Abstract
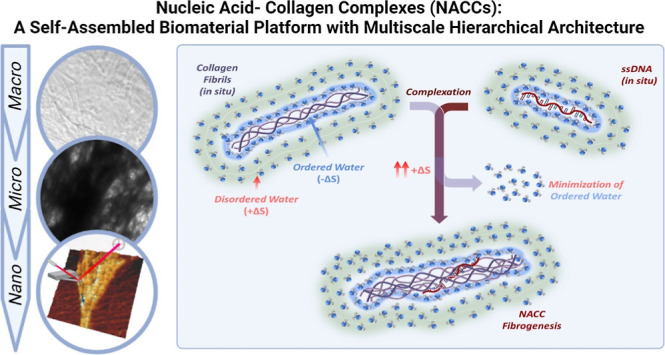
Nucleic acid–collagen complexes (NACCs) are a self-assembled biomimetic fibrillary platform arising from the spontaneous complexation of single-stranded DNA (ssDNA) oligonucleotides and collagen. NACCs merge the extracellular matrix functionality of collagen with the tunable bioactivity of ssDNA as aptamers for broad biomedical applications. We hypothesize that NACCs offer a hierarchical architecture across multiple length scales that significantly varies compared to native collagen. We investigate this using atomic force microscopy and electron microscopy (transmission electron microscopy and cryogenic electron microscopy). Results demonstrate key topographical differences induced by adding ssDNA oligonucleotides to collagen type I. NACCs form a dense network of intertwined collagen fiber bundles in the microscale and nanoscale while retaining their characteristic D-band periodicities (∼67 nm). Additionally, our exploration of thermodynamic parameters governing the interaction indicates an entropically favorable NACC formation driven by ssDNA. Thermal analysis demonstrates the preservation of collagen’s triple helical domains and a more stabilized polypeptide structure at higher temperatures than native collagen. These findings offer important insights into our understanding of the ssDNA-induced complexation of collagen toward the further establishment of structure–property relationships in NACCs and their future development into practical biomaterials. They also provide pathways for manipulating and enhancing collagenous matrices’ properties without requiring complex chemical modifications or fabrication procedures.
Introduction
To obtain bioactive materials for long-term efficacy and tissue regeneration, recent research efforts have focused on creating technologies that mimic the native extracellular matrix (ECM) environment and stimulate desired cell phenotypes. By closely replicating the ECM, biomaterial systems enable cells to model their native behavior, allowing biomedical researchers to study tissue-specific cells and regenerate complex tissues and organs. To achieve this, there is a need to develop innovative technologies that recapitulate the native microenvironment in a highly reproducible and user-friendly manner without harsh chemistries. Moreover, the complex multiscale structure that exists in living tissues must be introduced in cell-laden scaffolds to enable the hierarchical branching of cells, thereby enhancing their viability and function.1 Over the years, collagen-based hydrogels have emerged as a prominent biomaterial platform due to their ability to function as three-dimensional substrates for cell culture and scaffolds for engineered tissues, leveraging collagen’s natural abundance in the ECM, low immunogenicity, and biocompatibility.2 In parallel, the exploration of DNA aptamer-functionalized hydrogels has enabled targeted interactions with specific cellular entities.3 DNA aptamers are short single-stranded sequences (<100 nucleotides) that fold into tertiary structures and bind to targets like proteins with high affinity and specificity. They act as chemical equivalents of antibodies,4 with low to no immunogenicity, and can promote or inhibit receptor–ligand interactions.5 The emerging concept of “four-dimensional biomaterials” suggests the ability to replicate the three-dimensional spatial ECM network while directing biophysical or biochemical changes that ECM undergoes with time as a “fourth dimension.”6 Current strategies involve designing bioactive synthetic multifunctional peptide systems that promote the formation of multiscale fibrous structures through “click-chemistry” reactions.7 However, such four-dimensional biomaterials lack extensive biocompatibility (relying on metal ligands for stabilization),8 limit the number of cells incorporated within the topography, and fail to provide an entirely conducive environment for cell proliferation and ECM remodeling. Developing aptamer-functionalized, customizable collagen-based systems that mimic the ECM environment, stiffness, topography, and biochemistry can potentially promote and enhance reproducible native cellular functions by overcoming challenges linked to synthetic matrices’ native cellular functions.
A novel, fibrillary material platform utilizing collagen and single-stranded DNA (ssDNA) aptamers to render site-specific matrix bioactivity is under investigation by our group, referred to as nucleic acid–collagen complexes (NACCs). NACCs are characterized by their rapid self-assembly at room temperature upon the simple mixing of specific molar ratios of collagen type I and ssDNA oligonucleotides, independent of DNA base-pair sequence or G–C content.9 The unique interplay of DNA and collagen is positioned to be a next-generation biomaterial platform that leverages the benefits of both materials while addressing many of their individual limitations. NACCs rapidly form a three-dimensional network of fibrils (<1 μm) and larger fiber bundles (>10 μm), mimicking ECM architecture. Acting as a “molecular glue,” ssDNA spontaneously induces the microstructural rearrangement of collagen, forming hierarchical, interwoven fibers within seconds.10 NACCs are wholly native materials, potentially an ideal candidate to recapitulate a wide variety of ECM signals to cells.
