Abstract
Conversion of the normal protease-sensitive prion protein (PrP) to its abnormal protease-resistant isoform (PrP-res) is a major feature of the pathogenesis associated with transmissible spongiform encephalopathy (TSE) diseases. In previous experiments, PrP conversion was inhibited by a peptide composed of hamster PrP residues 109 to 141, suggesting that this region of the PrP molecule plays a crucial role in the conversion process. In this study, we used PrP-res derived from animals infected with two different mouse scrapie strains and one hamster scrapie strain to investigate the species specificity of these conversion reactions. Conversion of PrP was found to be completely species specific; however, despite having three amino acid differences, peptides corresponding to the hamster and mouse PrP sequences from residues 109 to 141 inhibited both the mouse and hamster PrP conversion systems equally. Furthermore, a peptide corresponding to hamster PrP residues 119 to 136, which was identical in both mouse and hamster PrP, was able to inhibit PrP-res formation in both the mouse and hamster cell-free systems as well as in scrapie-infected mouse neuroblastoma cell cultures. Because the PrP region from 119 to 136 is very conserved in most species, this peptide may have inhibitory effects on PrP conversion in a wide variety of TSE diseases.
The transmissible spongiform encephalopathy (TSE) diseases are fatal neurodegenerative diseases that include Creutzfeldt-Jakob disease, kuru, and Gerstmann-Sträussler-Scheinker syndrome in humans, scrapie in sheep and goats, and bovine spongiform encephalopathy in cattle (5, 28). TSE diseases are characterized by the accumulation of abnormal protease-resistant prion protein (PrP-res) which is derived from normal protease-sensitive prion protein (PrP-sen). PrP-res can be distinguished from PrP-sen by its increased β-sheet content, partial protease resistance, and tendency to form large aggregates (7, 24, 35). Although PrP-res by itself may induce functional damage to the central nervous system, the generation of spongiform pathology requires the presence of both PrP-res and PrP-sen (2).
PrP-res formation has been studied at three different levels: TSE-infected live animals, scrapie-infected tissue culture cells, and cell-free reactions in test tubes. At all three levels, certain restrictions between TSE agents from different species have been observed. One of these restrictions is the PrP amino acid sequence, which varies among species and appears to be important in PrP interactions involved in the species specificity of TSE diseases. For example, in transgenic mice expression of hamster PrP induces susceptibility to hamster scrapie (31, 32, 36). Conversely, coexpression of heterologous PrP molecules in transgenic mice can inhibit or delay onset of clinical TSE disease (27, 32), and expression of heterologous PrP in scrapie-infected mouse tissue culture cells can block the generation of mouse PrP-res (26). In the cell-free conversion system where incubation of PrP-res with PrP-sen leads to formation of new PrP-res, the species specificity of the conversion reaction appears to mirror the ability of various different TSE agents to cross from one species to another (1, 20, 33). In previous studies using chimeric recombinant PrP genes, sequences from the central region of PrP were found to be important in these species-specific effects (37). Furthermore, in recent studies a synthetic PrP peptide from the central portion of the hamster PrP molecule including residues 109 to 141 was able to directly inhibit the cell-free conversion of hamster PrP-sen to PrP-res (8). It was proposed that this peptide inhibited conversion possibly by substituting for PrP-sen or PrP-res in the binding interaction that leads to conversion (8).
In this study, using PrP-res derived from two mouse scrapie strains and one hamster scrapie strain in cell-free conversion reactions, we found formation of mouse and hamster PrP-res to be completely species specific. However, peptides corresponding to mouse PrP residues 108 to 140 (peptide MoP108-140) and hamster PrP residues 109 to 141 (peptide HaP109-141) were found to cross-inhibit cell-free conversion in both systems. Thus, the species-specific differences in these peptides were not important to the inhibition process. Strong inhibition was also obtained in analyses using a smaller PrP peptide from residue 119 to 136 with a sequence common to both mouse and hamster PrP. Furthermore, this peptide, P119-136, was found to inhibit the accumulation of PrP-res in a more physiological system using scrapie-infected murine neuroblastoma (Sc+MNB) cell cultures (30).
MATERIALS AND METHODS
Peptides.
Hamster peptides HaP119-128 (GAVVGGLGGY), HaP119-136 (GAVVGGLGGYMLGSAMSR), HaP121-141 (VVGGLGGYMLGSAMSRPMMHF), and HaP109-141 (MKHMAGAAAAGAVVGGLGGYMLGSAMSRPMMHF) and mouse peptide MoP108-140 (LKHVAGAAAAGAVVGGLGGYMLGSAMSRPMIHF) were synthesized by the Laboratory of Molecular Structure, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, Md. These peptides differ at three positions (underlined). Hamster PrP has a single amino acid insertion at position 54 compared with the mouse PrP; thus, mouse and hamster PrP amino acid residues differ by 1 in their numbering (21, 22, 34). Peptides were >95% pure, and analysis by high-pressure liquid chromatography revealed only a single peak. Alzheimer’s disease amyloid β protein fragment 1-40 (Aβ1-40) was purchased from Sigma. Lyophilized peptides were dissolved in deionized water at a concentration of 2 mM and stored at −20°C.
