Abstract
We have examined the nature of V3 sequence variability among subtype C human immunodeficiency virus type 1 (HIV-1) sequences from plasma-derived viral RNA present in infected men from Malawi. Sequence variability was assessed by direct sequence analysis of the V3 reverse transcription-PCR products, examination of virus populations by a subtype C V3-specific heteroduplex tracking assay (V3-HTA), and selected sequence analysis of molecular clones derived from the PCR products. Sequence variability in V3 among the subtype C viruses was not associated with the presence of basic amino acid substitutions. This observation is in contrast to that for subtype B HIV-1, where sequence variability is associated with such substitutions, and these substitutions are determinants of altered coreceptor usage. Evolutionary variants in subtype C V3 sequences, as defined by the V3-HTA, were not correlated with the CD4 level in the infected person, while such a correlation was found with subtype B V3 sequences. Viruses were isolated from a subset of the subjects; all isolates used CCR5 and not CXCR4 as a coreceptor, and none was able to grow in MT-2 cells, a hallmark of the syncytium-inducing phenotype that is correlated with CXCR4 usage. The overall sequence variability of the subtype C V3 region was no greater than that of the conserved regions of gp120. This limited sequence variability was also a feature of subtype B V3 sequences that do not carry the basic amino acid substitutions associated with altered coreceptor usage. Our results indicate that altered coreceptor usage is rare in subtype C HIV-1 isolates in sub-Saharan Africa and that sequence variability is not a feature of the V3 region of env in the absence of altered coreceptor usage.
Human immunodeficiency virus (HIV-1) isolates are phylogenetically clustered into distinct groups based on sequence analysis of the viral genome (reviewed in reference 45). These groups have been termed subtypes or clades and given alphabetical designations (e.g., subtype or clade A). Partial-sequence analysis of a portion of a viral genome derived from a tissue isolate taken in 1959 suggests that the different subtypes represent a fairly recent radiation (93), perhaps with each subtype representing the early establishment of independent focal infections. The major HIV-1 subtype found in the United States and western Europe, and the most extensively studied subtype, is subtype B. Subtype C virus is part of an expanding epidemic in sub-Saharan Africa and India and is now the most abundant subtype of HIV-1 worldwide (81).
A dramatic feature of subtype B HIV-1 infection is the de novo evolution of a more pathogenic variant in up to 50% of infected people (6, 10, 12, 20, 68, 70, 77). This variant represents an altered form of the virus whose appearance corresponds to an accelerated decrease in the number of circulating CD4+ T helper cells (42, 69). Typically, transmitted virus uses the CCR5 chemokine receptor as a coreceptor for entry into cells (3, 16, 26, 31, 32) and has also been characterized as nonsyncytium inducing (NSI), slow/low, and macrophage tropic (65, 84, 94); this type of virus has been named R5 (8). The new variant that has evolved from the initial R5 virus is able to use an alternate chemokine receptor, typically CXCR4 (36), and has been characterized as syncytium inducing (SI), rapid/high, and T-cell-line tropic (reviewed in reference 7). Variants that use CXCR4 are now called X4 (8). Primary X4 isolates can be dually tropic for CXCR4 and CCR5 (73).
Sequence alignments of the env gene revealed multiple variable regions (2, 76, 86), which were subsequently named V1 to V5 (57). The major determinant of specificity in coreceptor usage by subtype B isolates is within the V3 loop domain of the viral Env protein (16, 17, 75; reviewed in reference 71). Sequence changes within V3 are largely responsible for determining coreceptor specificity, probably through a direct interaction with surface residues of the chemokine receptor (9, 17, 79, 88).
There are distinctive sequence changes in V3 that are associated with the change in coreceptor usage. These changes include substitutions that result in the presence of increased numbers of basic amino acids at discrete positions within V3 (13, 24, 38, 55, 72). Other changes in the V3 sequence are strongly associated with the presence of the basic amino acid substitutions (56), and some of these changes play a direct role in coreceptor specificity (14). Overall, subtype B V3 sequences that contain basic amino acid substitutions display twice as much amino acid variability from the consensus sequence as do sequences without the basic amino acids (13, 55). This increased variability is in addition to the basic amino acid substitutions. The presence of this additional sequence variability has allowed the heteroduplex tracking assay (HTA) (25) to be used to rapidly identify V3 evolutionary variants that are strongly correlated with the more pathogenic (X4) form of the subtype B HIV-1 (60).
While the evolution of X4 variants among patients infected with subtype B virus is a striking and well-documented phenomenon, its impact on people infected with viruses of other subtypes is not known. Viruses that use the CXCR4 receptor (SI phenotype) have been identified among most of the HIV-1 subtypes (23, 28, 63, 80, 89–92). In cases where the X4 viruses of other subtypes have been examined, these variants encode increased numbers of basic amino acids in their V3 loops (23, 28, 63, 89, 92), suggesting that common mechanisms are determining changes in coreceptor interactions. However, the potential for differences in patterns of coreceptor usage among the different subtypes was suggested by the observation that subtype C SI/X4 variants were rare among a group of 16 people under care in Sweden (80) and among a group of 22 French military personnel infected during overseas deployment (63), although subtype C SI/X4 variants have been observed (78, 80, 91).
Understanding the differences in the evolution of subtype C virus is becoming increasingly important because the dominance of this subtype in the worldwide HIV epidemic will lead to the inevitable expansion of subtype C vaccine development. Because of our ongoing clinical studies in Malawi (18, 33), we had an opportunity to examine this question in a study of a large cohort of men infected primarily with subtype C HIV-1.
We adapted the V3-HTA (60) to detect V3 evolutionary variants of subtype C virus. In a group of plasma samples from 80 HIV-1-infected men from Malawi, we found that 31% of the samples showed evidence of evolutionary variants within the V3 region. However, the presence of these variants was not related to low levels of CD4+ T cells, as is seen in people infected with subtype B HIV-1. Sequence analysis of viral RNA revealed a virtual absence of basic amino acid substitutions in the V3 regions of the evolutionary variants. Virus isolates were established from a subset of the Malawi subjects, and all of them preferentially used the CCR5 receptor for virus entry. Total sequence variability in subtype C V3 sequences was comparable to the reduced variability seen with sequences from subtype B viruses that do not contain basic amino acid substitutions in the V3 loop and was similar in magnitude to the variability in the conserved regions of env. These results indicate that X4 variants are rare among men in sub-Saharan Africa infected with subtype C virus and that V3 variability is a feature of viruses with altered coreceptor usage.
MATERIALS AND METHODS
Source of patient samples.
Plasma and peripheral blood mononuclear cells (PBMC) were collected and processed from subjects in the sexually transmitted disease and dermatology clinics of the Lilongwe Central Hospital in Lilongwe, Malawi, as described previously (18). HIV-1 seropositivity was determined by two enzyme immunoassays (Genetic Systems HIV-1/HIV-2 EIA; Genetics Systems Corp., Redmond, Wash.; and Murex HIV-1+2; Murex Diagnostics Ltd., Dartford, United Kingdom) and Western blot analysis (Organon-Teknika, Durham, N.C.), after which the HIV-1 serotype was determined by an envelope V3 peptide immunoassay with antigens specific for clades A through F (33, 62). Plasma samples from subjects infected with HIV-1 subtype B were chosen from the samples that were collected as part of AIDS Clinical Trial Group Clinical Trial 201 (37) with Institutional Review Board approval. Subtype B samples were chosen from subjects with fewer than 400 CD4+ T cells/μl.
Virus isolation and test for coreceptor usage.
The following reagents were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program: the MT-2 cell line was from Douglas Richman, the U373-MAGI cell lines (85) were from Michael Emerman, the YU-2 molecular clone (51) was from Beatrice Hahn and George Shaw, and the HIV-189.6 viral isolate (19) was from Ronald Collman. Malcolm Martin provided the AD8 (ADA) (15) and NL4-3 molecular clones (1), and Nathaniel Landau provided the HIV-1 clone of HXB with the env gene derived from Ba-L (39). PBMC from infected individuals were cocultured with phytohemagglutinin-stimulated PBMC from uninfected donors in qualitative HIV cultures as described previously (82). Culture supernatants were tested twice a week for p24 antigen production (Organon-Teknika). Virus isolates were assayed in triplicate for syncytium formation in MT-2 cells by using a previously described method (41). Briefly, 50 μl of PBMC coculture (including the cells) was added to 5 × 104 MT-2 cells in 150 μl of medium in a 96-well plate. The cultures were monitored for syncytia twice a week; they were scored positive when there were three to five syncytia per high-power field under light microscopy.
