Abstract
Most details of the processing of the hepatitis A virus (HAV) polyprotein are known. Unique among members of the family Picornaviridae, the primary cleavage of the HAV polyprotein is mediated by 3Cpro, the only proteinase known to be encoded by the virus, at the 2A/2B junction. All other cleavages of the polyprotein have been considered to be due to 3Cpro, although the precise location and mechanism responsible for the VP1/2A cleavage have been controversial. Here we present data that argue strongly against the involvement of the HAV 3Cpro proteinase in the maturation of VP1 from its VP1-2A precursor. Using a heterologous expression system based on recombinant vaccinia viruses directing the expression of full-length or truncated capsid protein precursors, we show that the C terminus of the mature VP1 capsid protein is located near residue 764 of the polyprotein. However, a proteolytically active HAV 3Cpro that was capable of directing both VP0/VP3 and VP3/VP1 cleavages in vaccinia virus-infected cells failed to process the VP1-2A precursor. Using site-directed mutagenesis of an infectious molecular clone of HAV, we modified potential VP1/2A cleavage sites that fit known 3Cpro recognition criteria and found that a substitution that ablates the presumed 3Cpro dipeptide recognition sequence at Glu764-Ser765 abolished neither infectivity nor normal VP1 maturation. Altered electrophoretic mobility of VP1 from a viable mutant virus with an Arg764 substitution indicated that this residue is present in VP1 and that the VP1/2A cleavage occurs downstream of this residue. These data indicate that maturation of the HAV VP1 capsid protein is not dependent on 3Cpro processing and may thus be uniquely dependent on a cellular proteinase.
The genomes of all picornaviruses encode a single polyprotein which is co- and posttranslationally proteolytically cleaved by virus-encoded proteinase(s). These processing events are well characterized for most picornaviral genera, including the enteroviruses, rhinoviruses, cardioviruses, and aphthoviruses (reviewed in reference 23). The mechanism and location of the primary cleavage of the polyprotein differ between the genera. In viruses of the genera Enterovirus and Rhinovirus, the primary cleavage occurs at the VP1/2A junction, resulting in the release of the P1 capsid protein precursor from the P2-P3 nonstructural protein precursor. This cleavage occurs by cis action of the 2A proteinase at its N terminus. In viruses of the genera Cardiovirus and Aphthovirus, the primary cleavage event occurs at the 2A/2B junction, releasing the L-P1-2A (cardioviruses) or P1-2A (aphthoviruses) capsid protein precursor from the 2BC-P3 nonstructural protein precursor. This primary cleavage occurs in cis and is dependent on the sequence of the 2A protein, which is comprised of only 18 amino acid residues in the aphthoviruses. A similar sequence is located at the C terminus of the larger cardioviral 2A protein, and in both aphthoviruses and cardioviruses, there is complete conservation of the three C-terminal residues of 2A (Asn-Pro-Gly) as well as the N-terminal residue (Pro) of protein 2B (8). In all four of these picornaviral genera, all other cleavages within the polyprotein are carried out by the 3Cpro proteinase (or its precursor 3CDpro), with the exception of the L/P1 cleavage directed by the L proteinase in aphthoviruses, and the maturation cleavage of VP0 to VP4 and VP2, which occurs in all picornaviruses by a mechanism that has yet to be explained but which appears to be dependent on packaging of the viral RNA.
In contrast to these well-characterized events, the processing of the polyprotein has been difficult to study in the genus Hepatovirus. This genus of the family Picornaviridae is comprised of a single virus species, hepatitis A virus (HAV), which typically has a protracted and noncytolytic replication cycle in cell culture and fails to induce shutdown of cellular host cell protein synthesis in infected cells (reviewed in reference 18). It has recently been shown that the primary cleavage of the HAV polyprotein occurs at the 2A/2B junction, which has been mapped by the N-terminal sequencing of 2B. Unlike all other picornaviruses, this primary cleavage of the HAV polyprotein is carried out by the 3Cpro proteinase, which is the only proteinase known to be encoded by the virus (13, 21, 26). The P1-2A capsid protein precursor is most likely released from the nonstructural protein precursor (2BC-P3) as soon as 3Cpro is synthesized, as the full-length polyprotein has not been observed in these studies. A P1-2A precursor produced in a cell-free translation system has been shown to be readily cleaved in vitro by purified, recombinant 3Cpro to generate VP0 (VP4-VP2), VP3, and VP1-2A (also termed PX) (20). We have shown that similar cleavage events occur in vivo in cells infected with recombinant vaccinia viruses expressing HAV polypeptides (see Results). However, the eventual fate of the VP1-2A product remains controversial, and the N-terminal residue of 2A has not been defined.
The VP1-2A polypeptide is unique to the hepatoviruses. It associates with VP0 and VP3 to form pentamers, the first intermediate in the morphogenesis of HAV particles (2, 5). The mature capsid protein VP1 is subsequently derived from the VP1-2A precursor later in the morphogenesis process, although preparations of infectious virus particles often contain detectable quantities of VP1-2A (2, 5). The mechanism by which the 2A moiety is cleaved from the VP1-2A precursor is not known. However, purified recombinant 3Cpro has been shown to cleave relevant HAV substrates that were generated in cell-free translation reactions, suggesting that 3Cpro may be responsible for the VP1/2A cleavage (20, 25). More recently, Probst et al. (24) have presented data suggesting that 3Cpro directs the cleavage between VP1 and 2A at a Glu-Ser dipeptide sequence that is present in most HAV strains at residues 764 and 765 of the polyprotein (Glu764-Ser; amino acid numbering is from the first AUG). This would result in a VP1 protein of 273 amino acid residues, since the N terminus of the mature capsid protein VP1 has been isolated from virions, microsequenced, and shown to be located at residue 492 (Val) of the polyprotein (12, 19).
Here, however, we present data that argue strongly against the involvement of the HAV 3Cpro proteinase in the maturation of VP1 from its VP1-2A precursor. We show that the C terminus of the mature capsid protein VP1 is located near but downstream of, residue 764 of the polyprotein. Furthermore, we demonstrate that 3Cpro is incapable of directing the cleavage of VP1 from the VP1-2A precursor in vivo, using recombinant vaccinia viruses that express relevant HAV substrates, and show that a substitution that ablates the presumed 3Cpro dipeptide recognition sequence at positions 764-765 of the polyprotein neither abolish infectivity of the HAV nor eliminate the normal maturation of the VP1 capsid protein. These data strongly refute the hypothesis that the maturation of VP1 is dependent on 3Cpro processing of the VP1-2A precursor and suggest a novel role for an unknown cellular proteinase in processing of a picornavirus polyprotein.