Purification and analysis of PrP-res.
PrP-res was purified by detergent lysis and differential centrifugation (13). Hamster 263K PrP-res was obtained from brains of scrapie-infected Syrian golden hamsters. Mouse Obihiro and mouse 87V PrP-res preparations were obtained from Slc/ICR or VM mice and were the generous gifts of Motohiro Horiuchi (Rocky Mountain Laboratories, Hamilton, Mont.) and James Hope (University of Edinburgh, Edinburgh, United Kingdom), respectively. The Obihiro mouse scrapie strain was originally derived from a scrapie-infected sheep (38) and was propagated in Slc/ICR mice by more than 20 passages at near-limiting dilution. The yield of PrP-res was determined by Western blotting with a polyclonal rabbit antiserum (R27) raised against a synthetic PrP peptide (residues 89 to 103) (6). The purity of the preparations was estimated at 50 to 60% by silver staining of gels after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). PrP-res preparations were diluted to 1 mg/ml in phosphate-buffered saline containing 1% sulfobetaine 3-14 and stored at −20°C until use.
Labeling and purification of PrP-sen.
Recombinant hamster and mouse PrP-sen molecules without the glycophosphatidylinositol (GPI) anchor (GPI negative) (19) were radiolabeled with Tran35S methionine/cysteine (Dupont-NEN) and purified as previously described (33). Radiolabeled GPI-negative mouse PrP-sen was immunoprecipitated from cell culture medium with rabbit polyclonal antiserum R27. Radiolabeled GPI-negative hamster PrP-sen was immunoprecipitated from lysed cells as previously described (33) by using mouse monoclonal antibody 3F4, which recognizes an epitope within hamster PrP containing methionine at positions 109 and 112 (16). The radiolabeled PrP-sen molecules were bound to protein A-Sepharose beads, eluted with 0.1 M acetic acid, and stored at 4°C.
Cell-free conversion assay.
The in vitro conversion reaction was performed as previously described (19, 33). Briefly, purified PrP-res was partially denatured with 2.5 M guanidine hydrochloride for 30 min at 37°C. An aliquot of 200 ng of partially denatured PrP-res was incubated with 12,000 cpm of immunoprecipitated 35S-labeled PrP-sen (∼1 ng) for 36 h at 37°C in the presence or absence of peptide. At the end of the incubation time, the reaction mixtures were digested with 50 μg of proteinase K (PK) per ml for 45 min at 37°C. At the concentrations used, none of the peptides affected the PK digestion of PrP-res (data not shown). One-tenth of the reaction mixture was reserved as a non-PK-treated control. Following PK digestion, a mixture of thyroglobulin (4 mg/ml) and Pefabloc (20 mM) was added, and the samples were precipitated in 5 volumes of methanol. The resultant pellets were resuspended in sample buffer (65 mM Tris-HCl [pH 6.8], 5% glycerol, 5% SDS, 4 M urea, 5% β-mercaptoethanol, 0.5% bromophenol blue), boiled for 5 min, and analyzed by SDS-PAGE on 16% acrylamide precast NOVEX gels. The amount of 35S-labeled PrP seen in PK-treated or untreated reactions was quantified by using a Storm PhosphorImager and ImageQuant software (Molecular Dynamics), and the percent conversion was calculated as (volume of PK-resistant form × 100)/(volume of PK-sensitive form × 10). Relative percent conversion was calculated as (volume of PK-resistant form in presence of peptide × 100)/(volume of PK-resistant form in control condition).
Assay for PrP-res accumulation in Sc+MNB cultures.
Sc+MNB cells (30) were maintained in Opti-MEM medium (Life Technologies) supplemented with 10% fetal calf serum and were seeded at 5 to 10% confluent density into 35-mm-diameter dishes. Twelve hours after plating at 37°C, the cells were treated with the indicated concentrations of peptides for 3 to 4 days. The cells were lysed, and the cleared supernatants were treated with PK (20 μg/ml) for 20 min at 37°C. PK digestion was stopped by the addition of 0.1 M Pefabloc, and PrP-res was pelleted by centrifugation at 300,000 × g for 2 h at 4°C. Pellets were solubilized in SDS-PAGE sample buffer by sonication and boiled 5 min prior to running on 14% acrylamide precast NOVEX gels. PrP-res was assayed by Western blotting with rabbit polyclonal anti-PrP antiserum R27, and the blots were developed by using an enhanced chemifluorescence system (Vistra ECF; Amersham). PrP-res bands were quantified with a Storm PhosphorImager (Molecular Dynamics).