Coreceptor usage was determined by using three U373-MAGI cell lines that express no coreceptor (U373-MAGI), the CCR5 coreceptor (U373-MAGI-CCR5), or the CXCR4 coreceptor (U373-MAGI-CXCR4) (85). The cell lines were maintained and infected as described previously (85). Briefly, each of the cell lines was plated in separate wells of a 48-well plate 1 day prior to infection. An aliquot of 90 μl of diluted viral supernatant from PBMC coculture was added to the cells and adsorbed for 2 h at 37°C under 5% CO2. The subtype C viral isolates were used at a 1:3 dilution in medium or undiluted. Positive control viruses (YU-2, Ba-L, ADA, NL4-3, and 89.6) in infected-cell supernatants were used at a 1:6 dilution. An aliquot of 0.5 ml of culture medium was then added to each well, and the plate was incubated for 40 to 48 h at 37°C under 5% CO2. The medium was then removed, and the cells were fixed for 5 min with 0.5 ml of fixing solution (1% formaldehyde, 0.2% glutaraldehyde) per well. The cells were washed twice with phosphate-buffered saline, 200 μl of staining solution (4 mM potassium ferricyanide, 4 mM potassium ferricyanide, 2 mM MgCl2, and 0.4 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal] per ml in PBS) was added to each well, and the plate was incubated at 37°C for 2 h. The cells were then washed with phosphate-buffered saline twice, and the blue cells were counted.
Viral RNA isolation, RT-PCR, and DNA sequence determination.
Viral RNA was isolated from 140 μl of patient plasma with a QIAamp viral RNA kit (Qiagen); the RNA was eluted with 50 μl of RNase-free water. Primers for reverse transcription-PCR (RT-PCR) were designed to correspond to the HIV-1 subtype C V3 consensus sequence present in the Human Retroviruses and AIDS database (44). The upstream (C+V3) primer was 5′-ATAGTACATCTTAATCAATCTGTAGAAATT-3′, and the downstream (C−V3) primer was 5′-CCATTTATCTTTACTAATGTTACAATGTGC-3′; these primers generate a 159-bp product. RT-PCRs were performed by the method described by Nelson et al. (60) with the following modifications. RT reaction mixtures of 20 μl consisted of 5 μl of the viral RNA eluate, 1× Expand HF buffer (Boehringer Mannheim), 2.5 mM MgCl2, 1 μl of 10 mM deoxynucleoside triphosphate mix (U.S. Biochemical), 10 U of RNase inhibitor (Boehringer Mannheim), 15 pmol of primer C-V3, and 10 U of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim). Reverse transcription was done at 42°C for 30 min, followed by 2 min at 95°C to inactivate the enzyme. A 30-μl aliquot of PCR mix (1× Expand HF buffer, 2.5 mM MgCl2, 15 pmol of primer C+V3, 2 U of Expand High Fidelity enzyme mix [Boehringer Mannheim]) was added to each RT reaction mixture. PCR was carried out in a Stratagene Gradient-40 Robocycler with the following program: one cycle at 95°C for 2 min 45 s; then 40 cycles at 95°C for 45 s, 49°C for 45 s, and 68°C for 1 min (after the first 10 cycles, 1 min was added to the 68°C step for 10 cycles, 2 min was added for the next 10 cycles, and 3 min was added for the last 10 cycles). RT-PCR of subtype B samples was performed as described previously (60). The RT-PCR products were purified by using QIAquick PCR purification columns (Qiagen), and the PCR products were sequenced with an ABI PRISM dye terminator cycle-sequencing kit (Perkin-Elmer). Alternatively, the PCR product was cloned into the pT7Blue(R) vector (Novagen), individual clones were screened by HTA, and examples of each HTA-defined species were sequenced by using an ABI PRISM dye terminator cycle-sequencing kit.
V3-HTA.
Probe construction, probe labeling, and HTA conditions for subtype C samples were adapted from those described by Nelson et al. (60) and Delwart et al. (25). Based on direct sequencing of several RT-PCR products, the product from patient C128 was chosen for probe construction. The C128 RT-PCR product was cloned into the pT7Blue(R) vector. Several clones were sequenced, and a probe plasmid (D516-11) was chosen that had only three nucleotide differences from the subtype C V3 consensus. The subtype C V3 probe was labeled by first digesting 1 μg of D516-11 plasmid with BamHI. The plasmid was end labeled for 15 min at room temperature in a reaction mixture of 50 μl containing 12.5 μCi of 35S-dATP (1,250 Ci/mmol; NEN Life Science Products), unlabeled dGTP at final concentration 1 mM, and 2 U of the Klenow fragment of DNA polymerase I; this was followed by heat inactivation of the enzyme. The plasmid was further digested with SpeI to release the probe from the vector. The labeled probe was purified by using a QIAquick PCR purification column and recovered in a final volume of 50 μl. Heteroduplex formation reactions were done in 10-μl reaction mixtures consisting of 8 μl of purified RT-PCR product, 1 μl of 10× annealing buffer (1 M NaCl, 100 mM Tris-HCl [pH 7.5], 20 mM EDTA), 1 μM primer C+V3, and 0.8 μl of labeled D516-11 probe. The reaction mixtures were denatured at 95°C for 2 min and allowed to anneal at room temperature for 15 min. The heteroduplexes were separated in nondenaturing 12% polyacrylamide gels. V3-HTA of subtype B RT-PCR products was performed as described previously (60).
A unified linear regression model in rank scale was used to assess the relationship between the CD4 cell count and the HTA mobility ratio of the samples with single and multiple bands for both the subtype B and subtype C data sets. When multiple bands were present, the mobility ratio value of the slowest-migrating band was used. Calculations were done with SAS (version 6.12) programs rk0101.sas and rk0102.sas.
Analysis of sequence heterogeneity.
Total amino acid variability within the newly determined V3 sequences was analyzed as follows. The total number of times a nonconsensus sequence amino acid was present was tallied, this number was divided by the total number of V3 sequences times 35 (the number of amino acid positions in V3), and the final number was multiplied by 100 to give the percentage of amino acid substitutions.
For analysis of evolutionary distance, envelope sequences spanning V1 through V5 were obtained from the Los Alamos HIV-1 database (44). A total of 33 subtype C and 69 subtype B sequences were used. Subtype B sequences were classified into two groups, either R5-like or X4-like, based on known biological properties of the viruses or the charge characteristics of amino acids at position 11 or 25 in the V3 loop. Nucleotide sequences from all viruses were aligned by using DNA (Harvard University Molecular Biology Computer Resources). The alignments were optimized manually to ensure that codons remained intact and gaps were minimized (50). Sequences were divided into segments, which ranged in length from 78 to 150 nucleotides or 26 to 50 codons, based on the location of hypervariable and conserved domains. Phylogenetic analysis to assess the relationship among the sequences and between subtype C and subtype B sequences in V3 was performed by using the neighbor-joining method (66) in the PHYLIP package (34, 35). Estimation of total distance in each segment among the three groups of viruses was based on the method of Nei and Gojobori (59) in MEGA (48). Distances within each segment among the three groups of sequences were evaluated by a one-way analysis of variance (SigmaStat; Jandel). A pairwise multiple comparison procedure was used to analyze the significance of relationships between groups (SigmaStat). A P value of <0.05 was considered significant.
Nucleotide sequence accession numbers.
The nucleotide sequences for the V3 region described here have been assigned GenBank accession no. AF153129 to AF153190.
RESULTS
Application of V3-HTA to subtype C virus.
As a starting point to study the nature of V3 sequence variability in subtype C virus, we used a collection of plasma samples taken from 80 separate subjects participating in a clinical trial (18, 33). Each of the samples was taken from an HIV-1-infected subject from Malawi, and the virus from each subject was characterized as subtype C by using a V3 peptide immunoassay (33, 62). The viral RNA level in plasma and CD4+-T-cell count for each of the subjects are presented in Table 1. The range of values for the CD4+-T-cell count was 36 to 1,253 cells/μl, with the median value being 273 cells/μl; the range of values for viral RNA load in plasma was 1.6 × 103 by 5,700 × 103 copies/ml.