Previous research groups focused on exploring the influence of collagen fibrillogenesis with long (>1000 base pairs), double-stranded DNA (dsDNA), and various theoretical models for the dsDNA–collagen complex were developed.11,12 However, attempts to clarify the nanostructural topographies of DNA–collagen complexes have yet to succeed.13 Similarly, mechanistic insights into the specific interaction of collagen with other forms of DNA, such as ssDNA, remain to be explored.
Results and Discussion
Unique Topographical Features of NACCs
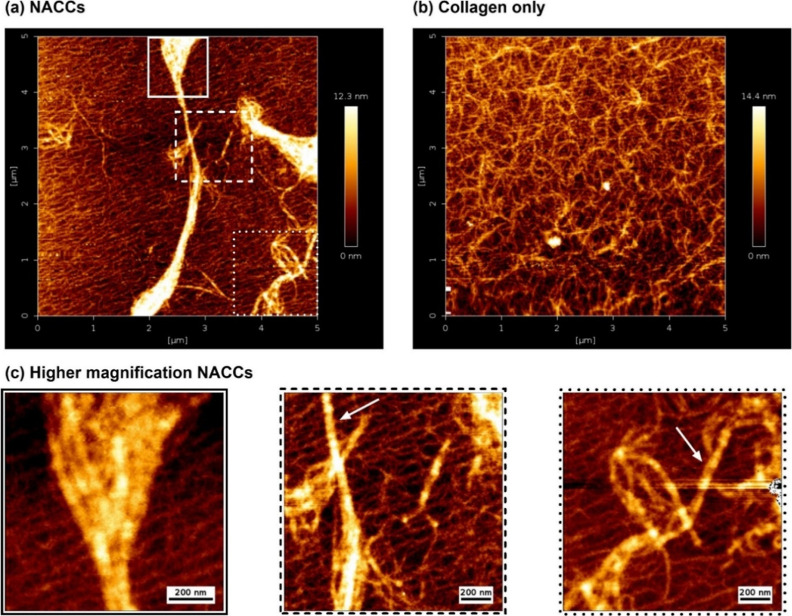
In this study, we successfully determine the topography of NACCs down to the nanoscale through quantitative imaging atomic force microscopy (AFM). The characterization of this unique material system with AFM aids in exploring the nanofiber morphology with a precision that surpasses the limits of other imaging modalities [such as optical or transmission electron microscopy (TEM)].14 In our investigation, we observed the presence of thick, aggregated bundles with variable diameters and lengths at random orientations in NACCs on top of a dense, carpet-like collagen mesh with overlapping fibrils (Figure 1a,c). Typically for collagen only, an even distribution of smaller collagen fibrils on a porous multilayer is expected,15 as also verified in our study, helping us establish direct visual comparisons between the collagen and NACCs (Figure 1b). For AFM, collagen film formation methodology and morphology can vary depending on the substrate (e.g., mica, glass, or polystyrene latex), opening pathways to manipulate the structure of naturally occurring collagen.16,17 For the purpose of this characterization, mica sheets were preferred as the substrate for sample immobilization due to their atomically flat nature, enabling better adsorption of fibers and retaining the fibrillary physiological characteristics of collagen.18,19 In NACCs, individual collagen fibrils were identified from the characteristic pattern of repeated D-band periods every ∼67 nm. This indicates that these structurally important bands remain unaffected upon ssDNA-induced complexation (Figures 2a, S1, and S2).
Figure 1.
Representative high-resolution AFM height topography images of DNA–collagen complexes after adsorption to mica sheets, compared with collagen only. (a) NACCs show the distribution of large fibril aggregates on top of root-like structures of self-assembled collagen fibrils. (b) For comparison, cross-linked collagen-only fibrils without any ssDNA present are shown. (c) Nanoscale observations focusing on the selected boxed regions of NACCs from (a). In the left image, we can see individual collagen fibrils complexed together. The central and right images show intertwined NACC fibrillary bundles. White arrows on representative individual collagen fibrils show retainment of the characteristic D-band periodicities.
Figure 2.
