microbiol. 1998;6:173–175. doi: 10.1016/s0966-842x(98)01267-0. [DOI] [PubMed] [Google Scholar]
- 37.Kurokawa R, Kalafus D, Ogliastro M H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 38.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 39.Lane D P, Crawford L V. T antigen is bound to a host protein in transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 40.Lane D P, Simanis V, Bartsch R, Yewdell J, Gannon J, Mole S. Cellular targets for SV40 large T-antigen. Proc R Soc Lond Ser B. 1985;226:25–42. doi: 10.1098/rspb.1985.0077. [DOI] [PubMed] [Google Scholar]
- 41.Lechner M S, Laimins L A. Inhibition of p53 DNA binding by human papillomavirus E6 proteins. J Virol. 1994;68:4262–4273. doi: 10.1128/jvi.68.7.4262-4273.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lechner M S, Mack D H, Finicle A B, Crook T, Vousden K H, Laimins L A. Human papillomavirus E6 proteins bind p53 in vivo and abrogate p53-mediated repression of transcription. EMBO J. 1992;11:3045–3052. doi: 10.1002/j.1460-2075.1992.tb05375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J S, Galvin K M, See R H, Eckner R, Livingston D, Moran E, Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 44.Lee S S, Weiss R S, Javier R T. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homologue of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 46.Linzer D I, Levine A J. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 47.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 48.Manser E, Huang H Y, Loo T H, Chen X Q, Dong J M, Leung T, Lim L. Expression of constitutively active α-PAK reveals effects of the kinase on actin and focal complexes. Mol Cell Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mietz J A, Unger T, Huibregtse J M, Howley P M. The transcriptional transactivation function of wild-type p53 is inhibited by SV40 large T-antigen and by HPV-16 E6 oncoprotein. EMBO J. 1992;11:5013–5020. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moran E. DNA tumor virus transforming proteins and the cell cycle. Curr Opin Genet Dev. 1993;3:63–70. doi: 10.1016/s0959-437x(05)80342-9. [DOI] [PubMed] [Google Scholar]
- 51.Munger K, Werness B A, Dyson N, Phelps W C, Harlow E, Howley P M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munger K, Yee C L, Phelps W C, Pietenpol J A, Moses H L, Howley P M. Biochemical and biological differences between E7 oncoproteins of the high- and low-risk human papillomavirus types are determined by amino-terminal sequences. J Virol. 1991;65:3943–3948. doi: 10.1128/jvi.65.7.3943-3948.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Myers G, Androphy E J. The E6 protein. In: Myers G, Bernard H U, Delius H, Icenogle J, Baker C C, Halpern A, Wheeler C, editors. Human papillomaviruses. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. pp. 47–57. [Google Scholar]
- 54.Nakagawa S, Watanabe S, Yoshikawa H, Taketani Y, Yoshiike K, Kanda T. Mutational analysis of human papillomavirus type 16 E6 protein: transforming function for human cells and degradation of p53 in vitro. Virology. 1995;212:535–542. doi: 10.1006/viro.1995.1511. [DOI] [PubMed] [Google Scholar]
- 55.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 56.O’Connor M, Bernard H U. Oct-1 activates the epithelial-specific enhancer of human papillomavirus type 16 via a synergistic interaction with NFI at a conserved composite regulatory element. Virology. 1995;207:77–88. doi: 10.1006/viro.1995.1053. [DOI] [PubMed] [Google Scholar]
- 57.O’Connor M J, Tan S H, Tan C H, Bernard H U. YY1 represses human papillomavirus type 16 transcription by quenching AP-1 activity. J Virol. 1996;70:6529–6539. doi: 10.1128/jvi.70.10.6529-6539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57a.O’Connor M J, Zimmermann H, Nielsen S, Bernard H U, Kouzarides T. Characterization of an E1A-CBP interaction defines a novel transcriptional adapter motif (TRAM) in CBP/p300. J Virol. 1999;73:3574–3581. doi: 10.1128/jvi.73.5.3574-3581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 59.Pan H, Griep A E. Altered cell cycle regulation in the lens of HPV-16 E6 or E7 transgenic mice: implications for tumor suppressor gene function in development. Genes Dev. 1994;8:1285–1299. doi: 10.1101/gad.8.11.1285. [DOI] [PubMed] [Google Scholar]
