Abstract
Infection with Japanese encephalitis virus (JEV), a mosquito-borne flavivirus, may cause acute encephalitis in humans and induce severe cytopathic effects in various types of cultured cells. We observed that JEV replication rendered infected baby hamster kidney (BHK-21) cells sensitive to the translational inhibitor hygromycin B or α-sarcine, to which mock-infected cells were insensitive. However, little is known about whether any JEV nonstructural (NS) proteins contribute to virus-induced changes in membrane permeability. Using an inducible Escherichia coli system, we investigated which parts of JEV NS1 to NS4 are capable of modifying membrane penetrability. We found that overexpression of NS2B-NS3, the JEV protease, permeabilized bacterial cells to hygromycin B whereas NS1 expression failed to do so. When expressed separately, NS2B alone, but not NS3, was sufficient to alter bacterial membrane permeability. Similarly, expression of NS4A or NS4B also rendered bacteria susceptible to hygromycin B inhibition. Examination of the effect of NS1 to NS4 expression on bacterial growth rate showed that NS2B exhibited the greatest inhibitory capability, followed by a modest repression from NS2A and NS4A, whereas NS1, NS3, and NS4B had only trivial influence with respect to the vector control. Furthermore, when cotransfected with a reporter gene luciferase or β-galactosidase, transient expression of NS2A, NS2B, and NS4B markedly reduced the reporter activity in BHK-21 cells. Together, our results suggest that upon JEV infection, these four small hydrophobic NS proteins have various modification effects on host cell membrane permeability, thereby contributing in part to virus-induced cytopathic effects in infected cells.
Among the medically important flaviviruses, Japanese encephalitis virus (JEV), which causes acute encephalitis in humans, has the highest mortality rate and remains as one of the major threats to public health in several parts of Asia (7, 46). Like other arthropod-borne flavivirus infections, JEV infection involves complex relationships among insect vectors, vertebrate reservoirs, and human subjects (9). Upon JEV infection, marked differences in cytopathogenecity are observed in different types of cultured cells. Infection of vertebrate cells is often cytocidal, resulting in drastic cytopathic effects (CPE) and ultrastructural changes, whereas infection of mosquito cells is noncytopathic, usually leading to persistent infection (reviewed in reference 34).
A wide variety of primary and continuous cell cultures of different origins can support the productive growth of JEV. Among them, Vero, LLC-MK2 (monkey kidney), and BHK-21 (baby hamster kidney) cells are frequently used for virus titer determination by plaque assays due to their apparent CPE induced by JEV infection (41). At the microscopic level, such infected cells display cell rounding, shrinkage, and dislodgment from the growth surface. At the ultramicroscopic level, the most prominent feature of flavivirus infection is a dramatic proliferation of intracellular membranous structures, including rough endoplasmic reticulum (RER) and Golgi complex, within which virus particles accumulate (20, 21). The exact molecular mechanism used by JEV to induce the infected-cell CPE is largely unknown.
Cytocidal viruses injure cells through a variety of mechanisms (reviewed in reference 25). There are at least two general pathways of cell death, i.e., necrosis and apoptosis; cell death due to viral infection could be the result of either or both pathways. JEV replication triggers apoptosis in various cell lines (31). Cytolytic viruses are known to cause their host cells to disintegrate by increasing plasma membrane permeability, causing a loss of cellular ion gradients and leakage of essential compounds from the cell (reviewed in reference 8), which leads to necrosis. The effects of viruses on cell membrane occur in at least two ways: by promoting membrane fusion between virus and cell and between cell and cell, and by altering the permeability of the plasma membrane (reviewed in reference 25). A growing body of evidence indicates that the expression of one single gene from certain animal viruses is sufficient to modify membrane permeability. These viral proteins are called viroporins. Viroporins are rather small polypeptides with a hydrophobic stretch of amino acids capable of forming an amphipathic helix; therefore, they possess activities like some ionophores or membrane-active toxins (reviewed in reference 8). Several viral proteins have been proven to be viroporins; these include poliovirus 2BC and 3AB proteins (1, 27, 29); human immunodeficiency virus gp41 (4); influenza virus M2 protein (19); togavirus 6K protein (40); human respiratory syncytial virus small hydrophobic protein (36); rotavirus NSP4 protein (44); hepatitis A virus 3A (37), 2B (23), and 2BC proteins (23); hepatitis C virus E1 protein (14); and coxsackievirus 2B protein (45).
As with other cytocidal viruses, JEV is likely to affect different host cellular processes at different steps of the viral replication cycle. The JEV genome is a single-stranded, positive-sense RNA of approximately 11 kb and contains an open reading frame of more than 10 kb encoding a polyprotein (reviewed in references 10 and 38). In the infected cells, the viral polyprotein is proteolytically cleaved by cellular and/or viral proteases into more than 10 structural and nonstructural (NS) proteins. The order of flavivirus proteins is 5′-C-prM(M)-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5-3′. The NS proteins in JEV-infected cells comprise a glycosylated NS1, two hydrophilic proteins NS3 and NS5, and four small hydrophobic proteins, NS2A, NS2B, NS4A, and NS4B. Among these proteins, NS3 and NS5, which are believed to be enzymatic components of the viral RNA replicase (11), localize in the cytoplasm and remain associated with intracellular membranes (16). In fact, NS3 is a multifunctional viral enzyme that contains helicase and NTPase activities in its central region (26) and a protease activity in its N terminus when associated with the viral cofactor NS2B (17, 22). NS2A, NS2B, NS4A, and NS4B, albeit less highly conserved in primary sequences among flaviviruses, have similar structural features consisting predominately of multiple hydrophobic, potential membrane-spanning domains (10). Except for the enzyme cofactor activity of NS2B for NS3 protease, little is known about the biological functions of these hydrophobic proteins in the flavivirus life cycle and the roles they may play in contributing to virus-induced CPE in infected cells.