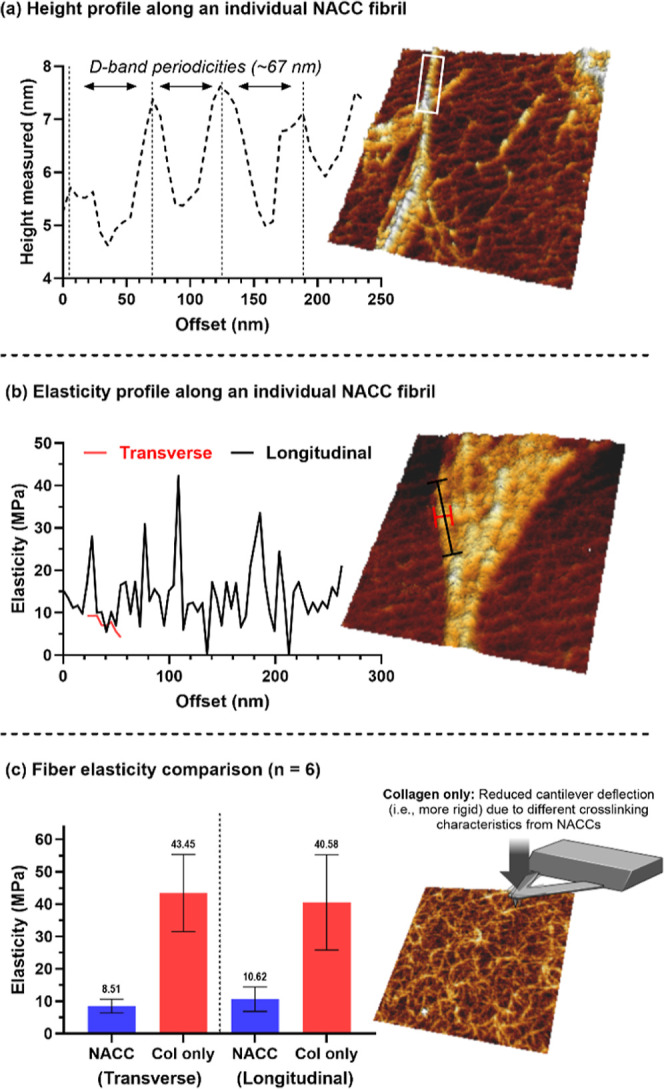
(a) Line height profile plotted along the white box shown on the fibril, confirming the presence of intervals every ∼67 nm, corresponding to the collagen D-band periodicities. Shown in 3D view is a NACC. (b) Elasticity profile showing a regular sinusoidal pattern along the lengthwise direction of a representative NACC fibril, plotted along the red and black lines shown on the 3D NACC image. (c) Bar chart comparison of average elasticity values (n = 6 fibrils) in NACCs and collagen only. The collagen-only sample is shown in 3D view.
We also examined the stiffness of NACC fibers in the lengthwise and transverse directions, with values of 10.62 ± 3.76 and 8.51 ± 1.94 MPa, respectively. Regular sinusoidal patterns of modulus variations were evident along the lengthwise but not the transverse direction—another indicator of D-band gap and overlap region retainment20 (Figure 2b). The average individual fiber stiffness value for collagen only was 40.57 ± 13.45 MPa lengthwise and 43.44 ± 10.89 MPa transversely (Figure 2c). This variation in stiffness between pure collagen samples and NACC formulations suggests that factors beyond the inherent properties of individual collagen fibers are at play.
The observed differences in the matrices’ mechanical properties might be attributed to structural reorganization aspects of the collagen fibrils, such as unique cross-linking patterns. Moreover, ssDNA strands, which exhibit a notably shorter persistence length than collagen fibers,21 are likely to influence the decreased elasticity observed in NACC samples. This aligns with topological images showing NACC samples with denser fiber accumulation, suggesting a more compact structure. The thermodynamic qualities linked to collagen’s strength also merit consideration, potentially explaining the observed mechanical property variations.
Collagen-only samples, characterized by a more uniform structure, display increased stiffness. This implies that stiffness variability along the longitudinal axis could signal features of matrix cross-linking. The material near cross-link points appears more rigid due to lesser deflection, but further from these points, the AFM cantilever detects increased deflection, indicating reduced stiffness—a hypothesis supported by uniform stiffness in the transverse direction, implying consistent proximity to cross-link points.
The structural topography, showcasing D-band periodicities and the fibrils’ overall morphology, alongside the stiffness mapping, offers profound insights into cross-linking dynamics. Our findings indicate that structures containing only collagen exhibit greater stiffness than NACCs, underscoring the complex relationship between structural details and mechanical behavior. This also highlights the importance of considering both molecular and structural factors in their analysis. The disorderly nature of NACC complexes appears to agree with the idea that hydrophobic and hydrophilic domain interactions drive the assembly of collagen peptides into hierarchical fibrillary materials.22,23 This can be of interest to researchers working on soft matter physics or sustainable biomaterials.24 Even though the orientation of collagen into various assemblies can be controlled by hydrodynamic flow,25 this was not controlled in our study to allow NACC fibers to form in their preferred, randomly distributed structures. Such fibrillary topographies can direct cellular processes in wound healing26 or tissue-mimetic templates27 and, as specifically demonstrated with NACCs, can provide contact guidance to endothelial cells and enhance the expression of angiogenic markers.28 Additionally, the hierarchical and curly nature of these rapidly self-assembled NACC nanofibrils enables their mineralization with hydroxyapatite toward directing osteocyte-like morphologies.29
NACCs Hierarchically Assemble across Multiple Length Scales
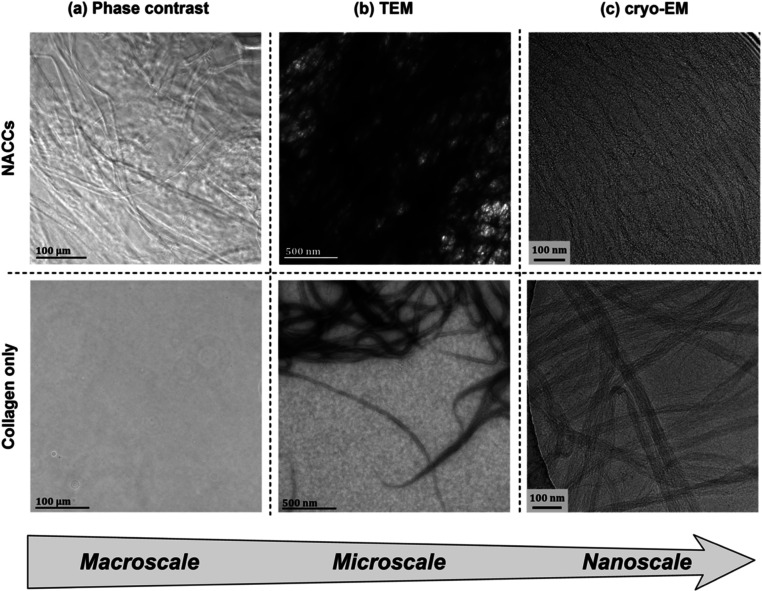
To further contribute to our understanding of NACC internal structure and multiscale hierarchy, we also looked at the organization of fibrils using phase contrast microscopy, TEM, and cryogenic electron microscopy (cryo-EM). We observed closely packed fiber bundles of collagen in NACCs, compared to randomly oriented and independently cross-linked fibrils in collagen only (Figures 3 and S3). Individual fibrils in NACCs are assembled into a mesh and run almost parallel.
