Abstract
Simian parainfluenza virus 5 (SV5) is a prototype of the Paramyxoviridae family of nonsegmented negative-sense RNA viruses. The single-stranded RNA genomes of these viruses contain a series of tandemly linked genes separated by intergenic (IG) sequences flanked by gene-end (GE) and gene-start (GS) sequences. The viral RNA polymerase (vRNAP) complex is thought to enter the genome at its 3′ end, and synthesis of mRNAs is thought to occur by a stop-start mechanism in a sequential and polar manner, with transcriptional attenuation occurring primarily at the intergenic regions. As a result, multiple nonoverlapping mRNA species are generated for each single entry of the vRNAP. To investigate the functions of GE, IG, and GS sequences in transcription, we constructed plasmids containing cDNAs of the full-length SV5 genome in which the gene junction sequences (GE, IG, and GS sequences) located between the hemagglutinin-neuraminidase (HN) and the polymerase (L) genes were replaced with the counterpart sequences from other gene junctions. By using reverse genetics, we recovered viable viruses from each cDNA construct, although their growth characteristics varied. Analysis of the HN and L mRNAs by quantitative RNase protection assay indicated that the ratios of HN to L mRNAs varied over a fourfold range. The alteration of the gene junction sequences also permitted examination of the hypothesized requirement for hexamer nucleotide position of the GS sites. The recovery of infectious viruses with transcription initiation sites that occurred at nucleotide positions 1, 2, 3, 5, and 6 of the hexamer suggest that the requirement is nonstringent.
The paramyxovirus family of nonsegmented negative-sense RNA viruses is classified in the order Mononegavirales, which contains many important human and animal pathogens, including rabies virus, vesicular stomatitis virus (VSV), human parainfluenza virus, Sendai virus, mumps virus, simian parainfluenza virus (SV5), Newcastle disease virus (NDV), measles virus, rinderpest virus, human respiratory syncytial virus (HRSV), and Ebola virus.
The single-stranded RNA genomes of the Mononegavirales range from ∼11,000 to 16,000 nucleotides in chain length and contain a series of tandemly linked genes separated by nontranscribed sequences. For paramyxoviruses the gene order is 3′-NP-P(V/C)-M-F-(SH)-HN-L-5′, where genes in parentheses are not found in all species (Fig. 1) (reviewed in references 9 and 20). The viral RNA-dependent RNA polymerase (vRNAP) that transcribes the nucleocapsid protein (NP)-encapsidated RNA into 5′-capped and 3′-polyadenylated mRNAs minimally consists of two proteins, phosphoprotein (P) and the large (L) polymerase protein (11). The vRNAP is thought to enter the genome RNA at a single 3′ entry site and to transcribe the genome by a sequential and polar process (1, 2, 10, 16).
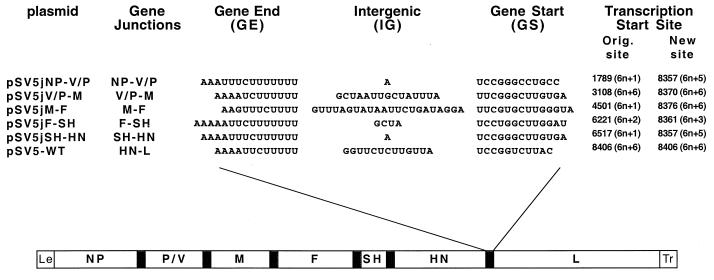
FIG. 1.
Gene organization and gene junctions of the paramyxovirus SV5. The three nucleotide sequence elements at the gene junction (GE, IG, and GS) are shown. Le, leader sequence which serves as the vRNAP entry site for transcription; Tr, trailer sequence which is important for RNA replication. SV5 encodes eight known viral proteins: nucleocapsid protein (NP), V protein (V), phosphoprotein (P), matrix protein (M), fusion protein (F), small hydrophobic protein (SH), hemagglutinin-neuraminidase (HN), and polymerase (L). P and V mRNAs are generated from the same V gene template via an RNA editing process: two nontemplated G residues are inserted into the primary V mRNA during transcription by a stuttering mechanism to generate the P mRNA (30). Plasmids containing different gene junctions are indicated and their GE, IG, and GS sequences are shown in vRNA sense. Transcription start sites are numbered from the 3′ end of the genome RNA. The position of the start site for each gene relative to a multiple of hexamers is shown as 6n + X (n is an integer). Nucleotide sequences from the SV5 complete genome sequence are available from GenBank under accession no. AF052755.
Although the exact details of mRNA production are not known, the process is currently believed to involve termination and reinitiation (stop and start) at each gene junction, and these junctions consist of three-nucleotide sequence elements. At the gene-end (GE) sequence, polyadenylation occurs through reiterative copying of a four- to seven-uridyl (U)-residue tract and transcription terminates releasing a polyadenylated RNA. At this juncture, the vRNAP either leaves the template (attenuation) or passes over an intergene (IG) region not found in mRNAs, and then reinitiates mRNA synthesis at a downstream transcriptional gene-start (GS) sequence. A failure to reinitiate transcription at a downstream site results in a gradient of mRNA production inversely proportional to the distance from the 3′ end of the genome, and this attenuation is not due to a stoichastic loss of vRNAP from the template (reviewed in references 9 and 20). For VSV it has been shown that the vRNAP spends more time crossing the gene junctions than in crossing the much larger encoding regions and that attenuation occurs at the IG regions (16). However, the degree of attenuation at each junction is not constant, occurring to the greatest extent at the junction between the glycoprotein gene (G or HN) and the L gene (reviewed in references 9 and 20).