In the present study, we observed that JEV infection can turn the originally insensitive cells into cells sensitive to the translational inhibitor hygromycin B or α-sarcine, indicating an increase in membrane permeability upon JEV infection. To further elucidate which JEV proteins are involved in this cytopathic process, we analyzed the changes in membrane permeability by overexpressing JEV NS proteins in an Escherichia coli inducible expression system (28). In addition, we investigated the effects of expression of such NS proteins on eukaryotic BHK-21 cells by assaying the gene expression of two reporter systems under different promoter controls. Our data illustrate that during JEV replication, some of its small hydrophobic NS proteins play a role in modifying membrane permeability of infected cells and thus might account in part for virus-induced CPE.
MATERIALS AND METHODS
Viruses and cell lines.
The plaque-purified Taiwanese local JEV RP-9 (12) was used for the cloning of JEV genes and for the infection of cells. Virus propagation was carried out with BHK-21 cells in RPMI 1640 medium containing 2% fetal bovine serum (GIBCO). Virus titers were determined by a plaque-forming assay on BHK-21 cells as previously described (32).
Construction of plasmids expressing JEV proteins.
For the expression of JEV proteins, cDNA fragments were reverse transcription-PCR amplified from JEV RP-9 (GenBank accession no. AF014161) as previously described (13, 33). The primer pairs used in the cloning procedures are listed in Table 1. The resulting cDNA fragments were cloned into TA vector pCR3.1, pCR3, or pCR3-uni (Invitrogen), in which the inserted gene is controlled by the enhancer-promoter sequences derived from the immediately-early gene of human cytomegalovirus (CMV), as well as by the bacteriophage T7 promoter. Standard recombinant DNA techniques (39) were used for plasmid construction. The inserted gene fragments, which were released from the TA vectors, were then subcloned into pET21 (Novagene) to fuse in frame with a T7 tag. After verification by sequencing, the resulting plasmids were used to transform E. coli BL21(DE3)pLysS (Novagene) for a membrane permeability assay. The plasmids expressing JEV NS proteins constructed in this study are listed in Table 2. In some experiments, to detect the viral proteins expressed in eukaryotic BHK-21 cells, a T7 tag was added in frame to the N terminus of each viral protein by the PCR cloning technique. A set of primers including a 5′ T7-tag primer derived from pET21 vector (Table 1) and a 3′ downstream primer for individual viral proteins was used to amplify the JEV genes with the above recombinant pET21 plasmids as templates. These PCR products were cloned into a eukaryotic expression vector, pCR3.1, and the resulting constructs were named Tag-NS2A, Tag-NS2B, Tag-NS4A, and Tag-NS4B.
TABLE 1.
Primers used for cloning JEV genes
| Gene | Position in JEV genomea | Primer sequence |
|---|---|---|
| NS1 | 2478–3533 | 5′-GCGGATCCbAGACACTGGATGTGCCA |
| 3′-GCGGATCCbTAAGCATCAACCTGTGA | ||
| NS2A | 3534–4214 | 5′-GCACCATGcTTTAATGGTGAA |
| 3′-GCCTAdTCTCTTCTTGTTTGGG | ||
| NS2B | 4215–4607 | 5′-GGGATGcGGGTGGCCAGCTACTGAGT |
| 3′-CTACTAdTCTTTTTGTTGT | ||
| NS2B-3 | 4215–6464 | 5′-GGGATGcGGGTGGCCAGCTACTGAGT |
| 3′-TTATTAdTCTCTTCCCTGCTGCGAAGTC | ||
| NS3 | 4608–6464 | 5′-ATGcGGGGGCGTGTTTTGGGACACGC |
| 3′-TTATTAdTCTCTTCCCTGCTGCGAAGTC | ||
| NS4A | 6465–6911 | 5′-CGACCATGcTCAGCCGTTAGC |
| 3′-GCCTAdTGCTGCCACCAC | ||
| NS4B | 6914–7676 | 5′-GCACCATGcAACGAGTACGGA |
| 3′-GCCTAdCCTTTTCAAGGAGGG | ||
| T7 tag | 5′-AATATGeGCTAGCATGACTGG |
According to the complete sequences of JEV, RP-9 (GenBank accession no. AF014161).
BamHI restriction enzyme recognition site created for cloning purposes.
Translational initiation codon added for proper protein expression.
Translational termination codon added for proper protein expression.
Starting codon of T7 tag in pET21 vector.
TABLE 2.
Plasmid constructs used in this study
| Region of JEV | Position in JEV genome | Vector(s) | Comments |
|---|---|---|---|
| NS1 | 2388–3533 | pcDNA3 | Signal peptide plus NS1 |
| NS1 | 2478–3533 | pET21b | NS1 full length, 352 aaa |
| NS2A | 3534–4214 | pCR3, pET21b | NS2A full length, 227 aa |
| NS2B | 4215–4607 | pCR3, pET21c | NS2B full length, 131 aa |
| NS2B (1–125) | 4215–4591 | pET21a | DraI fragment of NS2B |
| NS2B-NS3 (1–750) | 4215–6464 | pCR3, pET21a | NS2B-NS3 full length, 750 aa |
| NS2B-NS3 (1–373) | 4215–5334 | pCR3, pET21a | EcoRV fragment of NS2B-NS3 |
| NS2B-NS3 (127–373) | 4592–5334 | pET21b | DraI fragment of NS2B-NS3 |
| NS3 | 4608–6464 | pCR3.1, pET21c | NS3 full length, 619 aa |
| NS4A | 6465–6911 | pCR3-uni, pET21c | NS4A full length, 149 aa |
| NS4A (23–149) | 6530–6911 | pCR3, pET21c | HindIII fragment of NS4A |
| NS4B | 6914–7676 | pCR3.1, pET21a | NS4B full length, 255 aa |
| NS4B (1–219) | 6914–7570 | pET21b | EcoRI fragment of NS4B |
| NS4B (1–113) | 6914–7252 | pET21b | NcoI fragment of NS4B |
aa, amino acids.
Measurement of membrane modification by JEV infection.
