Abstract
The Rous sarcoma virus (RSV) Gag precursor polyprotein is the only viral protein which is necessary for specific packaging of genomic RNA. To map domains within Gag which are important for packaging, we constructed a series of Gag mutations in conjunction with a protease (PR) active-site point mutation in a full-length viral construct. We found that deletion of either the matrix (MA), the capsid (CA), or the protease (PR) domain did not abrogate packaging, although the MA domain is likely to be required for proper assembly. A previously characterized deletion of both Cys-His motifs in RSV nucleocapsid protein (NC) reduced both the efficiency of particle release and specific RNA packaging by 6- to 10-fold, consistent with previous observations that the NC Cys-His motifs played a role in assembly and RNA packaging. Most strikingly, when amino acid changes at Arg 549 and 551 immediately downstream of the distal NC Cys-His box were made, RNA packaging was reduced by more than 25-fold with no defect in particle release, demonstrating the importance of this basic amino acid region in packaging. We also used the yeast three-hybrid system to study avian retroviral RNA-Gag interactions. Using this assay, we found that the interactions of the minimal packaging region (Mψ) with Gag are of high affinity and specificity. Using a number of Mψ and Gag mutants, we have found a clear correlation between a reporter gene activation in a yeast three-hybrid binding system and an in vivo packaging assay. Our results showed that the binding assay provides a rapid genetic assay of both RNA and protein components for specific encapsidation.
Packaging of retroviral RNA is a precise process which leads to particles containing two copies of the viral genomic RNA and exclusion of viral and cellular mRNAs. Packaging of RNA is dependent only upon the Gag protein; the uncleaved Gag precursor protein is sufficient to allow RNA encapsidation (35, 48). Over the past decade, a number of studies have been undertaken to define the domains in the Gag proteins of murine, human, and avian retroviruses which interact with viral RNA and lead to specific encapsidation. These studies are complicated by the fact that assembly of Gag into particles is a prerequisite for encapsidation, and so regions of Gag which do not allow proper assembly could also be scored as packaging domains.
Previous work has shown that the avian retroviral Gag protein, consisting of matrix (MA), p2, p10, capsid (CA) (which includes the major homology region [MHR]), nucleocapsid (NC), and protease (PR) (see Fig. 1), contains three domains important for assembly. One is at the amino terminus of Gag and is involved in correct cellular localization of the protein (9, 60, 63). The second is a proline-rich region (PPPPPY) in the small p2 peptide, which is required for the late step in assembly (62). The third domain in the NC protein is important for protein-protein interactions (9, 61) and appears to be distinct from the two Cys-His motifs, which have been implicated in RNA packaging (1, 14, 20, 21, 28, 29, 45–47, 52). The basic residues of NC which surround the Cys-His motifs are also important for RNA packaging (18, 24, 30, 42, 52, 54). The study of chimeric proteins, which contain the NC domain of one retrovirus substituted for the cognate region of another retroviral Gag, indicates that NC mediates the specificity of RNA encapsidation (13, 22, 67). While all of these studies indicate that the NC domain plays important roles in RNA binding and RNA encapsidation, selection of RNA occurs through the Gag precursor protein, and NC itself can be either a rather nonspecific RNA binding protein which binds to RNA at a ratio of 4 to 7 nucleotides/protein (32, 33, 36, 66) or a specific binding protein under some conditions (2, 10, 18, 23).
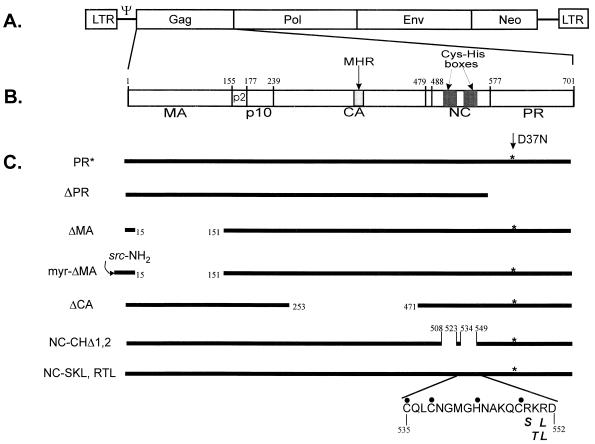
FIG. 1.
(A) Diagram of the RSV genome containing the selectable marker neo in the place of the src gene. LTR, long terminal repeat; Gag, viral structural protein; Pol, polymerase protein containing reverse transcriptase and integrase; Env, envelope protein. (B) Details of the Gag polyprotein. Amino acid numbering is from the amino terminus of Gag. (C) Gag constructs used to determine domains required for the specific viral RNA packaging into virions. End parts of the deletions are indicated by breaks flanked by amino acid numbers. PR* is a Gag polyprotein carrying an active-site mutation, D37N (indicated by asterisks), in the protease domain, which abolishes protease activity. In myr-ΔMA, the sequence encoded by the first 10 codons of p60v-src comprising a myristylation signal (64) were added to the N terminus of a Gag polyprotein lacking MA. Amino acids comprising the second Cys-His box in the NC domain are indicated by dots. The locations of the mutated residues in NC-SKL and RTL are indicated below the amino acid sequence, in italics.
RNA-Gag interactions have been assayed in vitro by using glutathione S-transferase (GST) fusion proteins or proteins containing NC and additional regions of Gag (12). Somewhat conflicting results have been reported in experiments involving GST fusions with human immunodeficiency virus type 1 (HIV-1) Gag proteins. One group showed that both GST-Gag protein and GST-NC protein bound to HIV-1 RNA with comparable specificity (11, 12). However, another group reported that while NC-p6 protein specifically bound to HIV-1 RNA, a GST-NC-p6 fusion protein was nonselective and bound equally to all transcripts, including antisense and non-HIV-1 RNA (43). Taken together, these studies seem to indicate that after fusion with extraneous protein domains such as GST, the secondary or tertiary structure of the NC domain might differ from that of the free NC protein. The altered conformation could lead to specific binding to RNA and to selective viral genomic RNA packaging into virions. In a different in vitro assay (25), it was shown that deletion of both MA and CA from the HIV-1 Gag proteins diminished the specificity of Gag binding to RNA. This again suggests that the conformation of NC is affected by upstream amino acid sequences.
To determine which regions of Gag are important for packaging in the context of normal avian retroviral assembly, we introduced a series of deletion mutations in Gag into a complete proviral vector, RCASBPneo (3), which also contains a PR active-site mutation, D37N (58). We demonstrate that while many of the individual domains in the Gag polyprotein, including MA, CA, or PR, are dispensable for RNA packaging, the NC protein plays a major role in specific packaging. Most strikingly, the small basic region in the NC domain plays important roles in the recognition and encapsidation of viral genomic RNA. In a complementary set of experiments, we used the yeast three-hybrid assay to detect Rous sarcoma virus (RSV) RNA-Gag interactions. In these studies, transcription of a reporter gene was rendered dependent on the interaction between Gag and RSV RNA. We found a clear correlation between the results of the rapid binding assay in yeast and the in vivo packaging assay. We also found that the binding affinity, in addition to the binding specificity, is an important component of packaging competence. Our results suggest that the simple and rapid yeast three-hybrid system is sensitive enough to replace the packaging assay for screening of Gag and some RNA mutations.
MATERIALS AND METHODS
Construction of RSV Gag mutations.