Figure 3.
(a) Phase contrast microscopy reveals the presence of fibers of various lengths in NACCs, in comparison to collagen alone, where the individual fibrils are indistinguishable. Such DNA–collagen fibers are also visible to the naked eye. (b) TEM (negative staining conditions: aq. uranyl acetate 1% w/v) demonstrates a dense network of complexed collagen fibrils and fiber bundles, in contrast to collagen only where fibrils do not assemble in that manner. (c) Cryo-EM images indicate a continuous mesh of collagen fibrils in NACCs. Since our cryogenic investigation was done without any staining, contrast variations are the direct result of electron densities. The small, electron-dense dark specks in NACC fibers lead us to believe that these are the single strands of DNA linking the fibers together, absent in collagen only.
Mechanistic Insights on NACC Fibrillogenesis
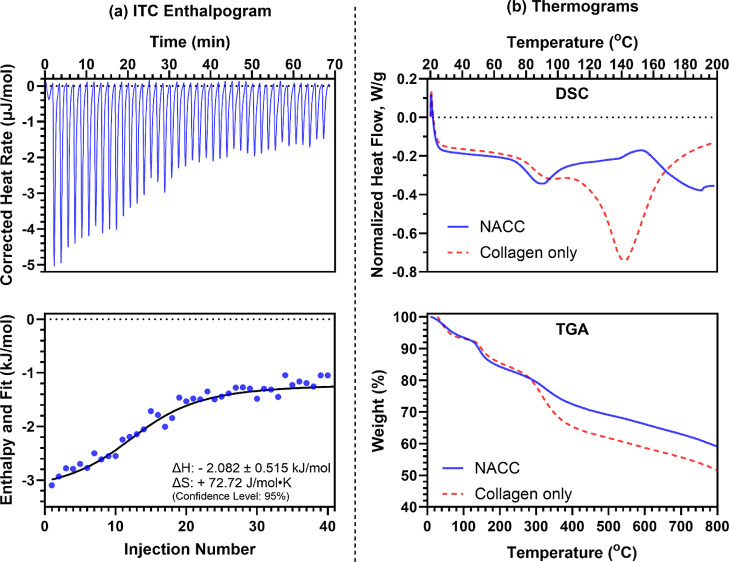
We utilized various thermoanalytical techniques to investigate the thermodynamic parameters governing the complexation and evaluate the thermal stability of NACCs. As a method used to reveal the fibrillogenic mechanisms of collagen,30 isothermal titration calorimetry (ITC) is a good starting point for monitoring the thermodynamics behind the ssDNA-induced complexation of collagen (i.e., NACCs). This provides initial critical guidelines for improving and controlling the design of energy-responsive and dynamic collagen-based materials, such as NACCs, in tissue engineering and regenerative medicine applications.31 The incremental titration of ssDNA in collagen solution has shown that this is an exothermic process, with the heat evolved during the addition of ssDNA to collagen yielding negative enthalpy (ΔH = −2.082 ± 0.515 kJ/mol as determined by an independent binding model fit). Moreover, we found that this type of complexation appears to be entropically favorable (ΔS = +72.72 J/mol·K), thereby driven by the spontaneous and increasing molecular disorder (Figure 4a).
Figure 4.
(a) Top panel: ITC enthalpogram of the titration of ssDNA into collagen (exotherm down). Bottom panel: Integrated data of the NACC ΔH and fit (kJ/mol) per injection. ITC reveals an initial high energy release and entropic favorability in the ssDNA-induced collagen complexation. (b) Top panel: differential scanning calorimetry (DSC) thermogram (exotherm up) shows a significant shift in thermal behavior upon NACC formation, with native collagen exhibiting an endothermic denaturation peak, while NACCs display an apparent exothermic event. Bottom panel: TGA thermogram confirms increased residual mass and elevated onset degradation temperatures in NACCs, indicating enhanced thermal stability.