BHK-21 cells in a six-well plate were infected with JEV at a multiplicity of infection of 5 as previously described (32). At the indicated time points postinfection (p.i.), the medium was removed and the cells were incubated for 30 min at 37°C with warm methionine (Met)- and cysteine (Cys)-free RPMI 1640 medium containing 2% dialyzed fetal bovine serum in the absence or presence of hygromycin B (500 μg/ml; Boehringer Mannheim) or α-sarcine (10 μg/ml; Sigma). The cells were then labeled with 100 μCi of [35S]Pro-mix (Amersham) per ml at 37°C for another 30 min. The culture fluids were removed, and the cell layers were rinsed with ice-cold phosphate-buffered saline and harvested in a lysis buffer (1% Nonidet P-40, 150 mM NaCl, 50 mM Tris-HCl [pH 7.5], 1 mM EDTA) containing a cocktail of protease inhibitors (20 μg of phenylmethysulfonyl fluoride per ml, 2 μg of leupeptin per ml, 2 μg of aprotinin per ml). The proteins were precipitated onto fiberglass discs (GF/C; Whatman) with a TCA solution (5% trichloroacetic acid, 20 mM sodium pyrophosphate). The discs were washed with 70% ethanol and dried at room temperature. To measure the total 35S label in the samples, the same amount of protein lysate was spotted on the disc, air dried, and counted in a β-counter (Beckman) with scintillation fluid (Biofluor; Dupont, NEN). The incorporation of [35S]Met was calculated as (incorporated/total) × 100% for each sample.
Induction of recombinant protein expression in E. coli.
A single colony of E. coli BL21(DE3)pLysS containing the indicated plasmid was grown overnight in Luria-Bertani medium in the presence of 100 μg of ampicillin per ml and 34 μg of chloramphenicol per ml. The cells were then diluted 100-fold in M9 medium supplemented with 0.2% glucose and antibiotics and grown at 37°C. Once the cultures reached an absorbance at 600 nm of 0.5 to 0.6, they were induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Rifampin (150 μg/ml; Boehringer Mannheim) was added 20 min after induction to inhibit the transcription of E. coli RNA polymerase.
Labeling and electrophoretic analysis of proteins.
To label the proteins synthesized by the transformed bacterial cells, after the indicated periods of IPTG induction 1-ml aliquots of cultures were collected and incubated with 10 μCi of [35S]Pro-mix per ml in the absence or presence of hygromycin B (1 mM) at 37°C. The labeled bacteria were pelleted, resuspended in a sample buffer (0.1 M dithiothreitol, 160 mM Tris-Cl [pH 6.8], 1% sodium dodecyl sulfate [SDS], 0.024% bromophenol blue, 10% glycerol), separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (10 to 15% polyacrylamide), and fluorographed at −70°C.
Radioimmunoprecipitation (RIP).
The JEV monoclonal antibodies (13) or T7 tag monoclonal antibody (Novagen) were first incubated with a mixture of protein A and protein G-Sepharose (Pharmacia) at room temperature for 1 h, and the 35S-labeled protein lysates were then added and incubated at room temperature for another 1 h. The resulting immunocomplexes were washed three times with RIPA buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate), analyzed by SDS-PAGE, and fluorographed at −70°C.
β-Galactosidase assay.
By using Lipofectamine PLUS (GIBCO-BRL) as described by the manufacturer, BHK-21 cells in a six-well plate were cotransfected with β-galactosidase reporter plasmid, pCMVβ (Clontech) (0.5 μg per well) and each of the plasmids expressing the various JEV proteins (2.5 μg per well). Two days posttransfection, the cell lysates were harvested and the enzyme activity was measured with a β-galactosidase assay system (Promega) as described by the manufacturer. The total amounts of protein in each sample were measured with a protein assay kit (Bio-Rad) based on the Bradford dye-binding procedure. The activity of reporter gene expression was normalized to the total amount of protein in each sample tested, and the relative enzyme activity of β-galactosidase was calculated as the ratio of enzyme activity from each sample to that from a negative vector control.
Luciferase assay.
BHK-21 cells were cotransfected with a luciferase reporter plasmid, pGL3-control (Promega), and each of the plasmids expressing the various JEV proteins as described above for the β-galactosidase assay. At 24 h posttransfection, the cells were lysed and assayed for their luciferase activity by using a luciferase assay system (Promega). The relative reporter gene expression was calculated as described above for the β-galactosidase assay.
RESULTS
Membrane modification by JEV infection.
To evaluate the membrane modification induced by JEV infection, we first assayed the permeabilization of infected BHK-21 cells to a translational inhibitor hygromycin B (Mr, 550) or α-sarcine (Mr, 16,800), neither of which is able to cross the unaltered membrane (3). As the data in Fig. 1A show, protein synthesis by infected cells, as measured by the amounts of [35S]methionine incorporated into total proteins, decreased in the presence of hygromycin B or α-sarcine at 18 h postinfection and further declined as the virus replication progressed. As a control, the mock-infected cells remained resistant to both inhibitors during the same period (Fig. 1B). Thus, similar to other cytolytic animal viruses (reviewed in reference 8), JEV modified the membrane permeability of infected cells during the infection process.
FIG. 1.
Membrane modification by JEV infection. The percentage of [35S]methionine incorporation was measured in JEV-infected (A) and mock-infected (B) BHK-21 cells at 6, 18, or 24 h p.i. in the absence or presence of the translation inhibitor hygromycin B (Hyg.) or α-sarcine (sar.) as described in Materials and Methods.
Expression of JEV protease NS2B-NS3 increased the membrane permeability.
Several viral proteins involved in the membrane modification have been identified by using an inducible E. coli expression system (reviewed in reference 8). A similar approach was used here to explore which JEV NS proteins possess the ability to alter membrane permeability. We first investigated the modification effect of JEV protease NS2B-NS3 on a bacterial membrane. The DNA fragment of full-length NS2B-NS3 was cloned in expression vector pET21 and transformed into E. coli BL21(DE3)pLysS as described in Materials and Methods. In the presence of the bacterial RNA polymerase inhibitor rifampin, expression of NS2B-NS3, which was under the control of the T7 promoter, was readily detected at 20, 40, and 60 min after IPTG induction (Fig. 2A, lanes 2, 4, and 6). The protein band indicated by the arrowhead in Fig. 2A was confirmed to be full-length NS2B-NS3, 83 kDa in size, by immunoprecipitation with monoclonal antibodies specific for JEV NS3 (reference 13 and data not shown). However, addition of hygromycin B greatly reduced the amount of NS2B-NS3 expression under the same condition (lanes 3, 5, and 7), indicating that NS2B-NS3 expression changed the bacterial membrane permeability, thus allowing the entry of the translational inhibitor. In contrast, inducible expression of JEV NS1 protein did not render the bacteria susceptible to hygromycin B inhibition (Fig. 2B), suggesting that NS1 did not have any effect on membrane modification.
