Abstract
Borna disease virus (BDV) causes acute and persistent infections in various vertebrates. During recent years, BDV-specific serum antibodies, BDV antigen, and BDV-specific nucleic acid were found in humans suffering from psychiatric disorders. Furthermore, viral antigen was detected in human autopsy brain tissue by immunohistochemical staining. Whether BDV infection can be associated with psychiatric disorders is still a matter of debate; no direct evidence has ever been presented. In the present study we report on (i) the detection of BDV-specific nucleic acid in human granulocyte cell fraction from three different psychiatric patients and (ii) the isolation of infectious BDV from these cells obtained from a patient with multiple psychiatric disorders. In leukocyte preparations other than granulocytes, either no BDV RNA was detected or positive PCR results were obtained only if there was at least 20% contamination with granulocytes. Parts of the antigenome of the isolated virus were sequenced, demonstrating the close relationship to the prototype BDV strains (He/80 and strain V) as well as to other human virus sequences. Our data provide strong evidence that cells in the granulocyte fraction represent the major if not the sole cell type harboring BDV-specific nucleic acid in human blood and contain infectious virus. In contrast to most other reports of putative human isolates, where sequences are virtually identical to those of the established laboratory strains, this isolate shows divergence in the region previously defined as variable in BDV from naturally infected animals.
Borna disease virus (BDV), a nonsegmented negative-strand RNA virus, belongs to the family Bornaviridae in the order Mononegavirales. The BDV antigenome exists of six open reading frames encoding the nucleoprotein (p40), the phosphoprotein (p24), a recently identified unglycosylated protein (p10), the glycosylated matrixprotein (p16→gp18), the glycoprotein (p57→gp84 or gp94), and the l-polymerase with a molecular mass of approximately 180 kDa (6, 12, 19, 20, 35–37, 44, 45, 47).
Borna disease (BD) is a virus-induced encephalomyelitis of horses and sheep. In recent years it was shown that BDV has a much broader host range than was anticipated so far, including naturally infected cats, dogs, birds, and cattle (7, 22, 23, 30, 48). Relevant to the present report, nonhuman primates have been experimentally infected, leading to various symptoms of neurological dysregulation (40, 50). The best-investigated experimental model is the infection of rats. In this model it could be shown that BD is a virus-induced immune system-mediated disease (15, 24, 30, 39) and that BDV-specific CD8+ T cells destroy virus-infected brain cells while CD4+ T cells function as helper cells (25–27, 38). Antibodies seem not to contribute to the immune system-mediated, destructive process, but neutralizing antibodies control virus tropism (13, 41).
The presence of antiviral antibodies (4, 31) and of BDV nucleic acid in blood of patients with psychiatric disease has been reported, and some authors even propose the involvement of BDV in mental illness (5, 16, 18). However, due to conflicting results obtained in attempts to detect viral nucleic acid consistently in human blood from psychiatric patients, this issue is still controversial (5, 16, 21, 28, 29). Recently, we demonstrated the persistence of BDV-specific nucleic acid in the blood of a psychiatric patient (28). This finding has enabled us to scrutinize the cell type carrying the viral nucleic acid. Here, we report the detection of BDV-specific nucleic acid and of infectious virus in the granulocytic cell fraction, a finding that is of great importance for the diagnosis, pathogenesis, and epidemiology of BDV infection in humans and animals.
MATERIALS AND METHODS
Processing of blood samples.
Blood was collected from patients with psychiatric diseases in the Klinik für Psychiatrie und Psychotherapie, Tübingen, Germany (see Results). Three different samples were collected from patients 1 (described previously [28]) and 2 at different time points, whereas only one sample was obtained from patient 3. A 5-ml volume of EDTA-blood was mixed with 5 ml of 0.9% ammonium chloride buffer for 30 min at 4°C to allow complete lysis of erythrocytes, and the remaining cells were then washed twice with phosphate-buffered saline (PBS). The cell pellet was used for RNA isolation as described below. For polymorphonuclear cell separation, 5 ml of EDTA-blood was layered on top of 5 ml of Polymorphprep (Life Technology, Freiburg, Germany) in a 12-ml centrifuge tube and centrifuged at 550 × g for 40 min in a swing-out rotor at 18°C. After centrifugation, two leukocyte bands became visible. The top band at the sample/medium interface consisted of mononuclear cells, and the lower band consisted of polymorphonuclear cells. The cell bands were harvested, washed twice with PBS, and counted, and equal numbers of cells (5 × 106) were treated with Trizol to extract total RNA.
Isolation of RNA.
To the cell pellets obtained from each of the two bands, 1.5 ml of Trizol (Life Technology) was added and mixed, and the mixture was incubated for 5 min at room temperature (RT). Thereafter, 400 μl of chloroform was added and mixed, and the mixture was incubated for 3 min at RT. Phase separation was performed in a 1.5-ml tube after centrifugation at 12,000 × g for 15 min at 4°C in an Eppendorf table centrifuge. The aqueous phase was transferred into a new tube, 500 μl of isopropyl alcohol was added, and the mixture was incubated for 10 min at RT and centrifuged at 12,000 × g for 15 min at 4°C. The RNA pellets was washed twice with 500 μl of 70% ethanol and centrifuged at 7,500 × g for 5 minutes at 4°C. The RNA was air dried and resolved in 30 μl of diethylpyrocarbonate-treated H2O. The RNA concentration was determined photometrically.
Reverse transcription.
RNA (1 μg) was transcribed into cDNA at 42°C for 1 h with 50 U of Expand reverse transcriptase (Boehringer, Mannheim, Germany)–100 mM dithiothreitol–20 U of RNase inhibitor (Pharmacia Biotech Products, Freiburg, Germany)–10 mM deoxynucleoside triphosphate mix (Pharmacia Biotech Products)–0.5 μg of BDV-p40 specific primer (BV829R) (33) or 0.2 μg of hexamer random primer (Boehringer).
Conditions for PCR.