RESULTS
Species specificity of the cell-free conversion reaction.
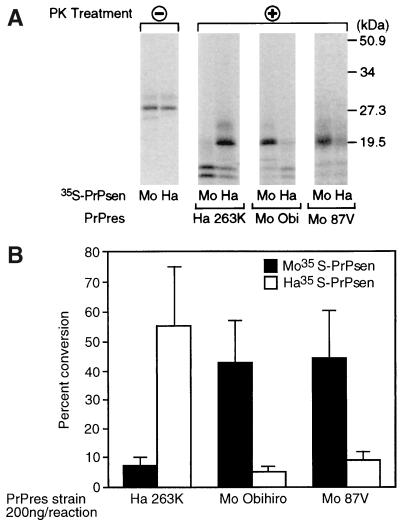
In previous studies, PrP-res derived from brain infected with the Chandler/RML isolate of mouse scrapie was able to convert both mouse and hamster PrP-sen to a protease-resistant form (20). This unexpected lack of species specificity might have been due to the fact that the Chandler scrapie isolate has not been cloned by limiting dilution (9) and may contain several different strains of scrapie agent with different abilities to interact with hamster PrP-sen. Therefore, in this study we used two different cloned strains of mouse scrapie, Obihiro and 87V (3, 18, 38), as sources of PrP-res to compare with the cloned hamster scrapie strain, 263K (17, 18). Purified PrP-res from each of these three scrapie strains was incubated with radiolabeled mouse or hamster PrP-sen molecules. PrP-res from strain 263K efficiently converted radiolabeled hamster PrP-sen to a ∼20-kDa PK-resistant form; however, by comparison, conversion of mouse PrP-sen was reduced by more than 90% (Fig. 1A and B). Similarly, PrP-res from both the Obihiro and 87V mouse strains was able to convert mouse PrP-sen to PK-resistant forms, but conversion of hamster PrP-sen was more than 90% lower (Fig. 1). Thus, when PrP-res from these three scrapie strains was used in the cell-free conversion system, strong species-specific differences were seen in the amount of conversion observed.
FIG. 1.
Species specificity of the cell-free conversion reaction. (A) Immunopurified mouse (Mo) and hamster (Ha) 35S-PrP-sen samples were incubated in the presence of hamster 263K (Ha 263K), mouse Obihiro (Mo Obi), or mouse 87V (Mo 87V) PrP-res. At the end of the incubation time, samples were treated (+) or not (−) with PK as described in Materials and Methods followed by SDS-PAGE analysis. Parallel experiments done using PrP-res from mouse Chandler scrapie gave results identical to those previously published (20). (B) Histogram representation of the cell-free conversion reactions induced by hamster 263K, mouse Obihiro, and mouse 87V PrP-res in the presence of immunopurified mouse or hamster 35S-Prp-sen. Only the PK-resistant bands (19 to 24 kDa) showing the 6- to 8-kDa downward size shift relative to the untreated 35S-PrP-sen precursor were quantified by PhosphorImager autoradiography. PK-resistant bands showing a downward size shift greater than 6 to 8 kDa (<19 kDa) likely represent partial conversion products and were not quantified. The results are expressed as the percent conversion of 35S-PrP-sen to 19- to 24-kDa PK-resistant forms, and each histogram represents the means of five independent experiments ± standard deviations (bars).
Kinetics of cell-free conversion reactions.
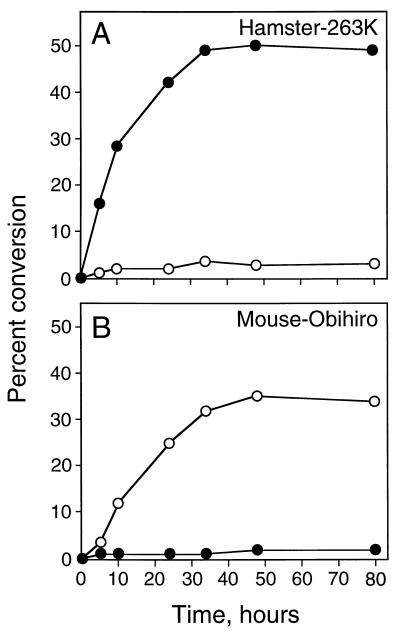
To assess whether the species specificity of the cell-free conversion reactions was a function of the incubation time, we incubated hamster 263K and mouse Obihiro PrP-res in the presence of mouse and hamster PrP-sen for various lengths of time. As shown in Fig. 2, the percent conversion for each reaction reached a plateau value after 36 h at 37°C and remained stable for up to 80 h. Less than 3% conversion was observed even after extended incubation of hamster PrP-res with mouse PrP-sen or mouse Obihiro PrP-res with hamster PrP-sen (Fig. 2). The specificity of these conversion reactions was clearly maintained throughout the 80-h time span of these experiments.