TABLE 1.
Subject profiles and V3-HTA results
| Samplea | HIV-1 RNA level (103 copies/ml) | CD4+-T-cell levelb | Mobility ratiosc | Samplea | HIV-1 RNA level (103) | CD4+-T-cell levelb | Mobility ratiosc | |
|---|---|---|---|---|---|---|---|---|
| C128 | 220 | 159 | 0.95 | S067 | 160 | ND | 0.92 | |
| C044 | 150 | 92 | 0.95 | S088 | 130 | 569 | 0.92 | |
| C120* | 1,000 | 268 | 0.95 | S100 | 150 | 210 | 0.92 | |
| S007* | 49 | 273 | 0.95 | S147 | 470 | 188 | 0.92 | |
| S036 | 110 | 631 | 0.95 | S173 | 15 | 36 | 0.92 | |
| S059 | 1,300 | 259 | 0.95 | C054 | 260 | 190 | 0.91 | |
| S080* | 850 | 212 | 0.95 | S005 | 720 | 29% | 0.91 | |
| S103* | 190 | 454 | 0.95 | S071 | 65 | 81 | 0.91 | |
| S111 | 17 | NDd | 0.95 | C087 | 470 | 284 | 0.90 | |
| S115 | 460 | 36 | 0.95 | S081 | 35 | 101 | 0.90 | |
| S116 | 220 | 628 | 0.95 | S171 | 180 | 441 | 0.90 | |
| S200 | 5,700 | 132 | 0.95 | S048 | 58 | 334 | 0.89 | |
| C012* | 380 | 863 | 0.94 | S002 | 10,000 | 41% | 0.88 | |
| C040 | 1.9 | 152 | 0.94 | S051 | 29 | 1,253 | 0.79 | |
| C065 | 260 | 171 | 0.94 | S018* | 580 | 476 | 0.69 | |
| C070 | 270 | 172 | 0.94 | C102 | 87 | 560 | 0.93/0.95 | |
| C096 | 320 | 350 | 0.94 | S083* | 810 | 166 | 0.92/0.95 | |
| C109 | 62 | 328 | 0.94 | S158 | 38 | 348 | 0.92/0.95 | |
| S003* | 240 | 21% | 0.94 | S101 | 610 | 235 | 0.92/0.94 | |
| S017* | 590 | 178 | 0.94 | S122 | ND | 359 | 0.92/0.94 | |
| S021* | 48 | 154 | 0.94 | S094 | 280 | 101 | 0.91/0.95 | |
| S119 | 530 | 364 | 0.94 | S077 | 63 | 153 | 0.90/0.95 | |
| S146 | 380 | 258 | 0.94 | C022 | 210 | 54 | 0.90/0.94 | |
| S148 | 66 | 344 | 0.94 | S176 | ND | 314 | 0.90/0.93 | |
| S166 | 48 | 274 | 0.94 | C007* | 78 | 190 | 0.90/0.92 | |
| S172 | 63 | 344 | 0.94 | S009* | 220 | 122 | 0.89/0.91 | |
| C011* | 540 | 67 | 0.93 | S040 | 17 | 480 | 0.88/0.91 | |
| C019 | 1,000 | 599 | 0.93 | S141 | 18 | 115 | 0.88/0.90 | |
| C047 | 140 | 291 | 0.93 | S191* | 640 | 187 | 0.87/0.91 | |
| C113 | 360 | 279 | 0.93 | C018* | 490 | 116 | 0.87/0.89/0.91 | |
| S047* | 760 | 469 | 0.93 | S134 | 340 | 534 | 0.83/0.87/0.91 | |
| S053 | 630 | 318 | 0.93 | C034 | 3,300 | 268 | 0.81/0.88/0.91 | |
| S070 | 55 | 670 | 0.93 | C030 | 580 | 322 | 0.80/0.95 | |
| S084* | 10 | 248 | 0.93 | S073 | 32 | 275 | 0.78/0.95 | |
| S128 | 140 | 11% | 0.93 | S123 | 590 | 399 | 0.78/0.92/0.94 | |
| S159 | 430 | 98 | 0.93 | C061 | 360 | 262 | 0.78/0.83/0.87 | |
| S174 | 170 | 529 | 0.93 | C111 | 69 | 210 | 0.77/0.79/0.88 | |
| S194 | 130 | 322 | 0.93 | S206 | 350 | 168 | 0.76/0.90/0.92 | |
| C045 | 710 | 479 | 0.92 | S180 | 170 | 37 | 0.74/0.93 | |
| S032 | 1.6 | 881 | 0.92 | S031* | 43 | 333 | 0.62/0.73/0.74/0.84 |
Samples for which a virus isolate was recovered are indicated by asterisks.
CD4+-T-cell count per microliter or percentage of CD4+ T cells among all T cells.
Mobility ratios are shown for all heteroduplexes in each sample.
ND, not determined.
Subtype C-specific primers were designed based on conserved regions of subtype C virus sequences in the Human Retroviruses and AIDS database (44) to amplify a 159-bp region of the env gene encompassing V3. Initially, RT-PCR products amplified from viral RNA in plasma from 20 subjects were analyzed by direct sequencing. Samples that gave largely unambiguous sequencing results (implying an absence of a significant mixture of V3 sequences) were used to construct a consensus V3 sequence for subtype C.
A clone from the RT-PCR product derived from the plasma of subject C128 was chosen for the V3-HTA probe because it was the closest to the consensus sequence; it varies from the consensus sequence at only three widely spaced positions, and all three of the differences are G-to-A transitions. The probe was labeled by first cleaving the V3 insert at one junction with the plasmid by using a unique restriction enzyme site in the plasmid. The overhanging end was filled in with a radioactive nucleotide to label one strand of the probe, allowing detection of heteroduplexes formed with only one of the probe strands. The probe insert was then released from the plasmid by cleavage at a second unique cleavage site at the other insert/plasmid junction. The inclusion of several flanking nucleotides from the plasmid in the probe results in distinct migrations for the reannealed probe strands versus the heteroduplexes formed between the labeled probe strand and complementary RT-PCR products (60).
Characterization of viral RNA in plasma by V3-HTA.
V3-HTA is able to identify many sequence variants within the region of env encoding V3 (60). The magnitude of sequence evolution that leads to a change in coreceptor usage is, in most cases, significant enough to score in the V3-HTA. Such sequence variants cause the heteroduplex formed between the PCR product and the probe to bend and therefore to be retarded during polyacrylamide gel electrophoresis. As a first step in characterizing subtype C V3 sequence variability, we applied this assay to the RT-PCR product from each of the 80 plasma samples. Such an analysis provides two pieces of information: first, the presence of variants with sequences distinct from the probe can be identified; second, the presence of mixtures of cocirculating viral sequences is revealed. Examples of V3-HTA patterns from subjects with homogeneous V3 sequences (single band) or heterogeneous V3 sequences (multiple bands) are shown in Fig. 1. A summary of the V3-HTA results for the plasma samples is shown in Table 1.
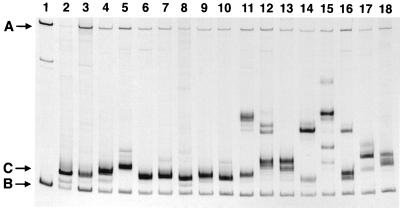
FIG. 1.
Examples of V3-HTA patterns obtained with the subtype C probe. RT-PCR products were generated from viral RNA isolated from the plasma of subjects infected with HIV-1 subtype C. Lanes: 1, probe without PCR product; 2, probe with PCR product generated from the D516-11 probe plasmid; 3 to 18, examples of RT-PCR products from subjects with either a single dominant heteroduplex band or multiple heteroduplex bands. Starting with lane 3, the order is as follows: S115, C011, S081, C044, S071, S200, S021, C065, S180, C111, C034, S073, S031, S123, S134, and S191. The positions in the gel of the single-stranded probe (A), probe homoduplex (B), and heteroduplexes with the most rapid migration (C) are shown.