Deletions were made in the context of a complete RSV-based provirus containing neo in the place of src gene (Fig. 1). Plasmid pASY155 was constructed by replacing the SacI255-HpaI2731 segment (numbering as described in references 19 and 55) of the avian leukosis virus-based vector RCASBPneo (3) with that of the RSV proviral clone pATV8R (5, 34). The PR active-site mutation, D37N, which has been described previously (58) was introduced into pASY155 by oligonucleotide-directed in vitro mutagenesis to create pASY165 (PR* in Fig. 1C). All deletion mutants were constructed by two rounds of DNA amplification by PCR technology. All mutants were moved to the pASY165 backbone. The PR deletion mutant, ΔPR, had complete deletion of the C-terminal PR domain in the Gag protein. The MA deletion mutation, ΔMA, leaves only 15 amino acids at the N terminus including ATG start codon. To construct the myrΔMA mutation, an oligonucleotide containing the first 10 codons of p60v-src, as previously reported in the construction of Pr76myr1 (64), was used to add a myristylation signal to the ΔMA mutation. The CA deletion mutation, ΔCA, lacks most of the CA domain including MHR. The RSV NC-CHΔ1,2 mutation is a deletion of both Cys-His boxes in the NC domain of Gag, and its construction has been described previously (46). The Arg549, Lys550, and Arg551 residues immediately downstream of the second Cys-His box in NC were altered by using appropriate mutant oligonucleotides to generate the NC-SKL and NC-RTL mutations.
Cell cultures and transfections.
The quail cell lines QT6 (5) and Q2bn-4D (59) were grown in GM+D+CK (Ham’s F10 medium containing 10% tryptose phosphate broth [Difco], 5% calf serum, 1% heat-inactivated chicken serum and 1% dimethyl sulfoxide). The modified calcium phosphate method (16) was used for DNA transfections on cells seeded in Dulbecco modified Eagle medium (DMEM) supplemented with 10% calf serum. Stably transfected mass cultures of G418-resistant cells at 0.15 mg/ml were obtained after 2 weeks of selection.
Protein labeling, density gradient analysis, and radioimmunoprecipitation assay (RIPA).
QT6 cultures containing the mutant proviruses were labeled with 250 μCi of [35S]methionine (EXPRESS 35S protein labeling mix; >1,000 Ci/mmol [NEN Research Products]) in 2 ml of serum-free DMEM minus methionine and cysteine (DMEM−Met−Cys) for 5 h and chased for 18 to 24 h with DMEM−Met−Cys medium supplemented with 10% fetal bovine serum. The supernatants were collected, and virus-like particles were harvested by clearing the supernatant of cell debris by low-speed centrifugation and then concentrated by high-speed centrifugation through a 20% sucrose cushion. The viral pellet was then resuspended in isotonic buffer as described previously (5). The labeled cells were washed twice with cold isotonic buffer, scraped from the plates, and then directly lysed with the lysis buffer (Direct Protect kit; Ambion Inc., Austin, Tex.).
For density gradient analysis, at 48 h after transfection of QT6 cells with plasmid DNA, the cells were labeled for 7 h in 2 ml of serum-free DMEM−Met−Cys supplemented with [35S]methionine. Viral pellets were layered onto 10 to 40% iodixanol (OptiPrep; Nycomed Pharma) and centrifuged at 36,000 rpm at 4°C for 4 h, using an L7 ultracentrifuge (Beckman). Fractions of 0.5 ml were collected, and the iodixanol density of each fraction was measured on a refractometer. Proteins in each fraction were precipitated with 10% trichloroacetic acid, and pellets were washed with acetone and then resuspended in 1% sodium dodecyl sulfate (SDS) plus TE buffer (10 mM Tris-Cl, 1 mM EDTA [pH 8.0]). Gag proteins in each fraction were immunoprecipitated, as described below.
For quantitation of intracellular and virion levels of Gag protein, RIPA were performed with either anti-avian leukosis virus polyclonal rabbit serum (anti-PrB) or antiserum specific for the p10 domain of RSV Gag polyprotein (anti-p10), obtained after immunization of a rabbit with a preparation of purified GST-p10 fusion protein. Concentrated [35S]methionine-labeled viral particles or labeled cellular extracts were incubated in 1.0 ml of Ab-buffer (20 mM Tris-HCl [pH 7.4], 50 mM NaCl, 0.5% Nonidet P-40 [NP-40], 0.5% deoxycholic acid [DOC], 0.5% SDS, 0.5% aprotinin, 1 mM EDTA [pH 8.0]) with 5 ml of anti-p10 serum and 30 ml of protein A-Sepharose beads (Pharmacia LKB Biotechnology, Inc.) for 90 min at room temperature. The antigen-antibody complexes were washed twice in RIPA buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1% DOC, 0.1% SDS, 0.5% aprotinin), once in high-salt buffer (10 mM Tris-HCl [pH 7.4], 2 M NaCl, 1% NP-40, 0.5% DOC), and then once more in RIPA buffer. The bound proteins were eluted in SDS sample buffer and loaded onto SDS–12.5% polyacrylamide gels. The gels were scanned directly with the Molecular Dynamics PhosphorImager. Radioactive bands were quantitated with ImageQuant software (Molecular Dynamics). The efficiency of particle release for each mutant was calculated as follows. The number of PhosphorImager machine units counted for each Gag band detected in the medium was divided by the number obtained for the respective Gag band detected in the cells. These ratios were then normalized to the ratio obtained for the intact full-length Gag precursor by correcting for the number of methionines in the respective Gag mutants to give a relative efficiency of particle release.
RPAs.
Purified viral and cellular RNAs were prepared as previously described (5). We used an antisense neo RNA as our riboprobe in RNase protection assays (RPAs). RPAs of purified RNAs were performed by the method specified for the RPA II kit from Ambion Inc., while RNA in crude cell lysates and viral particles was detected directly with the Direct Protect kit and the lysate RNase protection kit (Amersham-United States Biochemical, Cleveland, Ohio). Radioactively labeled protected RNA bands were quantitated with the PhosphorImager and expressed in machine units.
RT assay.
Each iodixanol density fraction containing unlabeled wild-type (wt) Gag particles was used to measure the reverse transcriptase (RT) activity by incorporation of [32P]TTP during synthesis of DNA on a poly(A) template as previously described (27). A 10-μl volume of the concentrated viral particles was added to 50 μl of reaction cocktails [50 mM Tris (pH 7.8), 75 mM KCl, 2 mM dithiothreitol, 5 mM MgCl2, 10 μg of polyA(dT)12 per ml, 0.05% NP-40] and the reaction mixtures were incubated at 37°C for 1 h. Then 4-μl volumes of the reaction mixtures were transferred to DE81 filters (Whatman), washed in 2× SSC (0.3 M sodium chloride, 30 mM sodium citrate) and in 95% ethanol, and dried. The filters were directly scanned with a PhosphorImager. Radioactive spots were quantitated with ImageQuant software.
RNA hybrid expression vector.