FIG. 2.
Kinetics of in vitro conversion reaction. The cell-free conversion reactions were performed by incubating hamster 263K (A) or mouse Obihiro (B) PrP-res with immunopurified mouse 35S-PrP-sen (open circles) or hamster 35S-PrP-sen (closed circles) at 37°C. At the indicated incubation times, the conversion reaction samples were digested with PK and then analyzed by SDS-PAGE and PhosphorImage autoradiography as described in the legend to Fig. 1. Percent conversion was plotted as a function of the incubation time. Each point represents the mean of two independent experiments.
PrP peptide inhibition of conversions induced by hamster and mouse PrP-res.
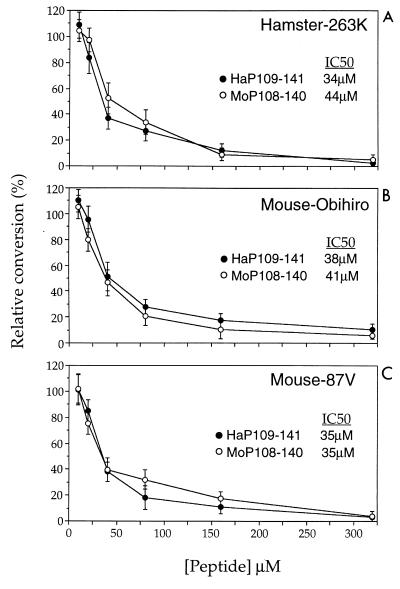
We previously described the inhibition of the hamster PrP-res-induced conversion by a hamster PrP peptide which contains the sequence from positions 109 to 141 in the central part of the PrP molecule (8). To investigate whether the inhibition of cell-free conversion by PrP peptides showed species specificity, hamster and mouse PrP peptides from this region (HaP109-141 and MoP108-140) were compared in the mouse and hamster conversion systems. Even though these peptides differed at three positions (see Materials and Methods), HaP109-141 and MoP108-140 were each able to inhibit all three hamster and mouse PrP-res-induced conversion reactions tested. The concentrations of each peptide required for 50% inhibition (IC50) were similar (34 to 44 μM) in all three reactions (Fig. 3). This was a somewhat surprising result in view of the distinct species specificity of the PrP-res–PrP-sen interactions in the cell-free conversion system, and it suggested that a PrP region with a common sequence in mouse and hamster might be involved in these reactions.
FIG. 3.
Dose response of the inhibition of conversion by hamster and mouse PrP peptides. The cell-free conversion reactions were performed by mixing PrP-res (200 ng) with immunopurified 35S-PrP-sen (∼1 ng) as described in Materials and Methods in the presence of various concentrations of HaP109-141 or MoP108-140. At the end of the incubation time, the samples were digested by PK and analyzed as described in Materials and Methods. (A) Hamster 263K strain PrP-res plus hamster PrP-sen. (B) Mouse Obihiro strain PrP-res plus mouse PrP-sen; (C) mouse 87V strain PrP-res plus mouse PrP-sen. Each point represents the mean of four independent experiments ± standard deviation (bars). The data were plotted as relative percent conversion in the presence of peptide compared to the control reaction in the absence of peptide.
Inhibition of the cell-free conversion by peptides P119-136 and P119-128.
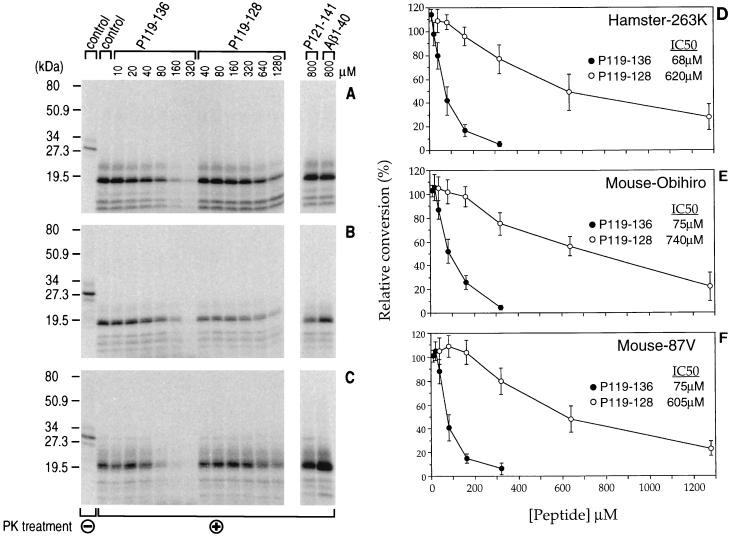
We previously found that the amino-terminal end of a hamster PrP peptide starting at residue 119 was required for peptide-induced inhibition of the cell-free conversion system (8). In the present studies, we synthesized two new hamster PrP peptides, P119-136 and P119-128, in an attempt to localize the sequence required for inhibiting the in vitro conversion reaction. From residues 119 to 136 there are no differences between the mouse and hamster PrP sequences. P119-136 gave strong inhibition of conversion with similar IC50 values (68, 75, and 75 μM) in the three PrP-res-induced conversion reactions tested (Fig. 4). The shorter peptide P119-128 partially inhibited the three conversion reactions and was found to be 10-fold less effective than P119-136 (IC50 = 605 to 740 μM). Control peptides P121-141 and Aβ1-40 showed no inhibition. In summary, PrP peptide P119-136 inhibited the conversion reaction induced by PrP-res from three different scrapie strains obtained from two different species. The fact that this peptide had the same sequence in both mouse and hamster PrP appeared to explain its ability to inhibit cell-free conversion reactions of both species.