- 60.Pim D, Massimi P, Banks L. Alternatively spliced HPV-18 E6* protein inhibits E6 mediated degradation of p53 and suppresses transformed cell growth. Oncogene. 1997;15:257–264. doi: 10.1038/sj.onc.1201202. [DOI] [PubMed] [Google Scholar]
- 61.Puri P L, Sartorelli V, Yang X J, Hamamori Y, Ogryzko V V, Howard B H, Kedes L, Wang J Y, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 62.Ronco L V, Karpova A Y, Vidal M, Howley P M. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roth J, Konig C, Wienzek S, Weigel S, Ristea S, Dobbelstein M. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J Virol. 1998;72:8510–8516. doi: 10.1128/jvi.72.11.8510-8516.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 65.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 66.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 67.Somasundaram K, El-Deiry W S. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene. 1997;14:1047–1057. doi: 10.1038/sj.onc.1201002. [DOI] [PubMed] [Google Scholar]
- 68.Song S, Gulliver G A, Lambert P F. Human papillomavirus type 16 E6 and E7 oncogenes abrogate radiation- induced DNA damage responses in vivo through p53-dependent and p53- independent pathways. Proc Natl Acad Sci USA. 1998;95:2290–2295. doi: 10.1073/pnas.95.5.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Steegenga W T, Riteco N, Jochemsen A G, Fallaux F J, Bos J L. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene. 1998;16:349–357. doi: 10.1038/sj.onc.1201540. [DOI] [PubMed] [Google Scholar]
- 70.Talis A L, Huibregtse J M, Howley P M. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J Biol Chem. 1998;273:6439–6445. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- 71.Thomas M, Banks L. Inhibition of Bak-induced apoptosis by HPV-18 E6. Oncogene. 1998;17:2943–2954. doi: 10.1038/sj.onc.1202223. [DOI] [PubMed] [Google Scholar]
- 72.Thomas M, Massimi P, Banks L. HPV-18 E6 inhibits p53 DNA binding activity regardless of the oligomeric state of p53 or the exact p53 recognition sequence. Oncogene. 1996;13:471–480. [PubMed] [Google Scholar]
- 73.Thomas M, Massimi P, Jenkins J, Banks L. HPV-18 E6 mediated inhibition of p53 DNA binding activity is independent of E6 induced degradation. Oncogene. 1995;10:261–268. [PubMed] [Google Scholar]
- 74.Thompson D A, Belinsky G, Chang T H, Jones D L, Schlegel R, Munger K. The human papillomavirus-16 E6 oncoprotein decreases the vigilance of mitotic checkpoints. Oncogene. 1997;15:3025–3035. doi: 10.1038/sj.onc.1201495. [DOI] [PubMed] [Google Scholar]
- 75.Tong X, Howley P M. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc Natl Acad Sci USA. 1997;94:4412–4417. doi: 10.1073/pnas.94.9.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Dyke T A. Analysis of viral-host protein interactions and tumorigenesis in transgenic mice. Semin Cancer Biol. 1994;5:47–60. [PubMed] [Google Scholar]
- 77.Vousden K H. Regulation of the cell cycle by viral oncoproteins. Semin Cancer Biol. 1995;6:109–116. doi: 10.1006/scbi.1995.0014. [DOI] [PubMed] [Google Scholar]
- 78.Vousden K H. HPV E6: ensuring all’s well at the end. Trends Microbiol. 1996;4:337–338. doi: 10.1016/0966-842x(96)30024-3. [DOI] [PubMed] [Google Scholar]
- 79.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 80.Whyte P, Buchkovich K J, Horowitz J M, Friend S H, Raybuck M, Weinberg R A, Harlow E. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 81.Yaciuk P, Carter M C, Pipas J M, Moran E. Simian virus 40 large-T antigen expresses a biological activity complementary to the p300-associated transforming function of the adenovirus E1A gene products. Mol Cell Biol. 1991;11:2116–2124. doi: 10.1128/mcb.11.4.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yaciuk P, Moran E. Analysis with specific polyclonal antiserum indicates that the E1A-associated 300-kDa product is a stable nuclear phosphoprotein that undergoes cell cycle phase-specific modification. Mol Cell Biol. 1991;11:5389–5397. doi: 10.1128/mcb.11.11.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 84.Zimmermann, H., and M. O’Connor. Unpublished data.
- 85.zur Hausen H. Human papillomaviruses in the pathogenesis of anogenital cancer. Virology. 1991;184:9–13. doi: 10.1016/0042-6822(91)90816-t. [DOI] [PubMed] [Google Scholar]