In contrast with, e.g., collagen-matrix metalloproteinase-1 complex formation, accompanied by positive entropy but endothermic enthalpy,32 we can infer that NACC formation does not involve any collagen triple helix unwinding.33,34 Initially, the titration indicated a high energy release because of NACC complexation, consistent with the literature suggesting increased enthalpic effects due to collagen aggregation.22 Subsequent addition of ssDNA resulted in a drop in the reaction heat rate, implying that less complexation can occur due to the sample cell reaching a critical mass concentration (Figure S4). We attribute these observations to be primarily due to the formation of collagen fibrillary bundles, triggered by the supramolecular interaction with ssDNA. ssDNA appears to drive collagen units to adopt specific quaternary conformations, eventually leading to the hierarchical formation of NACCs as higher-order structures. The association of the two species minimizes thermodynamically unfavorable interactions with water (local structuring of water molecules around the biomacromolecules). We believe that this largely entropic driving force enables the increase in the disorder of water molecules surrounding hydrophobic species (Figure S5), driven by the disruption of the structured water clathrates around individual collagen and ssDNA species in situ and minimizing such solvent cages through spontaneous complexation.35,36
ssDNA Enhances the Physicochemical Stability and Thermal Properties of Collagen
To investigate the consequence of NACC formation on the native properties of collagen, we performed DSC on type-I collagen alone and compared the resultant thermogram to the NACC counterpart. Techniques such as DSC enable the determination of intrinsic thermal properties of many protein-based biomaterials.37 Specifically, proteins or polypeptides and their secondary structures (often dictated by intramolecular hydrogen bonding) exhibit many complex thermal transitions, primarily dictated by the irreversible denaturation of such secondary structures.38 As expected, collagen specimens exhibit a significant endothermic transition, which we attribute to the denaturation of the α-helical secondary structure39 at approximately 130–140 °C (Figure 4b). Fascinatingly, this thermal denaturation event is no longer presented upon the formation of the NACCs; rather, an apparent exothermic event is displayed. We hypothesize that this significant alteration in thermal behavior could be attributed to a covalent cross-linking event between ssDNA and collagen. Regardless, this shift toward a higher temperature demonstrates greater thermodynamic stability of collagen upon complexation, allowing us to conclude that the triple helical domains of collagen are not only preserved but are also stabilized by adding ssDNA. These data highlight the potential of single-stranded oligonucleotides to not only impart augmented bioactivity to collagen-based materials but also act as stabilizers, conferring increased thermal stability within biomaterials without compromising their inherent structure.
Seeking to validate the thermal properties revealed by DSC, we utilized thermogravimetric analysis (TGA) to evaluate the consequence of ssDNA complexation and the exothermic event (vide supra) on the collagen materials’ thermal stability and decomposition profiles. The results appear to agree with the literature suggesting that the structural state of collagenous biomaterials affects the onset degradation temperatures;40 in this case, we indeed see evidence confirming the phenomenon observed in DSC studies. Namely, the NACC displays a patent increase in the residual mass following degradation (Figures 4d and S6), which is likely a result of covalent cross-linking between collagen fibers unique to the structure of the DNA–collagen complex. This covalent modification stabilizes the polypeptide structure at higher temperatures compared to native collagen, increasing the overall carbonization at significantly elevated temperatures (>350–400 °C). From our thermal investigations, we can infer that a prolonged in vivo longevity of DNA-complexed collagenous scaffolds may follow, given the stability under thermal stress enabled within NACCs, possibly offering more resistance to deterioration from collagenases or nucleases.41−43
Conclusions
The work we present herein is an updated report on our understanding of the dynamics of nucleic acid–collagen complexation and the formation of macro-, micro-, and nanofibers stemming from the unique union of these biomacromolecules. NACCs are composed of an ECM building block and a nucleotide building block that, when mixed, rapidly self-assemble into a three-dimensional network of insoluble and stable fibrils. Our results provide fundamental principles of this macromolecular interaction and can potentially be adapted to study similar self-assembling systems. We have shown that ssDNA modulates collagenous matrices’ internal structure and physicochemical properties. This adds to our previously published findings, which established that this interaction results in the robust reinforcement of the bulk matrix while retaining the secondary structure of individual collagen fibrils.
In summary, ssDNA acts as a driver for collagen reorganization, but it also gives us the ability to localize function-specific aptamers. NACCs present a compelling avenue toward establishing them as a highly reproducible, customizable, tunable, and bioactive aptamer-based material system for a wide range of biomedical applications. Being in an ideal blank state, they can be tailored to specific applications, both in terms of material properties as well as bioactivity. The spontaneous creation of bioactive fibrils resembling the native ECM unlocks the potential of NACCs to represent a groundbreaking advancement in biomaterials.
Methods
Extensive descriptions of methods are included in the Supporting Information. NACC complexation occurs at room temperature simply by mixing collagen type I (bovine, PureCol EZ Gel, Advanced Biomatrix) and ssDNA (IDT Technologies) solutions at desired ratios and gently pipetting. For AFM sample preparation, we developed a specialized protocol. Briefly, mica sheets were cleaved fresh using a razor blade, immediately followed by the deposition of the desired sample on the surface. The Bruker NanoWizard 4 was used with triangular pyramid probes in quantitative imaging mode. For TEM (Talos L120C G2), samples were placed on glow-discharged, carbon-coated Formvar copper grids, 400 mesh. Images with rat-tail collagen (Corning) were also taken for comparison. Cryo-EM images were recorded with a Tecnai G2 F20-TWIN TEM and a Gatan Inc. CCD camera. ITC was performed with a TA Instruments NanoITC. ssDNA oligonucleotide solution in the buret syringe (10 μM) was titrated over aqueous type-I collagen (950 μL, 1.5 mg/mL) in the sample cell at 25 ± 0.1 °C. 4.76 μL ssDNA injections were carried out at 100 s intervals, and the constant stirring speed in the sample cell was set to 250 rpm. For DSC and TGA, NACCs with collagen derived from an additional collagen species were evaluated (rat-tail, Corning), and similar trends were obtained as the bovine collagen. Samples were lyophilized for 24 h, and a final sample mass of around 5 mg was tested in aluminum containers. Measurements were carried out on a TA DSC2500 (20–200 °C range, ramp 10 °C/min) and TA TGA5500 (50–800 °C range).