FIG. 4.
Dose response of the inhibition of cell-free conversion by PrP peptides P119-136 and P119-128. (A to C) Representative results after PK digestion (+) of the samples. The first lane of each panel represents the PK-untreated sample (−). PrP-res was incubated with immunopurified 35S-PrP-sen in the absence (control) or presence of the indicated concentrations of PrP peptides P119-136, P119-128, and P121-141 and the Alzheimer’s protein peptide Aβ1-40. (A and D) Hamster 263K PrP-res incubated with hamster 35S-PrP-sen; (B and E) mouse Obihiro PrP-res incubated with mouse 35S-PrP-sen; (C and F) mouse 87V PrP-res incubated with mouse 35S-PrP-sen. Molecular mass markers are indicated on the left. (D to F) Dose-response curves of the inhibition of the cell-free conversion reactions induced by P119-136 (GAVVGGLGGYMLGSAMSR) and P119-128 (GAVVGGLGGY). The data represent the means of four independent experiments ± standard deviations (bars) and were plotted as relative percent conversion in the presence of peptide compared to the control reaction in the absence of peptide.
Inhibition of PrP-res formation by P119-136 in Sc+MNB cells.
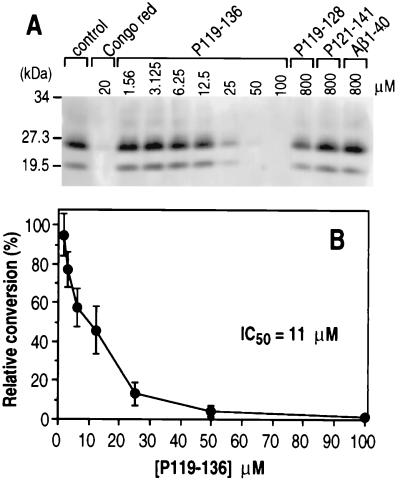
Because it remains unresolved whether scrapie infectivity can be generated in the cell-free conversion system, it was of interest to assess the inhibitory potency of various PrP peptides in scrapie-infected tissue culture cells where scrapie infectivity is known to be produced (29). Sc+MNB cells were incubated for 3 to 4 days in the presence of various peptides (Fig. 5A). In this system, results for peptides P119-136 and P119-128 were similar to those observed with cell-free conversion. Peptide P119-136 reduced the amount of PrP-res detectable in a concentration-dependent manner, with an IC50 of 11 μM (Fig. 5B). P119-128 gave only barely detectable inhibition at the highest concentration tested (800 μM). In contrast, P109-141 did not inhibit PrP-res formation under standard conditions but did give some inhibition when protease inhibitors were added to the culture medium (data not shown). The negative control peptides PrP P121-141 and Aβ1-40 showed no inhibition of the accumulation of PrP-res (Fig. 5A). In these studies, there was no evidence for cytotoxicity or reduced protein synthesis in cultures exposed to any of these peptides. In summary, P119-136 was an effective inhibitor of PrP-res generation in both the cell-free conversion and scrapie-infected cell culture systems.
FIG. 5.
Effect of PrP peptides P119-136 and P119-128 on the accumulation of PrP-res in Sc+MNB. (A) SDS-PAGE PhosphorImage of a representative experiment performed by treating Sc+MNB cells in the absence (control) or presence of indicated concentrations of Congo red, P119-136, P119-128, P121-141, or Aβ1-40. PrP-res was detected with a polyclonal rabbit anti-PrP serum and developed with the Amersham Vistra ECF system. (B) Dose-response curve of the inhibition of PrP-res accumulation induced by P119-136. The data represent the means of five independent experiments ± standard deviations (bars) and were plotted as relative percent conversion in the presence of peptide compared to the control culture in the absence of peptide.