Although the transcriptional processes for nonsegmented negative-sense RNA viruses are similar overall, comparisons of the gene junction sequences indicate important differences. For VSV, the gene junction sequences are highly conserved (29); for the paramyxoviruses Sendai virus and measles virus, the GS and GE sequences are invariant at each junction and the IG sequences are conserved (17). In contrast, for SV5 and HRSV the GE sequences are variable, and for SV5 the GS sequences are also variable. For SV5 and HRSV, the IG sequences vary from 1 to 22 nucleotides (SV5) or 1 to 52 nucleotides (HRSV). It was thought possible that the different gene junction sequences would lead to differential expression of the viral genes. By using a reverse genetics system and a dicistronic minigenome system, it has been found for HRSV that varying the IG sequences by using specific viral IG sequences does not affect transcriptional termination or reinitiation (19). Nonetheless, when the complete gene junction sequences of HRSV were inserted into a dicistronic minigenome system, modulation of HRSV transcriptional termination occurred (12). For SV5, analysis using the minigenome system revealed that specific (unnatural) mutations in the M-F junction sequences affect termination/reinitiation (26, 27).
The functional template for transcription of nonsegmented negative-sense RNA viruses is the helical nucleocapsid. For most members of the Paramyxovirinae, the nucleotide chain length is divisible by 6 (i.e., hexameric), and it is thought that each NP subunit interacts with precisely six nucleotides. For example, the SV5 RNA genome is 15,246 nucleotides, which is precisely 2,341 multiples of 6 nucleotides. It has been found that a hexameric length of the genome is important for efficient RNA replication (5, 21). This “rule of six” hypothesis has been extended to include the notion that transcription initiation for each gene start site occurs with similar phasing with respect to its position within the nucleotide hexamer (17). The rule of six does not apply to the Pneumovirinae subfamily of the Paramyxoviridae or the Rhabdoviridae.
To investigate the importance of the different gene junctions in controlling transcription, we substituted the junction sequences between HN and L genes with junction sequences from the other SV5 gene junctions in an infectious cDNA of SV5. By using reverse genetics, we recovered SV5 variants containing new gene junction sequences. This system has the advantage over dicistronic minigenome systems in that it permits comparison of different gene junctions at the same position in the viral genome during a natural viral infection without the complicating factors of using vaccinia virus infection or viral superinfection. We quantified the accumulation of the HN and L mRNAs in cells infected with the recovered SV5 variants (rSV5s). The data indicate that the different junction sequences have effects on mRNA production but the junction sequences are not responsible for the very low level of L mRNA found in virus-infected cells. The alteration of the gene junction sequences also permitted investigation of the hypothesized requirement for very similar hexamer nucleotide phasing of the GS sites (17). The recovery of infectious viruses with transcription initiation sites at nucleotide positions 1, 2, 3, 5, and 6 of the hexamer indicates the requirement is nonstringent.
MATERIALS AND METHODS
DNA and construction of plasmids.
A full-length infectious cDNA of the SV5 genome (pBH276) was described previously (13, 14). The sequences between an NcoI site located at the end of HN gene and the start codon for the L gene were replaced with sequences of identical length containing the GE, IG, and GS sequences from the NP-V/P, V/P-M, M-F, F-SH, and SH-HN junctions, using standard molecular cloning procedures (Fig. 1). Nucleotide sequences of the plasmids and details of the cloning procedures are available on request.
Plasmids expressing NP, P, and L proteins under the control of a bacteriophage T7 RNA polymerase promoter have been described previously (14). DNA oligonucleotides were synthesized by Macromolecular Resources (Colorado State University, Fort Collins).
Cells and viruses.
A549, MDBK, BHK 21F, and CV-1 cells were grown in Dulbecco modified Eagle medium (DMEM) containing 10% fetal calf serum (FCS). rSV5s were grown in MDBK or CV-1 cells in DMEM supplemented with 2% FCS. Plaque assays were performed in BHK 21F cells (24). MVA, a modified vaccinia virus containing a bacteriophage T7 RNA polymerase gene (6, 33), was grown in chicken embryo fibroblasts as described previously (6, 33).
Recovery of SV5 from cDNA.
To obtain infectious rSV5, A549 cells were infected with MVA, and then plasmids containing the full-length SV5 genomes were transfected into A549 cells together with plasmids encoding NP, P, and L under control of a T7 RNA polymerase promoter as described previously (14). At 24 h postinfection (p.i.), transfection media were changed to DMEM containing 10% FCS. The presence of virus released into the media was detected by infection of BHK 21F cells and the observation of syncytia. The recovered virus was plaque purified in BHK 21F cells and amplified in MDBK cells. The sequences of the rSV5 RNA between the HN and L junction regions were confirmed by reverse transcription-PCR (RT-PCR) sequencing using RNA purified from the virus-infected cells as described previously (14).