BDV cDNA was detected by first-round and nested PCR with primers located in the p40 gene of BDV as described by Sauder and de la Torre (33). First-round amplification was performed by hot-start PCR in a total volume of 50 μl containing 1 to 5 μl of cDNA, 50 ng of each primer, 20 mM deoxynucleoside triphosphate mix, 5 μl of 10× PCR buffer (Boehringer), and 0.5 μl of Taq polymerase (5 U/μl; Boehringer). Amplification was achieved by 40 cycles (95°C for 90 s, 58°C for 90 s, and 72°C for 90 s) in a Trio thermocycler (Biometra, Göttingen, Germany). Specific primers for p40, BV259F (5′-TTCATACAGTAACGCCCAGC-3′) and BV829R (5′-AACTACAGGGATTGTAAGGG-3′), were used. One hundred molecules or less of p40 INS (described by Sauder and de la Torre [33]) were detected to determine the sensitivity of the assay.
Nested PCR was performed identically to first-round PCR, using 1 μl of 1:10-diluted first-round PCR product as the template and the p40-specific primers BV277F (5′-GCCTTGTGTTTCTATGTTTGC-3′) and BV805R (5′-GCATCCATACATTCTGCGAG-3′). Amplification products were analyzed by electrophoresis in a 1.5% agarose gel containing 0.3 μg of ethidium bromide per ml.
For cloning and DNA sequencing, the five structural proteins of BDV were amplified with the primers and under the PCR conditions indicated in Table 1. The PCRs were performed in a total volume of 50 μl, as described for the p40 PCR, with primers BV259F and BV829R and under the various annealing conditions in Table 1. The specific PCR products were cloned into vector plasmid pcR-Topo as specified by the manufacturer (Invitrogen, Leek, The Netherlands). From each cloned PCR product, three different recombinant plasmids were DNA sequenced with the Big Dye terminator Cycle Sequencing Ready Reaction (Perkin-Elmer).
TABLE 1.
Primers used for sequencing of RW 98
| Protein | Designation (position)a | Sequence | Product size (bp) | Annealing conditions |
|---|---|---|---|---|
| p40 | p40-11 (11) | 5′-CAA CAA ACC ACT CAT CAT TC-3′ | 401 | 50°C, 90 s |
| p40-412 (412) | 5′-CCG TAA AAC TTC GCA GTC-3′ | |||
| p40 | p40-259 (259) | 5′-TTC ACA CAG TAA CGC CTA GC-3′ | 917 | 58°C, 60 s |
| p40-2 (1176) | 5′-TCA TTG TTT TTT AGT TTA GAC CAG T-3′ | |||
| p10 | p40-3 (1108) | 5′-GAG CTC TCA GGT GAG ATC TCT-3′ | 383 | 50°C, 90 s |
| p24 rev (1491) | 5′-GCT CAT CAT TCG ACA GCT GC-3′ | |||
| p24 | p24-A (1387) | 5′-TGA CCC AAC CAG TAG ACC A-3′ | 478 | 58°C, 60 s |
| p24-B (1865) | 5′-GTC CCA TTC ATC CGT TGT C-3′ | |||
| gp18 | gp18-1 (1882) | 5′-ATC GAA TCA CCA TGA ATT CAA AAC-3′ | 558 | 60°C, 90 s |
| gp18-2 (2340) | 5′-AGT GGA GTC AGT ATT GCA GCT AA-3′ | |||
| gp94 | p57-1 (2224) | 5′-AGC ACG CAA TCA ATG CAG CCT TC-3′ | 623 | 60°C, 90 s |
| p57-3 (2847) | 5′-GGA TGA GTT GCA GAG TAT GGT C-3′ | |||
| gp94 | p57-4 (2780) | 5′-CCC CGT ATA TCC TAC AGT CAG AAG-3′ | 980 | 60°C, 90 s |
| p57-2 (3760) | 5′-TGG TCG GTA CGG TTT ATT CCT GC-3′ |
The indicated nucleotide positions refers to the sequence of strain V (accession no. U04608).
Sequence data were analyzed with the Wisconsin package version 9.1 (Genetics Computer Group (GCG), Madison, Wis.) and the Basic Local Alignment Search Tool (BLAST) provided by the National Center for Biotechnology Information (24a).
Propagation of infectious BDV.
From patient 1, 107 cells from the granulocyte fraction were resuspended in 500 μl of sterile balanced salt solution (BSS) and sonicated on ice to disrupt the cells. A 50-μl volume of supernatant was incubated with 5 × 103 CRL 1405 cells in a total volume of 200 μl of cell culture medium. After 3 days the medium was changed, and 10 days later the cells were trypsinized and added to six-well plates. After this second culture passage, aliquots of cells were fixed and stained with BDV-specific monoclonal antibodies. After the fourth culture passage, 100% of the cells showed BDV-specific staining. BDV-infected cells were cryopreserved after each cell culture passage starting from the second passage.
Infectivity assay.
The infectivity of the BDV isolate (designated RW 98) was determined by using a guinea pig cell line (CRL 1405). A total of 5 × 103 cells were incubated for 7 days in 96-well plates containing sixfold dilutions of BDV RW 98 lysate, fixed with 4% formaldehyde–PBS, and treated with 0.5% Triton X-100–PBS. Nonspecific binding of immunological reagents was blocked by incubation of plates with PBS containing 10% fetal calf serum. Primary antibodies (BDV-specific mouse monoclonal antibodies [44]) were added, followed by a secondary anti-species peroxidase-labeled antibody (Dianova, Hamburg, Germany). The reaction was visualized with orthophenyldiamine and H2O2. Foci were counted, and the infectious titer was calculated.
Western blot analysis and immunofluorescence.