DISCUSSION
In this study, we investigated the species-specific formation of both hamster and mouse PrP-res and the inhibition of that process by PrP peptides. We found that although generation of PrP-res was highly species specific, inhibition of PrP-res formation by PrP peptides was not. This species-independent inhibition was mapped to a peptide sequence completely conserved between mouse and hamster PrP. Our data suggested that PrP peptides could be used as general inhibitors with therapeutic applications against a broad range of TSE diseases in different species.
The ability of an inhibitory peptide to survive in an in vivo environment and to be devoid of cell toxicity are critical properties for a therapeutically useful compound. Although P109-141 and P119-136 strongly inhibited PrP-res formation under cell-free conditions, only P119-136 was able to inhibit PrP-res formation in the context of a living cell. The reason for this difference is unclear but could be due to the stability of the peptide. P109-141 was more soluble than P119-136 (data not shown) and might be more rapidly degraded in the tissue culture medium. This hypothesis was supported by the fact that P109-141 could inhibit the accumulation of PrP-res in scrapie-infected cells if protease inhibitors were added to the medium (data not shown). Furthermore, no cytotoxicity was observed in cells exposed to P119-136. Therefore, P119-136 has possible therapeutic potential, and it will be of interest to determine whether this peptide is also able to reduce the level of scrapie infectivity in scrapie-infected animals.
Two of the peptides used in our experiments, P119-128 and P119-136, are comprised of sequences which are highly conserved in all the species in which the PrP gene has been sequenced. However, only P119-136 showed a strong inhibitory effect which was both species and strain independent. This difference in inhibitory action could be due to the eight amino acid residues present in P119-136 but absent in P119-128. This finding suggests the occurrence of a critical interaction which involves these eight residues. Interestingly, although residues 119 to 136 are highly conserved, some polymorphisms reside within the eight amino acids present in P119-136 but absent in P119-128. Furthermore, all of these polymorphisms are associated with resistance to TSE infection. For example, the PrP genotype at the codon for residue 129 in human PrP influences resistance to Creutzfeldt-Jakob disease (10, 23), while changes at position 136 in sheep (homologous to position 133 in P119-136) influence resistance to sheep scrapie (12, 14). Taken together, these observations suggest that amino acid residues 129 to 136 are important in PrP-PrP interactions in vivo and in vitro and that their presence influences the inhibitory action of peptide P119-136 on PrP conversion.
The mechanism of inhibition of PrP-res formation by P119-136 is not known. The peptide could act by binding PrP-sen and blocking any subsequent interactions with PrP-res. Conversely, it could bind to PrP-res and block binding to PrP-sen. The peptide could also disrupt interactions between PrP and other secondary molecules such as glycosaminoglycans which have been hypothesized to be involved in the conversion of PrP-sen to PrP-res (4). Any of these inhibitory actions might be dependent either on monomeric peptide or on peptide which has aggregated to form amyloid fibrils (11, 15).
Whatever the mechanism of the inhibition of PrP-res formation by P119-136, our current data differ from previous results for scrapie-infected cells which showed that expression of heterologous PrP interfered with PrP-res formation. This interference was sequence dependent and was mapped to amino acid residues 109, 112, and 139 in hamster PrP (25, 26, 37). These residues are outside the P119-136 region identified in the present report as important for inhibition of PrP-res formation. This finding suggests that compatibility or incompatibility involving different regions of PrP might result in different inhibitory mechanisms.
Our experiments clearly demonstrate that PrP-res derived from 87V- or Obihiro-infected mice showed a strong preference for conversion of mouse PrP-sen. These results are in contrast to previous results using PrP-res from the Chandler strain of mouse scrapie, which converted both mouse and hamster PrP-sen to PrP-res under similar conditions (20). The difference in specificity among the different strains of mouse scrapie may be a result of different PrP-res structures associated with each of these strains. Unlike the biologically cloned 263K, 87V, and Obihiro strains of scrapie, the mouse Chandler scrapie agent has not been cloned by limiting dilution and likely contains a heterogeneous mix of scrapie strains (3). Thus, the ability of Chandler-derived PrP-res to convert mouse and hamster PrP-sen to protease-resistant forms may be a reflection of heterogeneity of PrP-res structures associated with this isolate.
ACKNOWLEDGMENTS
We thank Gary Hettrick and Franck Aguila for help with preparation of the figures, Jan Lukszo for synthesis and purification of peptides, and Lynne Raymond and Gregory Raymond for cell culture advice and fruitful discussions. We greatly appreciate the help of Byron Caughey, Kim Hasenkrug, and John Portis for critical reading of the manuscript.
J.C. was supported by the Institut National de la Santé et de la Recherche Médicale and by the Bourses de l’Organisation du Traité Atlantique Nord.