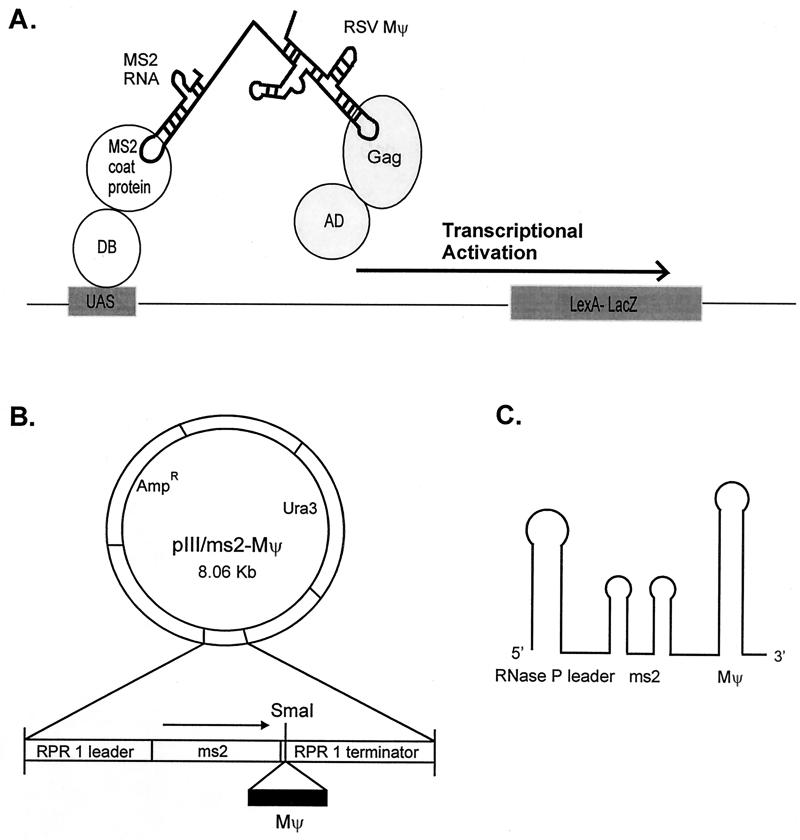
We have introduced the previously identified RSV minimal-packaging sequence, Mψ (7), into plasmid pIII/MS2-1 (56). The 160-nucleotide PCR-amplified Mψ sequence was subcloned into the unique SmaI site downstream of two copies of MS2 sequence. The orientation of the Mψ sequence relative to MS2 RNA was confirmed by sequencing. The MS2-Mψ hybrid RNA was expressed from the vector pIII/MS2-Mψ, which uses the RNA polymerase III promoter and terminator from the Saccharomyces cerevisiae RNase P RNA gene (RPR1). The plasmid also contains the selectable gene, URA3. To construct two copies of Mψ sequence in tandem at the 3′ end of MS2 sequence, two rounds of cloning were done because the SmaI site is no longer a unique restriction site in the pIII/MS2-Mψ plasmid. Two copies of the Mψ sequence were subcloned downstream of the MS2 sequence in plasmid pMS2-1 (56), and then the EcoRI fragment containing MS2-Mψ-Mψ sequence was swapped with that of plasmid pIII/MS2-1.
Protein hybrid expression vector.
Each RSV Gag mutant sequence shown in Fig. 1C was cloned into multicloning sites downstream of the activation domain (AD) of the Gal4 gene in plasmid pACTII (41). Each PCR-amplified Gag mutant sequence flanked with SfiI and EcoRI at the 5′ and 3′ ends of DNA, respectively, was annealed with the 7.5-kb SfiI-EcoRI fragment of plasmid pACTII. The hybrid protein is expressed from the ADH promoter in plasmid pACTII-Gag. The multicopy plasmid carries a selectable LEU2 gene.
Yeast transformation.
The yeast strain L40-coat (56), which stably expresses the LexA-MS2 coat protein fusion gene in the genome along with the TRP1 marker, was used to obtain double transformants expressing RNA hybrid plasmid and the AD hybrid protein plasmid. The genotype of this strain is MATa ura3-52 leu2-3,112 his3D200 trp1D1 ade2 LYS2::(lexA-op)-HIS3 ura3::(lexA-op)-lacZ lexA-MS2 coat (TRP1). Lithium acetate–single-stranded DNA–polyethylene glycol transformation was performed (26). Plates lacking uracil and leucine were used to select transformants carrying both the RNA hybrid plasmid and the protein hybrid plasmid.
Yeast β-gal activity assay.
The β-galactosidase (β-gal) activity of yeast double transformants was quantitatively assayed by directly measuring the enzyme activity with chlorophenol 5-bromo-4-chloro-3-indolyl-β-d-galactoside as a substrate (8). The enzymatic activity represents the average of three to four transformants, and independent assays were repeated four to six times.
RESULTS
Neither the MA nor the CA domain of Gag is required for RNA packaging.
To define the determinants and domains necessary for RNA packaging in the RSV Gag polyprotein, several deletion mutations were constructed in Gag in the context of the proviral vector RCASBPneo (3), which also contains a PR active-site mutation, D37N (58). Each deletion removed almost the entire MA, CA, or PR domain (Fig. 1). The first 10 codons of the src gene was added to the N termini of ΔMA mutant to supply a myristylation signal sequence, which is required for efficient viral assembly (64).
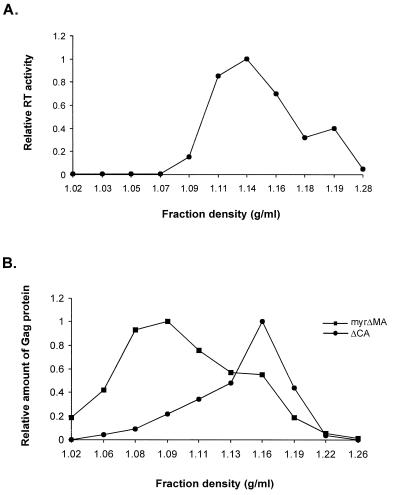
Viral mutant DNAs were transfected into QT6 cells, and after G418 selection, mass cultures with neo-expressing proviruses were obtained. The expression and stability of the mutant Gag precursor proteins in transfected cells were determined by RIPA. All mutants produced stable viral Gag precursors of the predicted molecular weight and at approximately the same level as the intact full-length Gag protein (Fig. 2A). Viral particles collected from the supernatants of metabolically labeled cells were concentrated and immunoprecipitated, as described in Materials and Methods. Moreover, all of the mutants except for ΔMA synthesized detectable amounts of viral particles (Fig. 2B). As expected, the mutant PR*, which lacks protease activity, produced viral particles assembled from unprocessed full-length Gag polyprotein precursor but no Gag cleavage products (Fig. 2A and B, lanes 2). Gag proteins were also detected in the supernatant of the ΔCA mutant (Fig. 2A and B, lanes 3). The efficiency of particle release was calculated for each mutant (Fig. 2C). The relative efficiency of the ΔCA mutant particles released into the medium was similar to that of the PR* protein. The density of ΔCA mutant particles detected in the supernatants was measured by the iodixanol gradient method. The reported densities for retroviruses generally fall between 1.14 and 1.18 g/ml (65). In our experiments, the measured density of wt RSV (as determined by an RT assay) was 1.14 g/ml (Fig. 3A), while that of the ΔCA mutant was 1.16 g/ml (Fig. 3B). This result indicates that the domain(s) necessary for particle assembly and release is still intact in the ΔCA mutant and that in avian retroviruses, the MHR is not required for the release of immature viral particles.
FIG. 2.