Of the 80 plasma samples, 55 had single bands in the V3-HTA analysis, indicating that the virus populations in these samples were homogeneous within V3. The remaining 25 samples had multiple bands in the V3-HTA analysis, revealing the presence of multiple virus species with different V3 sequences. Of the samples with single bands, 53 had bands that migrated near the bottom of the gel, indicating similarity to the consensus sequence. The other two samples (S018 and S051) had single bands that migrated more slowly, indicative of significant sequence differences with respect to the probe (Table 1). Sequence analysis of the bulk PCR product was done for 46 of the samples that gave a single band in V3-HTA and for 7 samples that gave multiple bands but for which there was a predominant band with rapid mobility and unambiguous sequence. In addition, seven samples that gave multiple bands were subjected to molecular cloning, and individual clones were screened by V3-HTA and sequenced. In total, sequence information was determined for 62 of the 80 samples.
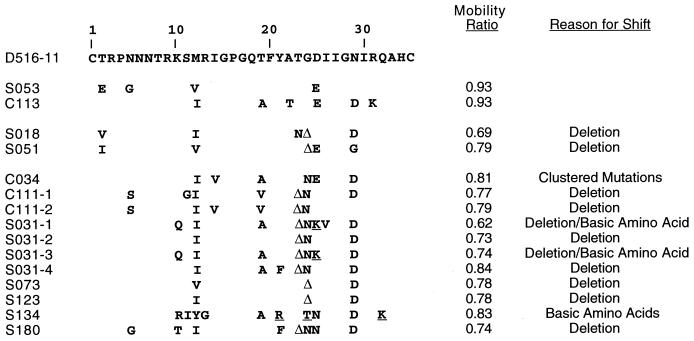
We previously found a strong correlation between subtype B HIV-1 samples that had slowly migrating bands in the V3-HTA analysis, the presence of sequence changes that would encode basic amino acid substitutions in the V3 loop, and the presence of SI viruses in the virus isolate (60). Therefore, we were interested in determining the nature of the sequence changes in the subtype C isolates that resulted in slow migration in the V3-HTA analysis. A summary of the amino acid substitutions encoded by the nucleotide changes for the shifted bands representing viruses from nine patients is shown in Fig. 2. This includes the two samples that had a single band with low mobility (S018 and S051) and seven of the samples with multiple bands. Bands were considered “shifted up” if the mobility ratio (the distance migrated by the heteroduplex divided by the distance migrated by the probe homoduplex) was less than 0.85, and by this definition only 10 of the samples with multiple bands included bands that were considered significantly shifted. Seven of the nine sequenced examples of shifted bands were due to three nucleotide deletions, usually corresponding to position 24 or 25 of V3. One of the sequences with a deletion (S031) and one other sequence (S134) had basic amino acid substitutions that are associated with the SI/X4 phenotype in subtype B viruses. The ninth sequence, C034, shifted because of clustered mutations away from the consensus, but these did not include basic amino acid substitutions. These results suggest that clustered mutations that include basic amino acid substitutions, which are characteristic features of subtype B X4 variants, are not a prominent feature of V3 sequence variability among subtype C sequences.
FIG. 2.
Alignment of inferred V3 amino acid sequences corresponding to V3-HTA bands with mobility ratios of less than 0.85. Each sequence represents a single clone from the RT-PCR product from that subject. The mobility of each clone was verified by V3-HTA. Basic amino acid substitutions at SI-associated positions are underlined; deletions are designated by Δ.
Comparison of evolutionary variants in subtype B and subtype C.
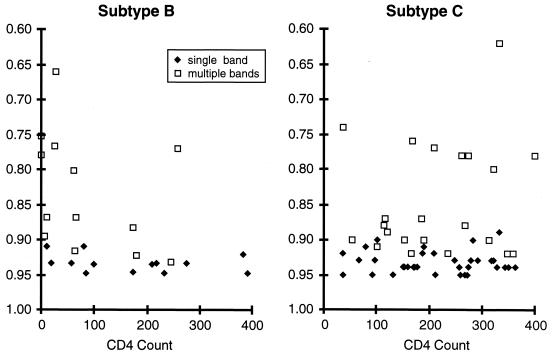
X4 viruses typically evolve late in the course of an HIV-1 infection, usually when the levels of CD4+ T helper cells fall below 400/μl (42). We carried out an analysis of the appearance of evolutionary variants among subtype B versus subtype C HIV-1 as a function of the patient CD4+-T-helper-cell level (Fig. 3). Samples were obtained from subjects infected with subtype B virus who were participants in an ACTG study. As expected, among the subtype B viruses, V3-HTA (with the subtype B probe) revealed slowly migrating bands and/or multiple bands with increasing frequency as patient CD4+-T-helper-cell levels declined. In contrast, the presence of slowly migrating bands derived from the subtype C viruses (detected with the subtype C probe) showed no relationship with the patient CD4+-T-helper-cell level. The association was tested by using a unified linear-regression model in rank scale (n = 83). The subtype B samples with multiple bands showed an association between decreasing mobility ratio and decreasing CD4 counts that approached statistical significance even with this small sample size (slope = 0.462; P = 0.06). This was not the case for the subtype B samples with single bands (slope = 0.158; P = 0.35) or the subtype C samples with either multiple bands (slope = 0.011; P = 0.94) or single bands (slope = 0.006; P = 0.96). Thus, there appears to be a difference between these two subtypes in the nature of V3 sequence variants as detected by V3-HTA. The difference was tested statistically by a comparison of slopes (0.426 versus 0.011; P = 0.13). A plausible explanation for the lack of statistical significance is the small sample size for subtype B (n = 26). The power of the test comparing slopes was approximately 0.324, or 32.4%.
FIG. 3.
V3-HTA mobility ratios plotted against the CD4+-T-cell count. Only the lowest mobility ratio was used for samples with multiple bands. The data for the subtype C viruses were taken from Table 1. The data from the subtype B viruses were generated from samples described in Materials and Methods.
Coreceptor usage of subtype C viruses.
Both the near absence of basic amino acid substitutions in samples scoring in the V3-HTA analysis and the lack of correlation between V3 sequence variants and CD4+-T-helper-cell levels suggested that subtype C viruses do not evolve to utilize different coreceptors in a manner analogous to that seen with subtype B viruses. To determine the nature of coreceptor usage among these viruses, PBMC corresponding to 79 of the 80 plasma samples were used in coculture assays to recover virus. Virus isolates were obtained from only 19 of the 79 samples. Fourteen of these isolates were from patients with less than 400 CD4+ T cells/μl, the point at which X4 viruses start appearing in people infected with subtype B viruses (42). The isolates were tested for coreceptor usage in U373-MAGI cell lines (85) and for syncytium induction in MT-2 cells, a frequent feature of subtype B X4 variants (43). As shown in Table 2, all of the isolates used CCR5 as a coreceptor, did not use CXCR4, and did not induce syncytia in MT-2 cells. An additional 12 subtype C isolates that were not associated with the 80 plasma samples were tested in the MT-2 assay, and all were NSI (data not shown), although this additional group has not been subjected to sequence, V3-HTA, or coreceptor usage analysis. To date we have not found a single example of CXCR4 coreceptor usage or SI phenotype among the 31 subtype C virus isolates in our collection.
TABLE 2.