REFERENCES
- 1.Bossers A, Belt P B G M, Raymond G J, Caughey B, de Vries R, Smits M A. Scrapie susceptibility-linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease-resistant forms. Proc Natl Acad Sci USA. 1997;94:4931–4936. doi: 10.1073/pnas.94.10.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandner S, Isenmann S, Raeber A, Fischer M, Sailer A, Kobayashi Y, Marino S, Weissmann C, Aguzzi A. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature. 1996;379:339–343. doi: 10.1038/379339a0. [DOI] [PubMed] [Google Scholar]
- 3.Bruce M E, McConnell I, Fraser H, Dickinson A G. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J Gen Virol. 1991;72:595–603. doi: 10.1099/0022-1317-72-3-595. [DOI] [PubMed] [Google Scholar]
- 4.Caughey B. Protease-resistant PrP accumulation and scrapie agent replication: a role for sulphated glycosaminoglycans? Biochem Soc Trans. 1994;22:163–167. doi: 10.1042/bst0220163. [DOI] [PubMed] [Google Scholar]
- 5.Caughey B, Chesebro B. Prion protein and the transmissible spongiform encephalopathies. Trends Cell Biol. 1997;7:56–62. doi: 10.1016/S0962-8924(96)10054-4. [DOI] [PubMed] [Google Scholar]
- 6.Caughey B, Raymond G J, Ernst D, Race R E. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caughey B W, Dong A, Bhat K S, Ernst D, Hayes S F, Caughey W S. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 8.Chabry J, Caughey B, Chesebro B. Specific inhibition of in vitro formation of protease-resistant prion protein by synthetic peptides. J Biol Chem. 1998;273:13203–13207. doi: 10.1074/jbc.273.21.13203. [DOI] [PubMed] [Google Scholar]
- 9.Chandler R L. Encephalopathy in mice produced by inoculation with scrapie brain material. Lancet. 1961;i:1378–1379. doi: 10.1016/s0140-6736(61)92008-6. [DOI] [PubMed] [Google Scholar]
- 10.Collinge J, Palmer M S, Dryden A J. Genetic predisposition to iatrogenic Creutzfeldt-Jakob disease. Lancet. 1991;337:1441–1442. doi: 10.1016/0140-6736(91)93128-v. [DOI] [PubMed] [Google Scholar]
- 11.Come J H, Fraser P E, Lansbury P T., Jr A kinetic model for amyloid formation in the prion diseases: importance of seeding. Proc Natl Acad Sci USA. 1993;90:5959–5963. doi: 10.1073/pnas.90.13.5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldmann W, Hunter N, Smith G, Foster J, Hope J. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J Gen Virol. 1994;75:989–995. doi: 10.1099/0022-1317-75-5-989. [DOI] [PubMed] [Google Scholar]
- 13.Hope J, Morton L J D, Farquhar C F, Multhaup G, Beyreuther K, Kimberlin R H. The major polypeptide of scrapie-associated fibrils (SAF) has the same size, charge distribution and N-terminal protein sequence as predicted for the normal brain protein (PrP) EMBO J. 1986;5:2591–2597. doi: 10.1002/j.1460-2075.1986.tb04539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter N, Goldmann W, Smith G, Hope J. The association of a codon 136 PrP gene variant with the occurrence of natural scrapie. Arch Virol. 1994;137:171–177. doi: 10.1007/BF01311184. [DOI] [PubMed] [Google Scholar]
- 15.Jarrett J T, Lansbury P T., Jr Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in Alzheimer’s disease and scrapie? Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 16.Kascsak R J, Rubenstein R, Merz P A, Tonna-DeMasi M, Fersko R, Carp R I, Wisniewski H M, Diringer H. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimberlin R H, Walker C A. Evidence that the transmission of one source of scrapie agent to hamsters involves separation of agent strains from a mixture. J Gen Virol. 1978;39:487–496. doi: 10.1099/0022-1317-39-3-487. [DOI] [PubMed] [Google Scholar]
- 18.Kimberlin R H, Walker C A, Fraser H. The genomic identity of different strains of mouse scrapie is expressed in hamsters and preserved on reisolation in mice. J Gen Virol. 1989;70:2017–2025. doi: 10.1099/0022-1317-70-8-2017. [DOI] [PubMed] [Google Scholar]
- 19.Kocisko D A, Come J H, Priola S A, Chesebro B, Raymond G J, Lansbury P T, Caughey B. Cell-free formation of protease-resistant prion protein. Nature. 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 20.Kocisko D A, Priola S A, Raymond G J, Chesebro B, Lansbury P T, Jr, Caughey B. Species specificity in the cell-free conversion of prion protein to protease-resistant forms: a model for the scrapie species barrier. Proc Natl Acad Sci USA. 1995;92:3923–3927. doi: 10.1073/pnas.92.9.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Locht C, Chesebro B, Race R, Keith J M. Molecular cloning and complete sequence of prion protein cDNA from mouse brain infected with the scrapie agent. Proc Natl Acad Sci USA. 1986;83:6372–6376. doi: 10.1073/pnas.83.17.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oesch B, Teplow D B, Stahl N, Serban D, Hood L E, Prusiner S B. Identification of cellular proteins binding to the scrapie prion protein. Biochemistry. 1990;29:5848–5855. doi: 10.1021/bi00476a029. [DOI] [PubMed] [Google Scholar]