MATERIALS AND METHODS
Cell cultures and viruses.
Fetal rhesus kidney (FRhK-4) cells were used for rescue of infectious HAV following transfection with synthetic, genome-length HAV RNA transcripts (7) and for the in vivo expression of HAV polypeptides following infection with recombinant vaccinia viruses. African green monkey kidney (BS-C-1) cells were used for radioimmunofocus assays (RIFA) (16) to characterize the replication phenotype of mutant HAVs and to determine the titer of virus stocks. These cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco BRL or Eurobio) supplemented with 5% fetal calf serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) (DMEM-5%) and a mixture of nonessential amino acids (Gibco BRL). Human 143B thymidine kinase-deficient cells were used for the isolation of recombinant vaccinia viruses, and monkey kidney (CV1) or human cervix carcinoma (HeLa) cells were used for propagation of these viruses. Both of these cell lines were maintained in DMEM-5%.
The wild-type vaccinia virus, Copenhagen strain, and its thermosensitive ts7 derivative (10) were propagated in HeLa cells and purified by centrifugation through a sucrose gradient. Titers of infectious virus were determined in CV1 cells. vTF7-3, a recombinant vaccinia virus expressing T7 DNA-dependent RNA polymerase (11), was obtained from B. Moss (National Institutes of Health, Bethesda, Md.). Infectious HAV was obtained by transfection of FRhK-4 cells with synthetic RNA derived from a chimeric cDNA (p5′P2P3-18f) (29) containing the P1 segment of a relatively low passage, cell culture-adapted variant of the HM175 strain (6, 7) in the background of a rapidly replicating, cytopathic HM175 variant, 18f (17).
HAV expression plasmids and generation of recombinant vaccinia viruses.
For expression of HAV polypeptides in eukaryotic cells, appropriate HAV cDNA fragments were cloned into plasmid pTM1 downstream of the T7 RNA polymerase promoter (22). Recombinant vaccinia viruses were generated from these plasmids by homologous DNA recombination as described previously (21).
Plasmid pTM/P1-2A was constructed by PCR amplification of nucleotides (nt) 748 to 3255 of the HAV sequence from p5′P2P3-18f (29) with a 5′ oligonucleotide primer complementary to nt 748 to 775 and a 3′ primer complementary to nt 3231 to 3255 and possessing an extension designed to create a SpeI restriction site at the 3′ end of the amplimer. The resulting DNA fragment was digested by SpeI, 5′ phosphorylated, and inserted between the NcoI and SpeI sites of the pTM1 polylinker.
To create pTM1 derivatives expressing a series of truncated HAV capsid protein precursors, nt 2221 to 2982, 2221 to 3039, 2221 to 3075, or 2221 to 3120 were PCR amplified from p5′P2P3-18f cDNA, using a series of 3′ primers containing a 3′ SpeI site and a 5′ primer complementary to nt 2221 to 2246. The resulting PCR amplimers were digested with NcoI (nt 2827) and SpeI and inserted in lieu of the corresponding full-length fragment (nt 2827 to 3255) of pTM/P1-2A. These plasmids encode truncated capsid protein precursors, with C termini corresponding to residue 745 (p-P1745), 764 (p-P1764), 776 (p-P1776), or 791 (p-P1791) of the polyprotein, or potential VP1 proteins of 254, 273, 285, or 300 residues, respectively.
Plasmid pTM/2BC-P3 was constructed from a dicistronic cDNA with an encephalomyocarditis virus internal ribosomal entry site insertion between 2A and 2B sequences of 5′P2P3-18f (pHAV-2AE2B) (2a). The Asp718-BamHI restriction fragment, which included the 132 3′-terminal nucleotides of the encephalomyocarditis internal ribosomal entry site, the initiation codon, HAV 2BC and P3 sequences, as well as HAV 3′ noncoding region and poly(A) sequence, was purified from pHAV-2AE2B and inserted between the Asp718 and BamHI sites of pTM1 to create pTM/2BC-P3, which expresses the HAV 2BC-P3 polypeptide.
Construction of mutated full-length HAV cDNAs by site-directed mutagenesis.
Potential 3Cpro cleavage sites which may represent the VP1/2A junction were altered in the infectious molecular clone, p5′P2P3-18f (29), using three different strategies for site-directed mutagenesis. For mutants 764Q, 764R, 776Q, 776R, and 791R (Table 1), the isolated SacI-EcoRI fragment (nt 3002 to 4990) of p5′P2P3-18f was inserted into the polylinker of phage M13mp18 DNA and subjected to site-directed mutagenesis according to the method of Taylor et al. (28). For mutants 745N and 745R (Table 1), the Bst1107I-SacI segment (nt 2037 to 3002) of p5′P2P3-18f was PCR amplified by using a 3′ oligonucleotide primer with nucleotide substitutions in the relevant codon. For mutants 791D and 791Q (Table 1), p5′P2P3-18f was used as a template for PCR-mediated mutagenesis by an adaptation of the method described by Stemmer and Morris (27). All mutated cDNA segments were sequenced to exclude spurious mutations prior to their reintroduction into the background of p5′P2P3-18f.
TABLE 1.
Infectivity of HAV mutant RNA transcripts in FRhK-4 cellsa
| Construct | aa 745 (nt 2980–82) | aa 764 (nt 3037–39) | aa 776 (nt 3073–75) | aa 791 (nt 3118–20) | Virus titer (RFU/ml)
|
|
|---|---|---|---|---|---|---|
| After transfection | After infectionb | |||||
| wt | Gln (UUG) | Glu (GAA) | Glu (GAG) | Glu (GAG) | 5 × 106–1 × 107 | 107 |
| 745N | Asn (AAC) | — | — | — | Lethal | NA |
| 745R | Arg (CGG) | — | — | — | Lethal | NA |
| 764Q | — | Gln (CAG) | — | — | 5 × 106–1 × 107 | ND |
| 764R | — | Arg (CGA) | — | — | 2 × 106 | 2 × 106 |
| 776Q | — | — | Gln (CAA) | — | Lethal | NA |
| 776R | — | — | Arg (CGG) | — | Lethal | NA |
| 791Q | — | — | — | Gln (CAG) | 1 × 103–1 × 104 | 6 × 104–3 × 105 |
| 791D | — | — | — | Asp (GAC) | 4 × 105 | 5 × 105–7 × 105 |
| 791R | — | — | — | Arg (CGG) | Lethal | NA |
wt, wild type (5′P2P3-18f RNA transcript); —, identical to wild type; NA, not applicable; ND, not done.
After infection with virus rescued from transfection lysates.
Transcription and transfection of full-length HAV RNAs.