Coreceptor usage determined by a U373-MAGI assay of viral isolates
| Isolate | No. of blue cells wella
|
CCR5/CXCR4 ratio | Coreceptor preference | Syncytia in MT-2 cells | ||
|---|---|---|---|---|---|---|
| U | U-CCR5 | U-CXCR4 | ||||
| YU2 | 2 | 132 | 10 | 13 | CCR5 | NDb |
| BAL | 3 | 301 | 53 | 5.7 | CCR5 | ND |
| AD8 | 1 | 153 | 15 | 10 | CCR5 | ND |
| 89.6 | 2 | 934 | 1,482 | 0.6 | CCR5/CXCR4 | ND |
| NL43 | 2 | 1 | 598 | 0.0017 | CXCR4 | + |
| C007 | 1 | 58 | 2 | 29 | CCR5 | − |
| C011 | 1 | 69 | 8 | 8.6 | CCR5 | − |
| C012 | 0 | 227 | 10 | 23 | CCR5 | − |
| C018 | 3 | 569 | 21 | 27 | CCR5 | − |
| C120 | 2 | 66 | 5 | 13 | CCR5 | − |
| S003 | 2 | 592 | 23 | 26 | CCR5 | − |
| S007 | 2 | 147 | 4 | 37 | CCR5 | − |
| S009 | 6 | 364 | 22 | 17 | CCR5 | − |
| S017 | 4 | 143 | 15 | 9.5 | CCR5 | − |
| S018 | 6 | 83 | 9 | 9.2 | CCR5 | − |
| S021 | 10 | 67 | 8 | 8.4 | CCR5 | − |
| S031 | 6 | 95 | 5 | 19 | CCR5 | − |
| S047 | 4 | 24 | 2 | 12 | CCR5 | − |
| S080 | 16 | 67 | 16 | 4.2 | CCR5 | − |
| S083 | 1 | 26 | 2 | 13 | CCR5 | − |
| S084 | 8 | 524 | 31 | 17 | CCR5 | − |
| S103 | 6 | 213 | 34 | 6.3 | CCR5 | − |
| S180 | 1 | 76 | 9 | 8.4 | CCR5 | − |
| S191 | 3 | 23 | 3 | 7.7 | CCR5 | − |
Assays were done in duplicate; the value given is the average from two wells. U, U373-MAGI; U-CCR5, U373-MAGI-CCR5; U-CXCR4, U373-MAGI-CXCR4.
ND, not determined.
Analysis of V3 sequence variability in subtype C HIV-1.
We combined the sequence data from the 62 samples analyzed in this study with 64 sequences available for subtype C viruses in the Human Retroviruses and AIDS database (44) to create a data set that would allow an estimate of total sequence variability within V3. Five subtype C sequences that had deletions were removed from the data set and not included in this analysis, leaving 121 sequences. Each deletion was of 3 nucleotides, i.e., one codon, and appeared at position 24 or 25, suggesting that these are positions that can accommodate a deletion. These deletions do not appear to be involved in altered coreceptor usage, since three of the viruses with these deletions were included in the analysis of coreceptor usage and all were specific for CCR5 (Fig. 2, Table 2).
The pattern of sequence variability for subtype C, as shown in Fig. 4, was compared to the patterns of sequence variability seen in the two sets of subtype B viruses, those without basic amino acid substitutions (NSI/R5-like) and those with basic amino acid substitutions (SI/X4-like) (56). We previously noted that NSI/R5-like sequences, presumed to represent R5 variants, have half as much sequence variability as do SI/X4-like sequences, presumed to represent X4 variants (56), and this pattern can be seen in Fig. 4. Total sequence variability among the subtype C viruses was nearly identical to that of the NSI/R5-like V3 loop sequences of the subtype B viruses and was approximately one-half of the variability of the subtype B viruses with SI/X4-like V3 loop sequences. Also, there was a virtual absence of nonconservative basic amino acid substitutions among subtype C sequences, with single examples at positions 11 and 25, the two positions most commonly substituted with a basic amino acid among subtype B sequences.
FIG. 4.
Sequence variability of subtype C V3 sequences compared to the variability of NSI/R5-like and SI/X4-like subtype B V3 sequences. The consensus sequence for subtype C was generated from a data set of 121 sequences. The NSI/R5-like and SI/X4-like variability patterns are from reference (56). Substitution percentages were calculated by dividing the sum of the substitutions away from the consensus by the total number of amino acids.
There are five amino acid positions within V3 where the subtype C consensus sequence differs substantially from the subtype B consensus sequence: positions 13, 18, 19, 22, and 25. At position 13, a basic amino acid appears in the subtype C consensus sequence while a basic amino acid is characteristic of the X4 variants among subtype B viruses (55). At position 19, threonine is the consensus amino acid in the subtype C virus, with alanine being a common substitution, while alanine is the consensus amino acid in the subtype B viruses. Similarly, at position 22, alanine is the consensus amino acid in the subtype C viruses while threonine is the consensus amino acid in the subtype B viruses, with alanine being a frequent substitution. In these last two cases, a pair of amino acids is allowed in one subtype while one of these is essentially fixed in the other subtype. Although the consensus amino acid at position 25 is different in these two subtypes, in both cases it is an acidic amino acid with both acidic amino acids being frequently represented in both viruses. Finally, the glutamine at position 18 represents the common amino acid at this position outside of the subtype B viruses (44).
Is V3 really variable?
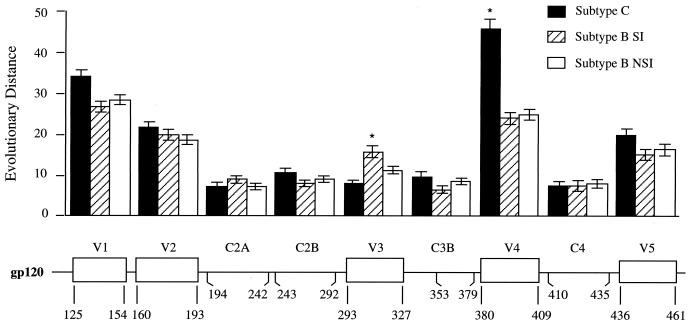
The original description of five variable regions flanked by conserved regions in the env gene was based on sequence comparisons of subtype B viruses that included R5-like and X4-like sequences (57). We considered the possibility that the excess variability in V3 is either the result of or associated with change in coreceptor usage. To determine the level of V3 variability in the absence of altered coreceptor use, we compared a collection of subtype C env sequences to a group of subtype B env sequences that were sorted into R5-like and X4-like groups by using a conservative criterion based on the presence or absence of a basic amino acid substitution at either position 11 or position 25. Between 30 and 39 env sequences spanning V1 through V5 were available for each group from the Los Alamos HIV-1 sequence database (44). The length of each sequence was divided into segments of 26 to 50 codons, representing discrete variable and conserved regions, and the evolutionary distance for each segment was determined. The distances between the groups of viral sequences within each segment and between segments were compared. The results are shown in Fig. 5. As expected, the V3 region of the X4-like subtype B sequences is more variable than the V3 region of the subtype B R5-like sequences (P < 0.01). The V3 region of subtype C sequences is similar in its variability to the R5-like sequences of subtype B and less variable than the subtype B X4-like sequences (P < 0.05). Finally, while the V3 domain of the subtype B X4-like sequences is more variable than the flanking conserved domains (P < 0.01), the V3 regions of both the subtype B R5-like sequences and the subtype C V3 sequences are no more variable than the flanking conserved regions. Thus, in the absence of sequence changes associated with change in coreceptor use (i.e., X4 viruses), the V3 domain is no more variable than the conserved regions of env.
FIG. 5.
Evolutionary distances of segments of gp120. Groups of sequences of the gp120 coding region for subtype C, subtype B R5-like, and subtype B X4-like viruses were assembled from the Los Alamos HIV-1 Sequence Database. A description of the sequences used is available on request. The sequences were aligned and then divided into segments of between 26 and 50 codons. These segments are indicated in the figure below the line, using the HIV-1JR-FL numbering. The region in C3 just downstream of V3 was omitted because of its recently described position as an external loop (loop E) in the protein structure with associated sequence variability (49). Evolutionary distance was calculated for each segment for each group of sequences. The distance of each segment within a group was compared to the equivalent distance in the other two groups to determine if they were significantly different. The two cases where a statistically significant difference was observed are noted with an asterisk. The vertical thin lines show standard error.
We extended the analysis of the extent of sequence variability between the groups of sequences to the other regions of env. With the exception of V4, each group of viral sequences (subtype C, subtype B X4-like, and subtype B R5-like) showed comparable levels of variability within each segment that was compared, including both the variable and conserved regions. However, within the V4 region, the subtype C sequences were significantly more variable than the comparable sequences from subtype B. A previous analysis of small regions immediately flanking V3 also found comparable levels of sequence variability among different subtypes (29). Thus, it is not the case that overall subtype C env genes are less divergent but, rather, that V3 is not more divergent.