FIG. 2.
Membrane modification in E. coli by JEV NS2B-NS3 proteins. E. coli BL21(DF3)pLysS harboring individual JEV NS genes cloned in pET21 plasmids was incubated without rifampin (no R.) or with rifampin plus IPTG (I.) in the absence or presence of hygromycin B (H.) after various periods as indicated at the top of the gels. Protein samples were labeled with [35S]methionine and analyzed by SDS-PAGE as described in Materials and Methods. (A) Full-length NS2B-NS3 (1 to 750); (B) full-length NS1; (C and D) partial NS2B-NS3 (amino acids 1 to 373 and 127 to 373, respectively). The radioimmunoprecipitation (RIP) of expressed protein is shown in panel D by using antibody against T7 tag (lane 6) or JEV NS3 (lane 7). The major protein bands as predicted for each construct are indicated by arrowheads, and the numbers on the sides of the gels denote the molecular masses of protein standards.
The ability of full-length NS2B-NS3 to permeabilize the E. coli membrane was further examined by expression and testing of tentative virus protease, NS2B plus the N-terminal one-third of NS3 (22). As shown in Fig. 2C, expression of pET/NS2B-3(1–373), encoding the full-length NS2B (131 amino acids) and the N-terminal 242 amino acids of NS3 (about 39% of NS3), still modified membrane permeability. On the other hand, expression of pET/NS2B-3(126–373) (Fig. 2D), containing the first 242 amino acids of NS3 plus the last 6 amino acids of NS2B, failed to trigger the entry of hygromycin B into bacterial cells. Moreover, expression of full-length NS3 alone also failed to modify the bacterial membrane (data not shown). These results strongly suggest that the membrane interacting domain of NS2B-NS3 is localized in the NS2B region.
Membrane modification by four small, hydrophobic proteins: NS2A, NS2B, NS4A and NS4B.
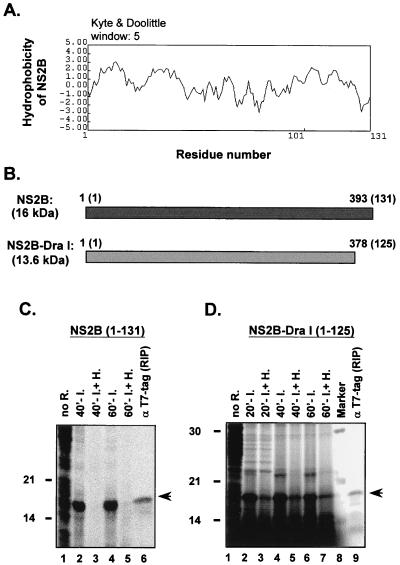
To further examine if NS2B expression alone was enough to alter membrane permeability, we cloned and evaluated NS2B in the bacterial pET system described above. The results shown in Fig. 3C indicate that expression of pET/NS2B(1–131), i.e., the full-length NS2B, resulted in a major protein band of 16 kDa as predicted for NS2B (Fig. 3C, lanes 2 and 4), which was immunoprecipitated by a monoclonal antibody against T7 tag present in this NS2B fusion protein (lane 6). Addition of hygromycin B markedly diminished the NS2B expression (lanes 3 and 5). These data clearly illustrate that when expressed separately, NS2B alone was sufficient to alter bacterial membrane permeability. Interestingly, inducible expression of pET/NS2B-DraI(1–125), a mutant NS2B with a deletion of the last 6 amino acids from its C terminus (Fig. 3B), entirely abolished the ability of NS2B to induce the influx of hygromycin B to cells (Fig. 3D). This result implicates the direct involvement of these 6 amino acids (LKTTKR), which are mainly positively charged and hydrophilic (Fig. 3A), in the membrane modification capability of NS2B. Besides, it is also possible that lack of these 6 amino acids will alter the conformation of the domain of NS2B involved in modifying membranes.
FIG. 3.
Membrane modification in E. coli by JEV NS2B. (A) Hydrophobicity plot of NS2B analyzed by the Kyte-Doolittle method with the DNAsis program (Hitachi). (B) Schematic diagram of NS2B regions expressed by plasmid constructs, shown in nucleotide numbers and (amino acid numbers are in parentheses). (C and D) Membrane permeability assays (described in the legend to Fig. 2).
The observation of the effect of NS2B on membrane modification suggests that the other three small, hydrophobic NS proteins, NS2A, NS4A, and NS4B, may also be involved in membrane permeabilization. To test this hypothesis, the JEV NS2A gene was cloned as described in Materials and Methods, and its expression from vector pCR3.1 or pET21 was readily detected by an in vitro transcription coupled with translation system (data not shown). However, no major NS2A protein bands could be identified when expressed by the bacterial pET inducible system. The NS2A protein derived from our JEV strain may have been extremely unstable or the codon usage may not have been suitable in the bacteria, such that this pET system was unable to detect NS2A. Hence, whether NS2A possesses the ability to modify the membrane permeability of E. coli cells by using the hygromycin B inhibition method as described above for NS2B remains unknown. In contrast, expression of NS4A (Fig. 4C) enhanced the influx of hygromycin B in bacteria, thereby blocking total-protein synthesis. Furthermore, expression of pET/NS4A-HindIII(23–149), a shorter mutant with a deletion of 22 residues from the N terminus of NS4A (Fig. 4B and D), maintained the membrane-modifying capability of its parental NS4A (Fig. 4B and C). Likewise, expression of NS4B (Fig. 5B and C) or its shorter constructs (Fig. 5B, D, and E) also exhibited the membrane-modifying capability, although at a lesser magnitude than that of NS2B (Fig. 3) or NS4A (Fig. 4). Together, these results indicate that overexpression of NS2B, NS4A, or NS4B permealizes E. coli cells to the translational inhibitor hygromycin B.
FIG. 4.