Acknowledgments
The authors would like to thank Bruker Nano Inc. (staff: Ming Ye and Jianjun Yao) for their technical support and expertise. Additionally, we express our gratitude to Dr. Nathalie Wall and Rachel Fontan (UF Department of Materials Science and Engineering) for the availability of ITC and the assistance of Rudy Alvarado and Paul Chipman from UF ICBR (University of Florida’s Interdisciplinary Center for Biotechnology Research, RRID: SCR_019146) for TEM and cryo-EM. The table of contents artwork was created with BioRender.com. This research was funded by the National Institutes of Health NIH R21 (#5R21GM146088-02) award to J.B.A..
Glossary
Nomenclature
- NACCs
nucleic acid–collagen complexes
- ssDNA
single-stranded DNA.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.4c04104.
Experimental protocols, materials, and methods (PDF)
Author Contributions
N. Pipis performed material preparation and primary data collection. K. A. Stewart and M. Tabatabaei assisted with experimental setup and data analysis. N. Pipis and Dr. J. B. Allen wrote the first draft of the manuscript, and Dr. L. N. Williams commented on previous versions. All authors have approved the final version of the manuscript.
This research was funded by the National Institutes of Health NIH R21 (#5R21GM146088-02) award to J.B.A.
The authors declare no competing financial interest.
Supplementary Material
References
- Li X.; Sun Q.; Li Q.; Kawazoe N.; Chen G. Functional Hydrogels With Tunable Structures and Properties for Tissue Engineering Applications. Front. Chem. 2018, 6, 499. 10.3389/fchem.2018.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrigiannidis S. O.; Rey J. M.; Dobre O.; González-García C.; Dalby M. J.; Salmeron-Sanchez M. A Tough Act to Follow: Collagen Hydrogel Modifications to Improve Mechanical and Growth Factor Loading Capabilities. Mater. Today Bio 2021, 10, 100098. 10.1016/j.mtbio.2021.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morya V.; Walia S.; Mandal B. B.; Ghoroi C.; Bhatia D. Functional DNA Based Hydrogels: Development, Properties and Biological Applications. ACS Biomater. Sci. Eng. 2020, 6 (11), 6021–6035. 10.1021/acsbiomaterials.0c01125. [DOI] [PubMed] [Google Scholar]
- Trausch J. J.; Shank-Retzlaff M.; Verch T. Replacing Antibodies with Modified DNA Aptamers in Vaccine Potency Assays. Vaccine 2017, 35 (41), 5495–5502. 10.1016/j.vaccine.2017.04.003. [DOI] [PubMed] [Google Scholar]
- Ramaswamy V.; Monsalve A.; Sautina L.; Segal M. S.; Dobson J.; Allen J. B. DNA Aptamer Assembly as a Vascular Endothelial Growth Factor Receptor Agonist. Nucleic Acid Ther. 2015, 25 (5), 227–234. 10.1089/nat.2014.0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilderbrand A. M.; Ovadia E. M.; Rehmann M. S.; Kharkar P. M.; Guo C.; Kloxin A. M. Biomaterials for 4D Stem Cell Culture. Curr. Opin. Solid State Mater. Sci. 2016, 20 (4), 212–224. 10.1016/j.cossms.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilderbrand A. M.; Ford E. M.; Guo C.; Sloppy J. D.; Kloxin A. M. Hierarchically Structured Hydrogels Utilizing Multifunctional Assembling Peptides for 3D Cell Culture. Biomater. Sci. 2020, 8 (5), 1256–1269. 10.1039/C9BM01894H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. B.; Jeon O.; Lee S. J.; Ding A.; Wells D.; Alsberg E. Induction of Four-Dimensional Spatiotemporal Geometric Transformations in High Cell Density Tissues via Shape-Changing Hydrogels. Adv. Funct. Mater. 2021, 31 (24), 2010104. 10.1002/adfm.202010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James B. D.; Saenz S.; van Gent A.; Allen J. B. Oligomer Length Defines the Self-Assembly of Single-Stranded DNA–Collagen Complex Fibers. ACS Biomater. Sci. Eng. 2020, 6 (1), 213–218. 10.1021/acsbiomaterials.9b01435. [DOI] [PubMed] [Google Scholar]
- Pipis N.; Duraivel S.; Subramaniam V.; Stewart K. A.; Angelini T. E.; Allen J. B.. Toward Bioactive Hydrogels: A Tunable Approach via Nucleic Acid-Collagen Complexation. Regenerative Engineering and Translational Medicine; Springer, 2024. 10.1007/s40883-024-00345-1 [DOI] [Google Scholar]
- Kitamura H.; Iwamoto C.; Sakairi N.; Tokura S.; Nishi N. Marked Effect of DNA on Collagen Fibrillogenesis in Vitro. Int. J. Biol. Macromol. 1997, 20 (3), 241–244. 10.1016/S0141-8130(97)00021-4. [DOI] [PubMed] [Google Scholar]