Human sera were tested by Western blot analysis and immunofluorescent staining. Briefly, for Western blot analysis, recombinant p40 and p24 (20) were separated by sodium dodecyl sulfate gel electrophoresis and proteins were blotted onto nitrocellulose. Human sera were diluted 1:500 and incubated for 1 h at RT. After being washed with 7 M urea for 4 min and then three times with PBS–0.2% Tween 20, the filter was incubated with goat anti-human peroxidase (Dianova) diluted 1:5,000 in 5% milk powder–PBS–0.2% Tween 20. The reaction was detected with the enhanced chemiluminescence system (Amersham, Pharmacia Biotech).
BDV-infected MDCK cells were used for immunofluorescence. Sera were diluted starting with a 1:2 dilution and incubated as described previously (26).
Fluorometric analysis.
Whole-blood cells were incubated with a mixture of phycoerythrin-conjugated mouse anti-human CD3 monoclonal antibody (clone HIT3a; Pharmingen, Hamburg, Germany) and a fluorescein isothiocyanate-conjugated mouse anti-human CD15 monoclonal antibody (clone HI98; Pharmingen) for 30 min. The CD15 monoclonal antibody recognizes eosinophilic and neutrophilic granulocytes and, to varying degrees, monocytes. The cells were washed twice, and erythrocytes were lysed with fluorescence-activated cell sorter (FACS) buffer (Becton-Dickinson). Mononuclear and polymorphonuclear cell preparations were treated with the same antibodies. The cells were analyzed in a Becton-Dickinson FACS Calibur fluorocytometer.
RESULTS
Detection of BDV-specific nucleic acid in the granulocytic cell fraction.
Usually, the presence of BDV is diagnosed by reverse transcription-PCR in lysates of peripheral blood mononuclear cells isolated from blood after Ficoll gradient separation. It is known that Ficoll gradient centrifugation results in an enrichment of peripheral blood mononuclear cells but that almost all polymorphonuclear cells are lost due to their higher density. To avoid the loss of particular cell types, Trizol was added to EDTA-blood to extract RNA. Alternatively, erythrocyte-free leukocytes were treated with Trizol to extract RNA. By these methods, we were able to detect BDV-specific nucleic acid in the blood of several patients with various psychological disorders (reference 28 and this report). The purpose of the present study was the identification of distinct peripheral blood cell populations, isolated from psychiatric patients, which harbor BDV-specific nucleic acid. Therefore, we used Polymorphprep gradient centrifugation of EDTA-blood, which allows the density separation of mononuclear and polymorphonuclear cells in two distinct bands almost free of erythrocytes. After centrifugation, the upper leukocyte band, migrating at the sample/medium interface, consisted of mononuclear cells and the lower band consisted of polymorphonuclear cells (manufacturer’s data). Both cell bands were harvested separately, any remaining erythrocytes were completely lysed, and the cells were washed twice with PBS and subsequently characterized by FACS analysis. We analyzed blood from 3 healthy blood donors and 15 selected patients with various psychiatric disease from the psychiatric clinic, including patients 1, 2, and 3. Patient 1 was diagnosed with schizophrenia of the undifferentiated type (DSM IV 295.90), patient 2 was diagnosed with recurrent major depressive disorder with severe mood-congruent psychiotic features (DSM IV 296.34), and patient 3 was diagnosed with recurrent major depressive disorder without psychiotic features (DSM IV 296.33). Anamneses revealed that patient 1 and 3 both had access to small farm animals during their youths whereas patient 2 was a permanent urban resident. From patients 1 and 2 we obtained three samples at different time points. As shown in Table 2, we were able to isolate polymorphonuclear cells (lower cell fraction) with a purity of 92 to 98% as determined by FACS analysis with the granulocyte-specific antibody anti-CD15. The contamination of the mononuclear cell fraction (upper band) with granulocytes was in the range of 5 to 22% (Table 2). Furthermore, blood samples from three healthy blood donors were tested, confirming the results obtained with blood samples from psychiatric patients (Table 2).
TABLE 2.
Summary of detection of viral nucleic acid and BDV-specific antisera in blood cell preparations from psychiatric patients and controls
| Patient | BDV RNA detected in:
|
BDV-specific antibodies by:
|
|||||
|---|---|---|---|---|---|---|---|
| Whole blood | Granulocyte fraction
|
Mononuclear cell fraction
|
WBa | IFb | |||
| % Purity | PCRc | % of residual granulocytes | PCR | ||||
| Psychiatric patients | |||||||
| 1, sample A | + | 92 | + | 15 | − | − | <1:2 |
| 1, sample B | + | 94 | + | 22 | + | − | <1:2 |
| 1, sample C | + | 98 | + | 12 | − | − | <1:2 |
| 2, sample A | + | 94 | + | 21 | + | − | <1:2 |
| 2, sample B | + | 96 | + | 11 | − | − | <1:2 |
| 2, sample C | + | 95 | + | 13 | − | − | <1:2 |
| 3, sample A | + | 97 | + | 5 | − | − | <1:2 |
| Control patients | |||||||
| 1 | − | 97 | − | 8 | − | − | <1:2 |
| 2 | − | 94 | − | 13 | − | − | <1:2 |
| 3 | − | 96 | − | 9 | − | − | <1:2 |
Western blot data.
Immunofluorescence data.
PCR, reverse transcription-PCR.
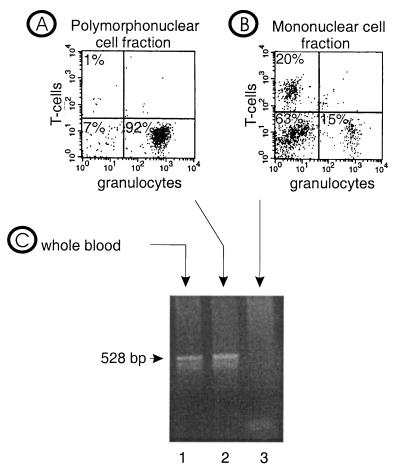
Equal numbers of polymorphonuclear and mononuclear cells (5 × 106) were treated separately to extract RNA. In parallel, whole blood from the patients was tested for the presence of BDV-specific nucleic acid. As shown representatively in Fig. 1 with sample A from patient 1, we were able to detect RNA encoding the nucleoprotein (p40) of BDV, the transcript mostly used for BDV diagnosis by nested reverse transcription-PCR (28, 33) (Fig. 1C). Next, the two cell fractions resulting from the Polymorphprep gradient were analyzed by the same p40 reverse transcription-PCR. BDV-specific amplification was found exclusively in the granulocyte fraction (Fig. 1A), and not in the mononuclear cell fraction (Fig. 1B). Blood from healthy donors was also tested after separation, and no BDV-specific amplification products were found (Table 2). Sequence analysis confirmed the BDV specificity of the reverse transcription-PCR and showed high homology to the prototype strains He/80 and strain V, originally isolated from horses (data not shown).