Membrane modification in E. coli by JEV NS4A. (A) Hydrophobicity plot of NS4A by the Kyte-Doolittle method. (B) Schematic diagram of NS4A regions expressed by plasmid constructs, shown in nucleotide numbers (amino-acid numbers are in parentheses). (C and D) Membrane permeability assays (described in the legend to Fig. 2).
FIG. 5.
Membrane modification in E. coli by JEV NS4B. (A) Hydrophobicity plot of NS4B by the Kyte-Doolittle method. (B) Schematic diagram of NS4B regions expressed by plasmid constructs, shown in nucleotide numbers (amino acid numbers are in parentheses). (C, D, and E) Membrane permeability assay (described in the legend to Fig. 2).
Effect of JEV NS proteins on E. coli cell growth.
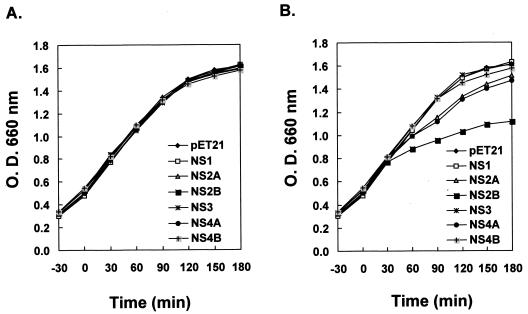
We next investigated whether expression of these JEV NS proteins, in the absence of translational inhibitors, affect bacterial cell growth. The growth rates of E. coli BL21(DE3)pLysS, transformed with various pET21 plasmids expressing each individual JEV NS protein, were analyzed spectrophotometrically by measurement of optical density. As shown in Fig. 6B, upon IPTG induction NS2B exhibited the greatest inhibitory effect on bacterial growth rate, followed by NS2A and NS4A, which gave a modest suppression, whereas NS1, NS3, NS4B, and the control pET21 had no influence. In contrast, all bacterial cells grew in a similar pattern without IPTG induction (Fig. 6A), illustrating that it was the expression of the various induced JEV NS proteins that caused the changes of bacterial cell growth kinetics.
FIG. 6.
Growth curves of E. coli BL21(DE3)pLysS transformed with pET21 recombinants expressing various JEV NS proteins without (A) or with (B) IPTG induction. IPTG for induction was added at time 0 in panel B. The cell density of bacterial growth was determined by measuring the optical density (O.D.) at 660 nm by spectrometry.
Effects of expression of small hydrophobic JEV NS proteins on eukaryotic gene expression.
The observation that some JEV NS proteins modify bacterial membranes and alter bacterial growth profiles prompted us to explore the effects of the expression of these NS proteins on mammalian cells. Two reporter gene systems, luciferase (controlled by the simian virus 40 [SV40] promoter) and β-galactosidase (governed by the CMV immediate-early promoter), were cotransfected with each JEV NS gene into BHK-21 cells. If the expression of JEV NS proteins could indeed modify cell membranes, including the plasma membrane and the intracellular organelle membranes, the changes in reporter activities should reflect the extent of physiological disturbance in the target cells examined. To conveniently identify NS proteins in the cells, a T7 tag was fused with the N terminus of each viral NS protein as described in Materials and Methods; expression of these NS fusion proteins was detected in vitro and in vivo by immunoprecipitation with monoclonal antibody against the T7 tag (data not shown). As shown in Fig. 7A, compared to the pcDNA3 control, luciferase activities decreased dramatically from the cells cotransfected with JEV NS2A, NS2B, or NS4B; in contrast, expression of NS4A or NS1 had only trivial effects on reporter activities. The levels of luciferase activity in culture supernatants were also determined, and no major difference was detected among these samples (data not shown). This result indicates that the decrease of luciferase activity in certain samples was due to the inhibition of reporter gene expression but not to the release of reporter gene product into culture supernatants. Therefore, the inhibition of reporter gene expression was more likely to result from the modification of intracellular organelle membranes, leading to disturbance of ion homeostasis (discussed below) or some other unknown reasons. Also, we observed an almost identical inhibitory pattern from the NS proteins without the T7 tag (Fig. 7B); that is, NS2A, NS2B, and NS4B, rather than NS4A, were able to cause potent suppression of luciferase activities compared to their opposite-orientation controls, NS2A-R and NS4A-R. The moiety of bacteriophage T7 tag in the fusion proteins appeared to play no role in altering the membrane integrity of BHK-21 cells.
FIG. 7.
Effects of expression of JEV NS proteins on reporter activity in BHK-21 cells. BHK-21 cells were cotransfected with a reporter gene and plasmids expressing various JEV NS proteins. The reporters include a luciferase gene (A and B) and a β-galactosidase gene (C and D). At 24 h (A and B) or 48 h (C and D) posttransfection, the cells were lysed and assayed for their luciferase or β-galactosidase activities. 2A-R and 4A-R in panels B and D are the constructs with inserts in opposite orientation.
We next tried to verify whether inhibition of reporter activity by expression of JEV NS genes was only associated with the genes under SV40 promoter control. The effect of expression of the various JEV NS genes was therefore also examined in BHK-21 cells cotransfected with another reporter gene, that encoding β-galactosidase, under the control of the CMV IE gene promoter. Although not as prominent as the results shown in Fig. 7A and B, there was still a similar inhibitory profile for each NS protein (Fig. 7C and D); i.e., expression of NS2A, NS2B, and NS4B, irrespective of the T7 tag, also repressed β-galactosidase activities, whereas expression of NS4A and NS1, just like the control pcDNA3, failed to affect the reporter activities. These results suggest that transient expression of certain small hydrophobic JEV NS proteins indeed displays global inhibition toward BHK-21 cells, presumably by modifying the integrity of intracellular organelle membranes or by some other unknown mechanisms.