Quantification of mRNA by RNase protection assay (RPA).
A DNA template from which a negative-sense RNA probe to the L mRNA could be synthesized was obtained from the L gene coding sequences by PCR. One DNA oligonucleotide used in the PCR contained a T7 RNA polymerase promoter and annealed to the positive-sense L strand; the other annealed to the negative-sense L strand. Both oligonucleotides contained irrelevant sequences to differentiate undigested probes from the RNase-protected fragment. The resulting PCR fragment contained a T7 RNA polymerase promoter, and from this template a 251-nucleotide-long negative-sense RNA probe could be generated; 230 of 251 nucleotides of the RNA probe were a perfect match to the 5′ end of the L mRNA. DNA templates from which probes specific for the 3′ end of the HN mRNA, the 5′ end of the V/P gene, and the 5′ end of the F gene could be generated were obtained by using a similar scheme; details are available on request. The DNA templates were transcribed by using T7 RNA polymerase in the presence of [α-32P]UTP by using a MaxiScript Kit (Ambion Inc., Austin, Tex.) according to the manufacturer’s instructions.
CV-1 cells were infected with SV5 at a multiplicity of infection (MOI) of 1 PFU per cell; at 48 h p.i., poly(A)-containing RNA species were isolated as described previously (23). Hybridization of the 32P-labeled RNA probes to mRNA was performed with a HySpeed RPA kit (Ambion). To minimize handling errors, 32P-labeled RNA probes were used in pairs in one hybridization reaction; e.g., to examine the HN/L mRNA ratios, both the HN and L probes were hybridized to the same aliquots of mRNA. Protected RNA fragments were analyzed on 6% polyacrylamide gels containing 8 M urea. Quantification of radioactivity was performed with a Storm Image system (Molecular Dynamics, Sunnyvale, Calif.).
RESULTS
Rescue of rSV5 containing altered HN/L gene junction sequences.
To examine the importance of different SV5 gene junctions in controlling transcription, we substituted the gene junction sequences between the HN and L genes with the junction sequences found between NP and V/P, V/P and M, M and F, F and SH, and SH and HN in cDNA of SV5 (Fig. 1). The junction between HN and L was chosen because it is the last gene junction, and we anticipated that any detrimental effect of its replacement would be minimized since there is only one gene downstream of the junction. The genome length of the expected rSV5 was kept the same as that of wild-type (wt) rSV5. To derive rSV5 from plasmid DNAs, the plasmids (Fig. 1) containing full-length SV5 genomes with the substituted junction sequences between HN-L, together with plasmids containing NP, P, and L genes (under control of a T7 promoter), were transfected into A549 cells which had been preinfected with MVA, a modified vaccinia virus encoding T7 RNA polymerase. All SV5s were recovered; the viruses were plaque purified, and their HN-L junction sequences were confirmed by RT-PCR sequencing (data not shown). Although frequency of rescue cannot be strictly quantified, no differences were observed in the ability to rescue any of the recombinant viruses; i.e., rescue was obtained from each of 6- by 6-cm-diameter plates for each virus on the first attempt. rSV5s were designated according to the new junction (j) sequence that they contained as indicated in Fig. 1.
Growth phenotypes of rSV5s.
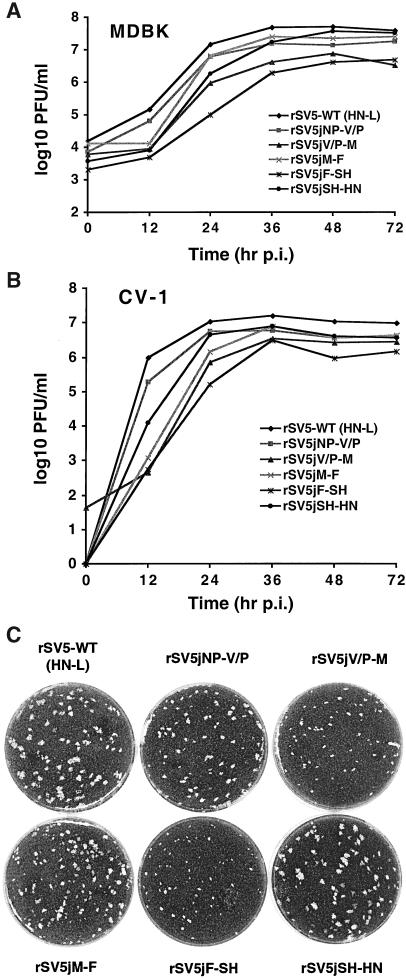
The growth characteristics of the rSV5s were examined in MDBK and CV-1 cells. MDBK cells were used because SV5 production from infected MDBK cells continues (after a plateau is reached) for many days with minimal cytopathic effect (7). In a single-step growth curve in MDBK cells infected with an MOI of 10 PFU/cell (Fig. 2A), it was found that from 12 to 24 h p.i., all rSV5s grew to a titer 1 to 2 logs lower than that of wt rSV5, but by 30 to 72 h p.i., their titers were only about 1 log lower than that of wt virus. In a single-step growth curve in CV-1 cells infected at 1 PFU/cell (Fig. 2B), all rSV5s grew more slowly than wt virus for the first 24 h but reached a titer that was within 1 log lower than that of wt SV5 by 36 to 72 h p.i. The plaque sizes of rSV5s in BHK 21F cells were also examined. SV5 does not form visible plaques in MDBK cells and forms only small plaques in CV-1 cells, but it does form readily visible plaques in BHK cells. The efficiencies of plaquing in BHK cells and CV-1 cells are comparable. It was found that among all rSV5s, rSV5jV/P-M and rSV5jF-SH yielded the smallest plaques (Fig. 2C), and thus the rSV5s with slowest growth in MDBK and CV-1 cells correspond to those with reduced plaque size in BHK cells.