Transcription of full-length HAV RNA and liposome-mediated RNA transfection of FRhK-4 cells were carried out essentially as described previously (21). Briefly, 1 μg of HaeII-digested plasmid DNA was transcribed by SP6 RNA polymerase (50 U; Promega) in a 50-μl reaction mix containing 0.5 mM each ribonucleoside triphosphate for 60 min at 37°C. The quality and approximate quantity of the transcription products were assessed by electrophoresis in 1% agarose gels. For transfections, approximately 5 μg of RNA transcript was mixed with 50 μl of DOTAP (Boehringer Mannheim) in a 150-μl total volume, incubated for 15 min at room temperature, and added to 3 ml of DMEM. This mixture was added dropwise to subconfluent monolayers of prewashed FRhK-4 cells (5 × 105 cells in 25-cm2 culture flasks). After overnight incubation at 37°C, the cells were washed and maintained in 5 ml of DMEM with 2% fetal calf serum for 14 days at 37°C. Cells were harvested mechanically, subjected to three freeze-thaw cycles, and extracted with an equal volume of chloroform.
HAV RIFA.
Lysates of transfected cells were assayed for infectious HAV by radioimmunofocus assay (RIFA) carried out in BS-C-1 cells as described previously (16). Infected cells were maintained at 37°C for 7 days before processing.
HAV polypeptide expression assays.
Segments of the HAV polyprotein were expressed in FRhK-4 cells (2 × 105 cells in 35-mm-diameter petri dishes) by coinfection of the cells with vTF7-3 and vaccinia virus-HAV recombinants expressing full-length or truncated capsid protein precursor (vv-P1-2A) and/or a 3Cpro precursor (vv-2BC-P3), each at a multiplicity of infection (MOI) of 5 PFU per cell.
Stocks of viable HAV mutants were used to infect FRhK-4 cells (2 × 105 cells in 35-mm-diameter petri dishes) at an MOI of 1 to 2 radioimmunofocus-forming units (RFU) per cell (16).
Immunoblot detection of HAV proteins.
Cytoplasmic extracts were prepared at 20 h postinfection (p.i.) (for vaccinia virus infections) or at 24, 48, 72, or 96 h p.i. (for HAV infections) by lysis of cells in 200 μl of 50 mM Tris-Cl (pH 7.5)–150 mM NaCl–1 mM EDTA–1% Nonidet P-40–0.1% sodium deoxycholate containing 25 μg of aprotinin per ml. A 10- to 20-μl aliquot was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by semidry transfer onto a polyvinylidene difluoride membrane (Amersham). Nonspecific binding sites were blocked in phosphate-buffered saline containing 0.1% Tween 20 (PBST) and 5% nonfat milk for 1 h at room temperature. The membrane was incubated overnight at 4°C with a mixture of HAV anti-VP1 and anti-VP2 guinea pig antibodies, diluted 1:4,000 and 1:8,000, respectively, in PBST containing 1% bovine serum albumin (PBST-BSA). After four washes with PBST, the membrane was incubated with anti-guinea pig antibodies conjugated to horseradish peroxidase (Sigma) diluted in PBST-BSA for 1 h at room temperature. After four washes with PBST, the HAV polypeptides were visualized by chemiluminescence (ECL Plus; Amersham).
RESULTS
Processing of the HAV P1-2A polypeptide by 3Cpro proteinase expressed by recombinant vaccinia viruses.
To study proteolytic cleavage of the HAV polyprotein by the 3Cpro proteinase of HAV, we expressed segments of the polyprotein in FRhK-4 cells in a hybrid T7-vaccinia expression system (11, 22). HAV cDNA sequences were cloned into plasmid pTM1 under control of the T7 promoter and recombinant vaccinia viruses were generated in vivo by homologous recombination with wild-type vaccinia virus DNA as described in Materials and Methods. One recombinant vaccinia virus (vv-P1-2A) expressed amino acids (aa) 1 to 836 of the HAV polyprotein, encompassing the entire capsid protein precursor P1-2A (Fig. 1). Another recombinant vaccinia virus (vv-2BC-P3) expressed aa 837 to 2226 of the HAV polyprotein, encompassing the 2BC-P3 polypeptide, as a precursor to proteinase 3Cpro. Coinfection of cells with one or more of these recombinant vaccinia viruses and with an additional recombinant vaccinia virus, vTF7-3, which expresses T7 RNA polymerase, resulted in expression of the HAV polypeptides.
FIG. 1.
Schematic representation of full-length or truncated capsid protein precursors expressed by recombinant vaccinia viruses. The identities of the amino acids located at the N and C termini of each capsid protein of the 5′P2P3-18f virus (29) as well as their positions within the polyprotein are indicated.
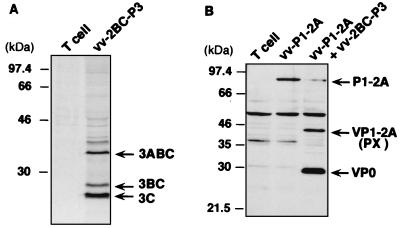
Self-cleavage of the 3Cpro precursor was assessed by [35S]methionine labeling of FRhK-4 cells coinfected with vTF7-3 and recombinant vaccinia virus vv-2BC-P3. Cell lysates were immunoprecipitated with anti-HAV 3C antibodies, and the products were separated by SDS-PAGE. This demonstrated that the 2BC-P3 polypeptide was efficiently processed to generate mature 3Cpro proteinase as a major product (Fig. 2A). Two additional products were precipitated by anti-3C antibodies; these were identified as 3ABC and 3BC on the basis of their electrophoretic mobilities. Thus, 3Cpro was active and capable of directing its cis cleavage from the 2BC-P3 precursor in this expression system.
FIG. 2.
(A) Autoproteolytic cleavage of the 2BC-P3 precursor of the 3Cpro proteinase. FRhK-4 cells were infected with vTF7-3 (T cell) or coinfected with vTF7-3 and vv-2BC-P3, each at an MOI of 5 PFU/cell. [35S]methionine-labeled HAV proteins were separated by SDS-PAGE (12% gel) following immunoprecipitation with anti-HAV 3C antibodies. (B) P1-2A is cleaved in trans by the 2BC-P3 precursor of the 3Cpro proteinase at the VP0/VP3 and VP3/VP1 junctions. FRhK-4 cells were coinfected with vTF7-3 and vv-P1-2A, with or without vv-2BC-P3 as a source of the 3Cpro proteinase, each at an MOI of 5 PFU/cell. Proteins were separated by SDS-PAGE (10% gel) and identified in an immunoblot using a mixture of anti-VP1 and anti-VP2 antibodies. Positions of molecular weight standards and of precursors and mature HAV polypeptides are shown at the left and right, respectively, of each panel.