FIG. 1.
Polymorphonuclear cell fraction (A) mononuclear cell fraction (B), and whole blood (C) was tested for the presence of BDV-specific nucleic acid by reverse transcription-PCR. Lanes: 1, polymorphonuclear cell fraction; 2, mononuclear cell fraction; 3, whole blood.
Isolation of infectious BDV.
In addition, infectious BDV could be isolated from the peripheral blood granulocyte cell fraction. To this end, 107 cells from the granulocytic fraction from patient 1 sample A were sonicated in BSS, and after centrifugation the supernatant was collected and incubated together with CRL 1405. As shown in earlier experiments, infection of these cells with BDV does not result in nucleic acid changes of the virus (10a). To monitor possible contamination with the BDV laboratory strain, noninfected CRL1405 cells were cultured in parallel on the same plates used for cocultivation and isolation. Upon testing for the presence of BDV-specific antigen, these cells were always found not to be infected. From persistently infected CRL1405 cells (seventh culture passage), the titer of this human BDV isolate was determined to be 3.9 log10 FFU/ml, and it was designated RW 98.
Sequence analysis of BDV RW 98.
Total RNA was extracted from the second and fourth cell culture passages on CRL1405 cells and amplified by reverse transcription-PCR with primers specific for BDV structural proteins. These PCR products were cloned, and three individual clones were sequenced for each PCR product (Table 3). Comparison between the PCR products obtained directly from the blood of patient 1, sample A, and the virus isolate revealed 100% sequence homology (data not shown). Furthermore, comparison of the human isolate RW 98 with the well-characterized animal strains He/80 and V revealed differences in single bases. As shown in Table 3, RW 98 is slightly more homologous to strain V (96.7 to 97.0%) than to strain He/80 used in our laboratory. The predicted amino acid sequences showed that the nucleoprotein p40 and the matrix protein gp18 are the most highly conserved proteins. In addition, RW 98 p40 showed one different amino acid compared to He/80 and V. For the matrix protein, single-base differences were found. Compared to He/80, all these changes are silent, and therefore RW 98 gp18 is identical to the amino acid sequence of gp18 from He/80. In contrast, compared to strain V, one amino acid change was found. Most divergent is the amino acid sequence homology for p10. Comparison of strain RW 98 with strain V revealed only 96.6% identity, and lower identity to He/80 was found (95.4%; Table 3). Single-base changes lead to four amino acid changes compared to He/80 and to three changes compared to strain V. RW 98 showed two amino acid changes compared to p24 of strain V and also two changes compared to He/80. For the glycoprotein gp84/94 of BDV isolate RW 98, it could be shown that it was less highly conserved than the matrix protein of He/80 and strain V. Single-base differences leading to seven amino acid changes compared to He/80 and to nine amino acid changes compared to strain V were found (Table 4). For the glycoprotein, two different sizes are predicted. For strain V, the start codon at position 2236 is used, resulting in a protein with a molecular mass of 94 kDa, while for He/80, the methionine at nucleotide position 2685 is used as translational start, resulting in a protein with a molecular mass of 84 kDa (12, 36).
TABLE 3.
Homology of the human BDV isolate to strain He/80 and strain V
| BDV protein | Identity (%) of nucleic acid sequence to:
|
Identity (%) of amino acid sequence to:
|
||||
|---|---|---|---|---|---|---|
| He/80 | V | He/80
|
V
|
|||
| Similarity | Identity | Similarity | Identity | |||
| p40 | 96.5 | 97.0 | 100 | 99.7 | 100 | 99.7 |
| p10 | 97.7 | 98.1 | 96.6 | 95.4 | 96.6 | 96.6 |
| p24 | 96.7 | 96.7 | 99.0 | 99.0 | 99.5 | 99.2 |
| gp18 | 96.7 | 96.7 | 100 | 100 | 100 | 99.3 |
| gp84/94 | 95.7 | 96.1 | 99.4 | 98.0 | 98.8 | 98.2 |
TABLE 4.
Amino acid changes between strains RW 98, He/80, and V
| BDV protein | Position | Amino acid in BDV strain
|
||
|---|---|---|---|---|
| RW 98 | He/80 | V | ||
| p40 | 275 | Glu | Glu | Asp |
| 361 | Lys | Arg | Lys | |
| p24 | 4 | Arg | Gly | Arg |
| 26 | Ser | Ser | Pro | |
| 34 | Ile | Ile | Val | |
| 194 | Thr | Ala | Thr | |
| p10 | 20 | Ala | Glu | Ala |
| 42 | Ile | Ile | Thr | |
| 50 | Asp | Asp | Glu | |
| 55 | Thr | Ile | Ile | |
| 59 | Ser | Glu | Ser | |
| 86 | Val | Ile | Val | |
| gp18 | 108 | Asp | Asp | Glu |
| gp84/94 | 8a | Pro | Leu | |
| 17a | Ala | Val | ||
| 28a | Pro | Leu | ||
| 193/43 | Arg | Lys | Lys | |
| 220/70 | Ala | Ala | Thr | |
| 234/84 | Val | Val | Ala | |
| 242/92 | Pro | Ser | Pro | |
| 243/93 | Arg | Lys | Arg | |
| 245/95 | Lys | Arg | Lys | |
| 282/132 | Val | Met | Val | |
| 296/246 | Gly | Gly | Ser | |
| 304/154 | Val | Ile | Ile | |
| 311/161 | Ile | Val | Val | |
The position for gp84/94 was determined at nucleic acid 2236 (start for strain V gp94). The start codon for gp84 from He/80 is located at position 2685 (amino acid position 151 compared to strain V).