DISCUSSION
In the present study, we demonstrated that not only does JEV infection render target cells susceptible to translational inhibitors (Fig. 1) but also separate expression of certain NS proteins of JEV changes membrane permeability and alters the cell growth rate in an E. coli system (Fig. 2 to 6). The JEV NS proteins responsible for hygromycin B inhibition in the bacterial system were the small hydrophobic proteins NS2B (Fig. 3C), NS4A (Fig. 4C), and NS4B (Fig. 5C); overexpression of NS2A, NS2B, or NS4A resulted in delayed bacterial growth (Fig. 6B) even in the absence of translational inhibitors. On the other hand, expression of NS2A, NS2B, or NS4B, but not NS1 or NS4A, suppressed reporter systems in mammalian BHK-21 cells (Fig. 7). Being capable of functioning in both prokaryotic and eukaryotic systems, NS2A, NS2B, and NS4B appear to be the viroporins of JEV. These results may thus provide a molecular explanation for JEV-induced CPE observed in infected cells. By contrast, expression of JEV NS4A exhibited an effect only on bacterial membrane permeability (Fig. 4) but not on the expression of reporter genes in mammalian cells (Fig. 7). Conceivably, such a discrepancy may be attributed to differences between the protein nature of NS4A synthesized in bacterial and mammalian cells or between the membrane composition derived from these two different types of cells. More experiments are needed to further study the exact role of NS4A on membrane modification.
The biological functions of small hydrophobic NS proteins of JEV are largely unexplored. Recent evidence has suggested that viroporins might play a crucial role in the release of viruses from infected cells (reviewed in reference 8). Theoretically, in the early phase of virus replication, viroporins of JEV in the cellular membrane compartments may form a hydrophilic pore by oligomerization, thus allowing ions and low-molecular-weight hydrophilic compounds to pass nonspecifically. As infection progresses and the amounts of viroporins in the membrane increase, other viral and/or cellular proteins may be recruited to widen the pore size. Consequently, macromolecules including cellular enzymes may leak from infected cells, resulting in CPE. This notion is supported by what we observed in JEV-infected cells. Starting from 6 h p.i., JEV gradually turned the infected BHK-21 cells, which were originally resistant to low-molecular-mass translational inhibitors (0.5 to 16.8 kDa), into cells sensitive to such a translational inhibition (Fig. 1); moreover, starting from 16 h p.i., a cytoplasmic enzyme, lactate dehydrogenase (140 kDa), also began to leak out of the cells (31). In addition, the kinetics of given infected cells sensitive to translational inhibitors seem to correlate with the virulence of JEV strains used; that is, the cells infected by attenuated strain RP-2ms (12) exhibited a delayed profile to translation inhibition compared to cells infected by its counterpart virulent strain, RP-9 (data not shown). This phenomenon is relevant because not only is the plaque size of RP-2ms on BHK-21 cells smaller than that of RP-9, but also RP-2ms virions are often accumulated intracellularly while RP-9 particles are readily released extracellularly (12). Presumably, different viroporins derived from these two JEV strains may account for such phenotypic differences. However, the amino acid sequences of small hydrophobic NS proteins are identical between RP2-ms (GenBank accession no. AF014160) and RP-9, implying that other unidentified viroporins, for example, virus structural proteins E and prM/M, may play an important role in virion release as well as in virus virulence. Thus, it will be of interest to identify such viroporins from other parts of JEV genome in the near future.
Theoretically, NS2A, NS2B, NS4A, and NS4B of flavivirus, processed posttranslationally by cellular signalase in the ER lumen together with viral NS2B-NS3 protease in the cytosol, are predicted to be able to span the ER membrane at least once (reviewed in reference 10). In fact, the association of these viral proteins with membranes has been demonstrated in a flavivirus, West Nile virus, in which stringently washed membranes derived from the infected BHK-21 cells contain virus-encoded NS2A, NS2B, and NS4B, although NS4A was not found in this study (47). In addition to the hydrophobic region that interacts with the membrane, viroporins usually contain a short stretch of basic amino acids flanked by a membrane-interacting domain, a unique feature seen in several membrane-active toxins (8). These basic amino acids are thought to participate in membrane permeabilization, probably by destabilizing the structural integrity of lipid bilayers. Consistent with this concept, JEV NS2B also comprises a region of 6 amino acids (3 of which are basic amino acids) at the carboxyl terminal, and removal of these 6 amino acids abolishes the membrane-modifying capability of NS2B (Fig. 3). NS2B (131 amino acids), the smallest of these four hydrophobic proteins, might behave in a manner similar to other reported viroporins (reviewed in reference 8) based on protein length, the requirement for basic amino acids for membrane modification (Fig. 3), and the ability to inhibit bacterial cell growth (Fig. 6).
Flaviviruses do not markedly inhibit the macromolecule synthesis of host cells until late in the infection (34). By analyzing a sensitive reporter system, we observed that JEV infection suppressed gene expression of luciferase or β-galactosidase in BHK-21 cells even before 24 h p.i. (data not shown). We further demonstrated that individual expression of JEV NS2A, NS2B, or NS4B blocked the activities of two different reporter systems in BHK-21 cells (Fig. 7). However, the way in which transient expression of these JEV NS proteins inhibits reporter functions in mammalian system remains obscure. The inhibition of reporter gene expression was more likely to result from the modification of intracellular organelle membranes which might lead to disturbance of ion homeostasis. Among viroporins identified so far, poliovirus 2BC protein (2), coxsackie B3 virus 2B protein (45), and rotavirus NSP4 (43) have been found to be capable of disrupting the intracellular Ca2+ homeostasis of infected cells. Intracellular Ca2+ appears to be an important regulatory signal for various biological events, and the ER serves as the major reservoir of Ca2+ in the cells (reviewed in reference 18). Since small hydrophobic NS proteins of JEV are associated primarily with the ER membranes once they are synthesized, it is possible that occurrence of these NS proteins in the ER membrane triggers the release of Ca2+ from the ER to the cytoplasm. Alteration of the intracellular Ca2+ distribution may conceivably activate certain signaling pathways that eventually lead to the inhibition of reporters under SV40 or CMV promoter control. Besides, some membrane interactive proteins, such as poliovirus 3A (30) and 2BC (6), Sindbis virus envelope glycoproteins (24), and rotavirus NSP4 (35), have all been reported to cause cytotoxicity. Whether JEV hydrophobic NS proteins are also directly involved in cell death induced by JEV infection remains to be determined. Other cellular events, such as proliferation and rearrangement of membrane structures (5, 42) and inhibition of glycoprotein trafficking through the membranous compartments (5, 15), have also been reported for other viroporins. Whether JEV hydrophobic NS proteins induce these cytopathic events also deserves further study. The results of these studies will enhance our understanding of the role of JEV hydrophobic NS proteins in the process of virus-induced CPE.