To determine whether the capsid protein precursor, P1-2A, could be cleaved in trans by 3Cpro in this expression system, FRhK-4 cells were coinfected with vTF7-3 and vv-P1-2A, with or without coinfection with vv-2BC-P3, which expresses the 3Cpro precursor. Cell lysates were subjected to SDS-PAGE, and HAV polypeptides visualized in immunoblots with a mixture of anti-VP1 and anti-VP2 antibodies. The 94-kDa P1-2A precursor was processed only in cells expressing the 2BC-P3 proteinase precursor, yielding VP1-2A (PX) and VP0 (Fig. 2B). This result demonstrates that 3Cpro expressed from vv-2BC-P3 is able to process P1-2A in trans at both the VP0/VP3 and VP3/VP1 junctions. However, no mature VP1 was identified in these cell lysates, indicating that the proteolytically active 3Cpro proteinase was not able to direct cleavage at the VP1/2A junction in this system.
Mature VP1 capsid protein from HAV-infected cells comigrates with a recombinant VP1 molecule of 273 amino acid residues (VP1764).
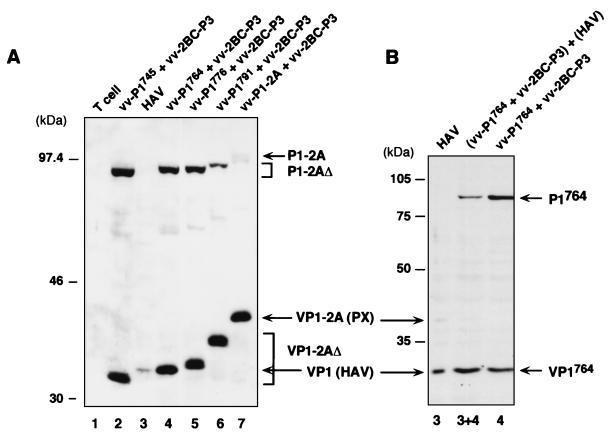
In an effort to determine the approximate location of the C terminus of the mature VP1 capsid protein, we constructed a series of recombinant vaccinia viruses expressing truncated P1-2A capsid protein precursors: aa 1 to 745 (vv-P1745), 1 to 764 (vv-P1764), 1 to 776 (vv-P1776), and 1 to 791 (vv-P1791). The C-terminal residues of these truncated precursors correspond to each of the four potential 3Cpro cleavage sites that may represent the VP1/2A junction: Gln745-Ser, Glu764-Ser, Glu776-Ser, and Glu791-Ser (Fig. 1). These sites were identified on the basis of the substrate recognition criteria of picornaviral 3Cpro proteinases, which include a requirement for a Gln or possibly a Glu residue at the P1 position immediately preceding the scissile peptide bond (3). We examined the processing of these truncated P1-2A substrates by 3Cpro expressed in cells that were coinfected with vv-2BC-P3 and vTF7-3 (Fig. 3A). Products containing VP1 residues were identified by immunoblotting with anti-VP1 antibodies. As expected, the proteolytic cleavage of these substrates by 3Cpro produced C-terminally truncated VP1-2A polypeptides representing aa 492 to 745, 492 to 764, 492 to 776, and 492 to 791 (Fig. 3A).
FIG. 3.
The mature capsid protein VP1 comigrates with VP1764. (A) FRhK-4 cells were infected with either HAV 5′P2P3-18f (lane 3) or vTF7-3 (lane 1) or coinfected with vTF7-3, the indicated recombinant vaccinia virus expressing a truncated P1-2A capsid protein precursor, and with vv-2BC-P3 as a source of 3Cpro proteinase (lanes 2, 4, 5, 6, and 7). Proteins were separated by SDS-PAGE (10% gel) and identified in an immunoblot with anti-VP1 antibodies. (B) A mixture of equal volumes of cellular extracts from HAV-infected cells and cells coinfected with vTF7-3, vv-P1764, and vv-2BC-P3 was loaded in lane 3+4. Positions of molecular weight standards and of precursors and mature HAV polypeptides are shown at the left and right, respectively, of each panel.
When subjected to SDS-PAGE, the electrophoretic mobilities of these truncated VP1-2A molecules bracketed that of mature VP1 isolated from HAV-infected cells (Fig. 3A, lanes 2 to 4). The natural VP1 protein (Fig. 3A, lane 3) roughly comigrated with the recombinant VP1764 product derived from the vv-P1764 precursor (lane 4) that contains 273 amino acid residues. To determine whether we could detect subtle differences in the electrophoretic mobilities of these two proteins, equal amounts of cytoplasmic extracts from HAV-infected cells and from cells coinfected with vv-P1764 and vv-2BC-P3 were mixed and loaded onto an SDS-polyacrylamide gel between lanes containing the individual extracts. As shown in Fig. 3B (lane 3+4), the mixture of the natural VP1 and recombinant VP1764 proteins migrated as a single band. This result suggests that the C terminus of the mature VP1 capsid protein lies close to, if not at, residue 764.
Similar to the results we obtained for cells expressing the entire P1-2A polyprotein segment as a substrate for 3Cpro (Fig. 2), neither of the two recombinant truncated P1-2A precursor proteins that contain the N terminus of 2A (P1776 and P1791) were completely processed into mature VP1 in the presence of the proteinase (Fig. 3A).
Infectivity of mutated HAV RNA transcripts.
To ascertain whether any of the four potential VP1/2A cleavage sites listed above constitute actual substrates for the 3Cpro proteinase within the context of the full-length HAV polyprotein, substitutions were introduced at the P1 residue of these sites within a genome-length, infectious HAV cDNA clone, p5′P2P3-18f. The P1 residue represents the most important determinant of 3Cpro specificity. Two types of mutations were engineered at each of these putative junctions (Table 1). One set of mutations was designed to preserve or even enhance 3Cpro recognition and cleavage (Gln745→Asn and Glu764,776,791→Gln), while a second set of substitutions were nonconservative and designed to abolish 3Cpro recognition of the dipeptide sequence (Gln745 and Glu764,776,791→Arg). In addition, we introduced a conservative substitution that maintained the charge of the Glu residue at position 791 (Glu791→Asp). For each mutation, we attempted to rescue infectious HAV from two independent clones by in vitro transcription of the cDNA with SP6 RNA polymerase, followed by liposome-mediated transfection of the RNA transcripts into permissive FRhK-4 cells (see Materials and Methods). Cell lysates were prepared 2 weeks after transfection, and the presence of virus was determined by RIFA in BS-C-1 cells.