Furthermore, the human isolate RW 98 was compared to sequences of other human isolates (8) and to various sequences of BDV-specific regions from various species reported in GenBank. Due to the available data for complete BDV protein sequences, the nucleic acid sequence and amino acid sequence of p24 were compared to those of three different human isolates, revealing single-base differences and also single-amino-acid changes; however, it cannot be concluded that these changes belong to distinct regions (Table 5). Thus, from the sequence data available and from data published recently (16), there is no indication for type, isolate, or strain specificity of different BDV isolates.
TABLE 5.
RW 98 p24 compared to other human isolatesa
| Isolate | Nucleic acid identity (%) | Amino acid sequence
|
Positionb | Change | |
|---|---|---|---|---|---|
| Identity (%) | Similarity (%) | ||||
| H1 | 96.2 | 98.5 | 99.5 | 4 | Glu→Arg |
| 34 | Ile→Val | ||||
| 127 | His→Tyr | ||||
| H2 | 96.5 | 99.0 | 99.5 | 4 | Arg→Gly |
| 34 | Ile→Val | ||||
| H3 | 96.5 | 99.0 | 99.5 | 4 | Glu→Arg |
| 34 | Ile→Val | ||||
Data from reference 8.
This refers to the sequence of RW 98.
Sequence divergence control.
Since single-base changes can be created artificially by reverse transcription-PCR, we controlled the mutation frequency by amplification of a 917-bp PCR product of BDV p40 (Table 1) of strain He/80. Three clones were sequenced, revealing 100% identity. In addition, for sequencing of the human BDV isolate RW 98, all the different clones showed identical sequences, except for gp94 (980 bp). Here, divergences between the individual clones were found, revealing 99.6% identity, but all mutations were silent.
Detection of BDV-specific antibodies.
Sera were obtained from all three patients and tested either in Western blot analysis or by immunofluorescence for the presence of BDV-specific antibodies. Interestingly, in these sera, no virus-specific antibodies could be detected, even at dilutions of 1:2 in immunofluorescence (Table 2).
DISCUSSION
Data from studies of human patients with various psychiatric disorders suggest that BDV is able to infect humans and may even represent a human pathogen. The potential role of BDV as a human pathogen, however, requires substantiation. Controversy has arisen about reports that blood mononuclear cells of psychiatric patients contain BDV nucleic acid. Whereas several researchers have demonstrated the presence of virus-specific RNA in these cells, others were not successful (5, 16, 21, 29, 34). Recently, we reported the presence of BDV nucleic acid in the blood of a patient with somatization disorders and schizophrenia (28). Blood from this patient and two additional psychiatric patients, found to be positive for BDV-RNA by reverse transcription-PCR, was scrutinized to define which cell population carries the viral nucleic acid. From the results presented above we conclude that the fraction of cells containing the majority of granulocytes represents a major carrier of BDV-specific nucleic acid in the peripheral blood, but we cannot formally rule out the presence of BDV in a subpopulation that sediments with the granulocytic cell fraction. Whether granulocytes are productively infected or whether they have simply taken up infected cells or free virus remains to be determined. Polymorphonuclear leukocytes can harbor viruses (2, 43, 46, 49), although it has rarely been shown that viruses infect these cells directly (reviewed in reference 1). On the other hand, it has been demonstrated that granulocytes take up virions; alternatively, infected cells or cell debris might be ingested by phagocytosing cells. In this context, it is worth mentioning that polymorphonuclear cells can be present in the brain.
The relatively small number of cells and the single fraction of blood cells harboring BDV might be the reason why detection of virus in blood is difficult. The presence of nucleic acid either in the polymorphonuclear blood cells, comprising up to 98% granulocytes, or in erythrocyte-free whole blood but not in the mononuclear fraction strengthens this assumption. When the mononuclear cell population contained less than 20% granulocytes, no BDV-specific reverse transcription-PCR product was amplified. This might indicate that not all granulocytes contain the virus. Alternatively, a small number of contaminating nongranulocyte cells still could be responsible for the presence of infectious BDV in the polymorphonuclear cell fraction; however, the only other known cells expressing the CD15 marker to various degrees are monocytes, which can be ruled out as virus carriers since these cells are distinguishable from granulocytes by FACS analysis.
So far, only little information is available on cell types outside the nervous system infected with BDV. Previous work with rats has revealed the presence of virus-specific antigen in organ-specific cells of immunocompromised animals (14, 41, 42). Furthermore, BDV RNA has been detected in immunocompromised but also in immunocompetent rats in the late chronic phase of the disease (32). These authors gave evidence that BDV might be present in stromal cells of the bone marrow and the thymus of infected rats. It is tempting to speculate on the situation in humans based on these findings, but the experiments as designed by Rubin et al. (32) are not possible in humans.
The identification of a patient with BDV-specific nucleic acid in blood and the finding of cells in the granulocyte fraction as carriers permitted us to try to isolate virus from these cells. The resulting virus (RW 98) could be identified as BDV and could be clearly distinguished from the virus strain used in this laboratory as well as from another animal strain (strain V). Sequence analysis of the RNA of the human isolate RW 98 showed 100% homology to the reverse transcription-PCR products obtained from the blood of this patient. In contrast, minor sequence differences were found between RW 98 and the prototype animal strains He/80 and V. Here, the nucleoprotein p40 was the most highly conserved protein whereas the majority of amino acid changes were found in p10.