ACKNOWLEDGMENTS
This work was supported by grants from the National Science Council (NSC), Taiwan, ROC (NSC-88-2314-B-001-044), and Academia Sinica, ROC, awarded to Y.-L.L.
We thank Douglas Platt for editorial correction of the manuscript.
REFERENCES
- 1.Aldabe R, Barco A, Carrasco L. Membrane permeabilization by poliovirus proteins 2B and 2BC. J Biol Chem. 1996;271:23134–23137. doi: 10.1074/jbc.271.38.23134. [DOI] [PubMed] [Google Scholar]
- 2.Aldabe R, Irurzun A, Carrasco L. Poliovirus protein 2BC increases cytosolic free calcium concentrations. J Virol. 1997;71:6214–6217. doi: 10.1128/jvi.71.8.6214-6217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso M A, Carrasco L. Molecular basis of the permeabilization of mammalian cells by ionophores. Eur J Biochem. 1982;127:3567–3590. doi: 10.1111/j.1432-1033.1982.tb06909.x. [DOI] [PubMed] [Google Scholar]
- 4.Arroyo J, Boceta M, Gonzalez M E, Michel M, Carrasco L. Membrane permeabilization by different regions of the human immunodeficiency virus type 1 transmembrane glycoprotein gp41. J Virol. 1995;67:4095–4102. doi: 10.1128/jvi.69.7.4095-4102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barco A, Carrasco L. A human virus protein, poliovirus protein 2BC, induces membrane proliferation and blocks the exocytic pathway in the yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3349–3364. doi: 10.1002/j.1460-2075.1995.tb07341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barco A, Carrasco L. Identification of regions of poliovirus 2BC protein that are involved in cytotoxicity. J Virol. 1998;72:3560–3570. doi: 10.1128/jvi.72.5.3560-3570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke D S, Leake C J. Japanese encephalitis. In: Monath T P, editor. The arboviruses: epidemiology and ecology. Vol. 3. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 63–92. [Google Scholar]
- 8.Carrasco L. Modification of membrane permeability by animal viruses. Adv Virus Res. 1995;45:61–112. doi: 10.1016/S0065-3527(08)60058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamberlain R W. Epidemiology of arthropod-borne togaviruses: The role of arthropods as hosts and vectors and of vertebrate hosts in natural transmission cycles. In: Schlesinger R W, editor. The togaviruses: biology, structure, replication. New York, N.Y: Academic Press, Inc.; 1980. pp. 175–228. [Google Scholar]
- 10.Chambers T J, Hanh C S, Galler R, Rice C M. Flavivirus genome, organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–698. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 11.Chen C-J, Kuo M-D, Chien L-J, Hsu S-L, Wang Y-M, Lin J-H. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J Virol. 1997;71:3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L-K, Lin Y-L, Liao C-L, Lin C-G, Huang Y-L, Yeh C-T, Lai S-C, Jan J-T, Chin C. Generation and characterization of organ-tropism mutants of Japanese encephalitis virus in vivo and in vitro. Virology. 1996;223:79–88. doi: 10.1006/viro.1996.0457. [DOI] [PubMed] [Google Scholar]
- 13.Chen L-K, Liao C-L, Lin C-G, Lai S-C, Liu C-I, Ma S-H, Huang Y-Y, Lin Y-L. Persistence of Japanese encephalitis virus is associated with abnormal expression of the nonstructural protein NS1 in host cells. Virology. 1996;217:220–229. doi: 10.1006/viro.1996.0109. [DOI] [PubMed] [Google Scholar]
- 14.Ciccaglione A R, Marcantonio C, Costantino A, Equestre M, Geraci A, Rapicetta M. Hepatitis C virus E1 protein induces modification of membrane permeability in E coli cells. Virology. 1998;250:1–8. doi: 10.1006/viro.1998.9380. [DOI] [PubMed] [Google Scholar]
- 15.Doedens J R, Kirkegaard K. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 1995;14:894–907. doi: 10.1002/j.1460-2075.1995.tb07071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edward Z, Takegami T. Localization and functions of Japanese encephalitis virus nonstructural proteins NS3 and NS5 for viral RNA synthesis in the infected cells. Microbiol Immunol. 1993;37:239–243. doi: 10.1111/j.1348-0421.1993.tb03206.x. [DOI] [PubMed] [Google Scholar]
- 17.Falgout B, Pethel M, Zhang Y-M, Lai C-J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of Dengue virus nonstructural proteins. J Virol. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill D L, Waldron R T, Rys-Sikora K E, Ufret-Vincenty C A, Graber M N, Favre C J, Alfonso A. Calcium pools, calcium entry, and cell growth. Biosci Reports. 1996;16:139–157. doi: 10.1007/BF01206203. [DOI] [PubMed] [Google Scholar]
- 19.Guinea R, Carrasco L. Influenza virus M2 protein modifies membrane permeability in E. coli cells. FEBS Lett. 1994;343:242–246. doi: 10.1016/0014-5793(94)80564-4. [DOI] [PubMed] [Google Scholar]
- 20.Hase T, Dubois D R, Summers P L. Comparative study of mouse brains infected with Japanese encephalitis virus by intracerebral or intraperitoneal inoculation. Int J Exp Pathol. 1990;71:857–869. [PMC free article] [PubMed] [Google Scholar]
- 21.Hase T, Summers P L, Ray P, Asafo-Adjei E. Cytopathology of PC12 cells infected with Japanese encephalitis virus. Virchows Arch B. 1992;63:25–36. doi: 10.1007/BF02899241. [DOI] [PubMed] [Google Scholar]
- 22.Jan L-R, Yang C-S, Trent D W, Falgout B, Lai C-J. Processing of Japanese encephalitis virus non-structural proteins: NS2B-NS3 complex and heterologous proteases. J Gen Virol. 1995;76:573–580. doi: 10.1099/0022-1317-76-3-573. [DOI] [PubMed] [Google Scholar]