Viral particle release of Gag mutants. (A) RIPA analysis of the expression of Gag polyproteins in transfected cells. Lysates from G418 mass culture of QT6 cells transfected with the mutant plasmids described in Fig. 1 were immunoprecipitated with anti-p10 antisera and electrophoresed on SDS-polyacrylamide gels, as described in Materials and Methods. The specific Gag polyproteins of the expected sizes are indicated by the asterisks. Abbreviations are same as in Fig. 1; untransf., untransfected cells. Two separate ΔMA transfectants are shown. (B) RIPA analysis of pelleted virus-like particles collected from supernatants. (C) Relative efficiency of particle release. The number of PhosphorImager machine units counted for each Gag band detected in the media was divided by the number obtained for the respective Gag band detected in the cells. These ratio were then normalized to the ratio obtained for the intact full-length Gag polyprotein by correcting for the number of methionines in the respective Gag mutants to give a relative efficiency of particle release. Each experiment was done four or five times, and the bars show the standard deviations.
FIG. 3.
Viral particle density analysis. (A) Viral particles obtained from stably transfected QT6 cells with wt RSV proviral DNAs were fractionated through 10 to 40% iodixanol gradients. Eleven fractions were collected, and the density of each fraction was measured on a refractometer. RT assays of each fraction were performed, as described in Materials and Methods. (B) QT6 cells transfected with mutant plasmid DNAs were labeled with [35S]methionine, and viral pellets were centrifuged in 10 to 40% iodixanol gradients. Ten fractions were collected, and the density of each fraction was measured on a refractometer. The labeled Gag proteins in each fraction were immunoprecipitated and electrophoresed on SDS-polyacrylamide gels, as described in Materials and Methods. The relative amount of Gag proteins in each fraction were quantitated by using ImageQuant software.
Although ΔMA was unable to direct the assembly of extracellular viral particles (Fig. 2A and B, lanes 7 and 8), this assembly defect can be suppressed by the addition of the myristylated amino terminus of the oncoprotein p60src, called Myr1 (64). We added the Myr1 sequence to the amino terminus of ΔMA Gag to obtain the myrΔMA mutant, which was as efficient as the full-length Gag polyprotein in particle release (Fig. 2A and B, lanes 5; Fig. 2C). Since the levels of cytoplasmic Gag proteins in both the ΔMA and myrΔMA mutants were similar to that of full-length Gag, the defect associated with the ΔMA mutation is likely to be primarily in the function(s) of particle release. These results are not unexpected, since the amino terminus of the MA domain has been shown to contain AD1, the assembly domain necessary for targeting and/or binding the Gag polyprotein to the membrane (63). However, extracellular pelletable proteins from the myrΔMA mutant were less dense than wild-type (wt) RSV particles, with a peak density of 1.09 g/ml (Fig. 3B). This suggests that the MA domain is also required for proper viral assembly.
Deletion of both Cys-His boxes in the NC domain of Gag reduced the relative particle release efficiency of the NC-CHΔ1,2 mutant by about fivefold (Fig. 2A and B, lanes 1; Fig. 2C). However, changing the sequence C-terminal of the second Cys-His box from RKR to SKL had no effect on particle assembly and release compared to the full-length intact Gag polyprotein (Fig. 2A and B, lanes 4; Fig. 2C).
Since the RCASBPneo vector contains an intact packaging region, we were able to examine the packaging of genomic RNA directly into particles. To accomplish this, we used an antisense neo RNA as the riboprobe in RNase protection assays. Since each transfected culture produced somewhat different levels of genomic viral RNA in the cells (data not shown), we normalized the extracellular RNA to the cellular RNA levels (Fig. 4B). Only in particles released from ΔCA did the viral RNA level approach that of PR* (Fig. 4A).
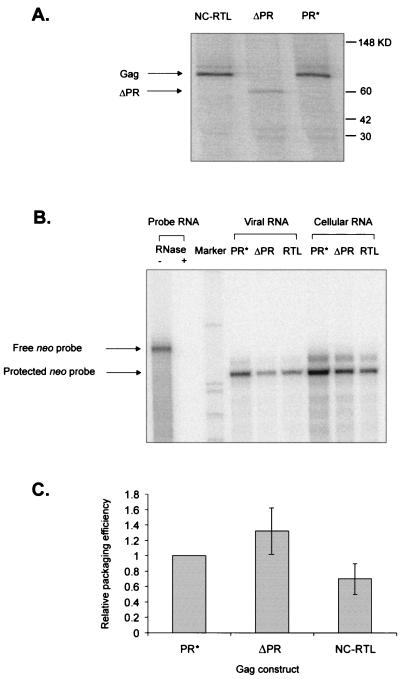
FIG. 4.
RNA packaging of Gag mutants. (A) RPA to measure RNA packaging. The probe used was an antisense neo RNA. The sample in lane 1 contained probe RNA with no RNase added. Abbreviations are same as in Fig. 1. (B) Ratio of viral to cellular RNA. The unit numbers from PhosphorImager analysis were obtained. The numbers for the packaged neo-specific RNA in virions were divided by those in the cells. The relative packaged viral RNAs of Gag mutants were normalized to PR*. The standard deviations are indicated by bars. (C) Relative packaging efficiencies of Gag mutants normalized to PR*. Packaging efficiency was calculated as the ratio of relative amount of neo-specific RNA packaged in particles, as measured by RPA (Fig. 4B), to the relative number of particles, as measured by RIPA (Fig. 2C). The standard deviations are indicated by bars.
To quantitate packaging of genomic RNA by the Gag mutants, the relative amounts of packaged RNA shown in Fig. 4B were corrected for the amounts of viral protein in the supernatants in Fig. 2C. The results, which summarize the packaging efficiency of each mutant provirus, are presented in Fig. 4C. Deletion of the CA domain did not affect the ability of the Gag polyprotein to efficiently package its own genomic RNA, indicating that in the context of the Gag precursor, the RSV CA domain is dispensable for RNA packaging as well as for particle assembly and release. The RNA packaging into myrΔMA pelletable proteins detected in the supernatants, which had lower density than wt particles, was reduced, but only about 2.5-fold, indicating that the MA domain is not necessary for packaging.
We also examined the effects of the PR domain deletion on viral assembly and RNA packaging. We found that the ΔPR mutation had no effect on the ability of virus to package Mψ RNA (Fig. 5).
FIG. 5.
Viral packaging by NC-RTL and ΔPR mutants. (A) RIPA to determine the number of viral particles released from cultures transfected with the indicated Gag mutant vectors. The bands of Gag polyproteins of the expected size are indicated by arrows. (B) RPA to determine the amount of neo-specific RNA expressed in the stably transfected cells and in viral particles. The samples in probe RNA had no viral RNA added. (C) Relative packaging efficiencies of ΔPR and NC-RTL mutants normalized to PR*. Packaging efficiency was calculated as the ratio of relative amount of neo-specific RNA packaged in particles, as measured by RPA (panel B), to the relative number of particles, as measured by RIPA (panel A). Each experiment was done three to five times, and the standard deviations are indicated by bars.
The basic region immediate downstream of the distal Cys-His box is important for the specific encapsidation of viral genomic RNA.