The finding that all our patients harbored virus in the blood in the absence of virus-specific antibodies might be important for the pathogenesis of BDV infection. Recently, we showed that virus is detectable in peripheral tissues of immunocompromised rats, which were unable to synthesize antiviral antibodies. However, transfusion of BDV-specific neutralizing antisera into these animals again resulted in the restriction of the virus to the central nervous system (41). Therefore, we hypothesize that infectious virus will be found predominantly in the blood of infected individuals lacking a potent humoral and/or cellular immune response against BDV. These might include patients undergoing treatment interfering with immune reactivity as well as immunological low- or non-responders or patients suffering from other diseases with an immunosuppressive effect; particular psychiatric diseases have been associated with poor general immunoreactivity (9, 17).
BDV infection of naturally infected hosts or experimentally infected species results in disease with different clinical features. At least some disturbances of functions such as sensory or behavioral abnormalities occur in the absence of a detectable local immune response (3, 10, 11). Symptoms of full-blown BD such as ataxia, paresis, and transient paralysis, however, appear to be clearly correlated with the local immune response in the brain (reviewed in reference 39).
The present observation raises the question whether infectious BDV might also be present or might even persist in blood and/or blood cells of animals, in which behavioral alterations are definitely much more difficult to diagnose than in humans. We therefore suggest a more intense search for infectious virus outside the nervous system of animals; furthermore, the results of those investigations might indicate whether animals represent a virus reservoir and whether BD should be regarded as a zoonosis. The remarkable similarity of BDV isolated from animals, from tissue culture, and from humans could indicate a common source of infection.
In conclusion, we were able to demonstrate the presence of infectious BDV in the cell fraction harboring granulocytes, a finding that is important for further strategies of diagnosis and also provides an explanation of controversial results obtained in the past. Moreover, this report on the presence of infectious virus in human blood is important for further investigations on the pathogenesis and epidemiology of human BD. In addition, we found that the infectious BDV isolated from the blood of a psychiatric patient differed from the two best-characterized laboratory BDV strains but, overall, showed an amazingly high conservation for an RNA virus. Although our sequence comparisons were not indicative of BDV strain-specific differences, more sequence data on other virus isolates are needed to finally clarify this point.
ACKNOWLEDGMENTS
We thank S. Herzog for testing antisera and Karl Heinz Adam, Silke Gommel, and Berthilde Bauer for technical assistance.
The work was supported in part by the Deutsche Forschungsgesellschaft (grant Sti 71/2-2) (to L.S. and O.P.), by the Bundesministerium für Gesundheit (through Paul-Ehrlich-Institut) and by the European Union (Pathogenesis of subacute and chronic inflammatory diseases of the central nervous system, grant CHRX-CT94-0670).
REFERENCES
- 1.Abramson J S, Mills E L. Depression of neutrophil function induced by viruses and its role in secondary microbial infections. Rev Infect Dis. 1988;10:326–341. doi: 10.1093/clinids/10.2.326. [DOI] [PubMed] [Google Scholar]
- 2.Abramson J S, Parce J W, Lewis J C, Lyles D S, Mills E L, Nelson R D, Bass D A. Characterization of the effect of influenza virus on polymorphonuclear leukocyte membrane responses. Blood. 1984;64:131–138. [PubMed] [Google Scholar]
- 3.Bautista J R, Schwartz G J, de la Torre J C, Moran T H, Carbone K M. Early and persistent abnormalities in rats with neonatally acquired Borna disease virus infection. Brain Res Bull. 1994;34:31–40. doi: 10.1016/0361-9230(94)90183-x. [DOI] [PubMed] [Google Scholar]
- 4.Bode L, Riegel S, Ludwig H, Amsterdam J D, Lange W, Koprowski H. Borna disease virus-specific antibodies in patients with HIV infection and with mental disorders. Lancet. 1988;ii:8612. doi: 10.1016/s0140-6736(88)90505-3. [DOI] [PubMed] [Google Scholar]
- 5.Bode L, Zimmermann W, Ferszt R, Steinbach F, Ludwig H. Borna disease virus genome transcribed and expressed in psychiatric patients. Nat Med. 1995;1:232–236. doi: 10.1038/nm0395-232. [DOI] [PubMed] [Google Scholar]
- 6.Briese T, Lipkin W I, de la Torre J C. Molecular biology of Borna disease virus. Curr Top Microbiol Immunol. 1995;190:1–16. doi: 10.1007/978-3-642-78618-1_1. [DOI] [PubMed] [Google Scholar]
- 7.Caplazi P, Waldvogel A, Stitz L, Braun U, Ehrensperger F. Borna disease in naturally infected cattle. J Comp Pathol. 1994;111:65–72. doi: 10.1016/s0021-9975(05)80112-4. [DOI] [PubMed] [Google Scholar]
- 8.de la Torre J C, Bode L, Durrwald R, Cubitt B, Ludwig H. Sequence characterization of human Borna disease virus. Virus Res. 1996;44:33–44. doi: 10.1016/0168-1702(96)01338-x. [DOI] [PubMed] [Google Scholar]
- 9.Dietrich D E, Schedlowski M, Bode L, Ludwig H, Emrich H M. A viro-psycho-immunological disease-model of a subtype affective disorder. Pharmacopsychiatry. 1998;31:77–82. doi: 10.1055/s-2007-979305. [DOI] [PubMed] [Google Scholar]
- 10.Dittrich W, Bode L, Ludwig H, Kao M, Schneider K. Learning deficiencies in Borna disease virus-infected but clinically healthy rats. Biol Psychiatry. 1989;26:818–828. doi: 10.1016/0006-3223(89)90122-4. [DOI] [PubMed] [Google Scholar]
- 10a.Furrer, E., et al. Unpublished data.