- 23.Palmer M S, Dryden A J, Hughes J T, Collinge J. Homozygous prion protein genotype predisposes to sporadic Creutzfeldt-Jakob disease. Nature. 1991;352:340–342. doi: 10.1038/352340a0. [DOI] [PubMed] [Google Scholar]
- 24.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, Prusiner S B. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion protein. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Priola S A, Caughey B, Race R E, Chesebro B. Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J Virol. 1994;68:4873–4878. doi: 10.1128/jvi.68.8.4873-4878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priola S A, Chesebro B. A single hamster amino acid blocks conversion to protease-resistant PrP in scrapie-infected mouse neuroblastoma cells. J Virol. 1995;69:7754–7758. doi: 10.1128/jvi.69.12.7754-7758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prusiner S B, Scott M, Foster D, Pan K M, Groth D, Mirenda C, Torchia M, Yang S L, Serban D, Carlson G A, Hoppe P C, Westaway D, DeArmond S J. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 28.Prusiner S B, Telling G, Cohen F E, DeArmond S J. Prion diseases of human and animals. Semin Virol. 1996;7:159–173. [Google Scholar]
- 29.Race R E, Caughey B, Graham K, Ernst D, Chesebro B. Analyses of frequency of infection, specific infectivity, and prion protein biosynthesis in scrapie-infected neuroblastoma cell clones. J Virol. 1988;62:2845–2849. doi: 10.1128/jvi.62.8.2845-2849.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Race R E, Fadness L H, Chesebro B. Characterization of scrapie infection in mouse neuroblastoma cells. J Gen Virol. 1987;68:1391–1399. doi: 10.1099/0022-1317-68-5-1391. [DOI] [PubMed] [Google Scholar]
- 31.Race R E, Priola S A, Bessen R A, Ernst D, Dockter J, Rall G F, Mucke L, Chesebro B, Oldstone M B A. Neuron-specific expression of a hamster prion protein minigene in transgenic mice induces susceptibility to hamster scrapie agent. Neuron. 1995;15:1183–1191. doi: 10.1016/0896-6273(95)90105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raeber A J, Race R E, Brandner S, Priola S A, Sailer A, Bessen R A, Aguzzi A, Oldstone M B A, Weissmann C, Chesebro B. Astrocyte-specific expression of hamster prion protein (PrP) renders PrP knockout mice susceptible to hamster scrapie. EMBO J. 1997;16:6057–6065. doi: 10.1093/emboj/16.20.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raymond G J, Hope J, Kocisko D A, Priola S A, Raymond L D, Bossers A, Ironside J, Will R G, Chen S G, Petersen R B, Gambetti P, Rubenstein R, Smits M A, Lansbury P T, Jr, Caughey B. Molecular assessment of the transmissibilities of BSE and scrapie to humans. Nature. 1997;388:285–288. doi: 10.1038/40876. [DOI] [PubMed] [Google Scholar]
- 34.Robakis N K, Sawh P R, Wolfe G C, Rubenstein R, Carp R I, Innis M A. Isolation of a cDNA clone encoding the leader peptide of prion protein and expression of the homologous gene in various tissues. Proc Natl Acad Sci USA. 1986;83:6377–6381. doi: 10.1073/pnas.83.17.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Safar J, Roller P P, Gajdusek D C, Gibbs C J., Jr Conformational transitions, dissociation, and unfolding of scrapie amyloid (prion) protein. J Biol Chem. 1993;268:20276–20284. [PubMed] [Google Scholar]
- 36.Scott M, Foster D, Mirenda C, Serban D, Coufal F, Walchli M, Torchia M, Groth D, Carlson G, DeArmond S J, Westaway D, Prusiner S B. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 37.Scott M, Groth D, Foster D, Torchia M, Yang S L, DeArmond S J, Prusiner S B. Propagation of prions with artificial properties in transgenic mice expressing chimeric PrP genes. Cell. 1993;73:979–988. doi: 10.1016/0092-8674(93)90275-u. [DOI] [PubMed] [Google Scholar]
- 38.Shinagawa M, Takahashi K, Sasaki S, Doi S, Sato G. Characterization of scrapie agent isolated from sheep in Japan. Microbiol Immunol. 1985;29:543–551. doi: 10.1111/j.1348-0421.1985.tb00856.x. [DOI] [PubMed] [Google Scholar]