In contrast to the lack of a role in packaging for the MA, CA, and PR domains, we found that the RNA packaging efficiencies of mutants in the NC protein were reduced markedly. The NC-CHΔ1,2 mutant reduced packaging about sixfold in these assays, but this deletion also reduced viral particle production (Fig. 2C). In contrast, there was no effect on viral assembly with the basic-region mutant, NC-SKL (Fig. 2C), but these two amino acid changes reduced packaging more than 25-fold in our assays (Fig. 4A and C). To test the importance of the basic region in packaging, a second mutant, in which RKR was mutated to RTL, was constructed (Fig. 1). In the context of PR*, the level of Gag synthesized by the RTL mutation was equivalent to that of wt Gag (Fig. 5A). Packaging of RNA into the mutant particles was also similar to that in the wt particles (Fig. 5B and C). This indicates that all three basic residues in this region are not necessary for packaging but that Arg549 is a critical residue.
The yeast three-hybrid system is a rapid genetic assay for Mψ RNA-protein interactions.
Since the initial interactions between RNA and Gag polyprotein are likely to be a prerequisite for packaging into virions, we established a rapid genetic assay for Mψ RNA-Gag protein interactions by using the yeast three-hybrid system (56). This system is similar to the two-hybrid approach for protein-protein interactions, based on the transcriptional activation of separable domains of regulatory proteins such as GAL4 and LexA, and is composed of three hybrid molecules (Fig. 6A). One protein hybrid consists of the DNA binding domain of a transcriptional activator fused to a known RNA binding domain, in our case the MS2 coat protein. The second hybrid is a RNA-RNA hybrid. The N-terminal MS2 RNA, which specifically binds to the MS2 coat protein, is fused at the C terminus to the target RNA. We fused the MS2 RNA to the minimal-packaging region of RSV, Mψ (7) (Fig. 6B). The third hybrid consists of the AD of Gal4 fused to the protein of interest, in our case the RSV Gag polyprotein. If interactions between the RSV minimal-packaging RNA (Mψ) and the Gag polyprotein occur, they will drive the transcriptional activation of the reporter gene, lacZ. Since the RSV protease is encoded within the Gag polyprotein, we used the active-site protease mutant PR* in our experiments.
FIG. 6.
(A) Schematic diagram of a yeast three-hybrid system. The specific binding of MS2-RSV Mψ RNA to the Gag polyprotein would reconstitute the activity of transcriptional activator and lead to the expression of a reporter gene, lacZ. DB, DNA binding domain of LexA; AD, activation domain of Gal4; UAS, binding site for the transcriptional activator upstream of the reporter gene. (B) Diagram of the RNA hybrid expression vector. The RSV Mψ was introduced into the SmaI site downstream of two copies of the MS2 RNA coat protein binding sequence. MS2-Mψ hybrid RNAs were expressed from an RNase P promoter by using RNA polymerase III in S. cerevisiae. Thus, RNA hybrid contains both 5′ RNase P RNA leader and 3′ terminator sequences. Ura3 is a selectable gene. (C) Schematic structure of the hybrid RNA. It retains the 5′ MS2 stem-loop structure(s) and the 3′ end of RNase P RNA.
To examine the specificy of the three-hybrid system for Mψ RNA-Gag interactions, we tested a variety of control plasmids by using a quantitative β-gal assay to monitor RNA-protein interactions. Interactions of an iron-regulatory protein and its structurally well-characterized RNA, the iron response element (IRE), were used as a positive control and showed a high level of β-gal activity (data not shown). Since our test system consists of sense Mψ (sMψ) fused to MS2 and GagPR* fused to the AD, we tested a variety of plasmid combinations lacking essential components, as well as the RNA hybrid plasmid containing antisense Mψ (αsMψ). In the absence of any one of the hybrid components, transformants showed little or no activity (data not shown), indicating that the hybrid RNA must be capable of binding simultaneously to both protein hybrids to allow transcription.
We next tested the effects of the Gag mutations which we previously analyzed in the in vivo packaging assay (Fig. 1 and 4). All of the mutants in Table 1 produced roughly equivalent amounts of Gag polyprotein with the predicted molecular weights in the yeast cells as measured by RIPA (data not shown). We found that binding of the full-length Gag polyprotein and binding of the Gag protein lacking the CA domain were at least as high as that of the IRE–iron-regulatory protein control (data not shown). Both PR* and ΔCA hybrids preferentially bound to the sMψ RNA (Table 1). The Gag protein did not bind to the IRE RNA (data not shown), indicating that this assay detects more than just binding to a highly structured vertebrate RNA motif. The NC-CHΔ1,2 deletion mutant showed only low-level binding to Mψ RNA. This is consistent with a low level of RNA packaging (Fig. 4A and C). Interestingly, the NC-CHΔ1,2 mutant did not show specific binding to Mψ RNA, since the β-gal activity was the same when the RNA hybrid containing αsMψ RNA was used. Thus, in this assay, the Cys-His boxes are required for binding specificity as well as affinity. Mutation of the basic region distal to the second Cys-His box (NC-SKL) yielded a drastically reduced packaging efficiency (Fig. 4C). In contrast to the results with NC-CHΔ1,2, NC-SKL still yielded specific binding to Mψ RNA, although the level of β-gal activity was only 30% that of wt (Table 1). An additional basic-region mutant, NC-RTL, gave 80% of the wt level of β-gal activity (Table 1). This mutant was wt for packaging (Fig. 5). These results indicate that the in vivo packaging assay and the three-hybrid assay correlate well but that the binding assay is much less sensitive to changes in proteins than is the packaging assay.
TABLE 1.
RSV Mψ-Gag interaction, binding specificity, and packaging efficiency
| Gag construct | β-gal activity (units) witha:
|
sMψ/αsMψ RNA ratio | Packaging efficiencyb | |
|---|---|---|---|---|
| sMψ RNA | αsMψ RNA | |||
| PR* | 2,650 ± 540 | 126 ± 40 | 21 | 1 |
| ΔCA | 3,620 ± 900 | 362 ± 80 | 10 | 0.95 ± 0.05 |
| NC-CHΔ1,2 | 480 ± 100 | 460 ± 80 | 1 | 0.16 ± 0.03 |
| NC-SKL | 860 ± 120 | 66 ± 40 | 13 | 0.04 ± 0.01 |
| NC-RTL | 2,120 ± 20 | 66 ± 10 | 32 | 0.7 ± 0.2 |
The enzymatic activity represents the means of four to six assays from three or four independent transformants and the standard deviations.
Packaging efficiency of mutant Mψ RNAs.