DISCUSSION
In up to 50% of people infected with subtype B HIV-1, a distinctive variant of the virus evolves with the ability to use an alternative coreceptor. We have used five criteria to evaluate the presence of equivalent variants among subtype C HIV-1. Based on the results of experiments examining all five criteria, we conclude that X4 variants of HIV-1 are rare among subtype C viruses, in contrast to their frequent appearance among subtype B viruses. First, basic amino acid substitutions were rare among a large collection of V3 sequences from subtype C viruses (Fig. 2 and 4). Such substitutions are the hallmark of X4 variants among subtype B viruses (13, 24, 38, 55, 72). Second, total sequence variability among subtype C V3 sequences was low, comparable to the V3 variability of sequences representative of subtype B R5 variants and lower than the V3 variability of sequences representative of X4 variants (Fig. 4 and 5). The absence of the variability in subtype C sequences that would be associated with X4 variants probably explains, at least in part, the previous observation that subtype C V3 sequences are more highly conserved than in other subtypes (29, 46). Third, there was a lack of correlation between the appearance of multiple V3 sequence variants and more extensive V3 sequence evolution as a function of decreasing CD4+-T-helper-cell levels (Fig. 3). Such a correlation is seen with subtype B viruses (Fig. 3), and the appearance of multiple divergent V3 species is correlated with the presence of SI variants (60). Fourth, when a subset (19 of 80) of the subtype C viruses were isolated by culture of primary PBMC, none of the viral isolates was able to form syncytia in MT-2 cells (Table 2); replication in transformed T-cell lines is another feature of SI/X4 variants (43). Fifth, a direct test of coreceptor usage demonstrated that these isolates preferentially used CCR5 and had not evolved to use CXCR4 for entry (Table 2). Thus, by all five criteria, subtype C viruses appear to be predominantly R5 variants of HIV-1 with, by comparison to subtype B HIV-1, a significant underrepresentation of X4 variants. X4 variants among different subtypes appear to select basic amino acid substitutions in V3 as a generalizable strategy for evolving to use CXCR4 as a coreceptor (23, 28, 63, 89, 92). Why, then, is evolution to use CXCR4 not a prominent feature of infection with subtype C HIV-1?
There is as yet too little information to answer this question, but there is a range of possibilities that can be considered. Although subtype C viruses can evolve to use CXCR4, perhaps the ability of a subtype C Env protein to accommodate the required amino acid changes is lower than that of a subtype B Env protein. Given the ability of HIV-1 to evolve, this possibility seems unlikely but remains unproved. The detection of at least some SI/X4 variants with a subtype C Env (78, 80, 91) demonstrates that there is no absolute block to the evolution of these variants. Another possibility is that subtype C viruses have not had sufficient time to evolve into X4 variants. HIV-1-infected people in developing countries can go through a more rapid disease course (5). X4 variants generally appear after a number of years of infection with a subtype B virus, typically when levels of CD4+ T helper cells are below 400/μl. Therefore, a shorter disease course in people living in developing countries may preclude X4 variants from appearing. A corollary of this scenario is the possibility that the evolution of X4 variants is comparable to the appearance of an opportunistic infection. However, if one succumbs to an initial opportunistic infection, subsequent opportunistic infections are precluded. The lack of linkage between low CD4+-T-helper-cell levels and the presence of V3 sequences characteristic of X4 variants suggests that it is not the absence of a longer disease course that precludes the appearance of subtype C X4 variants. One limitation of our study is that our subject population consisted of relatively healthy persons, although they did have a wide range of CD4+-T-helper-cell levels (Table 1). Also, it is clear that other subtypes, for example subtype D, are readily evolving SI/X4 variants in a similar population (reviewed in reference 29).
Several genetic polymorphisms affect the disease course and the evolution of SI/X4 virus in people infected with HIV-1. A deletion in the CCR5 gene (22, 27, 40, 52, 54, 67) and a genetic polymorphism in the CCR2 gene (4, 47, 64, 74, 83) have been associated with slowed disease progression in infected people, although it has been suggested that the CCR2 effect occurs predominantly in people of African descent (58). The absence of these allelic variants in a given population might increase the apparent rate of disease progression and perhaps influence the frequency of SI/X4 evolution. The CCR5 mutation is rare among African-American and African populations (22, 52, 53, 67), but the CCR2 polymorphism is common (4, 74), and its presence contrasts with the more rapid disease progression that is a feature of HIV-1 infection in sub-Saharan Africa (5). These mutations also select for an increased incidence of SI/X4 variants (21, 83), again in contrast to what is observed in the Malawi cohort. An allelic variant of the SDF-1 gene has been reported to reduce HIV-1 disease progression, perhaps through elevated levels of SDF-1 that protect against SI/X4 variants, but this allele is not common, at least among African-Americans (87), although the opposite effect has also been reported (58). Thus, the near absence of SI/X4 variants among the cohort of men in Malawi infected with subtype C HIV-1 cannot easily be ascribed to known genetic variation in the host population.
The reduced variability of R5-like viral sequences prompted the question whether this represented significant variability within the env gene (Fig. 5). We found that V3 in the absence of the sequence variability associated with changes in coreceptor use is no more variable than the conserved regions of gp120. V3 variability among X4-like viral sequences is about twice that among R5 sequences (13, 55). Half of this variability is linked to the presence of basic amino acid substitutions, and another quarter of the variability is represented by another set of substitutions that are biased in their presence in X4-like viral sequences (56). It is not clear what role this additional variability plays in the biology of the subtype B Env protein, i.e., whether it plays a direct role in altered coreceptor usage or whether this variability accumulates for other as yet unknown reasons that are linked to altered coreceptor usage. Similarly, simian immunodeficiency virus isolates most often use CCR5 (reviewed in reference 30), and where it has been examined, V3 is not variable (11, 61).
The lack of coreceptor switching in subtype C virus may affect transmission. Virus that is transmitted is almost always CCR5 dependent, regardless of the variants present in the transmitting donor (65, 84, 94). If the subtype C virus continues to use the CCR5 coreceptor throughout infection, persons carrying the subtype C virus may be more infectious throughout their entire infection than persons carrying subtype B virus who evolve X4 variants.
ACKNOWLEDGMENTS
We thank Malcolm Martin for molecular clones of the AD8 and NL4-3 HIV-1 genomes and Nathaniel Landau for the molecular clone of the Ba-L HIV-1 genome.