No infectious virus was recovered from any of the mutants with substitutions involving the potential Gln745-Ser or Glu776-Ser dipeptide cleavage sites: 745N, 745R, 776Q, or 776R (Table 1). The lethal nature of these mutations, even when the substitution was conservative in terms of the likelihood of preserving 3Cpro recognition (Table 1), suggests that these dipeptide sequences are unlikely to function as substrates for 3Cpro cleavage during virus replication. Alternatively, if these are sites of 3Cpro cleavage, they must possess highly stringent requirements for 3Cpro recognition.
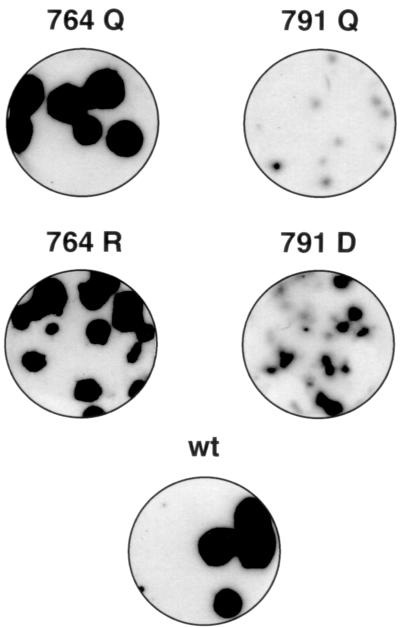
No virus was rescued from the 791R mutant (Table 1). The lethal nature of this mutant is consistent with what would be expected if Glu791-Ser were a 3Cpro cleavage site. In contrast, an HAV-specific cytopathic effect was observed after transfection with the 791D RNA transcripts, and virus was detected in lysates of these cells by RIFA (Fig. 4). No cytopathic effect was observed following transfection with the 791Q mutant, but small viral replication foci were detected by RIFA in BS-C-1 cells infected with lysates of these cells (Table 1 and Fig. 4). With both the 791Q and 791D mutants, the titers of infectious HAV were lower and RIFA foci were smaller than those of the parental virus (Table 1 and Fig. 4). However, these attributes were more pronounced with the 791Q mutant than with the 791D mutant. Taken together, these results suggest that conservation of the charge of the Glu791 residue (as in the 791D mutant) may be more important for viral replication than preserving a good context for 3Cpro recognition (as in the 791Q mutant). These data thus make the Glu791-Ser dipeptide an improbable 3Cpro substrate.
FIG. 4.
RIFA assay of mutant HAVs. Petri dish cultures of BS-C-1 cells were infected with wild-type (wt) virus (5′P2P3-18f) or with HAV mutants containing the indicated substitutions at residue 764 or 791. The inocula for the mutant viruses were lysates of infected FRhK-4 cells, collected 2 weeks after transfection with synthetic genome-length RNA. The BS-C-1 cell cultures were maintained for 1 week at 37°C before processing for detection of HAV radioimmunofoci as described in Materials and Methods.
Substitutions involving Glu764-Ser were of particular interest, since this dipeptide is located either at or very near the site of the VP1/2A cleavage (Fig. 3A). Thus, it is noteworthy that transcripts encoding either a Gln or an Arg at position 764 (764Q or 764R) yielded viable viruses. Both of these mutant viruses replicated to titers comparable to those for the parental virus (Table 1). Moreover, they produced RIFA foci only slightly smaller than (764R) or the same size as (764Q) those of the parental virus (Fig. 4). The latter observation is particularly important, since it indicates that the Glu→Arg substitution in 764R, which would be expected to abolish 3Cpro recognition of the Glu764-Ser dipeptide, had no appreciable effect on replication of the virus.
The viable HAV mutants that were rescued from transfected FRhK-4 cells were subjected to an additional passage in these cells. This led to an amplification of the titer of the 791Q mutant, which nonetheless remained 10- to 100-fold lower than that of parental virus (Table 1). For each of the mutants, however, the size of the RIFA foci remained unchanged after this additional passage (data not shown). In all cases, the RNA sequences of the viruses recovered, either directly from the transfected cells or following an additional passage in FRhK-4 cells, retained the nucleotide substitutions that had been introduced at the suspect 3Cpro cleavage sites by site-directed mutagenesis.
Cleavage at the VP1/2A junction during replication of virus with amino acid substitutions at Glu764 or Glu791.
Since the VP1/2A precursor is present in small quantities in some purified preparations of infectious HAV particles (2, 5), the VP1/2A cleavage might not be necessary for replication of the virus. If this were the case, then the viable mutant viruses with substitutions at Glu764 or Glu791 could still have a defect in their ability to fully process the VP1-2A precursor to mature VP1. To exclude this possibility, we infected FRhK-4 cells with these mutants and at various times p.i. prepared cytoplasmic extracts for immunoblot analysis (Fig. 5). VP1- and VP2-reactive proteins were detected with a mixture of anti-VP1 and anti-VP2 antibodies. VP0 and/or VP2 were detected in lysates of cells infected with each of these mutants with kinetics similar to those of the parental virus (Fig. 5).
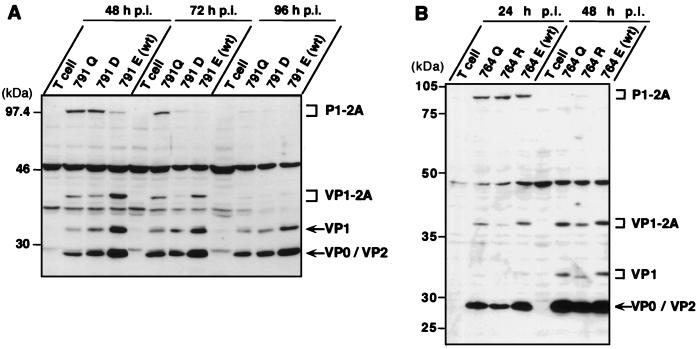
FIG. 5.
HAV 764 and 791 mutants produce a fully processed, mature VP1 capsid protein. FRhK-4 cells were mock infected (T cell) or infected with the indicated mutant at an MOI of 1 (A) or 2 (B) RFU/cell, and cytoplasmic extracts were prepared at the indicated times p.i. Proteins were separated by SDS-PAGE (10% gel) and identified in immunoblots with a mixture of anti-VP1 and anti-VP2 antibodies. VP0 and VP2 bands are indistinguishable in these gels. Positions of molecular weight standards and of precursors and mature HAV polypeptides are shown at the left and right, respectively, of each panel. wt, wild type.