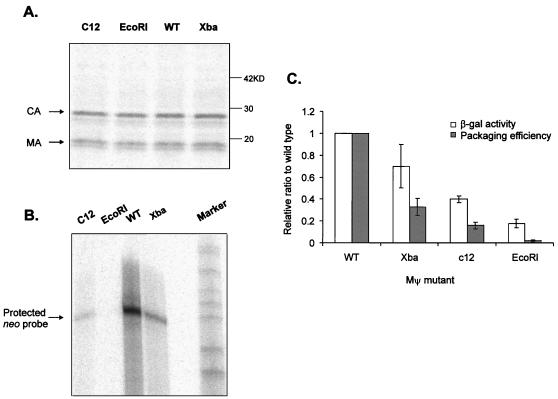
To further test whether the binding affinity of RNA to Gag polyprotein plays an important role in packaging, we constructed a series of RNA mutants with mutations in the Mψ region and compared their packaging efficiencies with their β-gal activities. The computer-generated putative secondary structure of Mψ RNA is shown in Fig. 7. The O3 stem, composed of S1 and S2, is known to be an important structure for packaging (7, 37). The mutations used in this analysis are highlighted in the figure. The β-gal activities of the mutants were in the range of 0.2- to 0.7-fold of wt Mψ RNA activities in the yeast three-hybrid system. Mψ confers efficient packaging when tethered to heterologous RNAs (7, 7a). To measure the ability of mutant Mψ RNAs to confer packaging in the heterologous assay, we placed mutant Mψ RNAs 3′ of the neo gene in plasmid pCMVneo (5, 44). The constructs were then transfected into the Q2bn-4D packaging cell line, and stably transfected mass cultures were obtained after selection with G418. Quantitative RIPA and RPAs were performed with viral particles collected from culture supernatants. Supernatants from packaging cells transfected with the three mutants or wt CMV-neo Mψ RNAs yielded equivalents amounts of pelletable capsid protein in the cell supernatants (0.8 to 1.2 relative to Mψ [Fig. 8A]). However, these mutants had different effects on packaging of neo RNA into particles (Fig. 8B). The efficiency of packaging was calculated and compared to the results of the three-hybrid assay (Fig. 8C). There was a clear correlation between the level of RNA packaging and β-gal activity. However, as seen with the Gag mutants, the three-hybrid assay was less sensitive to changes in RNA structure than was the packaging assay.
FIG. 7.
Putative secondary structure of minimal-packaging RNA in avian leukosis virus (RSV Mψ) (7). The location of the O3 stem is shown as S1 and S2. Nucleotides substituted for each Mψ mutations are shown as white nucleotides in black boxes in the appropriate regions of Mψ RNA. In the XbaMΨ mutant, GGGG was replaced by UCUAGA; in the c12 MΨ mutant, CUGCG was replaced by GAUUC; in the EcoRIMΨ mutant, GGGGG was replaced by GAAUUC.
FIG. 8.
Effects of Mψ mutations on RNA packaging. (A) RIPA to determine the number of viral particles released from cultures transfected with indicated plasmids. (B) RPA to measure the amount of neo-specific RNA packaged in virions. (C) Comparison of the β-gal activity measured in the yeast three-hybrid system and the packaging efficiency determined in vivo. The packaging efficiency was calculated as the ratio of the relative amount of neo-specific RNA packaged in particles, as measured by RPA (Fig. 8B), to the relative number of particles, as measured by RIPA (Fig. 8A). Both packaging efficiencies and β-gal activities were normalized to PR*. Each experiment was done three or four times, and the bars show the standard deviations.
A modified Gag-Mψ binding assay with increased sensitivity.
While we consistently observed a correlation between the binding in yeast and in vivo packaging, the differences in β-gal activity detected are not sufficient to predict the in vivo behavior of mutants in packaging, the key goal of these experiments. For example, there was only a 2.5-fold difference in β-gal activity between the NC-SKL and NC-RTL mutants whereas there was a 1.5-log difference in packaging (Table 1; Fig. 4 and 5). The hybrid RNA used in our assays contains two tandem MS2 binding sites but only one copy of Mψ. Thus, it was possible that the low sensitivity of our assay is a product of a single Mψ RNA in the RNA hybrid. To test this, we created a hybrid RNA containing two tandem Mψ structures. We found that the use of such an RNA had no effect on the efficiency of the assay with several different Gag proteins (data not shown). As another way to increase sensitivity, we exploited the finding that deletion of the protease domain of Gag (ΔPR; Fig. 1), rather than inactivation of the active site, led to a sixfold increase in β-gal activity compared to wt Gag (Table 2) as well as a moderate increase in packaging (Fig. 5). We examined several NC mutants (NC-SKL and RTL) in the context of ΔPR to determine if use of the ΔPR protein could increase the sensitivity of the assay. We found that the NC-RTL-ΔPR mutant had 10-fold more β-gal activity than the NC-SKL-ΔPR mutant did (Table 2), which is consistent with its 17-fold-greater packaging ability. These results indicate that the yeast three-hybrid assay with ΔPR Gag as one protein hybrid is nearly as sensitive as the packaging assay for screening of mutants.
TABLE 2.
β-Gal activity and packaging efficiency of ΔPR mutants
DISCUSSION
In this study, we have examined deletions of large portions of the Gag protein on both viral assembly and RNA packaging. We found that deletion of most of CA or PR had no effect on either. An MA deletion mutant with a myristylation signal at the amino terminus produces extracellular particles with an altered density but which still contain genomic RNA. In contrast to these results, mutation of several residues in the distal basic region of NC, in particular Arg549, has a profound effect on packaging but not assembly.
Three domains have been shown to be important for assembly of avian retroviral particles (50). These are AD1, located at the amino terminus of MA, which is required for membrane targeting; AD2, located in the p2 region between MA and p10, which is required for late budding; and AD3, located in NC, which is required for Gag-Gag interactions. In addition, the RSV Gag MHR has been reported to be involved in the late stage of virion maturation as well as in the entry of the viral core into a new host (17). All of our mutations contain AD2. However, the CA deletion lacks the MHR and still produces particles with the correct density. While deletion of AD1 in ΔMA prevented assembly, addition of a myristylation signal restored particle assembly. This is consistent with previous results (40, 63) which suggested that Myr is required to target Gag to the membrane and thus potentiate the frequency of Gag-Gag interactions. While the myrΔMA mutant produced particles with aberrant density, these particles still contained genomic RNA. This is consistent with the fact that MA is not a specific RNA binding protein in vitro (57). In RSV and HIV NC, AD3 overlaps with the Cys-His box zinc finger motifs, which are required for the formation of viral particles with proper density (15). Mutants lacking AD3 produce particles inefficiently (9, 53, 61). The fact that our NC-CHΔ1,2 mutant is defective in particle assembly is consistent with all of these findings.
Sakalian et al. (53) also looked at effects of CA deletions on assembly and packaging. However, these authors found that deletion of the MHR in the capsid reduced the packaging efficiency about fivefold in transiently transfected COS cells, while we found no effect on deletion of almost all of the CA region, including the MHR. One possibility to explain this discrepancy is that there is a much higher level of viral gene expression in transiently transfected Cos cells than in the cells we used, which contain low copies of integrated proviruses. In addition, our CA deletion was different from that used by Sakalian et al. These authors also showed that deletions in CA and the MHR region did not affect the content of gag or pol gene products or viral RNA but led to the production of noninfectious particles with size heterogeneity. We have not examined this for our ΔCA mutant.
There have been inconsistent reports of the effects of deletion of the PR domain on packaging. Our results are consistent with those of Sakalian et al. (53), who found that ΔPR Gag protein, in the context of Myr 1, can efficiently encapsidate RNA in COS cells. Our data disagree with other published reports of studies using somewhat different vector systems (4, 48). We cannot explain why the ΔPR mutant increases packaging efficiency. It is possible that the mutant particles contain fewer Gag proteins and thus increase the apparent ratio of RNA genomes to Gag proteins in virions.