- 11.Gies U, Bilzer T, Stitz L, Staiger J F. Disturbance of the cortical cholinergic innervation in Borna disease prior to encephalitis. Brain Pathol. 1998;8:39–48. doi: 10.1111/j.1750-3639.1998.tb00133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez-Dunia D, Cubitt B, Grasser F A, de la Torre J C. Characterization of Borna disease virus p56 protein, a surface glycoprotein involved in virus entry. J Virol. 1997;71:3208–3218. doi: 10.1128/jvi.71.4.3208-3218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatalski C G, Hickey W F, Lipkin W I. Evolution of the immune response in the central nervous system following infection with Borna disease virus. J Neuroimmunol. 1998;90:137–142. doi: 10.1016/s0165-5728(98)00076-9. [DOI] [PubMed] [Google Scholar]
- 14.Herzog S, Kompter C, Frese K, Rott R. Replication of Borna disease virus in rats: age-dependent differences in tissue distribution. Med Microbiol Immunol. 1984;173:171–177. doi: 10.1007/BF02122108. [DOI] [PubMed] [Google Scholar]
- 15.Hirano N, Kao M, Ludwig H. Persistent, tolerant or subacute infection in Borna disease virus-infected rats. J Gen Virol. 1983;64:1521–1530. doi: 10.1099/0022-1317-64-7-1521. [DOI] [PubMed] [Google Scholar]
- 16.Iwata Y, Takahashi K, Peng X, Fukuda K, Ohno K, Ogawa T, Gonda K, Mori N, Niwa S, Shigeta S. Detection and sequence analysis of borna disease virus p24 RNA from peripheral blood mononuclear cells of patients with mood disorders or schizophrenia and of blood donors. J Virol. 1998;72:10044–10049. doi: 10.1128/jvi.72.12.10044-10049.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khansari D N, Murgo A J, Faith R E. Effects of stress on the immune system. Immunol Today. 1990;11:170–175. doi: 10.1016/0167-5699(90)90069-l. [DOI] [PubMed] [Google Scholar]
- 18.Kishi M, Arimura Y, Ikuta K, Shoya Y, Lai P K, Kakinuma M. Sequence variability of Borna disease virus open reading frame II found in human peripheral blood mononuclear cells. J Virol. 1996;70:635–640. doi: 10.1128/jvi.70.1.635-640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kliche S, Briese T, Henschen A H, Stitz L, Lipkin W I. Characterization of a Borna disease virus glycoprotein, gp18. J Virol. 1994;68:6918–6923. doi: 10.1128/jvi.68.11.6918-6923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kliche S, Stitz L, Mangalam H, Shi L, Binz T, Niemann H, Briese T, Lipkin W I. Characterization of the Borna disease virus phosphoprotein, p23. J Virol. 1996;70:8133–8137. doi: 10.1128/jvi.70.11.8133-8137.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lieb K, Hallensleben W, Czygan M, Stitz L, Staeheli P. No Borna disease virus-specific RNA detected in blood from psychiatric patients in different regions of Germany. The Bornavirus Study Group. Lancet. 1997;350:1002. doi: 10.1016/s0140-6736(05)64066-4. [DOI] [PubMed] [Google Scholar]
- 22.Lundgren A L, Zimmermann W, Bode L, Czech G, Gosztonyi G, Lindberg R, Ludwig H. Staggering disease in cats: isolation and characterization of the feline Borna disease virus. J Gen Virol. 1995;76:2215–2222. doi: 10.1099/0022-1317-76-9-2215. [DOI] [PubMed] [Google Scholar]
- 23.Malkinson M, Weisman Y, Perl S, Ashash E. A Borna-like disease of ostriches in Israel. Curr Top Microbiol Immunol. 1995;190:31–38. doi: 10.1007/978-3-642-78618-1_3. [DOI] [PubMed] [Google Scholar]
- 24.Narayan O, Herzog S, Frese K, Scheefers H, Rott R. Pathogenesis of Borna disease in rats: immune-mediated viral ophthalmoencephalopathy causing blindness and behavioral abnormalities. J Infect Dis. 1983;148:305–315. doi: 10.1093/infdis/148.2.305. [DOI] [PubMed] [Google Scholar]
- 24a.National Center for Biotechnology Information. Basic Local Alignment Search Tool. [Online.] http://www.ncbi.nlm.nih.gov. [19 February 1999, last date accessed.]
- 25.Noske K, Bilzer T, Planz O, Stitz L. Virus-specific CD4+ T cells eliminate Borna disease virus from the brain via induction of cytotoxic CD8+ T cells. J Virol. 1998;72:4387–4395. doi: 10.1128/jvi.72.5.4387-4395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Planz O, Bilzer T, Sobbe M, Stitz L. Lysis of major histocompatibility complex class I-bearing cells in Borna disease virus-induced degenerative encephalopathy. J Exp Med. 1993;178:163–174. doi: 10.1084/jem.178.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Planz O, Bilzer T, Stitz L. Immunopathogenic role of T-cell subsets in Borna disease virus-induced progressive encephalitis. J Virol. 1995;69:896–903. doi: 10.1128/jvi.69.2.896-903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Planz O, Rentzsch C, Batra A, Rziha H, Stitz L. Persistence of Borna disease virus-specific nucleic acid in the blood of a psychiatric patient. Lancet. 1998;352:623. doi: 10.1016/S0140-6736(05)79577-5. [DOI] [PubMed] [Google Scholar]
- 29.Richt J A, Alexander R C, Herzog S, Hooper D C, Kean R, Spitsin S, Bechter K, Schuttler R, Feldmann H, Heiske A, Fu Z F, Dietzschold B, Rott R, Koprowski H. Failure to detect Borna disease virus infection in peripheral blood leukocytes from humans with psychiatric disorders. J Neurovirol. 1997;3:174–178. doi: 10.3109/13550289709015807. [DOI] [PubMed] [Google Scholar]