P.V. is supported by the Swiss National Science Foundation (3233-48902.96). In addition, this work was supported by the following grants from the National Institutes of Health: R01-AI44667 (to R.S.), R01-DK381 (to M.S.C.), R01-HD32259 (to M.M.G.), the UNC Center for STD Research (U01-AI31496), and the UNC Center For AIDS Research (P30-HD37260), including the help of Rakhi Kilaru and Paul Stewart.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alizon M, Wain-Hobson S, Montagnier L, Sonigo P. Genetic variability of the AIDS virus: nucleotide sequence analysis of two isolates from African patients. Cell. 1986;46:63–74. doi: 10.1016/0092-8674(86)90860-3. [DOI] [PubMed] [Google Scholar]
- 3.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1a, MIP-1b receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 4.Anzala A O, Ball T B, Rostron T, O’Brien S J, Plummer F A, Rowland-Jones S L University of Nairobi Collaboration for HIV Research. CCR2-64I allele and genotype association with delayed AIDS progression in African women. Lancet. 1998;351:1632–1633. doi: 10.1016/s0140-6736(05)77688-1. [DOI] [PubMed] [Google Scholar]
- 5.Anzala O A, Nagelkerke N J, Bwayo J J, Holton D, Moses S, Ngugi E N, Ndinya-Achola J O, Plummer F A. Rapid progression to disease in African sex workers with human immunodeficiency virus type 1 infection. J Infect Dis. 1995;171:686–689. doi: 10.1093/infdis/171.3.686. [DOI] [PubMed] [Google Scholar]
- 6.Asjo B, Morfeldt-Manson L, Albert J, Biberfeld G, Karlsson A, Lidman K, Fenyo E M. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;ii:660–662. [PubMed] [Google Scholar]
- 7.Berger E A. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11:S3–S16. [PubMed] [Google Scholar]
- 8.Berger E A, Doms R W, Fenyo E M, Korber B T, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 9.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S G, Caron M G, Cullen B R. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjorndal A, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyo E-M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns D P W, Desrosiers R C. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J Virol. 1991;65:1843–1854. doi: 10.1128/jvi.65.4.1843-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng-Mayer C, Seto D, Tateno M, Levy J A. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 13.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chesebro B, Wehrly K, Nishio J, Perryman S. Mapping of independent V3 envelope determinants of human immunodeficiency virus type 1 macrophage tropism and syncytium formation in lymphocytes. J Virol. 1996;70:9055–9059. doi: 10.1128/jvi.70.12.9055-9059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 17.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 18.Cohen M S, Hoffman I F, Royce R A, Kazembe P, Dyer J R, Daly C C, Zimba D, Vernazza P L, Maida M, Fiscus S A, Eron J J, Jr AIDSCAP Malawi Research Group. Reduction of concentration of HIV-1 in semen after treatment of urethritis: implications for prevention of sexual transmission of HIV-1. Lancet. 1997;349:1868–1873. doi: 10.1016/s0140-6736(97)02190-9. [DOI] [PubMed] [Google Scholar]
- 19.Collman R, Balliet J W, Gregory S A, Friedman H, Kolson D L, Nathanson N, Srinivasan A. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66:7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Aquila R T, Sutton L, Savara A, Hughes M D, Johnson V A NIAID AIDS Clinical Trials Group Protocol 241 Virology Team. CCR5/delta(ccr5) heterozygosity: a selective pressure for the syncytium-inducing human immunodeficiency virus type 1 phenotype. J Infect Dis. 1998;177:1549–1553. doi: 10.1086/515307. [DOI] [PubMed] [Google Scholar]
- 22.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S J. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 23.de Jong J, Simon F, van der Groen G, Baan E, Saragosti S, Brun-Vezinet F, Goudsmit J. V3 loop sequence analysis of seven HIV type 1 group O isolates phenotyped in peripheral blood mononuclear cells and MT-2 cells. AIDS Res Hum Retroviruses. 1996;12:1503–1507. doi: 10.1089/aid.1996.12.1503. [DOI] [PubMed] [Google Scholar]
- 24.de Jong J J, de Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 26.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major coreceptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 27.de Roda Husman A-M, Koot M, Cornelissen M, Keet I P, Brouwer M, Broersen S M, Bakker M, Roos M T, Prins M, De Wolf F, Coutinho R A, Miedema F, Goudsmit J, Schuitemaker H. Association between CCR5 genotype and the clinical course of HIV-1 infection. Ann Intern Med. 1997;127:882–890. doi: 10.7326/0003-4819-127-10-199711150-00004. [DOI] [PubMed] [Google Scholar]
- 28.De Wolf F, Hogervorst E, Goudsmit J, Fenyo E-M, Rubsamen-Waigmann H, Holmes H, Galvao-Castro B, Karita E, Wasi C, Sempala S D, Baan E, Zorgdrager F, Lukashov V, Osmanov S, Kuiken C, Cornelissen M the WHO Network For HIV Isolation and Characterization. Syncytium-inducing and non-syncytium-inducing capacity of human immunodeficiency virus type 1 subtypes other than B: phenotypic and genotypic characteristics. AIDS Res Hum Retroviruses. 1994;10:1387–1400. doi: 10.1089/aid.1994.10.1387. [DOI] [PubMed] [Google Scholar]
- 29.Dighe P K, Korber B T, Foley B T. Global variation in the HIV-1 V3 region. Human retroviruses and AIDS 1997: a compilation and analysis of nucleic acid and amino acid sequences, p. III–75 to III–207. Los Alamos, N.M: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1997. [Google Scholar]
- 30.Doms R W, Edinger A L, Moore J P. Coreceptor use by primate lentiviruses. Human retroviruses and AIDS 1998: a compilation and analysis of nucleic acid and amino acid sequences, p. III-20–III-35. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1998. [Google Scholar]
- 31.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 32.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 33.Dyer J R, Kazembe P, Vernazza P L, Gilliam B L, Maida M, Zimba D, Hoffman I F, Royce R A, Schock J L, Fiscus S A, Cohen M S, Eron J J., Jr High levels of human immunodeficiency virus type 1 in blood and semen of seropositive men in sub-Saharan Africa. J Infect Dis. 1998;177:1742–1746. doi: 10.1086/517436. [DOI] [PubMed] [Google Scholar]
- 34.Felsenstein J. PHYLIP version 3.5c. Seattle: University of Washington; 1993. [Google Scholar]
- 35.Felsenstein J. PHYLIP—phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 36.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 37.Fiscus S A, Heggem-Snow A, Troiani L, Wallmark E, Folds J D, Sheff B, van der Horst C M. Transient high titers of HIV-1 in plasma and progression of disease. J Acquired Immune Defic Syndr. 1995;9:51–57. [PubMed] [Google Scholar]
- 38.Fouchier R A, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gartner S, Markovits P, Markovitz D M, Kaplan M H, Gallo R C, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 40.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 41.Japour A J, Fiscus S A, Arduino J M, Mayers D L, Reichelderfer P S, Kuritzkes D R. Standardized microtiter assay for determination of syncytium-inducing phenotypes of clinical human immunodeficiency virus type 1 isolates. J Clin Microbiol. 1994;32:2291–2294. doi: 10.1128/jcm.32.9.2291-2294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koot M, Keet I P, Vos A H, de Goede R E, Roos M T, Coutinho R A, Miedema F, Schellekens P T, Tersmette M. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 43.Koot M, Vos A H, Keet R P, de Goede R E, Dercksen M W, Terpstra F G, Coutinho R A, Miedema F, Tersmette M. HIV-1 biological phenotype in long-term infected individuals evaluated with an MT-2 cocultivation assay. AIDS. 1992;6:49–54. doi: 10.1097/00002030-199201000-00006. [DOI] [PubMed] [Google Scholar]
- 44.Korber B, Hahn B, Foley B, Mellors J W, Leitner T, Myers G, McCutchan F, Kuiken C L. Human retroviruses and AIDS 1997: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.M: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1997. [Google Scholar]
- 45.Korber B T, Allen E E, Farmer A D, Myers G L. Heterogeneity of HIV-1 and HIV-2. AIDS. 1995;9:S5–S18. [PubMed] [Google Scholar]
- 46.Korber B T, MacInnes K, Smith R F, Myers G. Mutational trends in V3 loop protein sequences observed in different genetic lineages of human immunodeficiency virus type 1. J Virol. 1994;68:6730–6744. doi: 10.1128/jvi.68.10.6730-6744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kostrikis L G, Huang Y, Moore J P, Wolinsky S M, Zhang L, Guo Y, Deutsch L, Phair J, Neumann A U, Ho D D. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat Med. 1998;4:350–353. doi: 10.1038/nm0398-350. [DOI] [PubMed] [Google Scholar]
- 48.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. CABIOS. 1994;10:189–192. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 49.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamers S L, Sleasman J W, Goodenow M M. A model for alignment of Env V1 and V2 hypervariable domains from human and simian immunodeficiency viruses. AIDS Res Hum Retroviruses. 1996;12:1169–1178. doi: 10.1089/aid.1996.12.1169. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Hui H, Burgess C J, Price R W, Sharp P M, Hahn B H, Shaw G M. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J Virol. 1992;66:6587–6600. doi: 10.1128/jvi.66.11.6587-6600.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 53.Martinson J J, Chapman N H, Rees D C, Liu Y T, Clegg J B. Global distribution of the CCR5 gene 32-basepair deletion. Nat Genet. 1997;16:100–103. doi: 10.1038/ng0597-100. [DOI] [PubMed] [Google Scholar]