- 23.Jecht M, Probst C, Gauss-Müller V. Membrane permeability induced by hepatitis A virus proteins 2B and 2BC and proteolytic processing of HAV 2BC. Virology. 1998;252:218–227. doi: 10.1006/viro.1998.9451. [DOI] [PubMed] [Google Scholar]
- 24.Joe A K, Foo H H, Kleeman L, Levine B. The transmembrane domains of Sindbis virus envelope glycoproteins induce cell death. J Virol. 1998;72:3935–3943. doi: 10.1128/jvi.72.5.3935-3943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knipe D M. Virus-host cell interactions. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 273–299. [Google Scholar]
- 26.Kuo M-D, Chin C, Hsu S-L, Shiao J-Y, Wang T-M, Lin J-H. Characterization of the NTPase activity of Japanese encephalitis virus NS3 protein. J Gen Virol. 1996;77:2077–2084. doi: 10.1099/0022-1317-77-9-2077. [DOI] [PubMed] [Google Scholar]
- 27.Lama J, Carrasco L. Expression of poliovirus nonstructural protein in Escherichia coli cells. J Biol Chem. 1992;267:15932–15937. [PubMed] [Google Scholar]
- 28.Lama J, Carrasco L. Inducible expression of a toxic poliovirus membrane protein in Escherichia coli: comparative studies using different expression systems based on T7 promoters. Biochem Biophys Res Commun. 1992;188:972–981. doi: 10.1016/0006-291x(92)91327-m. [DOI] [PubMed] [Google Scholar]
- 29.Lama J, Carrasco L. Mutations in the hydrophobic domain of poliovirus protein 3AB abrogate its permeabilizing activity. FEBS Lett. 1995;367:5–11. doi: 10.1016/0014-5793(95)00523-c. [DOI] [PubMed] [Google Scholar]
- 30.Lama J, Sanz M A, Carrasco L. Genetic analysis of poliovirus protein 3A: characterization of a non-cytopathic mutant virus defective in killing vero cells. J Gen Virol. 1998;79:1911–1921. doi: 10.1099/0022-1317-79-8-1911. [DOI] [PubMed] [Google Scholar]
- 31.Liao C-L, Lin Y-L, Wang J-J, Huang Y-L, Yeh C-T, Ma S-H, Chen L-K. Effect of enforced expression of human bcl-2 on Japanese encephalitis virus-induced apoptosis in cultured cells. J Virol. 1997;71:5963–5971. doi: 10.1128/jvi.71.8.5963-5971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin Y-L, Huang Y-L, Ma S-H, Yeh C-T, Chiou S-Y, Chen L-K, Liao C-L. Inhibition of Japanese encephalitis virus infection by nitric oxide: antiviral effect of nitric oxide on RNA virus replication. J Virol. 1997;71:5227–5235. doi: 10.1128/jvi.71.7.5227-5235.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Y-L, Chen L-K, Liao C-L, Yeh C-T, Ma S-H, Chen J-L, Huang Y-L, Chen S-S, Chiang H-Y. DNA immunization with Japanese encephalitis virus nonstructural protein NS1 elicits a protective immunity in mice. J Virol. 1998;72:191–200. doi: 10.1128/jvi.72.1.191-200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monath T P, Heinz F X. Flaviviruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 961–1034. [Google Scholar]
- 35.Newton K, Meyer J C, Bellamy A R, Taylor J. Rotavirus nonstructural glycoprotein NSP4 alters plasma membrane permeability in mammalian cells. J Virol. 1997;71:9458–9465. doi: 10.1128/jvi.71.12.9458-9465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pérez M, Garcia-Barreno B, Melero J A, Carrasco L, Guinea R. Membrane permeability changes induced in Escherichia coli by the SH protein of human respiratory syncytial virus. Virology. 1997;235:342–351. doi: 10.1006/viro.1997.8696. [DOI] [PubMed] [Google Scholar]
- 37.Pisani G, Beneduce F, Gauss-Muller V, Morace G. Recombinant expression of hepatitis A virus protein 3A: interaction with membranes. Biochem Biophys Res Commun. 1995;211:627–638. doi: 10.1006/bbrc.1995.1859. [DOI] [PubMed] [Google Scholar]
- 38.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 931–959. [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Sanz M A, Pérez L, Carrasco L. Semliki Forest virus 6K protein permeability after inducible expression in Escherichia coli cells. J Biol Chem. 1994;269:12106–12110. [PubMed] [Google Scholar]
- 41.Stim T B. Arbovirus plaquing in two simian kidney cell lines. J Gen Virol. 1969;5:329–338. [Google Scholar]
- 42.Teterina N L, Bienz K, Egger D, Gorbalenya A E, Ehrenfeld E. Induction of intracellular membrane rearrangements by HAV proteins 2 and 2BC. Virology. 1997;237:66–77. doi: 10.1006/viro.1997.8775. [DOI] [PubMed] [Google Scholar]
- 43.Tian P, Yanfang H, Schilling W P, Lindsay D A, Estes M K. Rotavirus NSP4 affects intracellular calcium levels. J Virol. 1994;68:251–257. doi: 10.1128/jvi.68.1.251-257.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian P, Ball J M, Zeng C Q-Y, Estes M K. The rotavirus nonstructural glycoprotein NSP4 possesses membrane destabilization activity. J Virol. 1996;70:6973–6981. doi: 10.1128/jvi.70.10.6973-6981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Kuppeveld F J M, Hoenderop J G J, Smeets R L L, Willems P H G M, Dijkman H B P M, Galama J M D, Melchers W J G. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 1997;16:3519–3532. doi: 10.1093/emboj/16.12.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaughn D W, Hoke C H., Jr The epidemiology of Japanese encephalitis: prospects for prevention. Epidemiol Rev. 1992;14:197–221. doi: 10.1093/oxfordjournals.epirev.a036087. [DOI] [PubMed] [Google Scholar]
- 47.Wengler G, Wengler G, Nowak T, Castle E. Description of a procedure which allows isolation of viral nonstructural proteins from BHK vertebrate cells infected with the West Nile flavivirus in a state which allows their direct chemical characterization. Virology. 1990;177:795–801. doi: 10.1016/0042-6822(90)90552-3. [DOI] [PubMed] [Google Scholar]