FIG. 2.
Growth and plaque sizes of rSV5s. (A) Single-step growth curve of rSV5 in MDBK cells. MDBK cells were infected with rSV5s containing different junction sequences as indicated at an MOI of 10 PFU, and media were collected at 12-h intervals (24 h at the last point). (B) Single-step growth curves of rSV5 in CV-1 cells. CV-1 cells were infected with rSV5s containing different junction sequences as indicated at an MOI of 1 PFU. Viral titers were measured by plaque assay using BHK 21F cells. (C) Representative BHK 21F cell monolayers showing the plaques produced by rSV5s. Monolayers were stained with Giemsa stain as described elsewhere (24).
Expression of HN and L mRNAs.
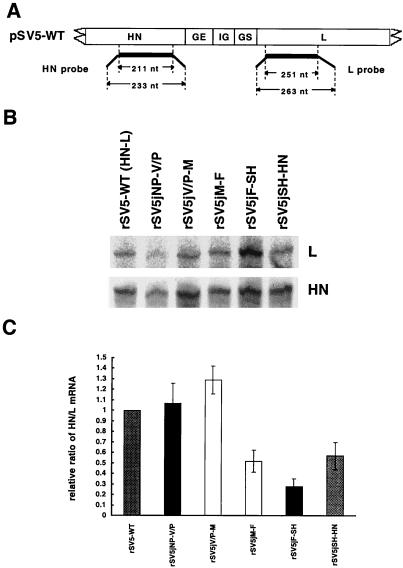
To investigate whether expression levels of the mRNAs for HN and L were altered by substitution of their gene junction sequences with those found for the other SV5 genes, the levels of HN and L mRNAs that accumulated in rSV5-infected cells were compared by RPA. RPA analysis was chosen over Northern blot analysis because an RPA yields data than can be meaningfully quantified. CV-1 cells were infected with rSV5s at an MOI of 1 PFU per cell. At 48 h p.i., no visible cytopathic effect was observed; at this time, poly(A)-containing mRNAs were isolated and annealed to 32P-labeled RNA probes specific for HN or L (Fig. 3A). A representative result of RPA using RNA probes specific for HN and L mRNAs is shown in Fig. 3B. Different exposure times were used to detect the HN and L protected RNA fragments. The radioactivity in the HN and L bands was quantified, and the relative ratio of HN mRNA to L mRNA that accumulated in wt rSV5-infected cells was set as 1.0. A histogram showing the quantification of five different experiments is shown in Fig. 3C. The relative ratio of HN/L mRNA accumulation for rSV5jV/P-M (1.29) was found to be slightly higher than those for rSV5jNP-V/P (1.07) and wt rSV5 (1.0). In contrast, HN/L mRNA ratios were lower for rSV5jM-F (0.52) and rSV5jSH-HN (0.57) than for wt rSV5. rSV5jF-SH had the lowest HN/L mRNA ratio, 0.28.
FIG. 3.
RPA analysis of the levels of HN and L mRNA. (A) Schematic diagram of the probes. The sizes (in nucleotides [nt]) of the RNA probes and the protected fragments are shown. The thin line at the end of each probe represents sequences that will not hybridize to the target mRNAs, enabling distinction of the probes and the protected fragments. (B) RNA protected fragments for HN and L mRNAs. [α-32P]UTP-labeled specific RNA probes as indicated in panel A were synthesized as described in Materials and Methods. RPAs were carried out with the HN and L probes simultaneously in the same reaction, and both protected fragments were resolved in the same lane of a polyacrylamide gel. To obtain similar levels of exposure for the two protected species, two different exposures of the gels were used. (C) The HN/L mRNA ratio for each experiment, quantified with a Storm Image system. To compare different experiments, the HN/L mRNA ratio of each rSV5 was normalized to the HN/L mRNA ratio of wt rSV5, set as 1. The relative ratio of HN/L mRNA from five different experiments ± standard error of mean was obtained and is shown in the histogram. Numerical values are 1.07 ± 0.43 for rSV5jNP-V/P, 1.29 ± 0.30 for rSV5jV/P-M, 0.52 ± 0.23 for rSV5jM-F, 0.28 ± 0.16 for rSV5jF-SH, and 0.57 ± 0.29 for rSV5jSH-HN.