- 30.Rott R, Becht H. Natural and experimental Borna disease in animals. Curr Top Microbiol Immunol. 1995;190:17–30. doi: 10.1007/978-3-642-78618-1_2. [DOI] [PubMed] [Google Scholar]
- 31.Rott R, Herzog S, Fleischer B, Winokur A, Amsterdam J, Dyson W, Koprowski H. Detection of serum antibodies to Borna disease virus in patients with psychiatric disorders. Science. 1985;228:755–756. doi: 10.1126/science.3922055. [DOI] [PubMed] [Google Scholar]
- 32.Rubin S A, Sierra-Honigmann A M, Lederman H M, Waltrip R W, Jr, Eiden J J, Carbone K M. Hematologic consequences of Borna disease virus infection of rat bone marrow and thymus stromal cells. Blood. 1995;85:2762–2769. [PubMed] [Google Scholar]
- 33.Sauder C, de la Torre J C. Sensitivity and reproducibility of RT-PCR to detect Borna disease virus (BDV) RNA in blood: implications for BDV epidemiology. J Virol Methods. 1998;71:229–45. doi: 10.1016/s0166-0934(98)00005-6. [DOI] [PubMed] [Google Scholar]
- 34.Sauder C, Muller A, Cubitt B, Mayer J, Steinmetz J, Trabert W, Ziegler B, Wanke K, Mueller-Lantzsch N, de la Torre J C, Grasser F A. Detection of Borna disease virus (BDV) antibodies and BDV RNA in psychiatric patients: evidence for high sequence conservation of human blood-derived BDV RNA. J Virol. 1996;70:7713–7724. doi: 10.1128/jvi.70.11.7713-7724.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneemann A, Schneider P A, Lamb R A, Lipkin W I. The remarkable coding strategy of borna disease virus: a new member of the nonsegmented negative strand RNA viruses. Virology. 1995;210:1–8. doi: 10.1006/viro.1995.1311. [DOI] [PubMed] [Google Scholar]
- 36.Schneider P A, Hatalski C G, Lewis A J, Lipkin W I. Biochemical and functional analysis of the Borna disease virus G protein. J Virol. 1997;71:331–336. doi: 10.1128/jvi.71.1.331-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwemmle M, Salvatore M, Shi L, Richt J, Lee C H, Lipkin W I. Interactions of the borna disease virus P, N, and X proteins and their functional implications. J Biol Chem. 1998;273:9007–9012. doi: 10.1074/jbc.273.15.9007. [DOI] [PubMed] [Google Scholar]
- 38.Sobbe M, Bilzer T, Gommel S, Noske K, Planz O, Stitz L. Induction of degenerative brain lesions after adoptive transfer of brain lymphocytes from Borna disease virus-infected rats: presence of CD8+ T cells and perforin mRNA. J Virol. 1997;71:2400–2407. doi: 10.1128/jvi.71.3.2400-2407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stitz L, Dietzschold B, Carbone K M. Immunopathogenesis of Borna disease. Curr Top Microbiol Immunol. 1995;190:75–92. doi: 10.1007/978-3-642-78618-1_5. [DOI] [PubMed] [Google Scholar]
- 40.Stitz L, Krey H, Ludwig H. Borna disease in rhesus monkeys as a models for uveo-cerebral symptoms. J Med Virol. 1981;6:333–340. doi: 10.1002/jmv.1890060408. [DOI] [PubMed] [Google Scholar]
- 41.Stitz L, Noske K, Planz O, Furrer E, Lipkin W I, Bilzer T. A functional role for neutralizing antibodies in Borna disease: influence on virus tropism outside the central nervous system. J Virol. 1998;72:8884–8892. doi: 10.1128/jvi.72.11.8884-8892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stitz L, Schilken D, Frese K. Atypical dissemination of the highly neurotropic Borna disease virus during persistent infection in cyclosporin A-treated, immunosuppressed rats. J Virol. 1991;65:457–460. doi: 10.1128/jvi.65.1.457-460.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Summerfield A, Hofmann M A, McCullough K C. Low density blood granulocytic cells induced during classical swine fever are targets for virus infection. Vet Immunol Immunopathol. 1998;63:289–301. doi: 10.1016/s0165-2427(98)00108-1. [DOI] [PubMed] [Google Scholar]
- 44.Thiedemann N, Presek P, Rott R, Stitz L. Antigenic relationship and further characterization of two major Borna disease virus-specific proteins. J Gen Virol. 1992;73:1057–1064. doi: 10.1099/0022-1317-73-5-1057. [DOI] [PubMed] [Google Scholar]
- 45.Thierer J, Riehle H, Grebenstein O, Binz T, Herzog S, Thiedemann N, Stitz L, Rott R, Lottspeich F, Niemann H. The 24K protein of Borna disease virus. J Gen Virol. 1992;73:413–416. doi: 10.1099/0022-1317-73-2-413. [DOI] [PubMed] [Google Scholar]
- 46.Van Strijp J A, Van Kessel K P, van der Tol M E, Fluit A C, Snippe H, Verhoef J. Phagocytosis of herpes simplex virus by human granulocytes and monocytes. Arch Virol. 1989;104:287–298. doi: 10.1007/BF01315550. [DOI] [PubMed] [Google Scholar]
- 47.Wehner T, Ruppert A, Herden C, Frese K, Becht H, Richt J A. Detection of a novel Borna disease virus-encoded 10 kDa protein in infected cells and tissues. J Gen Virol. 1997;78:2459–2466. doi: 10.1099/0022-1317-78-10-2459. [DOI] [PubMed] [Google Scholar]
- 48.Weissenbock H, Nowotny N, Caplazi P, Kolodziejek J, Ehrensperger F. Borna disease in a dog with lethal meningoencephalitis. J Clin Microbiol. 1998;36:2127–2130. doi: 10.1128/jcm.36.7.2127-2130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West B C, Eschete M L, Cox M E, King J W. Neutrophil uptake of vaccinia virus in vitro. J Infect Dis. 1987;156:597–606. doi: 10.1093/infdis/156.4.597. [DOI] [PubMed] [Google Scholar]
- 50.Zwick W, Seified O, Witte J. Weitere Untersuchungen über die Gehirn-Rückenmarksentzündung der Pferde (Bornasche Krankheit) Z Infekt Kr Haustiere. 1928;32:150. [Google Scholar]