- 54.Michael N L, Chang G, Louie L G, Mascola J R, Dondero D, Birx D L, Sheppard H W. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med. 1997;3:338–340. doi: 10.1038/nm0397-338. [DOI] [PubMed] [Google Scholar]
- 55.Milich L, Margolin B, Swanstrom R. V3 loop of the human immunodeficiency virus type 1 Env protein: interpreting sequence variability. J Virol. 1993;67:5623–5634. doi: 10.1128/jvi.67.9.5623-5634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milich L, Margolin B H, Swanstrom R. Patterns of amino acid variability in NSI-like and SI-like V3 sequences and a linked change in the CD4-binding domain of the HIV-1 Env protein. Virology. 1997;239:108–118. doi: 10.1006/viro.1997.8821. [DOI] [PubMed] [Google Scholar]
- 57.Modrow S, Hahn B H, Shaw G M, Gallo R C, Wong-Staal F, Wolf H. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J Virol. 1987;61:570–578. doi: 10.1128/jvi.61.2.570-578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mummidi S, Ahuja S S, Gonzalez E, Anderson S A, Santiago E N, Stephan K T, Craig F E, O’Connell P, Tryon V, Clark R A, Dolan M J, Ahuja S K. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med. 1998;4:786–793. doi: 10.1038/nm0798-786. [DOI] [PubMed] [Google Scholar]
- 59.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 60.Nelson J A E, Fiscus S A, Swanstrom R. Evolutionary variants of the human immunodeficiency virus type 1 V3 region characterized by using a heteroduplex tracking assay. J Virol. 1997;71:8750–8758. doi: 10.1128/jvi.71.11.8750-8758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Overbaugh J, Rudensey L M, Papenhausen M D, Benveniste R E, Morton W R. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J Virol. 1991;65:7025–7031. doi: 10.1128/jvi.65.12.7025-7031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pau C P, Kai M, Holloman-Candal D L, Luo C C, Kalish M L, Schochetman G, Byers B, George J R WHO Network for HIV Isolation and Characterization. Antigenic variation and serotyping of HIV type 1 from four World Health Organization-sponsored HIV vaccine sites. AIDS Res Hum Retroviruses. 1994;10:1369–1377. doi: 10.1089/aid.1994.10.1369. [DOI] [PubMed] [Google Scholar]
- 63.Peeters M, Vincent R, Perret J-L, Lasky M, Patrel D, Liegeois F, Courgnaud V, Seng R, Matton T, Molinier S, Delaporte E. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J Acquired Immune Defic Syndr. 1999;20:115–121. doi: 10.1097/00042560-199902010-00002. [DOI] [PubMed] [Google Scholar]
- 64.Rizzardi G P, Morawetz R A, Vicenzi E, Ghezzi S, Poli G, Lazzarin A, Pantaleo G Swiss HIV Cohort. CCR2 polymorphism and HIV disease. Nat Med. 1998;4:252–253. doi: 10.1038/nm0398-252. [DOI] [PubMed] [Google Scholar]
- 65.Roos M T, Lange J M, de Goede R E, Coutinho R A, Schellekens P T, Miedema F, Tersmette M. Viral phenotype and immune response in primary human immunodeficiency virus type 1 infection. J Infect Dis. 1992;165:427–432. doi: 10.1093/infdis/165.3.427. [DOI] [PubMed] [Google Scholar]
- 66.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 67.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 68.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyo E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 69.Schellekens P T, Tersmette M, Roos M T, Keet R P, de Wolf F, Coutinho R A, Miedema F. Biphasic rate of CD4+ cell count decline during progression to AIDS correlates with HIV-1 phenotype. AIDS. 1992;6:665–669. doi: 10.1097/00002030-199207000-00008. [DOI] [PubMed] [Google Scholar]
- 70.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E, van Steenwijk R P, Lange J M, Schattenkerk J K, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seillier-Moiseiwitsch F, Margolin B H, Swanstrom R. Genetic variability of the human immunodeficiency virus: statistical and biological issues. Annu Rev Genet. 1994;28:559–596. doi: 10.1146/annurev.ge.28.120194.003015. [DOI] [PubMed] [Google Scholar]
- 72.Shioda T, Levy J A, Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O’Brien T R, Jacobson L P, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner M W, O’Brien S J Hemophilia Growth and Development Study (HGDS); Multicenter AIDS Cohort Study (MACS); Multicenter Hemophilia Cohort Study (MHCS); San Francisco City Cohort (SFCC); ALIVE Study. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 75.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Starcich B R, Hahn B H, Shaw G M, McNeely P D, Modrow S, Wolf H, Parks E S, Parks W P, Josephs S F, Gallo R C, Wong-Staal F. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 77.Tersmette M, Lange J M, de Goede R E, de Wolf F, Eeftink Schattenkerk J K, Schellekens P T, Coutinho R A, Huisman J G, Goudsmit J, Miedema F. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet. 1989;i:983–985. doi: 10.1016/s0140-6736(89)92628-7. [DOI] [PubMed] [Google Scholar]
- 78.Tien P C, Chiu T, Latif A, Ray S, Batra M, Contag C H, Zejena L, Mbizvo M, Delwart E L, Mullins J I, Katzenstein D A. Primary subtype C HIV-1 infection in Harare, Zimbabwa. J Acquired Immune Defic Syndr. 1999;20:147–153. doi: 10.1097/00042560-199902010-00006. [DOI] [PubMed] [Google Scholar]
- 79.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its coreceptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 80.Tscherning C, Alaeus A, Fredriksson R, Bjorndal A, Deng H, Littman D R, Fenyo E M, Albert J. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology. 1998;241:181–188. doi: 10.1006/viro.1997.8980. [DOI] [PubMed] [Google Scholar]
- 81.UNAID/WHO. Report on the global HIV/AIDS epidemic. Geneva, Switzerland: World Health Organization; 1998. [Google Scholar]
- 82.U.S. Department of Health and Human Services. Division of AIDS. DAIDS virology manual for HIV laboratories. Washington, D.C: U.S. Department of Health and Human Services; 1997. pp. 46–52. [Google Scholar]
- 83.van Rij R P, de Roda Husman A-M, Brouwer M, Goudsmit J, Coutinho R A, Schuitemaker H. Role of CCR2 genotype in the clinical course of syncytium-inducing (SI) or non-SI human immunodeficiency virus type 1 infection in the time to conversion to SI virus variants. J Infect Dis. 1998;178:1806–1811. doi: 10.1086/314522. [DOI] [PubMed] [Google Scholar]
- 84.van’t Wout A B, Kootstra N A, Mulder-Kampinga G A, Albrecht-van Lent N, Scherpbier H J, Veenstra J, Boer K, Coutinho R A, Miedema F, Schuitemaker H. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Investig. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vodicka M A, Goh W C, Wu L I, Rogel M E, Bartz S R, Schweickart V L, Raport C J, Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 86.Willey R L, Rutledge R A, Dias S, Folks T, Theodore T, Buckler C E, Martin M A. Identification of conserved and divergent domains within the envelope gene of the acquired immunodeficiency syndrome retrovirus. Proc Natl Acad Sci USA. 1986;83:5038–5042. doi: 10.1073/pnas.83.14.5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Winkler C, Modi W, Smith M W, Nelson G W, Wu X, Carrington M, Dean M, Honjo T, Tashiro K, Yabe D, Buchbinder S, Vittinghoff E, Goedert J J, O’Brien T R, Jacobson L P, Detels R, Donfield S, Willoughby A, Gomperts E, Vlahov D, Phair J, O’Brien S J ALIVE Study; Hemophilia Growth and Development Study (HGDS); Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS); San Francisco City Cohort (SFCC) Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 88.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 89.Xiao L, Owen S M, Goldman I, Lai A A, de Jong J J, Goudsmit J, Lal R B. CCR5 coreceptor usage of non-syncytium-inducing primary HIV-1 is independent of phylogenetically distinct global HIV-1 isolates: delineation of consensus motif in the V3 domain that predicts CCR-5 usage. Virology. 1998;240:83–92. doi: 10.1006/viro.1997.8924. [DOI] [PubMed] [Google Scholar]
- 90.Zhang L, Carruthers C D, He T, Huang Y, Cao Y, Wang G, Hahn B, Ho D D. HIV type 1 subtypes, coreceptor usage, and CCR5 polymorphism. AIDS Res Hum Retroviruses. 1997;13:1357–1366. doi: 10.1089/aid.1997.13.1357. [DOI] [PubMed] [Google Scholar]
- 91.Zhang L, Huang Y, He T, Cao Y, Ho D D. HIV-1 subtype and second-receptor use. Nature. 1996;383:768. doi: 10.1038/383768a0. [DOI] [PubMed] [Google Scholar]
- 92.Zhong P, Peeters M, Janssens W, Fransen K, Heyndrickx L, Vanham G, Willems B, Piot P, van der Groen G. Correlation between genetic and biological properties of biologically cloned HIV type 1 viruses representing subtypes A, B, and D. AIDS Res Hum Retroviruses. 1995;11:239–248. doi: 10.1089/aid.1995.11.239. [DOI] [PubMed] [Google Scholar]
- 93.Zhu T, Korber B T, Nahmias A J, Hooper E, Sharp P M, Ho D D. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature. 1998;391:594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]
- 94.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]