Expression of V/P and F mRNAs.
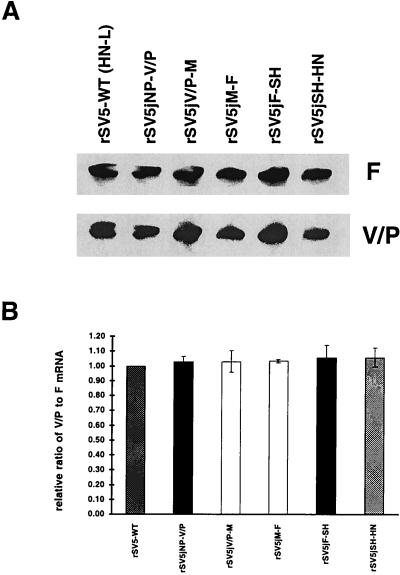
To ensure that the different ratios of HN/L mRNA accumulation observed were not due to experimental error, the ratio of V/P mRNA to F mRNA in rSV5-infected cells was examined. It was anticipated that transcription of upstream genes would not be affected by changes at the HN-L gene junction. A representative result of RPA using probes specific for V/P and F genes is shown in Fig. 4A, and a histogram showing the quantification of three different experiments is shown in Fig. 4B. The relative ratio of V/P mRNA to F mRNA that accumulated in wt rSV5-infected cells was set as 1.0. As expected, the relative ratios of V/P mRNA to F mRNA were found to be very similar for the different rSV5 viruses containing the altered HN-L gene junction sequences.
FIG. 4.
Relative ratios of V/P mRNA to F mRNA of rSV5s. (A) RPA of V/P mRNA and F mRNA of the rSV5s containing the different HN-L gene junction sequences. Probes were generated to the 5′-end regions of the V/P mRNA and F mRNA. (B) Relative ratios of V/P mRNA to F mRNA derived from three experiments ± standard error of the mean, calculated as described in the legend to the Fig. 3. Numerical values are 1.03 ± 0.04 for rSV5jNP-V/P, 1.03 ± 0.07 for rSV5jV/P-M, 1.03 ± 0.01 for rSV5jM-F, 1.061 ± 0.09 for rSV5jF-SH, and 1.06 ± 0.07 for rSV5jSH-HN.
Analysis of mRNA readthrough products.
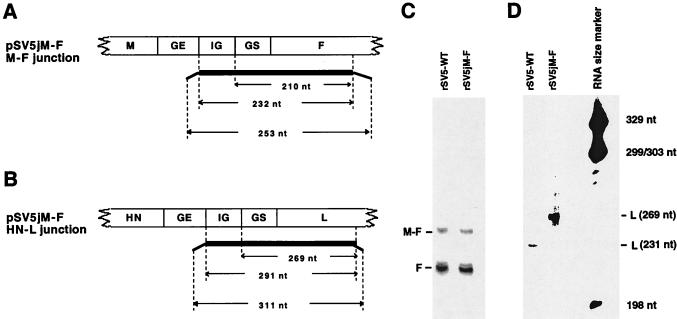
During mRNA transcription, inefficient polyadenylation/termination sometimes occurs. The result is bicistronic, and occasionally tricistronic, mRNAs containing IG sequences. Although readthrough transcription for VSV is a rare event, it occurs more frequently with some paramyxoviruses, e.g., Sendai virus and HRSV (reviewed in references 9, 20, and 31). For SV5, readthrough occurs more often at the SV5 M-F gene junction than at other gene junctions (23, 27). Interestingly, in SV5-infected cells the ratio of monocistronic to bicistronic M and F mRNAs decreases with time p.i. (27). As the probes used in the RPA analysis shown in Fig. 3 were against the coding sequences of the respective genes, the HN/L mRNA ratios measured are, theoretically, the ratios of HN mRNA (plus readthrough mRNA) to L mRNA (plus readthrough mRNA). The impact of the readthrough mRNAs from the junctions other than the M-F junction is negligible, as these junctions produce only small amounts of readthrough products. However, as readthrough products are detected most frequently at the SV5 M-F gene junction, we examined the amount of readthrough product produced at the HN-L junction in rSV5jM-F-infected cells by using probes (Fig. 5A and B) against both coding sequences and IG sequences in an RPA. As shown in Fig. 5C, an RNA probe (Fig. 5A) containing part of the F gene and the IG sequence between M and F was used to detect the readthrough products containing the M-F junction in poly(A)-containing RNA isolated from wt rSV5- and rSV5jM-F-infected cells. It was observed that the percentage of readthrough at the M-F junction for both viruses was similar to that found previously (∼20%) (27). To examine for HN-L readthrough products synthesize in rSV5jM-F-infected cells, a probe (Fig. 5B) that hybridized to the altered HN/L gene junction was made. This probe matched the 5′ end of the L mRNA, the (F) GS sequences and the (M-F) IG sequences of rSV5jM-F. The same probe matched only the 5′ end of the L mRNA of wt rSV5. When the probe was annealed to poly(A)-containing mRNA isolated from wt rSV5-infected cells in an RPA, the expected 231-nucleotide protected RNA species was observed (Fig. 5D). When the probe was annealed to poly(A)-containing mRNA isolated from rSV5jM-F-infected cells in an RPA, a 269-nucleotide protected RNA species was observed. Even though approximately 10 times more RNA was loaded on the gel for rSV5jM-F than the RNA for wt rSV5, a readthrough product (291 nucleotides) was not observed (Fig. 5D). As the HN/L readthrough product was not detected even for the M-F gene junction substitution of rSV5jM-F, a junction which has the highest readthrough products at its natural location, it appears reasonable to conclude that the HN/L mRNA ratios that we obtained from different viruses are fair reflections of the ratio of HN and L mRNAs. The unanticipated loss of the readthrough products at the HN and L gene junction of rSV5jM-F may be a result of a different RNA structural environment around the 3′ end (mRNA sense) of the HN gene and the 5′ end (mRNA sense) of the L gene on the viral RNA, low processivity of the vRNAP on the L gene template, or short half-life of the HN-L bicistronic mRNA.