- Pidaparti R. M.; Svintradze D. V.; Shan Y.; Yokota H. Optimization of Hydrogen Bonds for Combined DNA/Collagen Complex. J. Theor. Biol. 2009, 256 (2), 149–156. 10.1016/j.jtbi.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Svintradze D. V.; Mrevlishvili G. M.; Metreveli N.; Jariashvili K.; Namicheishvili L.; Skopinska J.; Sionkowska A. Collagen–DNA Complex. Biomacromolecules 2008, 9 (1), 21–28. 10.1021/bm7008813. [DOI] [PubMed] [Google Scholar]
- Kontomaris S. V.; Stylianou A.; Chliveros G.; Malamou A. Overcoming Challenges and Limitations Regarding the Atomic Force Microscopy Imaging and Mechanical Characterization of Nanofibers. Fibers 2023, 11 (10), 83. 10.3390/fib11100083. [DOI] [Google Scholar]
- Stylianou A.; Yova D. Surface Nanoscale Imaging of Collagen Thin Films by Atomic Force Microscopy. Mater. Sci. Eng. C 2013, 33 (5), 2947–2957. 10.1016/j.msec.2013.03.029. [DOI] [PubMed] [Google Scholar]
- Stylianou A.; Gkretsi V.; Stylianopoulos T. Atomic Force Microscopy Nano-Characterization of 3D Collagen Gels with Tunable Stiffness. MethodsX 2018, 5, 503–513. 10.1016/j.mex.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylianou A.; Politopoulos K.; Kyriazi M.; Yova D. Combined Information from AFM Imaging and SHG Signal Analysis of Collagen Thin Films. Biomed. Signal Process. Control. 2011, 6 (3), 307–313. 10.1016/j.bspc.2011.02.006. [DOI] [Google Scholar]
- Stylianou A. Atomic Force Microscopy for Collagen-Based Nanobiomaterials. J. Nanomater. 2017, 2017, 1–14. 10.1155/2017/9234627. [DOI] [Google Scholar]
- Leow W. W.; Hwang W. Epitaxially Guided Assembly of Collagen Layers on Mica Surfaces. Langmuir 2011, 27 (17), 10907–10913. 10.1021/la2018055. [DOI] [PubMed] [Google Scholar]
- Song Q.; Jiao K.; Tonggu L.; Wang L. G.; Zhang S. L.; Yang Y. D.; Zhang L.; Bian J. H.; Hao D. X.; Wang C. Y.; Ma Y. X.; Arola D. D.; Breschi L.; Chen J. H.; Tay F. R.; Niu L. N. Contribution of Biomimetic Collagen-Ligand Interaction to Intrafibrillar Mineralization. Sci. Adv. 2019, 5 (3), eaav9075 10.1126/sciadv.aav9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.; Weers B.; Stellwagen N. C. DNA persistence length revisited. Biopolymers: Original Research on Biomolecules 2002, 61 (4), 261–275. 10.1002/bip.10151. [DOI] [PubMed] [Google Scholar]
- Cejas M. A.; Kinney W. A.; Chen C.; Vinter J. G.; Almond H. R.; Balss K. M.; Maryanoff C. A.; Schmidt U.; Breslav M.; Mahan A.; Lacy E.; Maryanoff B. E. Thrombogenic Collagen-Mimetic Peptides: Self-Assembly of Triple Helix-Based Fibrils Driven by Hydrophobic Interactions. Proc. Natl. Acad. Sci. U.S.A. 2008, 105 (25), 8513–8518. 10.1073/pnas.0800291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs J. M.; Mant C. T.; Hodges R. S. Determination of Intrinsic Hydrophilicity/Hydrophobicity of Amino Acid Side Chains in Peptides in the Absence of Nearest-Neighbor or Conformational Effects. Pept. Sci. 2006, 84 (3), 283–297. 10.1002/bip.20417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurtsever A.; Wang P.-X.; Priante F.; Morais Jaques Y.; Miyazawa K.; MacLachlan M. J.; Foster A. S.; Fukuma T. Molecular Insights on the Crystalline Cellulose-Water Interfaces via Three-Dimensional Atomic Force Microscopy. Sci. Adv. 2022, 8 (41), eabq0160 10.1126/sciadv.abq0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F.; Hörber H.; Howard J.; Müller D. J. Assembly of Collagen into Microribbons: Effects of pH and Electrolytes. J. Struct. Biol. 2004, 148 (3), 268–278. 10.1016/j.jsb.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Liu G. Y.; Agarwal R.; Ko K. R.; Ruthven M.; Sarhan H. T.; Frampton J. P. Templated Assembly of Collagen Fibers Directs Cell Growth in 2D and 3D. Sci. Rep. 2017, 7 (1), 9628. 10.1038/s41598-017-10182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylianou A.; Kontomaris S. B.; Kyriazi M.; Yova D.. Surface Characterization of Collagen Films by Atomic Force Microscopy. In XII Mediterranean Conference on Medical and Biological Engineering and Computing 2010; Bamidis P. D., Pallikarakis N., Eds.; Springer: Berlin, Heidelberg, 2010; IFMBE Proceedings, pp 612–615. 10.1007/978-3-642-13039-7_154. [DOI]