Each of the VP1-containing polypeptides (P1-2A, VP1-2A, and the mature VP1) could be detected within 48 h of infection with the 791Q and 791D mutants at an MOI of 1 RFU/cell (Fig. 5A). Between 48 and 72 h p.i., the P1-2A precursor appeared to be progressively converted to VP1-2A and then fully processed into VP1 in cells infected with either mutant (Fig. 5A). The processing of the 791Q mutant may have been somewhat delayed, as a significant amount of the P1-2A product remained present as late as 72 h p.i. However, P1-2A and VP1-2A were almost completely processed into VP1 in cells infected with each of these mutant viruses by 96 h p.i. (Fig. 5A). Interestingly, the electrophoretic mobilities of the P1-2A and VP1-2A products were slightly altered by the Glu→Gln change introduced at residue 791 of the 791Q mutant compared to those of parental virus. However, the electrophoretic mobilities of the fully processed VP1 proteins were identical for both mutants and parental virus (Fig. 5A). This result indicates that the C terminus of VP1 lies upstream of residue 791.
The fully processed VP1 protein was also produced in FRhK-4 cells infected with the 764Q and 764R mutants at an MOI of 2 RFU/cell (Fig. 5B). There were no apparent delays in the processing of the VP1 intermediates in comparison to the wild-type virus. Importantly, however, the electrophoretic mobilities of the P1-2A, VP1-2A, and VP1 proteins were each significantly altered by the Glu→Gln substitution at residue 764 of the mutant 764Q and even more by the Glu→Arg change in 764R (Fig. 5B). This finding indicates that the mature VP1 protein contains residue 764 and that the VP1/2A cleavage must therefore be downstream of the potential 3Cpro cleavage site at Glu764/Ser.
DISCUSSION
Viruses of the genus Hepatovirus are unique among the Picornaviridae with respect to the primary cleavage of the viral polyprotein, which is carried out by the viral 3Cpro proteinase at the 2A/2B junction (13, 21). The capsid protein precursor, polypeptide P1-2A, is subsequently cleaved by 3Cpro to generate VP0, VP3, and VP1-2A (Fig. 2 and references 20, 24, and 26). In contrast to these well-documented events, considerable uncertainty has surrounded the identification of the junction between VP1 and 2A and the mechanism of this cleavage. One difficulty has been that we and others (24) have been unable to detect the 2A protein in HAV-infected cells. This has made it impossible to purify this protein and determine its N-terminal residue by microsequencing. Our inability to detect 2A could be because it is highly unstable and rapidly degraded, and/or because it is not released from VP1-2A as an intact protein. The role of the 2A polypeptide sequence in virus replication has also been elusive. The 2A proteins of hepatoviruses show no sequence homologies with the 2A proteinase of the enteroviruses and rhinoviruses nor with the cardiovirus 2A protein. The HAV 2A polypeptide is present as a C-terminal extension of VP1 (VP1-2A = PX) in viral pentamers but is generally absent or present in only small quantities in mature virions (2, 5). The mechanism of the VP1/2A cleavage remains unclear, as indicated by the lack of unambiguous reports concerning 3Cpro-mediated cleavage of VP1 from various substrates (13, 20, 24–26).
Several approaches were undertaken in this study in an effort to elucidate the process leading to maturation of the VP1 capsid protein. We determined the approximate C terminus of the mature VP1 protein that is present in infectious virions, by comparing its electrophoretic mobility in SDS-PAGE with those of a series of truncated HAV VP1-2A proteins expressed by recombinant vaccinia viruses (Fig. 3A). The C termini of these recombinant proteins corresponded to potential 3Cpro cleavage sites that may represent the VP1/2A junction: Gln745-Ser, Glu764-Ser, Glu776-Ser, or Glu791-Ser. Of these four potential 3Cpro cleavage sites, two are unlikely to be sites of a 3Cpro-directed VP1 maturation cleavage, given other information. First, the C terminus of VP1 is unlikely to be located at Glu791 since the mature VP1 protein isolated from HAV virions has been shown to have a mass less than that of a recombinant polypeptide representing aa 492 to 791 of the polyprotein (9). Similarly, the most N-terminal of these potential sites, Gln745-Ser, is also unlikely to be the site of VP1/2A cleavage, since a neutralizing monoclonal antibody, H7-C27, binds in enzyme-linked immunosorbent assays to nested synthetic octapeptides representing aa 747 to 759 (our unpublished results). This leaves Glu776-Ser, which is conserved among all HAV strains for which the nucleotide sequence has been determined, and Glu764-Ser, which is not fully conserved but is Val764-Ser in one cell culture-adapted HAV variant (6). It was of interest, therefore, that we found the mature VP1 protein isolated from virions to have an electrophoretic mobility indistinguishable from that of a recombinant VP1 protein with a C terminus at Glu764 (Fig. 3B). This result demonstrated that the VP1 C terminus is located close to, if not at, residue 764 of the polyprotein.
Crystallographic studies of the HAV 3Cpro proteinase suggest that a Glu-Ser dipeptide, such as that present at residues 764-765 of the HAV polyprotein, could constitute a substrate for this proteinase, even though 3Cpro has been shown to exhibit a marked preference for Gln at the P1 position immediately upstream of the scissile bond (1, 3, 14). In support of this argument, the predicted HAV 3A/3B cleavage site, which has not yet been confirmed by protein sequencing, is also thought to involve a Glu residue at the P1 position (in this case, a Glu-Gly dipeptide). However, although we have recently shown that a Glu-Ser dipeptide is partially cleaved in trans by 3Cpro when introduced in lieu of the normal Gln-Ala dipeptide at the 2A/2B site, we have found that a LeuP4-ProP3-ThrP2-GluP1-SerP′1 pentapeptide sequence (which corresponds to the sequence context of the putative 3Cpro cleavage of VP1/2A Glu764-Ser765) is not cleaved at all when introduced at the 2A/2B junction (our unpublished results). This is probably due to the additive deleterious effects of two unfavorable residues, the Pro at the P3 position as well as the Glu at the P1 position (2b). Furthermore, a Val residue, such as found at position 764 of the attenuated HM175 p35 strain (6), is not a suitable P1 residue for 3Cpro trans cleavage (our unpublished results). All of these data suggest that 3Cpro-mediated cleavage of the HAV polyprotein at the Glu764-Ser765 dipeptide is not a very likely scenario, despite the fact that Glu764 is close to or at the C terminus of VP1 (Fig. 3B). That 3Cpro is likely not to cleave this dipeptide is further indicated by the failure of the HAV proteinase to process VP1-2A into a mature VP1 protein when P1-2A and 2BC-P3 polypeptides were expressed in vivo by recombinant vaccinia viruses, whereas 3Cpro is fully active to process P1-2A at other junctions (Fig. 2).