FIG. 5.
Readthrough products at the HN-L junction of rSV5jM-F. (A and B) Schematic diagrams of the RNA probes used. The thin line at the end of each probe indicates nonhybridizing nucleotide sequences to allow distinction of undigested probes from the protected fragments. (A) Junction region of the M and F genes. The [α-32P]UTP-labeled RNA probe was 263 nucleotides (nt), the size of the protected fragment containing IG, GS, and F sequences was 232 nucleotides, and the size of protected F mRNA fragment was 210 nucleotides. (B) Junction region of the HN and L genes of rSV5jM-F. The RNA probe was 311 nucleotides, the size of the protected L mRNA fragment was 269 nucleotides, and the expected size of the protected readthrough product was 291 nucleotides. For wt rSV5-infected cells, the size of the protected L mRNA fragment was only 231 nucleotides due to a difference in IG, GS, and the 5′ nontranslated region sequences. (C) Readthrough at the M-F gene junctions for wt rSV5 and rSV5jM-F mRNAs. The RNA probe used in the experiment contains sequences covering the IG sequence between M and F and part of the F gene. F mRNA and readthrough fragments are indicated as F and M-F, respectively. (D) Lack of detection of the readthrough products at the HN-L junctions of rSV5jM-F mRNAs. The probe shown in panel B was used in the RPA against rSV5jM-F mRNA. The same probe was also used in an RPA using wt rSV5 mRNAs. The protected monocistronic L mRNA are indicated. A 32P-labeled RNA size marker was used. Note that approximately 10 times more RNA was loaded on the gel for rSV5jM-F than the RNA for wt rSV5.
The position of the transcription start site is independent of hexamer position.
The recovery of SV5s with the replaced gene junctions permitted examination of the hypothesis (17) that the relative position of the site for initiation of transcription of mRNAs within a hexamer is important. All transcription start sites for SV5 genes are 6n + 1, 6n + 2, or 6n + 6 (Fig. 1). In several of the rSV5s with altered HN-L gene junction sequences, the position within a hexamer of the transcription start site was changed from its original position or from the position of the L gene start site (Fig. 1). For example, the transcription start site for the V/P gene is 6n + 1, and that for the L gene is 6n + 6. In rSV5jNP-V/P, the transcription start site for the L gene was changed to 6n + 5, a position that is not utilized in SV5 and also is not used for other paramyxovirus genomes. Nonetheless, the relative ratios of HN/L mRNA for wt rSV5 and rSV5jNP-V/P are very similar. Thus, there does not appear to be an obvious correlation between the relative ratio of HN/L mRNA and the position of the transcription start site with respect to hexamer position.
DISCUSSION
The development of the scheme to recover infectious nonsegmented negative-sense viruses from molecularly cloned DNA (reviewed in references 22 and 28) has permitted analysis of many aspects of the function of viral proteins and cis-acting genome regulatory sequences. We have used the reverse genetics system that we developed for the paramyxovirus SV5 (13, 14) to recover novel SV5 genomes in which the junction sequences between the HN and L genes were replaced with the junction sequences of the other SV5 genes.
We had concerns over the likelihood of rescuing SV5 containing altered junction sequences. Earlier work had demonstrated that the ability to replicate VSV, Sendai virus, HRSV, and SV5 minigenomes and to rescue complete infectious genomes depended on a narrow range of the level of expression of the L and P proteins (21, 25). Although more detailed data are available only for VSV, NDV, and Sendai virus (1, 2, 8, 18), it is generally considered that transcriptional attenuation occurs to the greatest extent at the 5′-proximal gene junction, between the glycoprotein gene (G or HN) and the L protein gene (reviewed in reference 20). Thus, if replacement of the SV5 HN-L gene junction with that of another junction had altered dramatically accumulation of the L mRNA, the ability to rescue SV5 might have been compromised. Nonetheless, all SV5 genomes containing the altered HN-L junction sequences were recovered as viable viruses, although rSV5 containing the V/P-M and F-SH junction sequences substituted for the normal HN-L sequences showed a slower single-step growth curve and a smaller plaque size than wt virus. As the viruses were selected for growth in the rescue assay, the caveat has to be added that we have not determined if the rSV5s contain identical nucleotide sequences except for the newly introduced junction sequence. Nonetheless, the frequency of rescue of the viruses was not different from that of wt rSV5, and thus it does not appear that a selection pressure was unduly exerted. Even for VSV there may be more plasticity in the expression levels of viral proteins than originally anticipated, as it was found possible to rescue novel VSV genomes containing the N gene at several new locations in the gene order, although the new viruses did exhibit altered growth characteristics (32).