- James B. D.; Allen J. B. Self-Assembled VEGF-R2 Targeting DNA Aptamer-Collagen Fibers Stimulate an Angiogenic-like Endothelial Cell Phenotype. Mater. Sci. Eng. C 2021, 120, 111683. 10.1016/j.msec.2020.111683. [DOI] [PubMed] [Google Scholar]
- James B. D.; Guerin P.; Iverson Z.; Allen J. B. Mineralized DNA-Collagen Complex-Based Biomaterials for Bone Tissue Engineering. Int. J. Biol. Macromol. 2020, 161, 1127–1139. 10.1016/j.ijbiomac.2020.06.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Masís J.; Cubero-Sesin J. M.; Guerrero S.; González-Camacho S.; Corrales-Ureña Y. R.; Redondo-Gómez C.; Vega-Baudrit J. R.; Gonzalez-Paz R. J. Self-Assembly Study of Type I Collagen Extracted from Male Wistar Hannover Rat Tail Tendons. Biomater. Res. 2020, 24, 19. 10.1186/s40824-020-00197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S.; Yu X.; You J.; Yin T.; Lin Y.; Chen W.; Dao L.; Du H.; Liu R.; Xiong S.; Hu Y. Study of the Thermodynamics and Conformational Changes of Collagen Molecules upon Self-Assembly. Food Hydrocolloids 2021, 114, 106576. 10.1016/j.foodhyd.2020.106576. [DOI] [Google Scholar]
- Manka S. W.; Brew K. Thermodynamic and Mechanistic Insights into Coupled Binding and Unwinding of Collagen by Matrix Metalloproteinase 1. J. Mol. Biol. 2020, 432 (22), 5985–5993. 10.1016/j.jmb.2020.10.003. [DOI] [PubMed] [Google Scholar]
- Chung L.; Dinakarpandian D.; Yoshida N.; Lauer-Fields J. L.; Fields G. B.; Visse R.; Nagase H. Collagenase Unwinds Triple-helical Collagen Prior to Peptide Bond Hydrolysis. EMBO J. 2004, 23 (15), 3020–3030. 10.1038/sj.emboj.7600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I.; Fragai M.; Luchinat C.; Melikian M.; Toccafondi M.; Lauer J. L.; Fields G. B. Structural Basis for Matrix Metalloproteinase 1-Catalyzed Collagenolysis. J. Am. Chem. Soc. 2012, 134 (4), 2100–2110. 10.1021/ja208338j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke T. M.; Tsai J.; Levitt M. Quantification of the Hydrophobic Interaction by Simulations of the Aggregation of Small Hydrophobic Solutes in Water. Proc. Natl. Acad. Sci. U.S.A. 2001, 98 (11), 5965–5969. 10.1073/pnas.111158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter I.; Leeuw N. H. de. A Molecular Dynamics Study of the Interprotein Interactions in Collagen Fibrils. Soft Matter 2011, 7 (7), 3373–3382. 10.1039/C0SM01192D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samouillan V.; Delaunay F.; Dandurand J.; Merbahi N.; Gardou J.-P.; Yousfi M.; Gandaglia A.; Spina M.; Lacabanne C. The Use of Thermal Techniques for the Characterization and Selection of Natural Biomaterials. J. Funct Biomater 2011, 2 (3), 230–248. 10.3390/jfb2030230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mant C. T.; Chen Y.; Yan Z.; Popa T. V.; Kovacs J. M.; Mills J. B.; Tripet B. P.; Hodges R. S. HPLC Analysis and Purification of Peptides. Peptide Characterization and Application Protocols 2007, 386, 3–55. 10.1007/978-1-59745-430-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzyński S.; Sionkowska A.; Marciniak A. DSC Study of Collagen in Disc Disease. J. Biophys. 2009, 2009, 819635. 10.1155/2009/819635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi M.; Sawada T.; Hatakeyama W.; Taira M.; Hachinohe Y.; Takafuji K.; Kihara H.; Takemoto S.; Kondo H. Characterization of Five Collagenous Biomaterials by SEM Observations, TG-DTA, Collagenase Dissolution Tests and Subcutaneous Implantation Tests. Materials 2022, 15 (3), 1155. 10.3390/ma15031155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S.; Oh M. H.; Park J. Y.; Park T. G.; Nam Y. S. Protein-Resistant, Reductively Dissociable Polyplexes for in Vivo Systemic Delivery and Tumor-Targeting of siRNA. Biomaterials 2013, 34 (9), 2370–2379. 10.1016/j.biomaterials.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Mrevlishvili G. M.; Svintradze D. V. DNA as a Matrix of Collagen Fibrils. Int. J. Biol. Macromol. 2005, 36 (5), 324–326. 10.1016/j.ijbiomac.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Nguyen M. K.; Huynh C. T.; Gilewski A.; Wilner S. E.; Maier K. E.; Kwon N.; Levy M.; Alsberg E. Covalently Tethering siRNA to Hydrogels for Localized, Controlled Release and Gene Silencing. Sci. Adv. 2019, 5 (8), eaax0801 10.1126/sciadv.aax0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.