These observations led us to test whether the Glu764-Ser dipeptide was an actual substrate for 3Cpro in the context of the full-length polyprotein by introducing various amino acid substitutions at Glu764 that were designed to either maintain or abolish potential recognition by the 3Cpro proteinase. The transfection of FRhK-4 cells with an HAV RNA transcript encoding a Glu764→Arg substitution at this putative P1 residue resulted in the rescue of a mutant virus (764R) with replication properties similar to those of the parental virus (Table 1 and Fig. 4). This substitution should completely abolish 3Cpro substrate recognition, as we have shown previously in studies of the 2A/2B primary cleavage site (21). We ruled out the possibility that the 764R HAV mutant was viable in the absence of VP1/2A cleavage, by demonstrating that the maturation of VP1 proceeded with normal kinetics in cells infected with this 764R mutant (Fig. 5). Taken together, these results argue strongly against the hypothesis that maturation of the VP1 protein occurs by 3Cpro-mediated cleavage at the Glu764-Ser dipeptide.
This conclusion contradicts a recent report describing the processing of HAV polypeptides in a transient expression system involving Cos-7 cells (24). VP1 was reported to be cleaved in trans by 3Cpro from a P1-2A substrate containing a Glu764 residue, but much less efficiently when a Val764 residue was present (as in the cell culture-adapted HM175 p35 variant). The authors concluded that 3Cpro effects a VP1/2A cleavage at the 764-765 dipeptide on the basis of (i) the variation in the efficiency with which these substrates were processed and (ii) the similar electrophoretic mobilities of the VP1 product of these cleavages and a polypeptide corresponding to residues 492 to 764 of the polyprotein. The latter result obtained with polypeptides from the HAS-15 HAV strain is in agreement with our observations concerning the mass of the VP1 protein of the HM175 strain (Fig. 3). It is surprising that the inefficient 3Cpro processing of the P1-2A substrate containing Val764 that was observed by Probst et al. (24) in their transient expression system was not reflected in inefficient maturation of the VP1 protein in cells infected with the attenuated HM175 variant. Indeed, in contrast to the situation with transient expression of P1-2A, VP1 was efficiently cleaved from VP1-2A during the replication of this virus which contains a Val764 residue (24). This can be construed as further evidence that 3Cpro does not mediate the maturation of VP1 from its VP1-2A precursor during replication of the virus.
Considering that the C terminus of the mature VP1 protein is located just downstream of residue 764 (Fig. 3 and 5) and that the only apparent 3Cpro recognition sequence in the vicinity of this residue (Glu764-Ser765) is not cleaved by 3Cpro during replication of the virus, we suggest that 3Cpro is not directly responsible for the maturation of VP1. It is possible that a downstream 3Cpro cleavage of the VP1-2A precursor, at an alternative 3Cpro recognition site, may result in an intermediate protein that is subsequently subjected to C-terminal trimming by a cellular proteinase once pentamers or provirions are formed. Such a situation is found in the case of the mengovirus (cardiovirus) VP1 capsid protein, which is posttranslationally trimmed by three carboxy-terminal amino acids following an initial VP1/2A cleavage mediated by 3Cpro (4). In the case of HAV, the presence of minor anti-VP1-reactive polypeptides with molecular masses intermediate between those of VP1-2A and VP1 in HAV-infected cells (15) would support such a mechanism. However, the results of our experiments involving mutagenesis of the potential 3Cpro cleavage sites that are located downstream of residue 764 (i.e., Glu776-Ser and Glu791-Ser) do not provide support for this hypothesis (Table 1). Indeed, the lethal nature of the Glu776→Gln substitution, which would be expected to enhance 3Cpro recognition and cleavage, argues against 3Cpro cleavage at the Glu776-Ser dipeptide. The Glu791-Ser dipeptide also seems unlikely to constitute a substrate for 3Cpro. Even though a Glu791→Arg substitution proved lethal to replication, we found that conservation of the charge of the putative P1 residue (Glu791→Asp substitution) was more important than maintaining a 3Cpro recognition sequence (Glu791→Gln substitution) to preserve virus replication. Thus, it seems that there is no sequence downstream of residue 764 in the vicinity of the VP1-2A cleavage that is recognized by 3Cpro. This finding suggests that the cleavage of VP1 from its VP1-2A precursor is not dependent on 3Cpro, which is the only proteinase known to be expressed by HAV.
This observation raises the possibility that a cellular proteinase contributes to the mechanism of VP1/2A cleavage during the replication of HAV. If so, such a major role for a cellular proteinase in the processing of the polyprotein would be unique to HAV among all picornaviruses. This interpretation leaves unanswered the question as to why no maturation of the VP1 protein from its VP1-2A precursor was observed in FRhK-4 cells expressing P1-2A and 2BC-P3 (3Cpro) from recombinant vaccinia viruses (Fig. 2). The entire HAV protein complement is present in this system, and FRhK-4 cells are permissive for virus replication and thus must express any putative cellular proteinase involved in maturation of the HAV capsid protein. Perhaps such a proteinase is induced by HAV infection and not present in sufficient quantities in uninfected FRhK-4 cells to render its detection possible. A difference in the abundance of this unknown cellular proteinase in the different cell types used in our study and in that of Probst et al. (24) may explain the discrepancy relating to whether processing of a P1-2A substrate to a mature VP1 product can occur. Alternatively, this unknown proteinase may be sequestered within an isolated compartment of the FRhK-4 cells which is not accessible to vaccinia-expressed HAV polypeptides. However, it cannot be ruled out that VP1-2A becomes a competent substrate for this proteinase only after assembly of the virus particle.
ACKNOWLEDGMENTS
We are grateful to Nicolas Escriou and Czeslaw Wychowski for their interest in this work and helpful discussions and to Marc Girard for continuous support. We thank Bernard Moss for providing plasmid pTM1 and the recombinant virus vTF7-3, and we thank Stephen Feinstone for the gift of anti-HAV 3C antibodies.
This work was supported in part by the Pasteur Institute and CNRS and in part by grant AI32599 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Allaire M, Chernaia M M, Malcolm B A, James M N. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature. 1994;369:72–76. doi: 10.1038/369072a0. [DOI] [PubMed] [Google Scholar]
- 2.Anderson D A, Ross B C. Morphogenesis of hepatitis A virus: isolation and characterization of subviral particles. J Virol. 1990;64:5284–5289. doi: 10.1128/jvi.64.11.5284-5289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Beard, M., L. Cohen, A. Martin, and S. M. Lemon. Unpublished data.
- 2b.Bergmann, E. M. Personal communication.