Previously we had shown that two recombinant SV5 isolates which encode green fluorescent protein (GFP) between the HN and L genes showed large differences in the relative expression level of GFP when the GFP gene was flanked by the NP-V/P junction sequences (high expression of GFP) or a second copy of the HN-L junction sequences (low expression of GFP) (14). Seemingly these data differ from those reported here, which indicate very similar patterns of attenuation over the NP-V/P and HN-L gene junction sequences. However, preliminary investigation using RT-PCR of the rSV5 containing the GFP gene under the control of the HN-L gene junction sequences indicates the existence of two genomes, the GFP-expressing virus and a pseudo-wt virus genome that has lost the GFP gene and contains a modified HN-L gene junction sequence (12a).
Quantification of the relative ratio of HN/L mRNA accumulated in wt SV5-infected cells and the cells infected with the novel SV5s containing altered HN-L junctions sequences indicated that although substitution of the junction sequences affected the HN/L mRNA ratio over a fourfold range, there was no drastic change in attenuation level. rSV5jV/P-M and rSV5jF-SH had the highest and lowest, respectively, HN/L mRNA ratios, and it is interesting that the altered HN/L mRNA ratio correlates with the altered growth phenotype of these viruses.
The mechanism by which the vRNAP interacts with the GE, IG, and GS sequences is not fully understood. For VSV, mutational analyses indicated that the seven-U tract in the GE sequences plays a critical role in governing transcription termination/polyadenylation (3, 15). However, for SV5 the length of the U tract in the GE sequences does not seem to be a major factor in determining attenuation at the HN-L gene junction, as wt rSV5, which contains five U residues, has a level of attenuation similar to that of rSV5jNP-V/P, which contains seven U residues. For HRSV, Kuo et al. found no effect of the IG sequences on transcription termination/polyadenylation when they compared the effects of the sequences in the context of the same GE and GS sequences, using a dicistronic minigenome (19). Our data for SV5 confirm that the length of the IG sequence alone is not a determining factor in termination/polyadenylation. rSV5jNP-V/P and wt SV5 have IG sequences of different lengths (1 and 13 nucleotides, respectively) at the HN-L junction, yet both viruses have an attenuation level higher than that for rSV5jF-SH, whose IG sequence is only four nucleotides. Nonetheless, our data suggest that the SV5 GE, IG, and GS sequences as a whole do have some role in modulating attenuation; this observation is similar to the finding for HRSV, where it was found that the gene junction sequences modulate gene expression (12).
Replacement of the HN-L junction sequences with the junction sequences from either NP-V/P and V/P-M, compared to wt SV5, had only a small effect on transcriptional attenuation over the HN-L junction, which suggests that other factors contribute to the low level of L mRNA accumulation in SV5-infected cells. For example, there may be poor processivity of the vRNAP on the L gene perhaps due to specific sequence elements; alternatively, the L mRNA may have a short half-life. To date, all in vivo assays used to quantify L mRNA (Northern blot analysis or RPA) have depended on selection of poly(A)-containing mRNA species. Thus, the assays measure only mRNA accumulation and would detect neither loss of polymerase processivity nor mRNA degradation. To distinguish between these two possibilities, it will be necessary to perform in vitro transcription assays; however, the in vitro transcription reaction for SV5 RNA (4) gives a very low level of activity and needs to be reconfigured for use in mRNA analysis.
ACKNOWLEDGMENTS
We thank Anthony Schmitt for helpful discussions and Heather MacLennon, Margaret Shaughnessy, and Helen Mercer for excellent technical help.
This work was supported in part by research grant AI-23173 from the National Institute of Allergy and Infectious Disease. B.H. is an Associate and R.A.L. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Abraham G, Banerjee A K. Sequential transcription of the genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:1504–1508. doi: 10.1073/pnas.73.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball L A, White C N. Order of transcription of genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:442–446. doi: 10.1073/pnas.73.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr J N, Whelan S P J, Wertz G W. cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buetti E, Choppin P W. The transcriptase complex of the paramyxovirus SV5. Virology. 1977;82:493–508. doi: 10.1016/0042-6822(77)90021-6. [DOI] [PubMed] [Google Scholar]
- 5.Calain P, Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll M W, Moss B. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: propagation and generation of recombinant viruses in a nonhuman mammalian cell line. Virology. 1997;238:198–211. doi: 10.1006/viro.1997.8845. [DOI] [PubMed] [Google Scholar]
- 7.Choppin P W. Multiplication of a myxovirus (SV5) with minimal cytopathic effects and without interference. Virology. 1964;23:224–233. doi: 10.1016/0042-6822(64)90286-7. [DOI] [PubMed] [Google Scholar]
- 8.Collins P L, Hightower L E, Ball L A. Transcriptional map for Newcastle disease virus. J Virol. 1980;35:682–693. doi: 10.1128/jvi.35.3.682-693.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins P L, McIntosh K, Chanock R M. Respiratory syncytial virus. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1313–1351. [Google Scholar]
- 10.Emerson S U. Reconstitution studies detect a single polymerase entry site on the vesicular stomatitis virus genome. Cell. 1982;31:635–642. doi: 10.1016/0092-8674(82)90319-1. [DOI] [PubMed] [Google Scholar]
- 11.Emerson S U, Yu Y-H. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J Virol. 1975;15:1348–1356. doi: 10.1128/jvi.15.6.1348-1356.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy R W, Harmon S B, Wertz G W. Diverse gene junctions of respiratory syncytial virus modulate the efficiency of transcription termination and respond differently to M2-mediated antitermination. J Virol. 1999;73:170–176. doi: 10.1128/jvi.73.1.170-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.He, B. Unpublished data.