We found that while deletion of PR had little effect on RNA encapsidation, mutations in the Cys-His boxes and distal basic residues of NC did. Not surprisingly, we found that the RNA-packaging efficiency of the ΔCys-His box mutant was reduced markedly, consistent with many previous studies involving chimeric Gag proteins (13, 22, 67) and deletion mutants of Gag proteins (53). The NC-CHΔ1,2 mutant reduced packaging about sixfold in these assays, a result which is somewhere between those previously reported by others (46), in which deletion of both Cys-His boxes completely abrogated packaging measured by Northern blots, and those previously obtained in our laboratory (4). Since all retroviral NC proteins have a high basic amino acid content, a property shared with other RNA binding proteins (38), the effects of basic amino acid mutations on in vitro RNA binding, RNA packaging, and viral infectivity have been investigated by several groups (18, 24, 30, 49, 54). In an alanine-scanning mutagenesis on HIV-1 NC proteins, a set of mutants with multiple substitutions in the zinc binding domains, the basic region that links them, and the residues that flank the two zinc binding domains indicates that clusters of positively charged amino acids in three different subdomains are necessary for efficient RNA packaging (51). Using avian retrovirus NC proteins, we found a single basic region immediate downstream of the distal Cys-His box, which drastically reduces the packaging efficiency. It is likely that removal of positively charged residues either reduces the affinity of NC for the negatively charged RNA or causes deleterious conformational changes in the protein. The latter explanation is more likely, since we have mutated two positively charged amino acids in both RTL and SKL but only the latter affects packaging.
Because in vivo packaging assays are time-consuming, we have tested whether a rapid yeast three-hybrid assay can be used to detect and analyze the interaction between avian retroviral ψ RNA and Gag polyprotein. The RSV Mψ is a sufficient packaging signal in the context of heterologous RNA (7, 7a), and therefore the RSV system is a good one to explore the specificity of interactions in yeast. We found that the wt Gag and the CA domain deletion mutant (ΔCA) show specific binding to Mψ RNA, consistent with the results of in vivo packaging assays. However, we also found that the NC-SKL mutant, which is defective for packaging, bound to the Mψ RNA in the yeast assay in a specific manner with about 30% efficiency. Using an additional NC mutant, NC-RTL, as well as a series of mutants with mutations in Mψ RNA, we found that only interactions in yeast which are high in β-gal expression, approaching wt levels, correlate with packaging of ψ RNA into virions. Thus, both binding specificity and affinity measured by the level of β-gal expression in yeast are important indicators of in vivo packaging.
It has been reported that fusions of either HIV-Gag or NC with GST specifically bind to HIV-1 RNA in gel mobility shift assays (12). We examined whether Gal4AD-RSVNC can specifically bind to Mψ RNA in the yeast assay. We found that while transformants with the Gal4AD-Gag plasmid were formed after 3 days of incubation, yeast cells transformed with the Gal4AD-NC fusion plasmid were very sick, forming only tiny colonies (data not shown). The toxicity of NC fusion may be due to the nonspecific binding of NC protein to yeast cellular RNAs. Bacharach and Goff (6) described use of a yeast three-hybrid assay with HIV Gag and RNA. These workers found that HIV Gag bound specifically to HIV RNA but that the HIV NC fusion plasmid bound to both HIV RNA and heterologous RNAs such as IRE, resulting in equally strong activation of the lacZ reporter. In this case, the yeast assay reflects the in vivo situation better than the gel shift assays do. However, our assay goes beyond the previous work, since we have found a correlation between binding and packaging assays. Furthermore, by using a ΔPR Gag, our assay is quite sensitive.
In the three-hybrid system, the hybrid RNAs are probably localized primarily in the nucleus, since hybrid RNAs are linked to the 5′ leader sequence of RNase P RNA, an RNA not known to enter the cytoplasm (39). Even if transported to the cytoplasm, the RNAs would be returned to the nucleus after binding to their cognate hybrid proteins, which themselves carry a nuclear localization signal. While RNA-protein interactions important in transcriptional activation occur in the nucleus, it is likely that the encapsidation of genomic RNAs occurs at the cytoplasmic membrane. The yeast nuclear assay is unlikely to completely mimic packaging, where cytoplasmic or membrane cellular proteins might act as chaperones for both particle assembly and encapsidation. For example, hsp90 has been shown to be critical for hepatitis B virus assembly (31). Binding of Gag and Mψ RNA is probably only the first step in a complete pathway. Thus, a mutation in Gag or MΨ could have only a moderate effect on initial RNA-protein interaction but could destabilize downstream events. This is probably why the yeast assay does not fully recapitulate the sensitivity of the packaging assay.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Cancer Institute (CA 18282) to M.L.L. and by a grant from National Institutes of Health to M.W.
We thank Michael Emerman for a critical review of the manuscript.
REFERENCES
- 1.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging results in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen P, Collins B, Brown D, Hostomsky Z, Gold L. A specific RNA structural motif mediates high affinity binding by the HIV-1 nucleocapsid protein (NCp7) Virology. 1996;225:306–315. doi: 10.1006/viro.1996.0605. [DOI] [PubMed] [Google Scholar]
- 3.Anderson D J, Stone J, Lum R, Linial M L. The packaging phenotype of SE21Q1b provirus is related to high proviral expression and not trans-acting factors. J Virol. 1995;69:7319–7323. doi: 10.1128/jvi.69.11.7319-7323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aronoff R, Hajjar A M, Linial M L. Avian retroviral RNA encapsidation: reexamination of functional 5′ RNA sequences and the role of nucleocapsid Cys-His motifs. J Virol. 1993;67:178–188. doi: 10.1128/jvi.67.1.178-188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aronoff R, Linial M L. Specificity of retroviral RNA packaging. J Virol. 1991;65:71–80. doi: 10.1128/jvi.65.1.71-80.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacharach E, Goff S P. Binding of the human immunodeficiency virus type 1 Gag protein to the viral RNA encapsidation signal in the yeast three-hybrid system. J Virol. 1998;72:6944–6949. doi: 10.1128/jvi.72.8.6944-6949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banks J D, Yeo A, Green K, Cepeda F, Linial M L. A minimal avian retroviral packaging sequence has a complex structure. J Virol. 1998;72:6190–6194. doi: 10.1128/jvi.72.7.6190-6194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Banks, J. D., B. Kealoha, and M. L. Linial. Submitted for publication.