- 3.Bergmann E M, James M N G. Proteolytic enzymes of the viruses of the family Picornaviridae. In: Dunn B, editor. Proteinases of infectious agents. San Diego, Calif: Academic Press; 1999. pp. 139–163. [Google Scholar]
- 4.Boege U, Scraba D G. Mengo virus maturation is accompanied by C-terminal modification of capsid protein VP1. Virology. 1989;168:409–412. doi: 10.1016/0042-6822(89)90284-5. [DOI] [PubMed] [Google Scholar]
- 5.Borovec S V, Anderson D A. Synthesis and assembly of hepatitis A virus-specific proteins in BS-C-1 cells. J Virol. 1993;67:3095–3102. doi: 10.1128/jvi.67.6.3095-3102.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen J I, Rosenblum B, Ticehurst J R, Daemer R J, Feinstone S M, Purcell R H. Complete nucleotide sequence of an attenuated hepatitis A virus: comparison with wild-type virus. Proc Natl Acad Sci USA. 1987;84:2497–2501. doi: 10.1073/pnas.84.8.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen J I, Ticehurst J R, Feinstone S M, Rosenblum B, Purcell R H. Hepatitis A virus cDNA and its RNA transcripts are infectious in cell culture. J Virol. 1987;61:3035–3039. doi: 10.1128/jvi.61.10.3035-3039.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnelly M L, Gani D, Flint M, Monaghan S, Ryan M D. The cleavage activities of aphthovirus and cardiovirus 2A proteins. J Gen Virol. 1997;78:13–21. doi: 10.1099/0022-1317-78-1-13. [DOI] [PubMed] [Google Scholar]
- 9.Dotzauer A, Vallbracht A, Keil G M. The proposed gene for VP1 of HAV encodes for a larger protein than that observed in HAV-infected cells and virions. Virology. 1995;213:671–675. doi: 10.1006/viro.1995.0040. [DOI] [PubMed] [Google Scholar]
- 10.Drillien R, Spehner D, Kirn A. Complementation and genetic linkage between vaccinia virus temperature-sensitive mutants. Virology. 1982;119:372–381. doi: 10.1016/0042-6822(82)90096-4. [DOI] [PubMed] [Google Scholar]
- 11.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthetizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gauss-Muller V, Lottspeich F, Deinhardt F. Characterization of hepatitis A virus structural proteins. Virology. 1986;155:732–736. doi: 10.1016/0042-6822(86)90234-5. [DOI] [PubMed] [Google Scholar]
- 13.Gosert R, Cassinotti P, Siegl G, Weitz M. Identification of hepatitis A virus non-structural protein 2B and its release by the major virus protease 3C. J Gen Virol. 1996;77:247–255. doi: 10.1099/0022-1317-77-2-247. [DOI] [PubMed] [Google Scholar]
- 14.Jewell D A, Swietnicki W, Dunn B M, Malcom B A. Hepatitis A virus 3C proteinase substrate specificity. Biochemistry. 1992;31:7862–7869. doi: 10.1021/bi00149a017. [DOI] [PubMed] [Google Scholar]
- 15.Jia X Y, Summers D F, Ehrenfeld E. Primary cleavage of the HAV capsid protein precursor in the middle of the proposed 2A coding region. Virology. 1993;193:515–519. doi: 10.1006/viro.1993.1157. [DOI] [PubMed] [Google Scholar]
- 16.Lemon S M, Binn L N, Marchwicki R H. Radioimmunofocus assay for quantitation of hepatitis A virus in cell cultures. J Clin Microbiol. 1983;17:834–839. doi: 10.1128/jcm.17.5.834-839.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemon S M, Murphy P C, Shields P A, Ping L H, Feinstone S M, Cromeans T, Jansen R W. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J Virol. 1991;65:2056–2065. doi: 10.1128/jvi.65.4.2056-2065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemon S M, Robertson B H. Current perspectives in the virology and molecular biology of hepatitis A virus. Semin Virol. 1993;4:285–295. [Google Scholar]
- 19.Linemeyer D L, Menke J G, Martin-Gallardo A, Hughes J V, Young A, Mitra S W. Molecular cloning and partial sequencing of hepatitis A viral cDNA. J Virol. 1985;54:247–255. doi: 10.1128/jvi.54.2.247-255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malcolm B A, Chin S M, Jewell D A, Stratton-Thomas J R, Thudium K B, Ralston R, Rosenberg S. Expression and characterization of recombinant hepatitis A virus 3C proteinase. Biochemistry. 1992;31:3358–3363. doi: 10.1021/bi00128a008. [DOI] [PubMed] [Google Scholar]
- 21.Martin A, Escriou N, Chao S-F, Girard M, Lemon S M, Wychowski C. Identification and site-directed mutagenesis of the primary (2A/2B) cleavage site of the hepatitis A virus polyprotein: functional impact on the infectivity of HAV RNA transcripts. Virology. 1995;213:213–222. doi: 10.1006/viro.1995.1561. [DOI] [PubMed] [Google Scholar]
- 22.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 23.Palmenberg A C. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 24.Probst C, Jecht M, Gauss-Muller V. Proteinase 3C-mediated processing of VP1-2A of two hepatitis A virus strains: in vivo evidence for cleavage at amino acid position 273/274 of VP1. J Virol. 1997;71:3288–3292. doi: 10.1128/jvi.71.4.3288-3292.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schultheiss T, Kusov Y Y, Gauss-Muller V. Proteinase 3C of hepatitis A virus (HAV) cleaves the HAV polyprotein P2-P3 at all sites including VP1/2A and 2A/2B. Virology. 1994;198:275–281. doi: 10.1006/viro.1994.1030. [DOI] [PubMed] [Google Scholar]
- 26.Schultheiss T, Sommergruber W, Kusov Y, Gauss-Muller V. Cleavage specificity of purified recombinant hepatitis A virus 3C proteinase on natural substrates. J Virol. 1995;69:1727–1733. doi: 10.1128/jvi.69.3.1727-1733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stemmer W P C, Morris S K. Enzymatic inverse PCR: a restriction site independent, single-fragment method for high-efficiency, site-directed mutagenesis. BioTechniques. 1992;13:215–220. [PubMed] [Google Scholar]
- 28.Taylor J W, Ott J, Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate modified DNA. Nucleic Acids Res. 1985;13:8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Chao S-F, Ping L-M, Grace K, Clarke B, Lemon S M. An infectious cDNA clone of a cytopathic hepatitis A virus: genomic regions associated with rapid replication and cytopathic effect. Virology. 1995;212:686–697. doi: 10.1006/viro.1995.1526. [DOI] [PubMed] [Google Scholar]