- 13.He B, Leser G P, Paterson R G, Lamb R A. The paramyxovirus SV5 small hydrophobic (SH) protein is not essential for virus growth in tissue culture cells. Virology. 1998;250:30–40. doi: 10.1006/viro.1998.9354. [DOI] [PubMed] [Google Scholar]
- 14.He B, Paterson R G, Ward C D, Lamb R A. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology. 1997;237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- 15.Hwang L N, Englund N, Pattnaik A K. Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic junctions. J Virol. 1998;72:1805–1813. doi: 10.1128/jvi.72.3.1805-1813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iverson L E, Rose J K. Sequential synthesis of 5′-proximal vesicular stomatitis virus mRNA sequences. J Virol. 1982;44:356–365. doi: 10.1128/jvi.44.1.356-365.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolakofsky D, Pelet T, Garin D, Housmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolakofsky D, Vidal S, Curran J. Paramyxovirus RNA synthesis and P gene expression. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 215–233. [Google Scholar]
- 19.Kuo L, Fearns R, Collins P L. The structurally diverse intergenic regions of respiratory syncytial virus do not modulate sequential transcription by a dicistronic minigenome. J Virol. 1996;70:6143–6150. doi: 10.1128/jvi.70.9.6143-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1177–1204. [Google Scholar]
- 21.Murphy S K, Parks G D. Genome nucleotide lengths that are divisible by six are not essential but enhance replication of defective interfering RNAs of the paramyxovirus simian virus 5. Virology. 1997;232:145–157. doi: 10.1006/viro.1997.8530. [DOI] [PubMed] [Google Scholar]
- 22.Palese P, Zheng H, Engelhardt O G, Pleschka S, Garcia-Sastre A. Negative-strand RNA viruses: genetic engineering and applications. Proc Natl Acad Sci USA. 1996;93:11354–11358. doi: 10.1073/pnas.93.21.11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paterson R G, Harris T J R, Lamb R A. Analysis and gene assignment of mRNAs of a paramyxovirus, simian virus 5. Virology. 1984;138:310–323. doi: 10.1016/0042-6822(84)90354-4. [DOI] [PubMed] [Google Scholar]
- 24.Paterson R G, Lamb R A. The molecular biology of influenza viruses and paramyxoviruses. In: Davidson A, Elliott R M, editors. Molecular virology: a practical approach. Oxford, England: IRL Oxford University Press; 1993. pp. 35–73. [Google Scholar]
- 25.Pattnaik A K, Wertz G W. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J Virol. 1990;64:2948–2957. doi: 10.1128/jvi.64.6.2948-2957.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rassa J C, Parks G D. Highly diverse intergenic regions of the paramyxovirus simian virus 5 cooperate with the gene end U tract in viral transcription termination and can influence reinitiation at a downstream gene. J Virol. 1999;73:3904–3912. doi: 10.1128/jvi.73.5.3904-3912.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rassa J C, Parks G D. Molecular basis for naturally occurring elevated readthrough transcription across the M-F junction of the paramyxovirus SV5. Virology. 1998;247:274–286. doi: 10.1006/viro.1998.9266. [DOI] [PubMed] [Google Scholar]
- 28.Roberts A, Rose J K. Recovery of negative-strand RNA viruses from plasmid DNAs: a positive approach revitalizes a negative field. Virology. 1998;247:1–6. doi: 10.1006/viro.1998.9250. [DOI] [PubMed] [Google Scholar]
- 29.Rose J K. Complete intergenic and flanking gene sequences from the genome of vesicular stomatitis virus. Cell. 1980;19:415–421. doi: 10.1016/0092-8674(80)90515-2. [DOI] [PubMed] [Google Scholar]
- 30.Thomas S M, Lamb R A, Paterson R G. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988;54:891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner R R, Rose J K. Rhabdoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1121–1135. [Google Scholar]
- 32.Wertz G W, Perepelitsa V P, Ball L A. Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc Natl Acad Sci USA. 1998;95:3501–3506. doi: 10.1073/pnas.95.7.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyatt L S, Moss B, Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]