- 8.Bartel P L, Roecklein J A, SenGupta D, Fields S. A protein linkage map of Escherichia coli bacteriophage T7. Nat Genet. 1996;12:72–77. doi: 10.1038/ng0196-72. [DOI] [PubMed] [Google Scholar]
- 9.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berglund J A, Charpentier B, Rosbash M. A high affinity binding site for the HIV-1 nucleocapsid protein. Nucleic Acids Res. 1997;25:1042–1049. doi: 10.1093/nar/25.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 12.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berkowitz R D, Ohagen A, Hoglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowles N E, Damay P, Spahr P F. Effect of rearrangements and duplications of the Cys-His motifs of Rous sarcoma virus nucleocapsid protein. J Virol. 1993;67:623–631. doi: 10.1128/jvi.67.2.623-631.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowzard J B, Bennett R P, Krisina N K, Ernst S M, Rein A, Wills J W. Importance of basic residues in the nucleocapsid sequence for retrovirus gag assembly and complementation rescue. J Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C, Okayama H. High efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craven R C, Leure-duPree A E, Weldon R A, Jr, Wills J W. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis B R, Schwartz D H, Marx J C, Johnson C E, Berry J M, Lyding J, Merigan T C, Zander A. Absent or rare human immunodeficiency virus infection of bone marrow stem/progenitor cells in vivo. J Virol. 1991;65:1985–1990. doi: 10.1128/jvi.65.4.1985-1990.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorfman T, Luban J, Goff S P, Haseltine W A, Gottlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dupraz P, Oertle S, Meric C, Damay P, Spahr P F. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J Virol. 1990;64:4978–4987. doi: 10.1128/jvi.64.10.4978-4987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupraz P, Spahr P-F. Specificity of Rous sarcoma virus nucleocapsid protein in genomic RNA packaging. J Virol. 1992;66:4662–4670. doi: 10.1128/jvi.66.8.4662-4670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fisher R J, Rein A, Fivash M, Urbaneja M A, Casas F, JR, Medaglia M, Henderson L E. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu X, Katz R A, Skalka A M, Leis J. Site-directed mutagenesis of the avian retrovirus nucleocapsid protein, pp12. J Biol Chem. 1988;263:2140–2145. [PubMed] [Google Scholar]
- 25.Geigenmuller U, Linial M L. Specific binding of human immunodeficiency virus type 1 (HIV-1) Gag-derived proteins to a 5′ HIV-1 genomic RNA sequence. J Virol. 1996;70:667–671. doi: 10.1128/jvi.70.1.667-671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 27.Goff S, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorelick R J, Henderson L E, Hanser J P, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc Natl Acad Sci USA. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorelick R J, Nigida S M, Jr, Bess J W, Jr, Arthur L O, Henderson L E, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Housset V, de Rocquigny H, Roques B P, Darlix J L. Basic amino acids flanking the zinc finger of Moloney murine leukemia virus nucleocapsid protein NCp10 are critical for virus infectivity. J Virol. 1993;67:2537–2545. doi: 10.1128/jvi.67.5.2537-2545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu J, Toft D O, Seeger C. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 1997;16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jentoft J E, Smith L M, Fu X D, Johnson M, Leis J. Conserved cysteine and histidine residues of the avian myeloblastosis virus nucleocapsid protein are essential for viral replication but are not “zinc-binding fingers”. Proc Natl Acad Sci USA. 1988;85:7094–7098. doi: 10.1073/pnas.85.19.7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karpel R L, Henderson L E, Oroszlan S. Interactions of retroviral structural proteins with single-stranded nucleic acids. J Biol Chem. 1987;262:4961–4967. [PubMed] [Google Scholar]
- 34.Katz R A, Omer C A, Weis J H, Mitsialis S A, Faras A J, Guntaka R V. Restriction endonuclease and nucleotide sequence analyses of molecularly cloned unintegrated avian tumor virus on structure of large terminal repeats in circle junctions. J Virol. 1982;42:346–351. doi: 10.1128/jvi.42.1.346-351.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaye J F, Lever A M. trans-acting proteins involved in RNA encapsidation and viral assembly in human immunodeficiency virus type 1. J Virol. 1996;70:880–886. doi: 10.1128/jvi.70.2.880-886.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan R, Giedroc D P. Nucleic acid binding properties of recombinant Zn2 HIV-1 nucleocapsid protein are modulated by COOH-terminal processing. J Biol Chem. 1994;269:22538–22546. [PubMed] [Google Scholar]
- 37.Knight J B, Si Z H, Stoltzfus C M. A base-paired structure in the avian sarcoma virus 5′ leader is required for efficient encapsidation of RNA. J Virol. 1994;68:4493–4502. doi: 10.1128/jvi.68.7.4493-4502.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lazinski D, Grzadzielska E, Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989;59:207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- 39.Lee J Y, Rohlman C E, Molony L A, Engelke D R. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol Cell Biol. 1991;11:721–730. doi: 10.1128/mcb.11.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee P P, Linial M L. Efficient particle formation can occur if the matrix domain of human immunodeficiency virus type 1 Gag is substituted by a myristylation signal. J Virol. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Legrain P, Dokhelar M C, Transy C. Detection of protein-protein interactions using different vectors in the two-hybrid system. Nucleic Acids Res. 1994;22:3241–3242. doi: 10.1093/nar/22.15.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leis J, Jentoft J. Characteristics and regulation of interaction of avian retrovirus pp12 protein with viral RNA. J Virol. 1983;48:361–369. doi: 10.1128/jvi.48.2.361-369.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, Gu Z, Geleziunas R, Kleiman L, Wainberg M A, Parniak M A. Expression, purification, and RNA-binding properties of HIV-1 p15gag nucleocapsid protein. Protein Expression Purif. 1993;4:304–311. doi: 10.1006/prep.1993.1039. [DOI] [PubMed] [Google Scholar]
- 44.Linial M. Creation of a processed pseudogene by retroviral infection. Cell. 1987;49:93–102. doi: 10.1016/0092-8674(87)90759-8. [DOI] [PubMed] [Google Scholar]
- 45.Meric C, Goff S P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J Virol. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meric C, Gouilloud E, Spahr P-F. Mutations in Rous sarcoma virus nucleocapsid protein p12 (NC): deletions of Cys-His boxes. J Virol. 1988;62:3328–3333. doi: 10.1128/jvi.62.9.3328-3333.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meric C, Spahr P-F. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J Virol. 1986;60:450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oertle S, Spahr P F. Role of the Gag polyprotein precursor in packaging and maturation of Rous sarcoma virus genomic RNA. J Virol. 1990;64:5757–5763. doi: 10.1128/jvi.64.12.5757-5763.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ottmann M, Gabus C, Darlix J L. The central globular domain of the nucleocapsid protein of human immunodeficiency virus type 1 is critical for virion structure and infectivity. J Virol. 1995;69:1778–1784. doi: 10.1128/jvi.69.3.1778-1784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poon D T, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rein A, Harvin D P, Mirro J, Ernst S M, Gorelick R J. Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J Virol. 1994;68:6124–6129. doi: 10.1128/jvi.68.9.6124-6129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakalian M, Wills J W, Vogt V M. Efficiency and selectivity of RNA packaging by Rous sarcoma virus Gag deletion mutants. J Virol. 1994;68:5969–5981. doi: 10.1128/jvi.68.9.5969-5981.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmalzbauer E, Strack B, Dannull J, Guehmann S, Moelling K. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J Virol. 1996;70:771–777. doi: 10.1128/jvi.70.2.771-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwartz D E, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 56.Sengupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steeg C M, Vogt V M. RNA-binding properties of the matrix protein (p19gag) of avian sarcoma and leukemia viruses. J Virol. 1990;64:847–855. doi: 10.1128/jvi.64.2.847-855.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stewart L, Vogt V M. trans-acting viral protease is necessary and sufficient for activation of avian leukosis virus reverse transcriptase. J Virol. 1991;65:6218–6231. doi: 10.1128/jvi.65.11.6218-6231.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stoker A W, Bissell M J. Development of avian sarcoma and leukosis virus-based vector-packaging cell lines. J Virol. 1988;62:1008–1015. doi: 10.1128/jvi.62.3.1008-1015.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verderame M F, Nelle T D, Wills J W. The membrane-binding domain of the Rous sarcoma virus Gag protein. J Virol. 1996;70:2664–2668. doi: 10.1128/jvi.70.4.2664-2668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weldon R A, Jr, Wills J W. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 64.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wills J W, Craven R C, Weldon R A, Nelle T D, Erdie C R. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J Virol. 1991;65:3804–3812. doi: 10.1128/jvi.65.7.3804-3812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.You J C, McHenry C S. HIV nucleocapsid protein. Expression in Escherichia coli, purification, and characterization. J Biol Chem. 1993;268:16519–16527. [PubMed] [Google Scholar]
- 67.Zhang Y Q, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]